Abstract
Mitochondrial aldehyde dehydrogenase (ALDH2) plays a central role in the detoxification of reactive aldehydes generated through endogenous and exogenous sources. The biochemical regulation of enzyme activity through post-translational modification provides an intricate response system regulating mitochondrial detoxification pathways. ALDH2 is a known target of lysine acetylation, which arises as a consequence of mitochondrial bioenergetic flux and sirtuin deacetylase activity. The mitochondrial deacetylase Sirtuin 3 (SIRT3) has been reported to alter ALDH2 lysine acetylation status, yet the mechanism and consequence of this interaction remain unknown. The in vitro results presented here provide a novel biochemical approach using stable-isotope dilution mass spectrometry to elucidate which lysine residues are targeted by SIRT3 for deacetylation. Furthermore, HPLC-MS/MS and computational modeling elucidate a potential role for acetyl-Lys369 on ALDH2 in perturbing normal β-nicotinamide adenine dinucleotide (NAD+) cofactor binding.
TOC image

INTRODUCTION
Mitochondrial aldehyde dehydrogenase (ALDH2) is a key enzyme in the maintenance of mitochondrial fidelity through the detoxification of endogenous and exogenous cytotoxic reactive aldehydes.1, 2 Numerous studies have established the regulation of ALDH2 expression and activity as a key target for therapeutic intervention towards ameliorating a host of disease pathologies, including neurotoxicity, heart failure, and atherosclerosis.3–5
Recent evidence suggests that protein acetylation may play a critical role in regulating metabolic and antioxidant pathways.6 The acetylation of N-ε-lysine residues impacts both protein structure and function and it was recently reported that treatment of human aortic endothelial cells with a low dose of ethanol resulted in both ALDH2 acetylation and a concomitant increase in its enzymatic activity.7–10 Although evidence for the existence of a mitochondrial acetyltranferase is not yet definitive, the primary mitochondrial deacetylase is the HDAC class III, NAD+-dependent sirtuin family member Sirtuin 3 (SIRT3).6 A number of publications have demonstrated that ALDH2 is a target of SIRT3 deacetylation.7, 11–13 Accordingly, Xue and colleagues investigated the effect of ethanol on SIRT3. No change in SIRT3 protein expression was observed in their in vitro system, which is consistent with another in vivo report in mice following a chronic ethanol diet study.14 However, SIRT3 activity was decreased for a short period of time concurrent with an observed increase in ALDH2 activity in their cell culture model. Another report, published earlier in that same year, also demonstrated the interaction of SIRT3 with ALDH2.13 SIRT3-deficient mice exhibited increased acetylation of ALDH2, but with no observed change in ALDH2 activity.
In aggregate, the mechanism by which lysine acetylation affects ALDH2 activity remains unclear. Therefore, we present a fundamental in vitro characterization of the impact of non-enzymatic acetylation on ALDH2 activity. A basic biochemical approach was utilized to assess the consequences of acetylation dynamics and SIRT3 activity on the structural, functional, and electrostatic properties of ALDH2. These results suggest that deacetylation of ALDH2 by SIRT3 partially recovered the hyperacetylation-mediated inhibition of ALDH2 activity. Additionally, our results indicate that SIRT3 differentially deacetylates lysine residues on ALDH2 and, importantly, deacetylates acetyl-Lys369 on ALDH2. Lys369 plays a role in NAD+ cofactor binding, which is abrogated by acetylation and elucidates an unreported consequence of SIRT3-mediated ALDH2 regulation. A novel stable-isotope dilution tandem mass spectrometry method was utilized to quantify SIRT3-directed deacetylation of specific lysine residues on ALDH2. These data were combined to develop a new metric to prioritize lysine residues targeted by SIRT3. Computational molecular modeling was then applied to define the acetyl-directed modulation of NAD+ binding to ALDH2. Modeling also demonstrates the capacity for SIRT3 to deacetylate Lys369 on ALDH2. This study presents an integrated in vitro and in silico approach towards characterizing how acetylation regulates protein activity, in general, and demonstrates the specificity of SIRT3 deacetylase activity towards numerous ALDH2 lysine residues.
EXPERIMENTAL PROCEDURES
Immunoblotting
Samples containing equivalent amounts of protein were acetylated and then resolved by non-reducing polyacrylamide gel electrophoresis. The proteins were then transferred to a HyBond-P membrane (GE Healthcare) and blocked using a tris-buffered solution containing 1% (v/v) Tween 20 and 5% (w/v) non-fat dry milk. Membranes were incubated overnight at a primary antibody concentration of 1:1000 for anti-acetyllysine (#9441; Cell Signaling). A tris-buffered solution containing 1% (v/v) Tween 20 was used to wash the membranes prior to and following one-hour incubation with 1:10000 goat anti-rabbit horseradish peroxidase conjugated secondary antibody. Bands were visualized using enhanced chemiluminescence substrate and a Bio-Rad ChemiDoc MP system.
Native gel electrophoresis
Samples were prepared as described below and subjected to non-reducing, non-denaturing polyacrylamide gel electrophoresis. The gels were then incubated overnight with Imperial Protein Stain (Thermo Scientific) and subsequently rinsed with distilled water to improve contrast.
ALDH2 activity assay
Human recombinant ALDH2 protein was purchased from MyBioSource (MBS203287) where it was expressed and isolated in E. Coli with a purity greater than 90% by SDS-PAGE. The enzyme was aliquoted, flash frozen and stored at −80° C. Upon use each aliquot was thawed on ice and any remaining enzyme never underwent freeze/thaw more than twice with no observable detriment to activity. The purchased protein is missing the first 17 amino acids corresponding to the mitochondrion transit peptide and all residues are reported according to the full 517 amino acid length protein. Dilutions of acetic anhydride were performed in methanol and subsequently incubated with human recombinant ALDH2 protein at room temperature for 30 minutes. Organic content was never greater than 10% (v/v) for acetic anhydride treatments. The samples were then diluted with 200 mM sodium phosphate (pH 7.4) and 1 mM β-nicotinamide adenine dinucleotide (NAD+) was added. For kinetic parameter experiments NAD+ concentrations ranged from 31.25 μM to 1.5 mM and the IC25, IC50 and IC75 concentrations of acetic anhydride were 1.3, 3.4 and 10 mM respectively. Samples were run in triplicate in a 96-well quartz plate on a Molecular Devices SpectraMax 190 spectrophotometer. Propionaldehyde (1 mM) was added immediately before gathering data points at 30 second intervals for 20 minutes at 340 nanometers (nm) to measure the conversion to NAD(H). An Eadie-Hofstee plot was created in addition to nonlinear fitting utilization to determine kinetic parameters (GraphPad Prism 6).
Stable isotope dilution mass spectrometry
These experiments were adapted from a previously published protocol.15 Human recombinant ALDH2 protein was treated with either acetic anhydride or isotope labeled (deuterium) acetic anhydride (Cambridge Isotope Laboratories) for 30 minutes at room temperature. Samples treated with 10 mM acetic anhydride were diluted in 200 mM sodium phosphate containing NAD+ and were then incubated with human recombinant SIRT3 (Cayman Chemical) at 37° C for 45 minutes to allow for any potential removal of acetyl modifications. Protein was precipitated using ice-cold acetone, reduced with 10 mM dithiothreitol (DTT) in 50 mM ammonium bicarbonate (NH3HCO3), alkylated with 30 mM iodoacetamide in 50 mM NH3HCO3 and alkylation was quenched with another bolus of 10 mM DTT. Protein was digested with trypsin overnight at 37 °C. The resulting peptides were desalted using a ZipTip C18 tip (EMD Millipore). Samples treated with acetic anhydride or deuterated acetic anhydride were combined 1:1 and analyzed with nanoliquid chromatography and a Bruker maXis Impact quadrupole time-of-flight mass spectrometer. By mixing the samples, it is possible to detect an acetyl modification removed by SIRT3 as such a peptide would still contain the heavy acetylation (Figure S1). Lysine acetylation events were determined by a mass shift of +42.01 daltons or +45.01 daltons resulting from the reaction with the normal acetic anhydride (H) or the deuterated (D) acetic anhydride, respectively.
Data analysis and protein identification of the LC-MS/MS spectra by MASCOT was used to identify the initial peptides containing an acetylated lysine. A manual search of the spectral data was then performed in order to find all the charge states for acetylated peptides. Verification to correctly identify the unacetylated and acetylated peptides was performed by manual peptide fragmentation analysis. During the manual searches, it was also discovered that some peptides had potassium adducts corresponding to a mass shift of +38.96 daltons when compared to the unadducted peptides. Also, these peptides containing potassium adducts were found at different charge states. The combination of multiple charge states and potassium adducts resulted in as many as 5 different chromatograms for a given acetyl-peptide and complicates integration of the peaks. An isotope subtraction of a small percentage of the integral for the samples treated with the deuterated anhydride was required as a result of an overlap of the fourth isotope peak of the spectra from the samples treated with the normal anhydride. The mass to charge and integral data for all peptides can be found in Table S1. Peptides containing an unmodified lysine with multiple missed cleavages as well as other acetylated lysine residues, were included for calculation for that unmodified lysine. This is an important variable, due to the study of lysine modification and trypsin/Lys-C proteolytic cleavage, which may go overlooked when quantifying lysine acetylation. Based on the MS/MS quantification data, a novel metric was calculated [SIRT3 Target Affinity Ratio (STAR)] to show the specificity of SIRT3 for specific acetyllysine residues on ALDH2.
SIRT3 rescue assays
Human recombinant SIRT3 containing a N-terminal hexahistidine tag was purchased from Cayman Chemical (#10011194) where is was expressed and purified from E. Coli at a purity of 60% by SDS-PAGE. The storage conditions mirrored those of ALDH2. SIRT3 was dialyzed overnight in phosphate-buffered saline and subsequently concentrated. Acetic anhydride was diluted in methanol and allowed to react with human recombinant ALDH2 at varied concentrations at room temperature for 30 minutes. Samples were then diluted in 200 mM sodium phosphate containing 5 mM NAD+. The samples were then incubated with SIRT3 for 45 minutes at room temperature. Propionaldehyde was added to the triplicate samples in the quartz plate. Measurement of NAD(H) was performed at 340 nm every 30 seconds for 30 minutes at 37° C.
Computational-based molecular modeling of human ALDH2
All computational modeling was performed using Accelrys Discovery Studio 4.0 (Biovia, Inc). The crystal structure coordinates for human ALDH2 and human SIRT3 were downloaded from the Protein Data Bank (www.pdb.org; PDB IDs: 1O01 & 3GLR, respectively).16, 17 The acetylated ALDH2 structure was generated by modifying Lys369 to acetyl-Lys369 in Discovery Studio. Proteins were typed with the CHARMM forcefield and energy minimized with the smart minimizer protocol within Discovery Studio using the Generalized-Born with simple switching implicit solvent model to an RMS convergence <0.01 kcal/mol.18, 19 The theoretical ALDH2-SIRT3 complex was generated by molecular superimposition of the acetylated Lys369 of ALDH2 over the acetyllysine in the co-crystallized peptide contained within the 3GLR structure, followed by energy minimization of the entire resulting complex as described above. Figures were rendered using Lightwave 2015.3 (NewTek, Inc).
Statistical analysis
Significance was calculated using a Student’s t-test. Statistical significance was determined if p<0.05. The activity of ALDH2 was calculated by determining the amount of NAD(H) produced when compared to a standard curve. This value was then divided by the length of the assay in minutes and the nanomoles of ALDH2 used. The SIRT3 Target Affinity Ratio (STAR) was calculated as follows:
- STA for particular lysine residue
RESULTS
In vitro lysine acetylation alters ALDH2 structure and decreases activity
Acetyl modification of lysine residues can modify protein charge state dynamics and hydrogen bonding capacity (Figure 1). A dose-dependent increase in acetylation of recombinant ALDH2 was observed when samples were run on a non-reducing native gel and visualized with a pan anti-acetyllysine primary antibody (Figure 2A, top). Protein migration was altered as a result of lysine acetylation. The change in migration can be largely attributed to the decreased positive charge of the acetylated lysine residues, as well as mass and conformational effects. Interestingly, the highest concentration of acetic anhydride appeared to impact tetrameric holoenzyme formation of ALDH2, as evidenced by laddering in the native gel (Figure S2). Acetic anhydride is highly reactive and may induce artefactual adducts, potentially confounding the data interpretation for this experiment. Therefore, these results were confirmed by incubation with sulfo-NHS-acetate to acetylate ALDH2, which resulted in a similar effect on native-PAGE migration and quaternary enzyme structure (Figure S3A). These results alleviate concerns regarding the use of acetic anhydride as an acetylating reagent.
Figure 1. Effect of acetylation on lysine dynamics.
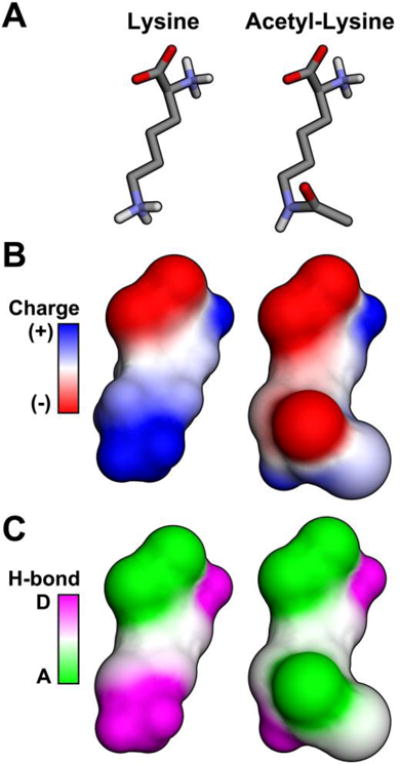
Shown are diagrams depicting an unmodified lysine or an acetylated lysine using (A) a line model, (B) surface electron density, and (C) hydrogen bonding. Acetylation neutralizes the positively charged lysine and decreases its capacity to function as an H-bond donor.
Figure 2. Acetylation inhibits ALDH2 activity in vitro.
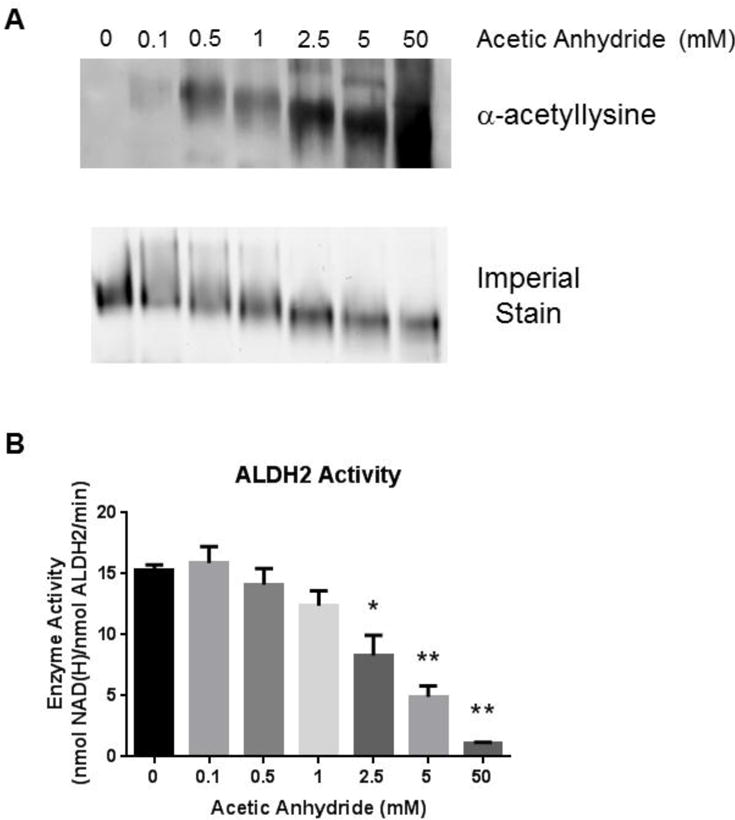
(A) A dose dependent increase in acetylated recombinant ALDH2 coincides with increasing concentrations of acetic anhydride when a non-reducing, non-denaturing native gel is immunoblotted using a pan-acetyllysine antibody. Increasing concentrations of acetic anhydride also caused an increased migration of the acetylated protein through the native gel. (B) Recombinant ALDH2 activity subsequent to treatment with acetic anhydride as measured by the absorbance of NAD(H) at 340 nm. Significance is compared against untreated (0 mM acetic anhydride) enzyme (* p<0.05, ** p<0.01). Error bars depict the standard deviation (n=4).
We hypothesized that ALDH2 activity would be diminished as a result of the structural changes to the homotetramer induced by acetylation. Accordingly, an ALDH2 activity assay was performed following incubation with increasing concentrations of acetic anhydride. Figure 2B depicts a concentration-dependent decrease in ALDH2 activity resulting from increased acetylation. The sigmoidal dose response curve is presented in Figure S4. Incubation with sulfo-NHS-acetate was employed to confirm that lysine-specific acetylation of ALDH2 decreased activity (Figure S3B). The reaction with the anhydride and acetate resulted in roughly the same decrease in ALDH2 activity with IC50 values of 3.37 mM for AA and 4.57 mM for sulfo-NHS-acetate. Regardless of the reagent, in vitro acetylation of ALDH2 results in significantly reduced enzymatic activity and is likely a result of several different factors, including tetramer formation and cofactor binding. An Eadie-Hofstee plot was generated from the data to determine the type of inhibition resulting from hyperacetylation (Figure 3). The slope of the lines increased with a concomitant increase in acetic anhydride. This concurrent increase in Km (slope) as acetylation of ALDH2 increases suggests acetylation affects NAD+ binding. However, the decreasing kcat value with increasing acetylation suggests a mixed inhibition, instead of a purely competitive inhibitory effect of acetylation (Table 1). These findings support the native gel results showing tetramer degradation as well as inhibition through cofactor disruption.
Figure 3. ALDH2 acetylation results in mixed inhibition.
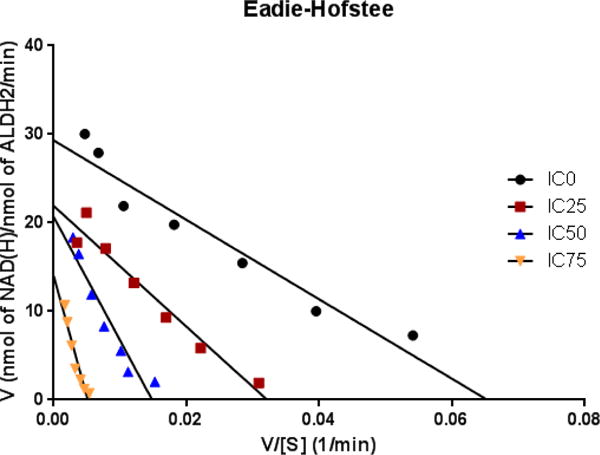
An Eadie-Hofstee plot of ALDH2 activity following untreated (IC0) or IC25, IC50 or IC75 concentrations of acetic anhydride further characterizes the acetylation-induced inhibition of ALDH2. The shift in both the slope of the lines (-Km) as well as the kcat values listed in Table 2 suggests that acetylation acts in a mixed inhibitory fashion.
Table 1. Kinetic parameters of recombinant ALDH2 following in vitro acetic anhydride treatment.
The decrease in kcat along with an increase in Km concurrent with increasing acetylation suggests mixed inhibition. Values listed are the average ± standard deviation.
| [Acetic Anhydride] | kcat (min−1) | Km (μM) |
|---|---|---|
| IC0 | 15.8 ± 1.2 | 213 ± 54 |
| IC25 | 14.1 ± 1.2 | 285 ± 75 |
| IC50 | 12.2 ± 1.0 | 715 ± 129 |
| IC75 | 11.9 ± 1.3 | 1530 ± 267 |
Deacetylation of ALDH2 by SIRT3 partially restores activity
The primary deacetylase in the mitochondrion is SIRT3. Along with previously published reports, the results from the current study confirm that ALDH2 is a target of SIRT3 and may be able to directly alter ALDH2 activity by deacetylating specific lysine residues; however, these specific residues have yet to be elucidated.7, 12, 13 Incubation of recombinant ALDH2 with 10 mM acetylating reagent results in a 65% decrease in activity. Interestingly, acetic anhydride treatment at 10 mM did not approach saturation of lysine acetylation. The addition of SIRT3 was able to restore 22% of the lost activity of ALDH2 (Figure 4). The similar observation that SIRT3 provides only a partial recovery to an acetylated protein in vitro has also been published using long-chain acyl-CoA dehydrogenase as a SIRT3 deacetylation target.20 However, these two examples demonstrate the ability of SIRT3 to recover an enzyme’s activity through deacetylation in a simple in vitro system. A similar recovery in ALDH2 activity was found utilizing ALDH2 acetylated via sulfo-NHS-acetate (data not shown) and further supports a mixed inhibition through tetramer breakdown and cofactor disruption.
Figure 4. SIRT3 partially restores ALDH2 activity.
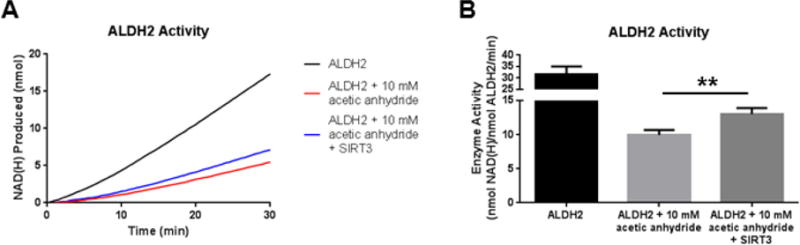
(A) Absorbance of NAD(H) at 340 nm is measured to assess ALDH2 activity. SIRT3 was added to acetylated ALDH2 and was able to partially recover ALDH2 activity. The lines plot the average NAD(H) produced in the triplicate wells. (B) The bar graph depicts the calculated activity of ALDH2 over 30 minutes for the activity assay (n = 3, standard deviation). (** p<0.01)
Identification of ALDH2 residues targeted by SIRT3 deacetylase activity
A number of ALDH2 lysine residues are known targets of SIRT3 (Compendium of Lysine Modification database). In order to thoroughly interrogate all known sites, stable isotope dilution mass spectrometry was employed. An overview of the methodology is depicted in Figure 5. By utilizing 1:1 mixed samples of ALDH2 treated with either normal or deuterated acetic anhydride, the first comprehensive quantitative analysis for acetylation removal by SIRT3 was performed (MS data in Table S1). This stable-isotope dilution approach is key, as the 3 dalton mass shift resulting from the three deuteriums of the acetyl modification allowed for accurate identification of lysine acetylation, regardless of removal by SIRT3 (Figure S1). The adjusted integral values were then used to calculate the percentage of acetylation occurring at a particular lysine residue and the percentage of acetylation removed by SIRT3 (Figure 6A). These two pieces of data were used in calculating the SIRT3 Target Affinity Ratio (STAR). The STAR values for the 16 lysine residues identified in ALDH2 are reported in Table 2.
Figure 5. Workflow for determining acetylation dynamics.
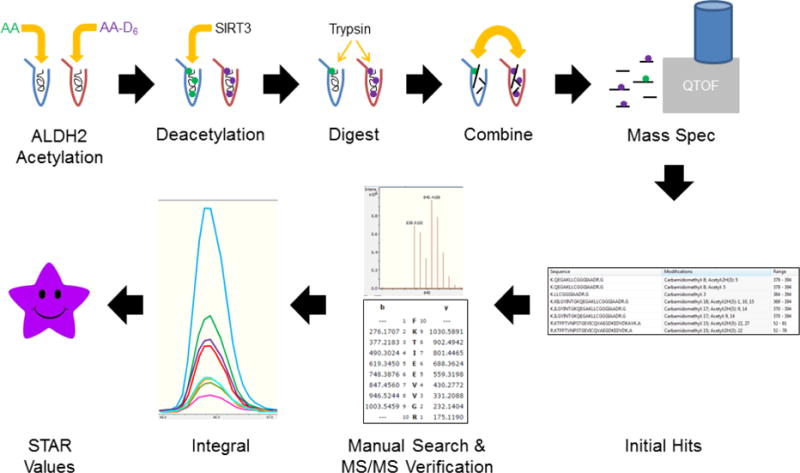
AA and AA-D6 denote samples treated with “non-labeled” and “deuterated” acetic anhydride, respectively. The SIRT3 Target Affinity Ratio (STAR) for each lysine residue was calculated as described in the Materials and Methods section.
Figure 6. ALDH2 acetylation dynamics determined via stable isotope dilution mass spectrometry and visualized via 3D model.
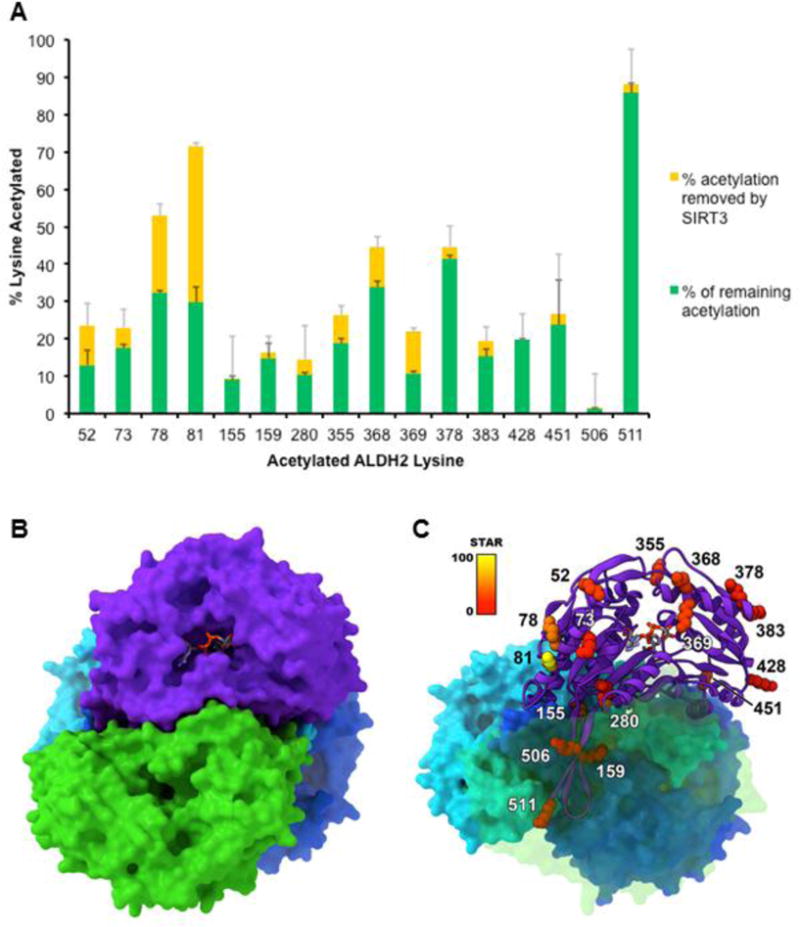
(A) The amount of total acetylation for each lysine residue was determined and is compared with the extent of deacetylation by SIRT3. Over a fixed period, SIRT3 displays a wide range of activity towards acetylated lysine residues of ALDH2. (B) The four monomers of the ALDH2 tetramer are shown in different colors with NAD+ shown in its binding pocket. (C) One monomer of ALDH2 is shown using a ribbon model with the lysine residues colored according to their STAR value.
Table 2. SIRT3 Target Affinity Ratio (STAR) values for acetylated lysines of ALDH2.
Lysines were identified using stable isotope dilution mass spectrometry and STAR values were calculated from the integrals of the corresponding chromatograms (n ≥ 3). Numbering is for the full 517 amino acid long human protein. (A higher STAR value denotes relatively greater SIRT3 deacetylation activity towards that residue.)
| Amino Acid of ALDH2 | STAR |
|---|---|
| Lys52 | 25.5 |
| Lys73 | 13.0 |
| Lys78 | 50.1 |
| Lys81 | 100.0 |
| Lys155 | 0.9 |
| Lys159 | 3.6 |
| Lys280 | 9.8 |
| Lys355 | 18.4 |
| Lys368 | 25.2 |
| Lys369 | 26.5 |
| Lys378 | 8.1 |
| Lys383 | 9.7 |
| Lys428 | 0.0 |
| Lys451 | 7.0 |
| Lys506 | 0.3 |
| Lys511 | 5.7 |
Structural analysis of acetylated lysines of ALDH2
Computational-based molecular analysis was employed to assist in determining the potential effects of lysine acetylation on ALDH2 structure and function. The STAR values were utilized to assist in helping to visualize the potential importance of the acetylation events of the 16 identified lysines (Figure 6). The majority of the lysine residues found to be acetylated (Lys52, Lys73, Lys78, Lys81, Lys280, Lys355, Lys378, Lys383, Lys428 and Lys451) are on the solvent accessible periphery of the protein. Interestingly, relative accessibility does not necessarily reflect deacetylation affinity by SIRT3 (Figure 6). The function of many of these lysines is unclear and further investigation would be required to illuminate what functionally-relevant role, if any, their acetylation has on ALDH2 structure and activity. Four amino acids (Lys155, Lys159, Lys506 and Lys511) are present at the interaction interface critical to ALDH2 dimerization. Acetylation of these residues would likely impact dimer and tetramer formation, thus impairing tetramer formation and/or stability. These modifications could provide an explanation for the multiple bands detected on the native gels, representing degradation of tetramer and dimer structures (Figure S2). These residues would also be inaccessible to SIRT3, which would further explain their low STAR values. In contrast, the acetylation of Lys280, Lys368 and Lys369 have the potential to have an effect on NAD+ binding based on their relative positioning within the protein structure. Lysine 368 is in very close proximity to the NAD+ binding pocket and Lys369 lies directly in the NAD+ binding pocket (Figures 6B & 6C). Lysine 280 participates in internal hydrogen bonding that is involved in maintaining the interactions between two helices at the opposite end of the NAD+ binding pocket from Lys369. Acetylation of Lys280 would disrupt these hydrogen-bonding interactions and potentially lead to a loss of cohesiveness of the NAD+ binding site, therefore negatively affecting NAD+ binding via more indirect means. The low to moderate STAR values for these three lysine residues important in cofactor binding may explain why only a partial recovery was observed in ALDH2 activity as a result of SIRT3 deacetylation.
To further examine the potential structural and functional effects of Lys369 acetylation, we performed computational modeling simulations to compare the native structure with acetylated ALDH2. In the energy minimized native structure, Lys369 constitutes part of the cofactor binding site and directly participates in hydrogen bond interactions with NAD+ (Figure 7A). Acetylation of this residue abolished this interaction, which both altered the binding orientation of the NAD+ in the pocket (Figure 7B) and the overall topology of the binding site itself (Figure 7 bottom panels). Given the changes in the binding site, we hypothesized that acetylation of Lys369 would negatively impact NAD+ binding directly. To test this, we performed NAD+ docking simulations to compare the predicted binding with the native and acetylated Lys369 structures. Although the results with the native structure were highly similar to the binding orientation of NAD+ in the crystal structure, we were unable to obtain any substantive predicted binding poses in the acetylated structure. Further investigation of the inhibitor effect of acetylation with NAD+ binding was interrogated by measuring the kinetic parameters of ALDH2 following exposure to different concentrations of acetic anhydride. The results are summarized in Table 1. The increasing Km values as acetylation increases is indicative of competitive inhibition at the NAD+ binding site. However, the decreasing kcat is a characteristic of noncompetitive inhibition. The combined data of the computational modeling and kinetic parameters both suggest that in vitro acetylation of ALDH2 acts through a mixed inhibitory fashion.
Figure 7. Modeling demonstrates that acetylation of Lys369 on ALDH2 prevents normal NAD+ cofactor binding into the binding pocket.
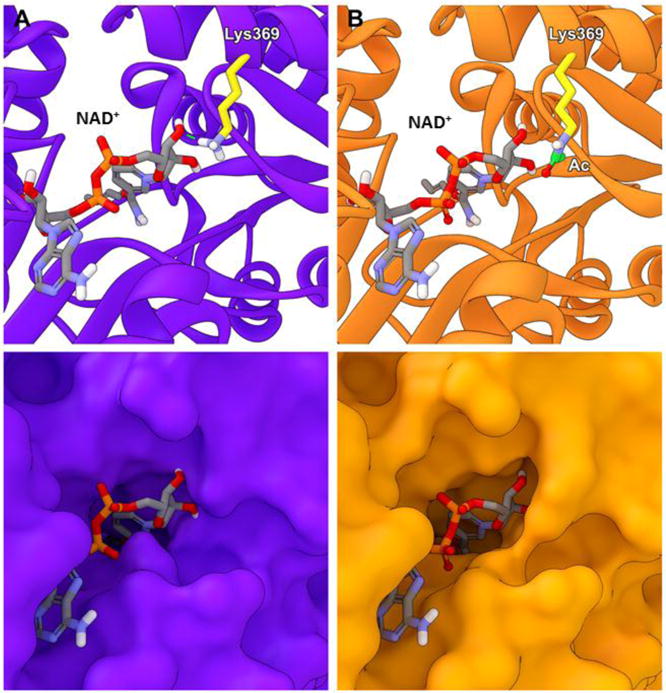
(A) Solvent-based energy minimization of the native structure (purple) and (B) acetylation on Lys369 (orange). Lys369 is highlighted in yellow with the acetyl group highlighted in green. The dashed green line indicates hydrogen bond interactions between Lys369 and NAD+, which is blocked by Lys369 acetylation.
Taken together, these results suggest that acetylation of these residues is functionally and/or catalytically relevant, likely resulting primarily from quaternary protein structure degradation and a reduction in NAD+ binding affinity. In order to gauge the potential protein-protein interactions between SIRT3 and ALDH2 (acetyl-Lys369) leading to the deacetylation of ALDH2, we performed a molecular overlay of the acetylated residue over the acetylated peptide contained with the SIRT3 crystal structure followed by solvent-based energy minimization of the theoretical SIRT3-ALDH2 complex. The overall interaction appeared highly likely, with favorable predicted interactions around the periphery of the SIRT3 active site as well as the ability to maintain the expected contacts between the acetyl modification, NAD+, and the catalytic His248 of SIRT3 (Figure 8).
Figure 8. SIRT3 deacetylates Lys369 on ALDH2, allowing NAD+ access into the cofactor binding pocket.
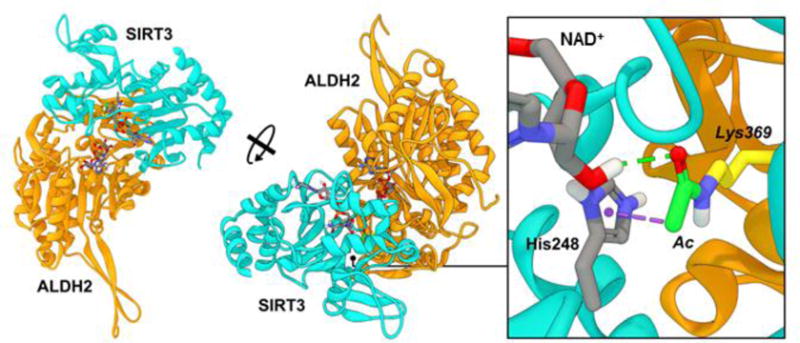
SIRT3 (cyan) binds to and deacetylates acetyl-Lys369 of ALDH2 (orange). Dashed lines indicate predicted non-bond interactions (green = hydrogen bonds, purple = pi-sigma interactions).
DISCUSSION
The modulation of protein activity via acetylation is emerging as a key mechanism of epiproteomic signaling resulting from altered metabolic status. The first report of acetylation of ALDH2 examined mitochondria isolated from mice to identify acetylation at Lys370.12 Subsequent publications have reported acetylation in mice at an additional 17 lysines.11, 21–25 These discoveries used a variety of models to identify lysine acetylation, particularly from mice that were sirtuin deficient in combination with caloric restriction, a high fat diet, or a chronic ethanol diet.11, 22, 23 The overall implications for lysine acetylation on protein structure and function, whether on an isolated residue or encompassing every lysine of a protein, remains a salient area of investigation.
Given the numerous implications for ALDH2 activity in biochemical toxicology and the number of acetylation sites reported to date, we provide a detailed in vitro characterization of SIRT3-ALDH2 interaction. Lu et al., previously expanded on the functional consequences of ALDH2 hyperacetylation while assessing acetaminophen hepatotoxicity in SIRT3 deficient mice.13 A direct interaction between SIRT3 and ALDH2 was demonstrated using bidirectional immunoprecipitation. This interaction was disrupted in the SIRT3 knockout mice with an accompanying increase in hepatic mitochondrial ALDH2 acetylation. Despite the robust increase in acetylated ALDH2, they reported no difference in ALDH2 activity between wild-type and SIRT3 knockout mice. Another publication demonstrated acetylation of ALDH2 upon exposure of human aortic endothelial cells to short-term ethanol.7 Interestingly, a concomitant increase in ALDH2 activity was found with increased ALDH2 acetylation; however, this increased activity, as well as the accompanying decrease in SIRT3 activity reported, was short lived with a return to basal levels 30 minutes after the peak. The work presented here represents a more canonical result of acetylation decreasing enzymatic activity by using human recombinant ALDH2 and two different acetylating reagents, acetic anhydride and sulfo-NHS-acetate. The decreased activity likely resulted from the effect acetylation has on electrostatic and structural properties. As such, the ALDH2 homotetramer appeared to be destabilized and the protein dissociated into individual monomers when hyperacetylated. These possible changes in quaternary protein structure provide further evidence that hyperacetylation decreases ALDH2 activity. Whether acetylation impacts protein quaternary structure in vivo remains to be determined. Subcellular fractionation and the presence of alternate ALDH isoforms are potentially confounding variables that hinder an accurate capture of in vivo ALDH2 activity. Here, the in vitro deacetylation of ALDH2 by SIRT3 restores activity by a modest 22%. A possible explanation for the observed decrease in activity is provided by the results of both the stable-isotope LC-MS/MS and computational analyses. Off-target effects, such as cysteine acetylation, may also play a potential role; however, these modifications were not observed in our spectral data.
The percentage of peptides containing unacetylated lysines following acetic anhydride-mediated acetylation varied substantially. While the in vivo stoichiometry of lysine acetylation remains a topic of debate, the ability of lysine acetylation to impact enzyme structure and function appears to be well accepted.6, 26 The novel methodology employed here allows for an accurate determination regarding which lysine residues are targeted by SIRT3. Indeed, lysine acetylation plays an important role in the ever-growing epiproteomic landscape as lysine residues are competitive targets for dozens of modifications, including trimethylation, ubiquitination, glycation, carbonylation, propionylation, succinylation, and malonylation. An important first step in understanding the complexity of lysine post-translational modification crosstalk is to elucidate which residues are targets for acetylation, how robustly these residues are deacetylated, and how these changes impact protein structure and function. Traditionally, amino acid substitution has been employed to characterize the functional relevance of PTMs. In the case of lysine acetylation, the high number of acetyllysine targets for each protein confound interpretation and may lead to false representation of lysine residue importance. Indeed, an insightful report highlights the numerous “tools to tackle protein acetylation”.27 Here, the authors detail the difficulty researchers face when attempting to characterize protein acetylation; including, knowing which deacetylases are relevant, lack of deacetylase specificity, how much of a given lysine is modified, and how many lysines are modified. Amino acid substitution falsely imparts saturating levels of acetyllysine, which we did not observe with our optimized in vitro assay. The authors also note the pitfalls of lysine mutations, as arginine (lysine) and glutamine (acetyllysine) mimics are far from perfect due to differences in their structure. Another additional report found that glutamine substitution may overestimate the potential effects of in vivo acetylation28. Therefore, our in vitro approach utilizes SIRT3-ALDH2 interactions coupled to stable-isotope dilution mass spectrometry to elucidate potential mechanisms of interaction.
An important aspect of this study is the novel method in assessing protein-specific acetylation and interpreting multiple-acetylated lysine residues on a given peptide by mass spectrometry. Including multi-acetylated peptides (one peptide with more than one acetyl-Lys) would result in the inability to analyze the acetylation flux at certain residues. Therefore, each tryptic variation of lysine-containing peptides must be taken into consideration. Interestingly, peptide coverage of ALDH2 also verified Lys355 and Lys368, the only hyperacetlyated lysines reported in humans, as a target of SIRT3.29 The majority of acetylated lysines on ALDH2 have been discovered in murine tissue. Unfortunately, when comparing the murine peptide sequence to the human peptide sequence, Lys355 is not conserved, which complicates interpretation towards its relevance. However, a comparison of SIRT3-targeted lysines between human and murine ALDH2 results in 10 homologous acetyllysine residues including Lys368. This homology includes a number of residues critical for ALDH2 protein activity.
The degree of acetylation and deacetylation occurring on ALDH2 lysines, as characterized by the STAR values, varies greatly. By integrating the data from our computational modeling and the STAR values, we were able to gain insight towards the functional relevance of the identified acetyllysine targets. The 5 top targets of lysine acetylation and SIRT3 removal were scrutinized. The two residues with the highest STAR values, Lys78 and Lys81, are on the periphery of the protein and may be involved in tetramer formation. This high-degree of accessibility likely aids acetylation reaction kinetics and also readily allows deacetylation by SIRT3. These two lysine residues are conserved among human, rat, and mouse species, but not yeast. A novel discovery is that the third and fifth residues with the highest STAR values are much more functionally relevant. Lysine 368 and Lys369 comprise part of the NAD+ binding pocket, suggesting that their acetylation status can alter cofactor binding. The finding that SIRT3 deacetylated three lysines (Lys280, Lys368, and Lys369) that may play a role in NAD+ binding suggests that SIRT3 modulates ALDH2 activity through altered cofactor binding. In particular, Lys369 is involved in direct hydrogen bond interactions with NAD+, which would be eliminated upon acetylation. Interestingly, Lys369 is conserved across human, mouse, rat, bovine, pig, and yeast, among others. This high-degree of conservation correlates with regulatory preservation across species and implicates Lys369 as a highly conserved site of ALDH2 regulation. While Lys 369 is a known target of acetylation, in vivo susceptibility of Lys369 for acetylation remains difficult to predict, as mitochondrial pH, acetyl-CoA levels, and deacetylase expression and activity are widely variable depending on the animal model applied.
In summary, in vitro acetylation of ALDH2 reduced its activity. Sixteen acetylated lysines on ALDH2 resulted in a change in protein structure and electrophilic profile. Incubating acetylated protein with SIRT3 resulted in a significant, albeit modest, recovery of ALDH2 activity. Enzyme assays suggest a mixed inhibition of ALDH2 due to lysine acetylation. Through the use of stable isotope dilution mass spectrometry, the level of lysine acetylation, and the degree to which SIRT3 deacetylates individual lysine resides of ALDH2 was quantified. These results suggest that acetylation of Lys369 may modulate ALDH2 activity. Future studies will aim to elucidate mechanisms of in vivo ALDH2 acetylation dynamics, including a role for sirtuin activity towards enhancing NAD+-ALDH2 binding. Overall, protein acetylation remains an interesting target for global characterization; however, the highly sensitive and specific in vitro characterization of individual proteins, as demonstrated here, is required to infer mechanistic implications for disease pathogenesis and therapeutic targeting.6
Supplementary Material
Acknowledgments
The authors would like to thank James J. Galligan, Thomas D. Hurley, and Matthew D. Hirschey for critical reading of the manuscript and thoughtful discussion. Special thanks to the Skaggs School of Pharmacy and Pharmaceutical Sciences Mass Spectrometry Core Facility for access to MS instrumentation.
FUNDING SOURCES
This work was supported by a National Institute on Alcohol Abuse and Alcoholism Grant R01AA022146 (KSF). Molecular modeling was performed at the University of Colorado Computational Chemistry and Biology Core Facility, which is funded in part by NIH/NCATS Colorado CTSI Grant Number UL1TR001082.
ABBREVIATIONS
- ALDH2
aldehyde dehydrogenase 2
- NAD+
(oxidized) β-nicotinamide adenine dinucleotide
- NAD(H)
(reduced) β-nicotinamide adenine dinucleotide
- nm
nanometer
- SIRT3
sirtuin 3
- STAR
SIRT3 target affinity ratio
- sulfo-NHS-acetate
sulfosuccinimidyl acetate
Footnotes
SUPPORTING INFORMATION
Mass spectrometry spectra illustrating SIRT3 deacetylating Lys511 of ALDH2, effects of acetylation by acetic anhydride or sulfo-NHS-acetate on ALDH2 migration and activity; Table S1 and S2, containing mass spectrometry data for STAR value calculations and acetylated ALDH2 peptides. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Muzio G, Maggiora M, Paiuzzi E, Oraldi M, Canuto RA. Aldehyde dehydrogenases and cell proliferation. Free Radic Biol Med. 2012;52:735–746. doi: 10.1016/j.freeradbiomed.2011.11.033. [DOI] [PubMed] [Google Scholar]
- 2.Chen CH, Ferreira JC, Gross ER, Mochly-Rosen D. Targeting aldehyde dehydrogenase 2: new therapeutic opportunities. Physiological reviews. 2014;94:1–34. doi: 10.1152/physrev.00017.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bai J, Mei Y. Overexpression of aldehyde dehydrogenase-2 attenuates neurotoxicity induced by 4-hydroxynonenal in cultured primary hippocampal neurons. Neurotox Res. 2011;19:412–422. doi: 10.1007/s12640-010-9183-1. [DOI] [PubMed] [Google Scholar]
- 4.Gomes KM, Campos JC, Bechara LR, Queliconi B, Lima VM, Disatnik MH, Magno P, Chen CH, Brum PC, Kowaltowski AJ, Mochly-Rosen D, Ferreira JC. Aldehyde dehydrogenase 2 activation in heart failure restores mitochondrial function and improves ventricular function and remodelling. Cardiovasc Res. 2014;103:498–508. doi: 10.1093/cvr/cvu125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stachowicz A, Olszanecki R, Suski M, Wisniewska A, Toton-Zuranska J, Madej J, Jawien J, Bialas M, Okon K, Gajda M, Glombik K, Basta-Kaim A, Korbut R. Mitochondrial aldehyde dehydrogenase activation by Alda-1 inhibits atherosclerosis and attenuates hepatic steatosis in apolipoprotein E-knockout mice. J Am Heart Assoc. 2014;3:e001329. doi: 10.1161/JAHA.114.001329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baeza J, Smallegan MJ, Denu JM. Mechanisms and Dynamics of Protein Acetylation in Mitochondria. Trends Biochem Sci. 2016;41:231–244. doi: 10.1016/j.tibs.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xue L, Xu F, Meng L, Wei S, Wang J, Hao P, Bian Y, Zhang Y, Chen Y. Acetylation-dependent regulation of mitochondrial ALDH2 activation by SIRT3 mediates acute ethanol-induced eNOS activation. FEBS letters. 2012;586:137–142. doi: 10.1016/j.febslet.2011.11.031. [DOI] [PubMed] [Google Scholar]
- 8.Sauve AA. Sirtuin chemical mechanisms. Biochimica et biophysica acta. 2010;1804:1591–1603. doi: 10.1016/j.bbapap.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verdin E, Hirschey MD, Finley LW, Haigis MC. Sirtuin regulation of mitochondria: energy production, apoptosis, and signaling. Trends Biochem Sci. 2010;35:669–675. doi: 10.1016/j.tibs.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimazu T, Hirschey MD, Hua L, Dittenhafer-Reed KE, Schwer B, Lombard DB, Li Y, Bunkenborg J, Alt FW, Denu JM, Jacobson MP, Verdin E. SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell Metab. 2010;12:654–661. doi: 10.1016/j.cmet.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fritz KS, Galligan JJ, Hirschey MD, Verdin E, Petersen DR. Mitochondrial acetylome analysis in a mouse model of alcohol-induced liver injury utilizing SIRT3 knockout mice. J Proteome Res. 2012;11:1633–1643. doi: 10.1021/pr2008384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Molecular cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 13.Lu Z, Bourdi M, Li JH, Aponte AM, Chen Y, Lombard DB, Gucek M, Pohl LR, Sack MN. SIRT3-dependent deacetylation exacerbates acetaminophen hepatotoxicity. EMBO Rep. 2011;12:840–846. doi: 10.1038/embor.2011.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fritz KS, Green MF, Petersen DR, Hirschey MD. Ethanol metabolism modifies hepatic protein acylation in mice. PLoS One. 2013;8:e75868. doi: 10.1371/journal.pone.0075868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fritz KS. Chemical acetylation and deacetylation. Methods Mol Biol. 2013;1077:191–201. doi: 10.1007/978-1-62703-637-5_13. [DOI] [PubMed] [Google Scholar]
- 16.Perez-Miller SJ, Hurley TD. Coenzyme isomerization is integral to catalysis in aldehyde dehydrogenase. Biochemistry. 2003;42:7100–7109. doi: 10.1021/bi034182w. [DOI] [PubMed] [Google Scholar]
- 17.Jin L, Wei W, Jiang Y, Peng H, Cai J, Mao C, Dai H, Choy W, Bemis JE, Jirousek MR, Milne JC, Westphal CH, Perni RB. Crystal structures of human SIRT3 displaying substrate-induced conformational changes. J Biol Chem. 2009;284:24394–24405. doi: 10.1074/jbc.M109.014928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brooks BR, Brooks CL, 3rd, Mackerell AD, Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. CHARMM: the biomolecular simulation program. Journal of computational chemistry. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feig M, Onufriev A, Lee MS, Im W, Case DA, Brooks CL., 3rd Performance comparison of generalized born and Poisson methods in the calculation of electrostatic solvation energies for protein structures. Journal of computational chemistry. 2004;25:265–284. doi: 10.1002/jcc.10378. [DOI] [PubMed] [Google Scholar]
- 20.Bharathi SS, Zhang Y, Mohsen AW, Uppala R, Balasubramani M, Schreiber E, Uechi G, Beck ME, Rardin MJ, Vockley J, Verdin E, Gibson BW, Hirschey MD, Goetzman ES. Sirtuin 3 (SIRT3) protein regulates long-chain acyl-CoA dehydrogenase by deacetylating conserved lysines near the active site. J Biol Chem. 2013;288:33837–33847. doi: 10.1074/jbc.M113.510354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y, Zhao W, Yang JS, Cheng Z, Luo H, Lu Z, Tan M, Gu W, Zhao Y. Quantitative acetylome analysis reveals the roles of SIRT1 in regulating diverse substrates and cellular pathways. Molecular & cellular proteomics: MCP. 2012;11:1048–1062. doi: 10.1074/mcp.M112.019547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hebert AS, Dittenhafer-Reed KE, Yu W, Bailey DJ, Selen ES, Boersma MD, Carson JJ, Tonelli M, Balloon AJ, Higbee AJ, Westphall MS, Pagliarini DJ, Prolla TA, Assadi-Porter F, Roy S, Denu JM, Coon JJ. Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome. Molecular cell. 2013;49:186–199. doi: 10.1016/j.molcel.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rardin MJ, Newman JC, Held JM, Cusack MP, Sorensen DJ, Li B, Schilling B, Mooney SD, Kahn CR, Verdin E, Gibson BW. Label-free quantitative proteomics of the lysine acetylome in mitochondria identifies substrates of SIRT3 in metabolic pathways. Proc Natl Acad Sci U S A. 2013;110:6601–6606. doi: 10.1073/pnas.1302961110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simon GM, Cheng J, Gordon JI. Quantitative assessment of the impact of the gut microbiota on lysine epsilon-acetylation of host proteins using gnotobiotic mice. Proc Natl Acad Sci U S A. 2012;109:11133–11138. doi: 10.1073/pnas.1208669109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Still AJ, Floyd BJ, Hebert AS, Bingman CA, Carson JJ, Gunderson DR, Dolan BK, Grimsrud PA, Dittenhafer-Reed KE, Stapleton DS, Keller MP, Westphall MS, Denu JM, Attie AD, Coon JJ, Pagliarini DJ. Quantification of mitochondrial acetylation dynamics highlights prominent sites of metabolic regulation. J Biol Chem. 2013;288:26209–26219. doi: 10.1074/jbc.M113.483396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baeza J, Dowell JA, Smallegan MJ, Fan J, Amador-Noguez D, Khan Z, Denu JM. Stoichiometry of site-specific lysine acetylation in an entire proteome. J Biol Chem. 2014;289:21326–21338. doi: 10.1074/jbc.M114.581843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamieniarz K, Schneider R. Tools to tackle protein acetylation. Chem Biol. 2009;16:1027–1029. doi: 10.1016/j.chembiol.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 28.Fujimoto H, Higuchi M, Koike M, Ode H, Pinak M, Bunta JK, Nemoto T, Sakudoh T, Honda N, Maekawa H, Saito K, Tsuchida K. A possible overestimation of the effect of acetylation on lysine residues in KQ mutant analysis. Journal of computational chemistry. 2012;33:239–246. doi: 10.1002/jcc.21956. [DOI] [PubMed] [Google Scholar]
- 29.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, Li Y, Shi J, An W, Hancock SM, He F, Qin L, Chin J, Yang P, Chen X, Lei Q, Xiong Y, Guan KL. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


