Summary
Stem cells undergo extensive metabolic rewiring during reprogramming, proliferation and differentiation, and numerous studies have demonstrated a significant role of metabolism in controlling stem cell fates. Recent applications of metabolomics, the study of concentrations and fluxes of small molecules in cells, have advanced efforts to characterize and maturate stem cell fates, assess drug toxicity in stem cell tissue models, identify biomarkers, and study the effects of environment on metabolic pathways in stem cells and their progeny. Looking to the future, combining metabolomics with other -omics approaches will provide a deeper understanding of the complex regulatory mechanisms of stem cells.
Introduction
Pluripotent stem cells (PSCs), including embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) have the ability to self-renew indefinitely and differentiate to any of the three germ layers [1,2]. In addition to potential applications in regenerative therapies, PSCs provide opportunities to model developmental progression and disease phenotypes, and can be used for drug screening and toxicity testing applications [3]. Major challenges to achieving the full potential of PSCs include identifying conditions that maintain their stemness and developing processes that effectively differentiate and mature PSCs to desired specialized cell types [3]. Another challenge is the development of phenotypic assays that employ stem cell-derived cells and tissues to expedite the process of drug screening and also provide mechanistic understanding of drug effects on human systems.
Metabolism and associated epigenetic remodeling have been found to play a crucial role in maintaining human PSC (hPSC) stemness and regulating differentiation [4–9••]. Metabolomics, the study of the complete set of small molecules or metabolites in a cell, is a reproducible, accurate and sensitive tool to analyze metabolic changes [10]. Metabolomics can also classify different cell types based on their molecular signatures [11], identify metabolite biomarkers in biological samples [12], and assess the effects of different drugs on cells and tissues [13]. In this review, we will highlight advances in applications of metabolomics in the PSC field in the past five years and discuss the challenges and future directions in employing metabolomics to advance in vitro and in vivo applications of PSCs.
Metabolite regulation of stem cell fate
Metabolism plays a crucial role in PSC survival, proliferation, and differentiation. Alterations in energy requirements during PSC homeostasis and differentiation can lead to significant changes in metabolic pathway utilization [14]. Specifically, during and after reprogramming to iPSCs, glycolysis is the predominant pathway for ATP generation and this is essential for hPSC maintenance and self-renewal [14,15]. Aerobic glycolysis is also common in other rapidly proliferating cells including cancer cells [16]. A hypotheses for utilization of aerobic glycolosis for energy generation in cancer cells is that glycolytic intermediates are used for nucleotide and protein synthesis to support increased proliferation [16]. Whether this is true for PSCs is still an open question. Importantly, the switch of energy generation from glycolysis to oxidative phosphorylation is sufficient to induce differentiation in PSCs [8], suggesting a role of glycolysis in PSC self-renewal. Other metabolic pathways, including lipid metabolism, have also shown to be enriched in PSCs and have been modulated to enhance iPSC reprogramming efficiency [17•].
One of the mechanisms by which metabolites can directly control PSC fate is by altering the epigenetic landscape [4–7•,14,18–20••]. S-adenosyl methionine (SAM) donates methyl groups for histone and DNA methylation, and levels of intracellular SAM can regulate methylation potential. Several metabolites have been shown to affect SAM levels, including methionine and threonine. Deprivation of methionine [5••] or threonine (in mouse ESCs) [18,19] in culture medium led to a rapid decrease in SAM and triggered histone and DNA demethylation, thereby increasing hPSC differentiation. Extended culture in methionine-deprived medium resulted in increased apoptosis [5••].
Histone methylation potential of naive human embryonic stem cells (hESCs) is reduced by increased activity of nicotinamide N-methyltransferase (NNMT) [6••]. NNMT catalyzes the conversion of SAM to 1-methylnicotinamide, which acts as a methyl sink [21] and is responsible for low levels of SAM. Histone/DNA demethylation is an equally important process in epigenetic regulation and it was shown that the intracellular α-ketoglutarate (α-KG) to succinate ratio regulates ten-eleven translocation (Tet)-dependent DNA demethylation, which is crucial for maintaining pluripotency in mouse ESCs [20••]. Directly altering this ratio of α-KG/succinate by supplementation of α-KG supported self-renewal while supplementation of succinate promoted differentiation, providing further evidence for metabolic regulation of pluripotency in mouse ESCs [20••].
Moussaieff et al. [7•] provided evidence that glycolytic acetyl-CoA affects histone acetylation in hPSCs. The authors showed that glycolytic production of acetyl-CoA promoted histone acetylation in PSCs and that modulation of glycolysis was sufficient to regulate pluripotency [7•]. There are several other metabolites which affect histone acetylation [22] and abundances of these metabolites can, therefore, affect PSC fates. Metabolites that affect histone post-translational modifications, such as acetylation, methylation, and phosphorylation, will also likely regulate global protein modifications that modulate stem cell pluripotency and differentiation. Onjiko et al. [9•] used single cell capillary electrophoresis-electrospray ionization mass spectrometry to show that different cell types in the 16-cell embryos of the South African clawed frog contained different quantities of metabolites. Additionally, changing metabolite concentrations altered cell migration during gastrulation, which in turn influenced the differentiation fates of these cells, indicating the importance of the balance of metabolites in determining the fates of stem cells during development [9•]. Overall, these studies highlight the important role metabolite concentrations play in regulation of protein post-translational modifications, epigenetics, and pluripotency and differentiation fates in stem cells.
Metabolism affects maturation of PSC-derived cells
In addition to regulation of stem cell fate, recent studies have also highlighted an important role of metabolism in maturation of hPSC-derived cell types [23••,24•]. For example, adult cardiomyocytes (CMs) primarily utilize fatty acid metabolism while immature hPSC-CMs rely on oxidative phosphorylation for energy generation [25]. A recent study by Kuppusamy et al. [23••] identified that the let-7 family of miRNAs (let-7i and let-7g) are upregulated in adult CMs. They showed that overexpression of let-7 miRNAs, whose targets involve genes in fatty acid metabolism and oxidative phosphorylation pathways, promoted maturation of hESC-CMs accompanied by a metabolic switch to fatty acid oxidation [23••].
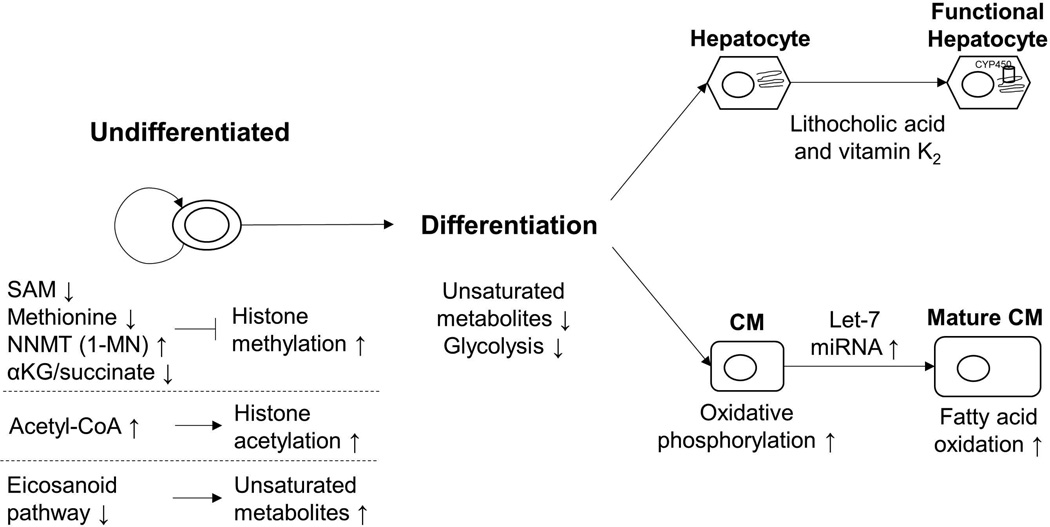
Similarly, hPSC-derived hepatocytes exhibit different toxicity responses than adult human hepatocytes [26]. For example, hPSC-derived hepatocytes express lower levels of cytochrome P450 (CYP450), which is important for metabolic transformation of lipids and for xenobiotic transformation, than adult hepatocytes [27]. Avior et al. [24•] observed metabolic maturation driven by microbial-derived metabolites, including lithocholic acid and vitamin K2, in hPSC-derived hepatocytes. They observed greater than 8-fold induction of CYP450 expression and were able to predict the TC50 (concentration causing 50% cell death) for several toxins with very high accuracy. These studies highlight the significant role metabolites and metabolic pathways play in maturation of hPSC-derived cell types. A summary of the effects of metabolites and pathways on altering the PSC fate is shown in Figure 1.
Figure 1. Regulation of PSC fate and maturation by metabolites and metabolic pathways.
Epigenetic regulation plays a significant role in maintaining the pluripotent state of PSCs. For example, histone methylation and acetylation is affected by specific metabolites. Stem cells also show increased abundance of unsaturated metabolites and inhibition of the eicosanoid pathway can assist in maintenance of pluripotency. During differentiation, metabolism shifts from glycolysis to oxidative phosphorylation or fatty acid oxidation accompanied by a reduction in unsaturated metabolites. Metabolic pathway regulation can drive maturation of hPSC-CMs and hPSC-derived hepatocytes. Abbrev. SAM: S-adenosyl methionine, NNMT: Nicotinamide N-Methyltransferase, 1-MN: 1-methylnicotinamide, CoA: coenzyme A, CM: cardiomyocyte
Identifying metabolic signatures and biomarkers via metabolomics
Cell types can be characterized by distinct and unique metabolite profiles. Several studies have profiled metabolite changes in hPSCs and hPSC-derived cells [8,11,28–30]. Yanes et al. [8] showed distinct metabolic changes in hESCs on differentiation to ectoderm and mesoderm and identified metabolic pathways that regulate hPSC differentiation. For example, eicosanoid pathway inhibition maintained pluripotency while substrates for oxidative metabolism, including fatty acids and acyl carnitines, promoted neurogenesis and cardiogenesis [8]. Panopoulos et al. [11], Varum et al. [28] and Meissen et al. [30] identified metabolic differences between ESCs, iPSCs and their somatic derivatives. Despite the epigenetic and functional similarities of ESCs and iPSCs [31], Panopoulos et al. [11] observed significant metabolic changes in unsaturated fatty acid metabolites, SAM, hypoxanthine and inosine. Based on gene expression analysis of glucose metabolism pathways, oxygen consumption rates and lactate production, Panopoulos et al. [11] and Varum et al. [28] concluded that hPSCs primarily relied on glycolysis to meet their energy demands. Meissen et al. [30] identified significant metabolic differences in mouse ESCs and iPSCs, mainly in amino acids and suggested differences in polyamine pathway activity due to significant differences in putrescine and 5-amthylthioadenosine (Table 1).
Table 1.
Summary of application of metabolomics to analyze the effects of culture conditions, drugs, and teratogens on PSCs and PSC-derived cells.
| Effect studied |
Metabolites/pathways affected (Sample type) |
PSC lines | Analytical technique |
Ref. |
|---|---|---|---|---|
| ESC vs. iPSC vs. somatic cells |
Unsaturated fatty acids ↓; SAM ↑, inosine ↑, hypoxanthine ↑ in iPSCs vs. ESCs (intracellular) |
hESCs, mESCs, iPSCs |
LC-MS, GC- MS |
[8,11,28,30] |
| hESCs vs. hECCs |
Octadecenoic acid ↑, glycerol-3- phosphate ↑, 4-hydroxyproline ↑, glutamic acid ↓, mannitol ↓, malic acid ↓, GABA ↓ in hESCs compared to hECCs (intracellular) |
H9 (hESC), NTERA2cl.D1 (hECC) |
GC-MS | [29] |
| Teratogens vs. non- teratogens |
Arginine to asymmetric dimethylarginine (ADMA) between 0.9 and 1.1 for non- teratogens (except for ascorbic acid and caffeine), GABA and malate are increased while succinate is reduced due to teratogens (supernatant) |
H9 (hESC) | LC-MS | [13,38] |
| Ethanol |
Embryoid bodies: MTA ↓ at higher dose and succinyladenosine ↑ at both low and high dose, thyroxine ↑ at lower dose Neural Progenitors: Kynurenine ↑ at lower dose of EtOH Neurons: Indoleacetaldehyde ↑ at lower dose (supernatant) |
H1, H9 (hESCs) |
LC-MS | [41] |
| Steroid hormones |
Estrogen: Lactate ↑, aspartate ↓, lysine ↓, phospholipids ↓, threonine ↓, valine ↓ Testosterone: Glycerol ↑, glycogen ↑, valine ↓ Progesterone: Organic acids ↓, phenylalanine ↓, proline ↓, tyrosine ↓ Common: Glucose ↑ and fatty acids ↓, inositol ↓ in germ-like cells (intracellular) |
BG01 (hESC), IMR90- derived iPSC |
GC-MS | [39] |
| Cisplatin | Oxidized and reduced glutathione ↑, urea ↑, proline ↑, putrescine ↑, spermine ↑, SAM ↑, several nucleotides were also altered (intracellular) |
HM1 (mESC) | NMR, LC- MS |
[40] |
| Passage difference of conditioned media |
Higher lactate, alanine, formate and lower tryptophan in HFF conditioned media which supported hESC maintenance (supernatant) |
H9 (hESC) | NMR | [42] |
| 5% vs. 20% oxygen |
Increased glucose consumption and lactate production at 5% oxygen (supernatant) |
Hues7, Shef3 (hESCs) |
Biochemistry Analyzer |
[45•] |
| 2% oxygen vs. 20% oxygen |
In addition to glycolysis, also utilized glutamine and amino acids for energy generation using oxidative phosphorylation and citric acid cycle (supernatant) |
MEL-2 (hESC) |
HPLC | [44] |
| Lipid and nutrient availability |
Metabolic rewiring takes place depending on nutrient availability and in addition to glycolysis, hPSCs also utilize oxidative phosphorylation (intracellular) |
HUES9, H9 (hESCs), iPS(IMR90)- c4 |
GC-MS | [43••] |
| 2D vs. 3D | Higher lactate-glucose ratios in small hESC colonies than large colonies (supernatant) |
H9 (hESC) | Biochemistry Analyzer |
[46] |
| Passaging methods |
Enzymatic passaging led to reduction of lipogenesis and glucose utilization (intracellular) |
H9 (hESC) | GC-MS | [47] |
Dawud et al. [29] found cell type-specific metabolic signatures in hESCs and human embryonal carcinoma cells, including differences in cell membrane components, despite similarities in concentrations of glycolysis pathway components. Recent studies have also reported metabolic signatures of hPSC-derived cell types including hPSC-derived vascular endothelial and smooth muscle cells [32], and CMs [33•].
Identifying molecular signatures can be useful in applications including purification of specific cell types based on their metabolic pathways [33•], assessing the efficiency of generating a target cell type based on the activity of specific pathways [17•] or comparing hPSC-derived cells with their primary counterparts [34]. Tohyama et al. [33•] developed an approach for purification of mouse and human PSC-derived CMs based on the significant differences in the glucose and lactate metabolism between CMs and other cell types. Only CMs were able to metabolize lactate in glucose-depleted culture medium and authors were able to achieve up to 99% CM purity. Pei et al. [17•] used lipid droplet abundance as a marker for reprogramming and determined that Rab32 improved iPSC reprogramming efficiency by enhancing lipid biosynthesis.
Metabolomics in iPSC-based disease modeling
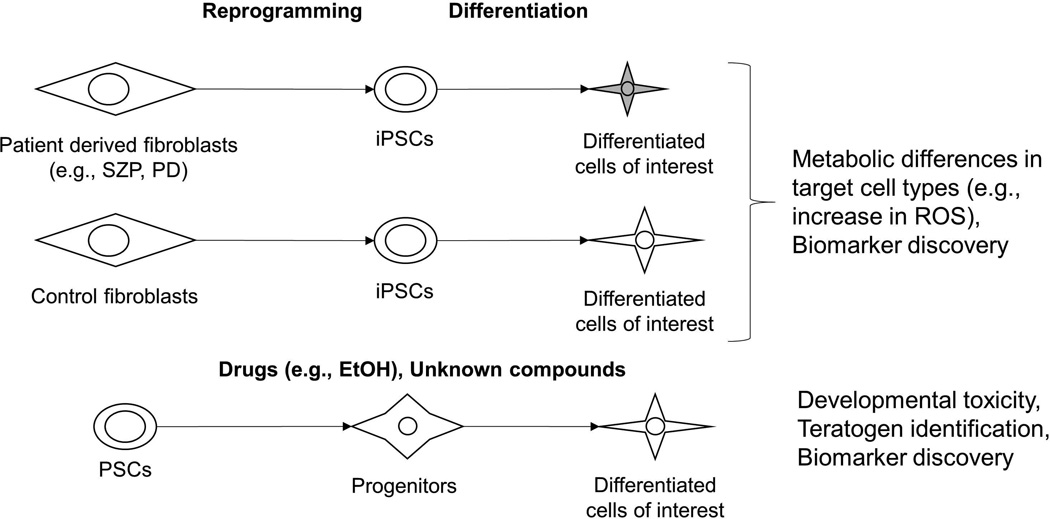
Recent studies have employed disease-specific iPSC models to study the metabolic changes the disease state imparts on specific cell populations [35–37]. For example, Paulsen et al. [35] demonstrated that neural progenitor cells (NPCs) differentiated from iPSCs reprogrammed from schizophrenia (SZP) patients generated more reactive oxygen species (ROS) and consumed more oxygen compared to NPCs derived from control iPSC lines. Importantly, this difference was only evident in NPC-SZP and not the patient fibroblasts or undifferentiated iPSCs. Interestingly, valproate treatment was able to restore the ROS levels similar to control, however extramitochondrial oxygen consumption was significantly increased due to valproate treatment in both NPC-control and NPC-SZP.
Imazumi et al. [36] and Cooper et al. [37] generated iPSCs from Parkinson’s disease (PD) patients harboring mutations in PARK2 [36] or PINK1 and LRRK2 [37] and observed several metabolic phenotypes, including increased ROS-mediated stress which led to reduced glutathione levels exclusively in iPSC-derived neurons (and not iPSCs or iPSC-derived fibroblasts) as compared to control iPSCderived neurons [36,37]. The PD iPSC-neurons also exhibited aberrations in mitochondrial morphology and impaired mitochondrial homeostasis [36,37], which was rescued by coenzyme Q10, rapamycin, or the LRRK2 kinase inhibitor GW5074 [37], further highlighting the value of metabolomics in assessing phenotypes in iPSC-based disease models and identifying potential therapeutic targets (Figure 2).
Figure 2. Applications of metabolomics in drug discovery using PSCs.
Patient-derived iPSCs can serve as in vitro models for drug screening and identifying the metabolic consequences of disease-related genetic mutations. PSCs can also serve as models to assess developmental toxicity of several drugs and quantify the teratomagenicity of compounds. Abbrev. iPSCs: induced pluripotent stem cells, SZP: schizophrenia, PD: Parkindon’s disease, PSCs: pluripotent stem cells, ROS: reactive oxygen species
Applications of metabolomics in identifying the effects of teratogens on PSCs
Several studies have used metabolomics to understand the effects of different drugs on stem cell metabolism [13,38–41]. West et al. [13] and Kleinstreuer et al. [38] detected teratogens and also identified biomarkers of developmental toxicity by quantifying metabolic changes in the hESC secretome in response to drug dosing. Palmer et al. [41] quantified the secretome of hESC-derived embryoid bodies, neural progenitors, and neurons in response to different doses of ethanol (EtOH) exposure to identify metabolic changes and biochemical pathways which play a role in alcohol-induced developmental neurotoxicity. They observed statistically significant changes due to EtOH exposure in all the three cell types, although none of the responses were common to all cell types. Based on these results, the authors suggested 5’-methylthioadenosine (MTA) and thyroxine as potential biomarkers for alcohol toxicity during early stages of development (Table 1).
West et al. [39] used GC-MS based metabolomics to assess the effects of several steroid hormones on the metabolism of hESC-derived germ-like cells and developed models to distinguish the effects of different hormones on metabolism. Combining transcriptomic and metabolomic analysis, Stechow et al. [40] identified cisplatin-regulated pathways in human PSCs, including nucleotide metabolism, the urea cycle, and arginine and proline metabolism. Several anti-oxidant associated metabolites and p53-regulated enzymes also showed significant enrichment due to genotoxic stress induced by cisplatin. These studies highlight the sensitivity of metabolism to drugs and hormones and the potential of hPSCs in drug toxicology screening (Figure 2).
Assessing the effect of environment on stem cell metabolism
In addition to studying the effects of drugs, metabolomics has also been used to understand the effect of medium components [42,43••], physiological and atmospheric oxygen concentrations [44,45•], 2D vs. 3D culture [46], and enzymatic passaging [47] on stem cell metabolism. Batch-to-batch variation in media can negatively impact the reproducibility of stem cell culture and differentiation. MacIntyre et al. [42] correlated metabolite concentrations in conditioned medium by human foreskin fibroblasts (HFFs) at different passages to their ability to maintain hESCs in culture (Table 1). HFF metabolism changed with extended culture, metabolite content of the conditioned media at different passages may account for differences in maintenance capability of hESCs by HFF-conditioned media [42].
Forristal et al. [45•] reported that hESCs cultured at 5% oxygen showed increased glucose consumption and lactate production as compared to hESCs cultured at atmospheric conditions (20% oxygen). A comprehensive analysis of metabolic fluxes was performed by Turner et al. [44] in hESCs cultured in atmospheric vs. hypoxic (2% oxygen) conditions. Although hESCs utilized glucose via aerobic and anaerobic glycolysis in both atmospheric and hypoxic conditions, they also utilized glutamine as the carbon source for oxidative phosphorylation to maximize the ATP production in atmospheric conditions, with amino acids as the major substrates for tricarboxylic acid (TCA) cycle. A recent study by Zhang et al. [43••] also reported the ability of hPSCs to utilize the TCA cycle in addition to glycolysis for energy generation depending on the availability of lipids in the media (Table 1).
Azarin et al. [46] studied the impact of 2D vs. 3D culture of hESCs on their cell cycle and metabolism and observed negligible changes in lactate-glucose ratios due to 3D culture, although they did report that 100 μm diameter hESC colonies contained higher lactate-glucose ratios than larger 3D colonies; the smaller colonies also exhibited less spontaneous differentiation than larger colonies [48], perhaps indicating a relationship between lactate-glucose ratio and hPSC self-renewal. Similarly, different methods of enzymatic passaging of hPSCs resulted in reduction of lipogenesis and glucose utilization in the central carbon metabolism as compared to non-enzymatic dissociation [47] (Table 1). Together, these studies demonstrate that identifying hPSC metabolic responses to culture conditions can be used to improve culture efficiency and robustness.
Conclusions and Future Perspectives
Recent studies have proven that metabolic changes and metabolite abundances can regulate PSC fates by affecting the epigenetic landscape in stem cells [4,5,18]. Patient-derived iPSCs are increasingly being used as in vitro models [49] to identify these metabolic alterations and predict new therapeutic targets to reverse or rescue the metabolic dysfunction in diseases. Advances in genome editing technologies like CRISPR-Cas9 can also be used to introduce mutations to study the effects of genetic disease on metabolism [50]. Recent studies have shown that understanding unique metabolic requirements can be used to develop strategies for efficient purification of specific cell types [33•] and that metabolic pathways play critical role in regulating cell maturation [23••]. Combining metabolomics with metabolic flux analysis (MFA) has the potential to provide deeper insight into how differentiation affects metabolism in stem cells and should prove useful in developing strategies to enhance pluripotency or differentiation fates in stem cells by modulating metabolic pathways [20••,43••,47,51].
There are numerous successful demonstrations of the application of metabolomics for toxicological screening in PSCs and PSC-derived cells, especially in studying developmental disorders [13]. Metabolomics has also been used to understand and optimize the impact of culture conditions on stem cell maintenance and differentiation. Different aspects of stem cell biology can also be assessed using metabolomics, including profiling the secretome and investigating interactions between stem cells and other cell types or substrates, and probing effects of geometry, organization, and density of stem cells in 2D and 3D culture. Advances in spectroscopic techniques have improved the sensitivity and reproducibility of metabolomics data, yet several challenges remain [10]. Even though we are able to measure metabolites to picomolar sensitivity using mass spectrometry approaches, accurate identification of these metabolites is still a challenge [10]. Differences in experimental methods, cell status, profiling approaches, and data processing and analysis have to be considered when comparing metabolic signatures. In the stem cell field, where the media and culture conditions can vary from lab to lab and are continually evolving, comparing results from different studies is particularly difficult. Detailed guidelines for sample preparation and analysis can overcome some of these challenges [52]. When combined with other -omics approaches, metabolomics promises to offer unprecedented insight into interactions between metabolic and signaling pathways that regulate stem cell fate.
Highlights.
Metabolism regulates stem cell fate by altering epigenetic markers
Metabolite signatures can be used to characterize differences between cell types
Metabolomics can be used to identify biomarkers of developmental toxicity to drugs
Metabolomics has also been used to optimize stem cell culture conditions
Acknowledgments
This work was supported by the National Institutes of Health grants R01CA164492 and R01EB007534.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mitalipov S, Wolf D. Engineering of Stem Cells. Springer Berlin Heidelberg: 2009. Totipotency, Pluripotency and Nuclear Reprogramming; pp. 185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Trounson A, McDonald C. Stem Cell Therapies in Clinical Trials: Progress and Challenges. Cell Stem Cell. 2015;17:11–22. doi: 10.1016/j.stem.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 4.Avitabile D, Magenta A, Lauri A, Gambini E, Spaltro G, Vinci MC. Metaboloepigenetics: the Emerging Network in Stem Cell Homeostasis Regulation. Curr. Stem Cell Res. Ther. 2016;11:352–369. doi: 10.2174/1574888x11666151203223839. [DOI] [PubMed] [Google Scholar]
- 5. Shiraki N, Shiraki Y, Tsuyama T, Obata F, Miura M, Nagae G, Aburatani H, Kume K, Endo F, Kume S. Methionine Metabolism Regulates Maintenance and Differentiation of Human Pluripotent Stem Cells. Cell Metab. 2014;19:780–794. doi: 10.1016/j.cmet.2014.03.017. •• This study demonstrated that hPSCs require high levels of methionine for survival and maintenance of the pluripotent state. Short term deficiency of methionine led to histone demethylation and differentiation while long term deprivation led to apoptosis.
- 6. Sperber H, Mathieu J, Wang Y, Ferreccio A, Hesson J, Xu Z, Fischer KA, Devi A, Detraux D, Gu H, et al. The metabolome regulates the epigenetic landscape during naive-to-primed human embryonic stem cell transition. Nat. Cell Biol. 2015;17:1523–1535. doi: 10.1038/ncb3264. •• The authors showed that upregulation of H3K27me3 repressive epigenetic marks during the naive-to-primed hESC transition is controlled by the metabolic enzyme nicotinamide N-methyltransferase which converts SAM to 1-methylnicotinamide.
- 7. Moussaieff A, Rouleau M, Kitsberg D, Cohen M, Levy G, Barasch D, Nemirovski A, Shen-Orr S, Laevsky I, Amit M, et al. Glycolysis-mediated changes in acetyl-CoA and histone acetylation control the early differentiation of embryonic stem cells. Cell Metab. 2015;21:392–402. doi: 10.1016/j.cmet.2015.02.002. • The authors provide evidence for the role of glycolysis in maintaining pluripotency via acetyl-CoA-mediated changes in histone acetylation.
- 8.Yanes O, Clark J, Wong DM, Patti GJ, Sánchez-Ruiz A, Benton HP, Trauger Sa, Desponts C, Ding S, Siuzdak G. Metabolic oxidation regulates embryonic stem cell differentiation. Nat. Chem. Biol. 2010;6:411–417. doi: 10.1038/nchembio.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Onjiko RM, Moody Sa, Nemes P. Single-cell mass spectrometry reveals small molecules that affect cell fates in the 16-cell embryo. Proc. Natl. Acad. Sci. U. S. A. 2015;112:6545–6550. doi: 10.1073/pnas.1423682112. •• Using single cell technology, the authors provided direct evidence for differential abundance of metabolites within the cells of the embryo with different tissue fates. Altering the abundance of several metabolites such as threonine, serine, methionine and acetylcholine in the blastomeres affected development and cell fate.
- 10.Mathew AK, Padmanaban VC. Metabolomics: The apogee of the omics trilogy. Int. J. Pharm. Pharm. Sci. 2013;5:45–48. [Google Scholar]
- 11.Panopoulos AD, Yanes O, Ruiz S, Kida YS, Diep D, Tautenhahn R, Herrerías A, Batchelder EM, Plongthongkum N, Lutz M, et al. The metabolome of induced pluripotent stem cells reveals metabolic changes occurring in somatic cell reprogramming. Cell Res. 2012;22:168–177. doi: 10.1038/cr.2011.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Armitage EG, Barbas C. Metabolomics in cancer biomarker discovery: current trends and future perspectives. J. Pharm. Biomed. Anal. 2014;87:1–11. doi: 10.1016/j.jpba.2013.08.041. [DOI] [PubMed] [Google Scholar]
- 13.West PR, Weir AM, Smith AM, Donley ELR, Cezar GG. Predicting human developmental toxicity of pharmaceuticals using human embryonic stem cells and metabolomics. Toxicol. Appl. Pharmacol. 2010;247:18–27. doi: 10.1016/j.taap.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 14.Zhang J, Nuebel E, Daley GQ, Koehler CM, Teitell MA. Metabolic regulation in pluripotent stem cells during reprogramming and self-renewal. Cell Stem Cell. 2012;11:589–595. doi: 10.1016/j.stem.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Folmes CDL, Nelson TJ, Martinez-Fernandez A, Arrell DK, Lindor JZ, Dzeja PP, Ikeda Y, Perez-Terzic C, Terzic A. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 2011;14:264–271. doi: 10.1016/j.cmet.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pei Y, Yue L, Zhang W, Wang Y, Wen B, Zhong L, Xiang J, Li J, Zhang S, Wang H, et al. Improvement in Mouse iPSC Induction by Rab32 Reveals the Importance of Lipid Metabolism during Reprogramming. Sci. Rep. 2015;5:16539. doi: 10.1038/srep16539. • The authors describe upregulation of lipid biosynthesis during iPSC reprogramming mediated by Oct4, Sox2, Klf4, and Rab32. Inhibition of lipid biosynthesis reduced the reprogramming efficiency highlighting the importance of lipid metabolism during reprogramming.
- 18.Shyh-Chang N, Locasale JW, Lyssiotis CA, Zheng Y, Teo RY, Ratanasirintrawoot S, Zhang J, Onder T, Unternaehrer JJ, Zhu H, et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 2013;339:222–226. doi: 10.1126/science.1226603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J, Alexander P, Wu L, Hammer R, Cleaver O, McKnight SL. Dependence of Mouse Embryonic Stem Cells on Threonine Catabolism. Science. 2009;325:435–439. doi: 10.1126/science.1173288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Carey BW, Finley LWS, Cross JR, Allis CD, Thompson CB. Intracellular α-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature. 2014;518:413–416. doi: 10.1038/nature13981. •• This study shows that manipulation of intracellular abundances can regulate the switch between self-renewal or differentiation in mouse ESCs. This is the first study to identify the regulation of Tet-dependent DNA demethylation by intracellular α-KG to succinate ratio.
- 21.Ulanovskaya OA, Zuhl AM, Cravatt BF. NNMT promotes epigenetic remodeling in cancer by creating a metabolic methylation sink. Nat. Chem. Biol. 2013;9:300–306. doi: 10.1038/nchembio.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choudhary C, Weinert BT, Nishida Y, Verdin E, Mann M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat. Rev. Mol. Cell Biol. 2014;15:536–550. doi: 10.1038/nrm3841. [DOI] [PubMed] [Google Scholar]
- 23. Kuppusamy KT, Jones DC, Sperber H, Madan A, Fischer K a, Rodriguez ML, Pabon L, Zhu W-Z, Tulloch NL, Yang X, et al. Let-7 family of microRNA is required for maturation and adult-like metabolism in stem cell-derived cardiomyocytes. Proc. Natl. Acad. Sci. 2015 doi: 10.1073/pnas.1424042112. •• This study exemplifies the potential of metabolic pathway induction of maturation in PSC-derived cell types. A switch from oxidative phosphorylation to fatty acid oxidation by overexpression of let-7 miRNAs resulted in maturation of hPSC-derived cardiomyocytes.
- 24. Avior Y, Levy G, Zimerman M, Kitsberg D, Schwartz R, Sadeh R, Moussaieff A, Cohen M, Itskovitz-Eldor J, Nahmias Y. Microbial-derived lithocholic acid and vitamin K 2 drive the metabolic maturation of pluripotent stem cells-derived and fetal hepatocytes. Hepatology. 2015;62:265–278. doi: 10.1002/hep.27803. • Microbial metabolites can induce expression of CYP450 in hPSC-derived hepatocytes, improving their capacity to predict compound toxicity.
- 25.Robertson C, Tran DD, George SC. Concise review: Maturation phases of human pluripotent stem cell-derived cardiomyocytes. Stem Cells. 2013;31:829–837. doi: 10.1002/stem.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szkolnicka D, Farnworth SL, Lucendo-Villarin B, Storck C, Zhou W, Iredale JP, Flint O, Hay DC. Accurate prediction of drug-induced liver injury using stem cell-derived populations. Stem Cells Transl. Med. 2014;3:141–148. doi: 10.5966/sctm.2013-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hannan NRF, Segeritz C-P, Touboul T, Vallier L. Production of hepatocyte-like cells from human pluripotent stem cells. Nat. Protoc. 2013;8:430–437. doi: 10.1038/nprot.2012.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Varum S, Rodrigues AS, Moura MB, Momcilovic O, Easley CA, Ramalho-Santos J, Van Houten B, Schatten G. Energy Metabolism in Human Pluripotent Stem Cells and Their Differentiated Counterparts. PLoS One. 2011;6:e20914. doi: 10.1371/journal.pone.0020914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abu Dawud R, Schreiber K, Schomburg D, Adjaye J. Human Embryonic Stem Cells and Embryonal Carcinoma Cells Have Overlapping and Distinct Metabolic Signatures. PLoS One. 2012;7:e39896. doi: 10.1371/journal.pone.0039896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meissen JK, Yuen BTK, Kind T, Riggs JW, Barupal DK, Knoepfler PS, Fiehn O. Induced Pluripotent Stem Cells Show Metabolomic Differences to Embryonic Stem Cells in Polyunsaturated Phosphatidylcholines and Primary Metabolism. PLoS One. 2012;7:1–9. doi: 10.1371/journal.pone.0046770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stadtfeld M, Hochedlinger K. Induced pluripotency: history, mechanisms, and applications. Genes Dev. 2010;24:2239–2263. doi: 10.1101/gad.1963910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patsch C, Challet-Meylan L, Thoma EC, Urich E, Heckel T, O’Sullivan JF, Grainger SJ, Kapp FG, Sun L, Christensen K, et al. Generation of vascular endothelial and smooth muscle cells from human pluripotent stem cells. Nat. Cell Biol. 2015;17:994–1003. doi: 10.1038/ncb3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tohyama S, Hattori F, Sano M, Hishiki T, Nagahata Y, Matsuura T, Hashimoto H, Suzuki T, Yamashita H, Satoh Y, et al. Distinct Metabolic Flow Enables Large-Scale Purification of Mouse and Human Pluripotent Stem Cell-Derived Cardiomyocytes. Cell Stem Cell. 2013;12:127–137. doi: 10.1016/j.stem.2012.09.013. • The authors identified the ability of cardiomyocytes to utilize lactate in absence of glucose which allowed them to purify cardiomyocytes by exposing them to lactate rich and glucose free medium.
- 34.Manganas Louis N, Zhang Xueying, Li Yao, Hazel Raphael D, Smith S. David, Wagshul Mark E, Henn Fritz, Benveniste Helene, Djuric Petar M, Grigori Enikolopov MM-S. Magnetic Resonance Spectroscopy Identifies Neural Progenitor Cells in the Live Human Brain. Science. 2007;318:980–985. doi: 10.1126/science.1147851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paulsen BdaS, Maciel RdeM, Galina A, da Silveira MS, Souza CdosS, Drummond H, Pozzatto EN, Junior HS, Chicaybam L, Massuda R, et al. Altered Oxygen Metabolism Associated to Neurogenesis of Induced Pluripotent Stem Cells Derived From a Schizophrenic Patient. Cell Transplant. 2012;21:1547–1559. doi: 10.3727/096368911X600957. [DOI] [PubMed] [Google Scholar]
- 36.Imaizumi Y, Okada Y, Akamatsu W, Koike M, Kuzumaki N, Hayakawa H, Nihira T, Kobayashi T, Ohyama M, Sato S, et al. Mitochondrial dysfunction associated with increased oxidative stress and α-synuclein accumulation in PARK2 iPSC-derived neurons and postmortem brain tissue. Mol. Brain. 2012;5:35. doi: 10.1186/1756-6606-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cooper O, Seo H, Andrabi S, Guardia-Laguarta C, Graziotto J, Sundberg M, McLean JR, Carrillo-Reid L, Xie Z, Osborn T, et al. Pharmacological rescue of mitochondrial deficits in iPSC-derived neural cells from patients with familial Parkinson’s disease. Sci. Transl. Med. 2012;4:141ra90. doi: 10.1126/scitranslmed.3003985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kleinstreuer NC, Smith AM, West PR, Conard KR, Fontaine BR, Weir-Hauptman AM, Palmer JA, Knudsen TB, Dix DJ, Donley ELR, et al. Identifying developmental toxicity pathways for a subset of ToxCast chemicals using human embryonic stem cells and metabolomics. Toxicol. Appl. Pharmacol. 2011;257:111–121. doi: 10.1016/j.taap.2011.08.025. [DOI] [PubMed] [Google Scholar]
- 39.West FD, Henderson WM, Yu P, Yang JY, Stice SL, Smith Ma. Metabolomic response of human embryonic stem cell-derived germ-like cells after exposure to steroid hormones. Toxicol Sci. 2012;129:9–20. doi: 10.1093/toxsci/kfs185. [DOI] [PubMed] [Google Scholar]
- 40.von Stechow L, Ruiz-Aracama A, van de Water B, Peijnenburg A, Danen E, Lommen A. Identification of Cisplatin-Regulated Metabolic Pathways in Pluripotent Stem Cells. PLoS One. 2013;8:e76476. doi: 10.1371/journal.pone.0076476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palmer JA, Poenitzsch AM, Smith SM, Conard KR, West PR, Cezar GG. Metabolic Biomarkers of Prenatal Alcohol Exposure in Human Embryonic Stem Cell-Derived Neural Lineages. Alcohol. Clin. Exp. Res. 2012;36:1314–1324. doi: 10.1111/j.1530-0277.2011.01732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.MacIntyre Da, Melguizo Sanchís D, Jiménez B, Moreno R, Stojkovic M, Pineda-Lucena A. Characterisation of human embryonic stem cells conditioning media by 1H-nuclear magnetic resonance spectroscopy. PLoS One. 2011;6:e16732. doi: 10.1371/journal.pone.0016732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang H, Badur MG, Divakaruni AS, Parker SJ, Jäger C, Hiller K, Murphy AN, Metallo CM. Distinct Metabolic States Can Support Self-Renewal and Lipogenesis in Human Pluripotent Stem Cells under Different Culture Conditions. Cell Rep. 2016;16:1–12. doi: 10.1016/j.celrep.2016.06.102. •• hESCs possess the ability to rewire metabolic pathways depending on the nutrient availability. Lipid-depleted medium upregulated lipid biosynthesis and redox pathways and downregulated oxidative phosphorylation.
- 44.Turner J, Quek L-E, Titmarsh D, Krömer JO, Kao L-P, Nielsen L, Wolvetang E, Cooper-White J. Metabolic Profiling and Flux Analysis of MEL-2 Human Embryonic Stem Cells during Exponential Growth at Physiological and Atmospheric Oxygen Concentrations. PLoS One. 2014;9:e112757. doi: 10.1371/journal.pone.0112757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Forristal CE, Christensen DR, Chinnery FE, Petruzzelli R, Parry KL, Sanchez-Elsner T, Houghton FD. Environmental Oxygen Tension Regulates the Energy Metabolism and Self-Renewal of Human Embryonic Stem Cells. PLoS One. 2013;8:e62507. doi: 10.1371/journal.pone.0062507. • The authors showed significant changes occur due to enzymatic passaging in hESCs, specifically in glucose metabolism, lipid biosynthesis and abundance of glycan moieties.
- 46.Azarin SM, Larson EA, Almodóvar-Cruz JM, de Pablo JJ, Palecek SP. Effects of 3D microwell culture on growth kinetics and metabolism of human embryonic stem cells. Biotechnol. Appl. Biochem. 2012;59:88–96. doi: 10.1002/bab.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Badur MG, Zhang H, Metallo CM. Enzymatic passaging of human embryonic stem cells alters central carbon metabolism and glycan abundance. Biotechnol. J. 2015;10:1600–1611. doi: 10.1002/biot.201400749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mohr JC, de Pablo JJ, Palecek SP. 3-D microwell culture of human embryonic stem cells. Biomaterials. 2006;27:6032–6042. doi: 10.1016/j.biomaterials.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 49.Robinton DA, Daley GQ. The promise of induced pluripotent stem cells in research and therapy. Nature. 2012;481:295–305. doi: 10.1038/nature10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hsu PD, Lander ES, Zhang F. Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oburoglu L, Tardito S, Fritz V, de Barros SC, Merida P, Craveiro M, Mamede J, Cretenet G, Mongellaz C, An X, et al. Glucose and Glutamine Metabolism Regulate Human Hematopoietic Stem Cell Lineage Specification. Cell Stem Cell. 2014;15:169–184. doi: 10.1016/j.stem.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 52.Lindon JC, Nicholson JK, Holmes E, Keun HC, Craig A, Pearce JTM, Bruce SJ, Hardy N, Sansone S-A, Antti H, et al. Summary recommendations for standardization and reporting of metabolic analyses. Nat. Biotechnol. 2005;23:833–838. doi: 10.1038/nbt0705-833. [DOI] [PubMed] [Google Scholar]