Abstract
Background
Photosynthetic organisms utilize carotenoids for photoprotection as well as light harvesting. Our previous study revealed that high-intensity light increases the expression of the gene for phytoene synthase (EgcrtB) in Euglena gracilis (a unicellular phytoflagellate), the encoded enzyme catalyzes the first committed step of the carotenoid biosynthesis pathway. To examine carotenoid synthesis of E. gracilis in response to light stress, we analyzed carotenoid species and content in cells grown under various light intensities. In addition, we investigated the effect of suppressing EgcrtB with RNA interference (RNAi) on growth and carotenoid content.
Results
After cultivation for 7 days under continuous light at 920 μmol m−2 s−1, β-carotene, diadinoxanthin (Ddx), and diatoxanthin (Dtx) content in cells was significantly increased compared with standard light intensity (55 μmol m−2 s−1). The high-intensity light (920 μmol m−2 s−1) increased the pool size of diadinoxanthin cycle pigments (i.e., Ddx + Dtx) by 1.2-fold and the Dtx/Ddx ratio from 0.05 (control) to 0.09. In contrast, the higher-intensity light treatment caused a 58% decrease in chlorophyll (a + b) content and diminished the number of thylakoid membranes in chloroplasts by approximately half compared with control cells, suggesting that the high-intensity light-induced accumulation of carotenoids is associated with an increase in both the number and size of lipid globules in chloroplasts and the cytoplasm. Transient suppression of EgcrtB in this alga by RNAi resulted in significant decreases in cell number, chlorophyll, and total major carotenoid content by 82, 82 and 86%, respectively, relative to non-electroporated cells. Furthermore, suppression of EgcrtB decreased the number of chloroplasts and thylakoid membranes and increased the Dtx/Ddx ratio by 1.6-fold under continuous illumination even at the standard light intensity, indicating that blocking carotenoid synthesis increased the susceptibility of cells to light stress.
Conclusions
Our results indicate that suppression of EgcrtB causes a significant decrease in carotenoid and chlorophyll content in E. gracilis accompanied by changes in intracellular structures, suggesting that Dtx (de-epoxidized form of diadinoxanthin cycle pigments) contributes to photoprotection of this alga during the long-term acclimation to light-induced stress.
Electronic supplementary material
The online version of this article (doi:10.1186/s12870-017-1066-7) contains supplementary material, which is available to authorized users.
Keywords: Euglena gracilis, Light-induced stress, Carotenoid, Phytoene synthase, crtB, Thylakoid, HPLC, Transmission electron microscopy, RNA interference, Double-stranded RNA
Background
Euglena gracilis is a microalga that has attracted much attention as a potential feedstock for biodiesel production. In outdoor cultivation for biofuel production, direct sunlight of high intensity can cause photoinhibition in microalgae and decrease the algal cell productivity [1, 2]. In photosynthesis of oxygenic phototrophs, excess light energy can generate various reactive oxygen species (ROS), such as superoxide radical (O2 −), hydrogen peroxide (H2O2), and hydroxyl radical (·OH) in the electron transport chain [3, 4] and singlet oxygen (1O2 *) in antenna complexes [5, 6]. ROS (such as 1O2 * and H2O2) have been shown to cause the cleavage of D1 protein in photosystem II (PSII) in vitro [7–9]. In addition, several studies [10, 11] have shown that ROS inhibit the repair of photodamaged PSII in vivo. When the reaction rate of photodamage to PSII exceeds the rate of repair, photoinhibition of photosynthesis occurs. To minimize this photoinhibition, plants have evolved several protective mechanisms such as chloroplast movement, screening of radiation, ROS scavenging, thermal energy dissipation, cyclic electron flow, and photorespiration [12].
In addition to their light-harvesting function, carotenoids contribute to photoprotection. They dissipate excess excitation energy of singlet-state chlorophylls as heat in xanthophyll-dependent non-photochemical quenching in oxygenic phototrophs [13]. Carotenoids also quench triplet-state chlorophylls in the antenna complex and singlet oxygen in the reaction center of PSII [6, 14, 15]. In general, PSII contains β-carotene in reaction center complexes [16, 17]. Lutein, 9′-cis neoxanthin and xanthophyll cycle pigments (violaxanthin and zeaxanthin) are components of antenna complexes of PSII [18, 19].
More than 750 structurally defined carotenoids have been identified in various photosynthetic and non-photosynthetic organisms including bacteria, archaea, fungi, algae, land plants, and animals [20]. Algae have evolved diverse pathways for carotenoid biosynthesis, and some algae synthesize division/class-specific carotenoids; e.g., the allenic carotenoids fucoxanthin in brown algae and diatoms, 19′-acyloxyfucoxanthin in Haptophyta and Dinophyta, and peridinin in dinoflagellates and the acetylenic carotenoids alloxanthin, crocoxanthin and monadoxanthin in Cryptophyta, and diadinoxanthin (Ddx) and diatoxanthin (Dtx) in Heterokontophyta, Haptophyta, Dinophyta and Euglenophyta [21]. The order Euglenida, which includes E. gracilis, synthesizes β-carotene and xanthophylls such as zeaxanthin, 9′-cis neoxanthin, Ddx, and Dtx [21–24].
Phytoene synthesis, the first step of carotenoid biosynthesis, by phytoene synthase (CrtB, also called Psy) is one of the rate-limiting steps in carotenoid biosynthesis [21, 25]. Steinbrenner and Linden [26, 27] reported that the expression of the phytoene synthase gene (psy) in Haematococcus pluvialis is induced in response to increased illumination. In addition, several studies have demonstrated light-induced accumulation of carotenoids in certain green algae, such as H. pluvialis [26, 27], Dunaliella salina [28, 29], and Chlorella zofingiensis [30, 31]. Consistent with these reports, our previous studies [32] revealed that high-intensity light (continuous illumination at 920 μmol m−2 s−1) increased the expression of the phytoene synthase gene in E. gracilis (EgcrtB), and this finding suggested that high-intensity light induces the accumulation of pigments assumed to be carotenoids in this alga.
To elucidate changes in carotenoid accumulation in E. gracilis in response to light stress, we analyzed the content and molecular species of carotenoids in cells grown under various light intensities. We found that the total carotenoid content in E. gracilis cells increased in response to light-induced stress. In particular, we found that light-induced stress resulted in an increase in the pool size of diadinoxanthin cycle pigments (Ddx and Dtx) and caused changes in intracellular structures, including chloroplasts. In addition, we transiently silenced EgcrtB expression using RNA interference (RNAi) in E. gracilis cells and found that the suppression of EgcrtB markedly decreased the proliferation and chlorophyll and carotenoid content accompanied by changes in intracellular structures under continuous illumination, even at a standard light intensity. Furthermore, we found that the Dtx/Ddx ratio was significantly increased by both light-induced stress and suppression of EgcrtB, suggesting that Dtx (de-epoxidized form of diadinoxanthin cycle pigment) contributes to photoprotection of E. gracilis during the long-term acclimation to light-induced stress.
Results
Effects of high-intensity light on the content of chlorophyll a and b in E. gracilis cells
E. gracilis cells were grown under continuous illumination in a range of 27–920 μmol m−2 s−1 for 7 days (Fig. 1a). Growth under 240 μmol m−2 s−1 yielded cells that looked pale green compared with control cells illuminated at a standard light intensity (55 μmol m−2 s−1). Indeed chlorophyll a and b content in these cells was 69% and 70%, respectively, of control cells, although the cell concentration did not differ significantly from control cells (Table 1). Similarly, after cultivation for 7 days under 460 μmol m−2 s−1, the cellular chlorophyll a and b content decreased to 61% and 59%, respectively, of control cells, whereas cell concentration increased as much as the control (Table 1). Cultivation under continuous light at 920 μmol m−2 s−1 for 7 days significantly decreased the cell concentration by 75% compared with control cells; moreover, this high-intensity light decreased chlorophyll a and b content by 58% and 55%, respectively, relative to the control.
Fig. 1.
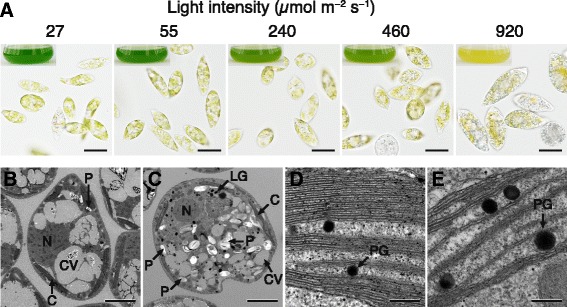
Effects of light intensity on the physical appearance of E. gracilis cells. a Algal cells and appearance of culture medium (insets) after cultivation for 7 days at 25 °C under continuous light at the indicated intensities. Scale bar, 20 μm. b and c Internal structure of cells grown under illumination at 55 (b) or 920 μmol m−2 s−1 (c) for 7 days. Scale bar, 5 μm. d and e Sections of chloroplasts of cells illuminated at 55 (d) or 920 μmol m−2 s−1 (e). Scale bar, 200 nm. C, chloroplast; CV, contractile vacuole; LG, lipid globule; N, nucleus; P, paramylon; PG, plastoglobule
Table 1.
Effect of high-intensity light on the growth and chlorophyll content of E. gracilis
| Treatment (μmol m−2 s−1) | Final cell concentration (×106 cells ml−1) | Cell weight (mg FW 106 cells−1) | Chlorophyll content (nmol 106 cells−1) | Chlorophyll a/b | |
|---|---|---|---|---|---|
| a | b | ||||
| 27 | 1.9 ± 0.1a | 2.2 ± 0.2a | 9.2 ± 0.8a | 1.3 ± 0.3a | 7.1 ± 0.7a |
| 55 | 1.9 ± 0.1a | 2.6 ± 0.1a | 8.9 ± 0.4a | 1.3 ± 0.1a | 6.9 ± 0.3a |
| 240 | 2.0 ± 0.0a | 3.0 ± 0.1a | 6.1 ± 0.1b | 0.9 ± 0.0b | 6.7 ± 0.2a |
| 460 | 2.0 ± 0.0a | 2.7 ± 0.1a | 5.4 ± 0.3b | 0.8 ± 0.1b | 7.0 ± 0.2a |
| 920 | 0.5 ± 0.1b | 5.5 ± 0.6b | 3.7 ± 0.3c | 0.6 ± 0.1b | 6.5 ± 0.7a |
Data represent the mean ± SD of biological triplicates. Different letters in each column indicate a significant differences (P < 0.05, Tukey’s test)
Cultivation for 7 days under 920 μmol m−2 s−1 yielded cells that appeared much larger than those illuminated at the standard light intensity, and the fresh weight of the cells was twice that of the control cells (Fig. 1a and Table 1). Furthermore, in contrast to cells grown under other light intensities, these cells appeared yellow-orange or reddish-orange and accumulated greater numbers of grayish granules thought to be composed of paramylon (~1–2 μm in diameter) in the cells.
Ultrastructure of E. gracilis cells grown under high intensity light
Figure 1c and e show the internal structure of cells and chloroplasts of E. gracilis grown under illumination at 920 μmol m−2 s−1; transmission electron microscopy (TEM) revealed a decrease in the number of thylakoid membranes in chloroplasts by approximately half compared with control cells grown under standard light intensity (Fig. 1b and d). TEM also revealed that the algal cells grown under the high-intensity light contained more plastoglobules (lipid globules in the interthylakoid space of chloroplasts) than control cells and that the plastoglobules of those cells were obviously larger than those in the control (Fig. 1d and e). The high-intensity light also markedly increased the number of osmium-philic droplets (lipid globules) in the cytoplasm compared with control (Fig. 1c).
Effects of high-intensity light on the relative content of carotenoids in E. gracilis cells
To identify carotenoid species in E. gracilis, we subjected cell extracts to high-performance liquid chromatography (HPLC) and measured absorption of the effluent at 445 nm (Fig. 2a). For control cells grown under illumination with 55 μmol m−2 s−1, HPLC analyses indicated that β-carotene, neoxanthin, Ddx and Dtx were the major carotenoids and accounted for 4, 6, 86, and 4%, respectively, of the total carotenoids (Fig. 2b). These four carotenoids were also the major species in cells grown under light of higher intensities (Additional file 1), and Fig. 3 shows the relative content of the major carotenoids in those cells. For cells illuminated at 240, 460, or 920 μmol m−2 s−1, neoxanthin content per cell significantly decreased by 19, 28, and 40%, respectively, relative to control cells; illumination with 27, 240, or 460 μmol m−2 s−1 had no obvious effect on the content of β-carotene, Ddx and Dtx relative to the control. In contrast, illumination at 920 μmol m−2 s−1 substantially increased the β-carotene, Ddx and Dtx content per cell by 2.6, 1.2, and 2.1-fold, respectively, compared with control cells, and the total major carotenoids per cell increased by 25% (Fig. 3).
Fig. 2.
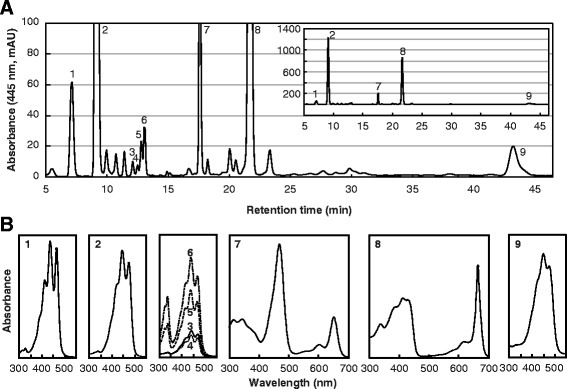
Analysis of carotenoid species in E. gracilis with HPLC. a HPLC chromatogram (445 nm) of extracts from E. gracilis. (Inset) The same chromatogram with an expanded y axis. mAU, milli-absorbance units. b Absorbance spectrum of individual peaks of major carotenoids (peaks 1–6 and 9). 1, neoxanthin; 2, diadinoxanthin; 3, all trans-diatoxanthin; 4–6, cis-diatoxanthin; 7, chlorophyll b; 8, chlorophyll a; 9, β-carotene
Fig. 3.
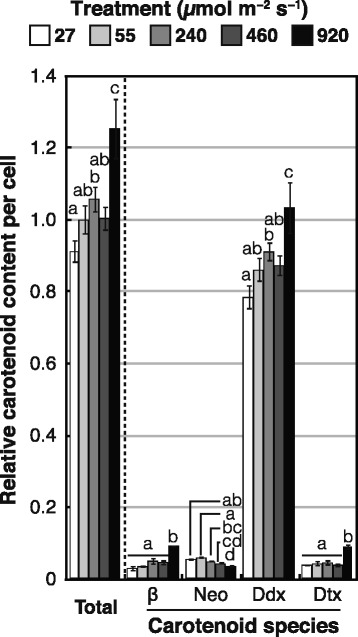
Effects of light intensity on carotenoid content of E. gracilis cells. Cells were grown under the indicated light intensities for 7 days. Relative carotenoid content per cell was calculated by normalizing molar ratios of major carotenoid species to chlorophyll a content. β, β-carotene; Neo, neoxanthin; Ddx, diadinoxanthin; Dtx, diatoxanthin. Levels of carotenoids per cell are expressed relative to total carotenoids in the cells illuminated at the standard light intensity of 55 μmol m−2 s−1. Error bars indicate ± SD of biological triplicates. Bars labeled with the same letter are not significantly different (Tukey’s multiple range test, P < 0.05)
Suppression of EgcrtB expression
EgcrtB expression was suppressed using RNAi mediated by double-stranded RNA (dsRNA). Figure 4a shows expression levels of EgcrtB in E. gracilis cells treated with dsRNA directed toward a partial sequence of EgcrtB. Treatment without EgcrtB-dsRNA (electroporation alone) had no obvious effect on EgcrtB expression. In contrast, expression of EgcrtB in cells cultured for 3 days was markedly decreased by the EgcrtB-dsRNA treatment. Although EgcrtB expression in EgcrtB-dsRNA-treated cells gradually recovered during the full 7-day cultivation period, expression was lower than that in non-electroporated cells. These results indicated that EgcrtB expression could be transiently suppressed by treating cells with EgcrtB-dsRNA.
Fig. 4.
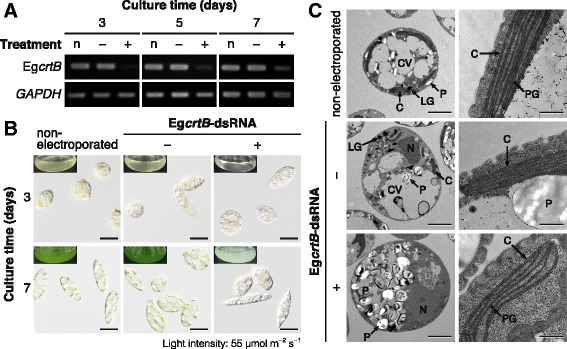
Effects of suppressing EgcrtB on E. gracilis cells. Cells treated with or without EgcrtB-dsRNA were cultured for 7 days at 25 °C under continuous illumination at 55 μmol m−2 s−1. a Semi-quantitative reverse transcription-PCR analysis of EgcrtB expression in cells with (+) or without (−) EgcrtB-dsRNA. Non-electroporated cells (n) were used as the control. GAPDH expression was used as an internal control for the semi-quantitative RT-PCR analysis. b Cells treated with or without EgcrtB-dsRNA and visual appearance of culture medium (insets) after cultivation for 3 and 7 days. Scale bar, 20 μm. c TEM images of intracellular structures (left column: scale bar, 5 μm) and sections of chloroplasts (right column: scale bar, 500 nm) of cells treated with or without EgcrtB-dsRNA. C, chloroplast; CV, contractile vacuole; LG, lipid globule; N, nucleus; P, paramylon; PG, plastoglobule
When control cells were grown under continuous light (55 μmol m−2 s−1) at 25 °C for 3 days, treatment with EgcrtB-dsRNA decreased the cell concentration to 43% and 61% compared with cells treated without electroporation or EgcrtB-dsRNA, respectively (Table 2). Electroporation alone decreased the cell concentration by 29%, but EgcrtB-dsRNA-mediated EgcrtB suppression caused a further marked decrease in cell concentration after cultivation for 3 days. After cultivation for 7 days, the number of cells treated without EgcrtB-dsRNA had increased as much as the non-electroporated cells, whereas the concentration of cells treated with EgcrtB-dsRNA had decreased by ~82%.
Table 2.
Effects of suppressing EgcrtB on cell concentration and chlorophyll content of E. gracilis
| Treatment | Cell concentration | Chlorophyll content (nmol 106 cells−1) | Chlorophyll a/b | ||
|---|---|---|---|---|---|
| Cultured for 3 days (×104 cells ml−1) | Cultured for 7 days (×106 cells ml−1) | a | b | ||
| non-electroporated | 7.7 ± 0.7a | 1.9 ± 0.0a | 7.7 ± 0.9a | 1.1 ± 0.1a | 6.9 ± 0.3a |
| EgcrtB-dsRNA(−) | 5.5 ± 0.7b | 1.8 ± 0.1a | 8.5 ± 1.6a | 1.2 ± 0.2a | 7.3 ± 0.3a |
| EgcrtB-dsRNA(+) | 3.3 ± 0.2c | 0.3 ± 0.0b | 1.3 ± 0.1b | 0.2 ± 0.0b | 6.1 ± 0.5b |
Data represent mean ± SD of biological replicates. The number of biological replicates was as follows: cell concentration, n = 3; chlorophyll content and chlorophyll a/b ratio, n = 4. Different letters in each column indicate a significant differences (P < 0.05, Tukey’s test)
Electroporation alone had no obvious effect on cell appearance (Fig. 4b) or chlorophyll a and b content (Table 2) compared with non-electroporated cells. In contrast, treatment with EgcrtB-dsRNA caused chlorosis in cells after cultivation for 3 days. After cultivation for 7 days, chloroplasts in these cells were still pale green, and the culture medium was mostly clear; moreover, the content of chlorophyll a and b in EgcrtB-suppressed cells was decreased to 17% and 20% of non-electroporated cells, respectively (Table 2).
TEM clearly revealed that EgcrtB-suppressed cells accumulated many more cytoplasmic paramylon granules compared with cells treated without electroporation or EgcrtB-dsRNA (Fig. 4c, left column). In contrast, EgcrtB-suppressed cells contained considerably fewer chloroplasts. When we examined 120–150 sections of individual cells, chloroplasts were found in <5% of sections of EgcrtB-suppressed cells, whereas almost all sections of cells treated without electroporation or EgcrtB-dsRNA contained several chloroplasts (data not shown). The number of thylakoid layers in EgcrtB-suppressed cells was slightly lower than in cells treated without electroporation or EgcrtB-dsRNA (Fig. 4c, right column).
Figure 5 shows the relative content of major carotenoid species in cells treated with or without EgcrtB-dsRNA. Treatment without EgcrtB-dsRNA had no significant effect on the content of the four major carotenoids in cells cultivated for 7 days. In contrast, treatment with EgcrtB-dsRNA drastically decreased the content of the total major carotenoids per cell by 86% relative to non-electroporated cells (Fig. 5). After cultivation for 7 days, the relative content of the four major carotenoids, namely β-carotene, neoxanthin, Ddx and Dtx, in the EgcrtB-suppressed cells was 12, 19, 13, and 21% of non-electroporated cells, respectively.
Fig. 5.
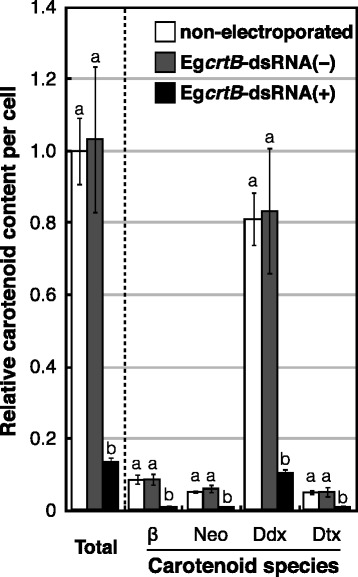
Effects of suppressing EgcrtB on carotenoid content of E. gracilis cells. Cells were treated with (+) or without (−) EgcrtB-dsRNA and cultured for 7 days at 25 °C under continuous light at 55 μmol m−2 s−1. Non-electroporated cells (n) were used as the control. Levels of carotenoids per cell are expressed relative to total carotenoids in the non-electroporated cells. β, β-carotene; Neo, neoxanthin; Ddx, diadinoxanthin; Dtx, diatoxanthin. Error bars indicate ± SD of biological quadruplicates. Bars labeled with the same letter are not significantly different (Tukey’s multiple range test, P < 0.05)
Discussion
Effects of high-intensity light on the content of chlorophyll a and b in E. gracilis cells
We previously reported that continuous illumination at an intensity of ~460 μmol m−2 s−1 appears to be a threshold of light stress that can be tolerated by E. gracilis grown at 25 °C [32]. In our present study, although the concentration of cells in cultures grown under illumination at 240 μmol m−2 s−1 was similar to that of control, chlorophyll content was significantly decreased in those cells after cultivation for 7 days (Table 1). This result suggests that illumination at 240 μmol m−2 s−1 can also induce light stress for this alga.
Steinbrenner and Linden [26] reported that illumination at 10–250 μmol m−2 s−1 decreases the total chlorophyll content in H. pluvialis cells in a light intensity-dependent manner. Similarly, Lamers et al. [29] reported that the total chlorophyll content of D. salina cells decreases in response to a stepwise increase in light intensity from 150 to 650 μmol m−2 s−1. These data are compatible with our findings.
Ultrastructure of E. gracilis cells grown under high intensity light
We found that treatment of E. gracilis cells with high-intensity light (920 μmol m−2 s−1) caused an increase in cell size and fresh weight (Fig. 1 and Table 1). Light-induced cell swelling has also been observed in D. salina. Lamers et al. [29] found that high-intensity light caused cell division arrest and increased the volume of D. salina cells immediately after the shift from 200 to 1400 μmol m−2 s−1. Our TEM study revealed that cells illuminated at 920 μmol m−2 s−1 accumulated more paramylon granules than control cells (Fig. 1b and c). This result is consistent with a report of light-induced starch accumulation in Dunaliella bardawil cells [33]. Accumulation of paramylon granules in E. graciilis might be due to the light stress-induced cell division arrest and would be the cause of cell swelling.
TEM revealed that the number of thylakoid layers clearly decreased in E. gracilis illuminated at 920 μmol m−2 s−1 (Fig. 1d and e). This decrease coincided with a significant decrease in chlorophyll content in these cells (Table 1). Considering that the chlorophyll a/b ratio of cells remained constant under various light intensities, the decrease in the chlorophyll a and b content in the cells was likely caused by a decrease in thylakoid layers in chloroplasts (Table 1 and Fig. 1). In contrast, exposure to high-intensity light induced the accumulation and enlargement of lipid globules in chloroplasts and the cytoplasm of these cells (Fig. 1). These findings are in accordance with previous reports of light-induced formation of lipid globules in D. salina [29] and D. bardawil [33].
Effects of high-intensity light on the relative content of carotenoids in E. gracilis cells
Our results revealed that the carotenoid content of E. gracilis increased in response to increasing light intensity (Fig. 3). This light-induced accumulation of carotenoids has been reported for several other green algae. In C. zofingiensis, for example, illumination at 150 μmol m−2 s−1 markedly increased the contents of zeaxanthin, canthaxanthin and astaxanthin [30]. Wang et al. [34] showed that illumination at 350 μmol m−2 s−1 induced astaxanthin accumulation and increased the total carotenoid content in H. pluvialis. Lamers et al. [29] reported that the β-carotene content of D. salina cells increased in response to an increase in light intensity from 150 to 650 μmol m−2 s−1, and they found that high-intensity light (1400 μmol m−2 s−1) led to an increase in the content of both lycopene and β-carotene.
Unlike most flagellated green algae, the eyespot apparatus (carotenoid-rich lipid globules) of E. gracilis is located in the cytoplasm [35]. Heelis et al. [36] reported that the major carotenoids in the eyespot globules in this alga are β-carotene, Ddx and Dtx. In addition, Cunningham and Schiff [37] observed that Euglena cells contain carotenoids in extraplastidic pools. Thus, the increase in these three carotenoid species in E. gracilis cells in response to high-intensity light was considered to be partly due to the accumulation of cytoplasmic lipid globules in addition to the accumulation of plastoglobules (Figs. 1 and 3).
Our HPLC analyses showed that, under the high-intensity light (920 μmol m−2 s−1), the pool size of diadinoxanthin cycle pigments (Ddx + Dtx) in E. gracilis cells increased by 1.2-fold relative to control cells, and the Dtx/Ddx ratio increased from 0.05 (control) to 0.09 (920 μmol m−2 s−1). In diatoms, it has been reported that a larger pool of Ddx promotes non-photochemical quenching [38], and the concentration of Dtx correlates directly with non-photochemical quenching in Phaeodactylum tricornutum [38, 39]. The increase in the Dtx/Ddx ratio of E. gracilis caused by illumination at 920 μmol m−2 s−1 suggests that Dtx participates in photoprotection of this alga during the long-term acclimation to high-intensity light.
Suppression of EgcrtB expression
Blocking carotenoid biosynthesis in E. gracilis by transient suppression of EgcrtB by RNAi caused chlorosis in cells and remarkably decreased the cell concentration and content of chlorophyll and carotenoid (Table 2 and Figs. 4 and 5). These results agree with previous studies showing that defects in phytoene synthase lead to a lack of or striking decrease in chlorophyll and carotenoid content in Chlamydomonas reinhardtii [40] and Scenedesmus obliquus [41]. McCarthy et al. [40] reported that the C. reinhardtii mutant lts1 with a defective PSY gene has a very pale-green phenotype and contained much less chlorophyll than the wild-type strain. A considerable decrease in chlorophyll concentration has also been observed in the C-6E mutant of S. obliquus with a defect in phytoene synthase [41]. Similarly, the S. obliquus mutant C-6E synthesizes only trace amount of carotenoids owing to a defect in the formation or function of phytoene synthase [41].
We did not observe any obvious difference in the shape of chloroplasts and thylakoid membranes between the non-electroporated cells and EgcrtB-suppressed cells in the TEM study, although the number of thylakoid layers of chloroplasts was slightly decreased by treatment with EgcrtB-dsRNA (Fig. 4c). These results suggest that the observed significant decrease in carotenoid content in EgcrtB-suppressed cells (Fig. 5) was likely due to the decrease in the number or size of chloroplasts.
The content of all major carotenoid species in E. gracilis markedly decreased in the same way by suppression of EgcrtB expression (Additional file 2 and Fig. 5). This result indicates that phytoene synthesis is considered to be the first committed and rate-limiting step of the carotenoid biosynthesis also in this alga. Our TEM study indicated that EgcrtB-suppressed cells accumulated paramylon granules in the cytoplasm (Fig. 4c), similar to cells illuminated at 920 μmol m−2 s−1 (Fig. 1c). In addition, although the pool size of diadinoxanthin cycle pigments (Ddx + Dtx) decreased by 86% with treatment with EgcrtB-dsRNA compared with non-electroporated cells, the Dtx/Ddx ratio increased from 0.06 (non-electroporated) to 0.10 in EgcrtB-suppressed cells. As mentioned above, a significant increase in the Dtx/Ddx ratio was observed under high-intensity light (Fig. 3). Hence, these results indicate that the EgcrtB-suppressed cells were light-stressed under illumination, even at 55 μmol m−2 s−1 because of carotenoid depletion. McCarthy et al. [40] proposed that chlorophylls in lts mutants of C. reinhardtii, which are unable to grow in the light–even very low-intensity light–would cause photooxidative stress in cells by acting as photosensitizers in the absence of carotenoids.
Conclusions
We found that the carotenoid content in E. gracilis cells increased in response to high-intensity light. Accumulation of carotenoids in these cells appeared to be associated with an increase in lipid globules in chloroplasts and the cytoplasm of this alga under the higher-intensity light conditions. Our results also revealed that suppression of EgcrtB resulted in a significant decrease in the content of carotenoids and led to an increase in the Dtx/Ddx ratio, as observed with the high-intensity light treatment. This study indicates that Dtx contributes to photoprotection of E. gracilis during long-term acclimation to light-induced stress.
Methods
Biological materials
Euglena gracilis Klebs (strain Z) was grown in 100 ml of Cramer-Myers medium [42] containing 0.1% ethanol at an initial cell concentration of 3 × 103 cells ml−1 in a 300-ml conical flask. The cells were cultured at 25 °C under continuous illumination at 27, 55 (control), 240, 460, and 920 μmol m−2 s−1 for 7 days as we reported previously [32]. Cell number was counted daily under a microscope using a plankton counter (MPC200, Matsunami Glass, Osaka, Japan). For the analysis of expression level of EgcrtB and determination of the content of carotenoids and chlorophyll, algal cells were harvested by centrifugation (3000×g, 2 min) and stored at −60 °C until the measurement.
Determination of chlorophyll a and b content in E. gracilis cells
For the determination of chlorophyll a and b in E. gracilis, pigments were extracted from cells three times with 1 ml of buffered aqueous 80% acetone [43]. Concentrations of chlorophyll a and b in the extracts were determined by the absorption with extinction coefficients reported by Porra et al. [43].
Extraction of carotenoids from E. gracilis cells and HPLC analysis
Under dim light, pigments were extracted twice from cells with 1 ml of acetone/methanol (7:2, v/v) immediately before HPLC analyses. After centrifugation, extracts were dried with a rotary evaporator. The residue was dissolved in chloroform/methanol (3:1, v/v) and then analyzed with an HPLC system equipped with Mightysil RP-18 GP analytical column (4.6 mm × 150 mm, 5 μm particles, Kanto Chemical, Tokyo, Japan) and guard column (4.6 mm × 5 mm, 5 μm particles, Kanto Chemical, Tokyo, Japan). The elution conditions were as follows: 0–10 min, linear gradient from 90% methanol/H2O (v/v) to 100% methanol; 10–50 min, isocratic 100% methanol at 1.0 ml min−1. Absorbance spectra (250–700 nm, 1.2 nm resolution) and retention times were recorded with an SPD-M20A Photodiode Array Detector (Shimadzu, Kyoto, Japan).
The composition of the major carotenoids was calculated from these molar absorption coefficients and areas under the peak in the chromatogram of absorbance at 445 nm. Relative carotenoid content per cell was calculated by normalizing molar ratios of major carotenoid species to chlorophyll a (carotenoids/chlorophyll a) based on absorbance at 445 nm in HPLC analysis with chlorophyll a content per cell (mol cell−1) determined as described above.
TEM
For our TEM study, cells were harvested by centrifugation (1000×g, 2 min), and fixed with 1.7% glutaraldehyde in 50 mM sodium cacodylate buffer (pH 7.0) for 2 h and then post-fixed in 2% osmium tetroxide in the same buffer for 2 h at room temperature. After dehydration in an ethanol series, fixed cells were embedded in Spurr’s resin. Ultrathin sections (80 nm thick) were cut with a diamond knife on an ULTRACUT E ultra-microtome (Leica, Wetzlar, Germany) and mounted on Formvar-coated grids. Sections were stained with 4% uranyl acetate for 18 min and 0.4% lead citrate solution for 7 min at room temperature and observed on a JEM-1400 instrument (JEOL, Tokyo, Japan) at 120 kV.
RNAi-mediated suppression of EgcrtB
Expression of EgcrtB in E. gracilis was transiently silenced with dsRNA-mediated interference as described by Iseki et al. [44]. For the synthesis of the template for EgcrtB-dsRNA, part of the EgcrtB cDNA (DDBJ accession No. LC062707) was amplified (472-bp fragment) by PCR with PrimeSTAR GXL Polymerase (Takara Bio, Shiga, Japan) and the primers 5′-TAATACGACTCACTATAGGGCAGCCGTACTACGACATGA-3′ and 5′-TAATACGACTCACTATAGGGGGATCTGGCTGTAGAGGTC-3′, which contain T7 RNA polymerase promoter sequence. The dsRNA of partial EgcrtB was synthesized with the MEGAscript T7 Transcription kit (Thermo Fisher Scientific, Massachusetts, USA).
The EgcrtB-dsRNA was introduced into E. gracilis cells with an electroporator (Micropulser, Bio-Rad, California, USA). Specifically, 2.0 × 106 cells were electroporated eight times at 0.4 kV with or without 15 μg of EgcrtB-dsRNA in 100 μl CM medium in a 0.2-cm gap cuvette (Bio-Rad, California, USA). Subsequently, cells were inoculated in 100 ml of CM medium containing 0.1% ethanol at an initial cell concentration of 3 × 103 cells ml−1 and cultivated for 7 days at 25 °C under continuous light at 55 μmol m−2 s−1 with agitation (90 rpm). Cells that had not been treated with EgcrtB-dsRNA or electroporated were used as non-electroporated cells.
Expression of EgcrtB in cells with or without EgcrtB-dsRNA was analyzed by semi-quantitative reverse transcription-PCR. Total RNA was extracted from cells using the RNAqueous kit (Thermo Fisher Scientific, Massachusetts, USA) and Plant RNA Isolation Aid (Thermo Fisher Scientific, Massachusetts, USA). First-strand cDNA was synthesized from total RNA with the QuantiTect Reverse Transcription kit (Qiagen, Hilden, Germany) and used as the template. GAPDH expression was used as an internal control for the semi-quantitative RT-PCR analysis. PCR was conducted with EmeraldAmp MAX PCR Master Mix (Takara Bio, Shiga, Japan). Primer sequences were as follows: GAPDH, 5′-GGTCTGATGACCACCATCCAT-3′ and 5′-CGACGACACGGTTGGAGTAT-3′; EgcrtB, 5′-CAGCCGTACTACGACATGATC-3′ and 5′-GGATCTGGCTGTAGAGGTCC-3′.
Additional files
Effects of light intensity on carotenoid composition of E. gracilis cells. (A–E) HPLC chromatogram (445 nm) of extracts from E. gracilis grown under illumination at 27 (A), 55 (B), 240 (C), 460 (D), or 920 μmol m−2 s−1 (E) for 7 days. (Insets) Same chromatograms with an expanded y axis. mAU, milli-absorbance units. 1, neoxanthin; 2, diadinoxanthin; 3, all trans-diatoxanthin; 4–6, cis-diatoxanthin; 7, chlorophyll b; 8, chlorophyll a; 9, β-carotene (PDF 96 kb)
Effects of suppressing EgcrtB on carotenoid composition of E. gracilis cells. (A–C) HPLC chromatogram (445 nm) of extracts from E. gracilis cells treated without electroporation or EgcrtB-dsRNA (non-electroporated) (A), or cells treated with (C) or without EgcrtB-dsRNA (B). (Insets) Same chromatograms with an expanded y axis. mAU, milli-absorbance units. 1, neoxanthin; 2, diadinoxanthin; 3, all trans-diatoxanthin; 4–6, cis-diatoxanthin; 7, chlorophyll b; 8, chlorophyll a; 9, β-carotene (PDF 69 kb)
Acknowledgements
We thank Ms. Megumi Kobayashi of Japan Women’s University for her help with TEM. We also thank Dr. Senji Takahashi for his excellent support in the analysis of carotenoids with HPLC.
Funding
This work was supported by grants from the JSPS KAKENHI (25450308 and 17K07945) and MEXT-supported Program for the Strategic Research Foundation at Private Universities (S1311014) to SK and TS.
Availability of data and materials
The data that support the findings of this study are included in this published article and its supplementary information files.
Abbreviations
- CrtB, PSY
Phytoene synthase
- Ddx
Diadinoxanthin
- dsRNA
Double-stranded RNA
- Dtx
Diatoxanthin
- HPLC
High-performance liquid chromatography
- RNAi
RNA interference
- ROS
Reactive oxygen species
- TEM
Transmission electron microscopy/microscopic
Authors’ contributions
SK designed the experiments, conducted the algal culture, chlorophyll determination, HPLC and gene expression analyses, and drafted the manuscript. ST and SK analyzed carotenoid species in cells with HPLC. TI established the RNAi method for the suppressing EgcrtB. MS and SK synthesized the dsRNA specific for EgcrtB and performed the EgcrtB suppression experiments in E. gracilis cells. MS and SK conducted the expression analysis of EgcrtB in EgcrtB-suppressed cells in cooperation with MA. NN conducted TEM of E. gracilis cells. TS conceived of the study, participated in its design and coordination, and helped to draft the manuscript. All authors read and approved the final manuscript.
Author’s information
ST present affiliation: Department of Molecular Microbiology, Faculty of Life Sciences, Tokyo University of Agriculture, 1-1-1 Sakuragaoka, Setagaya, Tokyo, 156-8502, Japan.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s12870-017-1066-7) contains supplementary material, which is available to authorized users.
Contributor Information
Shota Kato, Email: shota.kato.680@gmail.com.
Mika Soshino, Email: d.d.6xx9@gmail.com.
Shinichi Takaichi, Email: takaichi@nms.ac.jp.
Takahiro Ishikawa, Email: ishikawa@life.shimane-u.ac.jp.
Noriko Nagata, Email: n-nagata@fc.jwu.ac.jp.
Masashi Asahina, Email: asahina@nasu.bio.teikyo-u.ac.jp.
Tomoko Shinomura, Phone: +81-28-627-7724, Email: shinomura@nasu.bio.teikyo-u.ac.jp.
References
- 1.Richmond A, Lichtenberg E, Stahl B, Vonshak A. Quantitative assesment of the major limitations on productivity of Spirulina platensis in open raceways. J Appl Phycol. 1990;2:195–206. doi: 10.1007/BF02179776. [DOI] [Google Scholar]
- 2.Vonshak A, Guy R. Photoadaptation, photoinhibition and productivity in the blue-green alga, Spirulina platensis grown outdoors. Plant Cell Environ. 1992;15:613–616. doi: 10.1111/j.1365-3040.1992.tb01496.x. [DOI] [Google Scholar]
- 3.Edreva A. Generation and scavenging of reactive oxygen species in chloroplasts: a submolecular approach. Agric Ecosyst Environ. 2005;106:119–133. doi: 10.1016/j.agee.2004.10.022. [DOI] [Google Scholar]
- 4.Asada K. Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 2006;141:391–396. doi: 10.1104/pp.106.082040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krieger-Liszkay A. Singlet oxygen production in photosynthesis. J Exp Bot. 2005;56:337–346. doi: 10.1093/jxb/erh237. [DOI] [PubMed] [Google Scholar]
- 6.Telfer A. Singlet oxygen production by PSII under light stress: mechanism, detection and the protective role of β-carotene. Plant Cell Physiol. 2014;55:1216–1223. doi: 10.1093/pcp/pcu040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mishra NP, Ghanotakis DF. Exposure of a photosystem II complex to chemically generated singlet oxygen results in D1 fragments similar to the ones observed during aerobic photoinhibition. Biochim Biophys Acta. 1994;1187:296–300. doi: 10.1016/0005-2728(94)90003-5. [DOI] [Google Scholar]
- 8.Miyao M, Ikeuchi M, Yamamoto N, Ono T. Specific degradation of the D1 protein of photosystem II by treatment with hydrogen peroxide in darkness: implications for the mechanism of degradation of the D1 protein under illumination. Biochemist. 1995;34:10019–10026. doi: 10.1021/bi00031a025. [DOI] [PubMed] [Google Scholar]
- 9.Okada K, Ikeuchi M, Yamamoto N, Ono T, Miyao M. Selective and specific cleavage of the D1 and D2 proteins of photosystem II by exposure to singlet oxygen: factors responsible for the susceptibility to clevage of the proteins. Biochim Biophys Acta. 1996;1274:73–79. doi: 10.1016/0005-2728(96)00015-1. [DOI] [Google Scholar]
- 10.Nishiyama Y, Allakhverdiev SI, Yamamoto H, Hayashi H, Murata N. Singlet oxygen inhibits the repair of photosystem II by suppressing the translation elongation of the D1 protein in Synechocystis sp. PCC 6803. Biochemist. 2004;43:11321–11330. doi: 10.1021/bi036178q. [DOI] [PubMed] [Google Scholar]
- 11.Nishiyama Y, Yamamoto H, Allakhverdiev SI, Inaba M, Yokota A, Murata N. Oxidative stress inhibits the repair of photodamage to the photosynthetic machinery. EMBO J. 2001;20:5587–5594. doi: 10.1093/emboj/20.20.5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takahashi S, Badger MR. Photoprotection in plants: a new light on photosystem II damage. Trends Plant Sci. 2011;16:53–60. doi: 10.1016/j.tplants.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 13.Müller P, Li XP, Niyogi KK. Non-photochemical quenching. A response to excess light energy. Plant Physiol. 2001;125:1558–1566. doi: 10.1104/pp.125.4.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Telfer A. What is β-carotene doing in the photosystem II reaction centre? Phil Trans R Soc Lond B. 2002;357:1431–1440. doi: 10.1098/rstb.2002.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Triantaphylidès C, Havaux M. Singlet oxygen in plants: production, detoxification and signaling. Trends Plant Sci. 2009;14:219–228. doi: 10.1016/j.tplants.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 16.Kamiya N, Shen JR. Crystal structure of oxygen-evolving photosystem II from Thermosynechococcus vulcanus at 3.7-Å resolution. Proc Natl Acad Sci. 2003;100:98–103. doi: 10.1073/pnas.0135651100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferreira KN, Iverson TM, Maghlaoui K, Barber J, Iwata S. Architecture of the photosynthetic oxygen-evolving center. Sci. 2004;303:1831–1838. doi: 10.1126/science.1093087. [DOI] [PubMed] [Google Scholar]
- 18.Liu Z, Yan H, Wang K, Kuang T, Zhang J, Gui L, et al. Crystal structure of spinach major light-harvesting complex at 2.72 Å resolution. Nature. 2004;428:287–292. doi: 10.1038/nature02373. [DOI] [PubMed] [Google Scholar]
- 19.Standfuss J, van Scheltinga ACT, Lamborghini M, Kühlbrandt W. Mechanisms of photoprotection and nonphotochemical quenching in pea light-harvesting complex at 2.5 Å resolution. EMBO J. 2005;24:919–928. doi: 10.1038/sj.emboj.7600585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Britton G, Liaaen-Jensen S, Pfander H. Carotenoids Handbook. Birkhäuser Basel: Springer; 2004.
- 21.Takaichi S. Carotenoids in algae: distributions, biosyntheses and functions. Mar Drugs. 2011;9:1101–1118. doi: 10.3390/md9061101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bjørnland T. Chlorophylls and carotenoids of the marine alga Eutreptiella gymnastica. Phytochemistry. 1982;21:1715–1719. doi: 10.1016/S0031-9422(82)85046-2. [DOI] [Google Scholar]
- 23.Aitzetmüller K, Svec WA, Katz JJ, Strain HH. Structure and chemical identity of diadinoxanthin and the principal xanthophyll of Euglena. Chem Commun. 1968;1:32–3.
- 24.Takaichi S, Mimuro M. Distribution and geometric isomerism of neoxanthin in oxygenic phototrophs: 9′-cis, a sole molecular form. Plant Cell Physiol. 1998;39:968–977. doi: 10.1093/oxfordjournals.pcp.a029461. [DOI] [Google Scholar]
- 25.Ruiz-Sola MÁ, Rodríguez-Concepción M. Carotenoid biosynthesis in Arabidopsis: A colorful pathway. Arabidopsis Book. 2012;doi:10.1199/tab.0158. [DOI] [PMC free article] [PubMed]
- 26.Steinbrenner J, Linden H. Light induction of carotenoid biosynthesis genes in the green alga Haematococcus pluvialis: regulation by photosynthetic redox control. Plant Mol Biol. 2003;52:343–356. doi: 10.1023/A:1023948929665. [DOI] [PubMed] [Google Scholar]
- 27.Steinbrenner J, Linden H. Regulation of two carotenoid biosynthesis genes coding for phytoene synthase and carotenoid hydroxylase during stress-induced astaxanthin formation in the green alga Haematococcus pluvialis. Plant Physiol. 2001;125:810–817. doi: 10.1104/pp.125.2.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Król M, Maxwell DP, Huner NPA. Exposure of Dunaliella salina to low temperature mimics the high light-induced accumulation of carotenoids and the carotenoid binding protein (Cbr) Plant Cell Physiol. 1997;38:213–216. doi: 10.1093/oxfordjournals.pcp.a029155. [DOI] [Google Scholar]
- 29.Lamers PP, van de Laak CCW, Kaasenbrood PS, Lorier J, Janssen M, De Vos RCH, et al. Carotenoid and fatty acid metabolism in light-stressed Dunaliella salina. Biotechnol Bioeng. 2010;106:638–648. doi: 10.1002/bit.22725. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Huang J, Sandmann G, Chen F. High-light and sodium chloride stress differentially regulate the biosynthesis of astaxanthin in Chlorella zofingiensis (Chlorophyceae) J Phycol. 2009;45:635–641. doi: 10.1111/j.1529-8817.2009.00689.x. [DOI] [PubMed] [Google Scholar]
- 31.Campo JAD, Rodríguez H, Moreno J, Vargas MÁ, Rivas J, Guerrero MG. Accumulation of astaxanthin and lutein in Chlorella zofingiensis (Chlorophyta) Appl Microbiol Biotechnol. 2004;64:848–854. doi: 10.1007/s00253-003-1510-5. [DOI] [PubMed] [Google Scholar]
- 32.Kato S, Takaichi S, Ishikawa T, Asahina M, Takahashi S, Shinomura T. Identification and functional analysis of the geranylgeranyl pyrophosphate synthase gene (crtE) and phytoene synthase gene (crtB) for carotenoid biosynthesis in Euglena gracilis. BMC Plant Biol. 2016;4:1–12. doi: 10.1186/s12870-015-0698-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rabbani S, Beyer P, Lintig JV, Hugueney P, Kleinig H. Induced β-carotene synthesis driven by triacylglycerol deposition in the unicellular alga Dunaliella bardawil. Plant Physiol. 1998;116:1239–1248. doi: 10.1104/pp.116.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang B, Zarka A, Trebst A, Boussiba S. Astaxanthin accumulation in Haematococcus pluvialis (Chlorophyceae) as an active photoprotective process under high irradiance. J Phycol. 2003;39:1116–1124. doi: 10.1111/j.0022-3646.2003.03-043.x. [DOI] [Google Scholar]
- 35.Kivic PA, Vesk M. Structure and function in the Euglenoid eyespot apparatus: the fine structure, and response to environmental changes. Planta. 1972;105:1–14. doi: 10.1007/BF00385158. [DOI] [PubMed] [Google Scholar]
- 36.Heelis DV, Kernick W, Phillips GO, Davies K. Separation and identification of the carotenoid pigments of stigmata isolated from light-grown cells of Euglena gracilis strain Z. Arch Microbiol. 1979;121:207–211. doi: 10.1007/BF00425057. [DOI] [PubMed] [Google Scholar]
- 37.Cunningham FX, Schiff JA. Chlorophyll-protein complexes from Euglena gracilis and mutants deficient in chlorophyll b. I Pigment Composition Plant physiol. 1986;80:223–230. doi: 10.1104/pp.80.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lavaud J, Rousseau B, van Gorkom HJ, Etienne A-L. Influence of the diadinoxanthin pool size on photoprotection in the marine planktonic diatom Phaeodactylum tricornutum. Plant Physiol. 2002;129:1398–1406. doi: 10.1104/pp.002014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruban AV, Lavaud J, Rousseau B, Guglielmi G, Horton P, Etienne A-L. The super-excess energy dissipation in diatom algae: comparative analysis with higher plants. Photosynth Res. 2004;82:165–175. doi: 10.1007/s11120-004-1456-1. [DOI] [PubMed] [Google Scholar]
- 40.McCarthy SS, Kobayashi MC, Niyogi KK. White mutants of Chlamydomonas reinhardtii are defective in phytoene synthase. Genetics. 2004;168:1249–1257. doi: 10.1534/genetics.104.030635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sandmann G, Bishop NI, Senger H. The carotenoid-deficient mutant, C-6E, of Scenedesmus obliquus is blocked at the site of phytoene synthase. Physiol Plantarum. 1997;99:391–394. doi: 10.1111/j.1399-3054.1997.tb00552.x. [DOI] [Google Scholar]
- 42.Cramer M, Myers J. Growth and photosynthetic characteristics of Euglena gracilis. Arch Mikrobiol. 1952;17:384–402. doi: 10.1007/BF00410835. [DOI] [Google Scholar]
- 43.Porra RJ, Thompson WA, Kriedemann PE. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta. 1989;975:384–394. doi: 10.1016/S0005-2728(89)80347-0. [DOI] [Google Scholar]
- 44.Iseki M, Matsunaga S, Murakami A, Ohno K, Shiga K, Yoshida K, et al. A blue-light-activated adenylyl cyclase mediates photoavoidance in Euglena gracilis. Nature. 2002;415:1047–1051. doi: 10.1038/4151047a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Effects of light intensity on carotenoid composition of E. gracilis cells. (A–E) HPLC chromatogram (445 nm) of extracts from E. gracilis grown under illumination at 27 (A), 55 (B), 240 (C), 460 (D), or 920 μmol m−2 s−1 (E) for 7 days. (Insets) Same chromatograms with an expanded y axis. mAU, milli-absorbance units. 1, neoxanthin; 2, diadinoxanthin; 3, all trans-diatoxanthin; 4–6, cis-diatoxanthin; 7, chlorophyll b; 8, chlorophyll a; 9, β-carotene (PDF 96 kb)
Effects of suppressing EgcrtB on carotenoid composition of E. gracilis cells. (A–C) HPLC chromatogram (445 nm) of extracts from E. gracilis cells treated without electroporation or EgcrtB-dsRNA (non-electroporated) (A), or cells treated with (C) or without EgcrtB-dsRNA (B). (Insets) Same chromatograms with an expanded y axis. mAU, milli-absorbance units. 1, neoxanthin; 2, diadinoxanthin; 3, all trans-diatoxanthin; 4–6, cis-diatoxanthin; 7, chlorophyll b; 8, chlorophyll a; 9, β-carotene (PDF 69 kb)
Data Availability Statement
The data that support the findings of this study are included in this published article and its supplementary information files.


