Abstract
Mandibulofacial dysostosis with microcephaly (MFDM) is a multiple malformation syndrome comprising microcephaly, craniofacial anomalies, hearing loss, dysmorphic features, and, in some cases, esophageal atresia. Haploinsufficiency of a spliceosomal GTPase, U5–116 kDa/EFTUD2, is responsible. Here, we review the molecular basis of MFDM in the 69 individuals described to date, and report mutations in 38 new individuals, bringing the total number of reported individuals to 107 individuals from 94 kindreds. Pathogenic EFTUD2 variants comprise 76 distinct mutations and seven microdeletions. Among point mutations, missense substitutions are infrequent (14 out of 76; 18%) relative to stop-gain (29 out of 76; 38%), and splicing (33 out of 76; 43%) mutations. Where known, mutation origin was de novo in 48 out of 64 individuals (75%), dominantly inherited in 12 out of 64 (19%), and due to proven germline mosaicism in four out of 64 (6%). Highly penetrant clinical features include, microcephaly, first and second arch craniofacial malformations, and hearing loss; esophageal atresia is present in an estimated ~27%. Microcephaly is virtually universal in childhood, with some adults exhibiting late “catch-up” growth and normocephaly at maturity. Occasionally reported anomalies, include vestibular and ossicular malformations, reduced mouth opening, atrophy of cerebral white matter, structural brain malformations, and epibulbar dermoid. All reported EFTUD2 mutations can be found in the EFTUD2 mutation database (http://databases.lovd.nl/shared/genes/EFTUD2).
Keywords: EFTUD2, mandibulofacial dysostosis with microcephaly, MFDM, mandibulofacial dysostosis Guion-Almeida type, mandibulofacial dysostosis, microcephaly
Background
Mandibulofacial dysostosis with microcephaly (MFDM; MIM# 610536) is a multiple malformation syndrome comprising microcephaly, first and second branchial arch anomalies (Pierre-Robin sequence, malar hypoplasia, zygomatic clefting, microtia, middle ear malformations, choanal atresia, and/or auditory atresia), hearing loss, dysmorphic features, and variable systemic malformations (esophageal atresia, short stature, cardiac and/or genitourinary anomalies, and proximally-placed thumbs) [Guion-Almeida et al., 2006; Wieczorek et al., 2007; Wieczorek et al., 2009]. Haploinsufficiency of EFTUD2 (MIM# 603892), encoding U5–116 kDa, a spliceosomal GTPase, is responsible [Lines et al., 2012]. The majority of described mutations are de novo, with a variety of documented mutation types (missense, nonsense, frameshift, splice site, and complete or partial gene deletions, including cytogenetically-visible deletions); however, autosomal dominant inheritance and, less commonly, germline mosaicism have been reported [Lines et al., 2014].
Variants
Previous to this report, the literature includes 69 individuals (64 kindreds) with mutations of EFTUD2 (NM 004247.3), excluding individuals with cytogenetically visible chromosomal aberrations [Lines et al., 2012; Gordon et al., 2012; Need et al., 2012; Bernier et al., 2012; Luquetti et al., 2013; Voigt et al., 2013; Lehalle et al., 2014; Gandomi et al., 2015; Smigiel et al., 2015; Sarkar et al., 2015; Vincent et al., 2015]. Here, we summarize all published affected individuals and present genotypic and phenotypic data from a further 38 affected individuals belonging to 30 kindreds (Supp. Tables S1 and S2, Table 1, and Fig. 1). Across the entire series, the 83 distinct EFTUD2 mutations are classified as follows: seven large (whole-gene or multi-exon) deletions, 16 frameshift, 12 nonsense, 32 splice site, one small deletion/duplication, 14 missense, and one intronic change previously suggested to create a novel splice donor site. Of 64 individuals for whom parental genotypes were available, de novo inheritance was demonstrated in 48 (75%), whereas mutations were inherited from an affected parent in 12 (19%), or via germline mosaicism in four (6%; two sibling pairs with confirmed paternity but normal parental sequencing) (Supp. Fig. S1). Three of the splice-site mutations presented here have further been validated by RT-PCR (Supp. Fig. S2).
Table 1.
Penetrance of Clinical Findings in MFDM
| Feature | This study |
All reported individualsa |
Estimated penetrance (%) |
95% CI (%) |
|---|---|---|---|---|
| Craniofacial | ||||
| Micrognathia | 34/34 | 87/89 | 98 | 92–99 |
| Small or dysplastic pinna(e) | 31/34 | 84/87 | 97 | 90–99 |
| Malar hypoplasia | 34/36 | 78/84 | 93 | 85–97 |
| Hearing loss | 31/35 | 69/83 | 83 | 74–90 |
| Conductive | 16/27 | 32/51 | 63 | 49–75 |
| Mixed | 10/27 | 13/51 | 25 | 16–39 |
| Sensorineural | 1/27 | 6/51 | 12 | 6–23 |
| Auditory atresia/stenosis | 19/29 | 47/73 | 64 | 53–74 |
| Vestibular system abnormalities | 1/8b | 14/25b | 56b | 37–73b |
| Ossicular abnormalities | 4/8b | 8/15b | 53b | 30–75b |
| Facial asymmetry | nr | 25/47 | 53 | 39–67 |
| Preauricular tag(s) | 19/34 | 45/86 | 52 | 42–63 |
| Cleft palate | 20/35 | 41/88 | 47 | 37–57 |
| Choanal atresia | 6/34 | 27/83 | 33 | 23–43 |
| Neonatal resuscitation | 9/32 | 14/46 | 30 | 19–45 |
| Tracheostomy | 7/34 | 10/50 | 20 | 11–33 |
| Extracranial | ||||
| Thumb anomalies | 10/32 | 24/77 | 31 | 22–42 |
| Heart defects | 9/38 | 28/89 | 31 | 23–42 |
| Esophageal atresia/TEF | 5/35 | 23/85 | 27 | 19–37 |
| Development | ||||
| Developmental delay | 31/31 | 83/83 | 100 | 96–100 |
| Microcephaly | 27/33 | 78/89 | 88 | 79–93 |
| Congenital | 10/18 | 34/53 | 64 | 51–76 |
| Postnatal | 8/18 | 19/53 | 36 | 24–49 |
| Seizures | 9/30 | 21/77 | 27 | 19–38 |
Includes cases in: Lines et al. (2012), Need et al. (2012), Bernier et al. (2012), Gordon et al. (2012) (excluding individual #12), Voigt et al. (2013), Luquetti et al. (2013), Lehalle et al. (2014), Smigiel et al. (2015), and this study.
This feature has not been assessed in the majority of reported patients (estimate is likely inaccurate due to ascertainment bias).
Figure 1.
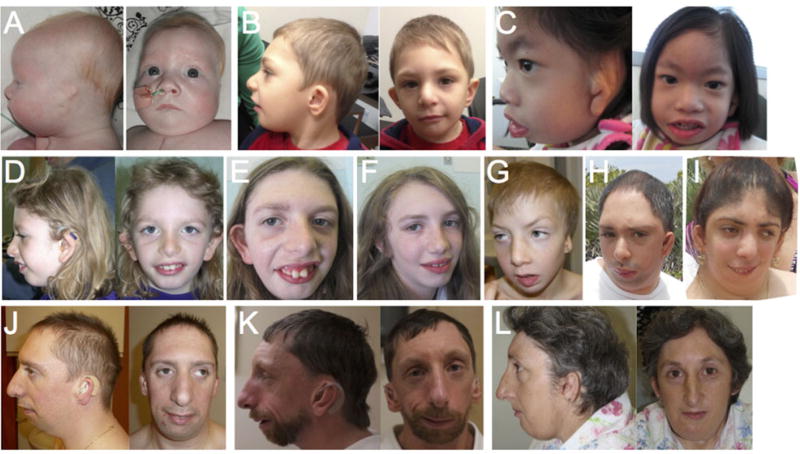
Craniofacial morphology in children and adults with mutations in EFTUD2. Panel A: Individual #62 (age 4 months); B: #52 (3.25 years); C: #60 (3.5 years); D: #10a (age 4.5 years); E: #10a (15 years); F: #10b (12 years); G: #57 (age not specified); H: #79a (28 years); I: #79b (25 years); J: #12 (31 years); K: #13 (43 years); L: #34 (47 years). The typical gestalt associated with MFDM (convex facial profile with micrognathia, midface hypoplasia, and sloping forehead, strong supraorbital ridges, high nasal bridge with a prominent ridge and rounded tip, ear anomalies including microtia, dysplastic pinnae, deficient superior helix, posteriorly “squared” earlobes, and/or preauricular tags), while seen in the majority of affected individuals, may occasionally be subtle (e.g., panels F and L).
Clinical Relevance
To better define the spectrum of EFTUD2-associated clinical symptoms, we reviewed the clinical features of all newly reported individuals in this study (Supp. Table S2), and tabulated the feature-specific penetrance of each of the major clinical findings across the entire cohort of all known affected individuals (Table 1) [Lines et al., 2012;Need et al., 2012; Bernier et al., 2012; Gordon et al., 2012; Voigt et al., 2013; Luquetti et al., 2013; Lehalle et al., 2014, Smigiel et al., 2015]. Highly penetrant features seen in >80% of individuals include developmental delay, microcephaly (here defined as occipitofrontal circumference (OFC) two or more standard deviations below mean), micrognathia, malar hypoplasia, small or dysplastic pinnae, and hearing loss (typically conductive, but occasionally sensorineural or mixed). Less frequent, but diagnostically useful, manifestations include preauricular tag(s) (52%), auditory canal atresia or stenosis (64%), tracheoesophageal fistula (TEF)/esophageal atresia or stenosis (27%), thumb abnormalities (31%), and choanal atresia or stenosis (33%; likely under-reported). Zygomatic arch hypoplasia/clefting and middle ear abnormalities (hypoplastic, fused, or absent ossicles, complete absence of middle ear structures, or hypoplasia/absence of semicircular canals) appear to be relatively frequent, although these features, which are best appreciated by computed tomography, have not been assessed systematically in most patients. The wide range of minor anomalies described in <10% of affected individuals includes: Renal anomalies (nineindividuals), cryptorchidism and/or small scrotum (seven), reduced mouth opening (seven), hemivertebrae or posterior dysraphism (six), cerebral and/or white matter atrophy (six), other CNS abnormalities (six; various including olfactory bulb agenesis; pontine hypoplasia; cerebellar hypoplasia, delayed gyration, delayed myelination, or exencephaly), absence of (naso-)lacrimal ducts (five), scoliosis/kyphosis (five), strabismus (four), absent 12th rib pair (four), epibulbar dermoid (three), branchial cleft and/or remnant (two), laryngeal cleft (one), midline mandibular defect (one), and gastric malrotation (one).
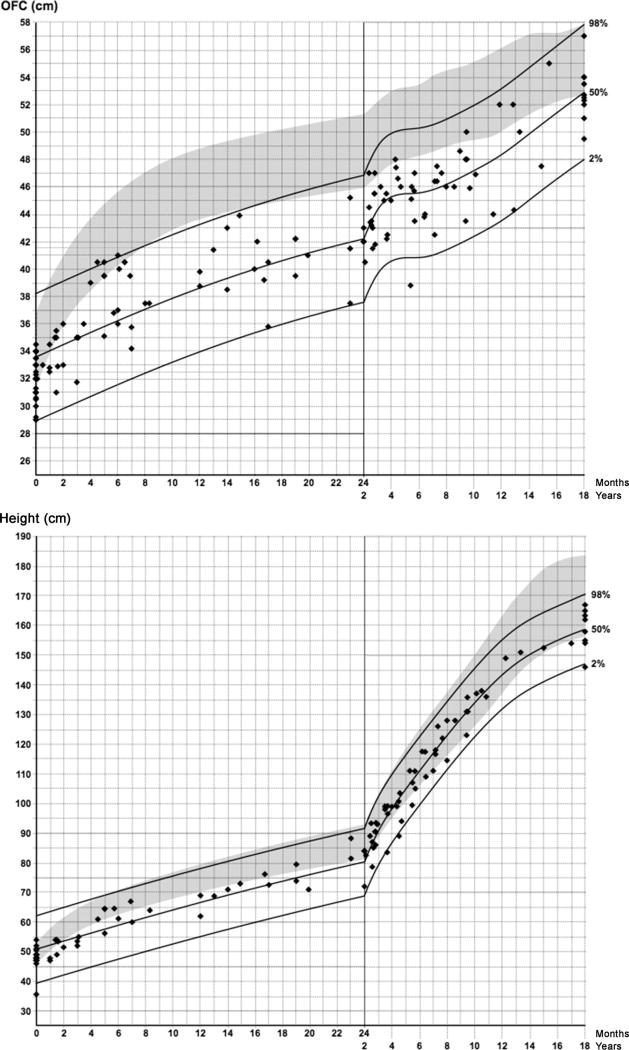
Although microcephaly was seen in all subjects in the original gene discovery cohort [Lines et al. 2012], several normocephalic individuals have since been reported [Gordon et al., 2012; Luquetti et al., 2013; Lehalle et al., 2014]. To study the penetrance of microcephaly in MFDM, we reviewed all available primary growth data in 57 affected individuals (Fig. 2). As described previously, microcephaly may be of either congenital or postnatal onset [Lines et al., 2012]. Despite the slower rate of cephalic growth during infancy and childhood, some affected individuals go on to exhibit gradual “catch-up” in OFC growth, with final adult OFC within the normal range. It may therefore be useful to review historical growth charts in normocephalic adults when considering the diagnosis of MFDM. Linear growth in MFDM may be normal or reduced (Fig. 2).
Figure 2.
Growth in MFDM. Primary growth data from 34 previously unpublished affected individuals, and 23 published affected individuals [Lines et al., 2012, Luquetti et al., 2013, Voigt et al., 2013, Sarkar et al., 2015] were modelled using the LMS method, based on a local generalized AIC criterion with smoothing penalty k = 10. The 2nd, 50th, and 98th percentile LMS curves (solid lines) are as shown. Each point represents an individual measurement from an affected individual; dates were corrected for prematurity if applicable. Top: Occipitofrontal circumference (OFC). Shaded area represents sex-averaged general population reference (−2SD to +2SD) [Nellhaus G, 1968]. Bottom: Linear growth versus general sex-averaged general population reference (3rd to 97th centile) from 2014 World Health Organization Growth Charts for Canada (http://www.whogrowthcharts.ca).
Although developmental delay is nearly uniform in MFDM, developmental outcomes vary significantly. The mean reported age of independent walking was 26.7 months in this cohort (n = 23), and 26.4 months for all patients (range: 13–60 months; n = 38). Among verbal individuals, the mean reported age of first meaningful word was 26.4 months (n = 24), and 27.4 months (range: 12 months to >5.5 years; n = 32) for all patients. Of adult patients in our cohort, one was a ward of the state, four lived with parents, one lived in a group home, and two lived in their own home, or with a spouse. At least one individual was able to carry out simple paid work outside of the home, and another was able to complete high school. Other individuals were nonverbal and dependent on caregivers for daily activities into adulthood, with significant behavioral difficulties. Of individuals for whom parental segregation analysis has been performed, dominant transmission was documented in 12 out of 64 meioses (19%), and germline mosaicism was proven in a further four out of 64 meioses (6%). These figures, which are higher than recognized previously, should be considered when counseling apparently unaffected parents with respect to recurrence risks.
Biological Relevance
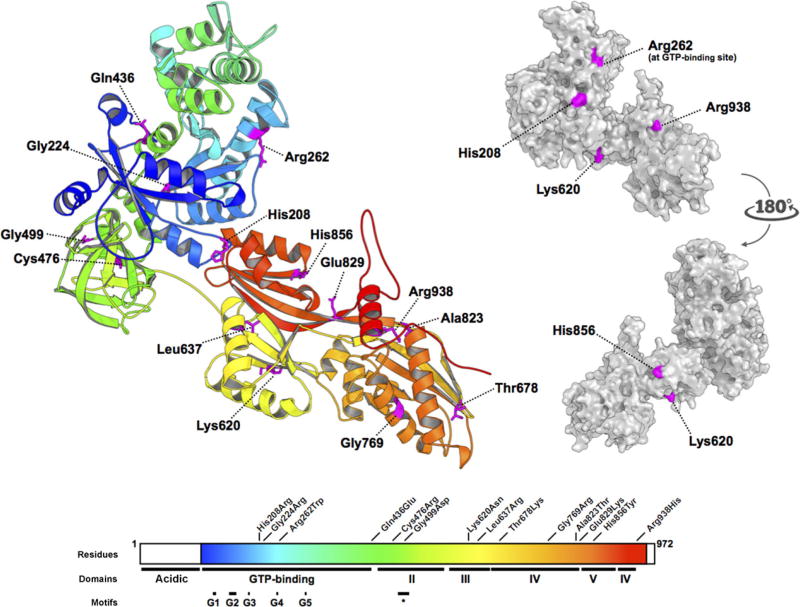
In the Saccharomyces cerevisiae U4/U5/U6 small nuclear ribonucleoprotein complex (“tri-snRNP”), the EFTUD2 orthologue Snu114p bridges the U5-snRNP helicases Brr2p and Prp8p, and several subunits of the U4/U6 snRNP complex [Häcker et al., 2008]. By analogy with the ribosomal elongation factor EF-2, Snu114p has been proposed to undergo a significant GTP-dependent conformational change, allowing the dissociation of U5 and U4/U6 snRNPs during splicing. Protein-protein and protein–GTP interactions are therefore both essential to EFTUD2’s function. In a structural model of EFTUD2, four [p.(His208Arg), p.(Lys620Asn), p.(His856Tyr), and p.(Arg938His)] of 14 described missense substitutions in MFDM alter basic, surface-forming residues that are potentially available for protein–protein interactions (Fig. 3). A fifth mutation, p.(Arg262Trp), is a recurrent mutation of a conserved residue in EFTUD2’s GTP-binding site observed in three unrelated families [Lines et al., 2012; Smigiel et al., 2015; this study]. The remaining nine missense substitutions, which are predicted to be interior to EFTUD2, could conceivably affect protein stability by any of several mechanisms, including effects on: protein stability, conformation, localization, and/or post-translational modifications.
Figure 3.
All EFTUD2 missense substitutions described to date. (Based on Fabrizio et al., 1997 and Lines et al., 2012). Residues 114–957 of EFTUD2 were modelled on the crystal structure of S. cerevisiae ribosomal elongation factor 2 (eEF2) (PDB: 1N0U). Missense substitutions are distributed throughout all domains of the protein. Conserved motifs identified by Fabrizio et al. (1997), including the GTP-binding domains G1 through G5, and the short conserved domain II motif (∗), are shown. The side chains of His208, Arg262, Lys620, and His856, and Arg938, all basic residues, are predicted to be surface-forming, whereas the sidechains of Gln436, Leu637, Thr678, and Glu829, Gly224, Cys476, Gly499, Gly769, and Ala823 are interior to the model.
Considering the entire allelic series for EFTUD2, the high proportion of truncating (frameshift, nonsense, or splice-site) mutations, and several instances of whole-gene or multi-exon deletions, suggests haploinsufficiency to be the predominant mechanism. How partial deficiency of a ubiquitously expressed spliceosomal subunit causes the specific pattern of malformations seen in MFDM is unknown. Craniofacial anomalies similar to those seen in MFDM are observed in other disorders of ribonucleoprotein metabolism and/or ribosome biogenesis, including: Nager syndrome (SF3B4) [Bernier et al., 2012], Cerebro-Costo-Mandibular syndrome (SNRPB) [Lynch et al., 2014], Richieri-Costa-Pereira syndrome (EIF4A3) [Favaro et al., 2014], Treacher Collins syndrome (TCOF1, POLR1C, POLR1D) [Treacher Collins Syndrome Collaborative Group 1996; Dauwerse et al., 2011], and Diamond-Blackfan anemia (multiple ribosomal genes) [Ruggero and Shimamura, 2014; Gripp et al., 2014]. In Treacher Collins syndrome, the prototype for this group of disorders, “nucleolar stress” as a result of impaired ribosome biogenesis causes p53-dependent attrition of a specific population of neural crest cells destined for the first and second arches [Dixon et al., 2006; Jones et al., 2008]. Similarly, in a Danio rerio morpholino model of Richieri-Costa-Pereira syndrome, deficiency of Eif4a3, an exon-junction protein deposited after splicing events, produces similar effects on the developing cranial neural crest [Singh et al., 2012; Favaro et al., 2014]. Craniofacial defects are therefore common to disorders of several different types of ribonucleoprotein complexes (ribosome, spliceosome, and exon–junction complex), and the nucleotide precursor defects Miller Syndrome (DHODH) and methotrexate toxicity [Ng et al., 2010], although the intermediate steps in this pathway are obscure at present.
Database
In order to establish a central repository for all known EFTUD2 mutations, we (D.B.) maintain the EFTUD2 Mutation Database (http://databases.lovd.nl/shared/genes/EFTUD2), according to the Leiden Open Variation Database (LOVD) system(currently version 3.0, build 12) [Fokkema et al., 2011]. As far as we are aware, all described mutations have been deposited in the EFTUD2 Mutation Database.
Diagnostic Relevance
The experience with EFTUD2 testing in MFDM suggests that sensitivity of sequencing and deletion testing approaches 100%.We are aware of only one described case of apparent nonpenetrance: the paternally-inherited deep-intronic change (c.2347+66A>G) reported in Gordon et al. (2012), for which he have neither a detailed clinical description of the parent in question, nor confirmatory RT-PCR studies to verify abnormal splicing. We consider individual #12 from the same publication to have another diagnosis due to the lack of characteristic features or a pathogenic EFTUD2 variant (see legend of Supp. Table S1 for details). Therefore, to our knowledge, there are no proven instances of nonpenetrance in an individual with a bona fide EFTUD2 mutation. The presence of an EFTUD2 mutation compatible with haploinsufficiency, particularly when de novo, is therefore highly compelling evidence to establish the diagnosis, although this can equally be determined based on clinical criteria [Lines et al., 2014].
Future Prospects
Because of the small number of total cases reported to date, and inherent clinical bias towards ascertaining “classic” (i.e., severe) cases, it is tempting to speculate that milder or attenuated EFTUD2 phenotypes, such as those with comparatively mild missense substitutions, may remain to be identified. The inclusion of EFTUD2 on next-generation-based sequencing panels should facilitate the recognition of the full clinical spectrum of MFDM. The main biological questions to be investigated include (i) which RNA transcripts are relevant to the developmental abnormalities seen in MFDM patients, and (ii) whether/how abnormal splicing produces “nucleolar stress” (impaired ribosome biogenesis) in the developing first- and second-arch-destined neural crest. Because this is a disorder characterized by haploinsufficiency, the phenotypic effect of pharmacologically or otherwise increasing the expression of the remaining EFTUD2 allele in a relevant animal model may warrant investigation.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the participation of the study participants and their families, without whom this work would not be possible. We thank Dr. Mark J. Stephan for his contribution to this work. We thank UCLA Clinical Genomics Center for their assistance with recruiting two of the individuals in this study.
Contract grant sponsors: Genome Canada; the Canadian Institutes of Health Research; the Ontario Genomics Institute; Ontario Research Fund; Genome Quebec; Children’s Hospital of Eastern Ontario Foundation; Resident Research Award from Physician’s Services Incorporated (PSI) Foundation.
Footnotes
Additional Supporting Information may be found in the online version of this article.
Ethical Compliance
Duly informed consent was obtained from all individuals participating in this study. The research protocol was approved by the Children’s Hospital of Eastern Ontario Research Ethics Board, and clinical data were obtained in a manner conforming with research ethics board and funding agency guidelines.
Disclosure statement: The authors declare no conflict of interest.
References
- Bernier FP, Caluseriu O, Ng S, Schwartzentruber J, Buckingham KJ, Innes AM, Jabs EW, Innis JW, Schuette JL, Gorski JL, Byers PH, Andelfinger G, et al. Haploinsufficiency of SF3B4, a component of the pre-mRNA spliceosomal complex, causes Nager syndrome. Am J Hum Genet. 2012;90:925–933. doi: 10.1016/j.ajhg.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauwerse JG, Dixon J, Seland S, Ruivenkamp CA, van Haeringen A, Hoefsloot LH, Peters DJ, Boers AC, Daumer-Haas C, Maiwald R, Zweier C, Kerr B, et al. Mutations in genes encoding subunits of RNA polymerases I and III cause Treacher Collins syndrome. Nat Genet. 2011;43:20–22. doi: 10.1038/ng.724. [DOI] [PubMed] [Google Scholar]
- Dixon J, Jones NC, Sandell LL, Jayasinghe SM, Crane J, Rey JP, Dixon MJ, Trainor PA. Tcof1/Treacle is required for neural crest cell formation and proliferation deficiencies that cause craniofacial abnormalities. Proc Natl Acad Sci USA. 2006;103:13403–13408. doi: 10.1073/pnas.0603730103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Laggerbauer B, Lauber J, Lane WS, Lührmann R. An evolutionarily conserved U5 snRNP-specific protein is a GTP-binding factor closely related to the ribosomal translocase EF-2. EMBO J. 1997;16:4092–4106. doi: 10.1093/emboj/16.13.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokkema IFAC, Taschner PEM, Schaafsma GCP, Celli J, Laros JFJ, den Dunnen JT. LOVD v.2.0: the next generation in gene variant databases. Hum Mutat. 2011;32:557–563. doi: 10.1002/humu.21438. [DOI] [PubMed] [Google Scholar]
- Gandomi SK, Parra M, Reeves D, Yap V, Gau CL. Array-CGH is an effective first-tier diagnostic test for EFTUD2-associated congenital mandibulofacial dysostosis with microcephaly. Clin Genet. 2015;87:80–84. doi: 10.1111/cge.12328. [DOI] [PubMed] [Google Scholar]
- Gordon CT, Petit F, Oufadem M, Decaestecker C, Jourdain AS, Andrieux J, Malan V, Alessandri JL, Baujat G, Baumann C, Boute-Benejean O, Caumes R, et al. EFTUD2 haploinsufficiency leads to syndromic oesophageal atresia. J Med Genet. 2012;49:737–746. doi: 10.1136/jmedgenet-2012-101173. [DOI] [PubMed] [Google Scholar]
- Gripp KW, Curry C, Olney AH, Sandoval C, Fisher J, Chong JX, UW Center for Mendelian Genomics. Pilchman L, Sahraoui R, Stabley DL, Sol-Church K. Diamond-Blackfan anemia with mandibulofacial dystostosis is heterogeneous, including the novel DBA genes TSR2 and RPS28. Am J Med Genet A. 2014;164A:2240–2249. doi: 10.1002/ajmg.a.36633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guion-Almeida ML, Zechi-Ceide RM, Vendramini S, Tabith Júnior A. A new syndrome with growth and mental retardation, mandibulofacial dysostosis, microcephaly, and cleft palate. Clin Dysmorphol. 2006;15:171–174. doi: 10.1097/01.mcd.0000220603.09661.7e. [DOI] [PubMed] [Google Scholar]
- Guion-Almeida ML, Vendramini-Pittoli S, Passos-Bueno MR, Zechi-Ceide RM. Mandibulofacial syndrome with growth and mental retardation, microcephaly, ear anomalies with skin tags, and cleft palate in a mother and her son: autosomal dominant or X-linked syndrome? Am J Med Genet A. 2009;149A:2762–2764. doi: 10.1002/ajmg.a.32816. [DOI] [PubMed] [Google Scholar]
- Häcker I, Sander B, Golas MM, Wolf E, Karagöz E, Kastner B, Stark H, Fabrizio P, Lührmann R. Localization of Prp8, Brr2, Snu114 and U4/U6 proteins in the yeast tri-snRNP by electron microscopy. Nat Struct Mol Biol. 2008;15:1206–1212. doi: 10.1038/nsmb.1506. [DOI] [PubMed] [Google Scholar]
- Jones NC, Lynn ML, Gaudenz K, Sakai D, Aoto K, Rey JP, Glynn EF, Ellington L, Du C, Dixon J, Dixon MJ, Trainor PA. Prevention of the neurocristopathy Treacher Collins syndrome through inhibition of p53 function. Nat Med. 2008;14:125–133. doi: 10.1038/nm1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehalle D, Gordon CT, Oufadem M, Goudefroye G, Boutaud L, Alessandri JL, Baena N, Baujat G, Baumann C, Boute-Benejean O, Caumes R, Decaestecker C, et al. Delineation of EFTUD2 haploinsufficiency-related phenotypes through a series of 36 patients. Hum Mutat. 2014;35:478–485. doi: 10.1002/humu.22517. [DOI] [PubMed] [Google Scholar]
- Lines MA, Huang L, Schwartzentruber J, Douglas SL, Lynch DC, Beaulieu C, Guion-Almeida ML, Zechi-Ceide RM, Gener B, Gillessen-Kaesbach G, Nava C, Baujat G, et al. Haploinsufficiency of a spliceosomal GTPase encoded by EFTUD2 causes mandibulofacial dysostosis with microcephaly. Am J Hum Genet. 2012;90:369–377. doi: 10.1016/j.ajhg.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lines M, Hartley T, Boycott K. Mandibulofacial dysostosis with microcephaly. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Dolan CR, Fong CT, Smith RJH, Stephens K, editors. GeneReviews® [Internet] Seattle, Washington: University of Washington, Seattle; 2014. pp. 1993–2015. [Google Scholar]
- Luquetti DV, Hing AV, Rieder MJ, Nickerson DA, Turner EH, Smith J, Park S, Cunningham ML. "Mandibulofacial dysostosis with microcephaly" caused by EFTUD2 mutations: expanding the phenotype. Am J Med Genet A. 2013;161A:108–113. doi: 10.1002/ajmg.a.35696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch DC, Revil T, Schwartzentruber J, Bhoj EJ, Innes AM, Lamont RE, Lemire EG, Chodirker BN, Taylor JP, Zackai EH, McLeod DR, Kirk EP, et al. Disrupted auto-regulation of the spliceosomal gene SNRPB causes cerebro-costo-mandibular syndrome. Nat Commun. 2014;5:4483. doi: 10.1038/ncomms5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Need AC, Shashi V, Hitomi Y, Schoch K, Shianna KV, McDonald MT, Meisler MH, Goldstein DB. Clinical application of exome sequencing in undiagnosed genetic conditions. J Med Genet. 2012;49:353–361. doi: 10.1136/jmedgenet-2012-100819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, Dent KM, Huff CD, Shannon PT, Jabs EW, Nickerson DA, Shendure J, Bamshad MJ. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nellhaus G. Head circumference from birth to eighteen years. Practical composite international and interracial graphs. Pediatrics. 1968;41:106–114. [PubMed] [Google Scholar]
- Favaro FP, Alvizi L, Zechi-Ceide RM, Bertola D, Felix TM, de Souza J, Raskin S, Twigg SR, Weiner AM, Armas P, Margarit E, Calcaterra NB, et al. A noncoding expansion in EIF4A3 causes Richieri-Costa-Pereira syndrome, a craniofacial disorder associated with limb defects. Am J Hum Genet. 2014;94:120–128. doi: 10.1016/j.ajhg.2013.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero D, Shimamura A. Marrow failure: a window into ribosome biology. Blood. 2014;124:2784–2792. doi: 10.1182/blood-2014-04-526301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A, Emrick LT, Smith EM, Austin EG, Yang Y, Hunter JV, Scaglia F, Lalani SR. Novel de novo mutations in EFTUD2 detected by exome sequencing in mandibulofacial dysostosis with Microcephaly syndrome. Am J Med Genet A. 2015;167A:914–918. doi: 10.1002/ajmg.a.36948. [DOI] [PubMed] [Google Scholar]
- Smigiel R, Bezniakow N, Jakubiak A, Błoch M, Patkowski D, Obersztyn E, Sasiadek MM. Phenotype analysis of Polish patients with mandibulofacial dysostosis type Guion-Almeida associated with esophageal atresia and choanal atresia caused by EFTUD2 gene mutations. J Appl Genet. 2015;56:199–204. doi: 10.1007/s13353-014-0255-4. [DOI] [PubMed] [Google Scholar]
- Singh G, Kucukural A, Cenik C, Leszyk JD, Shaffer SA, Weng Z, Moore MJ. The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell. 2012;151:750–764. doi: 10.1016/j.cell.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treacher Collins Syndrome Collaborative Group. Positional cloning of a gene involved in the pathogenesis of Treacher Collins syndrome. Nat Genet. 1996;12:130–136. doi: 10.1038/ng0296-130. [DOI] [PubMed] [Google Scholar]
- Vincent M, Geneviève D, Ostertag A, Marlin S, Lacombe D, Martin-Coignard D, Coubes C, David A, Lyonnet S, Vilain C, Dieux-Coeslier A, Manouvrier S, et al. Treacher Collins syndrome: a clinical and molecular study based on a large series of patients. Genet Med. 2015 doi: 10.1038/gim.2015.29. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Voigt C, Mégarbané A, Neveling K, Czeschik JC, Albrecht B, Callewaert B, von Deimling F, Hehr A, Falkenberg Smeland M, König R, Kuechler A, Marcelis C, et al. Oto-facial syndrome and esophageal atresia, intellectual disability and zygomatic anomalies - expanding the phenotypes associated with EFTUD2 mutations. Orphanet J Rare Dis. 2013;8:110. doi: 10.1186/1750-1172-8-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek D, Shaw-Smith C, Kohlhase J, Schmitt W, Buiting K, Coffey A, Howard E, Hehr U, Gillessen-Kaesbach G. Esophageal atresia, hypoplasia of zygomatic complex, microcephaly, cup-shaped ears, congenital heart defect, and mental retardation–new MCA/MR syndrome in two affected sibs and a mildly affected mother? Am J Med Genet A. 2007;143A:1135–1142. doi: 10.1002/ajmg.a.31752. [DOI] [PubMed] [Google Scholar]
- Wieczorek D, Gener B, González MJ, Seland S, Fischer S, Hehr U, Kuechler A, Hoefsloot LH, de Leeuw N, Gillessen-Kaesbach G, Lohmann DR. Microcephaly, microtia, preauricular tags, choanal atresia and developmental delay in three unrelated patients: a mandibulofacial dysostosis distinct from Treacher Collins syndrome. Am J Med Genet A. 2009;149A:837–843. doi: 10.1002/ajmg.a.32747. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.