Abstract
It has been well documented that genetic factors can influence predisposition to develop alcoholism. While the underlying genomic changes may be of several types, two of the most common and disease associated are copy number variations (CNVs) and sequence alterations of protein coding regions. The goal of this study was to identify CNVs and single-nucleotide polymorphisms that occur in gene coding regions that may play a role in influencing the risk of an individual developing alcoholism. Toward this end, two mouse strains were used that have been selectively bred based on their differential sensitivity to alcohol: the Inbred long sleep (ILS) and Inbred short sleep (ISS) mouse strains. Differences in initial response to alcohol have been linked to risk for alcoholism, and the ILS/ISS strains are used to investigate the genetics of initial sensitivity to alcohol. Array comparative genomic hybridization (arrayCGH) and exome sequencing were conducted to identify CNVs and gene coding sequence differences, respectively, between ILS and ISS mice. Mouse arrayCGH was performed using catalog Agilent 1 × 244 k mouse arrays. Subsequently, exome sequencing was carried out using an Illumina HiSeq 2000 instrument. ArrayCGH detected 74 CNVs that were strain-specific (38 ILS/36 ISS), including several ISS-specific deletions that contained genes implicated in brain function and neurotransmitter release. Among several interesting coding variations detected by exome sequencing was the gain of a premature stop codon in the alpha-amylase 2B (AMY2B) gene specifically in the ILS strain. In total, exome sequencing detected 2,597 and 1,768 strain-specific exonic gene variants in the ILS and ISS mice, respectively. This study represents the most comprehensive and detailed genomic comparison of ILS and ISS mouse strains to date. The two complementary genome-wide approaches identified strain-specific CNVs and gene coding sequence variations that should provide strong candidates to contribute to the alcohol-related phenotypic differences associated with these strains.
Introduction
According to a recent assessment of alcohol-related costs, excessive alcohol intake leads to approximately 79,000 deaths and $223.5 billion in spending in the US each year (Bouchery et al. 2011). Further, it was recently estimated that as high as 7 % of 18–29 year olds in the United States had alcohol abuse behaviors as defined by the DSM-IV (Grant et al. 2004) Alcoholism is a complex disease that has a heritability factor of 0.5 (Dick et al. 2009), which suggests a substantial genetic component that influences the risk of developing the condition. This study aims to apply two complementary genome-wide technologies to mouse strains developed as animal models of alcohol action. The two strains of mice, Inbred long sleep (ILS) and Inbred short sleep (ISS) (McClearn and Kakihana 1981), have been extensively studied (Bennet and Johnson 1998; Bennett et al. 2000a, b; Collins 1981; Deitrich 1990) in alcohol research. These two strains have been selectively bred based on their differential sensitivity to alcohol, including their phenotypic differences in their loss of righting reflex (LORR) (Markel et al. 1995, 1997; Bennet and Johnson 1998). ISS mice are able to right themselves much quicker than the ILS mice after receiving the same dose of ethanol. The regions linked to the loss of righting induced by ethanol (Lore) have been narrowed by quantitative trait loci (QTL) analysis, but no high-resolution strategy has been applied to determine exact genomic sequence variants and dosage changes (copy number variations; CNVs) that directly affect or cause the loss of righting phenotype. As genetic technologies and approaches have improved over recent years, some progress has been made in determining what genes underlie the phenotypic differences between ILS and ISS mice. Examples include the use of mouse QTL mapping (Bennet and Johnson 1998; Bennett et al. 2000a, b; Markel et al. 1995, 1997; Ehringer and Sikela 2002), gene expression profiling (Xu et al. 2001; MacLaren and Sikela 2005; MacLaren et al. 2006), comparative DNA sequencing (Ehringer et al. 2001, 2002) and transcriptome analysis (Darlington et al. 2013). However, these studies did not account for copy number variation, which contributes to many disorders (Weischenfeldt et al. 2013; Mefford et al. 2008; Brunetti-Pierri et al. 2008; Dumas et al. 2012). Additionally, mouse genome sequences contain many gaps, and complex duplication-rich regions are likely to be misassembled and ignored in most assays. The phenotypic affects of alcoholism, like many complex disorders with a genetic influence (Mefford et al. 2008; Brunetti-Pierri et al. 2008; International Schizophrenia Consortium 2008; Pinto et al. 2010; Jaillard et al. 2010; de Kovel et al. 2010), may be at least in part due to genomic structural variation such as CNVs. CNVs may influence alcohol action and metabolism by altering the dosage and possibly function of critical genes involved in these processes. Likewise, no genome-wide effort has yet been reported to identify strain-specific gene coding sequence differences between the ILS and ISS mice.
To better understand strain-specific genomic structural (CNVs) and gene coding changes that produce different alcohol-related phenotypes, arrayCGH (n = 6) and exome sequencing (n = 4) were performed on individuals from both strains. By allowing both structural variations and gene coding sequence differences to be identified, these combined approaches represent the most comprehensive genomic comparison of ILS/ISS mice so far reported.
Materials and methods
ArrayCGH
ArrayCGH was conducted using catalog Agilent 1×244 k mouse arrays. Each array contains 244,000 whole genome probes. The sample set included 4 ISS mice and 2 ILS mice, and the same reference DNA was used (C57BL/6J) so that we could compare all samples tested. The arrays were processed at Oxford Gene Technology in the UK.
ArrayCGH analysis
Cytosure version 3.3.2 was used to view and detect copy number variation across each sample. The criteria for calling either a duplication or a deletion required a log2 ratio greater than 0.3 or less than −0.3, respectively. Additionally, a minimum of five consecutive probes and more than 0.1 kb in length were required. The normalization was done in Cytosure version 3.3.2 using the chromosomal mean method. ILS and ISS mice were compared to the reference C57BL/6J to determine CNVs in each strain. These CNV counts and locations in turn were compared between strains and then compared within strains. This strategy allowed us to identify risk CNVs but also let us explore genomic variation that may exist in mice of the same strain. Aberration reports were generated using Cytosure version 3.3.2, and data were compiled in Xcel.
Exome sequencing
The exomes of individuals from both ILS (n = 2) and ISS (n = 2) strains were targeted and sequenced using the Agilent SureSelect XT Mouse Exome Kit. CSNPThe Illumina HiSeq 2000 was used with an average coverage of 30× and 2 × 100 bp paired-end reads.
Exome sequencing analysis
Sequence reads were trimmed of any poor quality sequence using a Perl script and then aligned to the mm10 reference genome using BWA (Li and Durbin 2009). ANNOVAR (Wang et al. 2010) was used with the mm10 RefSeq genes to determine the type of each cSNP (indel, synonymous, nonsynonymous, stop codon gain/loss, frameshift). A Perl script was used to parse the ANNOVAR variant call files to identify variants and genes that are found in both samples of one group but not any sample from the other group.
Independent cSNP verification using Sanger sequencing
11 ILS and 11 ISS mouse tail snips were obtained, and DNA was purified using a QIAGEN DNeasy Blood/Tissue extraction kit. PCR primers were designed to produce a 413 bp ampilicon encompassing exon 1 of the alpha-amylase 2B (AMY2B) gene. The primer sequences include forward: AGGTTTCACACATAACACTCACG and reverse: TCCATCACCTGTTCACATCC. Using the standard PCR protocol, the 22 purified DNAs were PCR amplified and the products were run on a 1 % agarose gel to verify amplification and size. After verification, 6 PCR products from each strain were purified using the Qiagen QIAquick PCR purification kit. Five of the purified PCR products from each strain were sequenced using the same Amy2b PCR primers and following standard Sanger sequencing protocol.
Results
Genome-wide arrayCGH was used to investigate the role of genome structural variation (e.g., CNVs) in the differential initial sensitivity to alcohol between ILS and ISS strains. Given the well-established phenotypic differences between the ILS and ISS strains, the most interesting CNVs identified in this study are those that are strain-specific, i.e., the CNV is found in either all ILS or all ISS mice tested. Using arrayCGH, 74 aberrations were detected that were common only to the ILS or to the ISS strains (Supplemental Table 1). Of these, ISS mice had 36 CNVs (6 gains, 30 losses), and ILS mice had 38 CNVs (10 gains, 28 losses). These aberrations ranged in size from 18,358 to 1,414,880 bp and were located on all autosomes except chromosome 9. Figures 1 and 2 shows an example of ILS-and ISS-strain-specific CNVs located on chromosome 4.
Fig. 1.
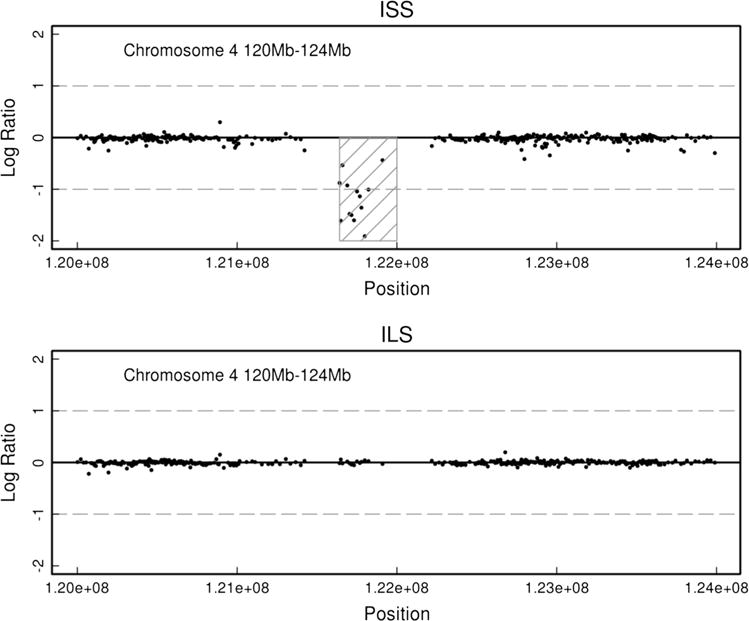
ISS-specific loss detected by arrayCGH. ArrayCGH determined log ratio profile of chromosome 4 from 120 to 124 Mb for both an ILS and ISS mouse. Note the deletion detected at approximately 121640501 bp to 122218644 bp in the ISS mouse and no apparent aberration in the same region of the ILS mouse
Fig. 2.
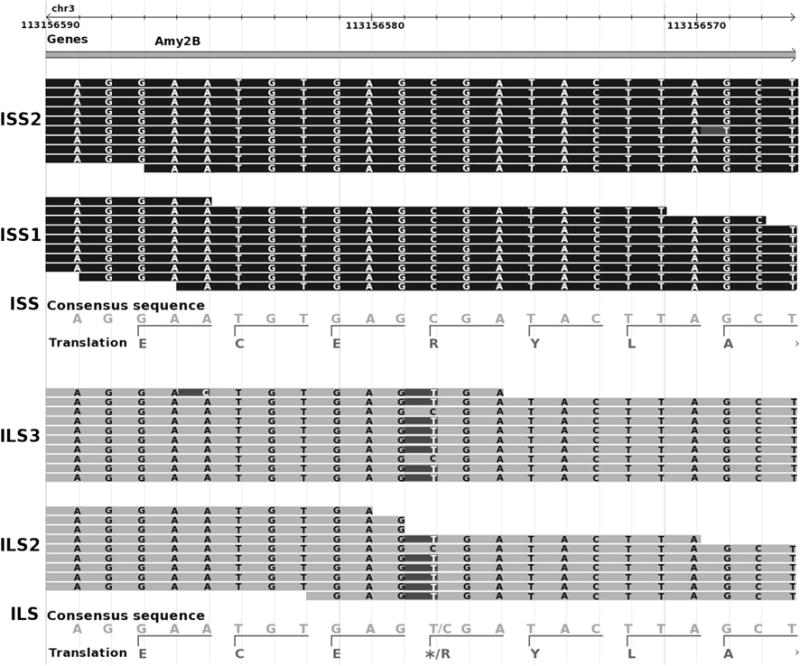
ILS-specific SNP causing a stop codon gain in AMY2B gene. Exome sequence reads from one ILS and one ISS mouse. Sequences are presented from all ILS and ISS mice tested. The ILS mice have a SNP that changes the base from a G (compared to reference C57BL/6J and ISS) to an A on the forward strand (top panel). Exome sequence reads for one ISS mouse are shown in middle panel. The bottom panel shows the consensus sequence from the reference genome (C57BL/6J). The AMY2B gene is on the negative strand, but the positive strand is shown. The SNP creates a premature TGA stop codon in the AMY2B gene specifically in ILS mice
More detailed analysis of these CNVs identified a number of genes that have been linked to biological processes potentially related to known phenotypic differences between ILS and ISS strains. Among ISS mice, several deletions were identified that contain genes that have been linked to brain function, a finding that may help explain why ISS mice show a more extreme phenotypic profile than the ILS mice. For example, an ISS-specific deletion on chromosome 7 contains the gene that encodes the alpha 1B subunit of the N type, voltage-dependent calcium channel, which controls neurotransmitter release from neurons. Similarly, an ISS-specific deletion on chromosome 14 contains the Erc2 gene that encodes a member of the ELKS/RAB6-interacting/CAST family thought to also be involved in neurotransmitter release. Finally, an ISS-specific deletion on chromosome 19 contains the Sorcs1 gene that encodes the sortilin-related VPS10 domain-containing receptor 1 that is strongly expressed in the central nervous system.
Among ILS mice, one of the most interesting CNVs was a duplication of a segment on chromosome 19 that included at least four genes, some of which have known, and important, biological functions. These include the NF-kappa-B (nfkb2) gene that has been implicated in immune-related functions, including inflammatory events, and the Psd gene, which is guanine-nucleotide exchange factor for ARF6 and encodes plekstrin and Sec7 domains.
Several other biologically interesting gene-based CNVs between the two mouse strains were detected by arrayCGH (Supplemental Tables 1, 2, 3). We identified CNVs in candidate genes including UDP-glucuronosyltransferase 1 family, polypeptide A1 (Ugt1a6a), major urinary protein (MUP) 1 and transmembrane protein 181 (Tmem181). More specifically, we found that ISS mice have a loss of Ugt1a6a which encodes a UDP-glucuronosyltransferase, an enzyme of the glucuronidation pathway important in xenobiotic metabolism. In humans, mutations in this gene result in Crigler–Najjar syndromes types I and II and in Gilbert syndrome. These syndromes are caused by a reduced ability to detoxify bilirubin, which is made in the liver (Sampietro and Iolascon 1999). Additionally, ILS has a loss of the MUP1 gene which encodes a protein produced by the liver and is involved in glucose and lipid metabolism (Hui et al. 2009; Zhou et al. 2009). Finally, ILS mice have a gain of the Tmem181 gene, which encodes a putative G-protein-coupled receptor expressed on the cell surface (Wollscheid et al. 2009) and is involved in toxin binding (Carette et al. 2009). All three of these genes have a plausible role in alcohol metabolism and warrant further investigation.
In addition, both strains shared common CNVs that were not found in the control mouse strain: C57BL/6J (Supplemental Table 2). Additionally, arrayCGH confirmed that within-strain variations do exist in both ILS and ISS strains, strengthening the evidence that mice from the same inbred strain are not genetically identical (Supplemental Table 3). (Watkins-chow and Pavan 2008; Cutler and Kassner 2009). Table 1 summarizes these findings.
Table 1.
Numbers of CNVs detected by ArrayCGH
| ISS-specific CNVs | ILS-specific CNVs | ILS/ISS shared CNVs compared to reference C57BL/6J | ILS within-strain CNVs | ISS within-strain CNVs |
|---|---|---|---|---|
| 36 | 38 | 22 | 120 | 127 |
The first two columns list the number of ISS- and ILS-strain-specific CNVs detected via arrayCGH compared to the reference C57BL/6J mouse. The middle column shows CNVs that the ILS and ISS mice share in contrast to the reference C57BL/6J. The right-most two columns show CNVs found within the ILS and within the ISS mouse strains
As a complement to arrayCGH, exome sequencing was applied to both strains to identify strain-specific gene coding sequence variations. Based on the exome sequences, a total of 4,365 unique exonic gene variants were detected. Of these, 961 and 585 were nonsynonymous gene changes for ILS and ISS mouse strains, respectively (Supplemental Table 4). Of all cSNPs, ILS mice had a total of 49 strain-specific frameshift indels, and ISS mice had 37 frameshift indels that were strain-specific. Table 2 lists the sequence polymorphisms detected by exome sequencing of the ILS and ISS strains. Additionally, 24 candidate genes were identified based on the presence of stop codons that were either gained or lost in a strain-specific manner (Table 3). One of the most interesting of these involved the Amy2b which contained a cSNP (on the negative strand) in the ILS strain that created a TGA stop codon. The Amy2b cSNP was sequenced via the Sanger method to verify that the cSNP causing the stop codon gain in the ILS mice was strain-specific. Five of each ILS and ISS mice underwent Sanger sequencing and confirmed a homozygous cSNP present in all ILS mice and absent in all ISS mice. The cSNP found in the ILS strain disrupts the entire translated protein because it is the first exon of ten exons in Amy2b.
Table 2.
Exome sequence differences between ILS and ISS mice
| Total strain-specific exonic mutationsa | Number of stop codons gaineda | Number of stop codons losta | Number of nonsynonymous changesa | Number of synonymous changesa | Number of splicing variant cSNPsa | Frameshift indelsa | |
|---|---|---|---|---|---|---|---|
| ILS strain-specific | 4,520 (2,597) | 12 (12) | 4 (4) | 1,530 (961) | 2,787 (1,462) | 18 (17) | 61 (49) |
| ISS strain-specific | 2,918 (1,768) | 6 (6) | 1 (1) | 829 (585) | 1,931 (1,035) | 12 (10) | 39 (37) |
The number of sequence alterations detected by whole exome sequencing is shown for ILS and ISS strains. Columns from left to right list the total number of unique exonic gene variants, number of stop codon gains, number of stop codon losses, number of nonsynonymous changes, number of synonymous changes, number of splicing variants and number of frameshift indels. For each column, the first number represents a strain-specific number of variants, and the second number in parentheses represents the number of strain-specific genes
The first number is number of strain-specific variants, and the second number in parentheses is the number of strain-specific genes
Table 3.
Genes with either stop codon gains or losses in either ILS or ISS strains
| Genes with stop codon gains | Genes with stop codon lost | Function | |
|---|---|---|---|
| ILS strain-specific | |||
| Ces1b | Carboxylesterase activity | ||
| Nck2 | Recruit and bind proteins involved in regulation of receptor protein kinases, and through these regulatory activities, this protein may be involved in cytoskeletal reorganization | ||
| Heatr7b1 | Molecular binding | ||
| Olfr411 | G-protein-coupled receptor activity and olfactory receptor activity | ||
| Mep1a | Hydrolase activity and metal ion binding | ||
| AMY2B | Hydrolyze 1,4-alpha-glucoside bonds in oligosaccharides and polysaccharides, and thus catalyze the first step in digestion of dietary starch and glycogen | ||
| Zbtb5 | Zinc finger and BTB domain-containing protein 5 | ||
| Aurkc | ATP binding, kinase activity, possible role in oocytes and microtubules organization with the cetrozome during mitosis, and is over expressed in cancer cell lines | ||
| Mrgprx2 | G-protein-coupled receptor activity and neuropeptide binding | ||
| Oca2 | Arsenite trasnsmembrane activity | ||
| Defb46 | Antimicrobial peptide implicated in the resistance of epithelial surfaces to microbial colonization | ||
| Vmn2r100 | G-protein-coupled receptor | ||
| Tmem100 | Developmental protein | ||
| lfi205 | Interferon activated gene 205 | ||
| Znhit3 | Thyroid receptor-interacting protein | ||
| Scnm1 | Plays a role in RNA splicing, may contribute to the recognition of nonconsensus donor sites | ||
| Vmn2r114 | G-protein-coupled receptor | ||
| ISS strain-specific | |||
| Krt42 | Structural molecule activity | ||
| Tubb4b | GTPase activity and binding | ||
| Sipa 1l2 | Gap activity for Ras-related regulatory protiens Rap1 and Rap2 | ||
| Prex1 | Guanine-nucleotide-releasing factor | ||
| Hbq1b | Heme binding, oxygen binding | ||
| Morn4 | Unknown | ||
| Mnda | Participates in blood cell-specific responses to interferons |
Gene symbols are listed for those genes that show either a stop codon gain or loss in either the ILS or ISS strains. Functions were obtained from GeneCards’ Weizmann Institute of Science, UniProt, and MGI Mouse Genome Informatic, Jackson Labs
Discussion
The ability to survey and compare entire genomes for structural and coding differences using the complementary methods of arrayCGH and whole exome sequencing represents a powerful tool for the study of the genes and genomic variations that underlie alcohol-related phenotypes. In this study, we identified mouse strain-specific genomic structural changes and novel gene sequence variations that have not been previously identified in mouse or human alcoholic populations.
Identification of strain-specific CNVs yielded several interesting findings. Among the most potentially relevant ISS-specific CNVs were three deletions that contained genes involved in neurotransmitter release and/or brain function, and include a neuronally expressed voltage-dependent calcium channel. Given that these gene-containing deletions presumably cause phenotypic effects related to important brain functions, they are excellent candidates for follow-up study in these mice as well as in human alcoholic populations.
This paper also represents the first whole exome sequencing of the ILS and ISS strains and identifies substantial sequence differences between the two strains. Overall, ILS mice had 4,925 more strain-specific cSNPs than the ISS mice (Table 2). This, in itself, may contribute to the more severe phenotype related to alcohol sensitivity, and further investigation examining associations of these mutations with alcoholism in larger populations will prove informative. ILS mice had 756 more nonsynonymous changes and 27 more cSNP that cause splicing variants when compared to the ISS strains. In general, the ILS mice harbor a larger burden of single base substitutions than ISS mice, and many of those cSNPs are in regions that are more likely to cause functional consequences.
Although exome sequencing identified several compelling gene coding variants that may play a role in alcohol response, one of the more biologically interesting candidates was Amy2b, which contained an ILS-specific premature stop codon. Genes that show a stop codon gain are likely to have a compromised function, and the fact that the ILS stop codon occurs near the N-terminus of the predicted protein suggests that the effect likely has a severe effect on the function of the protein. The Amy2b gene is a member of the amylase gene family, which is involved in starch metabolism. In humans, the amylase gene family is composed of five genes (AMY1A, AMY1B, AMY1C, AMY2A and AMY2B) which were generated during primate evolution from one ancestral gene copy (Samuelson et al. 1990). AMY2B is the only one of the five gene members that does not have a retroviral insertion site, which likely affects transcription (Samuelson et al. 1988a, b; Yokouchi et al. 1990), and suggests that it is an older version of the gene (Samuelson et al. 1990). Amylase 1 (AMY1) is produced in the salivary glands, and AMY1 CNVs have been identified in different human populations that correlate with starch intake in those geographic regions (Perry et al. 2007; Mandel et al. 2010).
To our knowledge, AMY2B copy number variation has not been studied in the human population. According to hg19, RefSeq identifies only one copy of the AMY2B in humans, while UCSC genes identify 4 AMY2B copies. It is plausible that extra copies have been missed or collapsed by the current genome assembly. However, in dogs, AMY2B was found to be of variable copy number ranging from 4 to 30 copies. Additionally, AMY2B copy number is greater in dogs than in wolves, and the increase in gene copy number corresponds to an increase in AMY2B expression level in dogs (Axelsson et al. 2013).
While alpha-amylase 2A and 2B are pancreatic enzymes, AMY2B has been found to be present in low levels in human and mouse liver (Koyama et al. 2001; Samuelson et al. 1988a, b), a primary organ of alcohol metabolism. Additionally, McGeachin and Potter tested several animals (rats, dogs, cats, New Zealand white rabbits, as well as tissue from hogs and cows) and found that amylase is present and produced in liver cells in all of these species. Further, animals with liver damage were associated with decreased amylase levels. They hypothesized that liver amylase (different than pancreatic and salivary amylase) is likely involved in glycogen synthesis (McGeachin and Potter 1960).
Perhaps, through differential expression of AMY2B in the liver, alcohol could be metabolized differently through a different level of starch metabolism. Plants store glucose as starch which undergoes a conversion to simple sugars in the brewing process to manufacture alcohol. Amylase facilitates this conversion (Jin et al. 2013). It is conceivable that the starches in plants used to make alcohol could have an effect on alcohol metabolism in the liver. To our knowledge, no literature has been published on alcohol metabolism and AMY2B. However, given the differing strain phenotypes with regards to alcohol metabolism, the major role that the liver plays in alcohol metabolism, and the connection to possibly functional and nonfunctional AMY2B, further investigation is warranted.
Taken together, the data presented here represent the most comprehensive genomic investigation of the ILS/ISS strains so far reported. The strain-specific CNVs and coding sequence alterations that were identified provide a rich source of candidates to help explain the alcohol-related phenotypes known to distinguish these strains and may be among the genetic factors that influence differential human risk to develop alcoholism and alcohol abuse.
Supplementary Material
Acknowledgments
NIAAA [2R01 AA011853 Genome variation underlying alcohol action (JMS), 3R01 AA011853-12S1 Genome variation underlying alcohol action (JMS), R01 AA016957 Genetic studies of alcohol tolerance (RAR)].
Contributor Information
Laura Dumas, Department of Biochemistry and Molecular Genetics, University of Colorado Denver School of Medicine, Aurora, CO 80045, USA, laura.dumas@ucdenver.edu.
C. Michael Dickens, Department of Biology, University of Memphis, Memphis, TN, USA.
Nathan Anderson, Department of Biochemistry and Molecular Genetics, University of Colorado Denver School of Medicine, Aurora, CO 80045, USA.
Jonathan Davis, Department of Biochemistry and Molecular Genetics, University of Colorado Denver School of Medicine, Aurora, CO 80045, USA.
Beth Bennett, Department of Pharmaceutical Sciences, University of Colorado, Denver School of Medicine, Aurora, CO 80045, USA.
Richard A. Radcliffe, Department of Pharmaceutical Sciences, University of Colorado, Denver School of Medicine, Aurora, CO 80045, USA
James M. Sikela, Department of Biochemistry and Molecular Genetics, University of Colorado Denver School of Medicine, Aurora, CO 80045, USA
References
- Axelsson E, Ratnakumar A, Arendt M-L, Maqbool K, Webster MT, Perloski M, Liberg O, Arnemo JM, Hedhammar A, Lindblad-Toh K. The genomic signature of dog domestication reveals adaptation to a starch-rich diet. Nature. 2013;495:360–364. doi: 10.1038/nature11837. [DOI] [PubMed] [Google Scholar]
- Bennet B, Johnson TE. Development of congenics for hypnotic sensitivity to ethanol by QTL-marker-assisted counter selection. Mamm Genome. 1998;9:969–974. doi: 10.1007/s003359900908. [DOI] [PubMed] [Google Scholar]
- Bennett B, Beeson M, Gordon L, Johnson TE. Reciprocal congenics defining individual quantitative trait loci for sedative/hypnotic sensitivity to ethanol. Alcohol Clin Exp Res. 2000a;26:149–157. [PubMed] [Google Scholar]
- Bennett B, Beeson M, Gordon L, Carosone-Link P, Johnson TE. Genetic dissection of quantitative trait loci specifying sensitivity to ethanol: mapping with interval-specific congenic recombinant lines. Alcohol Clin Exp Res. 2000b;26:1615–1624. doi: 10.1097/01.ALC.0000037136.49550.B3. [DOI] [PubMed] [Google Scholar]
- Bouchery EE, Harwood HJ, Sacks JJ, Simon CJ, Brewer RD. Economic costs of excessive alcohol consumption in the U.S., 2006. Am J Prev Med. 2011;41(5):516–524. doi: 10.1016/j.amepre.2011.06.045. [DOI] [PubMed] [Google Scholar]
- Brunetti-Pierri N, Berg JS, Scaglia F, Belmont J, Bacino CA, Sahoo T, Lalani SR, Graham B, Lee B, Shinawi M, Shen J, Kang SH, Pursley A, Lotze T, Kennedy G, Lansky-Shafer S, Weaver C, Roeder ER, Grebe TA, Arnold GL, Hutchison T, Reimschisel T, Amato S, Geragthy MT, Innis JW, Obersztyn E, Nowakowska B, Rosengren SS, Bader PI, Grange DK, Naqvi S, Garnica AD, Bernes SM, Fong CT, Summers A, Walters WD, Lupski JR, Stankiewicz P, Cheung SW, Patel A. Recurrent reciprocal 1q21.1–1q21.2 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nature Genet. 2008;40:1466–1471. doi: 10.1038/ng.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette JE, Guimaraes CP, Varadarajan M, Park AS, Wuethrich I, Godarova A, Kotecki M, Cochran BH, Spooner E, Ploegh HL, Brummelkamp TR. Haploid genetic screens in human cells identify host factors used by pathogens. Science. 2009;326(5957):1231–1235. doi: 10.1126/science.1178955. [DOI] [PubMed] [Google Scholar]
- Collins AC. A review of research using short-sleep and long-sleep mice. In: McClearn GE, Deitrich RA, Erwin VG, editors. Development of animal models as pharmacogenetic tools. U.S. Government Printing Office; Washington, DC: 1981. pp. 161–170. [Google Scholar]
- Consortium International Schizophrenia. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:178–179. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler G, Kassner PD. Copy number variation in the mouse genome: implications for the mouse as a model organism for human disease. Cytogenet Genome Res. 2009;123(1–4):297–306. doi: 10.1159/000184721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlington TM, Ehringer MA, Larson C, Phang TL, Radcliffe RA. Transcriptome analysis of Inbred Long Sleep and Inbred Short Sleep Mice. Genes Brain Behav. 2013;12:263–274. doi: 10.1111/gbb.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kovel CG, Trucks H, Helbig I, Mefford HC, Baker C, Leu C, Kluck C, Muhle H, von Spiczak S, Ostertag P, Obermeier T, Kleefuß-Lie AA, Hallmann K, Steffens M, Gaus V, Klein KM, Hamer HM, Rosenow F, Brilstra EH, K-N Trenité D, Marielle Swinkels MEM, Weber YG, Unterberger I, Zimprich F, Urak L, Feucht M, Fuchs K, Møller RS, Hjalgrim H, De Jonghe P, Suls A, Rückert I-M, Wichmann H-E, Franke A, Schreiber S, Nürnberg P, Elger CE, Lerche H, Stephani U, Koeleman BPC, Lindhout D, Eichler EE, Sander T. Recurrent microdeletions at 15q11.2 and 16p13.11 predispose to idiopathic generalized epilepsies. Brain. 2010;133:23–32. doi: 10.1093/brain/awp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitrich RA. Selective breeding of mice and rats for initial sensitivity to ethanol: contributions to understanding ethanol’s actions. In: Deitrich RA, Pawlowski AA, editors. Initial Sensitivity to Alcohol. National Institute of Alcohol Abuse and Alcoholism; Rockville: 1990. pp. 7–60. [Google Scholar]
- Dick DM, Prescott C, McGue M. The genetics of substance use and substance use disorders. In: Yong-Kyu K, editor. Handbook of behavior genetics. Springer; New York: 2009. pp. 433–453. [Google Scholar]
- Dumas LD, O’Bleness MS, Davis JM, Dickens CM, Anderson N, Keeney JG, Jackson J, Sikela M, Raznahan A, Giedd J, Rapoport J, Nagamani SSC, Erez A, Brunetti-Pierri N, Sugalski R, Lupski JR, Fingerlin T, Cheung SW, Sikela JM. DUF1220-Domain Copy Number Implicated in Human Brain-Size Pathology and Evolution. The American Journal of Human Genetics. 2012;91:444–454. doi: 10.1016/j.ajhg.2012.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehringer MA, Sikela JM. Genomic approaches to the genetics of alcoholism. Alcohol Res Health. 2002;26:181–192. [PMC free article] [PubMed] [Google Scholar]
- Ehringer MA, Thompson JT, Conroy O, Xu Y, Yang F, Canniff J, Beeson M, Gordon L, Bennett B, Johnson TE, Sikela JM. High-throughput sequence identification of gene coding variants within alcohol-related QTLs. Mam Genome. 2001;12:657–663. doi: 10.1007/s00335-001-1001-x. [DOI] [PubMed] [Google Scholar]
- Ehringer MA, Thompson J, Conroy O, Yang F, Hink R, Bennett B, Johnson TE, Sikela JM. Fine mapping of polymorphic alcohol-related quantitative trait loci candidate genes using interval-specific congenic recombinant mice. Alcohol Clin Exp Res. 2002;26:160–1603. doi: 10.1097/01.ALC.0000036921.33411.67. [DOI] [PubMed] [Google Scholar]
- Grant BF, Dawson DA, Stinson FS, Chou SP, Dufour MC, Pickering RP. The 12-month prevalence and trends in DSM-IV alcohol abuse and dependence: United States. Drug Alcohol Depend. 2004;74(3):223–234. doi: 10.1016/j.drugalcdep.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Hui X, Zhu W, Wang Y, Lam KS, Zhang J, Wu D, Kraegen EW, Li Y, Xu A. Major urinary protein-1 increases energy expenditure and improves glucose intolerance through enhancing mitochondrial function in skeletal muscle of diabetic mice. J Biol Chem. 2009;284(21):14050–14057. doi: 10.1074/jbc.M109.001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillard S, Drunat S, Bendavid C, Aboura A, Etcheverry A, Journel H, Delahaye A, Pasquier L, Bonneau D, Toutain A, Burglen L, Guichet A, Pipiras E, Gilbert-Dussardier B, Benzacken B, Martin-Coignard D, Henry C, David A, Lucas J, Mosser J, David V, Odent S, Verloes A, Dubourg C. Identification of gene copy number variations in patients with mental retardation using array-CGH: novel syndromes in a large French series. Eur J Med Genet. 2010;53:66–75. doi: 10.1016/j.ejmg.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Jin Z, Mu YW, Sun JY, Li XM, Gao XL, Lu J. Proteome analysis of metabolic proteins (pI 4–7) in barley (Hordeum vulgare) malts and initial application in malt quality discrimination. J Agric Food Chem. 2013;61(2):402–409. doi: 10.1021/jf3034418. [DOI] [PubMed] [Google Scholar]
- Koyama I, Komine S, Iino N, Hokari S, Igarashi S, Alpers DH, Komoda T. α-amylase expressed in human liver is encoded by the AMY-2B gene identified in tumorous tissues. Clin Chim Acta. 2001;309:79–83. doi: 10.1016/s0009-8981(01)00501-0. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler Transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLaren EJ, Sikela JM. Cerebellar gene expression profiling and eQTL analysis in inbred mouse strains selected for ethanol sensitivity. Alcohol Clin Exp Res. 2005;29:1568–1579. doi: 10.1097/01.alc.0000179376.27331.ac. [DOI] [PubMed] [Google Scholar]
- MacLaren EJ, Bennett B, Johnson TE, Sikela JM. Expression profiling identifies novel candidate genes for ethanol sensitivity QTLs. Mamm Genome. 2006;17:147–156. doi: 10.1007/s00335-005-0065-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel AL, Peyrot des Gachons C, Plank KL, Alarcon S, Breslin PAS. Individual differences in AMY1 gene copy number, salivary α-amylase levels, and the perception of oral starch. PLoS One. 2010;5(10):e13352. doi: 10.1371/journal.pone.0013352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markel PD, DeFries JC, Johnson TE. Use of repeated measures in an analysis of ethanol-induced loss of righting reflex in inbred long-sleep and short-sleep mice. Alcohol Clin Exp Res. 1995;19:299–304. doi: 10.1111/j.1530-0277.1995.tb01506.x. [DOI] [PubMed] [Google Scholar]
- Markel PD, Bennett B, Beeson M, Gordon L, Johnson TE. Confirmation of quantitative trait loci for ethanol sensitivity in long-sleep and short-sleep mice. Genome Res. 1997;7:92–99. doi: 10.1101/gr.7.2.92. [DOI] [PubMed] [Google Scholar]
- McClearn GE, Kakihana R. Selective breeding for ethanol sensitivity: short-sleep and long-sleep mice. In: McClearn GE, Deitrich RA, Erwin VG, editors. Development of animal models as pharmacogenetic tools. U.S. Government Printing Office; Washington, DC: 1981. pp. 147–159. (DHHS Publication No. (ADM) 81-113). [Google Scholar]
- McGeachin RL, Potter BA. Amylase in isolated liver cells. J Biol Chem. 1960;235:1354–1358. [Google Scholar]
- Mefford HC, Sharp AJ, Baker C, Itsara A, Jiang Z, Buysse K, Huang S, Maloney VK, Crolla JA, Baralle D, Collins A, Mercer C, Norga K, de Ravel T, Devriendt K, Bongers EMHF, de Leeuw N, Reardon W, Gimelli S, Bena F, Hennekam RC, Male A, Gaunt L, Clayton-Smith J, Simonic I, Park SM, Mehta SG, Nik-Zainal S, Woods CG, Firth HV, Parkin G, Fichera M, Reitano S, Lo Giudice M, Li KE, Casuga I, Broomer A, Conrad B, Schwerzmann M, Raber L, Gallati S, Striano P, Coppola A, Tolmie JL, Tobias ES, Lilley C, Armengol L, Spysschaert Y, Verloo P, De Coene A, Goossens L, Mortier G, Speleman F, van Binsbergen E, Nelen MR, Hochstenbach R, Poot M, Gallagher L, Gill M, McClellan J, King MC, Regan R, Skinner C, Stevenson RE, Antonarakis SE, Chen CF, Estivill X, Menten B, Gimelli G, Gribble S, Schwartz S, Sutcliffe JS, Walsh T, Knight SJL, Sebat J, Romano C, Schwartz CE, Veltman JA, de Vries BBA, Vermeesch JR, Barber JCK, Willatt L, Tassabehji M, Eichler EE. Recurrent rearrangements of chromosome 1q21.1–1q21.2 and variable pediatric phenotypes. N Engl J Med. 2008;359:1685–1699. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry GH, Dominy NJ, Claw KG, Lee AS, Fiegler H, Redon R, Werner J, Villanea FA, Mountain JL, Misra R, Carter NP, Lee Ch, Stone AC. Diet and the evolution of human amylase gene copy number variation. Nat Genet. 2007;39:1256–1260. doi: 10.1038/ng2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, Almeida J, Bacchelli E, Bader GD, Bailey AJ, Baird G, Battaglia A, Berney T, Bolshakova N, Bölte S, Bolton PF, Bourgeron T, Brennan S, Brian J, Bryson SE, Carson AR, Casallo G, Casey J, Chung BH, Cochrane L, Corsello C, Crawford EL, Crossett A, Cytrynbaum C, Dawson G, de Jonge M, Delorme R, Drmic I, Duketis E, Duque F, Estes A, Farrar P, Fernandez BA, Folstein SE, Fombonne E, Freitag CM, Gilbert J, Gillberg C, Glessner JT, Goldberg J, Green A, Green J, Guter SJ, Hakonarson H, Heron EA, Hill M, Holt R, Howe JL, Hughes G, Hus V, Igliozzi R, Kim C, Klauck SM, Kolevzon A, Korvatska O, Kustanovich V, Lajonchere CM, Lamb JA, Laskawiec M, Leboyer M, Le Couteur A, Leventhal BL, Lionel AC, Liu XQ, Lord C, Lotspeich L, Lund SC, Maestrini E, Mahoney W, Mantoulan C, Marshall CR, McConachie H, McDougle CJ, McGrath J, McMahon WM, Merikangas A, Migita O, Minshew NJ, Mirza GK, Munson J, Nelson SF, Noakes C, Noor A, Nygren G, Oliveira G, Papanikolaou K, Parr JR, Parrini B, Paton T, Pickles A, Pilorge M, Piven J, Ponting CP, Posey DJ, Poustka A, Poustka F, Prasad A, Ragoussis J, Renshaw K, Rickaby J, Roberts W, Roeder K, Roge B, Rutter ML, Bierut LJ, Rice JP, Salt J, Sansom K, Sato D, Segurado R, Sequeira AF, Senman L, Shah N, Sheffield VC, Soorya L, Sousa I, Stein O, Sykes N, Stoppioni V, Strawbridge C, Tancredi R, Tansey K, Thiruvahindrapduram B, Thompson AP, Thomson S, Tryfon A, Tsiantis J, Van Engeland H, Vincent JB, Volkmar F, Wallace S, Wang K, Wang Z, Wassink TH, Webber C, Weksberg R, Wing K, Wittemeyer K, Wood S, Wu J, Yaspan BL, Zurawiecki D, Zwaigenbaum L, Buxbaum JD, Cantor RM, Cook EH, Coon H, Cuccaro ML, Devlin B, Ennis S, Gallagher L, Geschwind DH, Gill M, Haines JL, Hallmayer J, Miller J, Monaco AP, Nurnberger JI, Jr, Paterson AD, Pericak-Vance MA, Schellenberg GD, Szatmari P, Vicente AM, Vieland VJ, Wijsman EM, Scherer SW, Sutcliffe JS, Betancur C. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampietro M, Iolascon A. Molecular pathology of Crigler–Najjar type I and II and Gilbert’s Syndrome. Haematologica. 1999;84(2):150–157. [PubMed] [Google Scholar]
- Samuelson LC, Keller PR, Darlington GJ, Meisler MH. Glucocorticoid and developmental regulation of amylase mRNAs in mouse liver cells. Mol Cell Biol. 1988a;8:3857–3863. doi: 10.1128/mcb.8.9.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelson LC, Wiebauer K, Gumuci DL, Meisler MH. Expression of the human amylase genes: recent origin of a salivary amylase promoter from an actin pseudogene. Nucleic Acids Res. 1988b;16:8261–8275. doi: 10.1093/nar/16.17.8261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelson LC, Biebauer K, Snow CM, Meisler MH. Retroviral and pseudogene insertion sites reveal the lineage of human salivary and pancreatic amylase genes from a single gene during primate evolution. Mol Cell Biol. 1990;10:2513–2520. doi: 10.1128/mcb.10.6.2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from next-generation sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins-chow DE, Pavan WJ. Genomic copy number and expression variation within the C57BL/6J inbred mouse strain. Genome Res. 2008;18(1):60–66. doi: 10.1101/gr.6927808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischenfeldt J, Symmons O, Spitz F, Korbel JO. Phenotypic impact of genomic structural variation: insights from and for human disease. Nat Rev Genet. 2013;14:125–138. doi: 10.1038/nrg3373. [DOI] [PubMed] [Google Scholar]
- Wollscheid B, Bausch-Fluck D, Henderson C, O’Brien R, Bibel M, Schiess R, Aebersold R, Watts JD. Mass-spectrometric identification and relative quantification of N-linked cell surface glycoproteins. Nat Biotechnol. 2009;27(4):378–386. doi: 10.1038/nbt.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Ehringer M, Yang F, Sikela JM. Comparison of global brain gene expression profiles between inbred long-sleep and inbred short-sleep mice by high-density gene array hybridization. Alcohol Clin Exp Res. 2001;25:810–818. [PubMed] [Google Scholar]
- Yokouchi H, Horii A, Emi M, Tomita N, Doi S, Ogawa M, Mori T, Matsubara K. Cloning and characterization of a third type of human α-amylase gene, AMY2B. Gene. 1990;90:281–286. doi: 10.1016/0378-1119(90)90191-s. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Jiang L, Rui L. Identification of MUP1 as a regulator for glucose and lipid metabolism in mice. J Biol Chem. 2009;284(17):11152–11159. doi: 10.1074/jbc.M900754200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


