Abstract
Though prostate cancer is often indolent, it is nonetheless a leading cause of cancer death. Defining the underlying molecular genetic alterations may lead to new strategies for prevention or treatment. Towards this goal, we performed array-based comparative genomic hybridization (CGH) on 86 primary prostate tumors. Among the most frequent alterations not associated with a known cancer gene, we identified focal deletions within 5q21 in 15 out of 86 (17%) cases. By high-resolution tiling array CGH, the smallest common deletion targeted just one gene, the chromatin remodeler chromodomain helicase DNA-binding protein 1 (CHD1). Expression of CHD1 was significantly reduced in tumors with deletion (P=0.03), and compared with normal prostate (P=0.04). Exon sequencing analysis also uncovered nonsynonymous mutations in 1 out of 7 (14%) cell lines (LAPC4) and in 1 out of 24 (4%) prostate tumors surveyed. RNA interference-mediated knockdown of CHD1 in two nontumorigenic prostate epithelial cell lines, OPCN2 and RWPE-1, did not alter cell growth, but promoted cell invasiveness, and in OPCN2-enhanced cell clonogenicity. Taken together, our findings suggest that CHD1 deletion may underlie cell invasiveness in a subset of prostate cancers, and indicate a possible novel role of altered chromatin remodeling in prostate tumorigenesis.
Keywords: prostate cancer, genomic profiling, tumor suppressor, chromatin remodeling, cell invasion
Introduction
Prostate cancer is the most frequently diagnosed cancer and the second leading cause of cancer death among men in the United States (Jemal et al., 2010); approximately one in six men will be diagnosed within their lifetime. Prostate cancer exhibits a range of clinical behaviors, from indolent growth to highly aggressive and metastatic disease. Key unmet clinical needs include distinguishing indolent from aggressive cancer (to determine whether and how aggressively to treat), and identifying effective therapies for later-stage castration-recurrent prostate cancer (Damber and Aus, 2008). Defining the full range of molecular genetic alterations in prostate cancer should provide improved understanding and new targets for prevention and treatment.
The molecular alterations underlying prostate cancer are partially understood (DeMarzo et al., 2003; Shen and Abate-Shen, 2010). Common events include deletion of tumor suppressors, including CDKN1B (p27/KIP1), RB1, TP53, PTEN and the prostate-specific homeobox transcription factor NKX3-1. Amplification of the MYC oncogene is also frequent. In addition, oncogenic fusions driving ETS-family oncogenic transcription factors (ERG, ETV1, ETV4 and ETV5), most commonly as TMPRSS2-ERG, have been identified in approximately half of prostate cancers. Androgen receptor alterations, including amplification and rearrangement, can also occur in castration-recurrent prostate cancer.
More recently, genomic profiling by comparative genomic hybridization (CGH) and single-nucleotide polymorphism arrays have provided comprehensive views of DNA copy number alterations in prostate cancer (Lapointe et al., 2004; Kim et al., 2007; Robbins et al., 2011), and have led to the nomination of new prostate cancer genes, for example, NCOA2 (Taylor et al., 2010). Next-generation genome sequencing is also now beginning to reveal the full landscape of somatic rearrangements (Berger et al., 2011).
Here, we have carried out genomic profiling by array CGH of a large collection of primary prostate tumors. Among our findings, we identify focal recurrent deletions within 5q21 targeting the chromatin remodeler chromodomain helicase DNA-binding protein (CHD1), whose loss we characterize to be a driver of cancer cell invasiveness.
Results and discussion
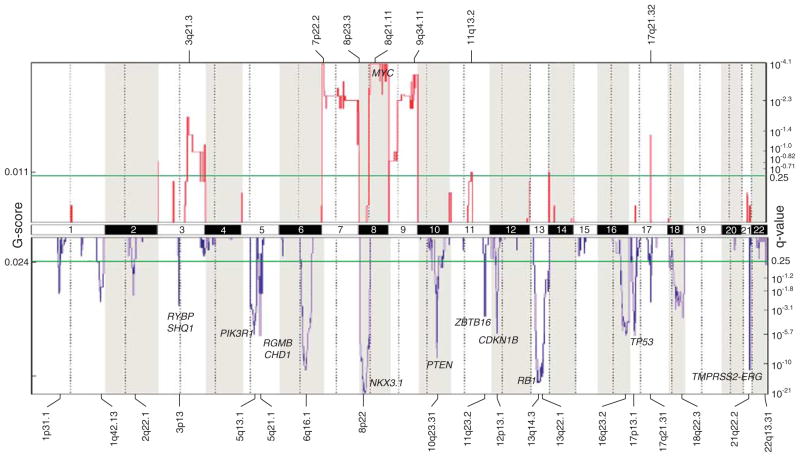
To survey the landscape of DNA copy number alterations in prostate cancer, we profiled a collection of 86 primary prostate tumors (clinicopathological characteristics summarized in Supplementary Table S1) using Agilent 44K CGH arrays (Agilent, Santa Clara, CA, USA). Genomic Identification of Significant Targets in Cancer analysis (Beroukhim et al., 2007) was applied to identify loci with significantly recurrent copy number alteration, which included 19 deletions and 7 amplifications (Figure 1). Deletions of known tumor suppressors included (ordered by chromosome) NKX3-1 (8p21.2) (occurring in 38% of tumors), PTEN (10q23.31) (22%), CDKN1B (12p13.1) (20%), RB1 (13q14.2) (40%) and TP53 (17p13.1) (21%). Deletion of 21q22.2 (29%) also defined intrachromosomal rearrangements generating TMPRSS2-ERG. Frequent gains included those on chromosomes 7 (12%) and 8 (9%), the latter spanning MYC (8q24.21), and 9 (10%).
Figure 1.
Landscape of copy number alterations in prostate cancer. Genomic Identification of Significant Targets in Cancer (GISTIC) plot identifies significant DNA copy number alterations (by consideration of both frequency and amplitude, and in comparison with randomly permuted data). Gains and losses are depicted in red and blue, respectively, and ordered by genome position. The significance threshold (false discovery rate, <0.25) is indicated, as are selected known cancer genes. Prostate tumors were obtained from radical prostatectomy cases performed at the Stanford University Hospital, with the Institutional Review Board approval and patient informed consent. Freshly-frozen specimens were cryostat sectioned and scalpel macrodissected to enrich tumor nuclei to >80%. Genomic DNA (and total RNA) were isolated using Qiagen Allprep DNA/RNA Mini Kits (Qiagen, Valencia, CA, USA). Tumor DNA and normal male reference DNA (pooled from eight donor leukocyte preparations) were respectively labeled with Cy5 and Cy3, as described (Kwei et al., 2008), then co-hybridized onto Agilent Human Genome CGH 44K arrays. Microarrays were scanned using an Agilent G2505C Microarray Scanner System, and normalized fluorescence ratios obtained using Feature Extraction 9.5.3 (Agilent). DNA gains and losses were called by Circular Binary Segmentation (Olshen et al., 2004) and significant alterations defined by GISTIC (Beroukhim et al., 2007). Array CGH data are accessible through the Gene Expression Omnibus (GSE29229).
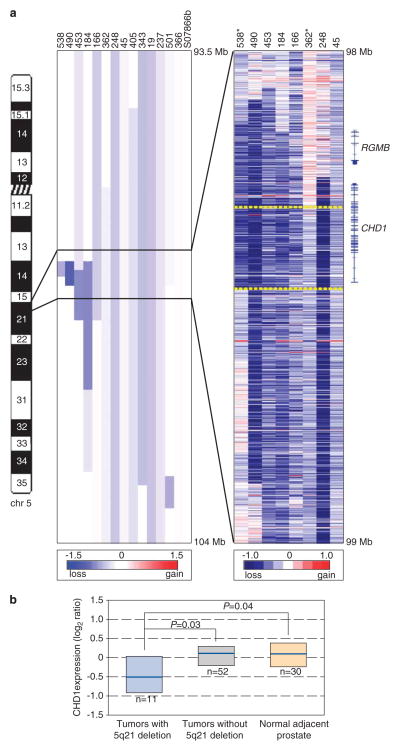
Among the most significant deletions not associated with a known tumor suppressor (and therefore presumably pinpointing a novel candidate), we noted deletions at 5q21.1 in 15 out of 86 (17%) prostate tumors. To more precisely map the boundaries of these deletions, we designed a custom high-density Agilent 15K CGH array, with probes covering 6.5Mb within 5q15–q21.1 and tiled every 500 bp. Profiling eight samples with informative deletions, we could delimit a smallest common region of deletion of 120 kb (Figure 2a). Remarkably, the commonly deleted region targeted just one gene, CHD1.
Figure 2.
The 5q21 deletion targets CHD1. (a) Left, Agilent 44K CGH array heatmap view of deletions spanning 5q21 in prostate tumors with deletion. Blue intensity (scale shown) identifies CBS-called deletions. Right, Agilent custom-tiling CGH array heatmap view of raw copy number ratios (scale indicated) for selected samples with informative deletions. Tumor samples no. 538 and no. 362 (asterisked) define the 120-kb smallest region of shared deletion (bracketed by yellow-dashed lines), which affects only CHD1. The exon structure of CHD1, and the only other nearby gene, RGMB, are shown. Note, for some samples, apparent deletion boundary discrepancies between the Agilent 44K CGH array and the custom tiling array data reflect the lower resolution and CBS-based smoothing in the former. The Agilent high-density custom CGH 15K array was designed using eArray 6.5 software, with probes spanning a subregion of 5q15–q21.1 (96.0–102.0 Mb; build 18) tiled on average every 500 bp. (b) Transcript levels of CHD1, measured by oligonucleotide microarray, are reduced in tumor samples with 5q21 deletion compared with samples without 5q21 deletion, and compared with normal prostate. Box plots show 25th, 50th (median) and 75th percentiles of sample-set expression. P-values (two-sided Mann–Whitney U-test) are shown. CHD1 transcript levels were obtained by HEEBO oligonucleotide microarray (http://www.microarray.org/sfgf/heebo.do) profiling, carried out in parallel on 63 of the same tumor samples, and, for a subset, matched adjacent-normal tissue. Values are reported as log2 ratios, normalized to the sample-set mean. The full gene-expression dataset will be detailed separately (Gulzar and Brooks; manuscript in preparation).
CHD1 is a member of the chromodomain helicase DNA-binding (CHD) family, a group of ATP-dependent chromatin remodelers that use the energy from ATP hydrolysis to alter histone–DNA contacts within nucleosomes to modulate gene expression (Hall and Georgel, 2007; Marfella and Imbalzano, 2007). All CHD family proteins (CHD1–9) have two tandem chromo (chromatin organization modifier) domains at the N-terminus and a SNF2-related helicase/ATPase domain in the central region; CHD1 and CHD2 also have a C-terminal DNA-binding domain (Hall and Georgel, 2007). Intriguingly, other CHD family members have been linked to cancer, most notably CHD5 deletions in neural-associated tumors (Bagchi and Mills, 2008).
Consistent with a possible tumor-suppressor role, CHD1 transcript levels (measured by microarray) were significantly decreased (1.4-fold on average) in the subset of prostate tumors with 5q21 deletion (P=0.03, two-tailed Mann–Whitney U-test), and in comparison with normal prostate tissue (P=0.04) (Figure 2b). Neither CHD1 deletion nor decreased expression was significantly associated with specific clinicopathological features, including preoperative serum prostate-specific antigen levels, tumor Gleason grade, tumor stage or biochemical recurrence (Supplementary Table S2). Among a collection of prostate cell lines, CHD1 protein levels were lower in established prostate cancer lines (DU145, LAPC-4, LNCaP, PC-3, PCA MDA 2a and PCA MDA 2b) compared with nontumorigenic prostate epithelial lines (OPCN2 and RWPE-1) (P<0.05, Mann–Whitney U-test) (Supplementary Figure S1). However, within the small set of prostate cancer lines, there was no significant relation between CHD1 copy number (by array CGH) and CHD1 protein levels (Supplementary Figure S1).
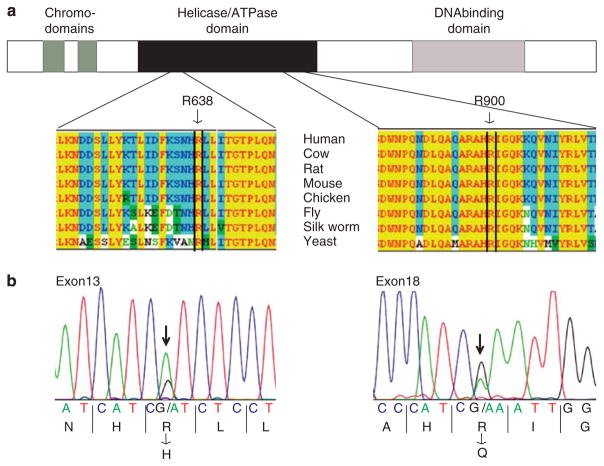
To obtain additional evidence for a tumor-suppressor role, we sequenced all 35 exons (coding regions and splice sites) of CHD1 in a sampling of 24 prostate tumors (Lapointe et al., 2007), including 5 with CHD1 deletion, as well as in 7 prostate cancer cell lines (ARCaP, DU145, LAPC-4, LNCaP, PC-3, PCA MDA 2a and PCA MDA 2b). Altogether, we identified two nonsynonymous mutations, an arginine (R638) to histidine mutation within the helicase/ATP-binding domain in 1 out of the 7 cell lines (14%), LAPC-4, and an arginine (R900) to glutamine mutation within the helicase/ATPase domain in 1 out of the 24 (4%) tumors (a sample without CHD1 deletion) (Figure 3). The latter mutation was confirmed to be somatic by its absence from the matched normal prostate DNA. Both mutations occur at amino-acid positions evolutionarily conserved from yeast to human (Figure 3a), and are strongly predicted to impair protein function (multivariate analysis of protein polymorphism P-values ≤0.001) (Binkley et al., 2010).
Figure 3.
DNA sequence mutations of CHD1. (a) Schematic illustration of the CHD1 protein, with functional domains annotated, along with identified mutations. Below, amino-acid sequence conservation is shown for the regions of CHD1 harboring identified mutations. (b) Sanger sequence traces of identified mutations, along with corresponding encoded amino acids. For sequencing, PCR primers covering all 35 exons (coding regions plus splice sites) of CHD1 were designed using ExonPrimer (http://ihg.gsf.de/ihg/ExonPrimer.html). Primer sequences and PCR conditions are available in Supplementary Table S3. PCR products were bidirectionally Sanger-sequenced (Geneway Research, Hayward, CA, USA), and mutations identified by BLAST alignment of sequence reads to the reference genome. All sequence traces were also manually reviewed. Amino-acid sequence conservation alignments were done using Jellyfish 3.3.1 software (Field Scientific, Lewisburg, PA, USA). Predictions of the functional impact of mutations were done using ProPhylER, based on evolutionary constraint and the physicochemical properties of the substituted amino acid (Binkley et al., 2010).
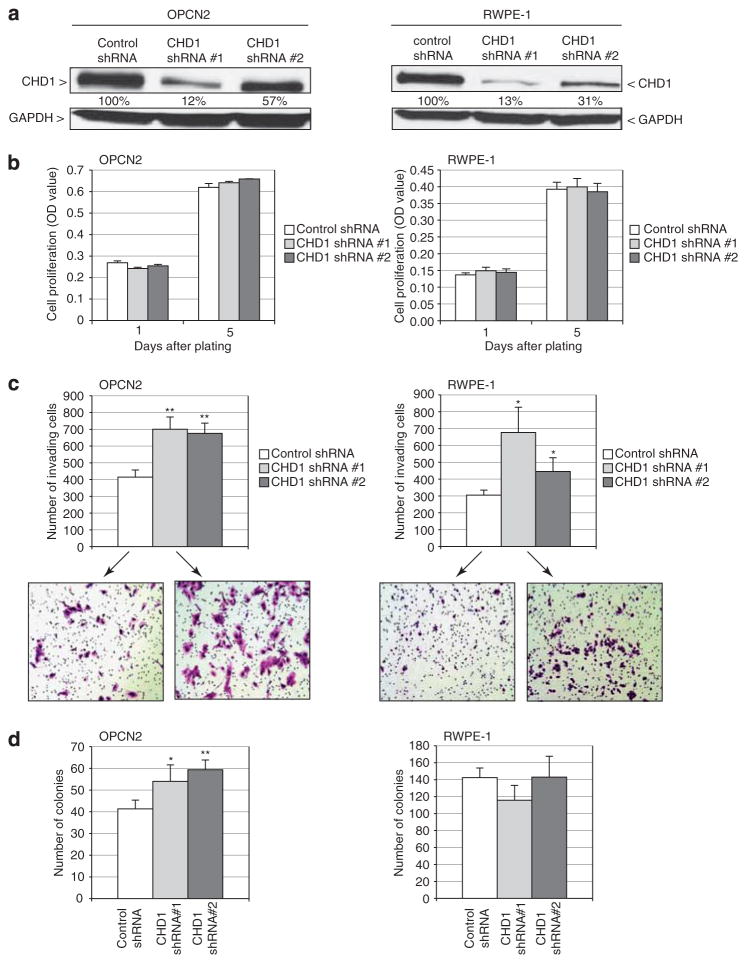
To more directly assess a tumor-suppressive function of CHD1, we used RNA interference to knock down its expression (simulating deletion) in the two immortalized but nontumorigenic prostate epithelial cell lines, OPCN2 and RWPE-1. Transduction of either of two different short hairpin RNAs targeting CHD1 led to reduced CHD1 protein levels, compared with a non-targeting short hairpin RNA control (Figure 4a). Resultant CHD1 knockdown did not significantly affect cell viability/proliferation (measured by mitochondrial activity; Figure 4b). In contrast, CHD1 knockdown in both OPCN2 and RWPE-1 resulted in significantly increased cell invasion through Matrigel (P<0.05, two-tailed Student’s t-test) (Figure 4c). In OPCN2 cells, CHD1 knockdown also enhanced cell clonogenicity (that is, colony formation on tissue culture plastic) (P<0.05) (Figure 4d); however, this finding was not apparent in RWPE-1 cells, suggesting invasiveness to be the more relevant and universal phenotype. Neither OPCN2 nor RWPE-1 parental cells were capable of anchorage-independent growth (that is, colony formation in soft agar), and CHD1 knockdown did not induce anchorage-independent growth (data not shown). Extending our findings to an established prostate cancer line, knockdown of CHD1 also increased invasiveness of LNCaP cells (P<0.01) (Supplementary Figure S2).
Figure 4.
CHD1 knockdown enhances cell invasiveness. (a) CHD1 knockdown by two independent short hairpin RNAs (shRNAs), in two different nontumorigenic prostate epithelial cell lines (OPCN2 and RWPE-1) verified by western blot analysis. Percent residual expression (relative to control nontargeting shRNA, and normalized to glyceraldehyde-3-phosphate-dehydrogenase) is indicated. OPCN2 cells were purchased from Asterand (Detroit, MI, USA) and grown in keratinocyte serum-free media (Invitrogen, Grand Island, NY, USA) supplemented with 2% fetal bovine serum (Hyclone, Logan, UT, USA). RWPE-1 cells were obtained directly from the American Type Culture Collection and grown in keratinocyte serum-free media supplemented with 0.05 mg/ml bovine pituitary extract and 5 ng/ml epidermal growth factor (Invitrogen). For RNAi-mediated knockdown, two different GIPZ shRNAs targeting CHD1 (112 969 and 112 972), along with a non-silencing-GIPZ shRNA, were obtained from Open Biosystems (Lafayette, CO, USA). The shRNAs were separately transfected (using Fugene HD reagent; Roche, Indianapolis, IN, USA) into 293TN cells, along with VSVG and pCMVdelta8.91 packaging plasmids, to generate replication-defective lentivirus. Viral supernatant was collected 48 h post transfection and used to infect OPCN2 and RWPE-1 cells. Pooled transductants were selected using 1 μg/μl puromycin for 14 days. Cells were lysed in RIPA buffer, and 30 μg whole-cell lysate was electrophoresed on a 4–15% Tris-HCl polyacrylamide gradient gel. Protein was transferred onto a polyvinylidene fluoride membrane, which was then blocked for 1 h with 5% dry milk in TBS-T buffer, and then incubated overnight with primary antibody at 4 °C. Detection was carried out using a horseradish peroxidase-conjugated secondary antibody and an enhanced chemiluminescence (ECL) kit (GE Healthcare, Piscataway, NJ, USA). Quantification was performed using ImageJ (http://imagej.nih.gov/ij). Antibodies used were anti-CHD1 rabbit polyclonal antibody (NB100-60411, 1:200; Novus biologicals, Littleton, CO, USA), anti-glyceraldehyde-3-phosphate-dehydrogenase (sc-25778, Santa Cruz Biotechnology, Santa Cruz, CA, USA), and horseradish-peroxidase conjugated anti-rabbit IgG (1:10 000; Pierce, Rockford, IL, USA). (b) CHD1 knockdown does not significantly alter cell viability/proliferation, measured by WST-1 assay 1 and 5 days after plating. WST-1 (Roche) is a colorimetric assay of mitochondrial activity in viable cells. (c) CHD1 knockdown promotes cell invasion through Matrigel. Photomicrographs show representative fields of crystal-violet stained traversed cells. *P<0.01; **P<0.01, two-sided Student’s t-test. Cell invasion was measured by Boyden chamber assay (BD Biosciences, Bedford, MA, USA). In all, 50 000 cells/24-well insert for OPCN2, or 100 000 cells for RWPE-1, were seeded onto pre-coated filters (8 μM pore size, Matrigel 100 μg/cm2), using a 0.5–10% fetal bovine serum gradient. After 24–48 h, cells traversing the filter were fixed with 10% buffered formalin, stained with crystal violet and manually counted. (d) CHD1 knockdown enhances cell clonogenicity (colony formation on tissue culture plastic) of OPCN2 cells. *P<0.01; **P<0.01, two-sided Student’s t-test. Clonogenicity was measured by sparse plating of 600 cells/15-cm plate. After 2 weeks, plates were stained with Giemsa and the number of colonies was counted. All the above assays were done in triplicate (mean±1 s.d. reported), and all experiments were replicated at least once.
To obtain additional mechanistic insight, we profiled the gene-expression changes occurring with CHD1 knockdown in each of the three cell lines (OPCN2, RWPE-1 and LNCaP) showing increased invasiveness. However, a common molecular link to invasiveness was not obvious, as none of the known effectors of invasion (for example, matrix metalloproteinases and the urokinase plasminogen pathway genes) (Mareel and Leroy, 2003; Kessenbrock et al., 2010; Mason and Joyce, 2011) showed consistently altered expression across the three cell line model systems (Supplementary Figure S3).
Other members of the CHD family have been implicated in human disease, including cancer (Hall and Georgel, 2007; Marfella and Imbalzano, 2007). Mice heterozygous for CHD2 knockout show reduced survival, due to widespread lymphoid hyperplasias and lymphomas (Nagarajan et al., 2009). Mice heterozygous for CHD5 deficiency are more prone to develop spontaneous tumors, and derived murine embryo fibroblasts exhibit enhanced proliferation and survival via the p19(Arf)/p53 pathway (Bagchi et al., 2007). Deletion of CHD5 (at 1p36.31) has been reported in various neural-associated and epithelial malignancies, the latter including colorectal, breast and cervical cancer (Bagchi and Mills, 2008). Mutation of CHD5 was also recently reported in a single prostate cancer sample (Robbins et al., 2011).
CHD1 itself had not been linked to cancer. Very recently, though, Berger et al., (2011) carried out paired-end whole-genome Illumina DNA sequencing of seven high-grade prostate cancers. From that analysis, they identified (among many other altered genes) a rearrangement breakpoint within CHD1 in two samples, and a splice-site mutation in a third sample, all predicted to truncate CHD1 protein. Our findings now define the census of structural alterations affecting CHD1, as well as provide key functional data supporting a tumor-suppressive role (where CHD1-loss promotes invasiveness). In the COSMIC catalog of somatic mutations in cancer (Forbes et al., 2010), CHD1 mutations are recorded infrequently in other cancers, including breast, lung, ovarian, and pancreatic cancers. The role of CHD1 in those and other cancer types remains to be determined.
In our study, deletion of CHD1 was far more common than DNA sequence mutation, perhaps reflecting the relatively low rate of DNA mutation in prostate compared with other cancers (Taylor et al., 2010; Berger et al., 2011). Furthermore, deletions occurred as heterozygous alterations, likely reflecting haploinsufficiency for tumor suppression, as has been documented for CHD5 (Bagchi et al., 2007). Previously, we had carried out array CGH on fewer prostate tumors and at lower resolution (Lapointe et al., 2007). In that cohort, 5q21 deletion was associated with ‘subtype-1’ prostate cancers, a gene-expression subtype itself linked to lower risk of biochemical (prostate-specific antigen) recurrence (Lapointe et al., 2004). Recently, however, Taylor et al. (2010) identified 5q21 deletion to be associated with increased risk of biochemical recurrence. In our current study, there was no statistically significant association. Future studies may clarify the relationship of 5q21 deletion with tumor aggressiveness and clinical outcome.
Members of the CHD family have been described as both transcriptional activators and repressors (Hall and Georgel, 2007; Marfella and Imbalzano, 2007). Recent studies have shown that CHD1 expression is critical for maintaining mouse embryonic stem cell pluripotency by maintaining open chromatin (Gaspar-Maia et al., 2009). The possible transcriptional programs linking CHD1 to invasiveness and tumor suppression remain to be elucidated, but may be complex, given that this chromatin remodeler likely has an impact on hundreds or even thousands of genes. Intriguingly, CHD8 has been shown to associate with and modulate androgen-receptor activity (Menon et al., 2010); however, we have so far been unable to identify a similar association for CHD1 (data not shown).
Recent studies have implicated other ATP-dependent chromatin remodeler families in human cancer, perhaps most notably the SWI/SNF multi-subunit complex. Next-generation DNA sequencing has uncovered ARID1A mutations in half of ovarian clear cell carcinomas (Jones et al., 2010; Wiegand et al., 2010), and PBRM1 mutations in ~40% of renal clear cell carcinomas (Varela et al., 2011). Given the known function of CHD1 in chromatin remodeling, our findings now suggest a possible novel role of altered chromatin remodeling in prostate cancer. CHD1 represents a new foothold to understand the disease, perhaps ultimately leading to new approaches for prevention or therapy.
Supplementary Material
Acknowledgments
We wish to thank the members of the Pollack lab for helpful discussion. Supported by grants from the NIH (CA122246 to JRP; CA111782 to JDB; CA130472 to SH), the Burroughs Wellcome Fund (1007519 to JRP), the American Cancer Society (PF-08-078-01-MGO to SH), Prostate Cancer Canada (to JL) and the Stanford Cancer Center (to JRP).
Footnotes
Conflict of interest
The authors declare no conflict of interest
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- Bagchi A, Mills AA. The quest for the 1p36 tumor suppressor. Cancer Res. 2008;68:2551–2556. doi: 10.1158/0008-5472.CAN-07-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagchi A, Papazoglu C, Wu Y, Capurso D, Brodt M, Francis D, et al. CHD5 is a tumor suppressor at human 1p36. Cell. 2007;128:459–475. doi: 10.1016/j.cell.2006.11.052. [DOI] [PubMed] [Google Scholar]
- Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci USA. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binkley J, Karra K, Kirby A, Hosobuchi M, Stone EA, Sidow A. ProPhylER: a curated online resource for protein function and structure based on evolutionary constraint analyses. Genome Res. 2010;20:142–154. doi: 10.1101/gr.097121.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damber JE, Aus G. Prostate cancer. Lancet. 2008;371:1710–1721. doi: 10.1016/S0140-6736(08)60729-1. [DOI] [PubMed] [Google Scholar]
- DeMarzo AM, Nelson WG, Isaacs WB, Epstein JI. Pathological and molecular aspects of prostate cancer. Lancet. 2003;361:955–964. doi: 10.1016/S0140-6736(03)12779-1. [DOI] [PubMed] [Google Scholar]
- Forbes SA, Tang G, Bindal N, Bamford S, Dawson E, Cole C, et al. COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Res. 2010;38:D652–D657. doi: 10.1093/nar/gkp995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar-Maia A, Alajem A, Polesso F, Sridharan R, Mason MJ, Heidersbach A, et al. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature. 2009;460:863–868. doi: 10.1038/nature08212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JA, Georgel PT. CHD proteins: a diverse family with strong ties. Biochem Cell Biol. 2007;85:463–476. doi: 10.1139/O07-063. [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- Jones S, Wang TL, Shih Ie M, Mao TL, Nakayama K, Roden R, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–231. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Dhanasekaran SM, Mehra R, Tomlins SA, Gu W, Yu J, et al. Integrative analysis of genomic aberrations associated with prostate cancer progression. Cancer Res. 2007;67:8229–8239. doi: 10.1158/0008-5472.CAN-07-1297. [DOI] [PubMed] [Google Scholar]
- Kwei KA, Kim YH, Girard L, Kao J, Pacyna-Gengelbach M, Salari K, et al. Genomic profiling identifies TITF1 as a lineage-specific oncogene amplified in lung cancer. Oncogene. 2008;27:3635–3640. doi: 10.1038/sj.onc.1211012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapointe J, Li C, Giacomini CP, Salari K, Huang S, Wang P, et al. Genomic profiling reveals alternative genetic pathways of prostate tumorigenesis. Cancer Res. 2007;67:8504–8510. doi: 10.1158/0008-5472.CAN-07-0673. [DOI] [PubMed] [Google Scholar]
- Lapointe J, Li C, Higgins JP, van de Rijn M, Bair E, Montgomery K, et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci USA. 2004;101:811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mareel M, Leroy A. Clinical, cellular, and molecular aspects of cancer invasion. Physiol Rev. 2003;83:337–376. doi: 10.1152/physrev.00024.2002. [DOI] [PubMed] [Google Scholar]
- Marfella CG, Imbalzano AN. The Chd family of chromatin remodelers. Mutat Res. 2007;618:30–40. doi: 10.1016/j.mrfmmm.2006.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason SD, Joyce JA. Proteolytic networks in cancer. Trends Cell Biol. 2011;21:228–237. doi: 10.1016/j.tcb.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon T, Yates JA, Bochar DA. Regulation of androgen-responsive transcription by the chromatin remodeling factor CHD8. Mol Endocrinol. 2010;24:1165–1174. doi: 10.1210/me.2009-0421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan P, Onami TM, Rajagopalan S, Kania S, Donnell R, Venkatachalam S. Role of chromodomain helicase DNA-binding protein 2 in DNA damage response signaling and tumorigenesis. Oncogene. 2009;28:1053–1062. doi: 10.1038/onc.2008.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- Robbins CM, Tembe WA, Baker A, Sinari S, Moses TY, Beckstrom-Sternberg S, et al. Copy number and targeted mutational analysis reveals novel somatic events in metastatic prostate tumors. Genome Res. 2011;21:47–55. doi: 10.1101/gr.107961.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen MM, Abate-Shen C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev. 2010;24:1967–2000. doi: 10.1101/gad.1965810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, Zeng T, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363:1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.