Abstract
Objectives
Heterogeneity in sepsis-related pathobiology presents a significant challenge. Resolving this heterogeneity presents an opportunity to understand pathobiology and improve patient care. Olfactomedin-4 (OLFM4) is a neutrophil subset marker and may contribute to sepsis heterogeneity. Our objective was to evaluate the expression of OLFM4 and characterize neutrophil heterogeneity in children with septic shock.
Design
Single-center, prospective cohort, as well as secondary analysis of existing transcriptomic and proteomic databases.
Setting
Tertiary care pediatric intensive care unit.
Patients
Patients from age 5 days to 18 years with septic shock were enrolled. Data collected included the expression of OLFM4 mRNA, serum protein concentrations and percentage of neutrophils that express OLFM4.
Interventions
None.
Measurements and Main Results
Secondary analysis of existing transcriptomic data demonstrated that OLFM4 is the most highly expressed gene in non-survivors of pediatric septic shock, compared to survivors. Secondary analysis of an existing proteomic database corroborated these observations. In a prospectively enrolled cohort, we quantified the percentage of OLFM4+ neutrophils in patients with septic shock. Patients with a complicated course, defined as ≥2 organ failures at day 7 of septic shock or 28-day mortality, had a higher percentage of OLFM4+ neutrophils, compared to those without a complicated course. By logistic regression, the percentage of OLFM4+ neutrophils was independently associated with increased risk of a complicated course (O.R. 1.09, 95% C.I. 1.01 to 1.17, p = 0.024).
Conclusions
OLFM4 identifies a subpopulation of neutrophils in patients with septic shock, and those with a high percentage of OLFM4+ neutrophils are at higher risk for greater organ failure burden and death. OLFM4 might serve as a marker of a pathogenic neutrophil subset in patients with septic shock.
Keywords: Neutrophils, Septic Shock, Pediatric, Heterogeneity
Introduction
Sepsis and septic shock are major causes of morbidity and mortality in all intensive care units throughout the world. Mortality rates from septic shock have improved over the past few decades, but remain unacceptably high (1, 2). Research has led to a better understanding of the pathologic mechanisms of sepsis, however clinical care has remained much the same for many years (3, 4). Virtually all recent clinical trials have failed to show improved outcomes, much of this because of the heterogeneity in the patients who are enrolled. This heterogeneity comes from many sources including initiating infection, host immune-competence, morbidities, and notable differences in immuno-pathobiology. There is a significant need for breakthroughs in understanding to identify novel therapeutic pathways that will also lead to increased ability to categorize patients with septic shock.
Neutrophils are the primary leukocyte in the blood and are the first line response to onset of infection. Neutrophils contain both pattern recognition immune receptors and Fc receptors that allow participation in both innate and adaptive immune responses. Characteristics of neutrophils in terms of count, maturity, migration, and granularity have been characterized in patients with septic shock, but only absolute neutrophils count and the presence of bands are used clinically (5). While lymphocytes have been subdivided into many subpopulations with unique immune roles, neutrophil heterogeneity that can associate different functions within neutrophil subsets has been postulated, but no definitive subpopulations or states have been characterized (6, 7).
Olfactomedin 4 (OLFM4) was originally identified as a granular protein induced with G-CSF and expressed in mature granulocytes in mice (8, 9). Subsequent work focused on the finding that OLFM4 was also expressed in human intestinal tumors, particularly gastric cancer where it was shown to have anti-apoptotic properties (10, 11). A mouse deficient of OLFM4 appeared to be phenotypically normal, but when challenged with intraperitoneal injection of Staphylococcus aureus, was protected from death when compared with wild type littermates (12, 13). OLFM4 mRNA has also been shown to be upregulated in the human disease such as RSV bronchiolitis in pediatric patients and acute respiratory distress syndrome (ARDS) in adults (14, 15). More recently, two groups reported OLFM4 expression in a subset of human neutrophils, but were unable to identify a phenotypic difference between the OLFM4 positive and negative neutrophil populations (16, 17).
We sought to identify novel pathways in immune cells during septic shock that may help explain the dynamic fluctuations and heterogeneity in patients. Review of a transcriptomic database reflecting whole blood derived RNA, revealed OLFM4 as significantly upregulated in these patients. Given the finding that OLFM4 is expressed in a subset of neutrophils, upregulated in several illnesses in adult and pediatric patients, and the OLFM4 null mouse had improved mortality when challenged with a lethal dose of bacteria, we set out to characterize the expression of OLFM4 in pediatric patients with septic shock.
Materials and Methods
Transcriptomic and Proteomic Data
OLFM4 mRNA expression data were extracted from an existing, microarray-based transcriptomic database. Details of the study protocol were previously published (18, 19) and the data are deposited in the Gene Expression Omnibus (Accession # GSE66099). Briefly, the data reflect children with septic shock and normal controls. The RNA used for microarray analyses was derived from whole blood obtained within the first 24 hours of meeting criteria for septic shock. The original consent form for this protocol allows for secondary analysis of clinical and biological data.
OLFM4 serum protein concentration data were extracted from an existing proteomic database generated during the derivation and validation of a panel of biomarkers to estimate the probability of sepis-associated acute kidney injury (20). The serum samples were obtained within the first 24 hours of meeting criteria for septic shock. The protocol for patient enrollment was previously published (20) and the original consent form allows for secondary analyses of clinical and biological data. For new patients (described below) and similar protocol was used except that OLFM4 was measured in the plasma rather than serum.
Prospective Enrollment of Study Subjects and Data Collection
The protocol for prospective collection and use of biological samples and clinical data was approved by the Institutional Review Board of Cincinnati Children’s Hospital Medical Center. Children ≤ 18 years of age admitted to the pediatric intensive care unit (PICU) and meeting pediatric-specific consensus criteria for SIRS, sepsis, or septic shock were eligible for enrollment (21). The only exclusion criteria were the presence of neutropenia, which precludes our flow cytometry protocol, and the inability to obtain informed consent, which was obtained from parents or legal guardians prior to any data or sample collection.
Whole blood samples for flow cytometry were obtained within 24 hours of first meeting the criteria for SIRS, sepsis, or septic shock in the PICU, which was typically at presentation to the PICU. These are referred to as “day 1” samples. Forty eight hours later, a second whole blood sample was obtained for flow cytometry, termed “day 3” samples. Clinical and laboratory data were collected daily while in the PICU. Organ failure data were tracked up to day 7 using previously published criteria (21). Mortality was tracked for 28 days after enrollment. Illness severity was estimated using the Pediatric Risk of Mortality (PRISM) score, which is based on laboratory values and clinical variables reflecting physiology (22). Control whole blood samples were obtained from children presenting to the same day surgery department for elective hernia repairs. The control subjects were otherwise healthy, and had no history of a febrile illness nor exposure to anti-inflammatory medications in the preceding two weeks.
Flow Cytometry
Flow cytometric staining of cells for OLFM4 expression was done as previously described by Welin and colleagues (17). Our protocol is described in the online supplemental data.
Statistical Analysis
Statistical procedures used SigmaStat Software (Systat Software, Inc., San Jose, CA). Initially, data are described using medians, interquartile ranges, frequencies, and percentages. Comparisons between groups used the Mann-Whitney U-test, Chi-square, or Fisher’s Exact tests as appropriate. For analysis of the flow cytometry data we combined the subjects with SIRS or sepsis into a single group referred to as “PICU Controls”. For logistic regression we used complicated course as the primary outcome variable. This is defined as persistence of two or more organ failures at day seven of septic shock or 28-day mortality (19, 23–28). We have used this outcome variable in previous studies because persistent, multiple organ failure is a major antecedent of death from septic shock. Using complicated course in the logistic regression modeling allows the exploration of association between percentage of OLFM+ neutrophils and a nuance of sepsis severity beyond the dichotomy of “alive” vs. “dead”. In addition to the percentage of OLFM4+ neutrophils, the logistic regression modeling considered age and illness severity as independent variables. Statistical methods for gene array data are described in the supplementary methods.
Results
OLFM4 mRNA Expression
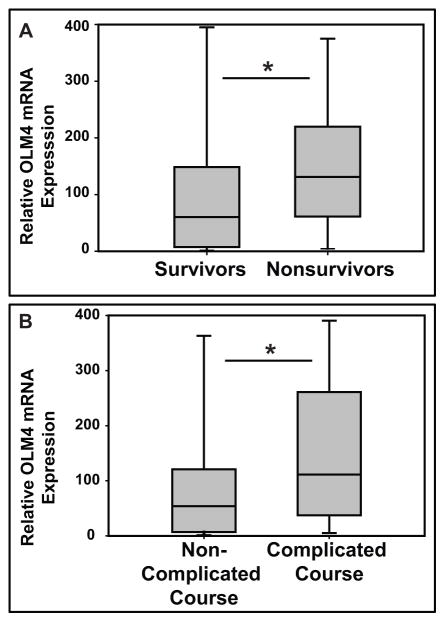
The clinical and demographic data for the study subjects in the transcriptomic database were previously published (18). Supplementary table 1 shows the top 5 most upregulated genes in the subjects with septic shock who did not survive (n = 29), compared to those who survived (n = 151). OLFM4 was the most highly expressed gene in the non-survivors based on a fold expression criterion and a Benjamini-Hochberg False Discovery Rate of 5%. OLFM4 mRNA expression was greater in the non-survivors, compared to the survivors (figure 1A). Similarly, figure 1B shows that OLFM4 mRNA expression was greater in the patients with septic shock and a complicated course (n = 53), compared to those without a complicated course (n = 127).
Figure 1. OLFM4 transcript is higher in non survivors and complicated course.
A. Graph showing OLFM4 mRNA expression is significantly higher in non-survivors (n=29) than survivors (n=151; * p=0.006) from septic shock. B. OLFM4 mRNA expression is also higher in those patients with complicated course (n=53) compared to non-complicated course (n=126; * p<0.001).
OLFM4 Protein in Serum
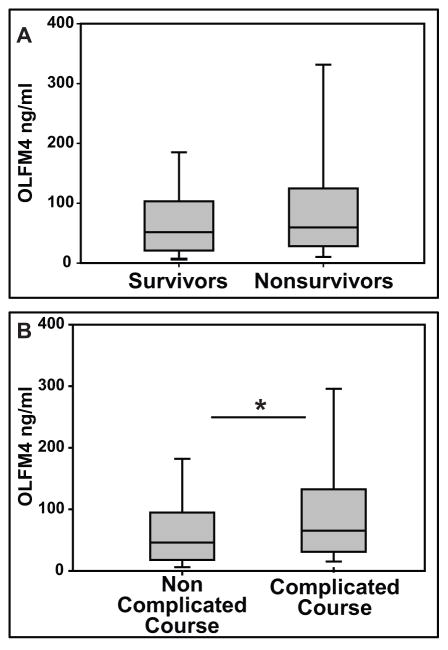
Given the increase in OLFM4 transcript expression we next evaluated the plasma protein concentration. The clinical and demographic data for the study subjects in the proteomic database were previously published (20). We measured baseline OLFM4 serum concentration in 22 healthy pediatric controls (mean= 0.93 ng/ml, range of 0.17 ng/ml to 3.0 ng/ml) and compared it to 450 patients with septic shock (mean= 86.3 ng/ml, range of 0.04 ng/ml to 867.4 ng/ml). Figure 2A compares OLFM4 serum concentrations between patients with septic shock who survived the 28 day study period (n = 388) and those who did not (n = 62). There was trend toward increased OLFM4 serum protein concentrations in the non-survivors, but this did not reach statistical significance (p = 0.099). However, when comparing OLFM4 serum concentrations between patients with septic shock without a complicated course (n = 316) and those with a complicated course (n = 134), the median OLFM4 serum concentrations was significantly higher in the patients with a complicated course (figure 2B).
Figure 2. OLFM4 Plasma protein expression is higher in patients with complicated course.
A. When comparing septic shock survivors (n=388) and non-survivors (n=62), there was no statistical difference (p=0.099). B. OLFM4 Plasma protein concentration is higher in those patients with complicated course (n=134) than without complicated course (n=316; * p=0.005).
Flow Cytometric Measurement of OLFM4+ Neutrophils
We next sought to evaluate if patients with septic shock had different percent of neutrophils that express OLFM4. Table 1 shows the clinical and demographic data for the prospectively enrolled study subjects who were evaluated by flow cytometry for OLFM4+ neutrophils. The subjects in the PICU control group were older than the subjects in the healthy control group. PRISM scores were greater in the septic shock group, compared to the PICU control group. No other differences were noted.
Table 1.
Demographic and clinical characteristics of the study subjects
| Characteristic | Healthy Control | PICU Control | Septic Shock |
|---|---|---|---|
| N | 33 | 29 | 41 |
| Median Age, Years (IQR) | 2.9 (0.7 – 6.4) | 6.6 (3.2 – 12.1)a | 4.4 (1.5 – 9.8) |
| Males, # (%) | 23 (70) | 15 (52) | 25 (61) |
| 28-day mortality, # (%) | --- | 2 (7) | 0 (0) |
| Complicated course, # (%) | --- | 2 (7) | 7 (17) |
| Median PRISM score (IQR) | --- | 4 (2 – 8) | 9 (5 – 12)b |
| Median WBC count, × 103 per mm3 (IQR) | --- | 11.8 (7.0 – 16.5) | 11.9 (6.3 – 17.8) |
| Median neutrophil count, × 103 per mm3 (IQR) | --- | 9.3 (5.4 – 13.9) | 6.8 (4.5 – 14.2) |
| # with gram negative bacteria (%) | --- | 0 (0) | 11 (27) |
| # with gram positive bacteria (%) | --- | 9 (31) | 2 (5) |
| # with other pathogen isolated (%) | --- | 1 (3) | 4 (10) |
| # with no pathogen isolated (%) | --- | 19 (66) | 24 (59) |
| # with comorbidity (%) | --- | 18 (62) | 33 (80) |
| # with malignancy (%) | --- | 3 (10) | 8 (20) |
| # with immune suppression (%) | --- | 2 (7) | 12 (29) |
| # with bone marrow transplantation (%) | --- | 0 (0) | 6 (15) |
p < 0.05 vs. Healthy Control, ANOVA on Ranks.
p < 0.05 vs. PICU Control, Rank Sum Test.
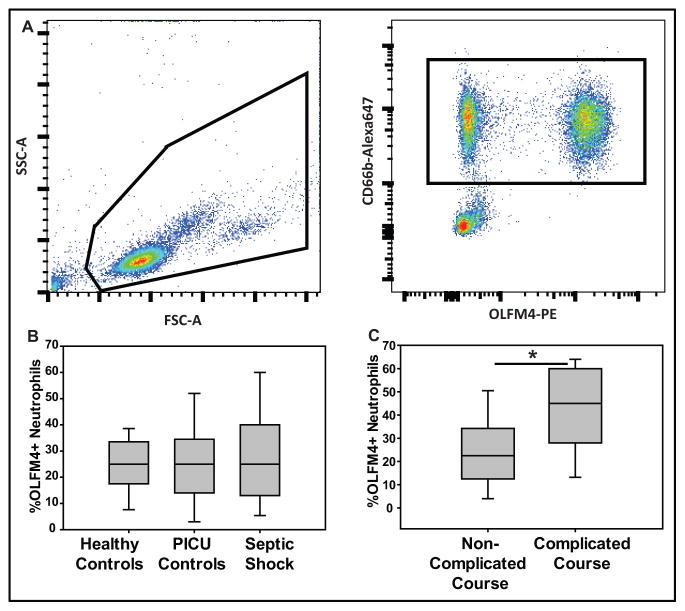
Figure 3A shows a representative flow cytometry analysis from a single patient with septic shock. The left panel shows the initial gating of the white blood cell population. The right panel shows two distinct populations of neutrophils based on negative or positive staining for OLFM4. We did not observe significant fluctuation in the mean fluorescence intensity (MFI), a surrogate measure of the amount of protein (data not shown).
Figure 3. Percentage OLFM4+ neutrophils is higher in those patients with complicated course.
A. Left figure depicts flow cytometric analysis showing forward and side scatter of leukocytes from a patient with septic shock with cells analyzed surrounded with black line; right shows the fluorescence markers used to identify neutrophils (CD66b positive, black box) and percentage of neutrophils that express OLFM4. B. Percentage of OLFM4+ neutrophils is not different between healthy controls (n=31), PICU controls (SIRS and sepsis; n=26), or patients with septic shock (n=43). C. When comparing just patients with septic shock, those with a complicated course (n=7) had a higher percentage OLFM4+ cells when compared to non-complicated course (n=34; * p=0.025).
We also evaluated the percentage of OLFM4+ neutrophils on day 1, between healthy controls, PICU controls, and subjects with septic shock (figure 3B). The median percentage of OLFM4+ neutrophils was not different between the three groups. Figure 3C compares the percentage of OLFM4+ neutrophils on day 1, between subjects with septic shock without a complicated course and those with a complicated course. The percentage of OLFM4+ neutrophils was higher in the subjects with a complicated course. Supplementary figures 1A and 1B show similar analyses, but represent day 3 samples. The day 3 observations were similar to that seen on day 1. We also evaluated if OLFM4 may be expressed by immature neutrophils, or bands, and found no correlation between band count and percent of OLFM4+ neutrophils (n=39; R value 0.100, p=0.58).
Logistic regression modeling to test for an association between the percentage of OLFM4+ neutrophils and complicated course among subjects with septic shock are shown in table 2. The percentage of OLFM4+ neutrophils on day 1 of septic shock was independently associated with increased odds of a complicated course (O.R. 1.09, 95% C.I. 1.01 to 1.17, p = 0.024), even when accounting for the age and degree of illness severity (PRISM score). A similar trend was observed when analyzing the percentage of OLFM4+ neutrophils on day 3 of septic shock, but this was marginally significant.
Table 2.
Multiple logistic regression to predict complicated course from septic shock
| Variable | Odds Ratio (95% CI) | P Value | Odds Ratio (95% CI) | P Value |
|---|---|---|---|---|
| Day 1 | Day 3 | |||
| Age | 0.619 (0.382–1.003) | 0.052 | 0.375 (0.127–1.103) | 0.075 |
| PRISM | 1.148 (0.945–1.394) | 0.165 | 1.198 (0.853–1.681) | 0.297 |
| % OLFM4+ Neutrophils | 1.088 (1.011–1.171) | 0.024 | 1.172 (0.998–1.376) | 0.053 |
To test if neutrophils are the source for serum OLFM4 we examined the correlation between the percentages of OLFM4+ neutrophils and serum OLFM4 concentrations (sup figure 2A). There was a significant trend for serum OLFM4 concentrations increasing relative to increasing percentages of OLFM4+ neutrophils, but there was substantial scatter about the regression line. This trend remained significant, with less scatter, when considering the absolute number of OLFM4+ neutrophils, instead of the percentage of OLFM4+ neutrophils (sup figure 2B).
Discussion
In these cohorts of children with septic shock, OLFM4 transcript was highly upregulated in those with a complicated course and was the most upregulated gene in those who died compared to those who survived. The mRNA upregulation was concomitant with a nearly 100-fold increase in serum protein concentration in patients with septic shock compared to healthy controls. Finally, in pediatric patients admitted to the ICU, there was no difference in the mean percentage of OLFM4+ neutrophils between healthy controls, PICU controls (SIRS and sepsis) and those with septic shock, however, a high percent of OLFM4+ neutrophils increased the likelihood of having a complicated course from septic shock (Figures 3B and 3C). This association was independent of illness severity and age. Collectively, these data indicate that OLFM4+ neutrophils are not associated with the risk of developing septic shock per se, but among those with septic shock they are associated with increased odds of having greater organ failure and death.
Neutrophils have long been recognized as the primary innate immune cell in the blood. While lymphocytes have been differentiated into many subpopulations with different effector function, neutrophils have been primarily classified by age (young vs old), activation status, or by location (marrow, blood, or extravasated into tissue) (7, 29, 30). Surface markers have greatly facilitated the differentiation of lymphocyte subpopulations, but there are few surface makers that differentiate neutrophils. CXCR4 and CD16 expression have been used to differentiate aged neutrophils (29); CD177 has been shown to have bimodal expression on neutrophils that differs between individuals with increased expression during certain inflammatory states (31, 32). To date, CD177 and OLFM4 remain the best markers for isolating bimodal neutrophil subpopulations, but there is little evidence for functional differences in these subpopulations. Two groups have started the characterization of the OLFM4+ neutrophils. Clemmensen et al. first showed the OLFM4 mRNA was expressed in all neutrophils during development, but only in a subpopulation of mature granulocytes (16). Weilin et al. found OLFM4 positive and negative neutrophils had similar activation, phagocytosis, and apoptosis. They also found that OLFM4 was located in neutrophils NETs and speculated it may play a role in NETosis (17). Here we present OLFM4 is highly upregulated in patients with septic shock and the degree of upregulation is associated with clinical outcomes, suggesting a potential role for this subpopulation of neutrophils.
When comparing patients who survived septic shock with those who did not, OLFM4 transcript had the highest fold increase of any gene from all leukocytes (neutrophils, monocytes and lymphocytes), yet OLFM4 is only expressed in a subset of neutrophils bases on flow cytometric analysis. We envision a few likely explanations for this observation. First, it could be that 25 to 50% of neutrophils account for the significant increase in mRNA transcriptional fold expression and other neutrophils never express OLFM4. Another explanation would be that all neutrophils express OLFM4 but that it is translated and rapidly secreted in some neutrophils or that the mRNA transcript never undergoes translation. Both interpretations suggest that there are separate subpopulations or at least states of neutrophils. In addition, there was no correlation between immature neutrophil bands and percentage of OLFM4+ neutrophils suggesting OLFM4 in not simply a marker of recently emigrated neutrophils. While there were changes in mean fluorescence intensity between patients and days 1 and 3, there was no consistent pattern to suggest an increase in secretion (decrease MFI; data not shown).
Increased transcriptional activity of OLFM4 was concurrent with a dramatic increase in plasma protein concentration. Under healthy conditions, OLFM4 is known to be expressed by the prostate, intestine and neutrophils. Patients with inflamed ulcerative colitis have been shown to produce OLFM4 in intestinal crypt cells and secrete it into the intestinal lumen (33). Other groups have attempted to identify a source of the serum OLFM4, but to date no definitive source has been identified (34). The expression of OLFM4 protein in the specific granules of neutrophils suggests that some plasma OLFM4 may be coming from neutrophil exocytosis. In patients with septic shock serum OLFM4 increased with increasing percentages of OLFM4+ neutrophils, but these was substantial scatter about the regression line, suggesting neutrophils contribute but are not the only source of OLFM4 serum concentrations. Further evidence that there is another source comes from patients with a low absolute neutrophil count that still have high serum OLFM4. An additional complicating factor is the recent finding that OLFM4 can serve as an autoantigen and some patients carry antibodies specific to OLFM4, which may also result in clearance of some of the plasma OLFM4 (35). Thus, the additional sources and function of a near 100 fold increase in plasma OLFM4 in patients with septic shock remains unknown at this time.
The question remains if the high concentration of OLFM4 in the plasma is simply a marker of cellular damage, or if there is a functional role for the OLFM4 protein. The strongest evidence for OLFM4 actually playing a role in pathology is the finding that OLFM4 deficient mice are protected from death when injected with Staphylococcus aureus and E. coli (13). The authors who generated this mouse also found that OLFM4 inhibited Cathepsin C, a serine protease important for activation of several proteases with putative-antimicrobial activity and postulated that the loss of OLFM4 increased Cathepsin C activity, thereby providing protection. However, crossing OLFM4 onto a Cathepsin C deficient mouse still provided protection, suggesting alternative mechanisms. Also, it is difficult to make physiologic sense that the most upregulated gene in neutrophils from patients with septic shock are those to inhibit antimicrobial activity, again suggesting there may be alternative functions for OLFM4.
Conclusions
The balance between an over exuberant immune response and one that controls the initiating infection remains a fundamental problem in the management of patients with septic shock. Massive stimulation of immune cells leads to a cytokine storm that often leads to the clinical presentation of septic shock with endothelial injury, coagulopathy, and vasodilation. Following initial presentation, patients are left in fluctuating state of chronic activation of some cells, while other immune cells are exhausted from over stimulation and rendered useless. Our goal is to identify specific immunologic pathways or cellular subpopulations that might be targeted to tone down an immune response to temper host damage but still allow for a pathogen clearing immune responses. Toward this direction, we identified a difference in neutrophil populations that correlate with disease severity, and may one day be eligible for targeting of specific neutrophil subpopulations, pathologic and non-pathologic.
Supplementary Material
A. Day 3 after admission to PICU, percentage OLFM4+ neutrophils in each of the subgroups. B. Day 3 after admission to PICU, percentage positive neutrophils again is associated with a complicated course (* p=0.033).
A. Linear regression showing correlation between percentage OLFM4+ cells and plasma OLFM4 concentration in patients with septic shock. B. Correlation is somewhat stronger when comparing absolute OLFM4+ neutrophil count and plasma OLFM4 concentration. For both graphs, outliers were placed on the border of the graph with their numerical value written beside them.
Acknowledgments
Source of support: Grant support is as follows: NIH K12 HD028827 (MNA), T32 GM008478 (MNA), R01GM099773 (HRW), and R01GM108025 (HRW)
Erin Frank, Kelli Howard and Laura Benken for consenting families and obtaining samples. All contributing hospitals who have provided patients samples in the PERSEVER study group.
Footnotes
Conflicts of interest
All authors report grant support from the NIH, no financial conflicts of interest.
Copyright form disclosures: Dr. Alder received support for article research from the National Institutes of Health (NIH). His institution received funding from the NIH. Dr. Lahni received support for article research from the NIH. Dr. Hildeman received support for article research from the NIH. Dr. Wong received support for article research from the NIH. His institution received funding from the NIH. Dr. Opoka disclosed that she does not have any potential conflicts of interest.
References
- 1.Martin GS, Mannino DM, Eaton S, et al. The epidemiology of sepsis in the United States from 1979 through 2000. The New England journal of medicine. 2003;348(16):1546–1554. doi: 10.1056/NEJMoa022139. [DOI] [PubMed] [Google Scholar]
- 2.Watson RS, Carcillo JA, Linde-Zwirble WT, et al. The epidemiology of severe sepsis in children in the United States. American journal of respiratory and critical care medicine. 2003;167(5):695–701. doi: 10.1164/rccm.200207-682OC. [DOI] [PubMed] [Google Scholar]
- 3.Opal SM, Dellinger RP, Vincent JL, et al. The next generation of sepsis clinical trial designs: what is next after the demise of recombinant human activated protein C?*. Critical care medicine. 2014;42(7):1714–1721. doi: 10.1097/CCM.0000000000000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanna W, Wong HR. Pediatric sepsis: challenges and adjunctive therapies. Critical care clinics. 2013;29(2):203–222. doi: 10.1016/j.ccc.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zonneveld R, Molema G, Plotz FB. Analyzing Neutrophil Morphology, Mechanics, and Motility in Sepsis: Options and Challenges for Novel Bedside Technologies. Critical care medicine. 2016;44(1):218–228. doi: 10.1097/CCM.0000000000001266. [DOI] [PubMed] [Google Scholar]
- 6.Kruger P, Saffarzadeh M, Weber AN, et al. Neutrophils: Between host defence, immune modulation, and tissue injury. PLoS Pathog. 2015;11(3):e1004651. doi: 10.1371/journal.ppat.1004651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beyrau M, Bodkin JV, Nourshargh S. Neutrophil heterogeneity in health and disease: a revitalized avenue in inflammation and immunity. Open Biol. 2012:2. doi: 10.1098/rsob.120134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenbauer F, Wagner K, Zhang P, et al. pDP4, a novel glycoprotein secreted by mature granulocytes, is regulated by transcription factor PU.1. Blood. 2004;103(11):4294–4301. doi: 10.1182/blood-2003-08-2688. [DOI] [PubMed] [Google Scholar]
- 9.Zhang JC, Liu WL, Tang DC, et al. Identification and characterization of a novel member of olfactomedin-related protein family, hGC-1, expressed during myeloid lineage development. Gene. 2002;283(1–2):83–93. doi: 10.1016/s0378-1119(01)00763-6. [DOI] [PubMed] [Google Scholar]
- 10.Grover PK, Hardingham JE, Cummins AG. Stem cell marker olfactomedin 4: critical appraisal of its characteristics and role in tumorigenesis. Cancer Metast Rev. 2010;29(4):761–775. doi: 10.1007/s10555-010-9262-z. [DOI] [PubMed] [Google Scholar]
- 11.Yu L, Wang L, Chen S. Olfactomedin 4, a novel marker for the differentiation and progression of gastrointestinal cancers. Neoplasma. 2011;58(1):9–13. doi: 10.4149/neo_2011_01_9. [DOI] [PubMed] [Google Scholar]
- 12.Liu WL, Yan M, Liu YQ, et al. Olfactomedin 4 down-regulates innate immunity against Helicobacter pylori infection. P Natl Acad Sci USA. 2010;107(24):11056–11061. doi: 10.1073/pnas.1001269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu WL, Yan M, Liu YQ, et al. Olfactomedin 4 Inhibits Cathepsin C-Mediated Protease Activities, Thereby Modulating Neutrophil Killing of Staphylococcus aureus and Escherichia coli in Mice. Journal of immunology. 2012;189(5):2460–2467. doi: 10.4049/jimmunol.1103179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brand HK, Ahout IML, de Ridder D, et al. Olfactomedin 4 Serves as a Marker for Disease Severity in Pediatric Respiratory Syncytial Virus (RSV) Infection. Plos One. 2015;10(7) doi: 10.1371/journal.pone.0131927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kangelaris KN, Prakash A, Liu KD, et al. Increased expression of neutrophil-related genes in patients with early sepsis-induced ARDS. Am J Physiol-Lung C. 2015;308(11):L1102–L1113. doi: 10.1152/ajplung.00380.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clemmensen SN, Bohr CT, Rorvig S, et al. Olfactomedin 4 defines a subset of human neutrophils. Journal of leukocyte biology. 2012;91(3):495–500. doi: 10.1189/jlb.0811417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Welin A, Amirbeagi F, Christenson K, et al. The Human Neutrophil Subsets Defined by the Presence or Absence of OLFM4 Both Transmigrate into Tissue In Vivo and Give Rise to Distinct NETs In Vitro. Plos One. 2013;8(7) doi: 10.1371/journal.pone.0069575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong HR, Shanley TP, Sakthivel B, et al. Genome-level expression profiles in pediatric septic shock indicate a role for altered zinc homeostasis in poor outcome. Physiol Genomics. 2007;30(2):146–155. doi: 10.1152/physiolgenomics.00024.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong HR, Cvijanovich NZ, Allen GL, et al. Corticosteroids Are Associated with Repression of Adaptive Immunity Gene Programs in Pediatric Septic Shock. American journal of respiratory and critical care medicine. 2014;189(8):940–946. doi: 10.1164/rccm.201401-0171OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong HR, Cvijanovich NZ, Anas N, et al. A Multibiomarker-Based Model for Estimating the Risk of Septic Acute Kidney Injury. Critical care medicine. 2015;43(8):1646–1653. doi: 10.1097/CCM.0000000000001079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldstein B, Giroir B, Randolph A, et al. International pediatric sepsis consensus conference: definitions for sepsis and organ dysfunction in pediatrics. Pediatr Crit Care Med. 2005;6(1):2–8. doi: 10.1097/01.PCC.0000149131.72248.E6. [DOI] [PubMed] [Google Scholar]
- 22.Pollack MM, Ruttimann UE, Getson PR. Pediatric Risk of Mortality (Prism) Score. Critical care medicine. 1988;16(11):1110–1116. doi: 10.1097/00003246-198811000-00006. [DOI] [PubMed] [Google Scholar]
- 23.Abulebda K, Cvijanovich NZ, Thomas NJ, et al. Post-ICU Admission Fluid Balance and Pediatric Septic Shock Outcomes: A Risk-Stratified Analysis. Critical care medicine. 2014;42(2):397–403. doi: 10.1097/CCM.0b013e3182a64607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mickiewicz B, Vogel HJ, Wong HR, et al. Metabolomics as a Novel Approach for Early Diagnosis of Pediatric Septic Shock and Its Mortality. American journal of respiratory and critical care medicine. 2013;187(9):967–976. doi: 10.1164/rccm.201209-1726OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong HR, Cvijanovich NZ, Anas N, et al. Developing a Clinically Feasible Personalized Medicine Approach to Pediatric Septic Shock. American journal of respiratory and critical care medicine. 2015;191(3):309–315. doi: 10.1164/rccm.201410-1864OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weiss SL, Cvijanovich NZ, Allen GL, et al. Differential expression of the nuclear-encoded mitochondrial transcriptome in pediatric septic shock. Critical care. 2014;18(6) doi: 10.1186/s13054-014-0623-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Atkinson SJ, Cvijanovich NZ, Thomas NJ, et al. Corticosteroids and Pediatric Septic Shock Outcomes: A Risk Stratified Analysis. Plos One. 2014;9(11) doi: 10.1371/journal.pone.0112702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wong HR, Cvijanovich NZ, Anas N, et al. Prospective Testing and Redesign of a Temporal Biomarker Based Risk Model for Patients With Septic Shock: Implications for Septic Shock Biology. Ebiomedicine. 2015;2(12):2087–2093. doi: 10.1016/j.ebiom.2015.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang DC, Chen G, Manwani D, et al. Neutrophil ageing is regulated by the microbiome. Nature. 2015;525(7570):528. doi: 10.1038/nature15367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pillay J, Ramakers BP, Kamp VM, et al. Functional heterogeneity and differential priming of circulating neutrophils in human experimental endotoxemia. Journal of leukocyte biology. 2010;88(1):211–220. doi: 10.1189/jlb.1209793. [DOI] [PubMed] [Google Scholar]
- 31.Hu N, Westra J, Huitema MG, et al. Coexpression of CD177 and Membrane Proteinase 3 on Neutrophils in Antineutrophil Cytoplasmic Autoantibody-Associated Systemic Vasculitis. Arthritis Rheum-Us. 2009;60(5):1548–1557. doi: 10.1002/art.24442. [DOI] [PubMed] [Google Scholar]
- 32.Stroncek D. Neutrophil-specific antigen HNA-2a (NB1, CD177): serology, biochemistry, and molecular biology. Vox Sang. 2002;83(Suppl 1):359–361. doi: 10.1111/j.1423-0410.2002.tb05334.x. [DOI] [PubMed] [Google Scholar]
- 33.Gersemann M, Becker S, Nuding S, et al. Olfactomedin-4 is a glycoprotein secreted into mucus in active IBD. J Crohns Colitis. 2012;6(4):425–434. doi: 10.1016/j.crohns.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 34.Clemmensen SN, Glenthoj AJ, Heeboll S, et al. Plasma levels of OLFM4 in normals and patients with gastrointestinal cancer. J Cell Mol Med. 2015;19(12):2865–2873. doi: 10.1111/jcmm.12679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amirbeagi F, Thulin P, Pullerits R, et al. Olfactomedin-4 autoantibodies give unusual c-ANCA staining patterns with reactivity to a subpopulation of neutrophils. Journal of leukocyte biology. 2015;97(1):181–189. doi: 10.1189/jlb.5A0614-311R. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. Day 3 after admission to PICU, percentage OLFM4+ neutrophils in each of the subgroups. B. Day 3 after admission to PICU, percentage positive neutrophils again is associated with a complicated course (* p=0.033).
A. Linear regression showing correlation between percentage OLFM4+ cells and plasma OLFM4 concentration in patients with septic shock. B. Correlation is somewhat stronger when comparing absolute OLFM4+ neutrophil count and plasma OLFM4 concentration. For both graphs, outliers were placed on the border of the graph with their numerical value written beside them.