Abstract
While years of investigation have elucidated many aspects of embryonic stem cell (ESC) regulation, the contributions of post-transcriptional and translational mechanisms to the pluripotency network remain largely unexplored. In particular, little is known in ESCs about the function of RNA binding proteins (RBPs), the protein agents of post-transcriptional regulation. We performed an unbiased RNAi screen of RBPs in an ESC differentiation assay and identified two related genes, NF45 (Ilf2) and NF90/NF110 (Ilf3), whose knockdown promoted differentiation to an epiblast-like state. Characterization of NF45 KO, NF90 + NF110 KO, and NF110 KO ESCs showed that loss of NF45 or NF90 + NF110 impaired ESC proliferation and led to dysregulated differentiation down embryonic lineages. Additionally, we found that NF45 and NF90/NF110 physically interact and influence the expression of each other at different levels of regulation. Globally across the transcriptome, NF45 KO ESCs and NF90 + NF110 KO ESCs show similar expression changes. Moreover, NF90 + NF110 RNA immunoprecipitation (RIP)-seq in ESCs suggested that NF90/NF110 directly regulate proliferation, differentiation, and RNA-processing genes. Our data support a model in which NF45, NF90, and NF110 operate in feedback loops that enable them, through both overlapping and independent targets, to help balance the push and pull of pluripotency and differentiation cues.
Keywords: NF45, NF90/NF110, RNA-binding protein, pluripotency, post-transcriptional regulation
INTRODUCTION
Early mammalian development is a complex phenomenon driven by a myriad of dynamic and precise molecular changes that enable cells to establish, maintain, and exit from the pluripotent state. Derived from the inner cell mass of the mammalian blastocyst and representing the early preimplantation epiblast (Boroviak et al. 2014), embryonic stem cells (ESCs) provide a valuable in vitro platform for dissecting these processes in detail, as they can both self-renew in culture indefinitely and differentiate into all of the tissue types of the body. Most ESC studies to date have focused on the roles of signaling pathways, transcriptional regulators, epigenetic modifications, and noncoding RNAs (Ng and Surani 2011; Young 2011; Watanabe et al. 2012; Greve et al. 2013; Flynn and Chang 2014). In contrast, post-transcriptional processes have only begun to receive attention recently, despite evidence that they are a major driver of the molecular changes that occur during ESC differentiation (Lu et al. 2009) and are important contributors to the efficiency of somatic cell reprogramming to induced pluripotent stem cells (Tahmasebi et al. 2014). These post-transcriptional events, which include alternative splicing, nuclear export, transcript stability, and translational efficiency, are directed by noncoding RNAs and RNA-binding proteins (RBPs), and the functions of RBPs in particular are poorly understood (Keene 2007; Van Nostrand et al. 2016). Building a more complete picture of the RBP-mediated molecular changes that regulate pluripotency is important, as it will enable the more informed use of stem cells for disease modeling, drug development, and regenerative medicine.
Until recently, most RBPs were largely predicted computationally based on homology with proteins with known RNA-binding domains. The development of the “mRNA interactome capture” technique, which uses in vivo UV crosslinking and oligo(dT) purification to identify proteins bound to poly-adenylated cellular RNAs (Baltz et al. 2012; Castello et al. 2012), enabled the generation of a catalog of experimentally assayed RBPs in different mammalian cells, including 555 RBPs in mouse ESCs (Kwon et al. 2013). To date, however, the functions of only a handful of RBPs have been carefully dissected in ESCs (Ye and Blelloch 2014). Thus, we elected to focus our efforts on identifying and characterizing pluripotency-associated RBPs in a well-defined, developmentally relevant in vitro system. Specifically, we examined the transition from an ESC to an epiblast-like cell (EpiLC) (Hayashi et al. 2011; Krishnakumar et al. 2016), a progression that parallels the earliest cell fate decision that occurs in the mammalian embryo proper.
Through an unbiased siRNA screen for putative RBPs that affect the ESC-to-EpiLC transition, we came to focus on two related genes, Ilf2 (NF45) and Ilf3 (NF90/NF110), the latter of which has a 90 kDa isoform (NF90) and a 110 kDa isoform (NF110). In addition to the EpiLC-promoting phenotype their knockdown produced in our screen, these genes were interesting to us for several reasons. First, they have been implicated in pluripotency and development (Shi et al. 2005; Wang et al. 2006; Lu et al. 2009; Horie et al. 2011). Second, although they were first identified in a complex together as a transcriptional activator of IL2 in Jurkat T cells (Shaw et al. 1988; Corthesy and Kao 1994; Kao et al. 1994), they were uncovered in all three mammalian “mRNA interactome capture” studies (Baltz et al. 2012; Castello et al. 2012; Kwon et al. 2013), which provides strong evidence that they have RNA-binding functions in vivo. Indeed, NF45 and NF90/NF110 have been found to participate in a diverse assortment of post-transcriptional processes (Shim et al. 2002; Shi et al. 2005; Merrill and Gromeier 2006; Sakamoto et al. 2009; Graber et al. 2010; Kuwano et al. 2010; Neplioueva et al. 2010; Lee et al. 2011; Singh et al. 2012; Chu et al. 2015; Wandrey et al. 2015; Guo et al. 2016), and their frequent co-occurrence therein suggests that they might interact with each other both physically and functionally. Importantly, none of the previous studies examining NF45 and NF90/NF110 molecular mechanisms were performed in ESCs, so we sought to investigate how NF45 and NF90/NF110 behave in the specific context of pluripotency and whether their activities are interconnected. Using genetic knockout models of these genes, we found that absence of NF45 or NF90 + NF110 impaired ESC proliferation and differentiation down embryonic lineages. Additionally, we found that NF45 and NF90/NF110 interact with one another both physically and functionally, controlling a number of genes involved in proliferation, differentiation, and post-transcriptional processes either alone or in concert. Our data suggest that, through feedback loops, NF45, NF90, and NF110 modulate the expression levels of one another, thereby coordinately influencing the regulatory programs that govern pluripotency and differentiation.
RESULTS
RNAi screen identifies NF45 and NF90/NF110 as promoters of pluripotency
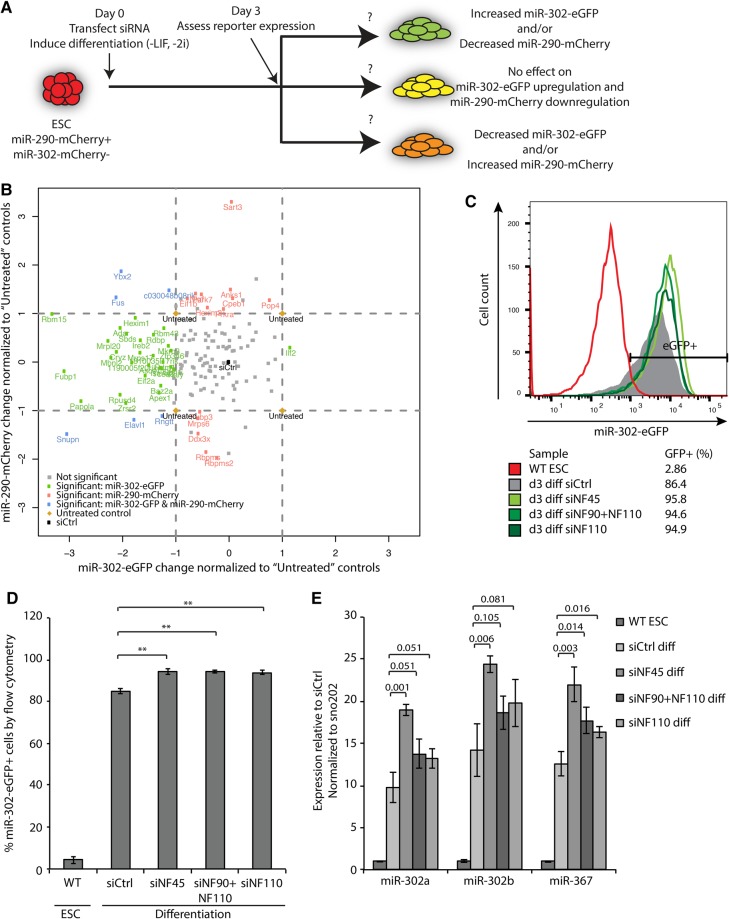
To discover RBPs important for regulating pluripotency and differentiation, we performed an siRNA knockdown screen of 356 putative RBPs in an in vitro mouse ESC differentiation system that monitors the ESC-to-EpiLC transition. The RBPs used in the screen (Supplemental Table S1) either have been annotated as such in the literature (McKee et al. 2005) or are associated with RNA-related gene ontology (GO) terms. Additionally, they are differentially expressed in a microarray study comparing ESCs with in vitro differentiated EpiLCs (Parchem et al. 2014). The ESC differentiation system uses fluorescent reporters to track the expression of two miRNA clusters associated with different stages of pluripotency. Specifically, mCherry was knocked into the mir-290∼295 (mir-290) locus and eGFP into the mir-302∼367 (mir-302) locus (Parchem et al. 2014). When maintained in naïve pluripotent conditions, these “dual reporter” ESCs express miR-290 exclusively and are mCherry+/eGFP−; when the ESCs are allowed to differentiate through the removal of leukemia inhibitory factor (LIF) and GSK and MEK inhibitors (2i), they turn on mir-302 and later shut off mir-290, thus becoming first mCherry+/eGFP+ and then mCherry−/eGFP+. Under optimized differentiation conditions, the initial activation of mir-302 is highly homogeneous and represents the acquisition of the primed pluripotent EpiLC state (Krishnakumar et al. 2016). Therefore, we focused on this phase of the transition to identify genes whose knockdown might disrupt the kinetics, direction, or extent of the differentiation process.
For the screen, dual reporter ESCs transfected with a pool of four siRNAs against each gene of interest were induced to differentiate and assessed for their miR-290-mCherry and miR-302-eGFP expression levels after 3 d using high-content, high-throughput microscopy (Fig. 1A). The screen yielded a number of hits (Supplemental Table S1). Focusing only on the 146 genes whose knockdown did not negatively affect cell proliferation and viability (defined in Supplemental Methods), we found 49 genes whose knockdown significantly changed the expression levels of miR-290-mCherry or miR-302-eGFP (Fig. 1B). Many of these genes (34 genes) led to a reduction in miR-302-eGFP expression, which represents an inhibition of the ESC-to-EpiLC transition. These genes were candidate promoters of differentiation, but we reasoned that the reduced miR-302-eGFP levels could also represent a general nonspecific manifestation of cellular toxicity that might occur when any important cellular gene is disrupted. More interestingly, there was only one gene, NF45 (Ilf2), whose knockdown resulted in increased miR-302-eGFP expression (Fig. 1B). This result therefore suggests that NF45 may normally inhibit premature differentiation out of the naïve state.
FIGURE 1.
An siRNA screen identifies NF45, NF90, and NF110 as promoters of pluripotency. (A) Schematic of siRNA screen design. (B) Scatterplot of siRNA screen hits with normalized t-statistics for miR-302-eGFP and miR-290-mCherry on the x and y axes, respectively. Coloring denotes genes whose knockdown significantly affected levels of miR-302-eGFP, miR-290-mCherry, or both (q < 0.001) after first z-score normalizing from a center of zero as defined by siCtrl-treated samples and then scaling by the t-statistic of the untreated samples. Genes whose knockdown negatively affected cell proliferation and viability (n = 210) were removed from this analysis. Note that some genes with large t-statistics are not called significant due to cutoffs described in the Supplemental Methods. (C) Flow cytometry of miR-302-eGFP expression of ESCs and ESCs differentiated for 3 d in the presence of control siRNA (siCtrl) or siRNAs against Ilf2/NF45 (siNF45), NF90 + NF110 (siNF90 + NF110), or NF110 only (siNF110). (D) Quantification of miR-302-eGFP+ cells from (C). (E) Transcript levels by qRT-PCR of mature miR-302 miRNAs in ESCs treated with the indicated siRNAs and differentiated for 3 d. Error bars represent SD of three biological replicates. (**) P < 0.005. Numbers above the graph in E indicate the P-values of the comparisons marked.
There is much literature on NF45 acting as a post-transcriptional regulator in addition to evidence of its involvement in the pluripotency network, as it was found to be complexed with Nanog (Wang et al. 2006) and is down-regulated in a Nanog-knockdown model of ESC differentiation (Lu et al. 2009). These pieces of data highlighted the potential significance of NF45; therefore, we chose to focus our subsequent investigations on this gene. To validate the NF45 knockdown phenotype, we performed flow cytometry on cells treated with siRNAs against NF45 and confirmed the increase in miR-302-eGFP expression compared with the cells treated with siCtrl nontargeting siRNA at 72 h of differentiation (Fig. 1C,D). Using qRT-PCR, we also confirmed the increase in the levels of processed miR-302 miRNAs (Fig. 1E). As NF45 is known to physically interact with NF90/NF110 (Guan et al. 2008; Wolkowicz and Cook 2012), which were not included in the original siRNA screen, we asked how depletion of NF90/NF110 would affect miR-302 expression. siRNAs against NF45, NF90 + NF110, and NF110 down-regulated their targets to similar levels (∼20%–50% of WT) (Supplemental Fig. S1A,B). Knockdown of either NF90 + NF110 or NF110 alone promoted miR-302 expression (Fig. 1C–E), although the increase in miRNA levels compared to siCtrl was less dramatic than that seen with NF45 knockdown (Fig. 1E). Expression of the miR-290 cluster miRNAs trended downward but did not reach a significance cutoff of P < 0.05 with NF45, NF90 + NF110, or NF110 knockdown (Supplemental Fig. S1C). Together, these data suggest that the reduction of NF45, NF90, and/or NF110 results in an accelerated shift toward the EpiLC state during ESC differentiation.
NF45 and NF90/NF110 regulate ESC proliferation and differentiation
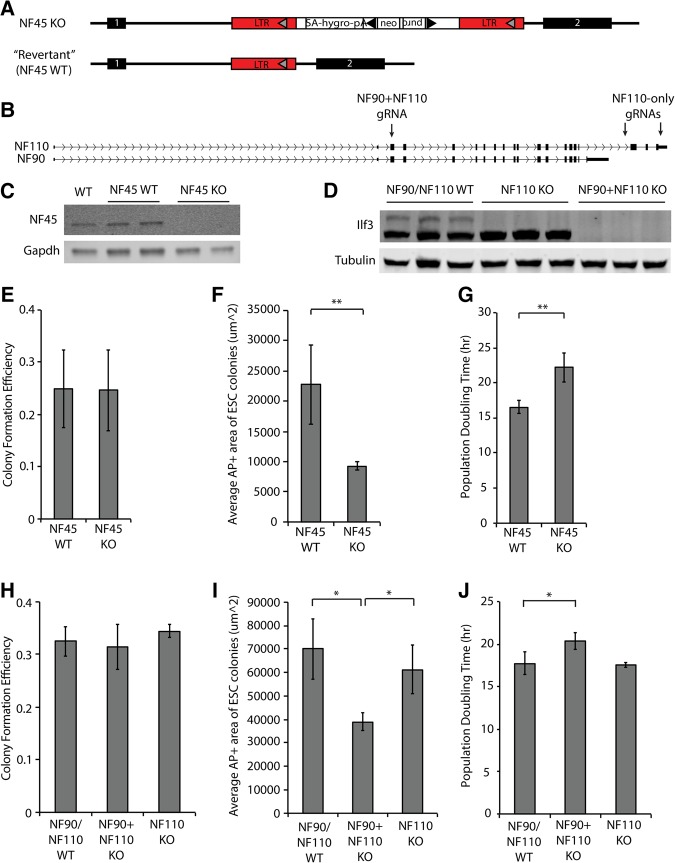
We next used genetic knockout models to further dissect the functions of NF45 and NF90/NF110 in pluripotency and differentiation. NF45−/− (NF45 KO) ESCs (Fig. 2A,C) created through a gene-trap system were obtained from Horie et al. (2011). As the wild-type (NF45 WT) control, we used their “revertant” counterpart, an ESC line in which the gene-trap construct was removed through FLP-FRT recombination. ESCs deficient for both NF90 and NF110 (NF90 + NF110 KO) or for NF110 alone (NF110 KO) (Fig. 2B,D, Materials and Methods) were generated by CRISPR-mediated mutagenesis; the parental line (NF90 + NF110 WT) served as their control.
FIGURE 2.
NF45 and NF90 promote ESC proliferation. (A) Schematic showing design of NF45 gene-trap allele. The gene trap construct is located in the intron between exons 1 and 2 of NF45. Transient expression of a Flippase results in removal of the construct in “revertant” cells, which we used as NF45 WT controls. (B) Schematic showing location of gRNAs for generating NF90 + NF110 and NF110 only KO ESCs by CRISPR. Details on CRISPR mutagenesis are in the Supplemental Methods. (C) Western blot showing protein expression of WT (dual reporter ESCs), NF45 WT, and NF45 KO ESCs. (D) Western blot showing protein expression in NF90 + NF110 WT, NF110 KO, and NF90 + NF110 KO ESCs. (E) Colony formation efficiency, (F) area of alkaline phosphatase staining per colony, and (G) population doubling time of NF45 WT and NF45 KO ESCs. (H) Colony formation efficiency, (I) area of alkaline phosphatase staining per colony, and (J) population doubling time of NF90/NF110 WT, NF90 + NF110 KO, and NF110 KO ESCs. Error bars represent SD of 3–4 biological replicates. (*) P < 0.05, (**) P < 0.005.
To determine whether NF45 and NF90/NF110 are required for ESC self-renewal and proliferation, we performed colony formation and proliferation assays. We found that NF45 KO, NF90 + NF110 KO, and NF110 KO ESCs have colony forming potentials similar to that of their WT counterparts (Fig. 2E,H), indicating that their loss does not affect ESC self-renewal. However, NF45 KO ESCs form smaller colonies (Fig. 2F) and have a longer doubling time (Fig. 2G), suggesting a decreased proliferative capacity consistent with a previous report (Horie et al. 2011). Notably, NF90 + NF110 KO ESCs also exhibit a proliferation defect of comparable magnitude, but NF110 KO ESCs are unaffected (Fig. 2I,J). These results show that both NF45 and NF90 promote ESC proliferation. It is possible that NF110 also promotes ESC proliferation but that NF90 can function redundantly to compensate for its loss; we were unable to test this possibility with our genetic knockout models.
We further investigated whether NF45 and NF90/NF110 are required for appropriate gene expression in the ESC state. We assayed the mRNA levels of a selection of ESC and EpiLC markers to ask whether the propensity of NF45- and NF90/NF110-deficient ESCs to differentiate to the EpiLC state begins with an inherent defect in the self-renewing state. NF45 KO ESCs expressed decreased levels of ESC markers like Klf4 and Rex1 (Supplemental Fig. S2A,B) and increased levels of EpiLC markers like Fgf5 and T/Brachyury (Supplemental Fig. S2C,D), suggesting that they are partially shifted to a primed state even under conditions that maintain naïve pluripotency, although levels of the EpiLC markers were still much lower than those seen with differentiation into EpiLCs (compare with Supplemental Fig. S3). In contrast, NF90/NF110 KO ESCs did not display obvious dysregulation of ESC and EpiLC markers (Supplemental Fig. S2E–H).
Having examined the roles of NF45, NF90, and NF110 in naïve pluripotency, we next sought to better characterize how loss of these genes affects the molecular network when ESCs are induced to differentiate. Using the 2D ESC-to-EpiLC differentiation assay used for the siRNA screen, we found that NF45 KO ESCs appropriately down-regulated ESC markers like Klf4 (Supplemental Fig. S3A) but overactivated EpiLC markers like Fgf5 and miR-302 (Supplemental Fig. S3B,C). Although NF90 + NF110 KO and NF110 KO ESCs showed a similar trend in gene dysregulation (Supplemental Fig. S3D–F), the degree to which EpiLC markers were up-regulated was milder, and only NF90 + NF110 KO but not NF110 KO ESCs exhibited an overactivation of miR-302.
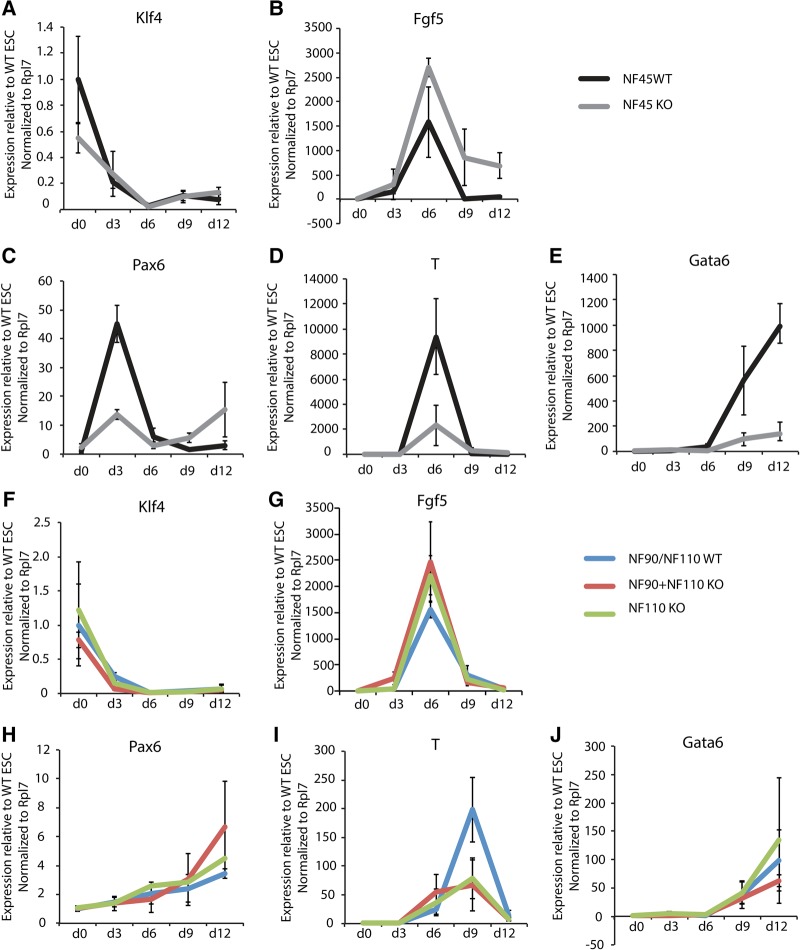
In order to more carefully dissect the impact of these genes upon exit from pluripotency, we studied the effects of NF45, NF90, and NF110 loss in a 3D embryoid body differentiation assay, which allows for undirected differentiation to the three embryonic germ layers. As in the 2D system, NF45 KO ESCs down-regulated ESC markers (Klf4) appropriately (Fig. 3A) while overactivating EpiLC markers (Fgf5, Fig. 3B). However, they were unable to activate markers of ectoderm (Pax6), mesoderm (T/Brachyury), or endoderm (Gata6) (Fig. 3C–E), suggesting that while NF45 KO ESCs are predisposed to achieving the primed EpiLC state, they cannot properly differentiate further to form the embryonic lineages. While lineage differentiation also showed signs of dysregulation in NF90 + NF110 KO and NF110 KO ESCs (Fig. 3F–J), the defects, especially in the NF110 KO ESCs, were different and less pronounced than those in the NF45 KO ESCs. Specifically, NF90 + NF110 KO ESCs appear to have a particular defect in activating the mesoderm program, which is indicated by their failure to activate T/Brachyury expression (Fig. 3I). In summary, our focused molecular and cellular analyses revealed that NF45 and NF90/NF110 have dissimilar effects on differentiation, where loss of NF45 resulted in more dramatic impairments in differentiation than loss of NF90 + NF110 or NF110 alone. These results are consistent with the observation that NF90/NF110 KO mice survive to birth (Shi et al. 2005), while NF45 KO mice exhibit embryonic lethality (http://www.informatics.jax.org/marker/MGI:1915031).
FIGURE 3.
Loss of NF45 and NF90/NF110 dysregulate differentiation down embryonic lineages. Transcript levels by qRT-PCR of (A,F) Klf4, (B,G) Fgf5, (C,H) Pax6, (D,I) T/Brachyury, and (E,J) Gata6 in NF45 WT, NF45 KO, NF90/NF110 WT, NF90 + NF110 KO, and NF110 KO ESCs (d0) and EBs (d3-d12). Error bars represent SD of 3–4 biological replicates.
NF45 and NF90/NF110 form complexes and influence expression levels of each other
Given the partially overlapping molecular and cellular phenotypes of their knockout ESC lines, we tested if and how NF45 and NF90/NF110 are physically or functionally interconnected. First, we examined the subcellular localization of NF45, NF90, and NF110 by fractionation. We found that all proteins are present in both the cytoplasm and the nucleus, although they are largely concentrated in the nucleus (Supplemental Fig. S4A). Expression and localization of these proteins did not change with differentiation to the EpiLC state (Supplemental Fig. S4A), and deletion of NF45 did not affect NF90/NF110 localization (Supplemental Fig. S4B). Their intracellular localization suggests that NF45 and NF90/NF110 are likely capable of participating in both nuclear and cytoplasmic processes.
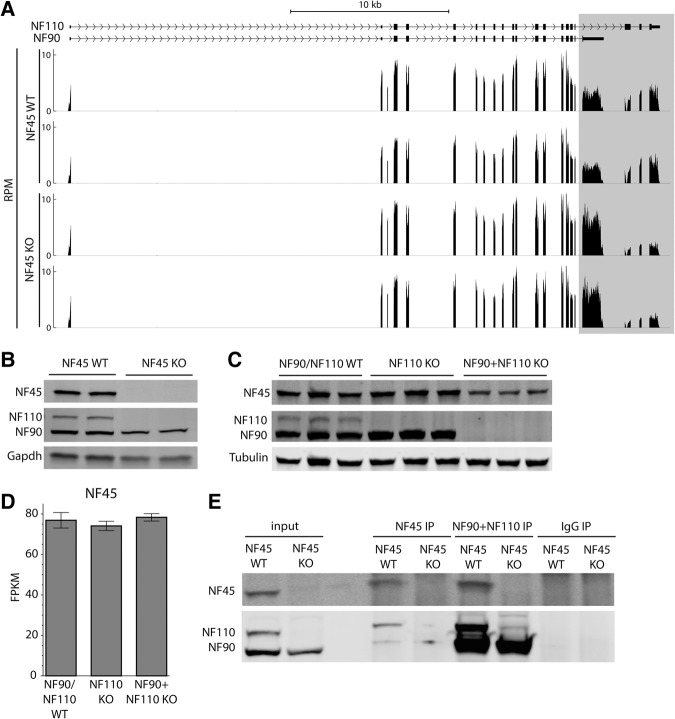
We then asked whether NF45 and NF90/NF110 interact with and influence the expression of each other in ESCs. Indeed, work in other cellular systems frequently discovers both NF45 and NF90/NF110 in the processes being studied, and X-ray crystallography has shown that these proteins can heterodimerize with each other through the dsRNA-binding domain associated with zinc fingers (DZF) motif that both proteins possess (Wolkowicz and Cook 2012). Intriguingly, we found that loss of either NF45 or NF90/NF110 impacts the expression of the other at different stages of molecular regulation. NF45 deletion leads to a splicing isoform switch of NF90/NF110, with transcripts of the NF90 isoform increasing relative to the NF110 isoform (Fig. 4A; Supplemental Fig. S4C). The effects of this isoform switch are further amplified at the protein level, where NF110 protein is entirely absent in NF45 KO cells (Fig. 4B), which suggests that in addition to controlling the balance of NF90 and NF110 mRNA levels, NF45 also positively regulates NF110 protein stability. NF90 protein levels are also reduced in the NF45 KO but to a much smaller degree than the NF110 isoform (Fig. 4B). Conversely, simultaneous loss of NF90 and NF110 but not loss of NF110 alone leads to a decrease in NF45 protein levels (Fig. 4C), while NF45 transcript levels are unaffected (Fig. 4D; Supplemental Fig. S4D). This observation suggests that NF90 promotes NF45 protein stability, although NF110 may also play a redundant role. Finally, we performed coimmunoprecipitations to determine whether NF45 and NF90/NF110 form protein complexes with each other in our ESCs. We found that NF45 and NF90/NF110 do indeed interact with each other (Fig. 4E). Together, these data show not only that NF45 and NF90/NF110 have overlapping phenotypes but also that they positively regulate each other's levels.
FIGURE 4.
NF45 and NF90/NF110 physically interact and influence the expression of each other at different levels of regulation. (A) Read coverage in RPM (reads per million mapped reads) of NF90/NF110 transcript in NF45 WT and NF45 KO ESCs. Differences between the NF90 and NF110 isoforms are highlighted. (B,C) Western blots showing NF45, NF90, and NF110 protein expression levels in the KO ESC lines indicated. (D) Mean FPKM (fragments per kilobase of transcript per million mapped reads) of NF45 transcript in NF90/NF110 WT, NF110 KO, and NF90 + NF110 KO ESCs. Error bars indicate standard error of the mean. (E) Western blots showing co-IP interactions between NF45 and NF90/NF110. Input lanes show 2% input.
Genome-wide expression analysis supports functional interactions between NF45 and NF90/NF110
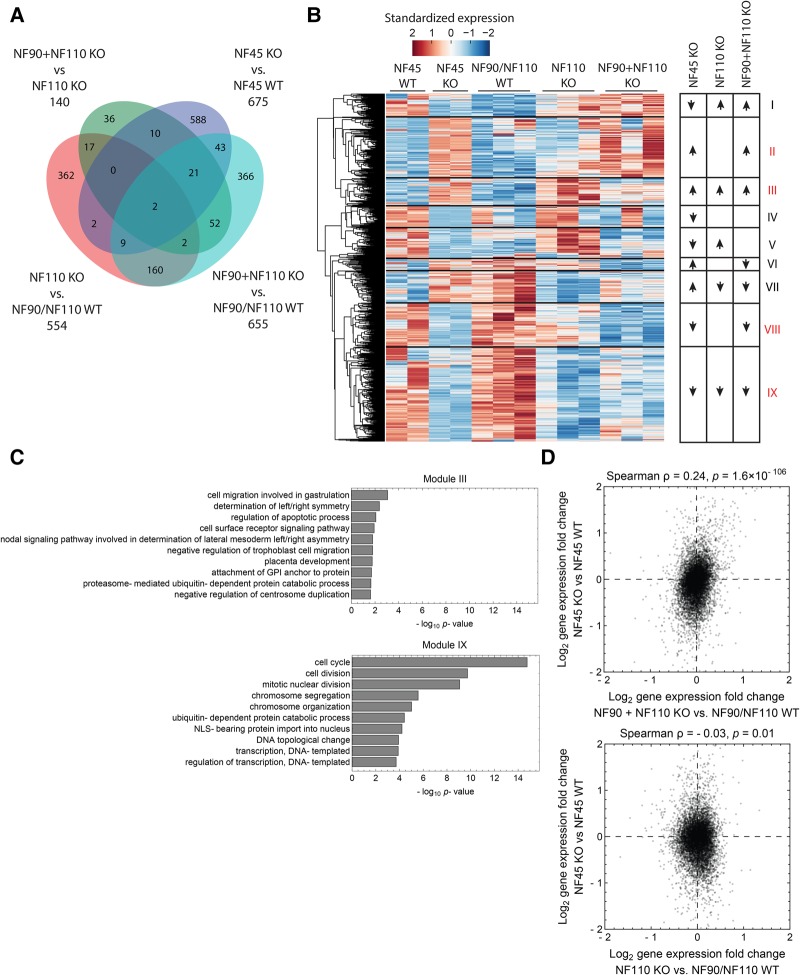
To further investigate the functional interactions of NF45, NF90, and NF110, as well as their roles in ESC proliferation and differentiation, we analyzed global gene expression in NF45 KO, NF90 + NF110 KO, and NF110 KO ESCs by RNA-seq (Supplemental Table S2). Comparing expression changes of select genes with transcript levels by qRT-PCR demonstrated consistency between RNA-seq data and qRT-PCR results (compare Supplemental Fig. S2A–H with Supplemental Fig. S5A–H). Examining the transcriptomes of these KO ESC lines in relation to their corresponding WT controls, we identified 1670 genes that are differentially expressed in at least one comparison (Fig. 5A; Supplemental Table S3; Materials and Methods). By requiring the genes to be robustly expressed (mean FPKM ≥ 10) in at least one condition, we retained 971 genes that represent candidate regulatory targets, both direct and indirect, of each protein.
FIGURE 5.
Global expression analysis is consistent with functional interactions among NF45, NF90, and NF110. (A) Venn diagram of differentially expressed genes in RNA-seq analysis of NF45 WT, NF45 KO, NF90 + NF110 WT, NF90 + NF110 KO, and NF110 KO ESCs. (B) Heat map of robustly expressed genes that are differentially expressed in at least one comparison considered in A. Genes are categorized through unsupervised clustering into nine modules indicating their pattern of expression in the different lines (Materials and Methods). (C) GO analysis of category III and IX genes as defined in B. (D) Correlation analysis of NF45 KO/NF45 WT versus NF110 KO/NF90 + NF110 WT or NF90 + NF110 KO/NF90 + NF110 WT transcriptomes at the gene level.
Grouping these 971 genes by unsupervised hierarchical clustering, we identified nine expression modules (Fig. 5B; Supplemental Table S4; Materials and Methods). We first focused on genes that were consistently up-regulated (Module III) or down-regulated (Module IX) in all three KO ESC lines, as they represent genes that NF45 and NF90/NF110 coordinately regulate either negatively or positively. Gene ontology analysis revealed that genes in Module III (up-regulated in all lines) show an overrepresentation of biological processes related to development and differentiation, such as the determination of left/right symmetry, placenta development, and cell migration, while genes in Module IX (down-regulated in all lines) are highly biased toward cell cycle and cell division related genes (Fig. 5C). Furthermore, genes up-regulated in NF45 KO and NF90 + NF110 KO but not in NF110 KO (Module II) showed enrichment in negative regulation of cell growth and proliferation, as well as positive regulation of differentiation (Supplemental Fig. S5I). Genes down-regulated in NF45 KO and NF90 + NF110 KO but not NF110 KO (Module VIII) showed enrichment in GO terms related to chromatin modification, nuclear division, and development (Supplemental Fig. S5I). These results are broadly consistent with our phenotypic data showing that deletion of NF45 or NF90/NF110 generally resulted in decreases in ESC proliferation and defects in differentiation.
The overall global transcriptomic changes associated with loss of NF45 resemble those associated with the loss of NF90 + NF110 KO as seen with correlation analyses performed at the individual gene level (Fig. 5D) and when aggregated into KEGG pathways (Supplemental Fig. S5J; Materials and Methods) or GO terms (Supplemental Fig. S5K; Materials and Methods). This similarity is absent when comparing NF45 KO with NF110 KO (Fig. 5D; Supplemental Fig. S5J,K), which suggests either that NF90 is the primary functional isoform or that NF90 and NF110 act redundantly. Combined with our phenotypic data, our results indicate that the molecular and cellular impairments of NF45 KO ESCs are similar yet not identical to that of NF90 + NF110 KO ESCs, suggesting both independent and collaborative functions among the three proteins.
NF90/NF110 RIP-seq in ESCs suggested that NF90/NF110 directly regulate proliferation, differentiation, and RNA-processing genes
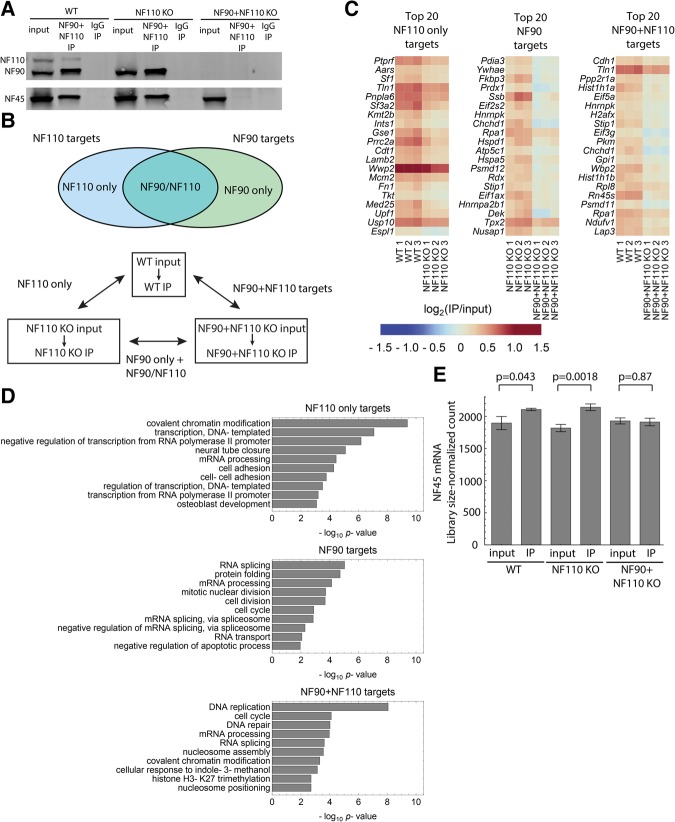
Although our transcriptomic analyses suggested overlapping roles for NF45 and NF90/NF110 in mRNA regulation, they could not distinguish between the primary and secondary effects of the proteins. Therefore, we sought to identify the combined direct RNA targets of NF45 and NF90/NF110. To do so, we performed NF90/NF110 RNA immunoprecipitation and sequencing (RIP-seq) in NF110 KO, NF90 + NF110 KO, and NF90 + NF110 WT ESCs, expecting that the immunoprecipitated NF90/NF110-bound RNAs would also include transcripts bound by NF45–NF90/NF110 complexes. Western blot analysis of input and RIP samples confirmed that immunoprecipitation of NF90/NF110 pulled down the expected proteins in each sample and that targets of NF45-NF90/NF110 complexes would also be included in our data sets (Fig. 6A). To distinguish between RNA targets of NF90 and NF110, we compared the RIP-seq data sets of NF110 KO, NF90 + NF110 KO, and WT ESCs with one another (Fig. 6B). In principle, the WT data set contains both NF90 and NF110 targets; the NF110 KO data set contains only NF90 targets, which consists of targets bound by NF90, including those that could be redundantly bound by NF110 or NF90 (Fig. 6B, top panel); and the NF90 + NF110 KO data set contains nonspecific targets of the NF90/NF110 antibody and experimental noise, both of which are also present in the WT and NF110 KO data. Thus, removing genes found in the NF110 KO RIP-seq data from the list of genes found in the WT RIP-seq data yields targets bound only by NF110 (“NF110 only targets”); a similar comparison of WT and NF90 + NF110 KO data yields targets bound by NF90 and/or NF110 (“NF90 + NF110 targets”), and a comparison of NF110 KO and NF90 + NF110 KO data yields targets bound by NF90 that could also be redundantly bound by NF110 (“NF90 targets,” Fig. 6B, bottom panel).
FIGURE 6.
Identification of NF90/NF110 RNA targets. (A) Representative RIP–Western blot showing immunoprecipitation of NF90, NF110, and NF45 in NF90/NF110 WT, NF110 KO, and NF90 + NF110 KO ESCs. (B) Identification of NF90, NF110, and NF90 + NF110 RNA binding proteins in ESCs by RIP-seq. Top panel defines NF110 and NF90 targets. Bottom panel defines the comparisons used to call RNA targets. (C) Heatmap of log2(IP/input) RIP-seq expression values of top 20 NF90, NF110 only, and NF90 + NF110 RNA targets as identified by a combination of the Poisson ratio test and the log ratio test (Supplemental Methods). (D) GO analysis of identified NF90, NF110 only, and NF90 + NF110 targets. (E) Mean library-size normalized read count (as calculated by DESeq2) of NF45 in the RIP-seq libraries. Error bar indicates standard error of the mean.
We developed two complementary computational approaches to identify NF90 and NF110 targets based on these comparisons (Supplemental Methods; Supplemental Fig. S6; Supplemental Table S5). One utilizes the stochastic nature of count data but treats the IP and input data as independent samples (Poisson ratio test, Supplemental Methods; Supplemental Fig. S6A), while the other takes into account the paired nature of the IP and input samples but does not make full use of the distributive properties of count data (log ratio test, Supplemental Methods; Supplemental Fig. S6B). We used a multiobjective optimization algorithm based on the concept of Pareto dominance (Diaz et al. 2015) to combine results from the two methods (Supplemental Fig. S6C), obtaining 208, 81, and 167 genes as NF110 only targets, NF90 targets, and NF90 + NF110 targets, respectively (Fig. 6C; Supplemental Table S6). Notably, NF90 targets previously identified in HeLa cells (Kuwano et al. 2010) also showed IP to input enrichment in our NF90 target identification method (Supplemental Fig. S7A), which demonstrates the robustness of our analysis and suggests that NF45, NF90, and NF110 likely regulate targets that are common across many cell types, as well as targets that are specific to certain cellular contexts.
Gene ontology analysis showed that the identified targets are broadly implicated in biological processes related to cell cycle, development, protein folding, and multiple steps in RNA metabolism, including transcription, splicing, and RNA transport (Fig. 6D), which is consistent with our molecular and cellular evidence that NF45 and NF90/NF110 are crucial for the regulation of ESC proliferation and differentiation, as well as published work associating NF45 and NF90/NF110 with different RNA-related activities. Indeed, NF45 and NF90/NF110 have been shown to participate in miRNA processing (Gregory et al. 2004; Sakamoto et al. 2009), mRNA splicing through its interactions with the components of the exon junction complex (Singh et al. 2012) and other splicing regulators (Damianov et al. 2016), protein translation through its association with precursors to the 60S ribosomal subunit (Wandrey et al. 2015), and lncRNA function through its interactions with Xist (Chu et al. 2015). Additionally, NF45 has been shown to regulate translation at cellular and viral internal ribosomal entry sites (Merrill and Gromeier 2006; Graber et al. 2010; Lee et al. 2011), and NF90 functions as an mRNA stabilizer and translational repressor, which, indeed, is perhaps its most established role (Shim et al. 2002; Shi et al. 2005; Kuwano et al. 2010; Neplioueva et al. 2010; Guo et al. 2016).
In order to find genes that are both direct targets of NF90/NF110 and are also regulated by NF90/NF110 at the mRNA expression level, we intersected identified NF90/NF110 targets with expression Modules III (up-regulated in all KO lines) and IX (down-regulated in all KO lines) as defined in Figure 5B. We found that genes up-regulated upon KO and directly targeted by NF90/NF110 consist predominantly of genes associated with cell development and differentiation (Supplemental Fig. S7B), while genes down-regulated upon KO and directly targeted by NF90/NF110 contain mostly genes related to cell division (Supplemental Fig. S7C). These results provide further evidence that NF90/NF110 regulate ESC differentiation and proliferation through direct targeting of downstream genes.
Interestingly, our data showed enriched NF45 mRNA in the IP compared to the input of both WT and NF110 KO but not NF90 + NF110 KO ESCs. The enrichment is small but statistically significant (Fig. 6E) and suggests that NF45 mRNA is bound by NF90/NF110. NF90/NF110 could potentially regulate NF45 post-transcriptionally, which could explain the reduced NF45 protein levels in the NF90 + NF110 KO (Fig. 4C). Together, these data uncover hundreds of potential targets of NF90 and NF110, many shared but others distinct, thus suggesting that NF110 is not simply redundant with NF90.
DISCUSSION
Post-transcriptional and translational processes and the RBPs that drive them have been historically understudied components of ESC regulatory circuitry. In this study, we use an RNAi screen for RBPs that affect the ESC-to-EpiLC transition to identify NF45 and NF90/NF110 as factors important for maintaining the ESC state. Using genetic knockout models, we found evidence for both physical and functional interactions of NF45, NF90, and NF110 with one another. Our data suggest that these proteins can function both independently and in complexes in ESCs to regulate RNA processing, cell proliferation, and cell differentiation. Because NF45 protein levels were decreased in NF90 + NF110 KO cells (Fig. 4C), it is possible that the NF90 + NF110-deficient phenotype is predominantly driven by the reduction in NF45 expression. However, even though the molecular and cellular dysregulation observed in NF90 + NF110 KO ESCs was generally similar and milder compared to that of NF45 KO ESCs, they also have distinct qualities. NF45 KO and NF90 + NF110 KO ESCs have different differentiation kinetics (Fig. 3; Supplemental Fig. S2), and they share only a limited number of differentially expressed genes globally (Fig. 5A). In addition, direct targets of NF90/NF110 identified through RIP-seq are involved in cell differentiation and division (Supplemental Fig. S7B,C). These data suggest that NF90/NF110 have at least some functions that are separate from its effects on NF45 expression levels.
Although we focused our screen analyses on genes that affected miR-302-eGFP activation, we also observed that some genes (20 genes) affected miR-290-mCherry expression, suggesting that they potentially influence the maintenance of naïve pluripotency (Fig. 1B). These genes should be interesting for future investigations. Of note, a similar screen of RBPs in ESCs recently identified components of the small subunit processome of rRNA biogenesis as important regulators of pluripotency (You et al. 2015), which highlights the significance of post-transcriptional and translational regulation of the ESC state. Due to differences in experimental design, however, there was only partial overlap with the RBPs tested here and no overlap in the hits identified.
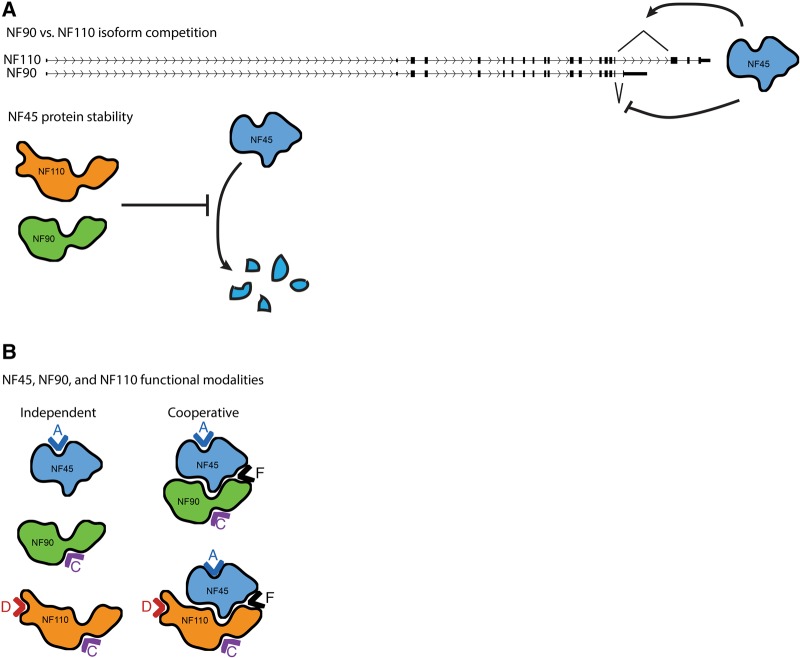
Our study design, which combined directed molecular experiments with genome-wide analyses, yielded rich data sets that allow us to propose theoretical models of NF45, NF90, and NF110 expression and function (Fig. 7). First, our results indicate that NF45, NF90, and NF110 influence the expression of one another through regulatory mechanisms diagrammed in Figure 7A. NF90 and NF110 are two alternative isoforms originating from the same gene and are thus in natural competition with each other. Loss of NF45 resulted in an isoform switch between NF90 and NF110, with NF90 transcripts increasing at the expense of NF110 transcripts (Fig. 4A), which suggests that NF45 normally promotes splicing of NF110 over NF90. Further, NF90 + NF110 KO but not NF110 KO ESCs showed decreased NF45 protein level (Fig. 4C), while NF45 transcript levels were unchanged (Fig. 4D), which indicates that NF90 promotes NF45 protein stability (Fig. 7A), although NF110 might be able to function redundantly. Taken together, our data support a model (Fig. 7A) in which NF45 and NF90 regulate each other through a negative feedback loop, with NF90 promoting NF45 protein levels, while NF45 indirectly decreases NF90 mRNA levels by promoting NF110 splicing. This negative feedback loop may help reduce stochastic fluctuations, ensuring proper downstream function of the complexes and individual components.
FIGURE 7.
Models of NF45, NF90, and NF110 regulation and function. (A) NF45, NF90, and NF110 expression levels are controlled through a homeostatic regulatory loop. NF45 causes an alternative splicing-driven isoform competition between NF90 and NF110, and NF90 (and possibly NF110) promote NF45 protein stability. (B) NF45, NF90, and NF110 can operate either independently or co-operatively. Carets indicate different functional modes: Modes A, C, and D represent regulatory groups of NF45, NF90 or NF110, and NF110, respectively; Mode F represent regulatory groups of NF45–NF90 or NF45–NF110 complexes (see main text and Supplemental Methods).
Second, our genomic analyses allowed us to dissect the functional interactions among NF45, NF90, and NF110 in unprecedented detail. In principle, these three proteins can function either on their own or co-operatively by forming complexes, which allows for a variety of possible regulatory modes. We were able to assess the actuality of each mode by using a combinatorial gene set expression analysis framework (Supplemental Methods; Supplemental Fig. S8A–D) based on the following observations: (i) There exists a set of genes that is regulated only by NF110, independent of both NF45 and NF90; (ii) genes regulated by the NF45-NF110 complex could also be redundantly regulated by the NF45–NF90 complex and vice versa; and (iii) there exists a set of genes that is regulated by either NF90 or NF110, independent of NF45. Applying this analysis framework to our transcriptomic data allowed us to identify four distinct regulatory modes that are supported by our experiments (Fig. 7B): Mode A, which consists of genes regulated by NF45 only; Mode C, which consists of genes regulated by NF90 or NF110 redundantly, but independent of NF45; Mode D, which consists of genes regulated by NF110 only, independent of both NF90 and NF45; and Mode F, which consists of genes regulated by either NF45–NF90 and/or NF45–NF110 complexes. NF45, NF90, and NF110 regulate ESC proliferation, pluripotency, and differentiation through these four distinct modes (Supplemental Fig. S8E), and the genes in each of these modes (Supplemental Table S7) should serve as a useful reference for future studies.
While some of the functions of NF45 and NF90/110 are probably shared across mammalian cells, others are likely specific to the unique needs of ESCs. For instance, the changes in NF45 and NF90/NF110 protein levels in the absence of each other have also been observed in HeLa cells (Guan et al. 2008), which suggests that establishing a proper balance of these factors could be important for maintaining cellular homeostasis generally. On the other hand, there was only partial concordance of our NF90/NF110 RIP-seq data with that of previously published NF90/NF110 targets, and indeed, NF90/NF110 RIP-chips performed in HeLa and 293T cells yielded completely nonoverlapping targets (Kuwano et al. 2010; Neplioueva et al. 2010), all of which suggests that NF90/NF110 RNA targets can be cell type-dependent. These differences could also be explained by technical variability under different experimental conditions. Indeed, capture of RBP-bound RNAs is inefficient, despite the development of a number of different UV crosslinking-based techniques that strive to increase the sensitivity and specificity of target identification (Wang et al. 2015; Van Nostrand et al. 2016). These challenges are compounded by the very nature of RNA binding as well, since many RNA-binding domains have limited sequence specificity (Singh and Valcarcel 2005; Lunde et al. 2007; Chen and Varani 2013; Jankowsky and Harris 2015). Additionally, double-stranded RNA (dsRNA) binding proteins like NF45 and NF90/NF110 (the DZF motif is dsRNA-binding, and NF90/NF110 also possesses dsRNA-binding motifs [Guan et al. 2008]) rely at least in part on recognition of the 3D RNA structure (Chen and Varani 2013). While these attributes make identifying RBP-binding motifs difficult, it could explain why a number of RBPs, including NF45 and NF90/NF110, have been found to participate in multiple layers of RNA metabolism (Ye and Blelloch 2014). With their relative binding sequence degeneracy, interactions with other protein partners is likely critical for establishing binding specificity. This could result in one RBP working with several other proteins to regulate different groups of transcripts in different ways in a single cell type.
In sum, our study uses novel experimental designs and analyses to establish a critical role for NF45 and NF90/NF110 in the ESC circuitry. Given the myriad regulatory activities with which NF45 and NF90/NF110 have been previously associated, as well as the different combinations of protein complexes they are capable of forming, it is likely that they play highly intricate and nuanced roles within the post-transcriptional network. Further work will be necessary to fully elucidate the mechanisms by which these proteins influence the cell fate decisions underlying early embryonic development.
MATERIALS AND METHODS
Cell culture and related assays
All ESCs were maintained on 0.2% gelatin-coated plates in FBS + LIF + 2i medium (DMEM, 15% FBS, nonessential amino acids, L-glutamine, penicillin/streptomycin, 55 µM β-mercaptoethanol, 1000 U LIF, 1 µM PD0325901, 3 µM CHIR99021). Dual reporter (mir-290-mCherry/mir-302-eGFP) ESCs (Parchem et al. 2014) and NF45 WT and NF45 KO ESCs (Horie et al. 2011) have been previously described. NF90 + NF110 and NF110 KO ESCs were generated using CRISPR (details in the Supplemental Methods). Colony formation, population doubling time, and differentiation assays are described in the Supplemental Methods.
RNAi screen
Dual reporter ESCs were plated into 96-well plates in FBS + LIF + 2i medium. LIF and 2i were removed the following day, and siRNAs were transfected. Plates were imaged 3 d later to assess miR-302-eGFP and miR-290-mCherry expression. Details on the screen development and the analysis methods are in the Supplemental Methods.
Western blot
For whole-cell extract, cells were lysed in protein lysis buffer (25 mM Tris pH 7.9, 150 mM NaCl, 0.1 mM EDTA, 0.1% Triton X-100, 10% glycerol) with protease inhibitors and resolved on 10% Mini-PROTEAN TGX gels (Bio-Rad). Cytoplasmic and nuclear fractions were collected using Thermo Fisher Scientific's Subcellular Fractionation Kit for Cultured Cells according to the manufacturer's protocol. Equal amounts of protein were loaded per sample. The following antibodies were used for blotting: 1:10,000 NF45 (Everest Biotech EB07784), 1:1000 NF90/NF110 (BD Biosciences clone 21/DRBP76), 1:10,000 Nanog (Calbiochem SC1000), 1:2000 GAPDH (Santa Cruz Biotechnology sc-25778), 1:10,000 Tubulin (Abcam ab6160).
Coimmunoprecipitation
ESCs were harvested, pelleted, and lysed in 10 pellet volumes of low NP40 lysis buffer (12.5 mM Tris pH 7.9, 150 mM NaCl, 0.1 mM EDTA, 0.1% Triton X-100, 10% glycerol) with cOmplete protease inhibitors (Roche). Lysates were freeze-thawed 3 times and then cleared by centrifugation at maximum speed for 15 min at 4°C. Antibodies (Ab) for immunoprecipitation (5 µg NF45 Ab/1 mL lysate [Everest Biotech EB07784], 5 µg NF90/NF110 Ab/1 mL lysate [BD Biosciences clone 21/DRBP76], 6 µg normal rabbit IgG/1 mL lysate [Thermo Fisher Scientific]) were conjugated to 40 µL Protein G Dynabeads/1 mL lysate (Thermo Fisher Scientific) in low NP40 lysis buffer for 2 h at room temperature. Lysates were incubated with Protein G Dynabead-conjugated antibodies overnight at 4°C. Immunoprecipitates were washed 2× with low NP40 lysis buffer and resolved and visualized by Western blot as described above.
qRT-PCR
Total RNA was collected using TRIzol (Thermo Fisher Scientific) according to the manufacturer's protocol. For cDNA synthesis, RNA was treated with DNase I (Invitrogen) and reverse-transcribed with oligo(dT) primers [or for RIP samples, a 1:1 mix of oligo(dT) and random hexamer primers] using the SuperScript III Kit (Invitrogen). Total cDNA was diluted 1:5, and qPCR was performed using gene-specific primer sets (listed in Supplemental Methods) and SensiFast SYBR Hi-ROX master mix (Bioline). MiRNA qRT-PCR was performed with the poly(A) and SYBR Green method as previously described using miRNA-specific forward primers and a 3′ RACE adaptor reverse primer (Shi and Chiang 2005). Primer specificity was verified through analysis of dissociation curves in experimental, no RT, and water-only samples. Primer sequences are in the Supplemental Methods.
RNA-seq library generation
For NF45 WT and NF45 KO ESCs, 1 µg of RNA per sample was processed using the Illumina TruSeq Stranded mRNA Sample Prep Kit according to the manufacturer's protocol. For NF90/NF110 WT, NF90 + NF110 KO, and NF110 KO ESCs, 1 µg of RNA per sample was processed using the KAPA Stranded RNA-seq with the RiboErase Kit according to the manufacturer's protocol.
RNA immunoprecipitation (RIP)
RIP was performed essentially as previously described (Keene et al. 2006; Peritz et al. 2006). ESCs were harvested, pelleted, and lysed in 10 pellet volumes of polysome lysis buffer (100 mM KCl, 5 mM MgCl2, 10 mM HEPES pH 7.5, 0.5% NP40 substitute, 1 mM DTT) with cOmplete protease inhibitors (Roche), 100 U/mL Superase-In (Thermo Fisher Scientific), and 400 µM vanadyl ribonucleoside complex (NEB). Lysates were freeze-thawed once, incubated with 100 U/mL Turbo DNase (Thermo Fisher Scientific) for 15 min at 37°C, and then cleared by centrifugation at maximum speed for 15 min at 4°C. Aliquots of the cleared lysate were taken at this step to serve as the input for Western blot and qRT-PCR analysis. Antibodies for immunoprecipitation (5 µg NF90/NF110 Ab/1 mL lysate [BD Biosciences clone 21/DRBP76], 6 µL normal rabbit IgG/1 mL lysate [Thermo Fisher Scientific]) were conjugated to 40 µL/1 mL lysate Protein G Dynabeads (Thermo Fisher Scientific) in polysome lysis buffer for 2 h at room temperature. Lysates were then incubated with Protein G Dynabead-conjugated antibodies for 4 h at 4°C. Immunoprecipitates were washed 3× with polysome lysis buffer and then 3× with polysome lysis buffer containing 1 M urea. An aliquot of the immunoprecipitate was taken for Western blot, while the rest of the immunoprecipitate, along with the qRT-PCR input lysate, was incubated with 1.2 mg/mL proteinase K (Roche) and 2× proteinase K buffer (100 mM Tris-HCl pH 7.5, 12.5 mM EDTA, 150 mM NaCl, 2% SDS) for 30 min at 55°C. Samples were then lysed in TRIzol LS (Thermo Fisher Scientific), and RNA was extracted and used for qRT-PCR or sequencing.
Mapping reads
Both RNA-seq and RIP-seq reads were mapped to the mm10 reference genome using TopHat2 (Kim et al. 2013) with options -g 1 --prefilter-multihits. Cuffnorm (Trapnell et al. 2012) was used with the gtf file from UCSC mm10 (Illumina iGenomes July 17, 2015 version) as transcript annotation to evaluate relative expression level of genes (fragments per kilobase of transcript per million mapped reads [FPKM]) (Supplemental Table S2). Genes belonging to the following categories were removed from subsequent analysis: tRNA, Mt-tRNA, rRNA, scRNA, snRNA, snoRNA, miRNA, and misc-RNA.
Differential expression
HTSeq-count (Anders et al. 2014) was used to summarize read counts for each gene. The resulting read count matrices were used in DESeq2 (Love et al. 2014) analysis of differential expression (Supplemental Table S3). Because paired-end 51 bp reads were used in NF45 RNA-seq experiments, while single-end 50-bp reads were used in NF90/NF110 RIP-seq experiments with much lower sequencing depth, NF45 RNA-seq data generally had higher sensitivity in calling differentially expressed genes compared to NF90/NF110 RIP-seq data. We thus used different thresholds for statistical significance in the two data sets for calling differentially expressed genes: adjusted P-value <0.001 for the NF45 RNA-seq data and adjusted P-value <0.05 for the NF90/NF110 RIP-seq data. In addition, we further required the log2 fold change (as calculated by DESeq2) to be at least 1 in NF45 RNA-seq data for a gene to be called differentially expressed.
Clustering analysis
The hierarchical clustering in Figure 5B was performed as follows. We identified 1670 genes differentially expressed in at least one of the following comparisons: NF45 KO versus NF45 WT, NF90 + NF110 KO versus NF110 KO, NF110 KO versus NF90/NF110 WT, and NF90 + NF110 KO versus NF90/NF110 WT. Note that the input samples from the NF90/NF110 RIP-seq experiments were used as RNA-seq data. By further requiring the genes to be robustly expressed (mean FPKM ≥ 10) in at least one condition, we retained 971 genes as candidate regulatory targets. These 971 genes were used for hierarchical clustering in Figure 5B. In order to remove batch effects and facilitate comparison between NF45 and NF90/NF110 RNA-seq data (RIP-seq input data), we separately standardized the FPKM value of each gene by subtracting the mean and dividing by the standard deviation across NF45 RNA-seq samples and NF90/NF110 RNA-seq samples. Hierarchical clustering of the genes was performed using the standardized expression values with Euclidean metric and average linkage. Expression modules were then extracted by first cutting the dendrogram at a fixed depth to obtain 20 clusters and then manually merging extremely small clusters with closest ones.
Gene ontology analysis
Gene ontology analysis was performed with the functional annotation tool from DAVID 6.8 (Huang et al. 2009a,b).
Pairwise expression correlation analysis
The pairwise expression correlation analysis is similar to the analysis performed as before (Bulut-Karslioglu et al. 2016). Figure 5D plots the log2 fold change of FPKM values between indicated conditions for 7995 genes that are robustly expressed (mean FPKM ≥ 10) in at least one of the NF45 KO, NF45 WT, NF90/NF110 WT, NF110 KO, and NF90 + NF110 KO RNA-seq samples. The Spearman correlation coefficients and associated P-values are also indicated. In Supplemental Figure S5B, we defined pathway expression as the mean FPKM values of genes associated with each of the 281 KEGG pathways (Kanehisa and Goto 2000; Kanehisa et al. 2016) that contain at least 10 genes. The log2 fold change of pathway expression values between indicated conditions was plotted, with Spearman correlation coefficients and associated P-values indicated. In Supplemental Figure S5C, we defined GO term expression as the mean FPKM values of genes associated with each of the 3516 GO terms (Huang et al. 2009a,b) that contain at least 10 genes. The log2 fold change of GO term expression values between indicated conditions were plotted, with Spearman correlation coefficients and associated P-values indicated.
RIP-seq analysis
NF90 and NF110 targets were called using a combination of the Poisson ratio test and log ratio test, which are described in the Supplemental Methods.
Combinatorial gene set expression analysis
This analysis aimed to infer the first-order approximation of the functional interactions between NF45, NF90, and NF110 by using the RNA-seq data from our genetic knockout models. Details are described in the Supplemental Methods.
DATA DEPOSITION
All raw sequencing data have been deposited in the Gene Expression Omnibus (GEO) under the accession code GSE95145.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Noelle L'Etoile, K. Mark Ansel, and Hiten Madhani for helpful discussions. We acknowledge the use of the UCSF Center for Advanced Technology sequencing core facility, as well as the UCSF Parnassus Flow Cytometry Core, which is run by the Diabetes Research Center (National Institutes of Health grant P30 DK063720). This work was supported by grants from the National Institutes of Health (R01GM101180 to R.B., R01CA163336 to J.S.S., and F30HD084120 to J.Y.), the National Science Foundation (1442504 to J.S.S., 1144247 to A.P.), and the University of California San Francisco Discovery Fellows Program (to J.Y.).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.061499.117.
REFERENCES
- Anders S, Pyl PT, Huber W. 2014. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltz AG, Munschauer M, Schwanhausser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M, et al. 2012. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell 46: 674–690. [DOI] [PubMed] [Google Scholar]
- Boroviak T, Loos R, Bertone P, Smith A, Nichols J. 2014. The ability of inner-cell-mass cells to self-renew as embryonic stem cells is acquired following epiblast specification. Nat Cell Biol 16: 516–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulut-Karslioglu A, Biechele S, Jin H, Macrae TA, Hejna M, Gertsenstein M, Song JS, Ramalho-Santos M. 2016. Inhibition of mTOR induces a paused pluripotent state. Nature 540: 119–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, et al. 2012. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149: 1393–1406. [DOI] [PubMed] [Google Scholar]
- Chen Y, Varani G. 2013. Engineering RNA-binding proteins for biology. FEBS J 280: 3734–3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Zhang QC, da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, Magnuson T, Heard E, Chang HY. 2015. Systematic discovery of Xist RNA binding proteins. Cell 161: 404–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corthésy B, Kao PN. 1994. Purification by DNA affinity chromatography of two polypeptides that contact the NF-AT DNA binding site in the interleukin 2 promoter. J Biol Chem 269: 20682–20690. [PubMed] [Google Scholar]
- Damianov A, Ying Y, Lin CH, Lee JA, Tran D, Vashisht AA, Bahrami-Samani E, Xing Y, Martin KC, Wohlschlegel JA, et al. 2016. Rbfox proteins regulate splicing as part of a large multiprotein complex LASR. Cell 165: 606–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz AA, Qin H, Ramalho-Santos M, Song JS. 2015. HiTSelect: a comprehensive tool for high-complexity-pooled screen analysis. Nucleic Acids Res 43: e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn RA, Chang HY. 2014. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell 14: 752–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graber TE, Baird SD, Kao PN, Mathews MB, Holcik M. 2010. NF45 functions as an IRES trans-acting factor that is required for translation of cIAP1 during the unfolded protein response. Cell Death Differ 17: 719–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. 2004. The microprocessor complex mediates the genesis of microRNAs. Nature 432: 235–240. [DOI] [PubMed] [Google Scholar]
- Greve TS, Judson RL, Blelloch R. 2013. microRNA control of mouse and human pluripotent stem cell behavior. Annu Rev Cell Dev Biol 29: 213–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan D, Altan-Bonnet N, Parrott AM, Arrigo CJ, Li Q, Khaleduzzaman M, Li H, Lee CG, Pe'ery T, Mathews MB. 2008. Nuclear factor 45 (NF45) is a regulatory subunit of complexes of NF90/NF110 involved in mitotic control. Mol Cell Biol 28: 4629–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Xue Y, Yang G, Yin S, Shi W, Cheng Y, Yan X, Fan S, Zhang H, Zeng F. 2016. Nanog RNA binding proteins YBX1 and ILF3 affect pluripotency of embryonic stem cells. Cell Biol Int 40: 847–860. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Ohta H, Kurimoto K, Aramaki S, Saitou M. 2011. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell 146: 519–532. [DOI] [PubMed] [Google Scholar]
- Horie K, Kokubu C, Yoshida J, Akagi K, Isotani A, Oshitani A, Yusa K, Ikeda R, Huang Y, Bradley A, et al. 2011. A homozygous mutant embryonic stem cell bank applicable for phenotype-driven genetic screening. Nat Methods 8: 1071–1077. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2009a. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2009b. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat Protoc 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Jankowsky E, Harris ME. 2015. Specificity and nonspecificity in RNA-protein interactions. Nat Rev Mol Cell Biol 16: 533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. 2016. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44: D457–D462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao PN, Chen L, Brock G, Ng J, Kenny J, Smith AJ, Corthésy B. 1994. Cloning and expression of cyclosporine A- and FK506-sensitive nuclear factor of activated T-cells: NF45 and NF90. J Biol Chem 269: 20691–20699. [PubMed] [Google Scholar]
- Keene JD. 2007. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet 8: 533–543. [DOI] [PubMed] [Google Scholar]
- Keene JD, Komisarow JM, Friedersdorf MB. 2006. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat Protoc 1: 302–307. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar R, Chen AF, Pantovich MG, Danial M, Parchem RJ, Labosky PA, Blelloch R. 2016. FOXD3 regulates pluripotent stem cell potential by simultaneously initiating and repressing enhancer activity. Cell Stem Cell 18: 104–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwano Y, Pullmann R Jr, Marasa BS, Abdelmohsen K, Lee EK, Yang X, Martindale JL, Zhan M, Gorospe M. 2010. NF90 selectively represses the translation of target mRNAs bearing an AU-rich signature motif. Nucleic Acids Res 38: 225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon SC, Yi H, Eichelbaum K, Föhr S, Fischer B, You KT, Castello A, Krijgsveld J, Hentze MW, Kim VN. 2013. The RNA-binding protein repertoire of embryonic stem cells. Nat Struct Mol Biol 20: 1122–1130. [DOI] [PubMed] [Google Scholar]
- Lee JW, Liao PC, Young KC, Chang CL, Chen SSL, Chang TT, Lai MD, Wang SW. 2011. Identification of hnRNPH1, NF45, and C14orf166 as novel host interacting partners of the mature hepatitis C virus core protein. J Proteome Res 10: 4522–4534. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Markowetz F, Unwin RD, Leek JT, Airoldi EM, MacArthur BD, Lachmann A, Rozov R, Ma'ayan A, Boyer LA, et al. 2009. Systems-level dynamic analyses of fate change in murine embryonic stem cells. Nature 462: 358–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunde BM, Moore C, Varani G. 2007. RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol 8: 479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee A, Minet E, Stern C, Riahi S, Stiles CD, Silver PA. 2005. A genome-wide in situ hybridization map of RNA-binding proteins reveals anatomically restricted expression in the developing mouse brain. BMC Dev Biol 5: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill MK, Gromeier M. 2006. The double-stranded RNA binding protein 76:NF45 heterodimer inhibits translation initiation at the rhinovirus type 2 internal ribosome entry site. J Virol 80: 6936–6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neplioueva V, Dobrikova EY, Mukherjee N, Keene JD, Gromeier M. 2010. Tissue type-specific expression of the dsRNA-binding protein 76 and genome-wide elucidation of its target mRNAs. PLoS One 5: e11710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Surani A. 2011. The transcriptional and signaling networks of pluripotency. Nat Cell Biol 13: 490–496. [DOI] [PubMed] [Google Scholar]
- Parchem RJ, Ye J, Judson RL, LaRussa MF, Krishnakumar R, Blelloch A, Oldham MC, Blelloch R. 2014. Two miRNA clusters reveal alternative paths in late-stage reprogramming. Cell Stem Cell 14: 617–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peritz T, Zeng F, Kannanayakal TJ, Kilk K, Eiríksdóttir E, Langel U, Eberwine J. 2006. Immunoprecipitation of mRNA-protein complexes. Nat Protoc 1: 577–580. [DOI] [PubMed] [Google Scholar]
- Sakamoto S, Aoki K, Higuchi T, Todaka H, Morisawa K, Tamaki N, Hatano E, Fukushima A, Taniguchi T, Agata Y. 2009. The NF90-NF45 complex functions as a negative regulator in the microRNA processing pathway. Mol Cell Biol 29: 3754–3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw JP, Utz PJ, Durand DB, Toole JJ, Emmel EA, Crabtree GR. 1988. Identification of a putative regulator of early T cell activation genes. Science 241: 202–205. [DOI] [PubMed] [Google Scholar]
- Shi R, Chiang VL. 2005. Facile means for quantifying microRNA expression by real-time PCR. Biotechniques 39: 519–525. [DOI] [PubMed] [Google Scholar]
- Shi L, Zhao G, Qiu D, Godfrey WR, Vogel H, Rando TA, Hu H, Kao PN. 2005. NF90 regulates cell cycle exit and terminal myogenic differentiation by direct binding to the 3′-untranslated region of MyoD and p21WAF/CIP1 mRNAs. J Biol Chem 280: 18961–18989. [DOI] [PubMed] [Google Scholar]
- Shim J, Lim H, Yates JR, Karin M. 2002. Nuclear export of NF90 is required for interleukin-2 mRNA stabilization. Mol Cell 10: 1331–1344. [DOI] [PubMed] [Google Scholar]
- Singh R, Valcarcel J. 2005. Building specificity with nonspecific RNA-binding proteins. Nat Struct Mol Biol 12: 645–653. [DOI] [PubMed] [Google Scholar]
- Singh G, Kucukural A, Cenik C, Leszyk JD, Shaffer SA, Weng Z, Moore M. 2012. The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell 151: 750–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahmasebi S, Alain T, Rajasekhar VK, Zhang JP, Prager-Khoutorsky M, Khoutorsky A, Dogan Y, Gkogkas CG, Petroulakis E, Sylvestre A, et al. 2014. Multifaceted regulation of somatic cell reprogramming by mRNA translational control. Cell Stem Cell 14: 606–616. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson D, Sauvageau M, Loyal G, Rinn JL, Pachter L. 2012. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31: 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand EL, Huelga SC, Yeo GW. 2016. Experimental and computational considerations in the study of RNA-binding protein-RNA interactions. Adv Exp Med Biol 907: 1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandrey F, Montellese C, Koos K, Badertscher L, Bammert L, Cook AG, Zemp I, Horvat P, Kutay U. 2015. The NF45/NF90 heterodimer contributes to the biogenesis of 60S ribosomal subunits and influences nucleolar morphology. Mol Cell Biol 35: 3491–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Rao S, Chu J, Shen X, Lavasseur DN, Theunissen TW, Orkin SH. 2006. A protein interaction network for pluripotency of embryonic stem cells. Nature 44: 364–368. [DOI] [PubMed] [Google Scholar]
- Wang T, Xiao G, Chu Y, Zhang MQ, Corey DR, Xie Y. 2015. Design and bioinformatics analysis of genome-wide CLIP experiments. Nucleic Acids Res 43: 5263–5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe A, Yamada Y, Yamanaka S. 2012. Epigenetic regulation in pluripotent stem cells: a key to breaking the epigenetic barrier. Philos Trans R Soc Lond B Biol Sci 368: 20120292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolkowicz UM, Cook AG. 2012. NF45 dimerizes with NF90, Zfr and SPNR via a conserved domain that has a nucleotidyltransferase fold. Nucleic Acids Res 40: 9356–9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Blelloch R. 2014. Regulation of pluripotency by RNA binding proteins. Cell Stem Cell 15: 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You KT, Park J, Kim VN. 2015. Role of the small subunit processome in the maintenance of pluripotent stem cells. Genes Dev 29: 2004–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RA. 2011. Control of the embryonic stem cell state. Cell 144: 940–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.