Abstract
The recombinant protein Ultrabithorax (Ubx), a Drosophila melanogaster Hox transcription factor, self-assembles into biocompatible materials in vitro that are remarkably extensible and strong. Here, we demonstrate that the strength of Ubx materials is due to intermolecular dityrosine bonds. Ubx materials auto-fluoresce blue, a characteristic of dityrosine, and bind dityrosine-specific antibodies. Monitoring the fluorescence of reduced Ubx fibers upon oxygen exposure reveals biphasic bond formation kinetics. Two dityrosine bonds in Ubx were identified by site-directed mutagenesis followed by measurements of fiber fluorescent intensity. One bond is located between the N-terminus and the homeodomain (Y4/Y296 or Y12/Y293), and another bond is formed by Y167 and Y240. Fiber fluorescence closely correlates with fiber strength, demonstrating that these bonds are intermolecular. To our knowledge, this is the first identification of specific residues that participate in dityrosine bonds in protein-based materials. The percentage of Ubx molecules harboring both bonds can be decreased or increased by mutagenesis, providing an additional mechanism to control the mechanical properties of Ubx materials. Duplication of tyrosine-containing motifs in Ubx increases dityrosine content in Ubx fibers, suggesting these motifs could be inserted in other self-assembling proteins to strengthen the corresponding materials.
Keywords: Ubx, protein-based materials, crosslinking, mechanical properties
1. Introduction
Protein-based materials have the potential to be customized for a variety of applications, including drug delivery, tissue engineering, surgical sealants, medical imaging, biosensors, biofabrication, and biomineralization.[1,2] However, realization of these innovations requires development of a variety of materials with different structural, mechanical, and functional properties.[3] For instance, macroscale materials in medical applications must be biodegradable,[4–6] biocompatible,[7,8] and have mechanical properties matching the tissues of interest;[9,10] whereas materials destined for biofabrication must form rigid nanoscale three-dimensional structures.[11,12] The methods used to generate and process protein-based materials can have a substantial impact on both the mechanical and functional properties of the products.[13–18]
Recombinant production of proteins provides a renewable supply of monomers for assembly whose sequences, and hence properties, can be easily engineered.[19] Multiple approaches to rationally engineer or control the mechanical properties of materials formed from recombinant proteins have been explored, including chemical crosslinking, oxidation to form disulfide or dityrosine bonds, and incorporation of nanoparticles and metal films.[14,20–27] Of these approaches, oxidation to form disulfide or dityrosine bonds is particularly attractive because these bonds do not always require additional steps for materials synthesis. The reversibility of disulfide bonds enables materials’ strength and stability to be responsive to external conditions.[28,29] In contrast, dityrosine bonds are useful when the mechanical properties must be consistent in a variety of chemical environments, reflecting their inclusion in many natural and engineered materials, including resilin, silk, fibrinogen, keratin, elastin, and collagen.[30–40] In many cases, photocrosslinking is used to rapidly form dityrosine bonds throughout a material.[37,39,41,42] Dityrosine crosslinks have also been used to drive assembly of proteins or peptides that would not otherwise form materials, or to covalently link multiple proteins for materials assembly.[39,42] However, the specific amino acids that form these bonds in protein-based materials have not been identified, information that is vital for engineering the sequence to control bond formation, and hence the structure and mechanical properties of the resulting materials. Although in some materials, dityrosine bonds have been attributed to a single tyrosine residue in a repeated motif,[42–44] each tyrosine motif is equally likely to participate in dityrosine bonds, resulting in monomer-to-monomer variation in the location of these bonds that would further complicate bond identification and sequence engineering.
In this study, we investigated the formation of dityrosine bonds in materials composed of Ultrabithorax (Ubx), a recombinant Drosophila melanogaster Hox transcription factor. In vitro, Ubx monomers coalesce in aqueous buffers near neutral pH to form globular aggregates, which further rearrange at the air-water interface to form nanoscale fibrils.[45] Fibrils associate laterally to generate macroscopic films, which are the building blocks for various macroscale Ubx architectures such as fibers, sheets, and bundles.[46] Ubx materials have many useful properties, including cytocompatibility, biocompatibility, and nonimmunogenicity.[47,48] Ubx materials can be functionalized i) with full-length proteins via gene fusion,[49,50] ii) with DNA by sequence-specific recognition,[51] and iii) with nanoparticles by non-covalent surface interactions.[45] Finally, Ubx materials are strong and remarkably extensible.[52]
Ubx contains 15 tyrosines that are embedded in distinct regions of the amino acid sequence (Figure 1A). Therefore, Ubx has the potential to form unique, and thus identifiable, dityrosine bonds in materials. In this study, we demonstrate that Ubx materials oxidize to form three dityrosine bonds, two of which are mutually exclusive, and we identify the participating tyrosine residues. In fibers pre-assembled in the absence of oxygen, exposure to oxygen rapidly triggers dityrosine formation, with biphasic bond formation kinetics. Because all Ubx monomers within the materials do not form both possible bonds, the dityrosine content can be increased by removing competing interactions. Dityrosine content directly correlates with the strength of the materials, suggesting these bonds are intermolecular and providing a mechanism to genetically tune the mechanical properties of the materials. These data illuminate the role of tyrosine residues in the formation and structure of Ubx materials, provide vital information for engineering the mechanical properties of Ubx fibers, and suggest approaches to insert specific dityrosine bonds into the sequence of other materials-forming proteins.
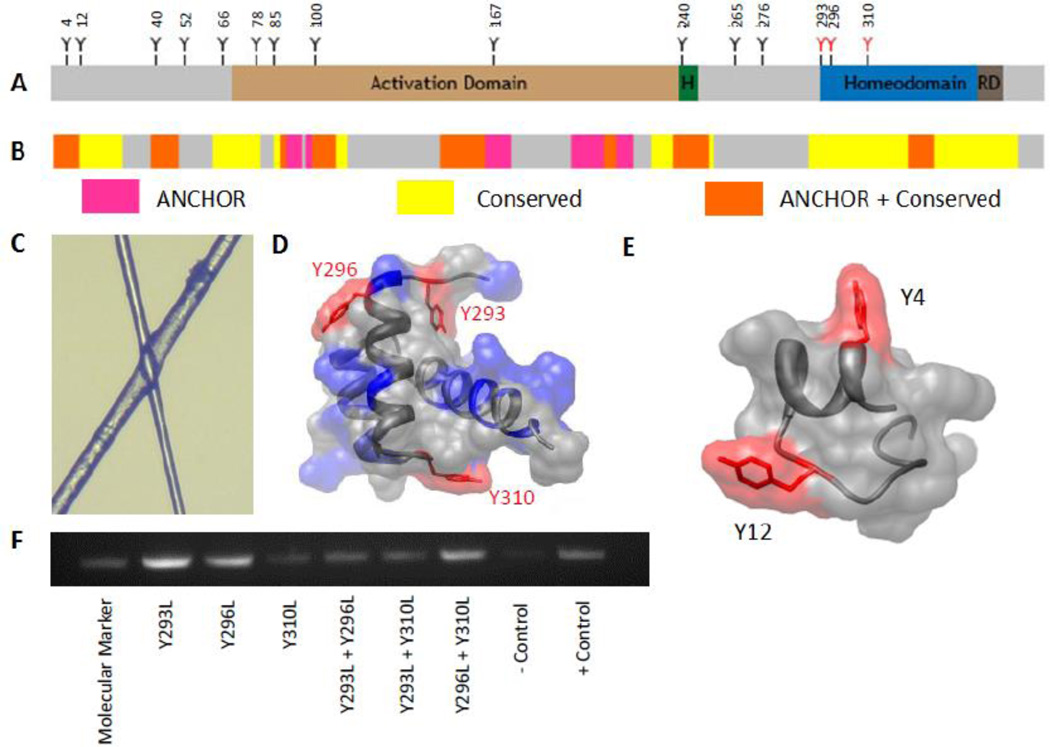
Figure 1. Location and functionality of Ubx tyrosine mutants.
A. Sequence schematic of Ubx, showing the location of tyrosine residues relative to functional domains and structural motifs. B. ANCHOR schematic showing areas of disorder in Ubx. C. Light microscopy of overlapping fibers shows that they are transparent and can diffract light. D. The three tyrosines in the Ubx homeodomain all lie on the surface of the domain (pdb: 1B8I).[68] E. Tyrosines 4 and 12 are not buried within this portion of Ubx. F. DNA binding data showing that the homeodomain remains functional in tyrosine mutants.
2. Results and Discussion
2.1. Ubx fibers are not amyloid
Elucidating the structure of protein-based materials is the first step toward understanding, and ultimately manipulating, the mechanical properties of these materials. In contrast to amorphous protein aggregates which often appear as white flocculates, Ubx materials are transparent (Figure 1C) and can diffract light,[45] suggesting a more regular structure. Since Ubx does not form materials as part of its natural function, one possibility is that Ubx fibers are amyloid, thus accounting for their transparency and strength.[53] However, X-ray diffraction and Thioflavin T binding studies of Ubx materials lack any indications of amyloid structure (data not shown). Furthermore, a large fraction of Ubx is extremely glycine-rich,[54] and thus is unlikely to form amyloid. The major structured region of Ubx is its DNA binding homeodomain, whose function is retained in the materials (Figure 1A, D),[51] suggesting that the helical structure of the homeodomain is likely intact as well. If both the unstructured and the structured regions of Ubx are unlikely to form amyloid, then amyloid structure cannot be responsible for the strength of Ubx materials.
2.2. Ubx materials contain dityrosine
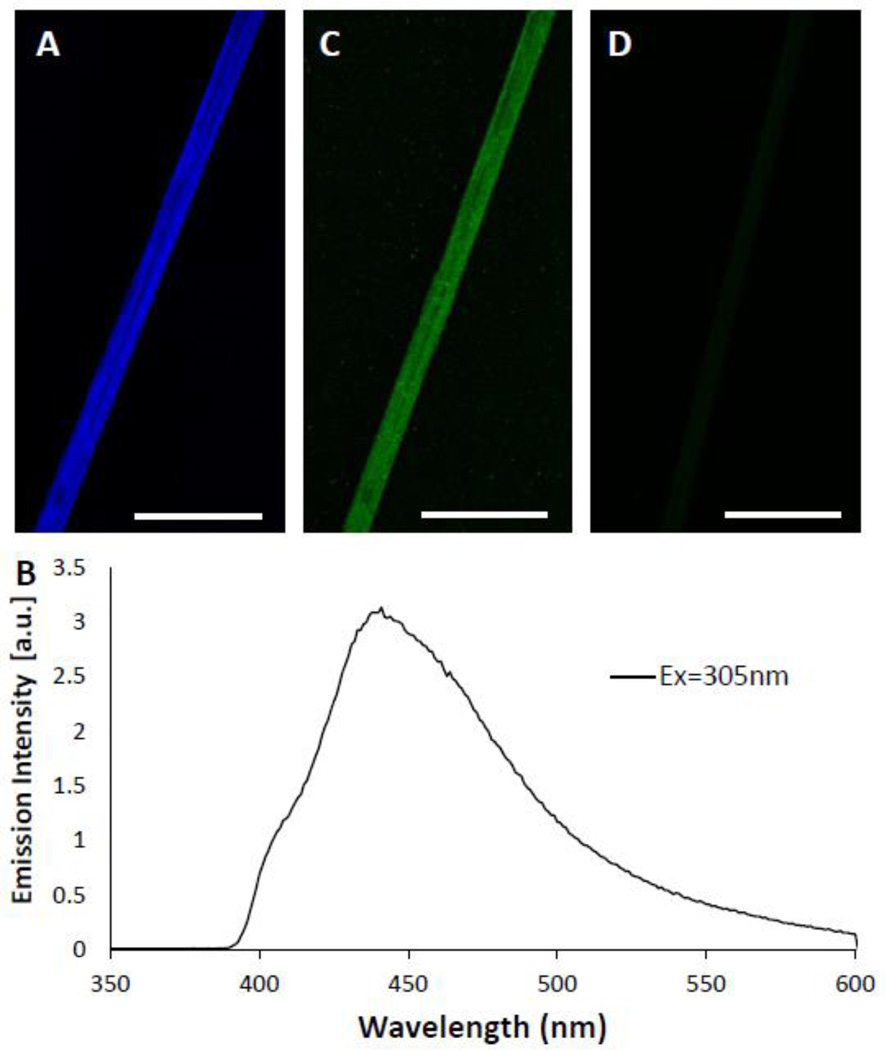
Intermolecular covalent crosslinks could also account for the strength of Ubx materials. Many natural materials rely on covalent crosslinks for strength,[31–36,55,56] and engineering covalent bonds into recombinant protein materials can dramatically improve both strength and assembly.[38] During fluorescent microscopy experiments, we observed that Ubx materials autofluoresce when excited at 305 nanometers (nm) (Figure 2A). The fluorescence emission spectrum (Figure 2B) corresponds with dityrosine, formed by oxidation of two tyrosine residues.[27,55,57,58] The emission maximum of dityrosine typically ranges from 410 nm to 430 nm.[58,59] For Ubx fibers, the emission peak is more red-shifted (438 nm). This difference may be due to proximity of the dityrosine bonds to positively charged amino acids, which can red-shift the emission spectra of aromatic amino acids by tens of nanometers.[60] Indeed, Ubx has a predicted net charge of +9, and the Ubx homeodomain, which contains 3 tyrosines, has a predicted net charge of +11 (Figure 1D).[45,50] Anti-dityrosine antibodies specifically recognize Ubx fibers in immunohistochemistry experiments, thus confirming the presence of dityrosine in Ubx materials (Figure 2C). The secondary antibodies alone are unable to bind fibers in the absence of primary antibodies (Figure 2D), supporting the specificity of the interaction. Together, the fluorescence and immunohistochemistry data demonstrate that dityrosine is present in Ubx fibers.
Figure 2. Ubx materials contain dityrosine.
A. Fibers autofluoresce blue. B. The Ubx emission spectrum, with a peak at 438 nm when excited at 305 nm is similar to other dityrosine containing proteins.[58–60] C. Immunofluorescence of Ubx fiber demonstrating anti-dityrosine primary antibodies recognize a Ubx fiber. D. A negative control experiment with the primary antibody omitted demonstrates that secondary antibodies do not adhere non-specificially to Ubx fibers. Scale bar equals 30 µm in all panels.
2.3. Measuring the kinetics of dityrosine bond formation
Tyrosine must oxidize to form dityrosine bonds; therefore, the kinetics of dityrosine bond formation can be monitored by assembling Ubx in a low oxygen environment and then exposing the resulting fibers to oxygen. To this end, Ubx was allowed to assemble in an argon-atmosphere glove box. Because films could be assembled and fibers could be drawn from films in this low oxygen environment, dityrosine bond formation is clearly not required for Ubx assembly. However, it is worth noting that these fibers were quite fragile and very difficult to handle.
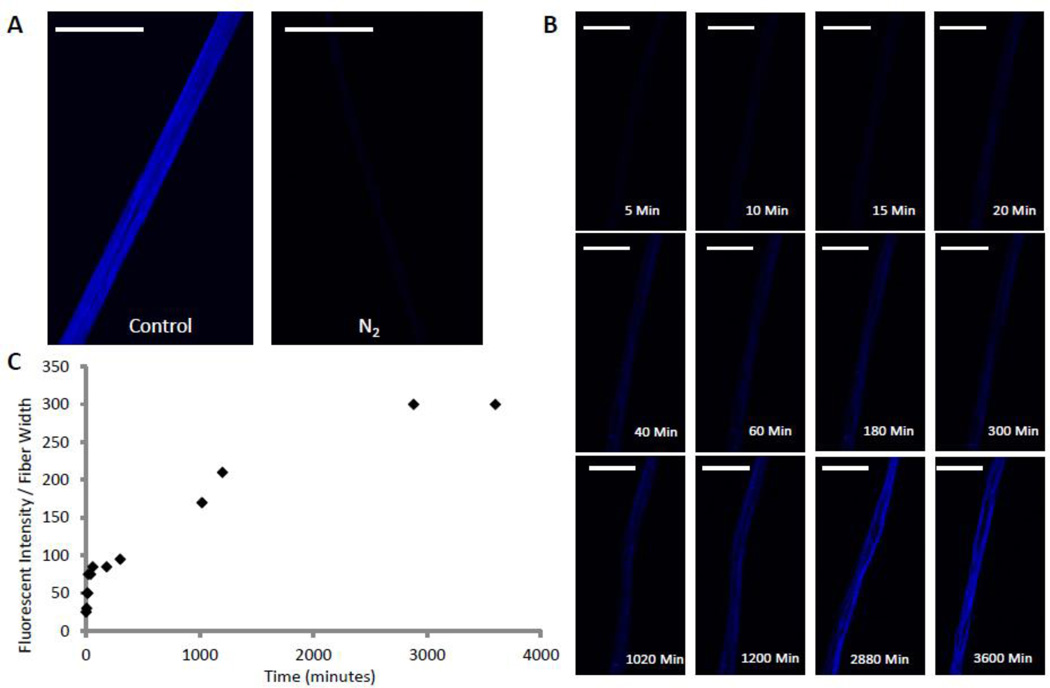
Fibers were placed in a custom imaging chamber (Figure S1) under nitrogen gas flow to maintain a low oxygen environment during transfer of the chamber from the glove box to a microscope. Cover slips on the top and bottom of the chamber allowed fibers inside the chamber to be analyzed by fluorescence microscopy. In a nitrogen environment, the blue dityrosine signal was nearly undetectable (Figure 3A). However, once the nitrogen gas in the chamber was replaced with air, the fibers gradually began to fluoresce blue (Figure 3B). Measurement of fluorescent intensity over time reveals two distinct transitions (Figure 3C): a fast initial transition occurring in a few minutes followed by a slow transition requiring days.
Figure 3. The kinetics of dityrosine bond formation reveals two transitions.
A. A fluorescent miscroscopy image of a Ubx fiber pulled and imaged in low oxygen environment N2 compared to a fiber pulled in normal atmosphere (control). B. Time-lapse images of a fiber pulled in low O2 after exposure to normal atmosphere. C. Graph of fluorescent intensity of the fiber shown in panel B over time showing 2 distinct transitions caused by dityrosine bond formation. Scale bar equals 30 µm in all panels.
2.4. Mutagenesis strategy
The presence of dityrosine provides an opportunity to manipulate the properties of Ubx materials by controlling dityrosine bond formation. To do so, the number of dityrosine bonds formed and the identity of the tyrosine residues that participate in these bonds must be determined. Because Ubx is produced as a recombinant protein in E. coli, we were able to use site-directed mutagenesis to identify participating tyrosines. This approach would be challenging to apply to many natural proteins that form materials. Because the amino acid sequence surrounding tyrosine residues impacts dityrosine bond formation,[59] tyrosines located in repeating motifs in natural materials should have equal probabilities of participating in a dityrosine bond.[43,44] In contrast, the unique sequences surrounding tyrosines in Ubx should lead to preferential interactions between specific tyrosines and thus consistent formation of the same dityrosine bonds. Furthermore, Ubx monomers rely on specific, long-range intramolecular interactions to regulate DNA binding.[54,61] These interactions involve regions of the protein containing tyrosine. Therefore, any intermolecular dityrosine bonds based on these interactions should form between specific residues.
A complication of the site-directed mutagenesis approach stems from the fact that Ubx has 15 tyrosine residues. The identity of tyrosines contributing to a single bond may vary, and more than one dityrosine bond may be present in the materials, creating an enormous array of possible bond arrangements. We narrowed our initial search based on interactions formed by Ubx monomers in DNA binding. When bound to DNA, Ubx can oligomerize in multiple orientations: side-to-side cooperative interactions when binding to linear DNA, and back-to-back interactions between clusters of cooperatively bound Ubx proteins to form the stem of a DNA loop.[62] Because Ubx fibers retain the ability to bind DNA (Figure 1F),[51] it is possible that interactions used on a small scale to enable cooperative DNA binding and DNA loop formation in vivo may also be applied on a much larger scale to form Ubx materials in vitro: side-to-side interactions to form nanoscale fibrils, and back-to-back interactions to allow the fibrils to interact to form films and fibers. Therefore, our first criterion for selecting tyrosines for mutagenesis was that the tyrosine should be located in a region important for regulating DNA binding.[54,61] Extending this logic, any tyrosine important for DNA binding is also expected to be evolutionarily conserved (Figure 1B), our second criterion. Next, for specific dityrosine bonds to form, the tyrosines would need to be embedded in regions of Ubx likely to participate in protein-protein interactions. The location of molecular recognition features, motifs capable of engaging in protein interactions, was predicted by the ANCHOR algorithm (Figure 1B).[63,64] It is important to note that this algorithm only identifies motifs located in intrinsically disordered regions; thus, it cannot provide information about the structured homeodomain. Finally, as demonstrated by the fragility of fibers drawn in a low-oxygen environment, dityrosine bonds significantly strengthen Ubx materials. Because fiber strength is one factor that determines the length of fibers that can be drawn from film, we reasoned that tyrosine residues, lost through truncation of the Ubx sequence, would shorten the average length of fibers produced by that Ubx variant. Fiber lengths were previously measured for a series of Ubx N-terminal and C-terminal truncation mutants.[46] This data provided the fourth criterion for selecting tyrosines for mutagenesis.
The ability of each of the 15 tyrosines in Ubx to meet these criteria is summarized in Table 1. Based on the logic described above, we hypothesized that tyrosines 4, 12, 100, 167, and 240 were most likely to be involved in dityrosine bonds. The three tyrosines on the surface of the homeodomain (HD) (293, 296, and 310; Figure 1A, D) were also selected because the homeodomain participates in long-range interactions with much of the rest of the protein,[54,61] and because the dityrosine spectrum is red-shifted. Conversely, tyrosines 40, 52, 66, 78, 85, 265, and 276 were deemed less likely candidates. The goal of our mutagenesis study was to remove the ability to form crosslinks, while retaining as much of the chemical nature of tyrosine as possible to prevent mutagenesis-induced structural rearrangements. Tyrosines in intrinsically disordered regions outside the homeodomain were mutated to serine, because the transfer coefficient of serine best mimics that of tyrosine as a free amino acid, leading to their similar values on the Kyte-Doolittle hydropathy scale.[65] Tyrosines within the homeodomain were mutated to leucine, because leucine most closely resembles the hydropathy of tyrosine on the surface of a protein.[66] These mutations do not alter the structure or function of the homeodomain, because fibers composed of homeodomain mutants can successfully bind DNA (Figure 1F). To confirm that hydrophobic patches created by the tyrosine to leucine mutants on the homeodomain surface were not causing a loss of fluorescence due to altered interactions with the rest of the protein, we also changed these three residues to serine. All Ubx variants carrying mutations in the homeodomain were able to form fibers which bound DNA (Figure 1F). Circular dichroism spectra of materials composed of wild-type Ubx and these mutants are similar (Figure S2), indicating the structure of the fibers was not significantly perturbed. Furthermore, for each position, the serine and leucine mutations had a similar impact on dityrosine fluorescence (Figure S3). Therefore, leucine mutations do not cause unanticipated effects on the structure of Ubx materials.
Table 1.
Criteria used to select tyrosines for mutagenesis studies.
| Tyrosine | Regulating DNA bindinga |
Sequence conservationb |
Anchorc | Fiber lengthd |
Selected? |
|---|---|---|---|---|---|
| 4 | Yes | Yes | Yes | Yes | Yes |
| 12 | Yes | Yes | No | Yes | Yes |
| 40 | Yes | Yes | Yes | No | No |
| 52 | No | No | No | No | No |
| 66 | No | Yes | No | No | No |
| 78 | No | Yes | No | No | No |
| 85 | No | No | No | No | No |
| 100 | Yes | Yes | Yes | Yes | Yes |
| 167 | Yes | No | Yes | Yes | Yes |
| 240 | Yes | Yes | Yes | N/A | Yes |
| 265 | Yes | No | No | N/A | No |
| 276 | Yes | No | No | N/A | No |
| 293 | Homeodomain | Yes | N/A | No | Yes |
| 296 | Homeodomain | Yes | N/A | No | Yes |
| 310 | Homeodomain | Yes | N/A | No | Yes |
2.5. Tyrosines that regulate DNA binding in Ubx monomers also participate in dityrosine bonds in Ubx fibers
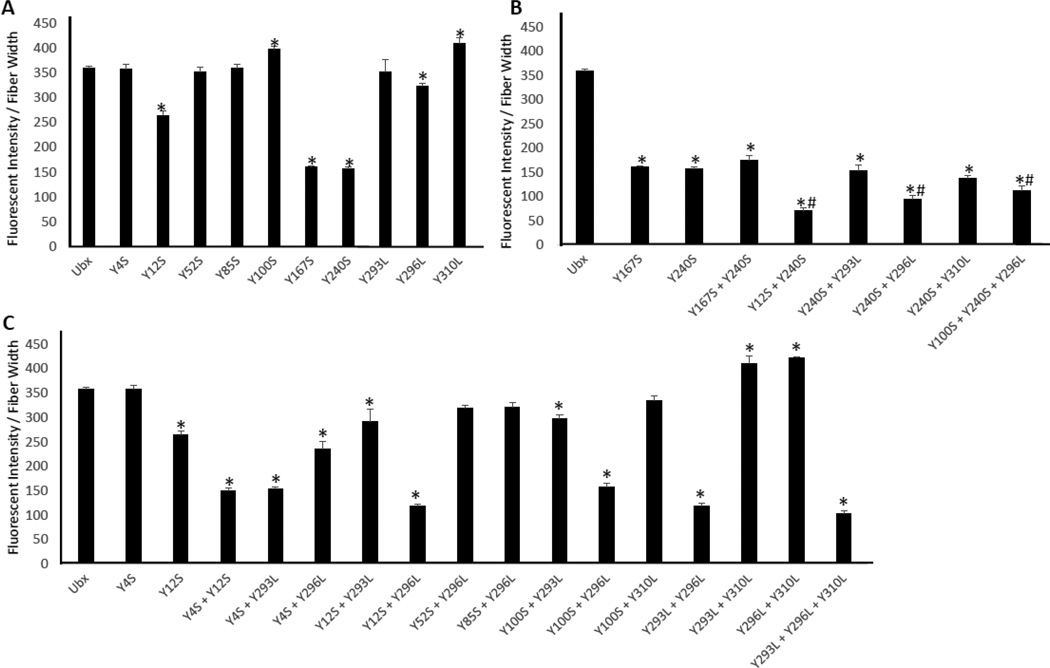
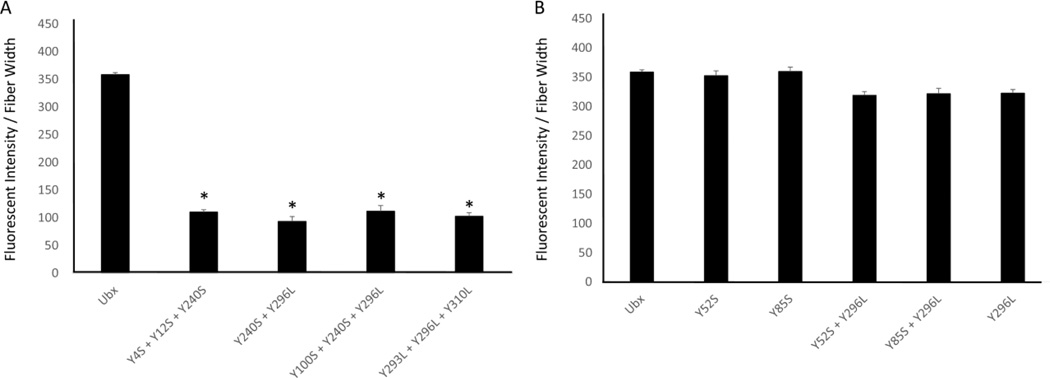
To find tyrosines involved in dityrosine bonds, we tested whether single mutations of the selected tyrosines reduce Ubx fiber fluorescence. Of these eight mutants, the fluorescence from Y12S, Y167S, Y240S, and Y296L mutants significantly decrease while the intensities from Y100S and Y310L mutants increase when compared to wild-type Ubx fiber fluorescence (Figure 4A; p ≤ 0.01 indicated by *). The intensity of blue fluorescence corresponds very well with immunostaining using the anti-dityrosine primary antibody (Figure S4, r2 = 0.99), confirming that changes in fiber fluorescence directly correspond to alterations in dityrosine content. The Y167S and Y240S mutations both reduce fluorescence to a similar degree, suggesting that Y167 and Y240 participate in the same dityrosine bond. To test this hypothesis, we created a Y167S + Y240S double mutant. If these residues participate in different dityrosine bonds, the loss of fluorescence should be additive. If Y167 and Y240 contribute to the same bond, then removing the second tyrosine should not cause an additional reduction in fluorescence. No further reduction in fluorescence was observed for the Y167S + Y240S double mutant (Figure 5B, S5, Table S1), suggesting that Y167 and Y240 form a single dityrosine bond (Figure 6A). This assignment is supported by the fact that double mutants combining Y240S with other affected tyrosines (for instance, Y12S + Y240S and Y240S + Y296L) all fluoresce less than the isolated Y240S mutant, indicating that Y12 and Y296 participate in a different bond than Y240 (Figure 4B; p ≤ 0.04 indicated by #). The Y167S + Y240S bond is responsible for a significant portion of the observed fluorescence in Ubx fibers (200/360 fluorescence units/µm). Finally, fibers composed of the Y293 mutant fluoresce to a similar extent as wild-type Ubx fibers. The fluorescence of the Y240S + Y293L mutant fibers is similar to that of Y240S mutant fibers. Thus, the Y240 mutant does not uncover any hidden contributions of Y293 to the Y167 + Y240 dityrosine bond.
Figure 4. Graph of fluorescent intensity divided by fiber width.
A. Single mutations of the 8 tyrosines predicted to be involved show that only Y12S, Y167S, Y240S, Y296L, and Y310L show a significant difference (p≤0.01 indicated by *) in fluorescent intensity when compared to Ubx. B. Comparison of mutants containing Y240S shows that Y240 binds Y167. Comparisons to Ubx are indicated by * (p=0.005) and to Y240 indicated by # (p=0.04) using t-tests. C. Comparison of mutants containing combinations of N-terminal (Y4 and Y12), Y100, and homeodomain mutants.
Figure 5.
A. The mutants Y4S + Y12S + Y240S, Y240S + Y296L, Y100S + Y240S + Y296L, and Y293L + Y296L + Y310L all show a loss of 250 fluorescent units per µm when compared to Ubx suggesting a loss of more than one tyrosine bond (p=0.005 indicated by *). B. Mutation of tyrosines Y52 and Y85 which were not predicted to be involved in dityrosine bonds has no effect, either in wild-type Ubx or in the Y296 background.
Figure 6.
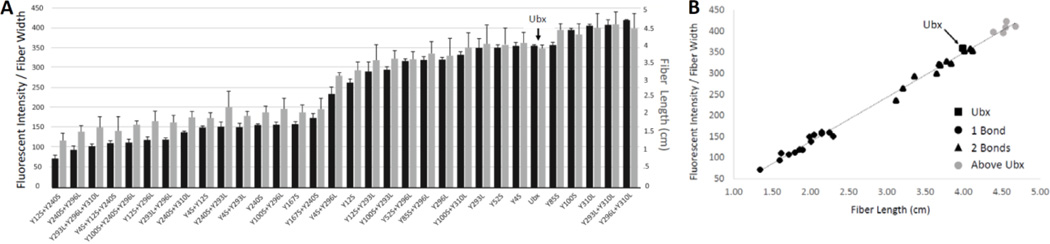
A. Graph of mutants from smallest to largest fluorescent intensity normalized to fiber width (black bars) and fiber length (grey bars). B. Scatter plot of fluorescent intensity compared to fiber length using linear regression with a coefficient of determination of 0.994.
Since all of the fluorescence cannot be attributed to the Y167/Y240 bond, at least one other bond is present. This additional bond(s) contributes less to the total fluorescence, indicating every monomer in the materials does not participate in this bond(s). This hypothesis is confirmed by the fact that Y12S and Y296L mutations decrease fluorescence while Y310L increases fluorescence when compared to wild-type Ubx (Figure 4A). The impact of mutagenesis varies between these three residues; therefore, either i) there are multiple additional bonds, ii) there is one bond, but tyrosines that do not engage in the bond contribute to a chemical environment that regulates bond formation, iii) there is one bond formed by different tyrosine residues in different monomers, or iv) some combination of these possibilities.
Mutation of tyrosines 12, 296, and 310 also alter fiber fluorescence. These residues are located in two regions of Ubx: the N-terminus and the homeodomain, suggesting a dityrosine bond may form between these two regions. Indeed, the sequence conservation of both the N-terminus and the homeodomain in Hox proteins[67] suggests these regions may interact. Furthermore, the N-terminus has a large impact on the DNA-binding affinity of the homeodomain.[48] Finally, the ANCHOR algorithm identifies the N-terminus as a region likely to engage in protein interactions.
Any bond or bonds between the N-terminus and the homeodomain could also involve two other tyrosines: Y4 and Y293. For any of these tyrosines to participate in dityrosine bond formation, they must be exposed to the solvent. X-ray crystallography data of the Ubx homeodomain[68] reveals Y293, Y296, and Y310 are all located on the homeodomain surface (Figure 1D). Likewise, a model of the structure of the N-terminus suggests both Y4 and Y12 are also solvent exposed (Figure 1E). Although any of these surfaces could potentially pack against other regions of Ubx, the extreme flexibility of the intervening intrinsically disordered regions suggests that these residues are likely to be occasionally exposed.
To determine which, if any, of these residues participate in dityrosine bond formation, we created a series of double and triple mutants involving these five amino acids. First, we assessed the role of the three tyrosines on the surface of the homeodomain (Y293, Y296, Y310). We have already established that the Y296L and Y310L mutants alter fluorescence (Figure 4A). In addition, we find that the Y293L mutation, when combined with Y296L, causes an additional loss of fluorescence (Figure 4C, S5). The difference in fluorescence between wild type fibers and Y293L + Y296L fibers is similar to the difference between wild type and Y240 fibers, and thus is equivalent to the loss of one bond. This suggests that either Y293 or Y296 can contribute one tyrosine to a single bond. This interpretation also explains why the Y293L mutation in isolation had no impact on fluorescence: Y296 provided an effective substitute.
Based on the logic described above, the other half of this bond may originate from the N-terminus of Ubx. Although the single Y4S mutation does not impact fiber fluorescence, Y4S in combination with Y12S significantly reduces fluorescence relative to Y12S fibers (Figure 4C). Thus, Y4 also impacts dityrosine content. The Y4 + Y12 scenario is similar to the one described above for Y293 + Y296: either Y4 or Y12 can participate in the dityrosine bond. Within a single fiber, different Ubx molecules may form a bond between the N-terminus and the homeodomain using different combinations of residues 4, 12, 293, and 296. However, the different chemical environments surrounding these residues should make some tyrosine pairs more likely to form a dityrosine bond. Comparison of pairs of double mutants should reveal if there are preferential interactions between 4 or 12 and 293 or 296. The Y4S + Y12S mutant removes all tyrosines from the N-terminus, and therefore prevents any possibility of forming a dityrosine bond with the homeodomain. The Y4S + Y293L double mutant had a similar level of fluorescence as Y4S + Y12S, suggesting neither possible N-terminus/homeodomain bond could form and thus Y4S does not bind Y293L. Therefore Y4 must bind Y296. Consistent with this conclusion, the fluorescence of Y4S + Y296L (one possible bond lost) was higher than Y4S + Y12S (both possible bonds lost), reflecting the fact that the Y12 and Y293 can still form a bond. Likewise, the Y12S + Y296 fibers fluoresce similar to Y4S + Y12S fibers, and with much less intensity than Y12S + Y293L fibers. Therefore Y12 binds Y293. Together, these results indicate that either a Y4/Y296 bond forms, or a Y12/Y293 bond forms.
Interestingly, the fluorescence of many variants involving Y310 (Y310L, Y293L + Y310L, and Y296L + Y310L) fibers increased relative to wild-type Ubx fibers. If Y310 quenched fluorescence of a dityrosine bond or induced a structure in which a dityrosine bond was quenched, then the same number of dityrosine bonds should be present in the wild type protein and the Y310 mutant. Consequently, immunofluorescence, using the anti-dityrosine antibody, should remain the same as for the fibers composed of wild-type Ubx. Instead, removal of Y310 increases immunofluorescence in proportion to the increase in dityrosine fluorescence. This increase in immunofluorescence was not only observed for Y310L fibers, but also for other pairs of Ubx variants in which the only difference is the presence or absence of the Y310 mutation. Therefore, removal of Y310 must increase the average number of dityrosine bonds formed per molecule of Ubx. These results suggest that Y310 acts as a decoy, in which tyrosines can interact with Y310, but not form a dityrosine bond. Removal of Y310 prevents Y4 and Y12 from forming unproductive interactions and thus increases the percentage of monomers that participate in a dityrosine bond.
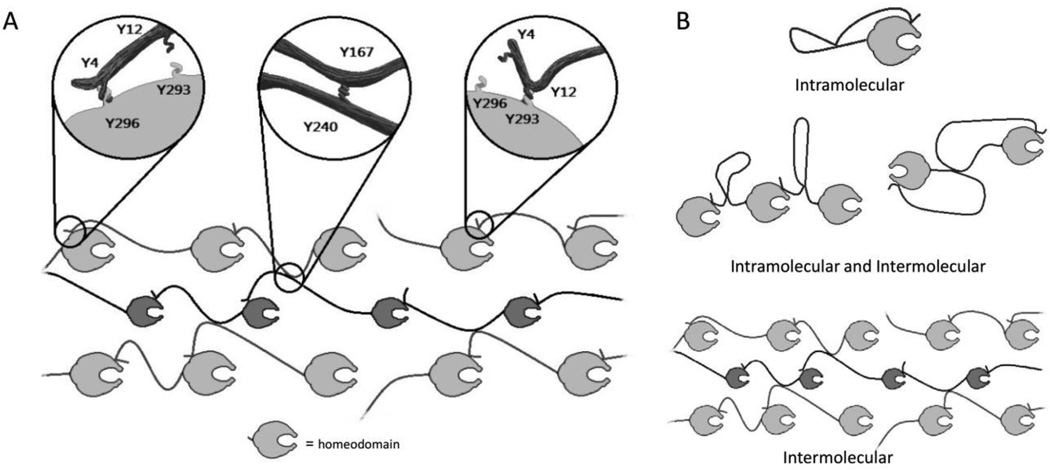
In the resulting model (Figure 6A), the N-terminus of Ubx (Y4 or Y12) interacts with the homeodomain (Y293, Y296, or Y310), but can only form a dityrosine bond with Y293 or Y296; while Y167/Y240 forms a second bond. Two separate bonds must form, because mutation of tyrosines from at least two groups results in a greater loss of fluorescence than removing multiple amino acids attributed to a single bond (Figure 5A; p<0.005 indicated by *).
Finally, plotting all mutants in order of increasing fluorescence clearly reveals two distinct transitions, corresponding to the presence of 0, 1, or 2 dityrosine bonds (Figure 7A). If this model is a complete description of the N-terminus/homeodomain interaction, then the fluorescence of Y4S + Y12S (150 units/µm) should equal that of Y293L + Y296L (100 units/µm). The discrepancy between these measurements may be due to differential contributions of Y100 to dityrosine bond formation. As a single mutant, Y100S increases fluorescence relative to wild-type Ubx, suggesting it is a decoy, like Y310, rather than a participant in dityrosine bond formation. However, double mutants of Y100S with Y293L, Y296L, or Y310L all fluoresce less than the corresponding Y293L, Y296, or Y310L single mutants (Figure 4D). Thus, Y100 may contribute to a chemical environment that can either aid dityrosine bond formation or act as a decoy, depending on the Ubx variant. In this model, differential contributions of Y100 and Y310 account for the differences in the fluorescence of Y4S + Y12S and Y293L + Y296L fibers.
Figure 7.
A. Artistic representation of proposed dityrosine bonds. B. Artistic representation options for intramolecular or intermolecular bonds.
2.6. Regions that do not regulate DNA binding in Ubx monomers also do not participate in dityrosine bonds in Ubx fibers
The data presented thus far only tested the tyrosines we selected based on involvement in DNA binding, conservation, predicted ability to participate in protein interactions, and location in a region that impacts fiber length (Table 1). To determine whether tyrosines outside of our selected group can also contribute to bond formation, we created the Y52S and Y85S mutants. Neither single mutant had any effect on fiber fluorescence (Figure 5B). However, the contributions of Y4 and Y293 were only apparent when mutated in combination with other tyrosines. Therefore we mutated Y52S and Y85S in conjunction with Y296L, a mutation that was able to uncover the contributions of both Y4 and Y293. The fluorescence of Y52S + Y296L and Y85S + Y296L mutant fibers was similar to the single Y296L mutant (Figure 5B). Therefore, Y296L does not reveal a hidden contribution of either Y52 or Y85, and these residues do not contribute to dityrosine bond formation.
2.7. Only two dityrosine bonds are formed by Ubx in materials
Both the mutant data and the kinetic data reveal two transitions, suggesting no more than two bonds are present (Figure 3, 6A). However, the mutants tested thus far do not eliminate absolutely all fluorescence from Ubx fibers. The remaining fluorescence could be due to random dityrosine bonds formed by the remaining amino acids, or it could be evidence of a third dityrosine bond. However, if our hypothesis is correct and Y167 and Y240 always participate in dityrosine bonds with each other, then one dityrosine bond contributes approximately 200 intensity units/µm, on our scale. Likewise, the Y4S + Y12S mutations completely remove the other dityrosine bond, also resulting in a loss of approximately 200 intensity units per µm. The maximum intensity observed, close to 400 intensity units/µm, was observed for the Ubx variants Y100L, Y310L, Y293L+Y310L, and Y296L+Y310L. Therefore, if a single bond is worth 200 units/µm and our maximum value for any of our mutants is 400 units/µm, then only 2 bonds can form. Thus, any remaining fluorescence is likely due to random bond formation. Interestingly, the fluorescent intensity of fibers composed of wild-type Ubx is only 359 units/µm, suggesting that monomers in these fibers only form approximately 1 ¾ bonds on average.
2.8. Dityrosine bonds are intermolecular and contribute to the strength of the materials
Although mutagenesis can identify the amino acids that participate in dityrosine bonds, this approach does not establish whether these bonds are intramolecular, intermolecular, or a mix of both types of bonds (Figure 7B). Many natural macroscale materials rely on intermolecular covalent crosslinks for strength, including disulfide and dityrosine bonds.[31–36,55,56] Consequently, adding covalent bonds to protein materials can dramatically improve both strength and assembly.[38,69] Therefore, if the dityrosine bonds in Ubx are intermolecular, they should impact the strength of the materials. The length to which a fiber can be pulled is dependent on protein assembly and the fibers’ inherent strength.[46,52] Since dramatic sequence changes, such as fusing large, charged proteins to Ubx monomers, do not impact assembly,[50] tyrosine point mutations are also unlikely to impact materials assembly. Therefore, changes in fiber length are expected to reflect changes in fiber strength. We observed that fibers formed in a low oxygen environment lack dityrosine bonds and are extremely short and fragile (data not shown). Plotting increasing values of both the normalized fluorescent intensity and the average fiber length reveals two transitions, corresponding to the formation of two bonds (Figure 6A). Comparison of fluorescence with fiber length using linear regression (Figure 6B) revealed a striking correlation of 0.994. This correlation suggests that fiber strength directly depends on dityrosine bond formation, and thus the bonds are intermolecular. It is important to note that a subset of mutants – those that remove decoy tyrosines - increase Ubx fluorescence. These point mutations also increase fiber strength. Thus, point mutations can either increase or decrease the average number of dityrosine bonds formed per monomer and consequently increase or decrease the strength of the resulting fibers.
2.9. Ubx tyrosine motifs as transferable motifs for strengthening protein-based materials
We have demonstrated that bonds are only formed between specific tyrosine residues in Ubx materials. These residues are located in conserved regions of the protein sequence which are separated by intrinsically disordered (unstructured) regions of the Ubx protein.[54] Therefore, it should be possible to add these conserved sequences to loops or unstructured regions of other self-assembling proteins, and thus add specific dityrosine bonds to increase the strength of those materials. As an example, the N-terminus-homeodomain bond yields less fluorescence than the Y167-Y240 bond due to competing interactions with the two decoy tyrosines, Y100 and Y310. Therefore, we reasoned that duplicating one of the tyrosines that can participate in this bond could allow both decoy binding and dityrosine bond formation. We created the 2×296 Ubx mutant in which amino acids G289-Q297, which includes Y296, were duplicated. In this variant, both the original Y293 and the duplicated Y293 were mutated to leucine. Fibers formed by this mutant were significantly more fluorescent than wild-type fibers (Figure S6, p = 0.003). Furthermore, the mutant protein created longer fibers, reflecting their increased strength. Therefore, the duplicated region was able to bind the decoy tyrosines and/or form a dityrosine bond, and thus can be considered active.
Since adding an entire region of the Ubx protein might not be feasible for some self-assembling proteins, we have identified shorter sequences likely to replace the large insertion. The sequences surrounding tyrosines that form dityrosine bonds must contribute to interaction specificity, and thus would need to be transferred to the heterologous system. Based on Ubx sequence conservation,[54] structure/disorder data,[54] and the predicted propensity to engage in protein interactions (Figure 1), we recommend the below sequences for transfer to other proteins to create dityrosine bonds. For proteins systems that can accommodate large insertions, the homeodomain (60 amino acids, RRRGR…LKKEI) and the N-terminus (MNSYFEQA) could be used. As an additional benefit, the solubility and stability of the homeodomain is expected to improve protein production when fused to a self-assembling protein.[50] For protein systems that can only tolerate small insertions or that self-assemble upon exposure to denaturing conditions, the conserved motifs surrounding residues 167 (VRPSACTPDSRVGGYLDTS) and 240 (FYPWMAIA) could be used. Thus the specific dityrosine-bond forming motifs in Ubx have the potential to be a useful tool for engineering the fluorescent and mechanical properties of other protein systems.
3. Conclusion
Although the extensibility of Ubx materials had previously been attributed to glycine-rich sequences resembling elastin,[46,52] the molecular interactions responsible for the strength of Ubx materials were unknown. We have shown that Ubx materials autofluoresce blue as a result of two intermolecular dityrosine bonds that rapidly and spontaneously form as the materials oxidize. The bonds, located between the N-terminus (Y4 or Y12) and the homeodomain (Y293 or Y296), and between Y167 and Y240 contribute to the strength of Ubx materials. Mutations that ablate one or both dityrosine bonds reduce fiber strength, whereas removing competing interactions or duplicating tyrosine-containing motifs similarly increase the strength of the materials.
4. Experimental Section
Production of Ubx materials
Protocols were used as established in the Bondos lab for expression, purification, and assembly of Ubx and Ubx fusion proteins into materials.[45,47–50] In brief, the ubx gene, cloned into pET-19b vector, was transformed into Rosetta (DE3) pLysS cells (Novagen). Single colonies were used to inoculate overnight liquid cultures. Protein expression was induced at mid-log phase with 1 mM IPTG for 4 hrs and cells were harvested by centrifugation and stored at −20 °C. Frozen cell pellets were lysed and cell debris was removed by centrifugation for 30 min at 35,000 × g (17,000 rpm by JA25.5 rotor). Ubx protein was purified from the clarified cell lysate by Ni-NTA chromatography and, as previously described, fibers were pulled from films produced in a “buffer reservoir”[49] using a buffer containing 50 mM sodium phosphate buffer, 500 mM NaCl, 5% glucose w/v, pH 8.0. Fibers were wrapped around a 5 mm sterile plastic inoculation loop and stored in a sterile tissue culture dish until use.
Measuring fluorescence in Ubx materials
Fluorescence resulting from diY or antibody binding (N=2, sample=15, replicates=45) was measured using identical DAPI and FITC settings on the Nikon Eclipse Ti A1R inverted confocal microscope and analyzed using Nikon Elements Imaging Software normalized to fiber width, which averaged approximately 15 µm. Z-stack images were captured using a 40X objective with a field depth of 1.1 µm and step sizes of 0.25 µm. Data in figures are displayed as average intensities +/− the standard deviation.
Quantitative measurement of dityrosine content based on fluorescence intensity requires tyrosine content to be the only variable. Since removal of specific tyrosine residues also prevents Ubx materials from being fluorescent, then there is clearly no other source of fluorescence that could interfere with our measurements. Since the materials can vary in size, the fluorescence intensity was always normalized to fiber diameter. This precaution allows us to quantitatively measure fluorescence, as previously demonstrated in measuring incorporation of different concentrations of Enhanced Green Fluorescent Protein-Ubx into Ubx materials.[50] Since the fluorescence intensity of the dityrosine signal is directly proportional to the fluorescence intensity of signal from anti-dityrosine antibodies (Figure S4), fluorescence intensity is a quantitative measure of dityrosine content. Data were analyzed for significance using univariate analysis of variance (ANOVA) with Tukey’s HSD test posthoc using Microsoft Excel (N=3, Replicates=50, p value < 0.05 was accepted as significant).
Immunofluorescence
Ubx fibers wrapped around inoculation loops were allowed to dry at room temperature for 2 hrs. Loops were placed in sterile 4 well cell culture plates and incubated in 250 µL of blocking solution (0.1% Triton X-100, 1% BSA, 0.2% sodium azide, and 5% goat serum in PBS) at room temperature for 1 hr. Primary antibodies raised against dityrosine (Genox) were diluted 1:500 in blocking solution and incubated with Ubx fibers for 1 hr. After two washes for 10 min each in 0.1% Triton X-100 in PBS (250 µL), loops were incubated with goat anti-rabbit Alexa 488 conjugated secondary antibodies (Molecular Probes, diluted 1:300 in blocking solution) for 1 hr. Loops were washed twice (10 min per wash) in 0.1% Triton X-100 in PBS (250 µL), placed on a 22 mm × 55 mm coverslip, and imaged immediately using a 40X objective Nikon Eclipse Ti A1R inverted confocal microscope equipped with NIS Elements AR 4.10.01 software to analyze fluorescence.
Measurement of absorption/emission spectra
A Ubx fiber was fractured and solvated in commercially available 10 mM PBS buffer solution. The dispersed Ubx solution was transferred into a four sided quartz cuvette for the photoluminescence measurement. Steady state emission spectra were recorded using a QuantaMaster 40 spectrofluorometer (Photon Technology International, Canada). Light from the excitation source, a Xenon arc lamp, was dispersed by a 1200 line/mm grating blazed at 500 nm and focused on the sample. A 380 nm long-pass filter was placed in the emission path to remove excitation light. The Ubx solution was excited at 325 nm. Emission was collected for 0.1 sec at each data point from 300 to 700 nm in steps of 1 nm.
Fiber Assembly and Imaging in Low Oxygen Atmosphere
Ubx protein was purified as previously described; however, Ubx monomers were assembled into films and drawn into fibers in an argon-atmosphere glovebox (MBraun Labmaster, ~ 2 ppm O2). Fibers were placed in a custom sealed imaging chamber (Figure S1) filled with N2 gas to capture any O2-independent autofluorescence. Ubx fibers were subsequently exposed to O2 by removing the flow of N2 gas and pushing room air into the chamber using a 50 mL syringe. The blue auto-fluorescence resulting from oxidation of Ubx fibers was analyzed over time using a Nikon Eclipse Ti A1R inverted confocal microscope equipped with NIS Elements AR 4.10.01 software.
Mutagenesis of Tyrosine
Tyrosines in the Ubx homeodomain region were mutated to leucine or serine using AccuPrime Pfx PCR kit (Invitrogen). Primers (Supplemental data Table 1) for each mutation were designed using the OligoCalc (northwestern.edu/biotools/oligocalc) and mfold (http://mfold.rna.albany.edu/?q=mfold) web servers. Mutated plasmids were transformed into DH5α competent cells (Zymo Research) and plated on LB agar with 50 µg/mL carbenicillin overnight at 37 °C. Colonies were selected and grown in 5 ml cultures of LB for plasmid purification using QIAprep miniprep (Qiagen) kit. Plasmids were sequenced to confirm each mutation prior to use. Ubx mutants were expressed in E. coli and purified as described above for the wild-type protein.
DNA Binding Assay
Ubx materials were produced using the drop method and DNA binding was measured as previously described.[51] Ubx was diluted in 250 µl of a solution containing 50 mM NaH2PO4 (pH 8.0), 300 mM NaCl, 10 mM β-mercaptoethanol, 5% glucose, and 200 mM imidazole, for a final protein concentration of 3 – 6 µM depending on the purification yield. Protein was carefully pipetted onto the surface of a siliconized glass slide, and covered with a screw cap from a 50 mL conical centrifuge tube (VWR International) and the slides were covered to prevent evaporation. After a 16 hr incubation at room temperature and humidity, a film, formed at the air-water interface, was drawn into fibers using a sterile inoculating loop. Fibers were subsequently washed three times in PBS buffer and dried for 1–2 hrs at room temperature. The DNA stock was diluted to a final concentration of 10 µg/mL in phosphate buffered saline. Fiber loops were then placed in a well (24 well culture plate); subsequently, 200 µL of the diluted DNA was pipetted in each well and allowed to incubate at room temperature (parafilm wrapped) overnight. Fibers were washed three times in PBS buffer (3–5 minutes each) to remove excess DNA from the fiber. The Ubx fiber was removed from the inoculating loop with micro-scissors and transferred to a PCR tube containing the following components: 1× PCR ThermoPol buffer (NEB), 50 µM each of dATP, dCTP, dGTP, dTTP, 0.5 µM of each primer, and 1 unit of Taq DNA polymerase (NEB) in a 50 µl reaction. The PCR reaction products were analyzed by electrophoresis through a 2% agarose gel, which was stained with ethidium bromide and visualized using a UV light source.
Fiber length measurements
Ubx protein was diluted to 1 mg in 590 mL of buffer in a shallow Teflon-coated tray (Nordic Ware), covered to prevent surface disruptions, and incubated 18 hrs at room temperature (approximately 25 °C) and 40 – 60% humidity. To measure changes in fiber production, which depends on both fiber assembly and fiber strength, the length of fibers drawn from the resulting films were measured. A minimum of eight measurements, produced from a minimum of two purifications, were made for each Ubx variant.
Supplementary Material
Acknowledgments
David W. Howell and Shang-Pu Tsai contributed equally to this work. The authors would like to thank the members of the Bayless and Bondos labs for helpful discussions and comments on the manuscript. We would like to especially thank Dr. Paul Lindahl for his expertise and use of his argon-atmosphere glovebox. Artwork for Figure 6 was generously donated by Bryan V. Frugé. J.T.A. is supported by the National Science Foundation Graduate Research Fellowship under grant number (R3E821). Funding was provided by NSF CAREER 1151394, the Ted Nash Long Life Foundation M1500779, and TAMHSC RDEAP to SEB as well as PHS grant HL095786 to KJB.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
David W. Howell, Department of Molecular and Cellular Medicine, Texas A&M Health Science Center, College Station, TX 77843, United States.
Shang-Pu Tsai, Department of Molecular and Cellular Medicine, Texas A&M Health Science Center, College Station, TX 77843, United States.
Kelly Churion, Department of Molecular and Cellular Medicine, Texas A&M Health Science Center, College Station, TX 77843, United States.
Jan Patterson, Department of Molecular and Cellular Medicine, Texas A&M Health Science Center, College Station, TX 77843, United States.
Colette Abbey, Department of Molecular and Cellular Medicine, Texas A&M Health Science Center, College Station, TX 77843, United States.
Joshua T. Atkinson, Systems, Synthetic, and Physical Biology Graduate Program, Rice University, Houston, TX 77005, United States
Dustin Porterpan, Department of Molecular and Cellular Medicine, Texas A&M Health Science Center, College Station, TX 77843, United States.
Yil-Hwan You, Department of Materials Science and Engineering, Texas A&M University, College Station, TX 77843, United States.
Kenith E. Meissner, Department of Materials Science and Engineering, Texas A&M University, College Station, TX 77843, United States Department of Biomedical Engineering, Texas A&M University, College Station, TX 77843, United States; Centre for Nanohealth, College of Engineering, Swansea University, Swansea, UK.
Kayla J. Bayless, Department of Molecular and Cellular Medicine, Texas A&M Health Science Center, College Station, TX 77843, United States
Sarah E. Bondos, Department of Molecular and Cellular Medicine, Texas A&M Health Science Center, College Station, TX 77843, United States Department of Biochemistry and Cell Biology, Rice University, Houston, Texas, 77005, United States.
References
- 1.Baneyx F, Schwartz DT. Curr. Opin. Biotechnol. 2007;18:312–317. doi: 10.1016/j.copbio.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 2.Deming TJ. Prog. Polym. Sci. 2007;32:858–875. [Google Scholar]
- 3.Maskarinec SA, Tirrell DA. Curr. Opin. Biotechnol. 2005;16:422–426. doi: 10.1016/j.copbio.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 4.Velema J, Kaplan D. Adv. Biochem. Eng. Biotechnol. 2006;102:187–238. doi: 10.1007/10_013. [DOI] [PubMed] [Google Scholar]
- 5.Woerdeman DL, Veraverbeke WS, Parnas RS, Johnson D, Delcour JA, Verpoest I, Plummer CJ. Biomacromolecules. 2004;5:1262–1269. doi: 10.1021/bm034530+. [DOI] [PubMed] [Google Scholar]
- 6.Grevellec J, Marquie C, Ferry L, Crespy A, Vialettes V. Biomacromolecules. 2001;2:1104–1109. doi: 10.1021/bm015525d. [DOI] [PubMed] [Google Scholar]
- 7.Rodríguez-Cabello JC, Prieto S, Requera J, Arias FJ, Ribeiro A. J. Biomater. Sci. Polym. Ed. 2007;18:269–286. doi: 10.1163/156856207779996904. [DOI] [PubMed] [Google Scholar]
- 8.Ong SR, Trabbic-Carlson KA, Nettles DL, Lim DW, Chilkoti A, Setton LA. Biomaterials. 2006;27:1930–1935. doi: 10.1016/j.biomaterials.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 9.Chilkoti A, Christensen T, MacKay JA. Curr. Opin. Chem. Biol. 2006;10:652–657. doi: 10.1016/j.cbpa.2006.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hollister SJ, Maddox RD, Taboas JM. Biomaterials. 2002;23:4095–4103. doi: 10.1016/s0142-9612(02)00148-5. [DOI] [PubMed] [Google Scholar]
- 11.Lagziel-Simis S, Cohen-Hadar N, Moscovich-Dagan H, Wine Y, Freeman A. Curr. Opin. Biotechnol. 2006;17:569–573. doi: 10.1016/j.copbio.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 12.Gazit E. FEBS J. 2007;274:317–322. doi: 10.1111/j.1742-4658.2006.05605.x. [DOI] [PubMed] [Google Scholar]
- 13.Huang L, McMillan RA, Apkarin RP, Pourdeyhimi B, Conticello VP, Chaikof EL. Macromolecules. 2000;33:2989–2997. [Google Scholar]
- 14.Qiu W, Teng W, Cappello J, Wu X. Biomacromolecules. 2009;10:602–608. doi: 10.1021/bm801296r. [DOI] [PubMed] [Google Scholar]
- 15.Liivak O, Blye A, Shah N, Jelinski LW. Macromolecules. 1998;31:2947–2951. [Google Scholar]
- 16.Leal Egaña A, Scheibel T. Biotechnol. Appl. Biochem. 2010;55:155–167. doi: 10.1042/BA20090229. [DOI] [PubMed] [Google Scholar]
- 17.Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 18.Georges PC, Hui J, Gombos Z, McCormick ME, Wang AY, Uemura M, Mick R, Janmey PA, Furth EE, Wells RG. Am. J. Physiol. Gastrointest. Liver. Physiol. 2007;293:G1147–G1154. doi: 10.1152/ajpgi.00032.2007. [DOI] [PubMed] [Google Scholar]
- 19.Gomes S, Leonor IB, Mano JF, Reis RL, Kaplan DL. Prog. Polym. Sci. 2012;37:1–17. doi: 10.1016/j.progpolymsci.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brooks AE, Strickler SM, Joshi SB, Kamersell TJ, Middaugh CR, Lewis RV. Biomacromolecules. 2008;9:1506–1510. doi: 10.1021/bm701124p. [DOI] [PubMed] [Google Scholar]
- 21.Grip S, Johansson J, Hedhammar M. Protein Sci. 2009;18:1012–1022. doi: 10.1002/pro.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teng W, Cappello J, Wu X. Biomacromolecules. 2009;10:3028–3036. doi: 10.1021/bm900651g. [DOI] [PubMed] [Google Scholar]
- 23.Lazaris A, Arcidiaono S, Huang Y, Zhou JF, Duguay F, Chretien N, Welsh EA, Soares JW, Karatzas CN. Science. 2002;295:472–476. doi: 10.1126/science.1065780. [DOI] [PubMed] [Google Scholar]
- 24.Kharlampieva E, Kozlovskaya V, Gunawidjaja R, Shevchenko VV, Vaia R, Naik RR, Kaplan DL, Tsukruk VV. Adv. Func. Mat. 2010;20:840–846. [Google Scholar]
- 25.Lee SM, Pippel E, Gösele U, Dresbach C, Qin Y, Chandran CV, Bräuniger T, Hause G, Knez M. Science. 2009;324:488–492. doi: 10.1126/science.1168162. [DOI] [PubMed] [Google Scholar]
- 26.Ding Y, Li Y, Qin M, Cao Y, Wang W. Langmuir. 2013;29:13299–13306. doi: 10.1021/la4029639. [DOI] [PubMed] [Google Scholar]
- 27.Souza JM, Giasson BI, Chen Q, Lee VM, Ischiropoulos H. J. Biol. Chem. 2000;275:18344–18349. doi: 10.1074/jbc.M000206200. [DOI] [PubMed] [Google Scholar]
- 28.Vaccaro E, Waite JH. Biomacromolecules. 2001;2:906–911. doi: 10.1021/bm0100514. [DOI] [PubMed] [Google Scholar]
- 29.Meng F, Hennink WE, Zhong Z. Biomaterials. 2009;30:2180–2198. doi: 10.1016/j.biomaterials.2009.01.026. [DOI] [PubMed] [Google Scholar]
- 30.Nairn KM, Lyons RE, Mulder RJ, Mudie ST, Cookson DJ, Lesieur E, Kim M, Lau D, Scholes FH, Elvin CM. Biophys. J. 2008;95:3358–3365. doi: 10.1529/biophysj.107.119107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sando L, Kim M, Colgrave ML, Ramshaw JA, Werkmeister JA, Elvin CM. J. Biomed. Mater. Res. A. 2010;95:901–911. doi: 10.1002/jbm.a.32913. [DOI] [PubMed] [Google Scholar]
- 32.Elvin CM, Brownlee AG, Huson MG, Tebb TA, Kim M, Lyons RE, Vuocolo T, Liyou NE, Hughes TC, Ramshaw JA, Werkmeister JA. Biomaterials. 2009;30:2059–2065. doi: 10.1016/j.biomaterials.2008.12.059. [DOI] [PubMed] [Google Scholar]
- 33.Chen WQ, Priewalder H, John JP, Lubec G. Proteomics. 2010;10:369–379. doi: 10.1002/pmic.200900624. [DOI] [PubMed] [Google Scholar]
- 34.Mullerova AM, Michlik I, Blazej A. Leder. 1974;5:85–155. [Google Scholar]
- 35.Qin G, Lapidot S, Numata K, Hu X, Meirovitch S, Dekel M, Podoler I, Shosyov O, Kaplan DL. Biomacromolecules. 2009;10:3227–3234. doi: 10.1021/bm900735g. [DOI] [PubMed] [Google Scholar]
- 36.Elvin CM, Brownlee AG, Huson MG, Tebb TA, Kim M, Lyons RE, Vuocolo T, Liyou NE, Hughes TC, Ramshaw JAM, Wermeister JA. Biomaterials. 2009;30:2059–2065. doi: 10.1016/j.biomaterials.2008.12.059. [DOI] [PubMed] [Google Scholar]
- 37.Ding Y, Li Y, Qin M, Cao Y, Wang W. Langmuir. 2013;29:13299–13306. doi: 10.1021/la4029639. [DOI] [PubMed] [Google Scholar]
- 38.Fang J, Li H. Langmuir. 2012;28:8260–8265. doi: 10.1021/la301225w. [DOI] [PubMed] [Google Scholar]
- 39.Lin Y, Wang S, Chen Y, Wang Q, Burke KA, Spedden EM, Stall C, Weiss AS, Kaplan DL. Nanomedicine. 2015;10:803–814. doi: 10.2217/nnm.14.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aparecido dos Santos-Pinto JR, Lamprecht G, Chen WQ, Heo S, Hardy JG, Priewalder H, Scheibel TR, Palma MS, Lubec G. J. Proteomics. 2014;105:174–185. doi: 10.1016/j.jprot.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 41.Vashi AV, Wekmeister JA, Vuocolo T, Elvin CM, Ramshaw JAM. J. Biomed. Mater. Res. Part A. 2012;100A:2239–2243. doi: 10.1002/jbm.a.34164. [DOI] [PubMed] [Google Scholar]
- 42.Fang J, Mehlich A, Koga N, Huang J, Koga R, Gao X, Hu C, Jin C, Rief M, Kast J, Baker D, Li H. Nature Commun. 4:2974. doi: 10.1038/ncomms3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elvin CM, Carr AG, Huson MG, Maxwell JM, Pearson RD, Vuocolo T, Liyou NE, Wong DC, Merritt DJ, Dixon NE. Nature. 2005;437:999–1002. doi: 10.1038/nature04085. [DOI] [PubMed] [Google Scholar]
- 44.Waffenschmidt S, Woessner JP, Beer K, Goodenough UW. Plant Cell. 1993;5:809–20. doi: 10.1105/tpc.5.7.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Majithia R, Patterson J, Bondos SE, Meissner KE. Biomacromolecules. 2011;12:3629–3637. doi: 10.1021/bm200889k. [DOI] [PubMed] [Google Scholar]
- 46.Greer AM, Huang Z, Oriakhi A, Lu Y, Lou J, Matthews KS, Bondos SE. Biomacromolecules. 2009;10:829–837. doi: 10.1021/bm801315v. [DOI] [PubMed] [Google Scholar]
- 47.Patterson JL, Abbey CA, Bayless KJ, Bondos SE. J. Biomed. Mater. Res. A. 2014;102:97–104. doi: 10.1002/jbm.a.34675. [DOI] [PubMed] [Google Scholar]
- 48.Patterson JL, Arenas-Gamboa AM, Wang TY, Hsiao HC, Howell DW, Pellois JP, Rice-Ficht A, Bondos SE. J. Biomed. Mater. Res. A. 2014 doi: 10.1002/jbm.a.35295. [DOI] [PubMed] [Google Scholar]
- 49.Huang Z, Salim T, Brawley A, Patterson J, Matthews KS, Bondos SE. Adv. Func. Mat. 2011;21:2633–2640. [Google Scholar]
- 50.Tsai SP, Howell DW, Huang Z, Hsiao HC, Lu Y, Matthews KS, Lou J, Bondos SE. Adv. Func. Mat. 2015;25:1442–1450. [Google Scholar]
- 51.Churion K, Howell DW, Ramasamy S, Catanese DJ, Tsai SP, Northern K, Zechiedrich L, Matthews KS, Bondos SE. Nat. Mater. 2015 submitted. [Google Scholar]
- 52.Huang Z, Lu Y, Majithia R, Shah J, Meissner K, Matthews KS, Bondos SE, Lou J. Biomacromolecules. 2010;11:3644–3651. doi: 10.1021/bm1010992. [DOI] [PubMed] [Google Scholar]
- 53.Knowles TP, Buehler MJ. Nat. Nanotechnol. 2011;6:469–479. doi: 10.1038/nnano.2011.102. [DOI] [PubMed] [Google Scholar]
- 54.Liu Y, Matthews KS, Bondos SE. J. Biol. Chem. 2008;283:20874–20887. doi: 10.1074/jbc.M800375200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Endrizzi BJ, Huang G, Kiser PF, Stewart RJ. Langmuir. 2006;22:11305–11310. doi: 10.1021/la0618216. [DOI] [PubMed] [Google Scholar]
- 56.Kelley FW. Fed. Proc. 1968;27:773. [PubMed] [Google Scholar]
- 57.Marquez LA, Dunford HB. J. Biol. Chem. 1995;270:30434–30440. doi: 10.1074/jbc.270.51.30434. [DOI] [PubMed] [Google Scholar]
- 58.Aeschbach R, Amado R, Neukom H. Biochim. Biophys. Acta. 1976;439:292–301. doi: 10.1016/0005-2795(76)90064-7. [DOI] [PubMed] [Google Scholar]
- 59.Harms GS, Pauls SW, Hedstrom JF, Johnson CK. J. Fluoresc. 1997;7:283–292. [Google Scholar]
- 60.Vivian JT, Callis PR. Biophys. J. 2001;80:2093–2109. doi: 10.1016/S0006-3495(01)76183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu Y, Matthews KS, Bondos SE. J. Mol. Biol. 2009;390:760–774. doi: 10.1016/j.jmb.2009.05.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beachy PA, Varkey J, Young KE, von Kessler DP, Sun BI, Ekker SC. Mol. Cell Biol. 1993;13:6941–6956. doi: 10.1128/mcb.13.11.6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dosztányi Z, Mészáros B, Simon I. Bioinformatics. 2009;25:2745–2746. doi: 10.1093/bioinformatics/btp518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mészáros B, Simon I, Dosztányi Z. PLoS Comput. Biol. 2009;5:e1000376. doi: 10.1371/journal.pcbi.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kyte J, Doolittle RF. J. of Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 66.Pace N, Scholtz JM, Grimsley GR. FEBS Lett. 2014;588:2177–2184. doi: 10.1016/j.febslet.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tour E, Hittinger CT, McGinnis W. Development. 2005;132:5271–5281. doi: 10.1242/dev.02138. [DOI] [PubMed] [Google Scholar]
- 68.Passner JM, Ryoo HD, Shen L, Mann RS, Aggarwal AK. Nature. 1999;397:714–719. doi: 10.1038/17833. [DOI] [PubMed] [Google Scholar]
- 69.Asai D, Xu D, Liu W, Garcia QF, Callahan DJ, Zalutsky MR, Craig SL, Chilkoti A. Biomaterials. 2012;33:5451–5458. doi: 10.1016/j.biomaterials.2012.03.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.