Abstract
miRNAs have emerged as key participants of p53 signaling pathways because they regulate or are regulated by p53. Here, we provide the first study demonstrating direct regulation of an oncogenic miRNA, miR-191-5p, by p53 and existence of a regulatory feedback loop. Using a combination of qRT-PCR, promoter-luciferase, and chromatin-immunoprecipitation assays, we show that p53 brings about down-regulation of miR-191-5p in breast cancer. miR-191-5p overexpression brought about inhibition of apoptosis in breast cancer cell lines (MCF7 and ZR-75) as demonstrated by reduction in annexin-V stained cells and caspase 3/7 activity, whereas miR-191-5p down-regulation showed the opposite. We further unveiled that SOX4 was a direct target of miR-191-5p. SOX4 overexpression was shown to increase p53 protein levels in MCF7 cells. miR-191-5p overexpression brought about down-regulation of SOX4 and thus p53 levels, suggesting the existence of a regulatory feedback loop. Breast cancer treatment by doxorubicin, an anti-cancer drug, involves induction of apoptosis by p53; we thus wanted to check whether miR-191-5p affects doxorubicin sensitivity. Interestingly, Anti-miR-191 treatment significantly decreased the IC50 of the doxorubicin drug and thus sensitized breast cancer cells to doxorubicin treatment by promoting apoptosis. Overall, this work highlights the importance of the p53-miR-191-SOX4 axis in the regulation of apoptosis and drug resistance in breast cancer and offers a preclinical proof-of-concept for use of an Anti-miR-191 and doxorubicin combination as a rational approach to pursue for better breast cancer treatment.
Keywords: breast cancer, apoptosis, miR-191, p53, SOX4
INTRODUCTION
Breast cancer is the most common invasive cancer in females that has shown improved survival owing to early detection and better therapeutic strategies. However, death due to breast cancer still shows a rising trend (Florea and Büsselberg 2013; American Cancer Society 2015). This has been attributed to multiple factors, including metastases and development of resistance to various therapies (Gonzalez-Angulo et al. 2007). Therefore, there is a need to understand breast tumor biology in its entirety and identify novel mediators involved in the pathogenesis.
Breast tumor development involves deregulated networks of specific protein coding genes, long noncoding RNAs, and miRNAs (Nana-Sinkam and Croce 2011). The key for breast cancer treatment lies in the identification and targeting of the crucial players among these complicated and huge networks. Recently, the significant role of an miRNA, miR-191-5p, in breast tumor biology was demonstrated by multiple groups (Di Leva et al. 2013; Nagpal et al. 2013, 2015). miR-191-5p was shown to be an estrogen and hypoxia responsive miRNA that promotes cell proliferation and migration in breast cancer (Nagpal et al. 2013, 2015). The oncogenic role of miR-191-5p has also been shown in other cancers (lung cancer, colon cancer, gastric cancer) (Xi et al. 2006; Elyakim et al. 2010; Patnaik et al. 2010; He et al. 2011; Shi et al. 2011; Zhou et al. 2013; Liu et al. 2014). miR-191-5p is up-regulated in breast cancer patients with a trend of higher expression in estrogen receptor positive patients as compared to the negative ones (Di Leva et al. 2013; Nagpal et al. 2013). We became interested in studying the regulation of miR-191-5p and identified multiple p53 binding sites in its promoter. Here, we report the regulation of miR-191-5p by p53 and the existence of a feedback loop wherein miR-191-5p emerges as an important regulator of p53 levels and consequently apoptosis and drug resistance in breast cancer. The work suggests the use of Anti-miR-191 therapy along with doxorubicin for effective breast cancer treatment.
RESULTS
P53 down-regulates miR-191-5p in breast cancer
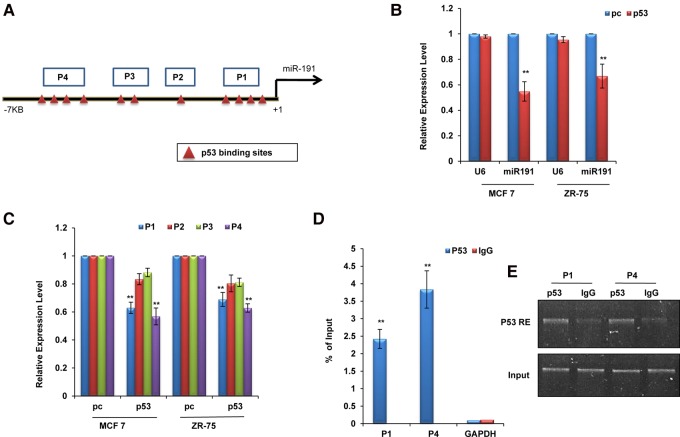
First, we looked for transcription factor binding sites in the upstream region of miR-191-5p. In silico analysis of regions ∼7 kb upstream of and 1 kb downstream from miR-191-5p using Promo 3.0 software showed the presence of multiple putative p53 binding sites showing <1% dissimilarity (Fig. 1A; Messeguer et al. 2002; Farré et al. 2003). To evaluate the role of p53 in the regulation of miR-191-5p, we overexpressed p53 in breast cancer lines and measured p53 and miR-191-5p levels 48 h post-transcription (Fig. 1B; Supplemental Fig. S1A,B). The miR-191-5p levels were significantly reduced in p53-transfected cells relative to cells transfected with the control (Fig. 1B). To determine whether p53-mediated miR-191-5p down-regulation is through the promoter, the regions bearing p53 binding sites (P1, four binding sites; P2, one binding site; P3, two binding sites; and P4, four binding sites) were cloned in luciferase promoter vector and scored for luciferase activity. The regions P1 and P4 showed a significant decrease in luciferase activity on p53 overexpression in MCF7 and ZR-75 cells, suggesting that p53 down-regulates miR-191-5p at the transcriptional level through the promoter (Fig. 1C). Further, in order to confirm direct binding of p53 to the predicted binding sites, chromatin immunoprecipitation (ChIP) assay was performed in MCF7 cells using p53 and IgG antibodies. We found that binding to P1 and P4 was more enriched in p53 as compared to the control (IgG) pull-down fractions (Fig. 1D,E). Based on these results, we confirm that p53 directly regulates the transcriptional expression of miR-191-5p through binding to the p53 response elements in its promoter.
FIGURE 1.
Transcriptional regulation of miR-191-5p. (A) Diagram showing putative p53 binding sites with <1% dissimilarity in the upstream region of pre-miR-191. P1–P4 refers to the four regions cloned in the pGL3 luciferase vector for studying the effect of p53 overexpression in breast cancer cells. (B) Real-time PCR data showing down-regulation of miR-191-5p levels in response to p53 overexpression as compared to the control vector (pc) in breast cancer cell lines (MCF7 and ZR-75). RNU6B (U6) has been used as a control for normalization. Graph was plotted using formula. (C) Luciferase activity of regions (P1–P4) was measured in MCF7 and ZR-75 48 h post-transfection with pc or p53. (D,E) p53 direct binding to the miR-191-5p upstream region was confirmed using ChIP analyses. qPCR (D) or semiquantitative (E) data showing enrichment of the p53 recognition elements (p53RE) in P1 and P4 regions on immunoprecipitation with p53 antibody as compared to the IgG control. The graphical data points represent mean ± SD of at least three independent experiments. (**) P < 0.01. Error bars denote ±SD.
miR-191-5p functions as an anti-apoptotic miRNA in breast cancer
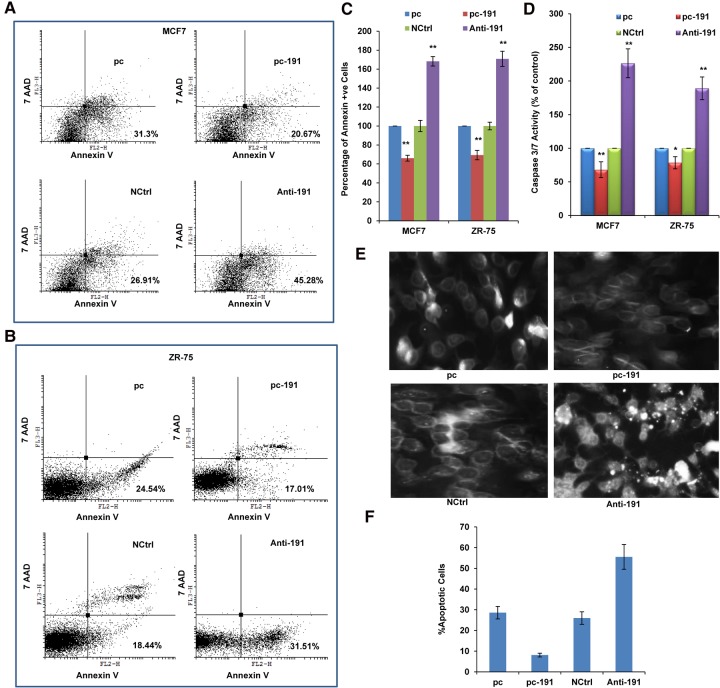
We next wanted to check whether miR-191-5p is involved in the regulation of apoptosis in breast cancer. The PE/Annexin V staining showed that the percentage of the early apoptotic cells was lower in the cells transfected with pc-191 when compared with the control (Fig. 2A–C). In contrast, the percentage of early apoptotic cells was higher in the cells transfected with Anti-191 than in NCtrl (Fig. 2A–C). Further, to know whether the effect of miR-191-5p on apoptosis is caspase dependent, we measured caspase 3/7 activity in the transfected MCF7 and ZR-75 cells. miR-191-5p transfected cells showed a decrease in caspase 3/7 activity, whereas Anti-191 treatment increased caspase 3/7 activity (Fig. 2D). Microscopic examination of DAPI (DNA binding fluorescent dye) stained cells revealed a higher number of cells showing shrinkage and the appearance of apoptotic bodies in Anti-191 treated cells as compared to control (Fig. 2E). In contrast, miR-191-5p overexpressing cells showed a lower number of apoptotic cells as compared to its control (Fig. 2F). Overall, these results demonstrate that miR-191-5p inhibits apoptosis in breast cancer cells.
FIGURE 2.
miR-191-5p functions as an anti-apoptotic miRNA in breast cancer. FACS analysis using PE-Annexin V and 7AAD was performed in MCF7 cells (A) and ZR-75 (B) transfected with pc-191 and Anti-191 along with their controls (pc and NCtrl, respectively). (C) Graph shows percentage of Annexin V positive cells. (D) Caspase 3/7 activity in MCF7 and ZR-75 cells 48 h post-transfection with pc-191 and Anti-191 along with their controls. (E) Evaluation of apoptosis by DAPI staining in MCF7 cells in response to miR-191-5p overexpression or inhibition. (F) Graph shows percentage of apoptotic cells. The graphical data points represent mean ± SD of at least three independent experiments. (*) P < 0.05, (**) P < 0.01. Error bars denote ±SD.
miR-191-5p targets genes in breast cancer
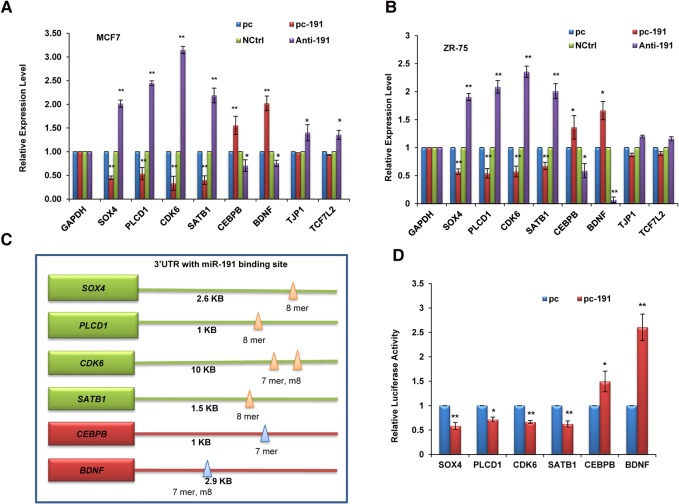
Next we wanted to evaluate the mechanism of miR-191-5p-mediated inhibition of apoptosis. We used online target prediction programs (TargetScan, miRanda) to investigate the potential targets of miR-191-5p and shortlisted eight target genes (SOX4, PLCD1, CDK6, SATB1, CEBPB, BDNF, TJP1, TCF7L2) that have been shown to be associated with breast cancer and also are related to regulation of apoptosis (Hoevel et al. 2004; John et al. 2005; Zahnow 2009; Hur et al. 2010; Li et al. 2010; Vanhecke et al. 2011; Chen et al. 2013; Agarwal et al. 2015; Mu et al. 2015; Tadesse et al. 2015). Interestingly, some of the targets (SOX4, PLCD1, CDK6, and SATB1) showed down-regulation; the others (CEBPB and BDNF) showed induction on pc-191 overexpression, while TJP1 and TCF7L2 remained unchanged (Fig. 3A,B). An opposite trend was seen on Anti-miR-191 treatment of MCF7 cells (Fig. 3A,B). To further confirm direct targeting of these transcripts by miR-191-5p, 3′ UTR-luciferase reporter assay was performed for six target transcripts (SOX4, PLCD1, CDK6, SATB1, CEBPB, and BDNF) containing miR-191-5p binding sites in their 3′ UTR (Fig. 3C). The 3′ UTR-luciferase assay results showed that SOX4, PLCD1, CDK6, and SATB1 were down-regulated by miR-191-5p, whereas CEBPB and BDNF were induced by miR-191-5p (Fig. 3D). The above results confirmed that all these genes (SOX4, PLCD1, CDK6, SATB1, CEBPB, and BDNF) are direct targets of miR-191-5p.
FIGURE 3.
Targets of miR-191-5p. Expression level of shortlisted target genes of miR-191-5p was determined by qRT-PCR in response to miR-191 overexpression or inhibition in MCF7 (A) and ZR-75 (B) cell lines. (C) Diagram showing miR-191-5p binding sites in the 3′ UTR of target genes. 7 mer, 8 mer, and m8 refer to seed match sites. (D) Luciferase activity of the 3′ UTR luciferase constructs bearing miR-191-5p binding sites in response to transfection with pc-191 or its control vector (pc) in MCF7 cells. The graphical data points represent mean ± SD of at least three independent experiments. (*) P < 0.05, (**) P < 0.01. Error bars denote ±SD.
miR-191-5p targets SOX4 and brings about down-regulation of p53 levels in breast cancer
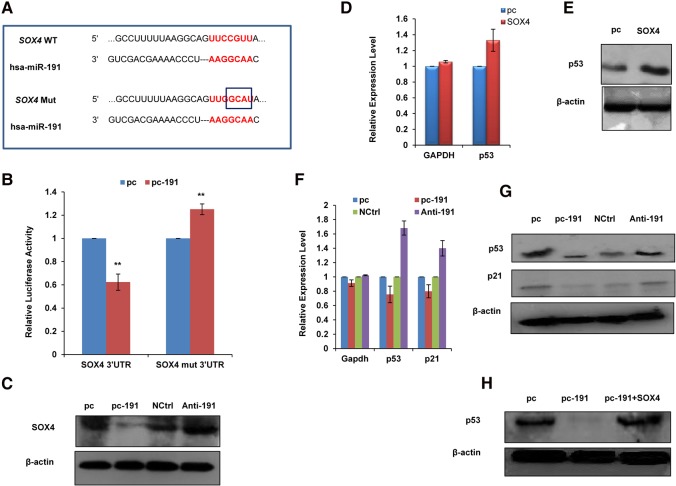
Pan et al. (2009) demonstrated that SOX4 regulates p53 protein stability in colon cancer. Therefore, we focused on SOX4 to explore the miR-191-5p-mediated effects on an apoptotic pathway. For this, we first confirmed the direct effect of miR-191-5p on SOX4 levels by performing luciferase assay with wild-type or mutated miR-191-5p binding site in the 3′ UTR of SOX4 (Fig. 4A). We found a significant decrease in the luciferase activity of wild-type SOX4 3′UTR but not the mutated one (Fig. 4B). Next, we measured SOX4 protein levels in response to miR-191-5p modulation. We found that miR-191-5p overexpression decreased the SOX4 protein levels, whereas Anti-191 treatment showed the opposite trend (Fig. 4C). To evaluate the effect of SOX4 on p53 protein stability in breast cancer, MCF7 cells were transfected with SOX4 or control plasmid, and 48 h post-transfection, the transcript as well as the protein level of p53 were measured (Fig. 4D,E; Supplemental Fig. S1C). We found that while the p53 transcript remained unaffected, the p53 protein levels were increased on SOX4 overexpression. Therefore, the regulation of p53 by SOX4 is more likely at the post-transcriptional level. This hinted at the existence of a feedback loop between miR-191-5p and p53, and we hypothesized that miR-191-5p by down-regulation of SOX4 may affect p53 levels. We found that overexpression of miR-191-5p in MCF7 cells resulted in a decrease of p53 protein levels with a minimal effect on transcript levels. A similar trend was seen on the levels of a well-known p53 target gene, p21 on miR-191-5p overexpression/inhibition (Fig. 4F,G). Replenishing SOX4 in miR-191-5p overexpressing cells brought back the p53 protein levels when compared to the control (Fig. 4H). Collectively, all these results demonstrated that miR-191-5p targets SOX4 and brings about down-regulation of p53 levels in breast cancer.
FIGURE 4.
miR-191-5p down-regulates SOX4 and p53 levels. (A) Diagram showing wild-type/mutated miR-191-5p binding site in the SOX4 3′ UTR. (B) Luciferase activity of the SOX4 wild-type/mutated miR-191-5p binding site bearing constructs was measured in response to pc or p53 overexpression. (C) SOX4 levels were measured at the protein level in MCF7 cells expressing pc-191 or Anti-191 along with their controls. The results show that miR-191-5p overexpression down-regulates SOX4 at the protein level. β-Actin was used for normalization of Western blotting data. (D,E) To evaluate the effect of SOX4 on p53 protein stability in breast cancer, MCF7 cells were transfected with SOX4 or control vector (pc), and 48 h post-transfection, p53 transcript as well as protein levels were measured by qRT-PCR and Western blotting. The real-time graph was plotted using formula. (F,G) The transcript and protein levels of p53 and p21 were evaluated by qRT-PCR and Western blotting in response to miR-191-5p overexpression or inhibition in MCF7 cells. The real-time graph was plotted using formula. (H) p53 protein level was determined by Western blotting in MCF7 cells transfected with pc-191 alone or in combination with SOX4. The graphical data points represent mean ± SD of at least three independent experiments. (*) P < 0.05, (**) P < 0.01. Error bars denote ±SD.
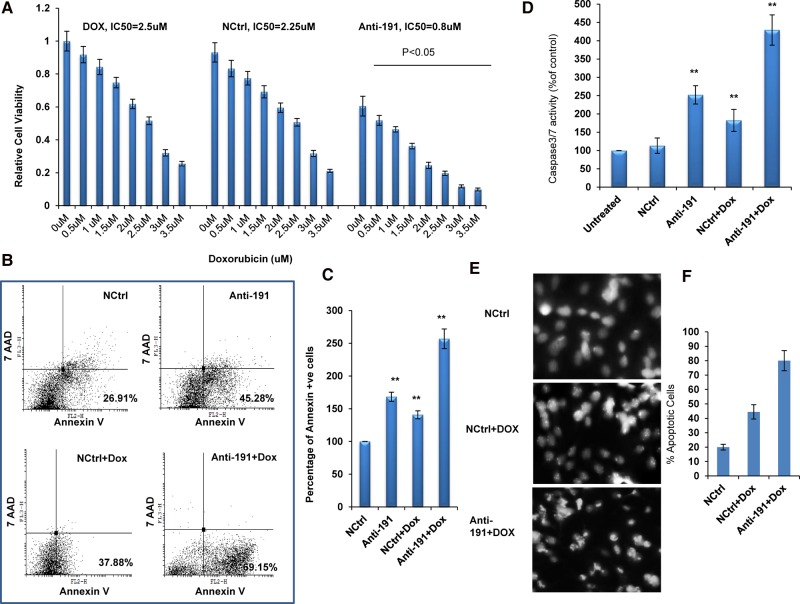
Anti-miR-191 treatment sensitizes breast cancer cells to doxorubicin-mediated apoptotic death
Apoptosis is a common and preferred target of several breast cancer treatment strategies. Since miR-191-5p is shown here to be anti-apoptotic miRNA, we wanted to evaluate the effect of using anti-miR-191 oligos (Anti-191) either alone or in combination with the chemotherapeutic drug doxorubicin for breast cancer treatment. Interestingly, we found that the IC50 value of doxorubicin was significantly decreased with the combination treatment of Anti-191 and doxorubicin when compared with the controls (NCtrl + Dox and Dox free) and displayed higher sensitivity to the treatment (Fig. 5A; Supplemental Fig. S2). This result suggested that inhibition of miR-191-5p sensitized breast cancer cells to chemotherapy. Similar results were obtained on using Anti-191 along with mitomycin C and paclitaxel, other chemotherapeutic drugs used for breast cancer treatment (Supplemental Fig. S3). To investigate whether the combination of Anti-191 with doxorubicin affects apoptosis in breast cancer cells, FACS analysis was done. The combination of Anti-191 with doxorubicin clearly increases the percentage of apoptotic cells (69.15%) as compared to its control (Fig. 5B,C). This was further confirmed by caspase 3/7 assay and DAPI-based microscopic analyses (Fig. 5D,E). These findings together demonstrate that inhibition of miR-191-5p promotes doxorubicin-induced apoptosis in breast cancer cells.
FIGURE 5.
The effects of miR-191-5p silencing on breast cancer chemotherapy. (A) MCF7 cells were treated with different concentrations of doxorubicin (free), or doxorubicin along with control oligo (NCtrl), or Anti-191. Subsequently, cell viability was evaluated by MTT assay. (B) FACS analysis using PE-Annexin V and 7AAD was performed in MCF7 cells treated with Anti-191 and doxorubicin. (C) Graph showing percentage of annexin V positive cells. (D) Caspase 3/7 activity was measured in MCF7 cells treated with Anti-191 and doxorubicin. (E) Microscopic images of DAPI-stained MCF7 cells treated with Anti-191 or its control and doxorubicin. (F) Graph shows percentage of apoptotic cells. The graphical data points represent mean ± SD of at least three independent experiments. (*) P < 0.05 and (**) P < 0.01. Error bars denote ±SD.
DISCUSSION
Apoptosis-mediated cancer cell death remains central to most treatment strategies. Thus, identification of miRNAs involved in the regulation of apoptosis holds immense importance. Here, we identify miR-191-5p as a negative regulator of apoptosis in breast cancer. We looked for miR-191-5p target genes to study its mechanism of action. We identified BDNF, CDK6, CEBPB, SATB1, SOX4, and PLCD1 as direct miR-191-5p targets. We have previously reported SATB1, CDK6, and BDNF as miR-191-5p targets but have included these here for their reported link with breast cancer apoptosis. miR-191-5p regulation of their levels has also been independently confirmed here in other breast cancer cell lines ZR-75 and T47D (Fig. 3B; Supplemental Fig. S4). BDNF has been shown to inhibit apoptosis in breast cancer cell lines MDA-MB-231 and MCF7 (Yang et al. 2012). SATB1 has also been shown to inhibit apoptosis and promote chemoresistance in MCF7 cells (Li et al. 2010). Similarly, high CDK6 protects cells from fulvestrant-mediated apoptosis in breast cancer (Alves et al. 2016). PLCD1 has been reported to function as a tumor suppressor in breast cancer that inhibits breast tumor cell formation in vivo by inducing apoptosis (Mu et al. 2015). CEBPB has been shown to promote tumorigenesis and its high levels are seen in breast cancer patients (Grimm and Rosen 2003). SOX4 has also been shown to affect apoptosis in breast cancer (Song et al. 2015). Overall, aberrations in miR-191-5p levels significantly affect the levels of proteins known to affect apoptosis.
We particularly focused on SOX4 because it has previously been shown to regulate the levels of p53, the master regulator of apoptosis (Hur et al. 2010; Zhou et al. 2015). There have been conflicting reports on the effect of SOX4 on p53 levels. While some studies demonstrate that SOX4 brings about the degradation of p53 levels, others show that it rather promotes p53 stabilization (Pan et al. 2009). Here, we found that SOX4 overexpression in MCF7 cells brought about a significant increase in p53 protein levels. This is in line with Pan et al. (2009), who reported that SOX4 is a novel mediator for p53 stabilization and activation in response to DNA damage (Pan et al. 2009). In our study, results revealed that miR-191-5p-mediated targeted down-regulation of SOX4 brings about down-regulation of p53, which further leads to inhibition of apoptosis in breast cancer. In turn, silencing of miR-191-5p by using Anti-191 brings about up-regulation of SOX4 and p53, which triggers apoptosis in breast cancer. This suggests that SOX4 may contribute to p53 protein stabilization in breast cancer cells, and by targeting SOX4, miR-191-5p brings about down-regulation of p53 in breast cancer cells. Several other miRNAs too have been shown to be associated with apoptosis and p53 pathways in breast cancer (Feng et al. 2011; Sharma et al. 2016). While miR-504 directly targets p53, other miRNAs were shown to indirectly affect p53 signaling by targeting upstream regulators or downstream mediators of p53 to affect apoptosis in breast cancer (Feng et al. 2011; Sharma et al. 2016). p53 has also been shown to regulate several miRNAs (miR-143, miR-145, miR-15, miR-16, miR-26a, and miR-34) involved in apoptosis in breast cancer (Feng et al. 2011). We found that p53 brings about down-regulation of miR-191-5p levels through binding to specific p53 binding sites in the promoter of miR-191-5p. Thus there exists an interesting p53-miR-191-SOX4 feedback loop that affects apoptosis in breast cancer. The role of miR-191-5p in regulation of apoptosis has also been demonstrated in hepatic and gastric cancer, but the mechanism was not elucidated (Elyakim et al. 2010; Shi et al. 2011). miR-191-5p has also been shown to target MDM4 in ovarian cancer and thus affect p53 levels (Wynendaele et al. 2010). Whether MDM4 is a target of miR-191-5p in breast cancer remains to be evaluated. miR-191-5p has been shown to be regulated by other transcription factors such as estrogen receptor and hypoxia-inducible factor-1 (Di Leva et al. 2013; Nagpal et al. 2013, 2015). The mutual regulation between p53 and ER-α or p53 and HIF-1 or HIF-1 and ER-α has been demonstrated by various groups in breast cancer (Berger et al. 2013; Obacz et al. 2013). However, what effect this cross-talk may have on miR-191-5p levels in tumors and consequently its impact on tumor aggressiveness and treatment outcome needs to be thoroughly investigated.
Doxorubicin is a commonly used effective drug for the treatment of breast cancer patients. However, patients developing resistance to this drug is common, representing a major hindrance to successful treatment (Smith et al. 2006; Raguz and Yagüe 2008). Here, we found that pretreatment of breast cancer cells with Anti-miR-191 sensitizes the cells to doxorubicin treatment and decreases its IC50 significantly. One of the mechanisms by which Anti-miR-191 treatment promotes doxorubicin sensitivity is by increasing p53 levels, leading to increased apoptosis in the cells, as demonstrated here. However, whether Anti-miR-191 also affects doxorubicin-mediated disruption of topoisomerase II-mediated DNA repair and generation of free radicals remains to be studied (Thorn et al. 2011). Latorre et al. (2012) previously demonstrated that HuR, an RNA binding protein, promotes doxorubicin sensitivity in breast cancer cells. HuR has also been shown to be a miR-191-5p target in breast cancer (Nagpal et al. 2015). Thus, miR-191-5p-mediated down-regulation of HuR can also be one of the mechanisms of doxorubicin resistance in breast cancer cells. Other miRNAs, such as miR-21, miR-34a, miR-181a, miR-17/20, and miR-218, also have been shown to affect doxorubicin resistance in breast cancer. The target genes of these miRNAs are involved in cell signaling or apoptotic pathways (miR-21-PTEN, miR-34a-NOTCH1, miR-181a-BCL2, miR-17/20-AKT1, and miR-218-SURVIVIN) (Wang et al. 2011; Li et al. 2012; Zhu et al. 2013; Yu et al. 2014; Hu et al. 2015). Considering that resistance to hormone therapy or chemotherapy drugs apart from doxorubicin is also partly due to inhibition of p53-mediated apoptosis, the effect of Anti-miR-191 on sensitizing cells to other therapies needs to be checked.
In summary, our findings add new insight into the role of miR-191-5p in the regulation of apoptosis in breast cancer. We also show the existence of a regulatory negative feedback loop between miR-191-5p and p53 in breast cancer. Considering previous reports of miR-191-5p as part of hypoxia and estrogen receptor signaling, it emerges as a critical mediator of multiple oncogenic and tumor suppressor pathways that are known to be deregulated in breast cancer. Thus, Anti-miR-191 treatment may lead to simultaneous inhibition of HIF and ER signaling and promotion of p53 signaling, leading to a multipronged attack on breast cancer. This, combined with the fact that Anti-miR-191 also promotes sensitivity toward doxorubicin treatment, makes it an attractive target for breast cancer treatment.
MATERIALS AND METHODS
Cell culture
The breast cancer cell lines (MCF7, ZR-75, and T47D) were obtained from Cell Repository of NCCS, Pune. All the cells were maintained in RPMI 1640 (GIBCO) medium. The media was supplemented with 100 U/mL penicillin, 100 µg/mL streptomycin, and 10% fetal bovine serum and incubated at 37°C in a 5% CO2 incubator.
Transfections
Cells were seeded (5 × 105 cells per well) in six-well plates. Of note, 2.5 µg of miR-191-5p plasmid (pc-191) or its control plasmid pcdna3.1 (pc) and 30nM anti-miR-191 (Anti-191) or the negative control (NCtrl) (Sigma-Aldrich) was transfected into six-well plates using lipofectamine 2000 (Invitrogen). After 5 h, media was changed, and after 48 h post-transfection the samples were assayed for transcript levels through quantitative reverse-transcription–PCR (qRT-PCR) or used for other cellular assays. Each experiment was repeated three times.
qRT-PCR
Total RNA was isolated using an RNA Extraction Kit (Fermentas) and reverse transcribed using a cDNA Synthesis Kit (Bio-Rad). The cDNA formed was then further amplified for the predicted genes with respective primers sets using SsoFast EvaGreen Master Mix (Bio-Rad). For miR191-5p detection, a set of stem–loop primers were used (see Supplemental Fig. S5 for a list of primers). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and RNU6B (U6) were used as controls for the normalization of the data. The expression fold change values were determined by formula.
Target prediction
The probable targets for miR-191-5p were selected by established target prediction programs. Two target prediction programs, TargetScan and miRanda, were used to select the miR-191-5p targets. The candidate targets were selected on the basis of published literature demonstrating their association with apoptosis.
Construction of SOX4 overexpression plasmid: pWPXL-SOX4 was a gift from Bob Weinberg (Addgene plasmid # 36984). To create the pc-SOX4 plasmid, a pWPXL-SOX4 plasmid was digested with Nde1 (Fermentas) and treated with Vent Polymerase (NEB) at 72°C for 15 min, then the plasmid was purified and digested with BamH1. Upon digestion with BamH1, the SOX4 insert was released from the pWPXL plasmid vector and ligated into the pcDNA3.1 plasmid (digested with BamH1 and EcoRV restriction enzymes [Fermentas]).
Construction of 3′ untranslated region- or promoter-luciferase constructs
To determine whether miR-191-5p down-regulates its target transcripts through its direct binding to the 3′ untranslated region (UTR), the region containing the miR-191-5p binding site in the 3′UTRs of the genes (SOX4, PLCD1, CDK6, SATB1, CEBPB, and BDNF) was amplified (using Taq polymerase; Fermentas) and cloned in the luciferase reporter vectors (pmiR-Report) downstream from a firefly luciferase gene (Supplemental Fig. S5). To further confirm whether the predicted target is the functional target of miR-191-5p, the miR-191-5p binding site in the 3′UTR of the gene (SOX4) was mutated through site-directed mutagenesis using inverse PCR and luciferase activity was observed (Supplemental Fig. S5). For promoter analysis, we amplified the miR-191-5p promoter fragments predicted to encompass p53 binding sites. The p53 binding sites upstream of miR-191-5p were cloned upstream of a luciferase gene in PGL3-tk-luciferase vector (Promega). All the clones were confirmed with PCR, restriction digestion, and sequence analysis.
Apoptosis assay
MCF7 cells were transiently transfected with pc-191 or anti-miR-191 (Anti-191) or respective control oligos (pc and NCtrl), and 48 h post-transfection apoptosis was assayed by fluorescence-activated cell sorting (BD Bioscience) using a PE Annexin V Staining Kit (BD Bioscience). The data obtained were analyzed using Cyflogic software. The experiment was repeated three times.
Western blot
MCF7 cells were transfected with miR-191-5p or SOX4 along with their controls and lysed using protein lysis buffer (Tris-50 mM, EDTA-5 mM, NaCl-150 mM and 1% Triton X-100 with 1 mM β-mercaptoethanol and protease inhibitor cocktail) 48 h post-transfection. The protein concentration was determined by using Bradford reagent (Sigma-Aldrich). An equal amount of protein lysates was separated with 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to nitrocellulose membrane (Bio-Rad). The membrane was then probed with a specific primary antibody at a dilution of 1:1000 (p53; Santa Cruz Biotechnology) or 1:3000 (β-actin; Santa Cruz Biotechnology) or 1:1000 (SOX4; Santa Cruz Biotechnology) followed by washing and incubation with the respective secondary antibody (p53 and β-actin–anti-mouse, horseradish peroxidase-linked, Santa Cruz Biotechnology; SOX4-anti-goat horseradish peroxidase-linked). The specific protein band was visualized by autoradiography using an ECL kit (Bio-Rad).
Chromatin immunoprecipitation
MCF7 cells were cultured in 100-mm culture dishes at 90%–100% confluency; then cells were harvested and crosslinked with formaldehyde, washed with phosphate-buffered saline, and resuspended in the RIPA buffer. Sonication was performed to obtain DNA fragment of 100- to 500-bp fragments (sonicated lysate was confirmed for fragment size through 2% agarose gel). For chromatin immunoprecipitation (CHIP), specific antibodies against p53 (Santa Cruz Biotechnology), salmon sperm DNA (Sigma-Aldrich), and protein A-sepharose beads (30 µL) were added to the chromatin extract and incubated overnight, washed and eluted with 0.5% w/v sodium dodecyl sulfate solution. Decrosslinking was done at 65° for 4–5 h and DNA was then purified with a PCR Purification Kit (HiMedia). Rabbit IgG was used as control antibody, whereas the chromatin extract without any antibody/beads treatment was used as positive control. For DNA sequence–specific quantification, qRT-PCR was done with an equal amount of chromatin extract using sequence-specific primers (Supplemental Fig. S5). The experiment was repeated twice.
Cell viability assay
Cell viability was determined by 3-(4,5-dimethylthiazole-2-yl)-2,5-diphenyl tetrazolium bromide (MTT). MCF7 and ZR-75 cells were seeded in a 96-well plate containing 100 μL of culture medium, and then MTT reagent was added at different time points. Subsequently, the plate was incubated for 2 h at 37°C in a humidified incubator containing 5% CO2 and quantified at 595 nm. The experiments were repeated three times.
Caspase-Glo 3/7 assay
Caspase-Glo 3/7 activity was determined by a Caspase-Glo 3/7 Assay Kit (Promega). Cells were seeded in a 96-well plate and transfections were done with miR-191-5p or its control. Forty-eight hours post-transfection, 50 μL of Caspase-Glo 3/7 reagent was added to the medium and the mixture was incubated at room temperature for 2 h, after which the luminescence was analyzed in a luminometer (Berthold).
DAPI staining
MCF7 cells were seeded in a 12-well plate and transfections were done with miR-191-5p or its control. Forty-eight hours post-transfection, cells were fixed with 4% paraformaldehyde for 20 min at room temperature. Fixed cells were washed with 1× PBS and permeabilized with 0.2% Triton X-100 for 10 min and again washed with PBS. Cells were stained with DAPI (1:1000, Sigma-Aldrich) and incubated in the dark for 20 min at room temperature. Cells were washed with PBS and analyzed under a fluorescent microscope.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grant SB/S0/BB-0088/2013 from the Science and Engineering Research Board, Department of Science & Technology, Government of India to R.K. S.S. thanks the Centre for Scientific and Industrial Research for a Senior Research Fellowship.
Author contributions: R.K. conceptualized and supervised the whole study. P.C.G. supervised and helped with the analyses of the FACS data. S.S. performed all the experiments. N.N. made some constructs for the luciferase assays.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.060657.117.
REFERENCES
- Agarwal V, Bell GW, Nam J, Bartel DP. 2015. Predicting effective microRNA target sites in mammalian mRNAs. eLife 4: e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves CL, Elias D, Lyng M, Bak M, Kirkegaard T, Lykkesfeldt AE, Ditzel HJ. 2016. High CDK6 protects cells from fulvestrant-mediated apoptosis and is a predictor of resistance to fulvestrant in estrogen receptor-positive metastatic breast cancer. Clin Cancer Res 22: 5514–5526. [DOI] [PubMed] [Google Scholar]
- American Cancer Society. 2015. Breast Cancer Facts & Figures 2015–2016. American Cancer Society, Atlanta, GA. [Google Scholar]
- Berger C, Qian Y, Chen X. 2013. The p53-estrogen receptor loop in cancer. Curr Mol Med 13: 1229–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Yuan T, Liu M, Chen P. 2013. Association between TCF7L2 gene polymorphism and cancer risk: a meta-analysis. PLoS One 8: e71730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Leva G, Piovan C, Gasparini P, Ngankeu A, Taccioli C, Briskin D, Cheung DG, Bolon B, Anderlucci L, Alder H, et al. 2013. Estrogen mediated-activation of miR-191/425 cluster modulates tumorigenicity of breast cancer cells depending on estrogen receptor status. PLoS Genet 9: e1003311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elyakim E, Sitbon E, Faerman A, Tabak S, Montia E, Belanis L, Dov A, Marcusson EG, Bennett CF, et al. 2010. hsa-miR-191 is a candidate oncogene target for hepatocellular carcinoma therapy. Cancer Res 70: 8077–8087. [DOI] [PubMed] [Google Scholar]
- Farré D, Roset R, Huerta M, Adsuara JE, Roselló L, Albà MM, Messeguer X. 2003. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res 31: 3651–3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Cen Zhang C, Wu R, Hu W. 2011. Tumor suppressor p53 meets microRNAs. J Mol Cell Biol 3: 44–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florea AM, Büsselberg D. 2013. Breast cancer and possible mechanisms of therapy resistance. J Local Glob Health Sci 2 10.5339/jlghs.2013.2 [DOI] [Google Scholar]
- Gonzalez-Angulo AM, Morales-Vasquez F, Hortobagyi GN. 2007. Overview of resistance to systemic therapy in patients with breast cancer. Adv Exp Med Biol 608: 1–22. [DOI] [PubMed] [Google Scholar]
- Grimm SL, Rosen JM. 2003. The role of C/EBPβ in mammary gland development and breast cancer. J Mammary Gland Biol Neoplasia 8: 191–204. [DOI] [PubMed] [Google Scholar]
- He Y, Cui Y, Wang W, Gu J, Guo S, Ma K, Luo X. 2011. Hypomethylation of the hsa-miR-191 locus causes high expression of hsa-mir-191 and promotes the epithelial-to-mesenchymal transition in hepatocellular carcinoma. Neoplasia 13: 841–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoevel T, Macek R, Swisshelm K, Kubbies M. 2004. Reexpression of the TJ protein CLDN1 induces apoptosis in breast tumor spheroids. Int J Cancer 108: 374–383. [DOI] [PubMed] [Google Scholar]
- Hu Y, Xu K, Yagüe E. 2015. miR-218 targets surviving and regulates resistance to chemotherapeutics in breast cancer. Breast Cancer Res Treat 151: 269–280. [DOI] [PubMed] [Google Scholar]
- Hur W, Rhim H, Jung CK, Kim JD, Bae SH, Jang JW, Yang JM, Oh ST, Kim DG, Wang HJ, et al. 2010. SOX4 overexpression regulates the p53-mediated apoptosis in hepatocellular carcinoma: clinical implication and functional analysis in vitro. Carcinogenesis 31: 1298–1307. [DOI] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. 2005. Human MicroRNA targets. PLoS Biol 3: e264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorre E, Tebaldi T, Viero G, Spartà AM, Quattrone A, Provenzani A. 2012. Downregulation of HuR as a new mechanism of doxorubicin resistance in breast cancer cells. Mol Cancer 11: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QQ, Chen ZQ, Xu JD, Cao XX, Chen Q, Liu XP, Xu ZD. 2010. Overexpression and involvement of special AT-rich sequence binding protein 1 in multidrug resistance in human breast carcinoma cells. Cancer Sci 101: 80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XJ, Ji MH, Zhong SL, Zha QB, Xu JJ, Zaoh JH, Tang JH. 2012. MicroRNA-34a modulates chemosensitivity of breast cancer cells to Adriamycin by targeting Notch1. Arch Med Res 43: 514–521. [DOI] [PubMed] [Google Scholar]
- Liu H, Xu XF, Zhao Y, Tang MC, Zhou YQ, Lu J, Gao FH. 2014. MicroRNA-191 promotes pancreatic cancer progression by targeting USP10. Tumor Biol 35: 12157–12163. [DOI] [PubMed] [Google Scholar]
- Messeguer X, Escudero R, Farré D, Núñez O, Martínez J, Albà MM. 2002. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 18: 333–334. [DOI] [PubMed] [Google Scholar]
- Mu H, Wang N, Zhao L, Li S, Li Q, Chen L, Luo X, Qiu Z, Li L, Ren G, et al. 2015. Methylation of PLCD1 and adenovirus-mediated PLCD1 overexpression elicits a gene therapy effect on human breast cancer. Exp Cell Res 332: 179–189. [DOI] [PubMed] [Google Scholar]
- Nagpal N, Ahmad HM, Molparia B, Kulshreshtha R. 2013. MicroRNA-191, an estrogen-responsive microRNA, functions as an oncogenic regulator in human breast cancer. Carcinogenesis 34: 1889–1899. [DOI] [PubMed] [Google Scholar]
- Nagpal N, Ahmad HM, Chameettachal S, Sundar D, Ghosh S, Kulshreshtha R. 2015. HIF-inducible miR-191 promotes migration in breast cancer through complex regulation of TGFβ-signaling in hypoxic microenvironment. Sci Rep 5: 9650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nana-Sinkam SP, Croce CM. 2011. Non-coding RNAs in cancer initiation and progression and as novel biomarkers. Mol Oncol 5: 483–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obacz J, Pastorekova S, Vojtesek B, Hrstka R. 2013. Cross-talk between HIF and p53 as mediators of molecular responses to physiological and genotoxic stresses. Mol Cancer 12: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Zhao J, Zhang WN, Li HY, Mu R, Zhou T, Zhang HY, Gong WL, Yu M, Man JH, et al. 2009. Induction of SOX4 by DNA damage is critical for p53 stabilization and function. Proc Natl Acad Sci 106: 3788–3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patnaik SK, Kannisto E, Yendamuri S. 2010. Overexpression of microRNA miR-30a or miR-191 in A549 lung cancer or BEAS-2B normal lung cell lines does not alter phenotype. PLoS One 5: e9219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raguz S, Yagüe E. 2008. Resistance to chemotherapy: new treatments and novel insights into an old problem. Br J Cancer 99: 387–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Patnaik PK, Aronov S, Kulshreshtha R. 2016. ApoptomiRs of breast cancer: basics to clinics. Front Genet 7: 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Su S, Long J, Mei B, Chen Y. 2011. MicroRNA-191 targets N-deacetylase/N-sulfotransferase 1 and promotes cell growth in human gastric carcinoma cell line MGC803. Acta Biochim Biophys Sin (Shanghai) 43: 849–856. [DOI] [PubMed] [Google Scholar]
- Smith L, Watson MB, O'Kane SL, Drew PJ, Lind MJ, Cawkwell L. 2006. The analysis of doxorubicin resistance in human breast cancer cells using antibody microarrays. Mol Cancer Ther 5: 2115–2120. [DOI] [PubMed] [Google Scholar]
- Song GD, Sun Y, Shen H, Li W. 2015. SOX4 overexpression is a novel biomarker of malignant status and poor prognosis in breast cancer patients. Tumor Biol 36: 4167–4173. [DOI] [PubMed] [Google Scholar]
- Tadesse S, Yu M, Kumarasiri M, Le BT, Wang S. 2015. Targeting CDK6 in cancer: state of the art and new insights. Cell Cycle 14: 3220–3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorn CF, Oshiro C, Marsh S, Hernandez-Boussard T, McLeod H, Klein TE, Altman RB. 2011. Doxorubicin pathways: pharmacodynamics and adverse effect. Pharmacogenet Genomics 21: 440–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhecke E, Adriaenssens E, Verbeke S, Meignan S, Germain E, Berteaux N, Nurcombe V, Le Bourhis X, Hondermarck H. 2011. Brain-derived neurotrophic factor and neurotrophin-4/5 are expressed in breast cancer and can be targeted to inhibit tumor cell survival. Clin Cancer Res 17: 1741–1752. [DOI] [PubMed] [Google Scholar]
- Wang ZX, Lu BB, Wang H, Cheng ZX, Yin YM. 2011. MicroRNA-21modulates chemosensitivity of breast cancer cells to doxorubicin by targeting PTEN. Arch Med Res 42: 281–290. [DOI] [PubMed] [Google Scholar]
- Wynendaele J, Böhnke A, Leucci E, Nielsen JS, Lambertz I, Hammer S, Sbrzesny N, Kubitza D, Wolf A, Gradhand E, et al. 2010. An illegitimate microRNA target site within the 3′ UTR of MDM4 affects ovarian cancer progression and chemosensitivity. Cancer Res 70: 9641–9649. [DOI] [PubMed] [Google Scholar]
- Xi Y, Formentini A, Chien M, Weir DB, Russo JJ, Ju J, Kornmann M, Ju J. 2006. Prognostic values of microRNAs in colorectal cancer. Biomark Insights 2: 113–121. [PMC free article] [PubMed] [Google Scholar]
- Yang X, Martin TA, Jiang WG. 2012. Biological influence of brain-derived neurotrophic factor on breast cancer cells. Int J Oncol 41: 1541–1546. [DOI] [PubMed] [Google Scholar]
- Yu Z, Xu Z, Disante G, Wright J, Wang M, Li Y, Zhao Q, Ren T, Ju X, Gutman E, et al. 2014. miR-17/20 sensitization of breast cancer cells to chemotherapy-induced apoptosis requires Akt1. Oncotarget 5: 1083–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahnow CA. 2009. CCAAT/enhancer-binding protein β: its role in breast cancer and associations with receptor tyrosine kinases. Expert Rev Mol Med 11: e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Zhang X, Li Z, Zhou C, Li M, Tang X, Lu C, Li H, Yuan Q, Yang M. 2013. Association of a genetic variation in a miR-191 binding site in MDM4 with risk of esophageal squamous cell carcinoma. PLoS One 8: e64331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Wang X, Huang Y, Chen Y, Zhao G, Yao Q, Jin C, Huang Y, Liu X, Li G. 2015. Down-regulated SOX4 expression suppresses cell proliferation, metastasis and induces apoptosis in Xuanwei female lung cancer patients. J Cell Biochem 116: 1007–1018. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Wu J, Li S, Ma R, Cao H, Ji M, Jing C, Tang J. 2013. The function role of miR-181a in chemosensitivity to adriamycin by targeting Bcl-2 in low-invasive breast cancer cells. Cell Physiol Biochem 32: 1225–1237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.