Abstract
Background
The Tohoku Medical Megabank project aims to create a next-generation personalized healthcare system by conducting large-scale genome-cohort studies involving three generations of local residents in the areas affected by the Great East Japan Earthquake. We collected medical and genomic information for developing a biobank to be used for this healthcare system. We designed a questionnaire-based pedigree-creation software program named “f-treeGC,” which enables even less experienced medical practitioners to accurately and rapidly collect family health history and create pedigree charts.
Results
f-treeGC may be run on Adobe AIR. Pedigree charts are created in the following manner: 1) At system startup, the client is prompted to provide required information on the presence or absence of children; f-treeGC is capable of creating a pedigree up to three generations. 2) An interviewer fills out a multiple-choice questionnaire on genealogical information. 3) The information requested includes name, age, gender, general status, infertility status, pregnancy status, fetal status, and physical features or health conditions of individuals over three generations. In addition, information regarding the client and the proband, and birth order information, including multiple gestation, custody, multiple individuals, donor or surrogate, adoption, and consanguinity may be included. 4) f-treeGC shows only marriages between first cousins via the overlay function. 5) f-treeGC automatically creates a pedigree chart, and the chart-creation process is visible for inspection on the screen in real time. 6) The genealogical data may be saved as a file in the original format. The created/modified date and time may be changed as required, and the file may be password-protected and/or saved in read-only format. To enable sorting or searching from the database, the file name automatically contains the terms typed into the entry fields, including physical features or health conditions, by default. 7) Alternatively, family histories are collected using a completed foldable interview paper sheet named “f-sheet”, which is identical to the questionnaire in f-treeGC.
Conclusions
We developed a questionnaire-based family tree-creation software, named f-treeGC, which is fully compliant with international recommendations for standardized human pedigree nomenclature. The present software simplifies the process of collecting family histories and pedigrees, and has a variety of uses, from genome cohort studies or primary care to genetic counseling.
Electronic supplementary material
The online version of this article (doi:10.1186/s12881-017-0433-4) contains supplementary material, which is available to authorized users.
Keywords: Pedigree, Family tree, Software, Questionnaire, Family health history, Interview sheet, Genetic counseling, Genome cohort studies, Primary care, Biobank
Background
Genealogical information is critical for accurate genetic diagnosis in clinical genetics. In 1995, the National Society of Genetic Counselors [1] introduced a standardized description method for presenting a family tree in genetic counseling, which was revised in 2008 to the current version [2]. Typically, pedigree charts are manually created via face-to-face personal interviews held on an individual basis. However, pedigree chart creation requires graphical skills and specialized knowledge of clinical genetics, and is thus a time- and labor-intensive process. A large amount of genetic data has been collected from numerous large-scale studies conducted in recent years, such as genome-cohort studies. The availability of accurate genealogical information is essential for performing comprehensive analysis of polymorphisms and associated diseases for use in genetic counseling, research, and diagnosis. Given the large amount of genetic information required and collected, it is currently not possible to create pedigree charts using the traditional method through interviews at a single recruiting event by an expert.
The Tohoku Medical Megabank (TMM) project [3] aims to restore community medical services that were negatively affected by the Great East Japan Earthquake, and to create a next-generation personalized healthcare system by conducting large-scale genome-cohort studies involving three generations of local residents in the disaster-stricken areas [4]. Specifically, we collected medical and genomic information, including family health history, for developing a biobank to be used in the planned healthcare system.
In this project, we designed a questionnaire-based pedigree-drawing software program named “f-treeGC”, which enables even less-experienced medical practitioners to accurately and rapidly collect genealogical information and create pedigree charts, in full compliance with international standards [2].
Implementation
Programming
f-treeGC is written in ActionScript 3.0, and may be run on Adobe AIR, which is a cross-platform runtime system.
Operating environment
f-treeGC is supported by both Windows (Windows 7, 8, and 10) and Macintosh (operating system (OS) X). Adobe AIR Runtime [5] must be installed before installation and use of f-treeGC, and Adobe Reader DC [6] is required for the printing function. Via these programs, the f-treeGC air file may be opened to install the software. The f-treeGC software program is available for use, at no monetary cost, at the Iwate Medical University Hospital website (http://www.iwate-med.ac.jp/hospital/clinics/medical/m26/).
Results
The method for the creation of pedigree charts is described in the following sections.
Confirmation of whether or not the client has a child
At system startup, the client is prompted to provide required information on the presence or absence of children (Additional file 1a). f-treeGC is capable of creating a pedigree that includes three generations (Fig. 1a). Couples with offspring are included in the 2nd generation of the family tree (Fig. 2a, Additional file 2), whereas clients with no children are included in the 3rd generation (Figs. 1a and 2b, Additional file 3). A representative correspondence table is shown in Table 1.
Fig. 1.
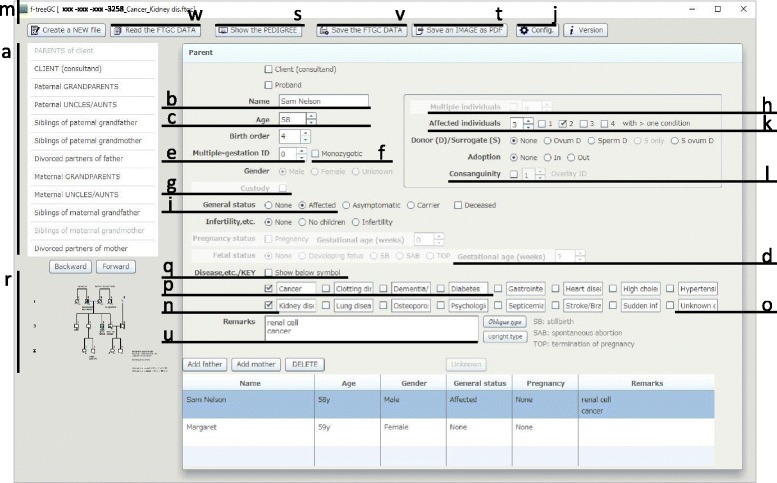
Information entry screen of f-treeGC; information for three generations is required to create the pedigree (a). The genealogical information requested includes name (b, u), age (c, d), gender, general status [affected (i, j, k), asymptomatic/presymptomatic carrier, carrier, or deceased], infertility status, pregnancy status, fetal status, and disease status (m, n, o, p, q, t) of individuals belonging to three generations. In addition, client, proband and birth order information, including multiple gestation (e, f), custody (g), multiple individuals (h), donor or surrogate, adoption, and consanguinity (l) information may be inserted. The chart-creation process is visible for inspection on the lower left side of the f-treeGC screen in real time (r). A multiple-choice questionnaire for genetic information is filled out by an interviewer or the client; then, f-treeGC automatically creates a pedigree chart (r, s, t, v, w)
Fig. 2.
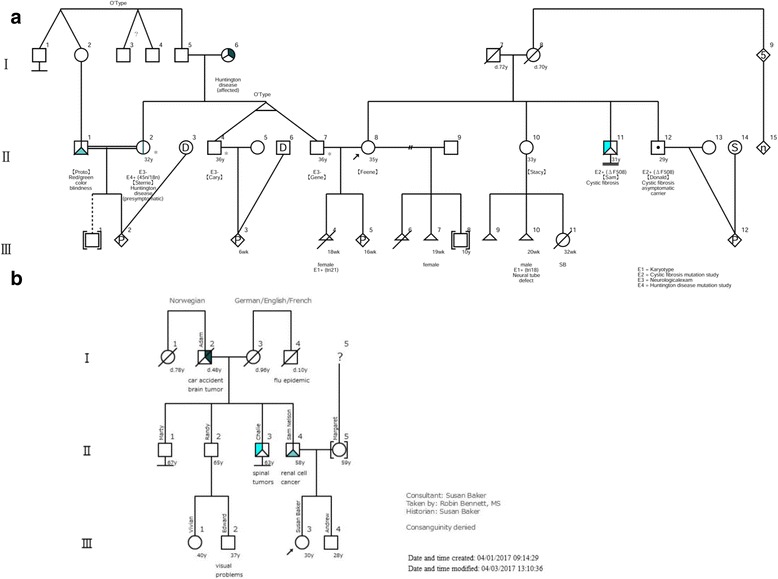
Fictitious pedigrees created by f-treeGC. a Fictitious ultimate pedigree, b A hypothetical pedigree representative of a family with von Hippel-Lindau syndrome shown in Fig. 1, partially modified from Bennett et al. [1, 7]. The clients are included in the 2nd generation (a) or in the 3rd generation (b) upon confirmation at system startup
Table 1.
Correspondence table of f-treeGC
| Client has a child | Client has NO children |
|---|---|
| Client (consultand) couple | Parents of client |
| Children of client couple | Client (consultand) |
| Husband’s (his) parents | Paternal grandparents |
| His siblings | Paternal uncles/aunts |
| His paternal uncles/aunts | Siblings of paternal grandfather |
| His maternal uncles/aunts | Siblings of paternal grandmother |
| His divorced partners | Divorced partners of father |
| Wife’s (her) parents | Maternal grandparents |
| Her siblings | Maternal uncles/aunts |
| Her paternal uncles/aunts | Siblings of maternal grandfather |
| Her maternal uncles/aunts | Siblings of maternal grandmother |
| Her divorced partners | Divorced partners of mother |
Multiple-choice questionnaire
The genealogical information requested includes name, age, gender, general status (affected, asymptomatic/presymptomatic carrier, carrier, or deceased), infertility status, pregnancy status, fetal status, and health status (occurrence of any diseases) of individuals in the three generations. In addition, information regarding client and proband, as well as birth order information such as multiple gestation, custody, multiple individuals, donor or surrogate, adoption, and consanguinity may be included (Fig. 1).
By default, the “Name” field refers to the type of relationship such as father or mother. The user should delete the relationship name before inputting the relevant name (Fig. 1b). In the “Age” box (Fig. 1c) and the “Gestational age (weeks)” box (Fig. 1d), the user may select “In blank”, “? (unknown)”, or the relevant number. Inputting the same number of individuals in the “Multiple-gestation ID” (Fig. 1e) indicates multiple gestation or pregnancy with multiple fetuses. The “Monozygotic” box (Fig. 1f) is for identical twins (pregnancy). The “Custody” box (Fig. 1g) is for the position of a break in a relationship line between divorced partners, and indicates the parent(s) with primary responsibility for the children following divorce. For multiple individuals, users may select “n (unknown)” or the relevant number(s) after checking the “Multiple individuals” box (Fig. 1h). The “Affected” button of general status (Fig. 1i) is for affected individuals, and users may set a key color for the affected status in the configuration (Fig. 1j). For affected individuals with two or more conditions, the user may check for the conditions (Fig. 1k). Considering the diversity in color perception and to enable distinction in subsequent black-and-white photocopies, f-treeGC shows multiple conditions using a color of a similar shade (Additional file 1b).
A printed paper version of the questionnaire
Alternatively, family health histories may be collected using the foldable interview sheet named “f-sheet” (Fig. 3, Additional file 4), which is a printed paper version of the questionnaire in f-treeGC. The f-sheet provides an overview of genetic relationships between families according to the manner in which the sheet is folded or developed. For example, the vertical line of the folded f-sheet corresponds to the left panel of the f-treeGC (Fig. 1a). Filling out a multiple-choice questionnaire on genetic information is easier for a medical practitioner with poor digital literacy. A skilled data entry clerk may subsequently input family health histories into f-treeGC from the f-sheet as a bundle.
Fig. 3.

Foldable interview sheet named “f-sheet”. a Large sheet, b gathered interview sheets for each individual, and c accordion-folded sheet. In diagram (a), the dashed-dotted lines, “-・-・-・-・-”, and broken lines, “- - - - -”, show the mountain fold and the valley fold, respectively
Overlay function for consanguinity
With respect to consanguinity, f-treeGC shows only marriages between first cousins using the overlay function. The “Consanguinity” box should be checked and the same “Overlay ID” should be entered for this function to be effective (Figs. 1l and 4, Additional file 5).
Fig. 4.
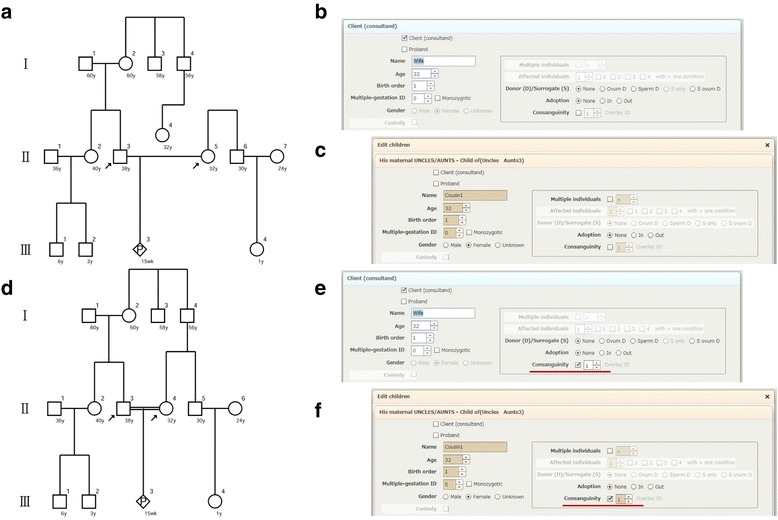
Overlay function of f-treeGC for consanguinity; a pedigree before (a) and after (d) overlay; the interview sheets of the wife (b, e) and a first cousin (c, f)
Entry fields and keys for physical features or diseases/conditions for genome cohort studies
By default, the file name automatically contains the first ten letters (Fig. 1m) that are typed into the sixteen entry fields of the “Disease, etc./KEY,” such as disease name (Fig. 1n), and for which the box is checked (Fig. 1o), to enable sorting or searching from the database. For example, the preliminary physical features entered are hair, ears, eyes, nose, philtrum, oral region, neck, hands/feet, chest, skin, abdominal, genitalia, and skeletal [7]. Diseases or conditions included in the My Family Health Portrait tool [8] include cancer, clotting disorder, dementia/Alzheimer’s disease, diabetes, gastrointestinal disorder, heart disease, high cholesterol, hypertension, kidney disease, lung disease, osteoporosis, psychological disorder, septicemia, stroke/brain attack, sudden infant death syndrome, and unknown disease (Fig. 1o). The first four out of sixteen entry fields may also be used as keys related to the multiple conditions of an affected individual (Fig. 1k, p). The “Show below symbol” box is for showing the entry below each symbol, only if the box on the left of the entry field is checked (Fig. 1q).
Pedigree chart creation
f-treeGC automatically creates a pedigree chart; the chart-creation process is visible for inspection on the screen in real time (Fig. 1r). The “Show the PEDIGREE” button (Fig. 1s) at the top of the screen displays a larger chart (Fig. 2b). The user may additionally edit a pedigree chart to visualize the larger chart with multiple displays.
A context menu appears when the family tree is right-clicked. The user may select “Add comment,” right-click on the comment box, and then type the appropriate text. Holding down the left mouse button allows the comment in the family tree to be moved and placed accordingly. The user may delete the comment, if required, by selecting “Delete comment”. By default, a pedigree chart does not include the name input (Fig. 1b, t) for privacy. The user may see names by selecting “Show names” in the context menu. To print or save an image of the pedigree, including names or other health conditions, the user may capture a screen shot (Fig. 2b, Additional file 3), or input the names manually in the “Remarks” box (Figs. 1u and 2a, Additional file 2).
Saving and reading the data
The genealogical information data may be saved as the original format file (FTGC file) by clicking the “Save the FTGC DATA” button (Fig. 1v), and shared between computers that have f-treeGC installed. The created/modified date and time may be changed as necessary before saving the data (Additional file 1c). In addition, the image of the pedigree chart may be saved in PDF format by clicking the “Save an IMAGE as PDF” button (Figs. 1t and 2a). The file may be password-protected and/or saved in a read-only format (Additional file 1c). A saved file may be read by clicking the “Read the FTGC DATA” button (Fig. 1w) at the top of the screen and selecting the file.
Comparison of the pedigree symbols used
A comparison of the pedigree symbols used in f-treeGC with those of several existing tools [8–11] is shown in Table 2. f-treeGC complies fully with the international recommendations of standardized human pedigree nomenclature [2], including common pedigree symbols, pedigree lines, assisted reproductive technology symbols, and pedigree symbols of genetic evaluation/testing information (Additional file 6).
Table 2.
Comparison of the pedigree symbols used
| Pedigree software packages | f-treeGC | PedigreeXP | Progeny Free Online Pedigree Tool | Genial Pedigree Draw | My Family Health Portrait |
|---|---|---|---|---|---|
| Version | 1.01 | 2.1.0.169 | Not described | 2.0 | 3.4 |
| Standard pedigree symbolsa | |||||
| Name of proband/consultand | Yes | Yes | Yes | Yes | No |
| Family names | Yes | Yes | Yes | Yes | Yes |
| Historian | Yesb | No | Yesb | No | No |
| Date of intake/update | Yes | No | No | No | Yes |
| Reason for taking pedigree | Yesb | No | Yesb | No | No |
| Ancestry of both sides of the family | Yes | Yes | Yes | Yes | No |
| Age | Yes | Yes | Yes | Yes | No |
| Generation numbered with Roman numerals I, II, III, etc. | Yes | Yes | Yesb | Yesb | No |
| Individual numbered 1, 2, 3, etc. | Yes | Yes | Yesb | Yesb | No |
| Individual | Yes | Yes | Yes | Yes | Yes |
| Affected individual | Yes | Yes | Yesd | Yes | Yes |
| Affected individual (> one condition) | Yesc | Yesc,d | Yes | Yes | No |
| Multiple individuals, number known | Yes | Yes | Yes | Yes | No |
| Multiple individuals, number unknown | Yes | Yes | Yes | Yes | No |
| Deceased individual | Yes | Yes | Yes | Yes | Yes |
| Consultand | Yes | No | No | Yes | No |
| Proband | Yes | Yesd | Yesd | Yes | No |
| Stillbirth (SB) | Yesb | No | No | No | No |
| Pregnancy (P) | Yes | Yes | Yes | Yes | No |
| Spontaneous abortion (SAB) | Yes | Yes | Yesd | Yes | No |
| Affected SAB | Yes | Yes | Yesd | No | No |
| Termination of pregnancy (TOP) | Yes | Yes | No | Yes | No |
| Affected TOP | Yes | Yes | No | No | No |
| Ectopic pregnancy | Yesb | Yesb | No | Yes | No |
| Relationship line | Yes | Yes | Yes | Yes | Yes |
| A break in relationship line | Yes | Yes | Yes | Yes | No |
| Divorced partner | Yese | Yes | Yes | Yes | No |
| Consanguinity | Yes | Yes | Yes | Yes | No |
| Multiple gestation, monozygotic | Yes | Yes | Yes | Yes | No |
| Multiple gestation, dizygotic | Yes | Yes | Yes | Yes | No |
| Multiple gestation, unknown | Yesb | No | Yesb | No | No |
| Multiple gestation, trizygotic | Yes | Yes | Yes | Yes | No |
| Family history not available/known for individual | Yes | No | No | No | No |
| No children by choice or reason unknown | Yes | No | Yes | Yesd | No |
| Infertility | Yes | No | Yes | Yesd | No |
| Adoption | Yes | Yes | Yes | Yes | Yesd |
| Ovum or sperm donor | Yes | No | No | Yes | No |
| Surrogate (gestational carrier) | Yes | No | No | Yes | No |
| Evaluation | Yesb | Yesb | Yes | Yesb | No |
| Carrier | Yes | Yesd | Yesf | Yes | No |
| Asymptomatic/presymptomatic carrier | Yes | No | No | Yes | No |
| Uninformative study | Yesb | Yesb | Yesb | Yesb | No |
| Affected individual with positive evaluation | Yesb | Yesb | Yes | Yesb | No |
aBennett et al., 2008 [2]
bBy using “Remarks”, “Comment”, “Notes”, or “Annotated Text”
cUp to four conditions
dDifferent symbols used from the recommended common pedigree symbols
eLimited to three individuals (consultand couple with children, or parents of consultand without children)
fBy using “Apply symbols”
System features and functions of f-treeGC as a family history collection tool
A well-designed online family history tool from the US Surgeon General, called My Family Health Portrait, is used for the collection and storage of family history data [11, 12]. In addition, the MeTree software program developed by the Duke Center for Applied Genomics and Precision Medicine enables collection of family health history and provides clinical decision-making support for more than 30 conditions such as cancer, cardiovascular diseases, liver, and diabetes [12, 13]. The Global Alliance for Genomics and Health (GA4GH) provides the GA4GH family history collection and clinical decision support tool inventory, and is open for submission. The information collected by f-treeGC is shown in Table 3 and in Additional file 7 derived from the submission form of the GA4GH family history tools catalog [12].
Table 3.
f-treeGC information
| Provenance | Academically developed |
| Tool URL | http://www.iwate-med.ac.jp/hospital/clinics/medical/m26/ |
| Target clinical population | Primary care and specialty |
| Family history information - source | Multiple sources |
| Family history information - data format | Both structured and free text |
| Family history information - analysis | Manual |
| Result recipients | Clinician |
| Data storage | Store in the original format file |
| Discrete data integration readiness | Application programing interfaces (API) |
| Consent documentation | Implied consent only |
| System features - platforms | Windows and Macintosh |
| Built in Adobe Air | |
| System features - architecture | Stand-alone |
| System features - security | Password security protection |
| Read-only mode | |
| Standard pedigree symbols | Fully compliant |
| Other functions | Custody |
| Pregnancy with twins or multiples |
Verification of the software and interview sheet
To verify that f-treeGC enables users without specialized knowledge of clinical genetics and graphical skills to easily create medical pedigrees, we provided nine subjects (six nurses and three clerks) with two scenarios (D, Duchenne muscular dystrophy; P, phenylketonuria) of fictitious family histories (Additional file 8), and compared the pedigrees obtained by f-treeGC (Additional file 9) with those derived manually. The pedigrees were scored according to a system of allocation points (Additional file 8) based on the international standard [2] to examine the performance and usability of f-treeGC. The creation time was indefinite, and we divided the trees into two groups with different orders of scenarios applied. We used Windows 7 as the OS for this test.
To verify that f-sheet improves the user experience for data input to f-treeGC, we provided 47 high school students from one high school (males, N = 28; females, N = 19; age range, 15–16 years; grade, the first year) with the software and data for two scenarios. Students were randomly assigned to two groups: students of one group created pedigrees for both scenarios (Additional file 8) using f-treeGC without f-sheet (group TT; males, N = 17; females, N = 6). Students of the other group first created pedigrees for scenario D using f-treeGC without f-sheet, and then created pedigrees for scenario P using f-treeGC with a completed f-sheet (group TS; males, N = 11; females, N = 13). The pedigrees with or without f-sheet were scored using our points allocation system (Additional file 8) based on the international standard [2]; then, the scores were compared to examine the efficacy and usability of f-sheet. The creation time was indefinite. The OS used for this purpose was Windows 7.
The Wilcoxon signed-rank test and Mann-Whitney U-test were performed for statistical analyses, using Statcel4 software (OMS Ltd. Publishing, Saitama, Japan). Significance was set at p < 0.05.
The family trees obtained using f-treeGC had higher scores than those that were manually created (p < 0.001) (Table 4). Moreover, the input time and family tree scores of trees created using f-treeGC were not affected by the difference in scenario content, order of application, or the qualifications of each user (Table 4). Furthermore, the scores of the family trees created using f-treeGC with a completed f-sheet were higher than those created using f-treeGC without f-sheet (p < 0.01) (Table 5).
Table 4.
Comparison of pedigree scores and creation time for each factor
| Factors | Median (Range) | P value | N | Statistics | ||||
|---|---|---|---|---|---|---|---|---|
| Method (manual method vs. f-treeGC software) | Manual | f-treeGC | Wilcoxon signed-rank sum test | |||||
| Score (%) of both scenarios | 59.5 | (23.7) | 86.4 | (25.8) | 0.00019 | *** | 18 | |
| Score (%) of scenario D | 58.8 | (14.7) | 86.8 | (16.2) | 0.00769 | ** | 9 | |
| Score (%) of scenario P | 60.2 | (23.7) | 86.0 | (25.8) | 0.00742 | ** | 9 | |
| Order of scenarios (D to P vs. P to D) | D to P | P to D | Mann-Whitney U-test | |||||
| Score (%) of scenario D by the manual method | 55.9 | (11.8) | 61.0 | (14.7) | 0.32312 | 5 × 4 | ||
| Time (sec/individuals) of scenario D by the manual method | 42.5 | (18.8) | 25.9 | (12.5) | 0.01390 | * | 5 × 4 | |
| Score (%) of scenario P by the manual method | 60.2 | (11.8) | 57.5 | (23.7) | 1.00000 | 5 × 4 | ||
| Time (sec/individuals) of scenario P by the manual method | 26.7 | (16.3) | 28.8 | (33.3) | 0.32516 | 5 × 4 | ||
| Score (%) of scenario D by f-treeGC | 83.8 | (16.2) | 87.5 | (13.2) | 0.61980 | 5 × 4 | ||
| Time (sec/individuals) of scenario D by f-treeGC | 65.0 | (49.4) | 61.9 | (23.4) | 0.46243 | 5 × 4 | ||
| Score (%) of scenario P by f-treeGC | 86.0 | (25.8) | 87.6 | (8.5) | 1.00000 | 5 × 4 | ||
| Time (sec/individuals) of scenario P by f-treeGC | 46.7 | (25.4) | 51.5 | (44.3) | 0.32719 | 5 × 4 | ||
| Order of scenarios (1st vs. 2nd) | 1st | 2nd | Wilcoxon signed-rank sum test | |||||
| Score (%) by the manual method | 55.9 | (23.7) | 60.3 | (14.7) | 0.44127 | 9 | ||
| Time (sec/individuals) by the manual method | 40.0 | (37.5) | 26.7 | (19.2) | 0.01086 | * | 9 | |
| Score (%) by f-treeGC | 83.9 | (18.9) | 86.8 | (25.8) | 0.67840 | 9 | ||
| Time (sec/individuals) by f-treeGC | 59.2 | (65.6) | 50.4 | (38.5) | 0.13864 | 9 | ||
| Qualification (nurse vs. clerk) | Nurse | Clerk | Mann-Whitney U-test | |||||
| Score (%) by the manual method | 61.0 | (23.7) | 54.1 | (17.2) | 0.13339 | 12 × 6 | ||
| Time (sec/individuals) by the manual method | 30.8 | (40.8) | 26.3 | (45.4) | 0.67310 | 12 × 6 | ||
| Score (%) by f-treeGC | 86.4 | (17.2) | 86.8 | (25.8) | 0.85103 | 12 × 6 | ||
| Time (sec/individuals) by f-treeGC | 57.1 | (53.1) | 39.4 | (47.9) | 0.18955 | 12 × 6 | ||
*p < 0.05
**p < 0.01
***p < 0.001
Table 5.
Comparison of pedigree scores using f-treeGC with or without f-sheet
| Factors | Median (Range) | P value | N | Statistics | ||||
|---|---|---|---|---|---|---|---|---|
| Group (TT vs. TS) | TT | TS | Mann-Whitney U-test | |||||
| Score (%) of scenario D | 89.7 | (10.3) | 87.5 | (26.5) | 0.0196 | * | 23 × 24 | |
| Score (%) of scenario P | 88.8 | (24.5) | 92.3 | (17.3) | 0.0109 | * | 23 × 24 | |
| Scenario (D vs. P) | D | P | Wilcoxon signed-rank sum test | |||||
| Score (%) of group TT | 89.7 | (10.3) | 88.8 | (24.5) | 0.6922 | 23 | ||
| Score (%) of group TS | 87.5 | (26.5) | 92.3 | (17.3) | 0.0018 | ** | 24 | |
*p < 0.05
**p < 0.01
Discussion
In the present study, we report the development of f-treeGC, a free stand-alone application built as a cross-platform runtime system. f-treeGC is capable of automatically creating a medical family tree compliant with international standards [2] by filling out available family tree information on a medical interview sheet (Fig. 1). Family histories are entered as both structured data and free text by the clinician or data entry clerk, and collected from patients or through f-sheet, which is a printed paper version of the questionnaire in f-treeGC (Fig. 3, Additional file 4). The family history data are stored in a computer in the original format file. f-treeGC may be used for collecting family health histories and creating pedigrees for individuals participating in situations such as primary care, genetic counseling, or genome cohort studies. The targeted clinical populations are recipients of primary and specialty health care facilities.
f-treeGC simplifies the process of creating pedigrees by confirming whether the client has offspring at system startup (Table 1, Additional file 1a) and by using the overlay function for confirming consanguinity (Fig. 4). Here, we show that f-treeGC, which is fully compliant with international recommendations for standardized human pedigree nomenclature (Table 2), is highly useful for creating pedigree charts for applications in genetic counseling.
However, the present study is not without limitations. As f-treeGC is only capable of creating a pedigree up to three generations, this software is currently unsuitable for creating large pedigrees. There are no auxiliary input functions for medical terms, pedigree-overlay function, nor a calculator for determining disease risk. f-treeGC is not adapted for compliance with Health-Level 7 (HL7) standards. Although numerous health and medical conditions exist [13], f-treeGC is limited to only sixteen medical conditions per person.
Low quality of family history data collected presents a challenge in pedigree analysis [14]. Before collecting family health histories, users should guide patients regarding what to inquire of relatives, as the amount and accuracy of the family history is limited. The Iwate Tohoku Medical Megabank Organization conducts genetics workshops mentored by medical geneticists or genetic counselors to highlight the importance of family health history before recruiting participants for cohort studies of the TMM project.
Family history, the ultimate genetic tool [15], is the most cost-effective and well known “genetic test” in clinical practice today [16]. However, recording family trees according to standard recommendations generally requires knowledge of graphical interfaces and clinical genetics [7]. In 2016, the National Human Genome Research Institute (NHGRI) convened a Family Health History Tool Meeting at the National Institute of Health (NIH) for identifying and sharing successful approaches to using family health history tools, and for identifying unresolved issues and potential solutions that may be addressed by policy, research, and/or collaborative efforts. The removal of barriers to health equality in populations with low levels of literacy, and exploration/expansion additional technological approaches for family health history collection was discussed in this meeting [14].
Six years have passed since the Great East Japan Earthquake and Tsunami. However, health and medical services has not been fully restored to date. The TMM project initiated two prospective cohort studies in the Miyagi and Iwate prefectures, which include the disaster-stricken areas: a population-based adult cohort study, in which 80,000 participants were recruited, and a birth and three-generation cohort study, in which 70,000 participants, included fetuses and their parents, siblings, grandparents, and extended family members, were recruited [4]. Collection of significant numbers of family health histories by conventional pedigree-drawing software programs is challenging in these regions owing to the lack of good internet service and personal computers. We used f-treeGC for genetic counseling at our institution, collecting approximately 100 patient histories and corresponding data, which would have taken a genetic counselor around twenty minutes in a clinical setting. In contrast, the software took about one minute per person to input two clinical scenarios at the verification experiment (Table 4). Since the use of f-treeGC in combination with f-sheet simplifies the process of collection of many numerous health histories and pedigrees (Table 5), its application is not limited to heredity clinics, but also to large-scale genome-cohort studies that handle large amounts of genetic information obtained through interviews at a single recruiting event.
The main advantages of f-treeGC are collection of several family histories for large-scale cohort studies in a short period of time, easing the burden of collection of genealogical information and creation of pedigree charts in remote medical practice by less experienced medical practitioners. In addition, the present tool facilitates online genetic counseling owing to its complete compliance with the international recommendations for standardized human pedigree nomenclature (Table 2).
Public awareness regarding the basic principles of genetics should be considered for the improvement of public health. Familial/pedigree information is valuable for variant filtering in high-throughput sequencing studies [17, 18]. Molecular approaches for the identification of disease-associated genes generally begin with pedigree-based methods, including positional cloning and founder gene approaches, prior to the use of pedigree-independent methods such as candidate gene approaches and genome-wide association studies [19]. With the recent explosion in whole-genome sequencing, linkage analysis has emerged as an important and powerful analytical method for the identification of genes involved in disease etiology, often in conjunction with whole-genome sequencing filtering approaches [20]. From this perspective, f-treeGC is a useful tool, not only for facile and accurate pedigree analysis, but also for conveniently collecting numerous family histories and pedigrees simultaneously. In future, we aim to add a calculator function for determining disease risk, an auxiliary input function for medical terms, a search function for family health conditions from free text, an adaption for HL7, and a pedigree-overlay function to the present f-treeGC software.
Conclusions
The f-treeGC software enables collection of family health history and automatically creates a medical family tree simply by filling out family tree information on a medical interview sheet, or by inputting the information in the questionnaire directly from the f-sheet.
Availability and requirements
Project name: TMM project
Project home page: http://www.amed.go.jp/en/program/list/04/01/042.html
Operating systems: Windows and Macintosh
Programing language: ActionScript 3.0
Licence: f-treeGC is a non-copylefted software, and is copyrighted by the Iwate Medical University and Holonic Systems, Ltd. The source code is not available.
Additional files
Dialog boxes of f-treeGC (a) Confirmation of whether or not the client has a child at system startup, (b) Configuration of the color for affected individuals, (c) File attribute setting for changing the created/modified date and time, setting a password, and converting to read-only. (PPTX 141 kb)
A pedigree file of Fig. 2a; fictitious ultimate pedigree, partially modified from Bennett et al. [1, 7]. (FTGC 20 kb)
A pedigree file of Fig. 2b; a hypothetical pedigree representative of a family with von Hippel-Lindau syndrome, partially modified from Bennett et al. [1, 7]. (FTGC 9 kb)
f-sheet; a printed paper version of the questionnaire in f-treeGC. (XLSX 57 kb)
A pedigree file of Fig. 4d; with respect to consanguinity, f-treeGC shows only marriages between first cousins using the overlay function. (FTGC 8 kb)
List of symbols used in f-treeGC; f-treeGC fully complies with the international recommendations of standardized human pedigree nomenclature [2]. (PDF 8 kb)
Current family history collection tools and f-treeGC; partially modified from GA4GH family history collection and clinical decision support tool inventory 6–9-16 v4.1 by Clinical Working Group of the Global Alliance for Genomics and Health. (XLSX 18 kb)
Two scenarios for the creation of pedigrees; scenario D (Duchenne muscular dystrophy) and scenario P (phenylketonuria). Any resemblance to real persons and pedigrees, living or dead, is purely coincidental. (XLSX 12 kb)
Model pedigrees of scenarios outlined (a) Scenario D, (b) scenario P. (PPTX 389 kb)
Acknowledgements
The authors thank the members of the Iwate Tohoku Medical Megabank Organization for suggestions and comments; Dr. Hiromasa Ono and Dr. Hidemasa Bono, Database Center for Life Science, for manual preparation; the members of Akita IP Firm for valuable suggestions; the members of Editage for English proofreading and valuable comments; Dr. Tohru Ohta, Dr. Rie Takai, and Dr. Norio Niikawa, Health Science University of Hokkaido, for experimental support and valuable comments; and Ms. Fumiyo Nakayama, Iwate Tohoku Medical Megabank Organization, for her assistance in preparing the manuscript; Mr. Yusuke Sakurai, Iwate Medical University Hospital, for generation of the website.
Funding
This research is supported by the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan Agency for Medical Research and Development (AMED), and Iwate Industry Promotion Center.
Availability of data and materials
The datasets supporting the conclusions are included in this article and in the additional files. The f-treeGC software program is available at no monetary charge at the Iwate Medical University Hospital website, http://www.iwate-med.ac.jp/hospital/clinics/medical/m26/, and the f-sheets are available at no monetary charge as additional files with this article.
f-treeGC is a non-copylefted software, and is copyrighted by the Iwate Medical University and Holonic Systems, Ltd. The source code is not available. f-treeGC is written in ActionScript 3.0, and may be run on Adobe AIR, which is a cross-platform runtime system. f-treeGC is supported by both Windows (Windows 7, 8, and 10) and Macintosh (OS X). An application for the f-treeGC software program is under review for the international Patent Cooperation Treaty (PCT/JP2016/078494).
f-sheet is copyrighted by the Iwate Medical University and registered as a utility model in Japan (3204531).
Abbreviations
- GA4GH
Global Alliance for Genomics and Health
- TMM project
Tohoku Medical Megabank project
Authors’ contributions
TT was responsible for the execution of the research project and for writing the first draft of the manuscript. AF designed, organized, and reviewed the research project and reviewed and critiqued the manuscript. KY was responsible for manual preparation, checking of software operation, and comparison of pedigree software packages. YB was responsible for computer programming. TH reviewed the manuscript. AS was responsible for manual preparation and reviewed the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Ethical approval for our study was obtained from the institutional review board of the Health Science University of Hokkaido (Approval ID: 174). This study was conducted in accordance with the Declaration of Helsinki, Japanese Act on the Protection of Personal Information, and Japan Ethical Guidelines for Medical and Health Research Involving Human Subjects. We obtained informed consent from parents of all the students enrolled in our study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s12881-017-0433-4) contains supplementary material, which is available to authorized users.
Contributor Information
Tomoharu Tokutomi, Email: tktmtmhr@iwate-med.ac.jp.
Akimune Fukushima, Email: akimunef@iwate-med.ac.jp.
Kayono Yamamoto, Email: kyamamot@iwate-med.ac.jp.
Yasushi Bansho, Email: bansho@holonic-systems.com.
Tsuyoshi Hachiya, Email: thachiya@iwate-med.ac.jp.
Atsushi Shimizu, Email: ashimizu@iwate-med.ac.jp.
References
- 1.Bennett RL, Steinhaus KA, Uhrich SB, O'Sullivan CK, Resta RG, Lochner-Doyle D, et al. Recommendations for standardized human pedigree nomenclature. J Genet Couns. 1995;4:267–279. doi: 10.1007/BF01408073. [DOI] [PubMed] [Google Scholar]
- 2.Bennett RL, French KS, Resta RG, Doyle DL. Standardized human pedigree nomenclature: update and assessment of the recommendations of the National Society of genetic counselors. J Genet Couns. 2008;17:424–433. doi: 10.1007/s10897-008-9169-9. [DOI] [PubMed] [Google Scholar]
- 3.Tohoku Medical Megabank project. http://www.amed.go.jp/en/program/list/04/01/042.html. Accessed 14 May 2017.
- 4.Kuriyama S, Yaegashi N, Nagami F, Arai T, Kawaguchi Y, Osumi N, et al. The Tohoku Medical Megabank project: design and mission. J Epidemiol. 2016;26:493–511. doi: 10.2188/jea.JE20150268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adobe AIR Runtime. http://get.adobe.com/jp/air/. Accessed 14 May 2017.
- 6.Adobe Reader DC. http://get.adobe.com/jp/reader/. Accessed 14 May 2017.
- 7.Bennett RL. The practical guide to the genetic family history. 2. Hoboken: Wiley-Blackwell; 2010. [Google Scholar]
- 8.PedigreeXP. PC PAL. https://www.pedigreexp.com/. Accessed 14 May 2017.
- 9.Progeny Free Online Pedigree Tool. Progeny Genetics LLC. http://www.progenygenetics.com/online-pedigree/. Accessed 14 May 2017.
- 10.Genial Pedigree Draw. Genial Genetic Solutions Ltd. http://www.pedigreedraw.com. Accessed 14 May 2017.
- 11.My Family Health Portrait. US Surgeon General. https://familyhistory.hhs.gov/FHH/html/index.html?setLng=en. Accessed 14 May 2017.
- 12.Global Alliance for Genomics and Health. http://genomicsandhealth.org/work-products-demonstration-projects/catalogue-global-activities-family-history-tools. Accessed 14 May 2017.
- 13.Orlando LA, Buchanan AH, Hahn SE, Christianson CA, Powell KP, Skinner CS, et al. Development and validation of a primary care-based family health history and decision support program (MeTree) N C Med J. 2013;74:287–296. [PMC free article] [PubMed] [Google Scholar]
- 14.The NHGRI Family Health History Tool Meeting 2016. https://www.genome.gov/27565264/the-nih-family-health-history-tool-conference-2016/. Accessed 14 May 2017.
- 15.Uhlmann WR, Scheuette JL, Yashar BM. A guide to genetic counseling. 2. Hoboken: Wiley-Blackwell; 2009. [Google Scholar]
- 16.Duke Center for Applied Genomic & Precision Medicine. https://precisionmedicine.duke.edu. Accessed 14 May 2017.
- 17.Wijsman EM. The role of large pedigrees in an era of high-throughput sequencing. Hum Genet. 2012;131:1555–1563. doi: 10.1007/s00439-012-1190-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bahlo M, Tankard R, Lukic V, Oliver KL, Smith KR. Using familial information for variant filtering in high-throughput sequencing studies. Hum Genet. 2014;133:1331–1341. doi: 10.1007/s00439-014-1479-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bakhtiar SM, Ali A, Baig SM, Barh D, Miyoshi A, Azevedo V. Identifying human disease genes: advances in molecular genetics and computational approaches. Genet Mol Res. 2014;13:5073–5087. doi: 10.4238/2014.July.4.23. [DOI] [PubMed] [Google Scholar]
- 20.Ott J, Wang J, Leal SM. Genetic linkage analysis in the age of whole-genome sequencing. Nat Rev Genet. 2015;16:275–284. doi: 10.1038/nrg3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dialog boxes of f-treeGC (a) Confirmation of whether or not the client has a child at system startup, (b) Configuration of the color for affected individuals, (c) File attribute setting for changing the created/modified date and time, setting a password, and converting to read-only. (PPTX 141 kb)
A pedigree file of Fig. 2a; fictitious ultimate pedigree, partially modified from Bennett et al. [1, 7]. (FTGC 20 kb)
A pedigree file of Fig. 2b; a hypothetical pedigree representative of a family with von Hippel-Lindau syndrome, partially modified from Bennett et al. [1, 7]. (FTGC 9 kb)
f-sheet; a printed paper version of the questionnaire in f-treeGC. (XLSX 57 kb)
A pedigree file of Fig. 4d; with respect to consanguinity, f-treeGC shows only marriages between first cousins using the overlay function. (FTGC 8 kb)
List of symbols used in f-treeGC; f-treeGC fully complies with the international recommendations of standardized human pedigree nomenclature [2]. (PDF 8 kb)
Current family history collection tools and f-treeGC; partially modified from GA4GH family history collection and clinical decision support tool inventory 6–9-16 v4.1 by Clinical Working Group of the Global Alliance for Genomics and Health. (XLSX 18 kb)
Two scenarios for the creation of pedigrees; scenario D (Duchenne muscular dystrophy) and scenario P (phenylketonuria). Any resemblance to real persons and pedigrees, living or dead, is purely coincidental. (XLSX 12 kb)
Model pedigrees of scenarios outlined (a) Scenario D, (b) scenario P. (PPTX 389 kb)
Data Availability Statement
The datasets supporting the conclusions are included in this article and in the additional files. The f-treeGC software program is available at no monetary charge at the Iwate Medical University Hospital website, http://www.iwate-med.ac.jp/hospital/clinics/medical/m26/, and the f-sheets are available at no monetary charge as additional files with this article.
f-treeGC is a non-copylefted software, and is copyrighted by the Iwate Medical University and Holonic Systems, Ltd. The source code is not available. f-treeGC is written in ActionScript 3.0, and may be run on Adobe AIR, which is a cross-platform runtime system. f-treeGC is supported by both Windows (Windows 7, 8, and 10) and Macintosh (OS X). An application for the f-treeGC software program is under review for the international Patent Cooperation Treaty (PCT/JP2016/078494).
f-sheet is copyrighted by the Iwate Medical University and registered as a utility model in Japan (3204531).


