Abstract
The role of many enzymes extends beyond their dedicated catalytic activity by fulfilling important cellular functions in a catalysis-independent fashion. In this aspect, little is known about 3′-end RNA-modifying enzymes that belong to the class of nucleotidyl transferases. Among these are noncanonical poly(A) polymerases, a group of evolutionarily conserved enzymes that are critical for gene expression regulation, by adding adenosines to the 3′-end of RNA targets. In this study, we investigate whether the functions of the cytoplasmic poly(A) polymerase (cytoPAP) GLD-2 in C. elegans germ cells exclusively depend on its catalytic activity. To this end, we analyzed a specific missense mutation affecting a conserved amino acid in the catalytic region of GLD-2 cytoPAP. Although this mutated protein is expressed to wild-type levels and incorporated into cytoPAP complexes, we found that it cannot elongate mRNA poly(A) tails efficiently or promote GLD-2 target mRNA abundance. Furthermore, germ cell defects in animals expressing this mutant protein strongly resemble those lacking the GLD-2 protein altogether, arguing that only the polyadenylation activity of GLD-2 is essential for gametogenesis. In summary, we propose that all known molecular and biological functions of GLD-2 depend on its enzymatic activity, demonstrating that polyadenylation is the key mechanism of GLD-2 functionality. Our findings highlight the enzymatic importance of noncanonical poly(A) polymerases and emphasize the pivotal role of poly(A) tail-centered cytoplasmic mRNA regulation in germ cell biology.
Keywords: poly(A) polymerase, poly(A) metabolism, translational regulation, germline development
INTRODUCTION
Gene expression programs are profoundly influenced by RNA-modifying enzymes. The catalytic activity of such proteins is often used to facilitate covalent RNA-structure changes, thereby providing a mechanistic basis for post-transcriptional gene regulation. Besides the obvious importance of the catalytic activity of enzymes per se, an increasing number of examples shows that enzymes also perform critical catalysis-independent functions. This is nicely exemplified in the field of signal transduction where fundamental noncatalytic roles are attributed to kinases such as scaffolding or allosteric regulation (Kung and Jura 2016). However, in the field of RNA research, it is currently unclear to which degree the molecular or biological functions assigned to many enzymes are dependent on their RNA-modifying activity.
A developmentally important class of RNA-modifying enzymes are noncanonical cytoplasmic poly(A) polymerases (cytoPAPs). The most prominent member of this class is germline development defective (GLD)-2 (Minasaki and Eckmann 2012). Originally discovered in the nematode Caenorhabditis elegans (Kadyk and Kimble 1998; Wang et al. 2002), numerous homologs of GLD-2 have been described in other invertebrates and vertebrates (Kwak et al. 2004; Rouhana et al. 2005; Benoit et al. 2008; Cui et al. 2008). Among the different species, GLD-2-type cytoPAPs are most commonly and predominantly expressed in germ cells (Rouhana et al. 2005), and in the case of nematodes and flies, it was found that this expression is essential for the formation of functional gametes (Kadyk and Kimble 1998; Benoit et al. 2008; Cui et al. 2008; Sartain et al. 2011). Therefore, this class of enzymes comprises important evolutionarily conserved regulators of germ cell development.
All members of the GLD-2-type cytoPAP family contain a nucleotidyl transferase domain (NTD) that is embedded in a large central domain. The crystal structure has been solved for C. elegans GLD-2 (Nakel et al. 2015). NTD and central domain are separated by a pronounced cleft that harbors the catalytically active site, whereby the catalytic domain itself is composed of a five-stranded β-sheet flanked by two α-helices (Nakel et al. 2015). Interestingly, the central domain, including its NTD, constitutes the main part of GLD-2 proteins in vertebrates (Kwak et al. 2004; Rouhana et al. 2005), whereas large protein extensions are found in nonvertebrates, such as flies and worms (Wang et al. 2002; Benoit et al. 2008; Cui et al. 2008), making nonvertebrate GLD-2 proteins prime candidates to investigate potential noncatalytic functions.
The isolated GLD-2 protein itself has only weak enzymatic activity in vitro. The purified human GLD-2 protein, although highly specific for the addition of adenosines, can only monoadenylate a wide set of RNA substrates (Chung et al. 2016). Interestingly, the activity of GLD-2 is stimulated by interacting proteins that bind to the catalytic domain (Wang et al. 2002; Kim et al. 2009). The crystal structure of worm GLD-2 in a complex with such binding partners shows that these interactions reduce the flexibility of the catalytic center, stabilize the fold of the catalytic domain, and extend its positively charged surface area for RNA interaction (Nakel et al. 2015, 2016). This suggests that GLD-2 has to be part of a protein complex in order to facilitate efficient polyadenylation.
It is envisioned that GLD-2-type cytoPAPs function as post-transcriptional activators by elongating mRNA poly(A) tails, thereby promoting mRNA stability in the cytoplasm. This idea is supported by several observations, made in different systems. In Caenorhabditis elegans, loss of GLD-2 protein correlates with a shortening of mRNA poly(A) tails as well as a decrease in abundance of many germline mRNAs (Kim et al. 2010; Nousch et al. 2014). Similar observations have been made in Drosophila melanogaster, where putative mRNA targets of the GLD-2 homolog, WISPY, are not polyadenylated in its absence, leading to inefficient protein synthesis during oocyte formation (Benoit et al. 2008; Cui et al. 2013).
The catalytic activity is not essential for the function of all noncanonical poly(A) polymerases. This is nicely illustrated by the functional requirements of Trf4-type PAPs. Most members of this protein family are involved in nuclear RNA quality control as part of the TRAMP complex, by oligoadenylating target RNAs that subsequently are degraded by the nuclear exosome (LaCava et al. 2005; Vanácová et al. 2005; Wyers et al. 2005). Loss of Trf4-type protein leads to an increase of many noncoding and nonfunctional RNAs (Kadaba et al. 2004; LaCava et al. 2005; Vanácová et al. 2005; Wyers et al. 2005; Davis and Ares 2006; Egecioglu et al. 2006). Interestingly, the overexpression of catalytically inactive Trf4 protein in yeast can restore many of these mRNAs back to wild-type levels (San Paolo et al. 2009), demonstrating that the enzymatic process of oligoadenylation is not essential for Trf4-mediated RNA degradation.
To date, all studies on gld-2 function were made in the absence of full-length GLD-2 proteins, making it impossible to distinguish between catalysis-dependent and -independent functions. In the work presented here, we investigate the importance of the poly(A) polymerase activity GLD-2 with regard to its molecular and biological functions by studying a missense mutation that resides in the catalytic domain of C. elegans GLD-2.
RESULTS AND DISCUSSION
Description of gld-2 point mutations
Several alleles of the gld-2 gene have been reported in C. elegans (Kadyk and Kimble 1998). We sequenced these alleles to identify mutations that are predicted to produce full-length protein but inhibit the enzymatic activity of GLD-2. Our search settled on the allele gld-2(q540), a missense mutation that results in the exchange of proline 652 to leucine (P652L), which is located within the nucleotidyl-transferase domain (NTD) (Fig. 1A). According to the recently described crystal structure of C. elegans GLD-2 (Nakel et al. 2015), proline 652 maps to β-sheet number 4 of the catalytic domain, which is part of the cleft that harbors the active site of the enzyme (Fig. 1B). Importantly, the high degree of evolutionary conservation from worm to humans of proline 652 (Fig. 1B) suggests structural and/or functional importance.
FIGURE 1.
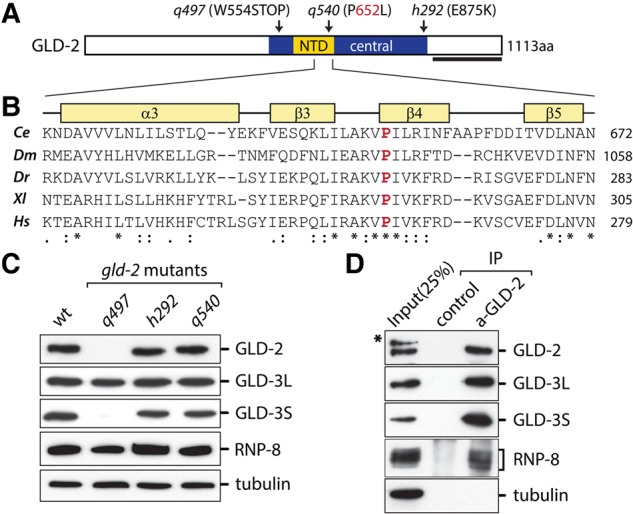
The GLD-2(P652L) protein is robustly expressed and interacts with GLD-3 and RNP-8. (A) Cartoon of GLD-2 protein domain structure. Position and impact of q497, q540, and h292 mutations on GLD-2 protein are indicated. The black, thick bar indicates the region that was used to raise all antibodies against this protein. (B) A protein sequence alignment comprising the second half of the nucleotidyl transferase domain (NTD) from Caenorhabditis elegans (Ce), Drosophila melanogaster (Dm), Danio renio (Dr), Xenopus laevis (Xl), and Homo sapiens (Hs) GLD-2. On top of the alignment, the secondary structure elements of the C. elegans protein are shown (α, α-helices; β, β-sheet). The conserved proline in β-sheet number 4 is highlighted in red. (C) Western blot analysis of the indicated proteins in the different gld-2 mutants. (D) Immunoprecipitation of GLD-2(P652L)-containing complexes from adult animal extracts, tested for a coenrichment of both known GLD-2-interacting proteins. The star indicates a background band that is detected by the mouse anti-GLD-2 antibody.
To test whether GLD-2(P652L) protein is stably expressed in gld-2(q540) worms, we performed Western blot analyses, comparing the mutant protein to wild-type and two other gld-2 alleles: gld-2(q497), which is considered to be a strong loss-of-function allele producing no full-length GLD-2 protein (Fig. 1A; Wang et al. 2002), and gld-2(h292), a missense mutation that replaces glutamic acid 875 with an arginine (E875R) in GLD-2 (Fig. 1A; Wang et al. 2002). Consistent with previous results, no full-length GLD-2 protein could be detected in q497, and an equally robust GLD-2 signal was present in wild-type as well as gld-2(h292) young adults (Fig. 1C; Wang et al. 2002). Importantly, a comparably strong GLD-2 signal was detected in extracts of gld-2(q540) animals, showing that in q540, full-length GLD-2(P652L) protein is present in similar amounts compared to wild-type GLD-2 or h292 full-length GLD-2(E875R).
To test whether germ cell-enriched GLD-2 mutant variants have an influence on the abundance of GLD-2-associated proteins, we analyzed the expression of GLD-3 and RNP-8 in the different gld-2 mutant backgrounds. Both proteins have previously been shown to physically bind to GLD-2 (Wang et al. 2002; Eckmann et al. 2004; Kim et al. 2009; Nakel et al. 2015, 2016). In germ cells, two variants of GLD-3 are expressed via alternative splicing: GLD-3 long (GLD-3L) and GLD-3 short (GLD-3S) (Eckmann et al. 2002). In the different GLD-2 mutants, the abundance of GLD-3L is unchanged compared to wild-type (Fig. 1C). Contrary to the long isoform, GLD-3S is nearly absent in q497 and slightly reduced in h292, which is also true for q540 (Fig. 1C). RNP-8 levels are comparable among all tested genotypes (Fig. 1C). Together, this shows that only the expression level of GLD-3S, but neither GLD-3L nor RNP-8, is dependent on the presence of GLD-2. The differential requirement of both GLD-3 isoforms on GLD-2 protein presence suggests that GLD-3S might be stabilized by its integration into a GLD-2 complex.
To test whether GLD-2(P652L) protein can still establish known protein interactions in vivo, we conducted immunoprecipitation experiments from hand-picked homozygous gld-2(q540) adult worms. GLD-2(P652L) was efficiently enriched in pull-down experiments with specific anti-GLD-2 antibodies, whereas no GLD-2 protein was detected in pull-downs from the same extract with unspecific antibodies (Fig. 1D). Importantly, both isoforms of GLD-3 as well as RNP-8 were specifically coimmunoprecipitated with GLD-2(P652L), whereas tubulin was not (Fig. 1D). As GLD-3 and RNP-8 occupy the same interaction surface on GLD-2, both proteins bind to GLD-2 in a mutually exclusive manner (Kim et al. 2009; Nakel et al. 2015, 2016), and our observations suggest that both proteins are able to bind to GLD-2(P652L), presumably forming two distinct GLD-2(P652L)-containing mRNP complexes in gld-2(q540). Although it cannot be excluded that the q540 mutation disturbs the association of unknown GLD-2–interacting factors, it is unlikely that, judging from the structural position of β-sheet number 4, Proline 652 participates in intermolecular interactions (Nakel et al. 2015). Regardless, based on the robust enrichment of GLD-3 and RNP-8 with the mutated enzyme, we concluded that GLD-2(P652L) cytoPAP is incorporated into protein complexes in vivo.
The enzymatic activity of GLD-2(P652L) cytoPAP is strongly compromised in vivo
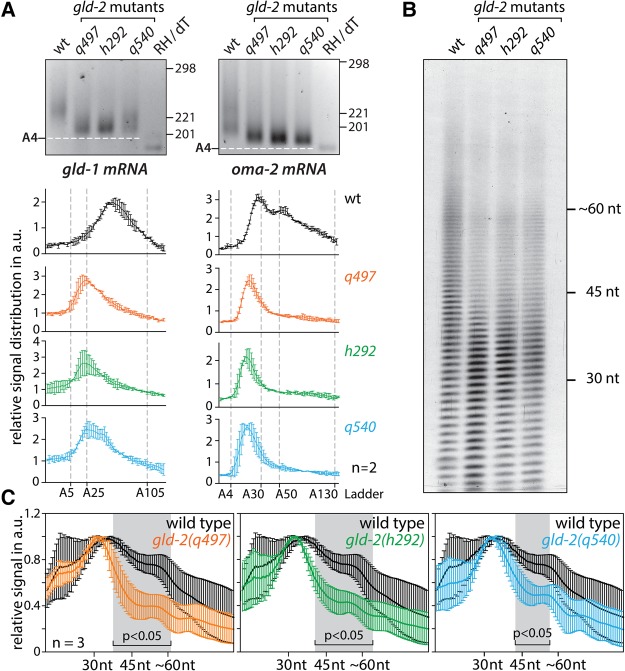
Loss of GLD-2 protein causes a decrease in mRNA poly(A)-tail lengths at the gene-specific as well as global level (Suh et al. 2006; Minasaki et al. 2014; Nousch et al. 2014). To test whether the GLD-2(P652L) enzyme polyadenylates mRNAs in vivo, we measured poly(A)-tail lengths of individual mRNAs and of bulk RNA in gld-2(q540) worms. Using the sPAT method (Minasaki et al. 2014), we concentrated our gene-specific analysis on two previously described GLD-2 target mRNAs: gld-1 and oma-2 (Suh et al. 2006; Schmid et al. 2009; Kim et al. 2010). Consistent with previous results (Suh et al. 2006; Kim et al. 2010; Minasaki et al. 2014), the average poly(A)-tail length of gld-1 and oma-2 mRNAs is shorter in the putative protein null allele gld-2(q497) than in wild-type (Fig. 2A). Interestingly, we made similar observations in gld-2(q540) animals, where both tested mRNAs also have shorter poly(A) tails (Fig. 2A). This suggests that gld-1 and oma-2 mRNAs are not efficiently polyadenylated by the GLD-2(P652L) enzyme. Still, small poly(A) tail differences are detected in gld-2(q497) and gld-2(q540) for gld-1 mRNA, which might reflect a differential sensitivity of different mRNAs toward GLD-2 activity. To test how prevalent the polyadenylation defect is in gld-2(q540) animals, we conducted bulk poly(A)-tail measurements. For this assay, we isolated total RNA, labeled it with radioactive cordycepin, removed everything that is not a poly(A) stretch by RNase digestion, and visualized poly(A) distribution on a sequencing gel. In wild-type, the majority of poly(A) tails is distributed between 30 and 60 nucleotides (nt) (Fig. 2B,C). In gld-2(q540) animals, poly(A) tails of 40 nt and longer are significantly less abundant, and the overall poly(A) profile is similar to the one detected in the protein null allele gld-2(q497) (Fig. 2B,C). However, with the assays used in this study, we cannot exclude that the GLD-2(P652L) protein still possesses some residual polyadenylation activity, as indicated by the small differences between gld-2(q497) and gld-2(q540) detected in the bulk poly(A) measurements (Fig. 2C). Nonetheless, our results show that a large number of mRNAs is inefficiently polyadenylated in gld-2(q540) animals, indicating that GLD-2(P652L) possesses a significantly reduced poly(A)-polymerase activity and cannot efficiently promote mRNA tail extension.
FIGURE 2.
Impact of different GLD-2 mutant proteins on mRNA polyadenylation. (A) sPAT analysis of gld-1 and oma-2 mRNA in wild-type (wt) and different gld-2 backgrounds. The lane RH/dT is a wild-type sample that was treated with oligo(dT) and RNase H prior to the sPAT assay to indicate the size of a completely deadenylated mRNA. Line scans from data generated by the sPAT assay are given below. Two independent biological repeats were analyzed. (B) The distribution of bulk poly(A) tails was analyzed in wild-type and the given gld-2 backgrounds. (C) Line scans from bulk poly(A) measurements. Three independent biological repeats were analyzed. Regions of statistically significant differences indicated with a gray stripe and black bracket are calculated via the Student's t-test.
The fold of the catalytic domain of GLD-2 is stabilized by interactions with GLD-3 or RNP-8 (Nakel et al. 2015, 2016), and interactions with either protein stimulate GLD-2's polyadenylase activity (Wang et al. 2002; Kim et al. 2009). As GLD-2(P652L) is still able to interact with both proteins (Fig. 1), it is unlikely that the loss of polyadenylation can be explained by an inability of the protein to interact with stimulating factors. It is more plausible that the P652L mutation has a direct effect on the catalytic efficiency of GLD-2 cytoPAP. In the structure of yeast nuclear poly(A) polymerase, PAP-1, the 3′-terminal residue of the poly(A) substrate is sandwiched between valine 141 and the base of the incoming ATP (Balbo and Bohm 2007). In the structure of GLD-2, this valine maps to position 651 and is thereby located one residue before the P652L mutation. With such an important nearby residue, it is plausible that the P652L mutation somehow interferes with the positioning of the substrate in the catalytic pocket and thus affects polyadenylation activity. Taken together, we therefore propose that GLD-2(P652L) is at best a catalytic null or at minimum a severely catalytically impaired enzyme.
We also included the h292 allele in our poly(A)-tail measurements. The h292 missense mutation has previously been described to abolish the polyadenylation activity of a GLD-2 fragment in vitro (Nakel et al. 2015). However, it was unclear whether similar negative effects would also manifest in vivo in the context of full-length GLD-2 (E875R) protein. Here, we find that both poly(A) tails of specific mRNAs (Fig. 2A) and bulk RNA poly(A)-tail profiles (Fig. 2B,C) are substantially reduced in gld-2(h292) animals, suggesting that also this mutation strongly affects the poly(A)-polymerase activity of GLD-2 in vivo. However, how this mutation, which is located outside of the catalytic domain, impairs GLD-2 activity is currently unclear. As part of the nucleotide recognition motif, E875 is likely to affect catalysis in a direct manner (Nakel et al. 2015). Moreover, in a yeast-two-hybrid assay, this mutation inhibits the interaction with GLD-3 (Wang et al. 2002). As the interaction with GLD-3 stabilizes the fold of the catalytic domain of GLD-2 (Nakel et al. 2015), the loss of polyadenylation in gld-2(h292) animals may also be due to an inability of GLD-2(E875R) to interact with GLD-3. However, a negative impact of the h292 mutation on the GLD-2/GLD-3 interaction could not be recapitulated in vitro upon coexpression and subsequent copurification of recombinant GLD-2 and GLD-3 fragments (Nakel et al. 2015). Therefore, future studies will be needed to provide a molecular explanation on how the h292 mutation impairs GLD-2 polyadenylation activity in vivo.
GLD-2-dependent polyadenylation is important for mRNA stability
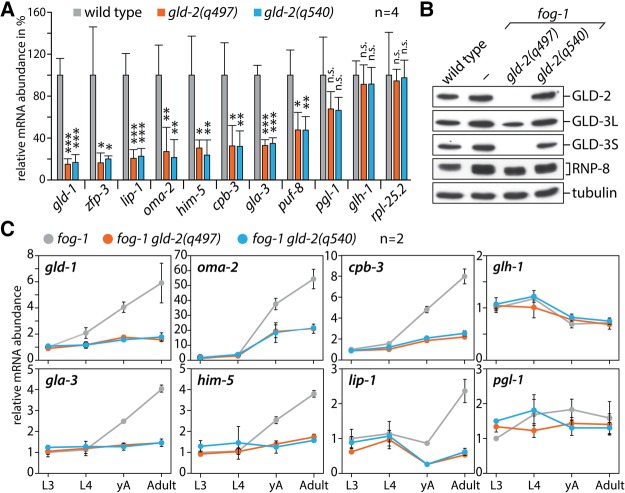
The presence of GLD-2 protein is important for a large number of germline mRNAs, as their abundance decreases in animals where GLD-2 is down-regulated or absent (Kim et al. 2010; Nousch et al. 2014). To test whether GLD-2(P652L) stabilizes mRNAs, we measured mRNA levels of previously described GLD-2 target mRNAs in young adults by qPCR, comparing the two gld-2 alleles, q540 and q497, to wild-type. The expression of each mRNA was normalized to rpl-11.1 levels, a germline-specific ribosomal protein and non-GLD-2 target mRNA (Maciejowski et al. 2005; Nousch et al. 2014). Consistent with previous results (Nousch et al. 2014), eight tested GLD-2 target mRNAs (gld-1, zfp-3, lip-1, oma-2, him-5, cpb-3, gla-4, puf-8) were significantly reduced in gld-2(q497) compared to wild-type, whereas three tested nontarget mRNAs (pgl-1, glh-1, rpl-25.2) remained unchanged (Fig. 3A). Interestingly, in gld-2(q540) animals, the abundance of all target mRNAs was comparable to gld-2(q497) animals (Fig. 3A). The observation that not even a partial rescue of GLD-2 targets could be detected strongly suggests that the polyadenylation-compromised GLD-2(P652L) enzyme is not able to stabilize mRNAs. This argues that the formation of polyadenylation-deficient cytoPAP complexes is not sufficient to promote target mRNA abundance, highlighting the importance of the enzymatic activity of GLD-2.
FIGURE 3.
Levels of GLD-2 target mRNAs are reduced in GLD-2(P652L)-expressing animals. (A) Abundance measurements of the indicated mRNAs via quantitative real-time PCR. Statistically significant differences are indicated and calculated via the Student's t-test. (***) P < 0.001, (**) P < 0.01, (*) P < 0.05; n.s., not significant. (B) Western blot analysis of the indicated proteins in fog-1 gld-2 double-mutant backgrounds. (C) mRNA abundance analysis during the development from L3 larvae to adults in given mutant backgrounds. L3, L3 larvae; L4, L4 larvae; earlyA, L4 + 12h; Adult, L4 + 24h.
In order to investigate the dynamics of GLD-2-mediated mRNA regulation, we decided to measure the accumulation of GLD-2 target mRNAs during the different stages of female germline development. C. elegans is a hermaphrodite that produces sperm during the L3 and L4 larval stages before switching to produce oocytes for the remainder of the animal's lifetime. In order to extend the time window of our analysis, we conducted our measurements in a fog-1 loss-of-function (lf) background. Removal of fog-1 activity feminizes the germline, and oogenesis already starts at the L3 larval stage (Barton and Kimble 1990). The absence of fog-1 has no influence on the expression of GLD-2(P652L) or its binding partners GLD-3L and RNP-8 (Fig. 3B). Also, in this genetic background, GLD-3S remained hardly detectable in the absence of functional GLD-2 protein (Fig. 3B). We measured six GLD-2 target mRNAs (gld-1, oma-2, gla-3, him-5, cpb-3, and lip-1) and two non-GLD-2 target mRNAs (glh-1 and pgl-1) at the L3 and L4 larval stage, as well as in younger (L4 + 12h) and older (L4 + 24h) adults. Each time point was normalized to the expression of rpl-11.1. In wild-type, most GLD-2 targets show a steady increase in expression following the L4 larval stage, with the exception of lip-1, which only increases during adulthood (Fig. 3C). On the contrary, these mRNAs show in both gld-2 mutant animals, q497 and q540, only a moderate increase in abundance, if any, during this time course (Fig. 3C). No such differences could be detected for glh-1 and pgl-1 (Fig. 3C). These data show that GLD-2 target mRNAs have similar levels and similar expression dynamics during oogenesis in the two tested gld-2 alleles, strongly arguing that GLD-2(P652L)-containing RNP complexes cannot stabilize mRNAs during any stages of oogenesis.
gld-2(q540) recapitulates known gld-2(0) phenotypes in the germline
GLD-2 protein expression is essential for germ cell development. In adult C. elegans hermaphrodites, female germ cells gradually mature with respect to their distal-to-proximal position within the tube-like gonad: Proliferative cells are located most distally and enter meiosis further proximally, starting with leptotene and eventually arresting in diakinesis at its proximal end (Fig. 4A). GLD-2 is important for at least two aspects of germ cell development: entry into and progression through meiosis (Kadyk and Kimble 1998; Eckmann et al. 2004). As a consequence, no functional gametes are produced in the absence of GLD-2 (Kadyk and Kimble 1998).
FIGURE 4.
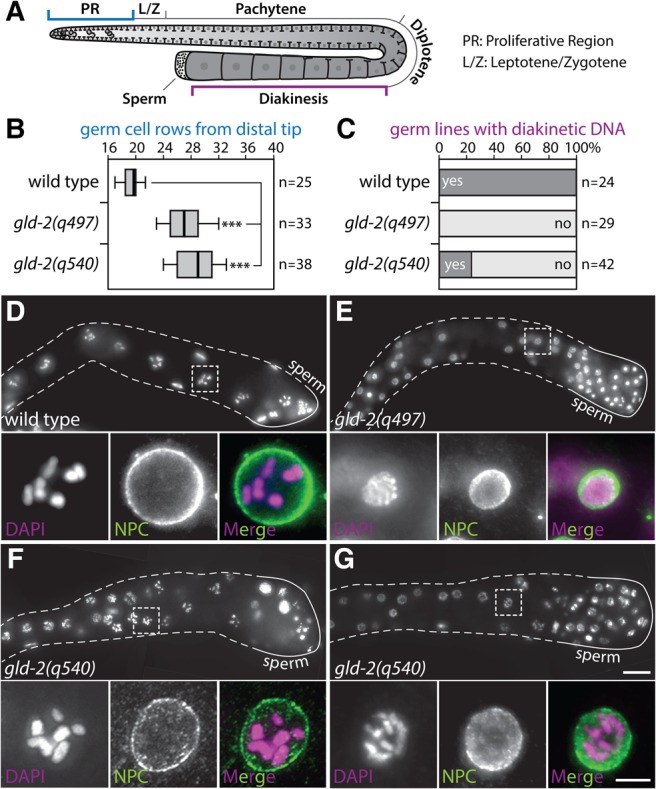
gld-2(q540) largely phenocopies the genetic null allele of gld-2. (A) Cartoon of the adult hermaphroditic syncytial germline tissue. Circles represent nuclei, T-structures are partial membranes. Important female germ cell stages are indicated; distal is top left, the position of sperm marks the most proximal end of the gonad. Color marked regions were analyzed in this work. (B) Analysis of the size of the proliferative region. Germ cell rows before meiotic entry were counted in the different genetic backgrounds as judged by DAPI staining. Significance was calculated by a two-tailed Student's t-test. (***) P ≤ 0.001. (C) Percentage of germlines that positively stain for diakinetic DNA in the proximal part, assessed by DAPI staining. (D–G) DAPI (purple in merge) staining of the indicated genetic backgrounds. NPC (green in merge), nuclear pore complex. Scale bar: (G) 15 µm, (blow-up) 5 µm.
It has previously been described that gld-2(q540) animals are sterile (Kadyk and Kimble 1998). However, it remained unclear whether all germ cell defects ascribed to the protein null allele q497 are also present in gld-2(q540) animals. We first concentrated on gld-2’s function in meiotic entry in the distal part of the gonad (Fig. 4A). In the absence of GLD-2 protein, the size of the proliferative region increases, which is interpreted as a delayed entry into meiosis (Eckmann et al. 2004). We analyzed the size of the proliferative region in wild-type and gld-2 mutants by DAPI staining. In wild-type gonads, germ cells enter meiosis around row 20 (germ cell distance measured from the distal tip) (Fig. 4B). Consistent with previous data (Eckmann et al. 2004), the size of the proliferative region is substantially increased in gld-2(q497) (Fig. 4B). Interestingly, the size of the proliferative region in gld-2(q540) is also larger than in wild-type, and no difference was detected between q497 and q540 (Fig. 4B), demonstrating that the gld-2(q540) missense mutation mimics the delayed meiotic entry phenotype of the genetic null allele gld-2(q497). Thus, we conclude that the polyadenylation-compromised GLD-2(P652L) protein is insufficient to compensate for meiotic entry defects, and that a timely transition from mitosis to meiosis is tightly linked to GLD-2's enzymatic activity.
The second major germ cell phenotype that we analyzed by DAPI staining occurs in the proximal part of the gonad (Fig. 4A). In this part of wild-type gonads, germ cells with highly condensed chromosomes are detected (Fig. 4C,D), marking them as cells that are in the diakinetic stage of meiosis. In the absence of GLD-2 protein, as seen in gld-2(q497), germ cells arrest in their meiotic program somewhere around the pachytene-diplotene stage, never reaching diakinesis (Fig. 4C,E). We made similar observations for the q540 allele; a large fraction of gonads (∼76%, n = 42) lacks diakinetic germ cell nuclei (Fig. 4C,G). Nonetheless, diakinetic chromosome configurations were detected in ∼24% of the analyzed gld-2(q540) germlines (Fig. 4C,F), showing that the q540 mutation mimics the meiotic arrest of germ cells of the q497 mutation to a large extent, but not completely. The overall phenotypic similarities between the two analyzed gld-2 alleles argue that polyadenylation is the most important function of GLD-2 in germ cells.
GLD-2 is broadly expressed during C. elegans germ cell development and forms distinct cytoPAP complexes with respect to its roles within the gonad. Based on the ubiquitous expression of GLD-3 and its phenotypic resemblance (Eckmann et al. 2002, 2004), a GLD-2/GLD-3 complex is most likely the main cytoPAP complex that regulates germ cell mRNAs in the distal part of the gonad. In combination with RNP-8, whose expression is limited to later stages of meiotic pachytene (Kim et al. 2009), at least two GLD-2 complexes are likely to operate in the proximal gonad. Interestingly, neither single mutant animals of rnp-8 or gld-3, nor rnp-8; gld-3 double mutants show the typical germ cell arrest seen in the loss of gld-2 (Eckmann et al. 2002; Kim et al. 2009), suggesting that additional GLD-2 interactors may exist for oogenesis. In either case, our data suggest that all GLD-2(P652L)-containing cytoPAP complexes are primarily compromised in their polyadenylation activity of target mRNAs.
Combined, our data suggest that the GLD-2 function is more diverse in later meiotic stages of germ cell development than during entry into meiosis. This may also reflect a different level of requirement or sensitivity for GLD-2-mediated polyadenylation. Although the expression of the catalytically compromised GLD-2(P652L) cytoPAP is most of the time insufficient for the progression through pachytene, germ cells occasionally develop further, suggesting that either GLD-2(P652L) protein has some remaining polyadenylation activity that we were unable to detect in our assays, or it might have polyadenylation-independent roles. Whether such minor roles exist and are connected to mRNA regulation needs to be addressed in the future. Nonetheless, we conclude that in strong contrast to Trf4-type nucleotidyl transferases, GLD-2-type poly(A) polymerases are likely to fulfill their cellular roles almost exclusively in a polyadenylation-dependent manner.
MATERIALS AND METHODS
Nematode strains and transgenesis
Worms were handled according to standard procedures and grown at 20°C (Brenner 1974). The N2 Bristol strain was used as a reference for wild-type. Strains used in this study: Linkage group (LG) I: gld-2(q497), gld-2(q540), gld-2(h292), and fog-1(q785). We noted that the original gld-2(q497) and gld-2(q540) strains (Kadyk and Kimble 1998) carried a background mutation in the mut-16 locus, which is on the same LG and corresponds to the allele mg461. To remove this mutation, both gld-2 stains were outcrossed with wild-type and rebalanced with hT2 [bli-4(e937) let-?(q782) qIs48] (I;III). Unless stated otherwise, adult germline phenotypes were scored 24 h past mid-L4 stage. For the time course experiment in Figure 3, worms were synchronized at the L1 stage via food deprivation, and developmental stages of the population were determined by DIC analysis of the developing germline of 5–10 animals.
Primary antibodies
Primary antibodies against the following proteins were used: rabbit anti-RNP-8 (Kim et al. 2009); mouse anti-NPC (Mab414, Covance), anti-tubulin (T5168, Sigma), and anti-GLD-2 (Millonigg et al. 2014); and rat anti-GLD-3 (Eckmann et al. 2002). The mouse anti-GLD-2 (Millonigg et al. 2014) recognized the C terminus of the protein. The rabbit anti-GLD-2 antibody used in the immunoprecipitation experiments was generated by immunizing New Zealand white rabbits with an epitope that covered amino acid 959–1113 of GLD-2, fused to a GST-affinity tag. The fusion protein had been expressed in BL21 bacteria and purified via a GST column to homogeneity. An analogously prepared MBP-fusion protein had been coupled to Hi-TRAP NHS columns (GE Healthcare) and used for affinity purification of the immune serum rb184.
Immunocytochemistry
Indirect immunocytochemistry of extruded and 1% PFA-fixed gonads was carried out in solution as described (Rybarska et al. 2009). Images were taken on a Zeiss Imager M1 equipped with an Axiocam MRm (Zeiss) and processed with AxioVision (Zeiss) and Photoshop CS5 (Adobe). Secondary antibodies coupled to fluorochromes FITC, CY3, and CY5 were purchased from Jackson ImmunoResearch (Dianova).
Western blotting and immunoprecipitations
For Western blotting experiments, we collected individual worms by hand and boiled them in Laemmli protein sample loading buffer prior to gel separation. Worm protein extracts for protein coimmunoprecipitations were made as previously described (Jedamzik and Eckmann 2009), with a minor modification of the procedure; to generate liquid nitrogen-frozen worm powder, we used a MR301 bead mill (Retsch) at 30 Hz. For the immunoprecipitation procedure, we coupled affinity-purified rabbit 184 anti-GLD-2 antibodies to Protein A Dynabeads (Invitrogen). All immunoprecipitates were analyzed by Western blotting with ECL detection of HRPO-coupled secondary antibodies (Jackson ImmunoResearch) as previously described (Jedamzik and Eckmann 2009).
RNA isolation and qPCR
Total RNA was isolated from hand-picked whole worms using TRIzol (Invitrogen). Total RNA (200 ng) was reverse-transcribed using random hexamer primers and ReverseAid Premium reverse transcriptase (Thermo Fisher Scientific), according to the manufacturer's protocols. Quantitative PCR (qPCR) was conducted on an iQ5 (Bio-Rad), using the ABsolute QPCR SYBR Green mix (Thermo Fisher Scientific) and gene-specific primers (sequences available upon request).
PCR-based poly(A)-tail measurement (sPAT assay)
For measuring poly(A) tails of specific mRNAs, we performed the previously described sPAT assay (Minasaki et al. 2014). In short, we isolated total mRNA from hand-picked worms, ligated an RNA anchor to the 3′ ends, performed gene-specific RT-PCR, and resolved the samples on high-resolution agarose gels. Lane quantifications were performed using Fiji (ImageJ).
Bulk poly(A)-tail length measurements
One microgram of total RNA was used to perform bulk poly(A)-tail measurements, following a previously described protocol (Temme et al. 2004), with the only exception that un-incorporated [α-32P]-Codycepin (PerkinElmer) was removed using mini Quick Spin Columns (Roche). Each sample was analyzed from three independent biological repeats; size markers were synthesized RNA oligos of 30 and 45 nt in length, and a loading dye band that corresponds to approximately 65 nt. Lane quantifications were performed using Fiji (ImageJ).
ACKNOWLEDGMENTS
We thank Elmar Wahle and Tosin Oyewale for critical reading of the manuscript, Judith Kimble for antibody reagents, the MPI-CBG for help with microscopy and its antibody facility for immunizing animals. This work was financially supported by the German Research Foundation (DFG) (EC369/2-3) and the DFG Heisenberg Program (EC369/3-1 and EC369/5-1) to C.R.E.
Author contributions: M.N. and C.R.E. designed the experiments. M.N. and R.M. performed the experiments. M.N., R.M., and C.R.E. analyzed the data. M.N. and C.R.E. wrote the manuscript.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.061473.117.
REFERENCES
- Balbo PB, Bohm A. 2007. Mechanism of poly(A) polymerase: structure of the enzyme-MgATP–RNA ternary complex and kinetic analysis. Structure 15: 1117–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton MK, Kimble J. 1990. fog-1, a regulatory gene required for specification of spermatogenesis in the germ line of Caenorhabditis elegans. Genetics 125: 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit P, Papin C, Kwak JE, Wickens M, Simonelig M. 2008. PAP- and GLD-2-type poly(A) polymerases are required sequentially in cytoplasmic polyadenylation and oogenesis in Drosophila. Development 135: 1969–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CZ, Jo DH, Heinemann IU. 2016. Nucleotide specificity of the human terminal nucleotidyltransferase Gld2 (TUT2). RNA 22: 1239–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Sackton KL, Horner VL, Kumar KE, Wolfner MF. 2008. Wispy, the Drosophila homolog of GLD-2, is required during oogenesis and egg activation. Genetics 178: 2017–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Sartain CV, Pleiss JA, Wolfner MF. 2013. Cytoplasmic polyadenylation is a major mRNA regulator during oogenesis and egg activation in Drosophila. Dev Biol 383: 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CA, Ares M Jr. 2006. Accumulation of unstable promoter-associated transcripts upon loss of the nuclear exosome subunit Rrp6p in Saccharomyces cerevisiae. Proc Natl Acad Sci 103: 3262–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckmann CR, Kraemer B, Wickens M, Kimble J. 2002. GLD-3, a bicaudal-C homolog that inhibits FBF to control germline sex determination in C. elegans. Dev Cell 3: 697–710. [DOI] [PubMed] [Google Scholar]
- Eckmann CR, Crittenden SL, Suh N, Kimble J. 2004. GLD-3 and control of the mitosis/meiosis decision in the germline of Caenorhabditis elegans. Genetics 168: 147–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egecioglu DE, Henras AK, Chanfreau GF. 2006. Contributions of Trf4p- and Trf5p-dependent polyadenylation to the processing and degradative functions of the yeast nuclear exosome. RNA 12: 26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedamzik B, Eckmann CR. 2009. Analysis of in vivo protein complexes by coimmunoprecipitation from Caenorhabditis elegans. Cold Spring Harb Protoc 10.1101/pdb.prot5299. [DOI] [PubMed] [Google Scholar]
- Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J. 2004. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev 18: 1227–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyk LC, Kimble J. 1998. Genetic regulation of entry into meiosis in Caenorhabditis elegans. Development 125: 1803–1813. [DOI] [PubMed] [Google Scholar]
- Kim KW, Nykamp K, Suh N, Bachorik JL, Wang L, Kimble J. 2009. Antagonism between GLD-2 binding partners controls gamete sex. Dev Cell 16: 723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KW, Wilson TL, Kimble J. 2010. GLD-2/RNP-8 cytoplasmic poly(A) polymerase is a broad-spectrum regulator of the oogenesis program. Proc Natl Acad Sci 107: 17445–17450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung JE, Jura N. 2016. Structural basis for the non-catalytic functions of protein kinases. Structure 24: 7–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JE, Wang L, Ballantyne S, Kimble J, Wickens M. 2004. Mammalian GLD-2 homologs are poly(A) polymerases. Proc Natl Acad Sci 101: 4407–4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121: 713–724. [DOI] [PubMed] [Google Scholar]
- Maciejowski J, Ahn JH, Cipriani PG, Killian DJ, Chaudhary AL, Lee JI, Voutev R, Johnsen RC, Baillie DL, Gunsalus KC, et al. 2005. Autosomal genes of autosomal/X-linked duplicated gene pairs and germ-line proliferation in Caenorhabditis elegans. Genetics 169: 1997–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millonigg S, Minasaki R, Nousch M, Eckmann CR. 2014. GLD-4-mediated translational activation regulates the size of the proliferative germ cell pool in the adult C. elegans germ line. PLoS Genet 10: e1004647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minasaki R, Eckmann CR. 2012. Subcellular specialization of multifaceted 3′end modifying nucleotidyltransferases. Curr Opin Cell Biol 24: 314–322. [DOI] [PubMed] [Google Scholar]
- Minasaki R, Rudel D, Eckmann CR. 2014. Increased sensitivity and accuracy of a single-stranded DNA splint-mediated ligation assay (sPAT) reveals poly(A) tail length dynamics of developmentally regulated mRNAs. RNA Biol 11: 111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakel K, Bonneau F, Eckmann CR, Conti E. 2015. Structural basis for the activation of the C. elegans noncanonical cytoplasmic poly(A)-polymerase GLD-2 by GLD-3. Proc Natl Acad Sci 112: 8614–8619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakel K, Bonneau F, Basquin C, Habermann B, Eckmann CR, Conti E. 2016. Structural basis for the antagonistic roles of RNP-8 and GLD-3 in GLD-2 poly(A)-polymerase activity. RNA 22: 1139–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nousch M, Yeroslaviz A, Habermann B, Eckmann CR. 2014. The cytoplasmic poly(A) polymerases GLD-2 and GLD-4 promote general gene expression via distinct mechanisms. Nucleic Acids Res 42: 11622–11633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhana L, Wang L, Buter N, Kwak JE, Schiltz CA, Gonzalez T, Kelley AE, Landry CF, Wickens M. 2005. Vertebrate GLD2 poly(A) polymerases in the germline and the brain. RNA 11: 1117–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybarska A, Harterink M, Jedamzik B, Kupinski AP, Schmid M, Eckmann CR. 2009. GLS-1, a novel P granule component, modulates a network of conserved RNA regulators to influence germ cell fate decisions. PLoS Genet 5: e1000494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Paolo S, Vanacova S, Schenk L, Scherrer T, Blank D, Keller W, Gerber AP. 2009. Distinct roles of non-canonical poly(A) polymerases in RNA metabolism. PLoS Genet 5: e1000555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartain CV, Cui J, Meisel RP, Wolfner MF. 2011. The poly(A) polymerase GLD2 is required for spermatogenesis in Drosophila melanogaster. Development 138: 1619–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Küchler B, Eckmann CR. 2009. Two conserved regulatory cytoplasmic poly(A) polymerases, GLD-4 and GLD-2, regulate meiotic progression in C. elegans. Genes Dev 23: 824–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh N, Jedamzik B, Eckmann CR, Wickens M, Kimble J. 2006. The GLD-2 poly(A) polymerase activates gld-1 mRNA in the Caenorhabditis elegans germ line. Proc Natl Acad Sci 103: 15108–15112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temme C, Zaessinger S, Meyer S, Simonelig M, Wahle E. 2004. A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. EMBO J 23: 2862–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanácová S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. 2005. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol 3: e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Eckmann CR, Kadyk LC, Wickens M, Kimble J. 2002. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature 419: 312–316. [DOI] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Régnault B, Devaux F, Namane A, Séraphin B, et al. 2005. Cryptic Pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121: 725–737. [DOI] [PubMed] [Google Scholar]