Abstract
Researchers increasingly visualize a significant role for artificial biochemical logical systems in biological engineering, much like digital logic circuits in electrical engineering. Those logical systems could be utilized as a type of servomechanism to control nanodevices in vitro, monitor chemical reactions in situ, or regulate gene expression in vivo. Nucleic acids (NA), as carriers of genetic information with well-regulated and predictable structures, are promising materials for the design and engineering of biochemical circuits. A number of logical devices based on nucleic acids (NA) have been designed to handle various processes for technological or biotechnological purposes. This article focuses on the most recent and important developments in NA-based logical devices and their evolution from in vitro, through cellular, even towards in vivo biological applications.
Keywords: DNA computation, DNA nanobiotechnology, logical systems, nucleic acids
Graphical Abstract
Introduction
Because of their unprecedented computing power, seamless coupling ability and incredible adaptability, silicon-based computers have been intensively applied in a vast array of areas. However, as a result of the physical limitations of conventional silicon chips, the development of more powerful microprocessors is heading towards a barrier.[1] Continued progress will require miniaturization of components, with a possible negative effect on instrumental performance. To address the challenge ahead, researchers have been pursuing the idea of constructing computers in which computations are performed by individual molecules, allowing for a continuous exponential increase in performance and decrease in size for microprocessors.
Nucleic acids (NA) have been found extremely effective for miniaturization of information storage, because only approximately 50 atoms are used for one bit of information.[2] In addition, easy chemical synthesis, combinatorial structures, and Watson–Crick complementarity principle provide sufficient theoretical and experimental bases for the rational design of logical devices. Therefore, researchers have challenged themselves to use DNA or RNA to build logical devices for molecular computation. As early as 1994, Adleman used DNA to solve a computational problem. He encoded the Hamiltonian path problem into different single-stranded DNA (ssDNA) sequences and applied biotechnologies (such as ligation, polymerase chain reaction, sequencing) to decode the answers to the correct Hamiltonian path.[3]
However, subsequent progress in constructing molecular computing devices has been frustratingly slow. Scientists have demonstrated that NA-based biocircuitry can perform logic gate operations, signal restoration, amplification, feedback, and cascading, all by distinct oligonucleotide strands. However, the chief objective is still to emulate or mimic the digital logic found in typical circuit boards, and so far the computing power cannot rival that of silicon computers in the execution of any algorithm.
It has become increasingly clear that the ability to interact with naturally occurring biomolecules, together with such unique properties as programmability, nanometric size, and autonomous operation, can be used for practical applications of NA-based logical devices. This has allowed biological properties to be interfaced to other materials, thereby opening up a novel and exciting direction in biological and biomedical applications. Accordingly, this article focuses on the most recent and important developments in NA-based logical devices and their evolution from in vitro, through cellular, even towards in vivo biological application.
In Vitro Nucleic Acid Based Logical Systems
A detailed understanding of nucleic acids, including specific connectivity of the nucleotides, Watson–Crick base pairing, and the double-helical structure of the double-stranded DNA (dsDNA), provides the theoretical basis of constructing NA-based logical devices. To engineer complex logical modules from molecules, the stereotypical structures of these modules should include three elements: input, processor, and output.[4] In addition, specific mechanisms should be employed to allow the signals to transfer through these elements. For NA-based devices, several common strategies will be introduced here to make circuit construction a predictive science.
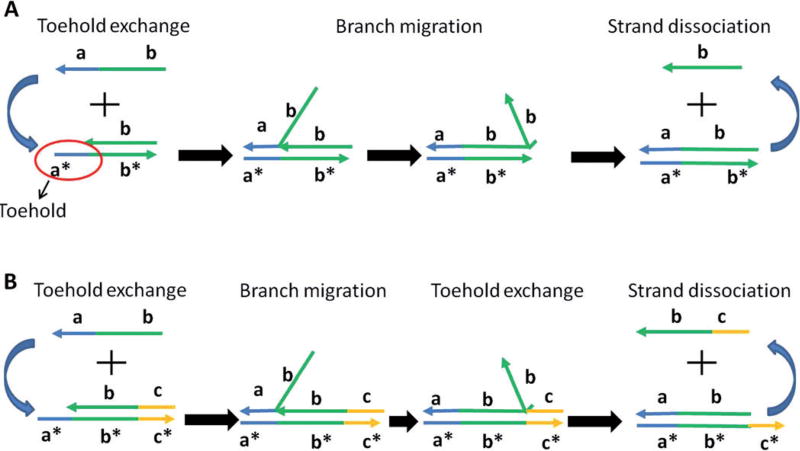
One significant mechanism to trigger signal transfer in the DNA-based logical network is called the toehold-mediated displacement reaction.[5] In typical toehold displacement reactions, a ssDNA (input) “reacts” with a dsDNA to displace one strand (output) of the dsDNA through binding to an overhang ssDNA region (toehold) of the dsDNA. Specifically, as shown in Figure 1A, this reaction is a three-step process including toehold binding, branch migration, and strand dissociation. Toehold binding occurs through the complementary sequences between input ssDNA and the toehold, and the displacement rate can be adjusted by changing the toehold base numbers.[6] Branch migration is a reversible process with the same binding sequences between input and output, except that the remaining sequences can be freely designed, allowing design diversity in the logical network. Seelig et al. have successfully implemented DNA-based logic gates including AND, OR, and NOT gates by applying this mechanism. These simple Boolean logic gates can also be cascaded into multilayer architectures to form more complex digital networks with an amplification strategy that prevents the damping of signals.[7] Li et al. also designed a three-input majority logic gate based on toehold-mediated displacement reaction. By smartly designing a circular DNA strand with identical joint domains but different recognition domains, the key features of a three-input majority gating, that is, the output will be true if any of the two inputs are true, were achieved (Figure 2 A).[8]
Figure 1.
A) Scheme of toehold-mediated displacement reaction. B) Scheme of toehold-exchange displacement reaction.
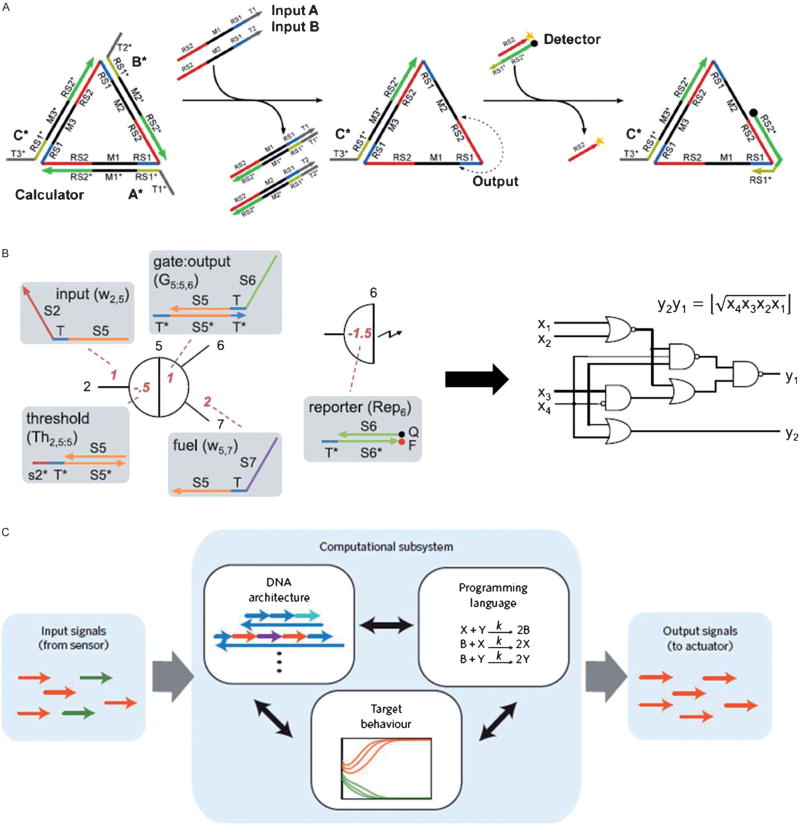
Figure 2.
DNA displacement reaction and its applications in constructing logical systems. A) Scheme of a three-input majority gate based on toehold-mediated DNA strand displacement reactions (reprinted by permission from reference [8], copyright 2013, American Chemical Society). B) Abstract diagram for a square-root circuit implemented with the logical DNA motif based on toehold-exchange DNA strand displacement reactions (from reference [12], reprinted with permission from AAAS). C) Scheme of programming chemical reactions using short single strands of DNA (reprinted by permission from reference [13], Macmillan Publishers Ltd., Nat. Nanotechnol., copyright 2013).
Although signal cascading by the classic toehold displacement provides considerable circuitry design programmability, this method is feasible only for constructing small-scale circuits that allow initial information-processing functions. One important restriction of the displacement reaction is the requirement that a longer strand must displace a shorter one, resulting in decreased sequence freedom for the downstream strands and limited numbers of circuit layer assembly. In order to improve the complexity of the logical circuit, Zhang and Winfree have developed a toehold-exchange displacement reaction (Figure 1 B).[9] The incoming single strand can be designed with a “shifted sequence” that is complementary to the bottom strand. The displacement reaction still occurs, but the displaced strand will also have a single-strand toehold region that may initiate the backward displacement reaction. This results in a reversible DNA displacement reaction with the input and output signals of the same sequence length. To drive the reaction to move only in the forward direction, a smart toehold exchange reaction has been developed in which a fuel strand with much higher concentration creates a catalytic cycle and allows the input to be transformed to output without being consumed.[10] This mechanism allows the entire circuit to be driven forward by the entropy gain of equilibration. With this mechanism in hand, Qian and Winfree constructed DNA-based logical circuits with much higher complexity including a neural network mimicry. This system consists of 110 DNA strands and demonstrates associative memory capable of answering 81 possible questions.[11] They also designed a 130-stranded DNA computer capable of computing the square root of a four-digit binary number (Figure 2 B).[12] Seelig et al. applied this principle to program chemical reactions, allowing the implementation of any reaction pattern that can be expressed mathematically (Figure 2 C).[13] Recent studies have also found ways to improve the reaction robustness of this toehold-exchange displacement reaction for detecting single nucleotide polymorphisms (SNP) in a DNA duplex. These results have the added benefit of performing normally under severe conditions, such as large changes in temperature or salt concentration.[14] The strategies described above provide more reasonable and effective guidelines for constructing complex nucleic acid circuits for biological applications.
As a catalytic molecule that can specifically cleave the RNA site of its substrate, deoxyribozymes (DNAzymes) provide another effective strategy to establish communication among the inputs and outputs in a logical circuit. In principle, the activation of one deoxyribozyme can lead to the cleavage of the precursor for downstream reactions, thus resulting in signal transfer between the input and output. In 2002, Stojanovic and colleagues first implemented AND, OR, and NOT logic gates by engineering the activity of two deoxyribozymes controlled by multiple oligonucleotide inputs.[15] Expanding on this, they further constructed double-layer deoxyribozyme-based circuits using gate-to-gate communication with the help of ligation reactions, wherein output from the ligating gates was sensed by phosphodiesterase gates in a cascade with ultimate fluorogenic output.[16] Later on, the scalability of this deoxyribozyme-based logical system was demonstrated by constructing a molecular automaton capable of playing “tic-tactoe” with a human player and even winning the game every time.[17] Another significant work completed by Willner and co-workers demonstrated the use of deoxyribozyme to construct input-guided assembly of computing circuits, such as half-adder/half-subtractor and fan-out gates.[18]
However, the logical systems mentioned above, that is, those based on DNA hybridization or a sequence-specific deoxyribozyme, are all irreversible reactions that are driven by thermodynamic equilibrium. These irreversible logical circuits may have limitations, such as one-time use for every new input and the accumulation of errors. Therefore, Turberfield et al. developed reversible DNA-based logical circuits (AND gates), the outputs of which are adjusted to changes in the inputs.[19] In other words, this system can stay close to an equilibrium state and continuously recompute outputs with different inputs.
To further expand the variety of NA-based logical circuits, Prokup et al. constructed a photochemically controlled DNA-based AND gate.[20] They used a caging group (6-nitropiperonyloxymethylene) to block the exposure of toeholds for circuit activation, thus allowing light as input to control the logical circuit with spatial and temporal resolution. Our group also designed a photon-induced toehold for formation of a pure duplex that can be used to trigger subsequent DNA hybridization chain reactions.[21] These photon-controllable devices provide the possibility for constructing reusable logical circuits on a larger scale to emulate and interface with electronic devices.
Expanding DNA to Functional NA Elements
While the development of pure NA-based logical systems has reached a sufficiently high level with elegant designs, the central molecules (i.e., inputs, transducers, and outputs) are still DNA strands, and the operational forces are controlled solely by DNA hybridization. Before expanding the applicability of logical systems from pure NA-based to multiple-component-based (e.g., proteins and small molecules), two significant questions need to be addressed. Can NA–protein or NA–small-molecule interactions be programmably enrolled in molecular circuits? Is NA circuitry capable of precise and smart manipulation of protein and other small-molecule functions?
A major challenge for direct manipulation of proteins and other small molecules by DNA logic circuitry is the need for a key component that can bridge nucleic acids with other molecules without influencing the programmability and versatility of logical circuits. Because they have the potential to interact specifically with proteins and small molecules, as well as control their functions, aptamers have been increasingly explored and developed as simple and effective ways to construct multiple- component-based logical devices. Aptamers also extend the recognition capabilities of nucleic acids in logical systems from Watson–Crick base-pairing to interactions with various targets through their unique secondary or tertiary structures. Therefore, aptamers can be directly used as building blocks to fabricate seamless logic-based aptamer circuits with enhanced capabilities and an extended scope of applications, from simple DNA base-pairing reactions to more complicated biomolecular reactions such as NA–protein or NA–small-molecule interactions, thus assigning the logical devices more functions for biological and biomedical applications.
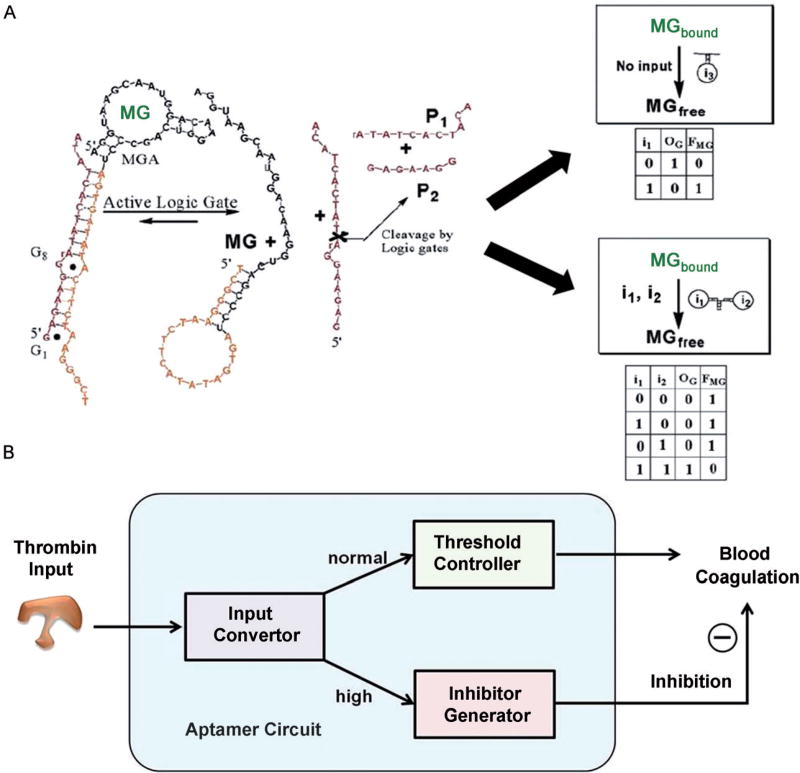
Stojanovic et al. were the first to include aptamers into DNA logical circuits (Figure 3A).[22] They constructed deoxyribozyme-based logic gates that could perform Boolean calculations on the input molecules to control the functional binding state of aptamers. Similar examples include the use of the binding between targets and aptamers to control the power release of biofuel cells according to the built-in Boolean NAND logic.[23] These two examples mainly addressed the first question mentioned above by enrolling NA–small-molecule interactions into molecular circuits.
Figure 3.
Examples of aptamer-based logical systems. A) The two conformations of malachite green aptamer and its incorporation into the construction of Boolean AND and NOT gates (reprinted by permission from reference [22], copyright 2005, American Chemical Society). B) Abstract diagram illustrating aptamer-based circuitry for logical control of protein activity. The circuit consists of three modules including input convertor, threshold controller, and inhibitor generator, which can be programmed with threshold control for smart manipulation of thrombin coagulation activity (reprinted by permission from reference [24], copyright 2012, American Chemical Society).
To address the second question of using NA-based circuits for precise control of molecular functions, our group has developed a logical circuit based on DNA–protein interactions with accurate threshold control, enabling autonomous, self-sustained, and programmable manipulation of protein activity in vitro (Figure 3B).[24] In general, a programmable and autonomous circuit with threshold control is constructed of three DNA modules: 1) an input convertor that converts the protein input to DNA input for downstream cascade reactions, 2) a threshold controller that sets the threshold concentration for the system to maintain regular protein activity, and 3) an inhibitor generator that inhibits excessively high protein activity once it surpasses the threshold. This circuit can intelligently sense the activity (i.e., the concentration) of protein and initiate the inhibitory function by means of a threshold control loop when excessively high protein activity occurs. By setting the threshold value according to each practical situation, the circuit may be usable as a smart drug delivery system in the design of personalized medicine.
In vitro hybrid systems composed of NA elements and purified enzymes further expand the applicability of NA-based logical devices. In this context, enzymes, with their exquisite catalytic properties, offer an effective experimental implementation for both signal generation (amplification) and waste product degradation, thus allowing the construction of pseudo-open systems on separate timescales.[25] In this respect, researchers have developed different dynamic logical systems with DNA or RNA as signal loaders and enzymes as mediators. Kim et al. first constructed an in vitro analogue of a gene regulation circuit in which two “genelets” (short DNA duplexes that contain an incomplete promoter) are able to control each other through the transcription of their RNA outputs mediated by T7 RNA polymerase and RNase H.[26] Consequently, this design behaved as a bistable switch and allowed the dynamic control of signals. Rondelez and co-workers further engineered an in vitro oscillator by using DNA as a template and enzymes as mediators and obtained predicted oscillation dynamics.[27] Specifically, a positive-feedback loop was designed using a DNA template (T1) that can self-duplicate in the presence of the DNA polymerase and nicking enzyme. A single-strand specific exonuclease is able to degrade the generated α-strand, while templates are protected by DNA backbone modifications. Then an inhibitory interaction that prevents T1 from amplification was incorporated by an inhibitor with stronger binding ability to T1. By connecting the production of the inhibitor to the presence of the α-strand and thereby closing the negative- feedback loop, this system exhibited continuous oscillation behavior that obeyed the quantitative mathematical analysis.
Applying NA-based Logical Systems for Cellular Applications
A blueprint of a NA-based logical system needs to be translated into an actual system operating in a living cell. To this end, the most intensive efforts have involved the harnessing of practical applications like sensing, regulating, and rewiring biological systems. Reproduction of the logical functions in biological systems is not trivial, especially in terms of device robustness and stability. On the one hand, researchers have focused on enhancing the reaction robustness to mutations or impurities in the sequences, as well as NA hybridization robustness over large ranges of temperature and salt conditions.[14, 28] On the other hand, increasing the stability of devices against complex cellular environments, such as enzymatic digestion, provides another possible solution.[29] The references cited here may provide solid experimental platforms for the possible implementation of complex NA-based logical systems in vivo.
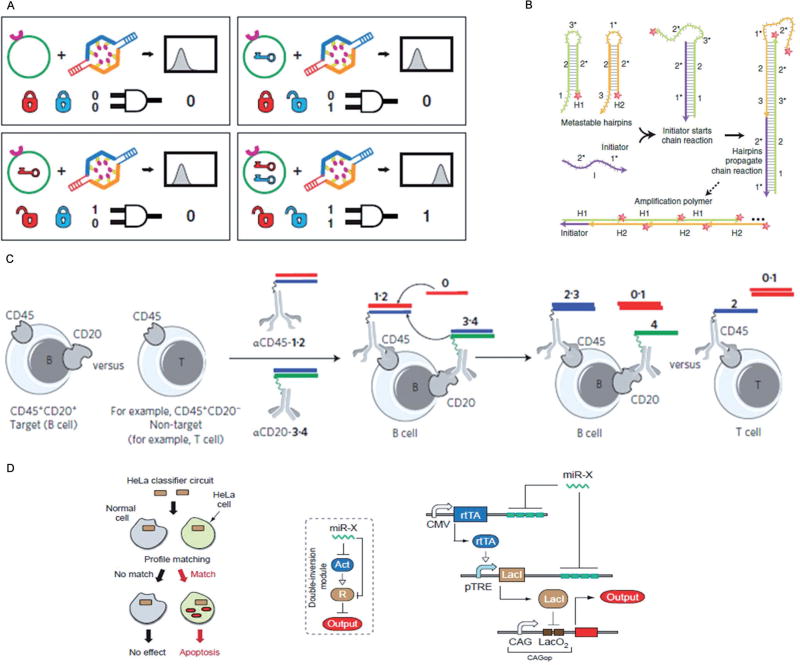
Pioneer work started with the application of intelligent sensing of cancer cells via biomarkers on cell membranes. Douglas et al. developed a DNA nanorobot with the ability to detect antigen information on cell membranes and autonomously release the payloads according to the different combinations of cell surface inputs (Figure 4 A).[30] Specifically, this nanorobot was assembled by the DNA origami method[31] in the shape of a hexagonal barrel with dimensions of 35 nm × 35 nm × 45 nm. The nanobarrel was noncovalently fastened in the front by staples modified with locks based on DNA aptamer that could be specifically opened through DNA–protein binding to cell surface proteins. Then the payloads, which were either labeling antibodies or gold nanoparticles, were selectively released to realize intelligent sensing and delivery. A similar idea was implemented by Stojanovic and co-workers, who built DNA-based logical devices on cell membranes for intelligent sensing (Figure 4C).[32] First, the DNA probes were conjugated with antibodies independently targeting specific antigens on cell membranes. The cell-bound probes would then undergo a cascade of reactions for fundamental logical operations including AND, OR, and NOT, through toehold-mediated displacement reaction.[33] This DNA-based logical device provides a reliable method for analysis of complex cell membrane information and the accurate identification of stem cells.
Figure 4.
NA-based logical circuits for cellular applications. A) Illustration and truth table showing how this aptamer-based nanorobot works (reprinted by permission from reference [30], copyright 2012, AAAS). B) Hybridization chain reaction mechanism (from reference [35a], reprinted with permission from Macmillan Publishers Ltd., Nat. Biotechnol., copyright 2010). C) Scheme of DNA-based logical circuit operating on a target B cell with a C45+CD20+ phenotype and on a nontarget cell with a CD45+CD20− phenotype (e.g., a T cell) (reference [32], reprinted by permission from Macmillan Publishers Ltd., Nat. Nanotechnol., copyright 2013). D) Schematic representation of a HeLa-specific classifier circuit operation (from reference [36], reprinted with permission from AAAS).
To further demonstrate the feasibility of assembling a DNA circuit on cell membranes and to broaden the utility of DNA circuits for cellular applications, our group also designed an aptamer-based DNA logical circuit constructed from cell membranes capable of selective recognition of cancer cells, controllable activation of a photosensitizer, and amplification of the photodynamic therapeutic (PDT) effect.[34] Here, the aptamers are able to selectively recognize target cancer cells and bind to the specific proteins on the cell membranes. An overhanging catalyst sequence on the aptamer then triggers a toehold-mediated catalytic strand displacement to activate the photosensitizer for amplified PDT. The specific binding-induced activation allows the DNA circuit to distinguish diseased cells from healthy cells, reducing damage to nearby healthy cells, while resulting in a high local concentration of singlet oxygen around diseased cells.
To go one step further from the cell surface to the interior, Choi and co-workers developed a programmable in situ amplification circuit based on a hybridization chain reaction (Figure 4 B).[35] With this approach, specific intracellular mRNAs can trigger a chain hybridization of metastable RNA hairpin molecules to self-assemble into tethered fluorescent amplification polymers. The programmability and sequence specificity of these amplification circuits enable multiple mRNA detection inside intact biological samples. Xie and co-workers designed an intracellular circuit that senses expression levels of a customizable set of endogenous microRNAs and classifies their expression patterns into a predetermined profile of cancer cells (Figure 4D).[36] Actually this idea originated in their previous approach, in which they designed an in vitro RNA computer that logically analyzed the levels of messenger RNA species and responded with a molecule capable of affecting levels of gene expression.[37] Also using microRNAs as manipulation elements, Deiters engineered a NA-based AND gate to respond to specific microRNA inputs and compute the presence or absence of multiple miRNAs in live mammalian cells.[38] These results may be useful for accurate and intelligent diagnosis in complex disease situations.
Going beyond logical sensing, the ability to rationally control and manipulate the functions of living cells will allow us to move a step further towards the use of NA-based logical systems in future biological and biomedical applications. In terms of constructing complex computation networks with numerous regulatory mechanisms, RNA interference (RNAi), antisense RNA, and exogenous regulatory elements, such as riboswitches, have been shown to function inside cells.[39] For example, Benenson et al. used RNA interference to assemble large-scale logical circuits in human kidney cells, thus allowing the cells to perform general Boolean logic to make decisions based on endogenous molecular inputs.[40] Smolke and Win rewired a yeast cell with the ability to compute two-input logical functions with small-molecule ligands as inputs. They fused ligand-binding aptamers to ribozyme moieties through a transmission linker that allows the binding event to activate or deactivate ribozyme.[41] These RNA-based intracellular devices may offer an exciting direction towards programmable cells and cell populations, and they promise to become a basis of large synthetic systems.
Summary and Outlook
The field of NA-based computation clearly has advanced to a stage at which arbitrary logical functions can be rationally designed in a predictable manner in vitro. With respect to device construction, logical systems can be integrated at the nucleic acid level, and inputs and outputs may be DNA, RNA, proteins, or small molecules. Operationally, the behaviors of these NA-based devices can be either static or oscillatory, in which the output may be produced, degraded, or modified reversibly or irreversibly in response to design principles. Furthermore, it should not be forgotten that the efforts of using programs to aid in device design greatly improve experimental implementation efficiencies. Combining these points together, both the quality and quantity of NA-based logical devices have experienced a dramatic increase in the last ten years. However, this does not mean that these devices have reached a mature state. Some limitations still need to be overcome prior to their translation into real-world applications. For example, the relatively slow reaction kinetics and high error rates may hinder the practical implementation of theoretical parallelism. Furthermore, the lack of biological stability or cooperative integration in complex environments may challenge in vivo applications.[42]
Despite these limitations, the unique feature of logical NA-based systems for interfacing living and nonliving matter still makes it important for various applications. On a fundamental level, NA-based logical systems can be used to control the temporal and spatial process of molecular operations or assembly reactions.[43] Moreover, logical circuitry may one day lead to the development of “smart drugs” that can sense and analyze multiple physiological cues and then perform logical operations to release drugs or regulate genetic expressions.[44] Another direction includes mimicking biological systems by using dynamic NA-based devices to gain more insight into naturally occurring systems, with concomitant translational applications, for example, in the design of artificial neural networks exhibiting autonomous brainlike behaviors.[11] It is difficult to predict how far these NA-based logical systems can go, as new technologies will continue to emerge. However, it is certain that highly sophisticated programmed architectures will continue to be designed, and they will find widespread applications in biology, biotechnology, and biomedicine.
Acknowledgments
D.H. acknowledges the ACS Division of Analytical Chemistry Fellowship sponsored by the Society for Analytical Chemists of Pittsburgh. H.K acknowledges the support from National Instrumentation Program (NIP) of China. This work is supported by the National Key Scientific Program of China (2011CB911000), NSFC grants (NSFC 21221003 and NSFC 21327009) and China National Instrumentation Program 2011YQ03012412, and by the National Institutes of Health (GM079359 and CA133086).
References
- 1.Ball P. Nature. 2000;406:118–120. doi: 10.1038/35018259. [DOI] [PubMed] [Google Scholar]
- 2.a) Goldman N, Bertone P, Chen SY, Dessimoz C, LeProust EM, Sipos B, Birney E. Nature. 2013;494:77–80. doi: 10.1038/nature11875. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Church GM, Gao Y, Kosuri S. Science. 2012;337:1628–1628. doi: 10.1126/science.1226355. [DOI] [PubMed] [Google Scholar]
- 3.Adleman LM. Science. 1994;266:1021–1024. doi: 10.1126/science.7973651. [DOI] [PubMed] [Google Scholar]
- 4.Benenson Y. Mol. Biosyst. 2009;5:675–685. doi: 10.1039/b902484k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yurke B, Turberfield AJ, Mills AP, Simmel FC, Neumann JL. Nature. 2000;406:605–608. doi: 10.1038/35020524. [DOI] [PubMed] [Google Scholar]
- 6.Turberfield AJ, Mitchell JC, Yurke B, Mills AP, Jr, Blakey MI, Simmel FC. Phys. Rev. Lett. 2003;90:118102. doi: 10.1103/PhysRevLett.90.118102. [DOI] [PubMed] [Google Scholar]
- 7.Seelig G, Soloveichik D, Zhang DY, Winfree E. Science. 2006;314:1585–1588. doi: 10.1126/science.1132493. [DOI] [PubMed] [Google Scholar]
- 8.Li W, Yang Y, Yan H, Liu Y. Nano Lett. 2013;13:2980–2988. doi: 10.1021/nl4016107. [DOI] [PubMed] [Google Scholar]
- 9.Zhang DY, Winfree E. J. Am. Chem. Soc. 2009;131:17303–17314. doi: 10.1021/ja906987s. [DOI] [PubMed] [Google Scholar]
- 10.Zhang DY, Turberfield AJ, Yurke B, Winfree E. Science. 2007;318:1121–1125. doi: 10.1126/science.1148532. [DOI] [PubMed] [Google Scholar]
- 11.Qian L, Winfree E, Bruck J. Nature. 2011;475:368–372. doi: 10.1038/nature10262. [DOI] [PubMed] [Google Scholar]
- 12.Qian L, Winfree E. Science. 2011;332:1196–1201. doi: 10.1126/science.1200520. [DOI] [PubMed] [Google Scholar]
- 13.Chen YJ, Dalchau N, Srinivas N, Phillips A, Cardelli L, Soloveichik D, Seelig G. Nat. Nanotechnol. 2013;8:755–762. doi: 10.1038/nnano.2013.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang DY, Chen SX, Yin P. Nat. Chem. 2012;4:208–214. doi: 10.1038/nchem.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stojanovic MN, Mitchell TE, Stefanovic D. J. Am. Chem. Soc. 2002;124:3555–3561. doi: 10.1021/ja016756v. [DOI] [PubMed] [Google Scholar]
- 16.Stojanovic MN, Semova S, Kolpashchikov D, Macdonald J, Morgan C, Stefanovic D. J. Am. Chem. Soc. 2005;127:6914–6915. doi: 10.1021/ja043003a. [DOI] [PubMed] [Google Scholar]
- 17.a) Stojanovic MN, Stefanovic D. Nat. Biotechnol. 2003;21:1069–1074. doi: 10.1038/nbt862. [DOI] [PubMed] [Google Scholar]; b) Pei RJ, Matamoros E, Liu MH, Stefanovic D, Stojanovic MN. Nat. Nanotechnol. 2010;5:773–777. doi: 10.1038/nnano.2010.194. [DOI] [PubMed] [Google Scholar]
- 18.Elbaz J, Lioubashevski O, Wang FA, Remacle F, Levine RD, Willner I. Nat. Nanotechnol. 2010;5:417–422. doi: 10.1038/nnano.2010.88. [DOI] [PubMed] [Google Scholar]
- 19.Genot AJ, Bath J, Turberfield AJ. J. Am. Chem. Soc. 2011;133:20080–20083. doi: 10.1021/ja208497p. [DOI] [PubMed] [Google Scholar]
- 20.Prokup A, Hemphill J, Deiters A. J. Am. Chem. Soc. 2012;134:3810–3815. doi: 10.1021/ja210050s. [DOI] [PubMed] [Google Scholar]
- 21.Huang FJ, You MX, Han D, Xiong XL, Liang HJ, Tan WH. J. Am. Chem. Soc. 2013;135:7967–7973. doi: 10.1021/ja4018495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolpashchikov DM, Stojanovic MN. J. Am. Chem. Soc. 2005;127:11348–11351. doi: 10.1021/ja051362f. [DOI] [PubMed] [Google Scholar]
- 23.Zhou M, Du Y, Chen CG, Li BL, Wen D, Dong SJ, Wang EK. J. Am. Chem. Soc. 2010;132:2172. doi: 10.1021/ja910634e. [DOI] [PubMed] [Google Scholar]
- 24.Han D, Zhu Z, Wu CC, Peng L, Zhou LJ, Gulbakan B, Zhu GZ, Williams KR, Tan WH. J. Am. Chem. Soc. 2012;134:20797–20804. doi: 10.1021/ja310428s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li YF, Qian H, Yi YF. J. Chem. Phys. 2008;129:154505. doi: 10.1063/1.2995855. [DOI] [PubMed] [Google Scholar]
- 26.Kim J, White KS, Winfree E. Mol. Syst. Biol. 2006;2:68. doi: 10.1038/msb4100099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montagne K, Plasson R, Sakai Y, Fujii T, Rondelez Y. Mol. Syst. Biol. 2011;7:466. doi: 10.1038/msb.2010.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.a) Chen SX, Zhang DY, Seelig G. Nat. Chem. 2013;5:782–789. doi: 10.1038/nchem.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zhang DY, Winfree E. Nucleic Acids Res. 2010;38:4182–4197. doi: 10.1093/nar/gkq088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.a) Wu YR, Sefah K, Liu HP, Wang RW, Tan WH. Proc. Natl. Acad. Sci. USA. 2010;107:5–10. doi: 10.1073/pnas.0909611107. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wang L, Yang CYJ, Medley CD, Benner SA, Tan WH. J. Am. Chem. Soc. 2005;127:15664–15665. doi: 10.1021/ja052498g. [DOI] [PubMed] [Google Scholar]
- 30.Douglas SM, Bachelet I, Church GM. Science. 2012;335:831–834. doi: 10.1126/science.1214081. [DOI] [PubMed] [Google Scholar]
- 31.Rothemund PWK. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 32.Rudchenko M, Taylor S, Pallavi P, Dechkovskaia A, Khan S, Butler VP, Rudchenko S, Stojanovic MN. Nat. Nanotechnol. 2013;8:580–586. doi: 10.1038/nnano.2013.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.See ref. [9].
- 34.Han D, Zhu GZ, Wu CC, Zhu Z, Chen T, Zhang XB, Tan WH. ACS Nano. 2013;7:2312–2319. doi: 10.1021/nn305484p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.a) Choi HM, Chang JY, Trinh le A, Padilla JE, Fraser SE, Pierce NA. Nat. Biotechnol. 2010;28:1208–1212. doi: 10.1038/nbt.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Dirks RM, Pierce NA. Proc. Natl. Acad. Sci. USA. 2004;101:15275–15278. doi: 10.1073/pnas.0407024101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie Z, Wroblewska L, Prochazka L, Weiss R, Benenson Y. Science. 2011;333:1307–1311. doi: 10.1126/science.1205527. [DOI] [PubMed] [Google Scholar]
- 37.Benenson Y, Gil B, Ben-Dor U, Adar R, Shapiro E. Nature. 2004;429:423–429. doi: 10.1038/nature02551. [DOI] [PubMed] [Google Scholar]
- 38.Hemphill J, Deiters A. J. Am. Chem. Soc. 2013;135:10512–10518. doi: 10.1021/ja404350s. [DOI] [PubMed] [Google Scholar]
- 39.Benenson Y. Curr. Opin. Biotechnol. 2009;20:471–478. doi: 10.1016/j.copbio.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rinaudo K, Bleris L, Maddamsetti R, Subramanian S, Weis R, Benenson Y. Nat. Biotechnol. 2007;25:795–801. doi: 10.1038/nbt1307. [DOI] [PubMed] [Google Scholar]
- 41.Win MN, Smolke CD. Science. 2008;322:456–460. doi: 10.1126/science.1160311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen X, Ellington AD. Curr. Opin. Biotechnol. 2010;21:392–400. doi: 10.1016/j.copbio.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Franco E, Friedrichs E, Kim J, Jungmann R, Murray R, Winfree E, Simmel FC. Proc. Natl. Acad. Sci. USA. 2011;108:E784–E793. doi: 10.1073/pnas.1100060108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gil B, Kahan-Hanum M, Skirtenko N, Adar R, Shapiro E. Nano Lett. 2011;11:2989–2996. doi: 10.1021/nl2015872. [DOI] [PubMed] [Google Scholar]