Abstract
Free-living animals must not only regulate the amount of food they consume, but also choose which types of food to ingest. The shifting of food preference driven by nutrient-specific hunger can be essential for survival, yet little is known about the underlying mechanisms. We identified a dopamine circuit that encodes protein-specific hunger in Drosophila. The activity of these neurons is increased following substantial protein deprivation. Activation of this circuit simultaneously promoted protein intake and restricted sugar consumption, via signaling to distinct downstream neurons. Protein starvation triggered branch-specific plastic changes in these dopaminergic neurons, thus enabling sustained protein consumption. These studies reveal a crucial circuit mechanism by which animals adjust their dietary strategy to maintain protein homeostasis.
Beyond satisfying caloric needs, animals must also ingest nutrients that cannot be biosynthesized. The “specific appetite” hypothesis posits that nutrient-specific hunger drives homeostatic consumption of substances such as protein and salt (1–3). Protein is an indispensable macronutrient (4, 5), and is particularly required in anabolic states, such as infancy and pregnancy (6, 7). Recent studies in Drosophila have identified signaling mechanisms regulating protein hunger, particularly in the context of post-mating effects (8–12). However, specific circuits encoding protein hunger remain unknown.
Yeast is an ethologically-relevant protein food source for Drosophila, containing a negligible amount of sugars. Studies in fruit flies demonstrated that mated females have greater drive for protein and amino acid intake (9, 11, 13) (fig. S1, A to D). We thus used this potent physiological drive for protein in mated females as an entry point to investigate the neural basis of protein hunger. We assayed yeast feeding in mated females following conditional silencing of different neuromodulatory cell groups (14). Dopamine (DA) neurons were specifically required for yeast consumption (fig. S1, E and F) and preference (Fig. 1A and fig. S1G). To identify the specific DA neurons mediating this effect, we used two restricted Gal4 drivers (TH-C-Gal4 and TH-D-Gal4) and found that conditional silencing of the neurons labeled with either driver led to significantly reduced protein preference (Fig. 1A and fig. S1G). These drivers label largely non-overlapping DA cells, aside from 2 cells in each PPM2 (Protocerebral Posterior Medial) subgroup (15) (fig. S2, A and B).
Fig. 1. DA-WED neurons are necessary and sufficient for protein hunger.
(A) Preference Index (PI) for yeast vs sucrose in mated female flies with indicated genotypes (n=5–15 trials). (B) Whole-mount brain immunostaining with anti-GFP (green) and anti-Bruchpilot (BRP, nc82, magenta). Scale bar: 100µm. High-magnification sections of DA-WED neuron cell bodies are shown on the right with anti-GFP (green) and anti-TH (blue). Scale bar: 10 µm. (C) Whole-mount brain immunostaining with anti-GFP (green) and anti-DsRed (magenta). “M” and “L” denote Medial (M) and Lateral (L) branches, respectively. Scale bar: 50 µm. (D and E) PI (D) and yeast intake per fly (E) showing that silencing DA-WED neurons suppressed protein hunger in yeast-deprived male flies (n=8–18 and 18–33 trials for D and E). (F and G) Conditional activation of DA-WED neurons increased preference for (F) and intake of (G) yeast in male flies (n=7–13 and 12–27 trials for F and G). Simplified box plots show 25th, 50th, and 75th percentiles. For bar graphs, mean ± SEM is shown. In this and subsequent figures “*”, “**”, “***”, and “ns” denote P<0.05, P<0.01, P<0.001, and not significant, respectively.
To isolate these PPM2 neurons, we used an intersectional approach (TH-C-FLP with TH-D-Gal4) to drive expression of FRT-stop-FRT-mCD8-GFP and identified two PPM2 neurons in each hemisphere that project ventrally to the “Wedge” neuropil (Fig. 1B). Based on this projection pattern, we named these two PPM2 neurons DA-WED cells. Next, using the FINGR system (TH-D-Gal4, TH-C-FLP, tub-FRT-Gal80-FRT (WED1-Gal4)) (16) (fig. S2D) to express dendritic vs terminal markers, we found that the Wedge area contains the dendritic field, whereas the two dorsally bifurcating branches contain pre-synaptic terminals (Fig. 1C). Intersection between TH-C-Gal4 and another restricted driver TH-F3-Gal4 (15) (WED2-Gal4) also revealed the two DA-WED neurons (fig. S2, C and E). We specifically inactivated these cells with Kir2.1 and found substantial inhibition of yeast preference and consumption (fig. S3, A and B). General hunger, thirst, and salt appetite were not affected when DA-WED neurons were silenced (fig. S3, C–E).
We next asked whether the DA-WED neurons play a general role in homeostatic regulation of protein intake, beyond mated females. Although male flies did not prefer yeast at baseline (fig. S1, A–D), they exhibited significant protein preference and consumption following yeast deprivation for 8 days (9), which was fully suppressed if an amino acid mix was provided during the deprivation period (fig. S4, A and B). Inactivating the DA-WED neurons significantly reduced yeast preference and consumption in protein-deprived male flies (Fig. 1, D and E). Conversely, conditional activation (17) of these cells induced yeast preference and consumption in males (Fig. 1, F, G and fig. S4, C and D). Similar data were obtained for virgin females (fig. S4, E to L). Conditional silencing (fig. S5, A and B) or activation (fig. S5, C and D) of DA-WED neurons reduced or increased protein consumption, respectively, over a range of internal protein hunger states in both mated female and male flies, and so we chose to focus on male flies for subsequent experiments to avoid post-mating effects. Reducing DA levels in DA-WED neurons by knocking down the neuronal specific isoform of Tyrosine Hydroxylase (TH) in these cells (fig. S6, A and B) decreased yeast intake in protein-starved male flies (fig. S6C).
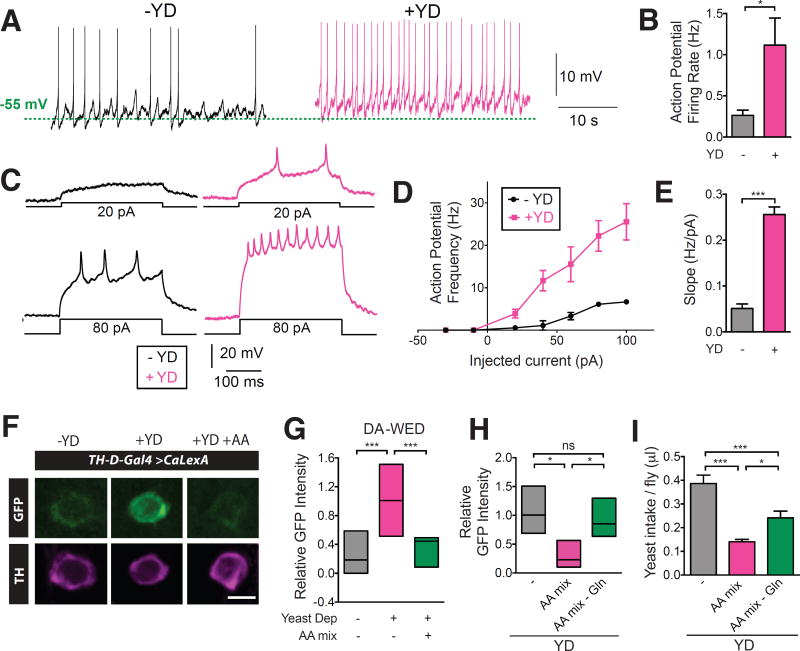
To investigate whether the activity of the DA-WED neurons correlates with protein need, we first performed perforated patch-clamp recordings (18) from these neurons (Fig. 2, A to E). Following yeast deprivation, the spontaneous action potential (AP) firing rate of DA-WED neurons increased ~4-fold compared to controls (Fig. 2, A and B). Moreover, evoked AP firing rates were higher at all measured depolarizing currents following yeast deprivation (Fig. 2, C to E). Protein starvation did not significantly alter the resting membrane potential or input resistance of these cells (fig. S7, A and B). Cytosolic Ca2+ levels within these cells (but not nearby PPM3 DA neurons), was substantially increased following yeast deprivation when monitored using the CaLexA (Calcium-dependent nuclear import of LexA) system (19) (Fig. 2, F and G, fig. S7, C and D). This increase in intracellular Ca2+ levels was suppressed when an amino acid mix was provided in the protein-deficient diet (Fig. 2, F and G). This effect was specific for protein, as general starvation did not affect CaLexA signal in the DA-WED neurons (fig. S7E). Similar data were obtained using GCaMP imaging, as an alternative method to measure cytosolic Ca2+ levels (fig. S7, F and G).
Fig. 2. Protein starvation increases activity of DA-WED neurons.
(A and B) Yeast deprivation increased spontaneous action potential (AP) firing rate from DA-WED neurons (n=7–9). (C to E), Mean AP frequency and f-I slope for evoked responses from DA-WED neurons was elevated following protein starvation (n=5). All recordings were from male flies. (F) Representative images of DA-WED neuron cell bodies from control, yeast-deprived, and yeast-deprived with amino acid supplemented TH-D-Gal4>CaLexA male flies. Scale bar: 5 µm. (G) CaLexA signal intensity from DA-WED cell bodies correlated with protein hunger status (n=12–18). (H and I) An amino acid mix lacking Gln failed to suppress the increase in DA-WED activity (n=4–5) and was less effective at inhibiting protein consumption following yeast deprivation (n=17–25 trials) in male flies.
What component of protein serves to signal protein satiety to the DA-WED neurons? We hypothesized that a specific amino acid may play this role. We first subjected flies to protein deprivation, but provided individual amino acids in the diet and then monitored the activity of DA-WED neurons using CaLexA. Tryptophan and glutamine (Gln) supplementation suppressed the enhanced activity of DA-WED neurons induced by protein deprivation, while proline and glutamate supplementation showed a trend towards this effect (fig. S8A). Next, we assessed whether supplementation of these 4 individual amino acids modulated homeostatic regulation of yeast intake. Yeast intake was significantly inhibited when Gln was added back to a protein-deficient diet (fig. S8B). Conversely, an amino acid mix lacking only Gln failed to suppress the increase in DA-WED activity and was less effective at inhibiting protein consumption following yeast deprivation (Fig. 2, H and I). We also found that Gln levels in the hemolymph were significantly reduced following protein deprivation (fig. S8C). Together, these data suggest that Gln in particular may be important for signaling protein deficiency.
Despite strong preference for sucrose at baseline, yeast deprivation caused flies to decrease their consumption of this nutrient (fig. S9A). Because DA-WED neurons are activated following substantial protein deprivation, we hypothesized that they also play a direct role in reducing sucrose intake under these conditions. Silencing DA-WED neurons using Kir2.1 significantly enhanced sugar intake after protein deprivation, whereas activating these neurons using dTrpA1 reduced sucrose consumption (Fig. 3, A and B and fig. S9C).
Fig. 3. Yeast and sucrose feeding are oppositely regulated by DA-WED neurons.
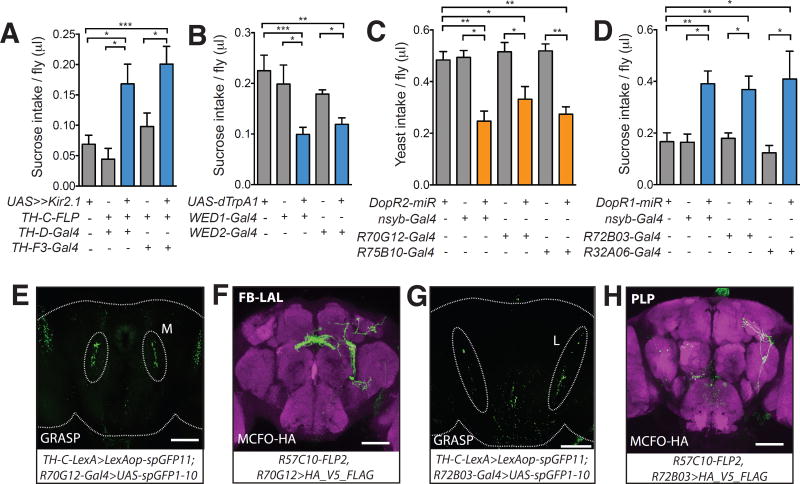
(A) Silencing DA-WED neurons increased sucrose intake in protein-deprived male flies (n=12–34 trials). (B) Conditional activation of DA-WED neurons reduced sucrose consumption in male flies (n=21–33 trials). (C) Knockdown of DopR2 using the drivers shown reduced yeast intake in protein-deprived male flies (n=18–47 trials). (D) Knockdown of DopR1 using the drivers shown elevated sucrose intake in protein-deprived male flies (n=15–40 trials). (E and G) Representative image from a GRASP experiment between DA-WED neurons and neurons labeled by R70G12-Gal4 or R72B03-Gal4. Native fluorescence from reconstituted GFP is shown. (F and H) Whole-mount brain image from Multicolor Flip-Out (MCFO) analysis for the driver lines shown stained with anti-HA (green) and anti-Bruchpilot antibodies (magenta). Brains in E–H are from male flies. Scale bar: 100µm.
We next investigated the mechanisms underlying the opposing effects of DA-WED neurons on sucrose and yeast intake. There are four DA receptors in Drosophila, DopR1, DopR2, D2R and DopEcR. Both DopR1 and DopR2 mutants exhibited a reduced preference for yeast following protein deprivation in a two-choice assay, which could reflect either a reduced preference for protein or a greater preference for sucrose (fig. S9B). Thus, we focused on these two receptors as potential targets acting downstream of the DA-WED neurons. In one-choice assays, DopR2 mutants exhibited significantly reduced yeast intake, while DopR1 mutants displayed a robust increase in sucrose feeding (fig. S9, D and E). Mutations in DopR2 and DopR1 also suppressed the increase in yeast consumption and decrease in sucrose feeding triggered by activating DA-WED neurons, respectively (fig. S9, F and G). We confirmed these findings by pan-neuronal knockdown of DopR1 or DopR2 (nsyb-Gal4>UAS-DopR1-miR or nsyb-Gal4>UAS-DopR2-miR) (fig. S9, H and I), which yielded similar results (Fig. 3, C and D). We therefore hypothesized that DopR2 and DopR1 act in discrete neuronal targets of the DA-WED cells to regulate protein and sugar feeding, respectively.
To identify putative candidate circuits acting downstream of the DA-WED cells, we performed a GRASP (GFP Reconstitution Across Synaptic Partners) (20) –based screen with Rubin Gal4 drivers derived from DopR2 and DopR1 enhancer sequences to assess connectivity with the DA-WED neurons (fig. S10A and S11A). Among DopR2-derived driver lines, only the R70G12-Gal4 line exhibited GRASP signal at the DA-WED terminal branches (specifically, the medial branch) (Fig. 3E). Moreover, knockdown of DopR2 using R70G12-Gal4 (fig. S10B) significantly reduced yeast intake following protein starvation (Fig. 3C), without affecting general hunger (fig. S12A) or sucrose consumption in yeast-deprived animals (fig. S12B). This reduction in protein appetite in R70G12-Gal4>UAS-DopR2-miR flies was observed over a range of protein hunger states in both mated female and male flies (fig. S12, C and D). To identify the specific cells in this driver that contact the DA-WED neurons, we conducted a MultiColor Flip Out (MCFO) experiment (21). We identified a neuron whose cell body is located in the posterior medial protocerebrum and sends its projections first to the fan-shaped Body (FB), before extending more anteriorly to the Lateral Accessory Lobe (LAL) area where the GRASP signal was observed (Fig. 3F). We therefore named these neurons “FB-LAL” cells based on this projection pattern. We identified an independent driver R75B10-Gal4 (fig. S13A) that also labeled the FB-LAL neuron and displayed GRASP signal at the medial branch of the DA-WED neurons (fig. S13B). Knockdown of DopR2 using R75B10-Gal4 decreased yeast intake in protein-starved flies (Fig. 3C). As expected, intersection between R70G12-Gal4 and R75B10-Gal4 drivers (R70G12-Gal4,R75B10-LexA>LexAop-FLP, UAS-FRT-stop-FRT-mCD8-GFP) revealed two pairs of FB-LAL neurons (fig. S13C). Silencing these cells reduced yeast intake in protein-starved flies, while activating these cells induced elevated yeast consumption in male flies (fig. S13, D–F). Activation of DA-WED neurons expressing ATP-gated P2X2 receptors (22) via ATP application induced a substantial increase in GCaMP signal in the cell bodies of downstream FB-LAL neurons (fig. S14A).
In contrast, among the DopR1-derived lines, R72B03-Gal4 driver demonstrated GRASP-signal at the DA-WED lateral branches (Fig. 3G). Knockdown of DopR1 using the R72B03-Gal4 (fig. S11B) driver triggered an increase in sucrose intake in protein-deprived flies (Fig. 3D), without affecting general hunger (fig. S12E) or yeast intake following protein starvation (fig. S12F). MCFO analysis using R72B03-Gal4 revealed a small subset of neurons in the Posterior Lateral Protocerebrum (PLP) that send their projections locally to the Ventrolateral and Superior neuropils, and so we named these cells PLP neurons (Fig. 3H). PLP neurons were also labeled by another driver line R32A06-Gal4 (fig. S15A). Similar to R72B03-Gal4, R32A06-Gal4 exhibited GRASP signal at the lateral branch of DA-WED neurons (fig. S15B). MCFO analysis using the new R32A06-Gal4 driver (fig. S15C) revealed cells whose cell body location and projection pattern match that seen with the PLP neuron identified in the R72B03-Gal4 driver (Fig. 3H). Knockdown of DopR1 using R32A06-Gal4 increased sucrose intake in protein-starved flies (Fig. 3D). Silencing or activating these PLP neurons using R72B03-Gal4 or R32A06-Gal4 led to a reduction or an increase, respectively, of sucrose intake (fig. S15, D–G).
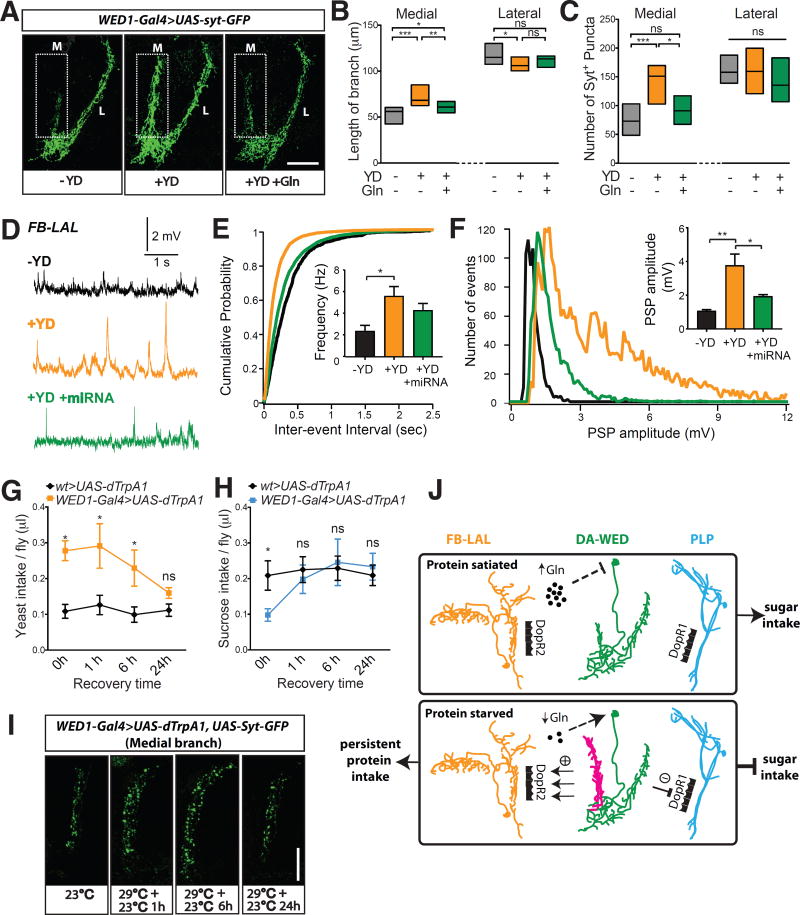
Following substantial deprivation of an essential nutrient, animals must engage in persistent behavior aimed at replenishing it. Plasticity of relevant neural circuits may underlie the persistent drive for motivated behaviors (18, 23). To address whether the DA-WED neurons undergo protein hunger-dependent plastic changes, we first assessed the morphology of their terminal branches using a membrane-tethered GFP (mCD8-GFP) (24) and GFP-tagged version of the synaptic vesicle-associated protein Synaptotagmin (Syt-GFP) (25). The medial, but not the lateral, branches were significantly elongated following protein deprivation, which was suppressed when Gln was selectively restored in the protein-deficient diet (fig. S16A and Fig. 4, A and B). Similar data were obtained for the number and total volume of Syt-GFP+ puncta (Fig. 4C and fig. S16B). To address whether the number of active zones also changed in the medial branches of DA-WED neurons following protein starvation, we expressed a truncated fragment of the active zone marker Bruchpilot (Brp-short) (26) in these cells. The number of BRP+ puncta in the medial, but not lateral, branch was significantly elevated when flies were protein-deprived, and this increase showed a trend towards being inhibited by Gln in the protein-deficient diet (fig. S16, C–E). We next used GRASP to determine whether protein starvation altered the connectivity between the DA-WED cells and their downstream targets. GRASP signal between the medial branch of the DA-WED neurons and the FB-LAL cells, but not the lateral branch and PLP neurons, was substantially elevated with protein starvation, and this increase was again suppressed if Gln was provided in the protein-deficient diet (fig. S16, F–H).
Fig. 4. Protein starvation triggers branch-specific plastic changes in DA-WED cells.
(A to C) DA-WED neuron medial, but not lateral, branch length and Syt+ puncta number were greater in yeast-deprived vs control male flies, and these effects were suppressed by Gln (n=17–20 for B and 25–56 for C). Scale bar: 50µm. (D to F) Sharp intracellular recordings of postsynaptic potentials (PSPs) from FB-LAL neurons in male flies (n=6–11). Yeast deprivation increased PSP frequency and induced a population of high amplitude PSPs, which was suppressed by knockdown of DopR2 in these neurons. (G to I) The increased yeast intake (G) and Syt+ puncta number in the medial branch (I), but not decreased sucrose intake (H), induced by activation of DA-WED neurons persisted for at least 6 hrs following cessation of heat treatment in male flies (n=7–25 trials for G and 9–16 trials for H). Scale bar: 25µm. (J) Model. At baseline (top), circulating Gln levels are high and act directly or indirectly on DA-WED neurons to suppress their activity, leading to greater sugar intake. Severe protein deprivation (bottom) reduces Gln levels, thus activating the DA-WED cells. Under these conditions, the medial branch (magenta) undergoes plastic changes, resulting in increased and persistent stimulation of the downstream FB-LAL neurons via DopR2 receptors to promote prolonged protein intake. Simultaneously, signaling from the lateral branch to downstream PLP neurons via DopR1 receptors induces a transient inhibition of sugar intake. Together, these changes result in a behavioral switch from consuming sugar to persistent intake of protein.
To characterize the functional consequences of the plastic changes in the DA-WED neurons, we performed sharp intracellular current-clamp recordings of the FB-LAL cells to measure frequency and amplitude of postsynaptic potentials (PSPs) (Fig. 4, D to F and fig. S17A). Protein deprivation induced a substantial increase in the frequency of PSPs in the FB-LAL neurons (Fig. 4E). Moreover, examination of the distribution of PSP amplitudes revealed the presence of high-amplitude PSPs solely in yeast-deprived animals (Fig. 4F). This population of high-amplitude PSPs likely reflects evoked responses seen only with protein deprivation and suggests that downstream signaling is markedly enhanced following protein starvation. Knockdown of DopR2 in FB-LAL cells significantly reduced the number of high-amplitude PSPs in yeast-deprived animals (Fig. 4, D–F), consistent with a model wherein the DA-WED neurons signal via DopR2 on the FB-LAL cells to promote protein consumption.
We next examined whether the plastic changes in the medial branches of the DA-WED cells led to a change in feeding behavior. We used dTrpA1 to activate the DA-WED cells and assessed yeast or sucrose consumption at different time points following cessation of heat treatment. dTrpA1 activation of the DA-WED neurons resulted in a prolonged increase in yeast consumption lasting at least 6 hrs (Fig. 4G) and induced persistent plastic changes in the medial, but not lateral branches, as revealed by Syt-GFP staining (Fig. 4I and fig. S17B). In contrast, while sucrose intake was reduced immediately after dTrpA1 activation, this response did not persist and was gone within 1h (Fig. 4H). After 24 hrs, both yeast consumption and terminal morphology of DA-WED neurons were indistinguishable from control flies (Fig. 4G and fig. S17B). Together, these findings demonstrate branch-specific plastic changes of the DA-WED neurons in response to substantial protein deprivation, providing a mechanism for the persistent hunger for protein, but not sugar, under these conditions (Fig. 4J).
Appropriate regulation of food consumption is essential for the survival of organisms that must navigate environments with variable and uncertain food availability and quality. A number of studies have investigated how animals reject diets devoid of essential amino acids (27, 28), and how monoaminergic and TOR/S6K signaling in Drosophila regulates mating-induced protein feeding (8, 9, 11, 12). However, little is known about the circuit mechanisms mediating the homeostatic regulation of protein intake. Our study suggests that protein hunger is encoded by the DA-WED neurons, providing a unique entree towards dissecting these mechanisms. Our findings suggest that glutamine, directly or indirectly, regulates the activity of these neurons, thus modulating behavioral responses to protein deprivation.
Our data suggest that the DA-WED neurons play a crucial role in homeostatic control of protein intake, functioning on multiple timescales to restore protein balance. Shortly after substantial protein deprivation, the DA-WED neurons act to mediate the behavioral switch between consumption of a food source preferred at baseline (sucrose) and the deprived nutrient (protein), by activating a dedicated “protein” feeding circuit, while simultaneously inhibiting a “sugar” feeding circuit. Over a longer timeframe, the DA-WED cells undergo branch-specific plastic changes that underlie the selective and persistent hunger for protein under these conditions. In the wild, these actions may correspond to promoting greater selectivity for protein in an initial foraging response, followed by maintenance of protein consumption after the protein food source has been identified.
Given that branch-specific plasticity has generally been described in post-synaptic dendrites (29, 30), our finding that the pre-synaptic terminals of the DA-WED neurons exhibit this phenomenon is highly unusual. The mechanisms underlying this process are currently unclear, but may depend upon extrinsic neuromodulatory influences that differentially regulate potential for plasticity of the distinct presynaptic terminals of the DA-WED cells. Further characterization of these and related circuit mechanisms should help delineate the fundamental principles governing protein-specific hunger. A better understanding of how animals choose to consume protein may also have implications for the treatment of obesity.
Supplementary Material
Acknowledgments
We thank Michael Pankratz, Hubert Amrein, Yun-Nung Jan, Christopher Potter, Bing Zhang, Barry Dickson, Mala Murthy, Stephan Sigrist, Kristin Scott, Serge Birman, Edward Kravitz, Jing Wang, Orie Shafer, and Gerald Rubin for kindly sharing reagents. We also thank the Bloomington Stock Center for fly lines. We thank John Schulze in the Proteomics Core of the Genome Center at the University of California, Davis for the amino acid analysis. We thank Audrey Branch for comments on the manuscript and Greg Suh and Makoto Kanai for discussion of unpublished data. This work was supported by NINDS Center Grant (NS05027) for use of the Core Machine Shop, a Japan Society for the Promotion of Science fellowship (M.T.), and NIH grants R01NS079584 and R21NS088521 and a Burroughs-Wellcome Career Award for Medical Scientists (M.N.W.). Data are stored on a password-protected computer and backed up on a hard drive located in the Department of Neurology at Johns Hopkins University. Data will be made available upon request.
References and Notes
- 1.Richter CP. Increased salt appetite in adrenalectomized rats. American Journal of Physiology. 1936;115:155–161. [Google Scholar]
- 2.Richter CP. Total self regulatory functions of animals and human beings. (Harvey Lecture Series, 1942–1943) 38 [Google Scholar]
- 3.Simpson SJ, Raubenbeimer D. The Nature of Nutrition. Princeton University Press; 2012. [Google Scholar]
- 4.Berthoud HR, Munzberg H, Richards BK, Morrison CD. Neural and metabolic regulation of macronutrient intake and selection. Proc Nutr Soc. 2012;71:390–400. doi: 10.1017/S0029665112000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rose WC. The Amino Acids in Nutrition. Yale J Biol Med. 1932;4:519–536. [PMC free article] [PubMed] [Google Scholar]
- 6.Deutsch JA, Moore BO, Heinrichs SC. Unlearned specific appetite for protein. Physiol Behav. 1989;46:619–624. doi: 10.1016/0031-9384(89)90341-7. [DOI] [PubMed] [Google Scholar]
- 7.White BD, Porter MH, Martin RJ. Effects of age on the feeding response to moderately low dietary protein in rats. Physiol Behav. 2000;68:673–681. doi: 10.1016/s0031-9384(99)00229-2. [DOI] [PubMed] [Google Scholar]
- 8.Corrales-Carvajal VM, Faisal AA, Ribeiro C. Internal states drive nutrient homeostasis by modulating exploration-exploitation trade-off. Elife. 2016;5 doi: 10.7554/eLife.19920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ribeiro C, Dickson BJ. Sex peptide receptor and neuronal TOR/S6K signaling modulate nutrient balancing in Drosophila. Curr Biol. 2010;20:1000–1005. doi: 10.1016/j.cub.2010.03.061. [DOI] [PubMed] [Google Scholar]
- 10.Ro J, et al. Serotonin signaling mediates protein valuation and aging. Elife. 2016;5 doi: 10.7554/eLife.16843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vargas MA, Luo N, Yamaguchi A, Kapahi P. A role for S6 kinase and serotonin in postmating dietary switch and balance of nutrients in D. melanogaster. Curr Biol. 2010;20:1006–1011. doi: 10.1016/j.cub.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walker SJ, Corrales-Carvajal VM, Ribeiro C. Postmating Circuitry Modulates Salt Taste Processing to Increase Reproductive Output in Drosophila. Curr Biol. 2015;25:2621–2630. doi: 10.1016/j.cub.2015.08.043. [DOI] [PubMed] [Google Scholar]
- 13.Toshima N, Tanimura T. Taste preference for amino acids is dependent on internal nutritional state in Drosophila melanogaster. J Exp Biol. 2012;215:2827–2832. doi: 10.1242/jeb.069146. [DOI] [PubMed] [Google Scholar]
- 14.Baines RA, Uhler JP, Thompson A, Sweeney ST, Bate M. Altered electrical properties in Drosophila neurons developing without synaptic transmission. J Neurosci. 2001;21:1523–1531. doi: 10.1523/JNEUROSCI.21-05-01523.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Q, Liu S, Kodama L, Driscoll MR, Wu MN. Two dopaminergic neurons signal to the dorsal fan-shaped body to promote wakefulness in Drosophila. Curr Biol. 2012;22:2114–2123. doi: 10.1016/j.cub.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bohm RA, et al. A genetic mosaic approach for neural circuit mapping in Drosophila. Proc Natl Acad Sci U S A. 2010;107:16378–16383. doi: 10.1073/pnas.1004669107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aso Y, et al. Mushroom body output neurons encode valence and guide memory-based action selection in Drosophila. Elife. 2014;3:e04580. doi: 10.7554/eLife.04580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu S, Liu Q, Tabuchi M, Wu MN. Sleep Drive Is Encoded by Neural Plastic Changes in a Dedicated Circuit. Cell. 2016;165:1347–1360. doi: 10.1016/j.cell.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masuyama K, Zhang Y, Rao Y, Wang JW. Mapping neural circuits with activity-dependent nuclear import of a transcription factor. J Neurogenet. 2012;26:89–102. doi: 10.3109/01677063.2011.642910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gordon MD, Scott K. Motor control in a Drosophila taste circuit. Neuron. 2009;61:373–384. doi: 10.1016/j.neuron.2008.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nern A, Pfeiffer BD, Rubin GM. Optimized tools for multicolor stochastic labeling reveal diverse stereotyped cell arrangements in the fly visual system. Proc Natl Acad Sci U S A. 2015;112:E2967–2976. doi: 10.1073/pnas.1506763112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lima SQ, Miesenbock G. Remote control of behavior through genetically targeted photostimulation of neurons. Cell. 2005;121:141–152. doi: 10.1016/j.cell.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 23.Hyman SE, Malenka RC. Addiction and the brain: the neurobiology of compulsion and its persistence. Nat Rev Neurosci. 2001;2:695–703. doi: 10.1038/35094560. [DOI] [PubMed] [Google Scholar]
- 24.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 25.Zhang YQ, Rodesch CK, Broadie K. Living synaptic vesicle marker: synaptotagmin-GFP. Genesis. 2002;34:142–145. doi: 10.1002/gene.10144. [DOI] [PubMed] [Google Scholar]
- 26.Fouquet W, et al. Maturation of active zone assembly by Drosophila Bruchpilot. J Cell Biol. 2009;186:129–145. doi: 10.1083/jcb.200812150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bjordal M, Arquier N, Kniazeff J, Pin JP, Leopold P. Sensing of amino acids in a dopaminergic circuitry promotes rejection of an incomplete diet in Drosophila. Cell. 2014;156:510–521. doi: 10.1016/j.cell.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 28.Efeyan A, Comb WC, Sabatini DM. Nutrient-sensing mechanisms and pathways. Nature. 2015;517:302–310. doi: 10.1038/nature14190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cichon J, Gan WB. Branch-specific dendritic Ca(2+) spikes cause persistent synaptic plasticity. Nature. 2015;520:180–185. doi: 10.1038/nature14251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fauth M, Tetzlaff C. Opposing Effects of Neuronal Activity on Structural Plasticity. Front Neuroanat. 2016;10:75. doi: 10.3389/fnana.2016.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.