Abstract
The hard tick Ixodes ricinus is an important disease vector whose salivary secretions mediate blood-feeding success on vertebrate hosts, including humans. Here we describe the expression profiles and downstream analysis of de novo-discovered microRNAs (miRNAs) expressed in I. ricinus salivary glands and saliva. Eleven tick-derived libraries were sequenced to produce 67,375,557 Illumina reads. De novo prediction yielded 67 bona fide miRNAs out of which 35 are currently not present in miRBase. We report for the first time the presence of microRNAs in tick saliva, obtaining furthermore molecular indicators that those might be of exosomal origin. Ten out of these microRNAs are at least 100 times more represented in saliva. For the four most expressed microRNAs from this subset, we analyzed their combinatorial effects upon their host transcriptome using a novel in silico target network approach. We show that only the inclusion of combinatorial effects reveals the functions in important pathways related to inflammation and pain sensing. A control set of highly abundant microRNAs in both saliva and salivary glands indicates no significant pathways and a far lower number of shared target genes. Therefore, the analysis of miRNAs from pure tick saliva strongly supports the hypothesis that tick saliva miRNAs can modulate vertebrate host homeostasis and represents the first direct evidence of tick miRNA-mediated regulation of vertebrate host gene expression at the tick–host interface. As such, the herein described miRNAs may support future drug discovery and development projects that will also experimentally question their predicted molecular targets in the vertebrate host.
Keywords: tick–vertebrate host interaction, deep-sequencing, microRNA, gene target prediction, interactomes/systems biology, disease biology
INTRODUCTION
Ixodes ticks are important arthropod disease vectors that transmit pathogens causing several human diseases, including Lyme disease and tick-borne encephalitis (for review, see Vayssier-Taussat et al. 2015; Nelder et al. 2016). An interesting and important aspect of their life cycle is their ability to feed on the vertebrate host for several days without being noticed. The secreted tick saliva mediates tick feeding success (Ribeiro 1987) through intrinsic antihemostatic, anti-inflammatory, and immunosuppressive actions on the host (Ribeiro et al. 1985). Many different tick salivary proteins and low-molecular-weight substances (e.g., prostaglandin E2) are known to modulate the host response (for review, see Kazimírová and Štibrániová 2013). Small RNAs, especially microRNAs (miRNAs), play critical pathophysiological roles, but nothing is known about if or how tick miRNAs participate in the tick–host feeding dyad. There are only some restricted data on arthropod miRNAs in arthropod physiology (Lucas and Raikhel 2013; Luhur et al. 2013; He et al. 2015), especially in mosquitoes (for review, see Asgari 2014; Blair and Olson 2015). Knowledge about tick miRNAs is sparse and limited to sequences, potential tissue localization, and evolution in only five tick species (Barrero et al. 2011; Zhou et al. 2013; Luo et al. 2015; Shao et al. 2015; Wang et al. 2015) and to their potential interactions with Flaviviruses (Tsetsarkin et al. 2016). Accordingly, miRNAs have been reported in the literature for the following tick species (alphabetically): Haemaphysalis longicornis (Zhou et al. 2013), Hyalomma anatolicum (Luo et al. 2015), Rhipicephalus haemaphysaloides (Wang et al. 2015), Rhipicephalus (Boophilus) microplus (Barrero et al. 2011), and Rhipicephalus sanguineus (Shao et al. 2015). miRNAs have yet to be described in Ixodes ticks. Ixodes ricinus is an important disease vector in Europe where they transmit viruses, bacteria, and protozoan parasites (Honig et al. 2015; Vayssier-Taussat et al. 2015; Brugger et al. 2016). Difficulties in genome analysis have restricted our knowledge of their miRNAs (Cramaro et al. 2015). Here we performed deep-sequencing analysis of I. ricinus salivary gland and midgut tissues which are critical to tick vectorial capacity. The de novo annotation of miRNAs expressed in these tissues and their temporal expression are described. Further, we present the first data on miRNAs in I. ricinus saliva, which we show possess nontemplated nucleotide modifications characteristic of exosomal secretion. More importantly, in silico target prediction reveals that these salivary miRNAs are likely to modulate host responses to tick attachment and feeding.
RESULTS
Taxonomic and read length distribution
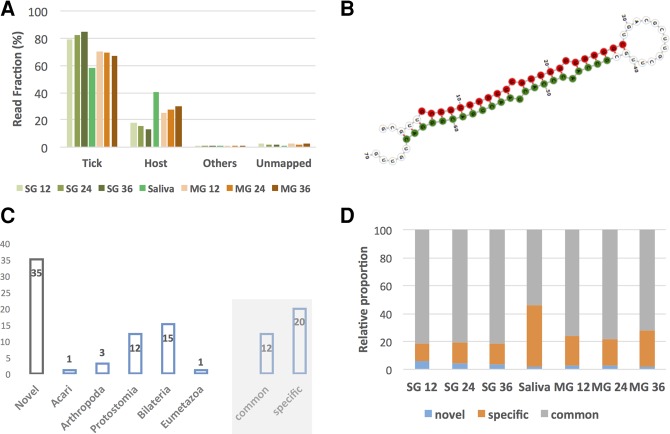
After adapter trimming and removal of short reads (≤15 nt), 67,375,557 reads were available for downstream analysis (Supplemental Table 2), 98.5% of which mapped to at least one genomic sequence. For libraries from adult ticks, most reads were of tick origin independent of tissue type, feeding time point, and developmental stage, followed by reads mapping to a host genome (Fig. 1A). The proportion of tick reads was higher in the salivary glands (between 76.9% and 82.5%) than in the midgut (between 62.7% and 65.5%) and, in the salivary gland (SG), the relative amount of tick reads increased over feeding time and the opposite in the midgut (MG). As expected, the opposite was true for host sequence reads. It should be noted that the MG samples serve as an out group as far as it concerns the study of the potential role of tick miRNAs in the interaction with the vertebrate host, which is the main objective of this study. A similar trend was observed with libraries originating from nymphal ticks (Supplemental Fig. 1A), albeit with less pronounced differences, presumably due to less blood being acquired by nymphs during feeding. A negligible number of reads could be assigned to viruses, bacteria, and endosymbionts.
FIGURE 1.
(A) Novel miRNAs and miRNA families in adult I. ricinus ticks. Most of the genetic material in the sequencing libraries is of tick origin. Sequence read distribution depending on organism of origin as a function of feeding time for adult ticks is shown. The MG represents up to 30% of host reads (at 36 h) while the percentage of host reads is lower in the SG. MG, midgut; SG, salivary glands; 12, 12 h of feeding; 24, 24 h of feeding; 36, 36 h of feeding. (B) Novel miRNA X1b is among the top 20 expressed miRNAs both in tick saliva and SG and a member of a novel miRNA family X1. The secondary structure and mature microRNA is shown (guide strand in red and passenger strand in green). (C) Number of miRNAs per taxonomic node. The gray shaded part shows the number of miRNAs in both the tick and putative hosts (common) and those that are only in the tick (specific). The novel miRNAs have not been used in this classification. Note that the taxonomic node of some miRNAs is bilateria, but the microRNA was lost and is not present in any of the possible hosts. (D) Relative expression levels as a function of miRNA presence (common, specific, and novel) under all experimental conditions.
The read length distribution (Supplemental Fig. 1B,C) provides an important “first look” of the types of small RNAs present in the different samples. The percentages of miRNAs ranged from 10% to 20%. Two main peaks were distinguishable in adult ticks: (i) a peak around 22 nt, constituting mainly mature miRNAs, and (ii) a broader peak between 26 and 30 nt with a clear local maximum at 28 nt of uncertain small RNA composition. The 22-nt peak was slightly higher in the MG samples, while the relative number of reads between 26 and 30 nt was similar in the MG and SG albeit decreasing as a function of feeding time. The relative frequencies decreased as a function of feeding time for the 22-nt peak (mature miRNAs) in the SG but not the MG. The read length distribution in nymphal ticks was qualitatively identical to that seen for adult ticks.
Sequence-based prediction of novel miRNAs
Given that the miRBase database (release 21) currently contains no I. ricinus miRNAs, we generated a miRNA catalog for I. ricinus using sRNAbench and miRanalyzer (Hackenberg et al. 2011) applied to the I. ricinus and I. scapularis genome assemblies (Cramaro et al. 2015; Gulia-Nuss et al. 2016) and the I. ricinus transcriptome (Kotsyfakis et al. 2015a). For all guide miRNAs from I. ricinus and their assigned homologs, we then searched for the last common ancestor using previously derived full lineages. The miRNA was deemed a “novel miRNA” if no homologous miRNAs were present in miRBase, a “common miRNA” if the miRNA was present in I. ricinus and also all putative hosts (the taxonomic node was in bilateria or before), and a “tick-specific” miRNA if it was not present in any putative host, i.e., the node was in protostomia (or more recent) or lost before vertebrates.
Sixty-seven different miRNAs (unique guide strand sequences) were detected, of which 35 were not in miRBase and are reported here for the first time as novel miRNAs. A putative important novel microRNA (iri-mir-X1b), a member of the novel miRNA family X1, was found in the top 20 expressed miRNAs in SG and saliva, so it was selected for further secondary structure presentation (Fig. 1B). This novel family has eight members in I. ricinus with conserved seeds (Supplemental Fig. 2A) in both the guide strand (5p arm, conserved seed length of 13 nt) and the passenger strand (3p arm, conserved seed length of 7 nt). The quality of our novel miRNAs and specifically for iri-mir-X1b was confirmed by the observation of (i) two stacks representing the two mature miRNAs; (ii) little fluctuation at the 5′ end of the guide strand; and (iii) no reads starting or ending within the loop of the hairpin structure (Supplemental Fig. 2B,C). In addition to this large family, a novel family containing two members (mir-X11a and mir-X11b) and two members of the mir-2 family (mir-2b and mir-2b2) were also detected (Supplemental Table 3).
Some of the 67 miRNAs had more than one genomic copy, so a total of 73 precursor sequences are reported. We defined two precursor sequences as copies of the same miRNA gene when they shared the same guide sequence. There were two copies in the genomic sequence for four miRNAs (iri-miR-100, iri-mir-X12, iri-mir-X13, iri-mir-X1c) and three copies for one miRNA (iri-mir-X16). Of the 73 precursor sequences, 35 were detected using the I. ricinus genome assembly, 31 the I. scapularis assembly, and seven in the I. ricinus transcriptome. As most of the detected miRNAs had both arms represented by reads in at least one sample, a total of 133 mature sequences are reported in Supplemental Table 3.
Evolutionary origin of novel miRNAs
As well as the 35 novel miRNAs, we found 16 miRNAs after the split of protostomia and deuterostomia (protostomia [12], arthropoda [three], and acari [one]) (Fig. 1C), i.e., these miRNAs could only belong to ticks but not to any of the possible hosts. Furthermore, 15 miRNAs had taxonomic nodes in bilateria and one in eumetazoa (mir-100, one of the conserved miRNAs present in all animals; Fig. 1C). Note that the taxonomic node of these miRNAs is before the last common ancestor of ticks and all putative hosts. However, as discussed below, not all of these 16 miRNAs were common to both ticks and hosts, as some were lost prior to the appearance of vertebrates. Specifically, 4/16 miRNAs (miR-2001-5p, miR-252b-5p, miR-278-3p, miR-981-3p; labeled bilateria and “specific”) were present in some deuterostomia, but seem to have been lost before vertebrates; they were therefore not present in any of the possible hosts.
Therefore, 20 specific miRNAs (four bilateria lost before vertebrates, 12 protostomia, three arthropoda, and one acari) and 12 common miRNAs (16 bilateria minus the four lost before vertebrates and being found in ticks and their possible hosts) were discovered. Note that the phylogenetic distribution of the 35 novel miRNAs was not considered since the range of species to which they belong could not be assessed due to their novelty. The 12 “common” miRNAs (present both in ticks and hosts) comprised 75.2% and 81.1% of total miRNA sequence reads in MG and SG, respectively (Fig. 1D). However, in saliva, the tick-specific miRNAs accumulated more reads (44.4% of total), prompting us to investigate them further.
miRNA expression analysis
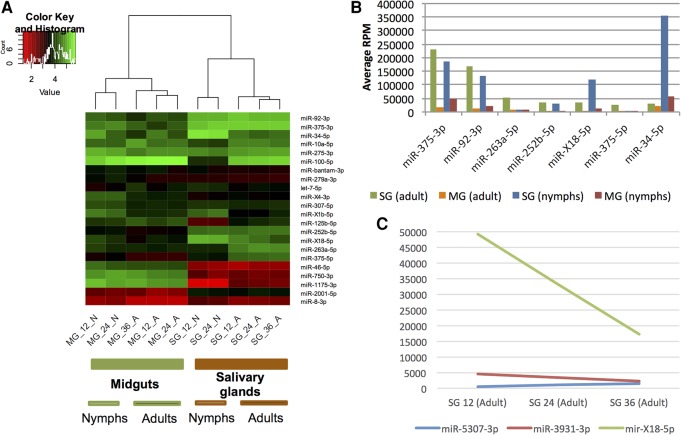
After mapping the reads to the predicted tick miRNAs, we generated a read count matrix for the miRNAs using sRNAbench (RPM-normalized values are presented in Supplemental Table 4). After normalizing to RPM expression values, highly expressed miRNAs constituting 95% of total miRNA reads were extracted and the RPM values were log10 transformed. There were two clearly separated clusters (Fig. 2A), one corresponding to the MG and one corresponding to the SG, indicating distinct miRNA expression profiles in the two tissues. Furthermore, the clusters clearly subdivided into nymph and adult samples. The miRNA expression profiles were indicative both of tissue and developmental stages, as detailed below.
FIGURE 2.
Expression analysis based on read counts for each miRNA. (A) Cluster analysis of the six adult and four nymphal samples from tick salivary glands and midgut. (B) Overexpressed miRNAs in the salivary gland compared to the midgut in both adults and nymphs. All miRNAs are over eightfold overexpressed with RPM values greater than 10,000. (C) Putative feeding-regulated miRNAs in adult tick salivary glands. MG, midgut; SG, salivary glands; 12, 12 h of feeding; 24, 24 h of feeding; 36, 36 h of feeding.
Salivary gland-regulated miRNAs in adults and nymphs
The adult and nymph read count expression matrices were used as an input for edgeR to calculate the miRNAs showing statistically significant differential expression (Supplemental Table 5 [adult ticks] and Supplemental Table 6 [nymphal ticks]). Data are represented as volcano plots (Supplemental Figs. 3A,B). A large number of miRNAs showed large and significant changes in both adult and nymphal ticks, and all miRNAs with corrected P-values (FDR) ≤ 0.05 were considered differentially expressed. SG- and MG-overexpressed miRNAs were next examined separately for adults and nymphs (Venn diagram, Supplemental Fig. 3C). Sixteen microRNAs were up-regulated in SG in both adults (28 up-regulated in total) and nymphs (23 up-regulated in total); that is, 57% (16/28) of all SG up-regulated miRNAs in adults were also up-regulated in nymphs, and 70% (16/23) of up-regulated miRNAs in nymphs were up-regulated in adults.
Regulation as a function of the developmental stage in salivary glands
Thirty-one miRNAs were overexpressed in adult SGs and seven in nymphal SGs (Supplemental Table 7). Of the differentially expressed miRNAs, four of those up-regulated in adult SGs had RPM values greater than 10,000, while only one miRNA was overexpressed in nymph SG (Supplemental Fig. 3D). Two highly expressed miRNAs showed particularly large changes in expression: miR-100-5p was over 60-fold higher in adults, while miR-34-5p is the most expressed miRNA in nymphal SG (RPM 353,965), ranking only ninth in adult SG (RPM 29,789; 12-fold higher in nymphs).
There was a large number of strongly regulated miRNAs in SG and MG with both high fold-changes and high expression. Expression levels are related to function (Mullokandov et al. 2012). miRNAs at least eightfold overexpressed with RPM values >10,000 RPM are shown in Figure 2B. Six miRNAs were at least eightfold overexpressed in SG compared to MG in adults, while five miRNAs were at least eightfold overexpressed in nymphs. However, four miRNAs (iri-miR-375-3p, iri-miR-92-3p, iri-mir-X18-5p, and iri-miR-252b-5p) were overexpressed in both adults and nymphs, two were “adult specific” (iri-miR-263a-5p, iri-miR-375-5p), and one was “nymph specific” (iri-miR-34-5p). Of these tick-specific miRNAs, iri-mir-X18-5p was a novel miRNA, iri-miR-252b-5p was only found in invertebrates but not in putative hosts, and one (iri-mir-375) had a highly conserved guide sequence (iri-miR-375-3p) but I. ricinus-specific passenger strand (iri-miR-375-5p).
Next, three putative feeding-regulated miRNAs were identified according to (i) at least twofold different RPM expression values at 12 and 36 h, and (ii) the 24 h RPM value lying in-between (Fig. 2C): iri-miR-5307-3p, the expression of which increased as feeding progressed; and miR-X18-5p and miR-3931-3p, which decreased. miR-X18-5p in particular seemed to be a good candidate for further analysis due to its high RPM values of nearly 50,000 at 12 h and 17,000 at 36 h. Remarkably, none of the feeding-regulated miRNAs were common, i.e., present in the tick and host: miR-X18-5p was novel (not in miRBase), miR-3931-3p was present in I. scapularis and the red spider mite Tetranychus urticae, and iri-miR-5307-3p was already described in I. scapularis and in the red flour beetle Tribolium castaneum in which it is called tca-mir-3477. The first 15 nt are identical between iri-miR-5307-3p and tca-mir-3477 and therefore they belong to the same family and should have the same name. We adopted miR-5307 as the name, given that this is the name of the microRNA in another tick—I. scapularis.
Saliva expression profiles
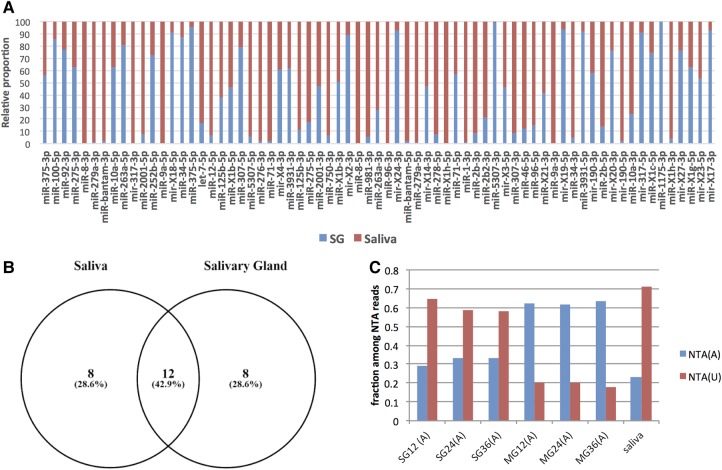
We next sought to establish (i) whether the read counts in pure tick saliva resembled those of the salivary glands; and (ii) the origin of the detected miRNAs. The vast majority of salivary gland miRNAs were also detected in saliva (Fig. 3A). Furthermore, among the top 20 expressed miRNAs from saliva and salivary glands, 12 were shared (Fig. 3B).
FIGURE 3.
Sequence reads from tick miRNAs were detected in pure tick saliva. (A) The relative proportion of RPM expression values between SG (blue) and saliva (red) for the top 70 expressed miRNAs. (B) Overlap between the top 20 expressed microRNAs in saliva and SG: 12 miRNAs are common to both SG and saliva. (C) Fraction of A and U nontemplated additions (NTAs) for SG, MG, and salivary samples. NTA(A), adenylation; NTA(U), uridylation. There are marked differences between SG and MG. Salivary miRNAs show an even higher proportion of uridylated reads compared to SG. MG, midgut; SG, salivary glands; 12, 12 h of feeding; 24, 24 h of feeding; 36, 36 h of feeding. (A) On the x-axis: adult ticks.
In general, miRNA read counts in saliva were more equally distributed (16 miRNAs with RPM values ≥10,000). In contrast, only 10 miRNAs in the SG had such high RPM values (see Supplemental Fig. 4). Interestingly, some of the highest expressed miRNAs in saliva were virtually absent in the salivary glands: miR-8-3p (RPM in saliva: 129,040) was 648-fold overrepresented in saliva, miR-bantam-3p (RPM in saliva: 87,987) was 45-fold overrepresented; miR- 317-3p (RPM in saliva: 59,401) was 249-fold overrepresented, and miR-279a-3p (RPM in saliva: 116,978) was 107-fold overrepresented in saliva compared to SG (Fig. 3A).
Moreover, it has been reported that exosome-secreted miRNAs show more uridylation than adenylation events at the 3′ ends of the mature sequences (Koppers-Lalic et al. 2014). The fraction of uridylation events was clearly more frequent in SG than in the midgut, with uridylation and adenylation the most frequent addition events (Fig. 3C). Most importantly, uridylation was even more frequent in saliva than in the SG, as expected for miRNAs of exosomal origin. Among the most abundant microRNAs in saliva, uridylation events are more frequent than adenylation (Supplemental Fig. 5A), and the percentage of uridylation decreases when the abundance decreases in saliva (Supplemental Fig. 5B).
In silico target analysis
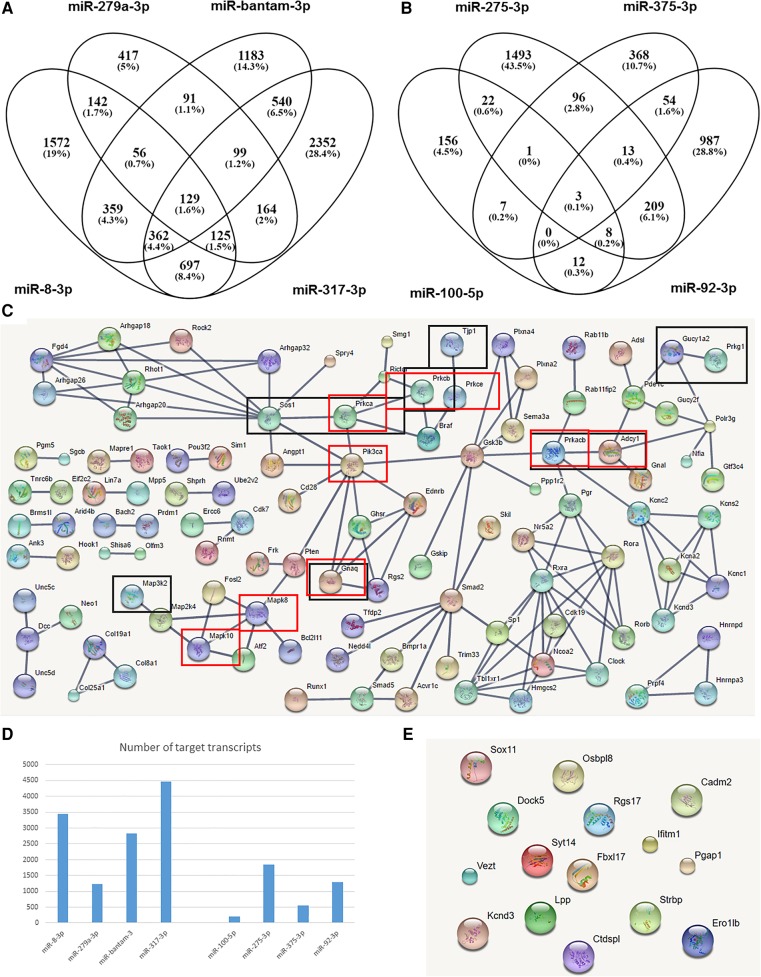
Given the sequence read differences for some miRNAs between SG and saliva, we next analyzed the putative functions of saliva-overexpressed miRNAs, namely miR-8-3p, miR-bantam-3p, mir-317-3p, and miR-279a-3p (together 39.3% of total miRNA expression in saliva; for expression in other samples, see Supplemental Fig. 6). The four most frequent miRNAs from saliva equally or slightly overrepresented in salivary glands were used as control: miR-375-3p, miR-100-5p, miR-92-3p, and miR-275-3p (32.3% of the total miRNA expression in saliva). All eight miRNAs were in the top 10 expressed miRNAs in saliva.
We first defined the putative host gene targets of all the tick miRNAs as those predicted by all three tested gene target prediction programs. The overlap in target prediction of the overrepresented miRNAs in saliva is shown in Figure 4A and the saliva control set in Figure 4B. There were considerable differences between the two sets of target transcripts. First, the overall number of putative targets was much lower in the control set (Fig. 4D), which is surprising as, with the exception of miR-275-3p, the other three miRNAs in the control set are common, evolutionarily conserved miRNAs, i.e., they are endogenous to the mouse genome. Conversely, all four tested saliva-overrepresented miRNAs should be considered tick-specific since miR-bantam-3p, mir-317-3p, and miR-279a-3p were not present in any of the putative host species, while miR-8-3p shared a seed sequence with only some miR-200 members present in some hosts. Regardless, the tick-specific miRNAs, in this case saliva-overrepresented miRNAs, had more predicted target genes in the host than those tick miRNAs with homologous sequences in the host genome.
FIGURE 4.
The four saliva-overrepresented miRNAs are predicted to have many more target genes in the host than the control group. (A,B) Venn diagrams that depict the degree of overlap in target host transcripts among the saliva-overrepresented miRNAs (A) and the control microRNAs (B). (C) A network built exclusively from host proteins targeted by saliva-overrepresented miRNAs. (Black boxes) Host genes from the “gap junction” KEGG pathway; (red boxes) genes from the “Inflammatory mediator regulation of TRP channels” KEGG pathway. (D) The total number of target transcripts for the eight miRNAs (the four saliva-overrepresented miRNAs on the left and the four control miRNAs on the right). (E) Target proteins for the four control miRNAs (no network formed).
To reduce the number of false-positive predictions, we next considered only host transcripts targeted by at least three of the four tested tick miRNAs. A clear surprise was that there were no significant KEGG pathways for the control salivary gland miRNAs (Fig. 4E). In contrast, for saliva-overrepresented miRNAs, 36 KEGG pathways were enriched for target genes (proteins) (Supplemental Table 8). Furthermore, the 460 target host proteins showed 147 physical interactions (expected value 43; P-value 0), even considering only interactions with the highest confidence (interaction scores >0.9 versus algorithm default of 0.4). All proteins targeted by at least three of four saliva-overrepresented miRNAs (predicted by all three target prediction programs) and that interacted physically with at least one other target protein with very high confidence are shown in Figure 4C. Many of the detected KEGG pathways were highly relevant to the tick–host interaction, such as “gap junction” and “inflammatory mediator regulation of TRP channels” (10 and nine target proteins, respectively). Note that of the 460 predicted target host proteins, 109 showed physical interactions with other target proteins (Fig. 4C) and all of the 10 gap junction and nine inflammatory mediator regulation of TRP channel proteins (P ≤ 8 × 10−7 and P ≤ 3.2 × 10−6, respectively).
DISCUSSION
Over half a century has passed since the first description of arthropod saliva as a major mediator of disease–vector life cycle success (Griffiths and Gordon 1952; Hudson et al. 1960). Prostaglandin was identified over 40 years ago as the first nonprotein constituent of tick saliva (Dickinson et al. 1976), and it was soon found to modulate host responses (Ribeiro et al. 1985). In 1988, prostacyclin was shown to be a nonprotein constituent of tick saliva related to host vasodilation (Ribeiro et al. 1988), and in 2003 endocannabinoids and related fatty acid amides in tick saliva were reported to have analgesic and anti-inflammatory activities in the vertebrate host (Fezza et al. 2003). More recently, the molecular mechanisms of host modulation by tick salivary prostaglandin E2 and the purine nucleoside adenosine have been elucidated (Sá-Nunes et al. 2007; Oliveira et al. 2011). Here we present the first high-throughput analysis of the miRNA repertoire in I. ricinus (or any other disease vector species) saliva. We also report for the first time that miRNAs represent another class of nonprotein molecules at the tick–host interface by reporting their detection through direct sequencing from pure tick saliva.
Our in silico downstream analysis shows that the saliva-specific microRNAs (those that are far more abundant in saliva compared to salivary glands) show combinatorial effects on the host targetome, i.e., many target genes in the same host pathway are regulated by more than one saliva-specific microRNA. This behavior cannot be observed in the control set of microRNAs. These combinatorial effects of pathogen microRNAs on host target genes that we report here for the first time, may be of importance in evolutionary terms to maintain a stable and secured regulation of certain host genes and pathways that apparently are important in tick–host interaction; single targets on the vertebrate host may get lost rapidly because the tick microRNA target sites on the vertebrate host are not protected by negative selection. Accordingly, from a tick's perspective, only combinatorial effects on the action of tick saliva microRNAs and redundant target genes on the host can guarantee evolutionary robustness of the tick feeding process. In addition, our in silico analysis suggests that most saliva-specific microRNAs are not endogenously present in the vertebrate host and can therefore regulate a completely different set of host genes, which are not under the control of host microRNAs in physiological conditions. Finally, our analysis of nontemplated nucleotide additions found in saliva versus those in salivary glands and in the midgut suggest that they are secreted in exosomes (Koppers-Lalic et al. 2014). Functional validation is now required to characterize the miRNA secretion mechanism and their exact gene targets in the vertebrate host. These future experiments should include a detailed analysis of each individual salivary miRNA and their molecular targets and mechanism(s) of action in the vertebrate host.
Research has hitherto focused on tick salivary proteins, not small RNA species. Indeed, thousands of proteins are predicted or experimentally confirmed as being present in tick saliva (Mudenda et al. 2014; Schwarz et al. 2014; Tirloni et al. 2014, 2015), and the functional analysis of a long list of tick recombinant proteins with respect to their host modulatory activity is elegantly reviewed elsewhere (Kazimírová and Štibrániová 2013; Wikel 2013). An emerging paradigm is that tick salivary proteins show pluripotency and redundancy in their action on the vertebrate host (Chmelař et al. 2016), i.e., the same tick salivary protein can target more than one host homeostatic mechanism, and the same host homeostatic mechanism may be targeted by more than one tick salivary protein. Our analysis strongly suggests that tick salivary miRNAs act in the same way, since each one is predicted to target more than one vertebrate host biological pathway and each pathway of the host is predicted to be targeted by more than one tick salivary miRNA. However, tick miRNAs are nonproteinaceous so no host immune responses can be directed against them.
Some of the—predicted as—targeted host KEGG pathways such as “gap junction” and “inflammatory mediator regulation of TRP channels” play a role in host homeostatic response. Gap junctions are intercellular channels made of connexin proteins, mediating both electrical and biochemical signals between cells. Damage to peripheral nerves or the spinal cord is often accompanied by neuropathic pain, which is a complex, chronic pain state (Jeon and Youn 2015). The ability of gap junction proteins to regulate immune responses, cell proliferation, migration, apoptosis, and carcinogenesis makes them attractive therapeutic targets for treating inflammatory and neoplastic disorders in different organ systems (Wong et al. 2017). TRP channels can be modulated indirectly by inflammatory mediators such as PGE2, bradykinin, ATP, NGF, and proinflammatory cytokines that are generated during tissue injury (Vay et al. 2012), and they play a role in neuropathic pain (Marwaha et al. 2016). Our systems-based approach will support many future research projects that will aim to characterize the molecular mechanisms mediating the interaction between the vertebrate host and the tick disease vector. These future studies should establish how saliva-specific tick miRNAs modulate host responses to support the long-lasting hematophagy of ticks on the vertebrate host; but it is equally important to characterize these novel small molecules as far as it concerns their interference with miRNA-regulated pathways of the vertebrate host for drug development and clinical application.
MATERIALS AND METHODS
Ethics statement
All animal experiments were carried out in accordance with the Animal Protection Law of the Czech Republic (§17, 246/1992Sb) and with the approval of the Akademie Ved Ceské Republiky Ethics Committee (approval 161/2010).
Ticks, tissue dissection, total RNA isolation, and sequencing
The same starting material (total RNA) as described in Kotsyfakis et al. (2015b) was used. Briefly, 1080 nymphs and 420 adult females and males were attached to experimental animals for feeding. Total RNA was extracted from pooled tissue dissected from female adult ticks and nymphal ticks feeding for 3 h periods up to 24 h (nymphs) or 36 h (adults) to produce 10 samples: four samples at 0–12 h and 12–24 h for nymphal salivary glands (SG) and midguts (MG), respectively, and six samples at 0–12 h, 12–24 h, and 24–36 h for adult SG and MG, respectively. Tick saliva was produced as described in Horká et al. (2009), and ticks were fed on rabbits for 6 d. It is technically impossible to produce a sufficient amount of pure tick saliva (for direct total RNA extraction from pure saliva) from ticks fed on animals for up to 36 h.
All the samples were prepared and sequenced using a 36-bp single-end standard sequencing protocol on a Genome Analyzer IIx (Illumina) following the manufacturer's instructions. The sequencing libraries were prepared according to the TrueSeq Small RNA Sample (Illumina) instructions. Clusters were generated using the Illumina cluster station. Fluorescent images were processed to sequences using Illumina Genome Analyzer Pipeline Analysis software v1.8. All sequences were submitted to the NCBI, and the corresponding Object IDs and URLs can be found in Supplemental File 1.
Bioinformatics
After an initial sequencing quality control step in FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc), preprocessing, mapping, and annotation were mainly conducted in sRNAbench (Barturen et al. 2014) with customized scripts as necessary. Briefly, the obtained sequence reads were 36 nt in length, but many small RNAs are between 27 and 33 nt. When forcing the detection of at least 10 nt of adapter as a typically used minimum length, only RNA molecules of up to 26 nt can be resolved (read length plus minimum adapter length). To detect all RNAs <36 nt, we implemented iterative adapter detection and trimming. First, the adapter was detected in the whole read, and, if not found, it was then searched using iteratively shorter minimum adapter lengths at the 3′ end. After adapter trimming, the reads were collapsed into unique reads followed by read count assignment, i.e., counting the number of times that each unique read was sequenced.
Unique reads were then aligned simultaneously to 99 different genome assemblies or sequences representing all species that might have contributed to our samples. Sequences included ticks (I. ricinus and I. scapularis), several putative host species, tick-borne pathogens, and putative tick endosymbionts, viruses, and other bacteria found in the I. ricinus microbiome (see Supplemental Table 1). Read alignment was conducted in Bowtie (Langmead et al. 2009), allowing one mismatch within the 20-nt seed region.
Based on the alignments, our goal was to assign each read to its putative species of origin. We defined the “best mapping” as the alignments with the lowest number of mismatches. If several mappings had the same number of mismatches within the seed, we attempted to disambiguate using the “longest alignment maintaining the number of observed mismatches within the seed” as a second criterion as described in Hackenberg et al. (2011). Even so, the assigned reads sometimes best mapped to several species, so we first labeled all reads that mapped to one of the two tick genomes (I. scapularis and I. ricinus) (Cramaro et al. 2015; Gulia-Nuss et al. 2016) or the available I. ricinus transcriptome assemblies (Kotsyfakis et al. 2015a) as of tick origin. To assign reads to other organisms, we only allowed reads that mapped uniquely to another category such as host, microbiome, parasites, endosymbionts, and viruses. After assigning all reads to putative species of origin, we classified them into different RNA categories using the following annotations: tRNAs predicted in I. ricinus using Aragorn (Laslett and Canback 2004); I. ricinus transcriptome (Kotsyfakis et al. 2015a); tRNAs predicted in I. scapularis and the noncoding RNA and gene transcript annotations for I. scapularis, Pediculus humanus, Rhodnius prolixux, Anopheles gambiae, Aedes aegypti, and Culex quinquefasciatus downloaded from VectorBase (Giraldo-Calderón et al. 2015); all eukaryotic tRNAs from a genomic tRNA database (Chan and Lowe 2016); RNA sequences and categories from RNAcentral (The RNAcentral Consortium 2015); and repetitive and transposon sequences from RepBase (Bao et al. 2015).
Novel miRNAs
Given that I. ricinus miRNA annotations do not exist, we used sRNAbench and miRanalyzer (Hackenberg et al. 2011) to de novo predict I. ricinus miRNAs using our merged sequence reads. We followed this approach because, given the large number of experimental conditions, it would be nearly impossible to first predict the miRNAs in each individual sample and then merge or unify the results in a second step. Accordingly, in the first step, reads were pooled from all experimental samples, and then only high-confidence predictions/annotations in the complete sequence data set were accepted. In a second annotation round, we successively used all 10 samples for miRNA prediction, removing in each round the previously predicted miRNAs so that they were not detected again.
Novel miRNAs were also predicted based on the I. scapularis genome; however, in some cases, the I. scapularis pre-miRNA sequence was not identical to the I. ricinus sequence, given the separate evolution from their last common ancestor. However, we determined the correct I. ricinus mature miRNA sequence as the one determined by our sequence reads. Therefore, in cases of sequence variation (mismatches) in the mature sequence between I. scapularis and I. ricinus, we “corrected” the I. scapularis-derived pre-miRNA sequence. In practice, this meant that for some novel miRNAs predicted on the I. scapularis genome, the pre-miRNA sequences were hybrids (part of the mature miRNA sequence was always the I. ricinus sequence but the remaining pre-miRNA sequence [not covered by our sequence reads] corresponded to I. scapularis). Secondary novel RNA structures were predicted using RNAfold and illustrated using the “forna” web server in the same ViennaRNA package (Lorenz et al. 2011).
miRNA families and taxonomy
We assigned each novel miRNA to the taxonomic node of its first appearance (based on phylogram analysis). We first downloaded the NCBI taxonomy database to generate the full lineage for each species and then assigned all predicted miRNAs to putatively homologous miRNAs in miRBase if (i) they had the same seed sequences defined as the sequences from nucleotides 2–8; (ii) the overall alignment had fewer than four mismatches. Note that for all novel miRNAs, both arms were supported by reads, so we defined the functional arm (guide strand) as the one with a higher number of homologous miRNAs in miRBase (more conserved sequence). Also note that for most miRNAs the guide and passenger strand were very different.
Differential expression and normalization
Our experimental design resulted in several possible comparisons: (i) SG versus MG in adults, (ii) SG versus MG in nymphs, and (iii) adults versus nymphs in SG. We used edgeR to determine differential expression between conditions (Robinson et al. 2010). Note that in these comparisons, the different feeding time points are treated as biological replicates. Briefly, using sRNAbench's differential expression module, we generated an expression matrix with the raw read counts for input into edgeR to obtain differential miRNA expression. edgeR normalizes the data using the trimmed mean of M-values (TMM) method. We also generated an expression matrix with reads per million (RPM)-normalized expression values using the “single assignment” procedure in sRNAbench. As a result, each read mapping multiple times was only assigned once to the miRNA with the highest expression and only affected reads mapping to several different reference sequences, i.e., normally miRNA sequences from the same family. The RPM values were obtained by dividing the read count of a given miRNA by the total number of reads mapped to the miRNA library. Finally, feeding-regulated miRNAs were defined as (i) at least twofold different RPM expression values at 12 and 36 h; and (ii) a 24-h RPM value lying in-between.
Cluster analysis
Hierarchical clustering was performed using log10 transformed RPM-normalized expression matrices as input data. The plots were generated with the heatmap.2 function of the “gplots” R package (https://cran.r-project.org/web/packages/gplots/gplots.pdf) maintaining the default Euclidian distance metric and average linkage clustering.
Target prediction of tick small RNAs and functional in silico analysis
To predict the host genes regulated by tick miRNAs, we applied TargetSpy (Sturm et al. 2010), MIRANDA (John et al. 2004), and PITA (Kertesz et al. 2007) implemented in the miRNAconsTarget program from sRNAtoolbox (Rueda et al. 2015). Only mRNA/miRNA interactions predicted by all three programs were further used. miRNA target prediction normally yields a high number of false positives, but cross-species comparisons and combinatorial effects can decrease this number (Min and Yoon 2010). It cannot be assumed that tick miRNA target sites are conserved in the host (due to tick–host adaptations), so we applied only combinatorial effects to increase target prediction quality. As a result, we analyzed the effect of sets of four miRNAs, considering only those transcripts targeted by at least three out of four of these miRNAs. Lists of target host genes were functionally characterized using the STRING web server (Franceschini et al. 2013). To comply with the input format, mouse transcript names were first converted to protein names with the “Retrieve/ID mapping” tool (UniProt consortium) (Apweiler et al. 2004). The networks of target genes and the KEGG pathways significantly enriched for target genes were extracted using the STRING output.
DATA DEPOSITION
All sequences were submitted to the NCBI (GenBank accession numbers MF061606-MF061678) and the corresponding object IDs and URLs can be found in Supplemental File 1.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Professor Jose M.C. Ribeiro for assistance in the collection of Ixodes ricinus transcriptome sequences and Nextgenediting (www.nextgenediting.com) for editorial assistance. We also thank the anonymous reviewers for their constructive comments. This work was supported by the intramural institutional support of the Institute of Parasitology, Biology Center of the Czech Academy of Sciences (RVO 60077344), and the Spanish Government (AGL2013-49090-C2-2-R).
Author contributions: M.H. and M.K. conceived and designed the study. A.S. and J.E. performed experiments. M.H., D.L., and M.K. analyzed data. M.H. and M.K. wrote the manuscript.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.061168.117.
REFERENCES
- Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, et al. 2004. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res 32: D115–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asgari S. 2014. Role of microRNAs in arbovirus/vector interactions. Viruses 6: 3514–3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W, Kojima KK, Kohany O. 2015. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA 6: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero RA, Keeble-Gagnere G, Zhang B, Moolhuijzen P, Ikeo K, Tateno Y, Gojobori T, Guerrero FD, Lew-Tabor A, Bellgard M. 2011. Evolutionary conserved microRNAs are ubiquitously expressed compared to tick-specific miRNAs in the cattle tick Rhipicephalus (Boophilus) microplus. BMC Genomics 12: 328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barturen G, Rueda A, Hamberg M, Alganza A, Lebron R, Kotsyfakis M, Shi BJ, Koppers-Lalic D, Hackenberg M. 2014. sRNAbench: profiling of small RNAs and its sequence variants in single or multi-species high-throughput experiments. Methods Next Generation Seq 1: 21–31. [Google Scholar]
- Blair CD, Olson KE. 2015. The role of RNA interference (RNAi) in arbovirus-vector interactions. Viruses 7: 820–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugger K, Boehnke D, Petney T, Dobler G, Pfeffer M, Silaghi C, Schaub GA, Pinior B, Dautel H, Kahl O, et al. 2016. A density map of the tick-borne encephalitis and Lyme Borreliosis vector Ixodes ricinus (Acari: Ixodidae) for Germany. J Med Entomol 53: 1292–1302. [DOI] [PubMed] [Google Scholar]
- Chan PP, Lowe TM. 2016. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res 44: D184–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmelař J, Kotál J, Kopecký J, Pedra JH, Kotsyfakis M. 2016. All for one and one for all on the tick-host battlefield. Trends Parasitol 32: 368–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramaro WJ, Revets D, Hunewald OE, Sinner R, Reye AL, Muller CP. 2015. Integration of Ixodes ricinus genome sequencing with transcriptome and proteome annotation of the naive midgut. BMC Genomics 16: 871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson RG, O'Hagan JE, Schotz M, Binnington KC, Hegarty MP. 1976. Prostaglandin in the saliva of the cattle tick Boophilus microplus. Aust J Exp Biol Med Sci 54: 475–486. [DOI] [PubMed] [Google Scholar]
- Fezza F, Dillwith JW, Bisogno T, Tucker JS, Di Marzo V, Sauer JR. 2003. Endocannabinoids and related fatty acid amides, and their regulation, in the salivary glands of the lone star tick. Biochim Biophys Acta 1633: 61–67. [DOI] [PubMed] [Google Scholar]
- Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, et al. 2013. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res 41: D808–D815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo-Calderón GI, Emrich SJ, MacCallum RM, Maslen G, Dialynas E, Topalis P, Ho N, Gesing S, the VectorBase Consortium, Madey G, et al. 2015. VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res 43: D707–D713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths RB, Gordon RM. 1952. An apparatus which enables the process of feeding by mosquitoes to be observed in the tissues of a live rodent; together with an account of the ejection of saliva and its significance in Malaria. Ann Trop Med Parasitol 46: 311–319. [DOI] [PubMed] [Google Scholar]
- Gulia-Nuss M, Nuss AB, Meyer JM, Sonenshine DE, Roe RM, Waterhouse RM, Sattelle DB, de la Fuente J, Ribeiro JM, Megy K, et al. 2016. Genomic insights into the Ixodes scapularis tick vector of Lyme disease. Nat Commun 7: 10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackenberg M, Rodríguez-Ezpeleta N, Aransay AM. 2011. miRanalyzer: an update on the detection and analysis of microRNAs in high-throughput sequencing experiments. Nucleic Acids Res 39: W132–W138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Ju C, Zhang X. 2015. Roles of small RNAs in the immune defense mechanisms of crustaceans. Mol Immunol 68: 399–403. [DOI] [PubMed] [Google Scholar]
- Honig V, Svec P, Halas P, Vavruskova Z, Tykalova H, Kilian P, Vetiskova V, Dornakova V, Sterbova J, Simonova Z, et al. 2015. Ticks and tick-borne pathogens in South Bohemia (Czech Republic)—spatial variability in Ixodes ricinus abundance, Borrelia burgdorferi and tick-borne encephalitis virus prevalence. Ticks Tick Borne Dis 6: 559–567. [DOI] [PubMed] [Google Scholar]
- Horká H, Cerná-Kýcková K, Skallová A, Kopecký J. 2009. Tick saliva affects both proliferation and distribution of Borrelia burgdorferi spirochetes in mouse organs and increases transmission of spirochetes to ticks. Int J Med Microbiol 299: 373–380. [DOI] [PubMed] [Google Scholar]
- Hudson A, Bowman L, Orr CW. 1960. Effects of absence of saliva on blood feeding by mosquitoes. Science 131: 1730–1731. [DOI] [PubMed] [Google Scholar]
- Jeon YH, Youn DH. 2015. Spinal gap junction channels in neuropathic pain. Korean J Pain 28: 231–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. 2004. Human MicroRNA targets. PLoS Biol 2: e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazimírová M, Štibrániová I. 2013. Tick salivary compounds: their role in modulation of host defences and pathogen transmission. Front Cell Infect Microbiol 3: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. 2007. The role of site accessibility in microRNA target recognition. Nat Genet 39: 1278–1284. [DOI] [PubMed] [Google Scholar]
- Koppers-Lalic D, Hackenberg M, Bijnsdorp IV, van Eijndhoven MA, Sadek P, Sie D, Zini N, Middeldorp JM, Ylstra B, de Menezes RX, et al. 2014. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep 8: 1649–1658. [DOI] [PubMed] [Google Scholar]
- Kotsyfakis M, Kopáček P, Franta Z, Pedra JH, Ribeiro JM. 2015a. Deep sequencing analysis of the Ixodes ricinus haemocytome. PLoS Negl Trop Dis 9: e0003754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotsyfakis M, Schwarz A, Erhart J, Ribeiro JM. 2015b. Tissue- and time-dependent transcription in Ixodes ricinus salivary glands and midguts when blood feeding on the vertebrate host. Sci Rep 5: 9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res 32: 11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R, Bernhart SH, Höner Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. 2011. ViennaRNA Package 2.0. Algorithms Mol Biol 6: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas K, Raikhel AS. 2013. Insect microRNAs: biogenesis, expression profiling and biological functions. Insect Biochem Mol Biol 43: 24–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luhur A, Chawla G, Sokol NS. 2013. MicroRNAs as components of systemic signaling pathways in Drosophila melanogaster. Curr Top Dev Biol 105: 97–123. [DOI] [PubMed] [Google Scholar]
- Luo J, Liu GY, Chen Z, Ren QY, Yin H, Luo JX, Wang H. 2015. Identification and characterization of microRNAs by deep-sequencing in Hyalomma anatolicum anatolicum (Acari: Ixodidae) ticks. Gene 564: 125–133. [DOI] [PubMed] [Google Scholar]
- Marwaha L, Bansal Y, Singh R, Saroj P, Bhandari R, Kuhad A. 2016. TRP channels: potential drug target for neuropathic pain. Inflammopharmacology 24: 305–317. [DOI] [PubMed] [Google Scholar]
- Min H, Yoon S. 2010. Got target? Computational methods for microRNA target prediction and their extension. Exp Mol Med 42: 233–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudenda L, Pierlé SA, Turse JE, Scoles GA, Purvine SO, Nicora CD, Clauss TR, Ueti MW, Brown WC, Brayton KA. 2014. Proteomics informed by transcriptomics identifies novel secreted proteins in Dermacentor andersoni saliva. Int J Parasitol 44: 1029–1037. [DOI] [PubMed] [Google Scholar]
- Mullokandov G, Baccarini A, Ruzo A, Jayaprakash AD, Tung N, Israelow B, Evans MJ, Sachidanandam R, Brown BD. 2012. High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nat Methods 9: 840–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelder MP, Russell CB, Sheehan NJ, Sander B, Moore S, Li Y, Johnson S, Patel SN, Sider D. 2016. Human pathogens associated with the blacklegged tick Ixodes scapularis: a systematic review. Parasit Vectors 9: 265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira CJ, Sá-Nunes A, Francischetti IM, Carregaro V, Anatriello E, Silva JS, Santos IK, Ribeiro JM, Ferreira BR. 2011. Deconstructing tick saliva: non-protein molecules with potent immunomodulatory properties. J Biol Chem 286: 10960–10969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro JM. 1987. Role of saliva in blood-feeding by arthropods. Annu Rev Entomol 32: 463–478. [DOI] [PubMed] [Google Scholar]
- Ribeiro JM, Makoul GT, Levine J, Robinson DR, Spielman A. 1985. Antihemostatic, antiinflammatory, and immunosuppressive properties of the saliva of a tick, Ixodes dammini. J Exp Med 161: 332–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro JM, Makoul GT, Robinson DR. 1988. Ixodes dammini: evidence for salivary prostacyclin secretion. J Parasitol 74: 1068–1069. [PubMed] [Google Scholar]
- The RNAcentral Consortium. 2015. RNAcentral: an international database of ncRNA sequences. Nucleic Acids Res 43: D123–D129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueda A, Barturen G, Lebrón R, Gómez-Martín C, Alganza A, Oliver JL, Hackenberg M. 2015. sRNAtoolbox: an integrated collection of small RNA research tools. Nucleic Acids Res 43: W467–W473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sá-Nunes A, Bafica A, Lucas DA, Conrads TP, Veenstra TD, Andersen JF, Mather TN, Ribeiro JM, Francischetti IM. 2007. Prostaglandin E2 is a major inhibitor of dendritic cell maturation and function in Ixodes scapularis saliva. J Immunol 179: 1497–1505. [DOI] [PubMed] [Google Scholar]
- Schwarz A, Tenzer S, Hackenberg M, Erhart J, Gerhold-Ay A, Mazur J, Kuharev J, Ribeiro JM, Kotsyfakis M. 2014. A systems level analysis reveals transcriptomic and proteomic complexity in Ixodes ricinus midgut and salivary glands during early attachment and feeding. Mol Cell Proteomics 13: 2725–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao CC, Xu MJ, Chen YZ, Tao JP, Zhu XQ. 2015. Comparative profiling of microRNAs in male and female Rhipicephalus sanguineus. Appl Biochem Biotechnol 176: 1928–1936. [DOI] [PubMed] [Google Scholar]
- Sturm M, Hackenberg M, Langenberger D, Frishman D. 2010. TargetSpy: a supervised machine learning approach for microRNA target prediction. BMC Bioinformatics 11: 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirloni L, Reck J, Terra RM, Martins JR, Mulenga A, Sherman NE, Fox JW, Yates JR III, Termignoni C, Pinto AF, et al. 2014. Proteomic analysis of cattle tick Rhipicephalus (Boophilus) microplus saliva: a comparison between partially and fully engorged females. PLoS One 9: e94831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirloni L, Islam MS, Kim TK, Diedrich JK, Yates JR III, Pinto AF, Mulenga A, You MJ, Da Silva Vaz I Jr. 2015. Saliva from nymph and adult females of Haemaphysalis longicornis: a proteomic study. Parasit Vectors 8: 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsetsarkin KA, Liu G, Kenney H, Hermance M, Thangamani S, Pletnev AG. 2016. Concurrent micro-RNA mediated silencing of tick-borne flavivirus replication in tick vector and in the brain of vertebrate host. Sci Rep 6: 33088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vay L, Gu C, McNaughton PA. 2012. The thermo-TRP ion channel family: properties and therapeutic implications. Br J Pharmacol 165: 787–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vayssier-Taussat M, Kazimirova M, Hubalek Z, Hornok S, Farkas R, Cosson JF, Bonnet S, Vourch G, Gasqui P, Mihalca AD, et al. 2015. Emerging horizons for tick-borne pathogens: from the ‘one pathogen-one disease’ vision to the pathobiome paradigm. Future Microbiol 10: 2033–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Gong H, Zhang H, Zhou Y, Cao J, Zhou J. 2015. Lipopolysaccharide-induced differential expression of miRNAs in male and female Rhipicephalus haemaphysaloides ticks. PLoS One 10: e0139241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong P, Laxton V, Srivastava S, Chan YW, Tse G. 2017. The role of gap junctions in inflammatory and neoplastic disorders. Int J Mol Med 39: 498–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikel S. 2013. Ticks and tick-borne pathogens at the cutaneous interface: host defenses, tick countermeasures, and a suitable environment for pathogen establishment. Front Microbiol 4: 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Zhou Y, Cao J, Zhang H, Yu Y. 2013. Distinctive microRNA profiles in the salivary glands of Haemaphysalis longicornis related to tick blood-feeding. Exp Appl Acarol 59: 339–349. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.