ABSTRACT
Plasmodium falciparum relies on monoallelic expression of 1 of 60 var virulence genes for antigenic variation and host immune evasion. Each var gene contains a conserved intron which has been implicated in previous studies in both activation and repression of transcription via several epigenetic mechanisms, including interaction with the var promoter, production of long noncoding RNAs (lncRNAs), and localization to repressive perinuclear sites. However, functional studies have relied primarily on artificial expression constructs. Using the recently developed P. falciparum clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 system, we directly deleted the var2csa P. falciparum 3D7_1200600 (Pf3D7_1200600) endogenous intron, resulting in an intronless var gene in a natural, marker-free chromosomal context. Deletion of the var2csa intron resulted in an upregulation of transcription of the var2csa gene in ring-stage parasites and subsequent expression of the PfEMP1 protein in late-stage parasites. Intron deletion did not affect the normal temporal regulation and subsequent transcriptional silencing of the var gene in trophozoites but did result in increased rates of var gene switching in some mutant clones. Transcriptional repression of the intronless var2csa gene could be achieved via long-term culture or panning with the CD36 receptor, after which reactivation was possible with chondroitin sulfate A (CSA) panning. These data suggest that the var2csa intron is not required for silencing or activation in ring-stage parasites but point to a subtle role in regulation of switching within the var gene family.
KEYWORDS: CRISPR/Cas9, Plasmodium falciparum, antigenic variation, transcriptional regulation, var genes
IMPORTANCE
Plasmodium falciparum is the most virulent species of malaria parasite, causing high rates of morbidity and mortality in those infected. Chronic infection depends on an immune evasion mechanism termed antigenic variation, which in turn relies on monoallelic expression of 1 of ~60 var genes. Understanding antigenic variation and the transcriptional regulation of monoallelic expression is important for developing drugs and/or vaccines. The var gene family encodes the antigenic surface proteins that decorate infected erythrocytes. Until recently, studying the underlying genetic elements that regulate monoallelic expression in P. falciparum was difficult, and most studies relied on artificial systems such as episomal reporter genes. Our study was the first to use CRISPR/Cas9 genome editing for the functional study of an important, conserved genetic element of var genes—the intron—in an endogenous, episome-free manner. Our findings shed light on the role of the var gene intron in transcriptional regulation of monoallelic expression.
INTRODUCTION
The pathogenesis of Plasmodium falciparum, the most virulent malaria parasite, relies on the expression of a family of clonally variant adhesion proteins on the surface of infected red blood cells (iRBCs) (1). These proteins, which are members of the P. falciparum erythrocyte membrane protein 1 (PfEMP1) family, are encoded by a family of ~60 var virulence genes. Similarly to other parasites, P. falciparum uses clonal antigenic variation to evade the immune system and to maintain an infection. Thus, only a single var gene is expressed at a time, but occasional switching occurs to allow expression of a different PfEMP1 (2). Monoallelic expression of var genes is believed to be under epigenetic control, as histone H3 lysine 9 trimethylation (H3K9me3) and H3K36me3 are enriched in the heterochromatin of silent var genes whereas transcriptionally permissive histone modifications such as H3K4me3 and H3K9ac are enriched at the single active var gene (3–5). It is unclear how one specific var gene is targeted for transcriptional activation while all others are maintained in a silent state; however, evidence suggests that underlying genetic elements play a key role in this regulation (reviewed in reference 6).
All var genes have a similar structure consisting of a 5′ upstream promoter sequence followed by two exons that flank a conserved intron (Fig. 1A). Studies using reporter genes and drug-selectable markers have demonstrated that a “pairing” of the 5′ upstream promoter with the var intron is required for both silencing and maintenance of the counting mechanism involved in monoallelic expression (reviewed in reference 7). The 5′ upstream sequence constitutively drives expression of reporter genes; however, this promoter activity is silenced in an S-phase-dependent manner when a var intron is placed at the 3′ end of the reporter gene (8–10). In an episomal context, the ability of the var intron to silence transgene transcription depends on its own promoter activity, and pairing of a var 5′ upstream promoter sequence with any downstream active promoter is able to silence transgenes and maintain monoallelic expression (11, 12). However, it has been shown that transcriptional activation of an episomal or integrated var promoter is able to silence the members of the var gene family, suggesting that the var intron is not required for maintenance of monoallelic expression (13).
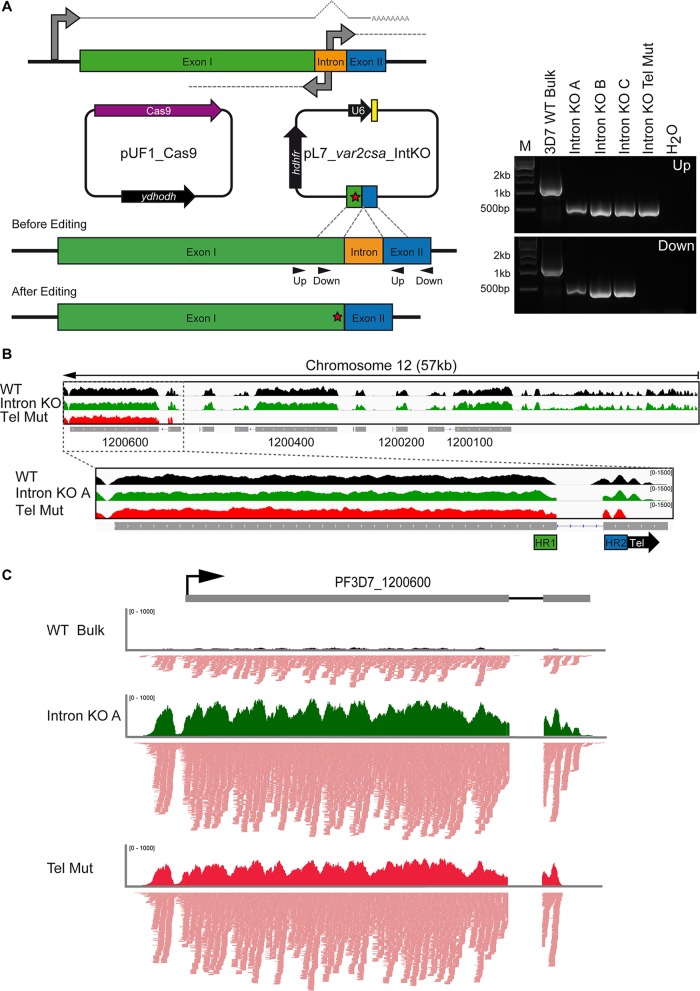
FIG 1 .
Generation of the intronless var gene mutant. (A) Schematic of a representative var gene and all associated RNA: full-length mRNA originating from the 5′ promoter and sense and antisense sterile transcripts originating from the intron (solid gray and dotted gray lines, respectively). var2csa intron knockout mutants were created by transfection with the pUF1_Cas9 and pL7_var2csa_IntKO plasmids. The pL7 plasmid contains the guide RNA (yellow) and DNA homology region KO construct corresponding to the 500 bp of exon I (green) immediately upstream of the intron (orange) fused in frame to the homology region corresponding to the 500 bp of exon II (blue) immediately downstream of the intron. A shield mutation (red) is included in the KO construct. Homologous recombination of the pL7 KO construct with the var2csa gene results in the deletion of the intron. Gel electrophoresis of PCRs performed on genomic DNA from 3D7 WT bulk culture (3D7 WT Bulk), three var2csa intron KO mutant clones (Intron KO A, Intron KO B, and Intron KO C), and one var2csa intron KO telomere mutant clone (Intron KO Tel Mut) demonstrated deletion of the intron. Primers (shown at left as arrowheads labeled “Up,” and “Down”) were designed to detect the presence of the var2csa intron. The marker is indicated (M), and a PCR using only water (H2O) served as a control. (B) Next-generation sequencing of genomic DNA from WT 3D7 parasites (black), var2csa intron KO clone A (green), and a telomere mutant clone (red) showed that the intron was deleted successfully with CRISPR/Cas9. The top panel shows 57 kb of DNA adjacent to the chromosome 12 telomere (at the right). The bottom panel is an enlargement of the var2csa gene (Pf3D7_1200600). DNA read enrichment is indicated on the y axis, and gene models are shown in gray at the bottom. A schematic of the positions of the telomere repeats (black “Tel”) and homology regions 1 and 2 (HR1 in green and HR2 in blue) within the telomere mutant is shown at the bottom. (C) Stranded sequencing of RNA harvested 12 hpi from WT 3D7 parasites (black), var2csa intron KO clone A (green), and a telomere mutant clone (red) showed that var2csa full-length transcripts were produced in 3D7 WT parasites and the intron KO clone. The telomere mutant clone produced var2csa transcript up to, but not past, the end of the second homology region used for CRISPR/Cas9 editing. Coverage plots are shown at the top of each panel, and read stacks are shown at the bottom, where red lines indicate sense transcription reads, and blue lines indicate antisense transcription reads. Numbers of reads are indicated on the y axis, and the gene model is shown in gray at the top.
Interestingly, sense and antisense sterile transcripts have been shown to originate from the introns of var genes, suggesting that the var intron has bidirectional promoter activity that could be involved in transcriptional activation or silencing (Fig. 1A) (14–16). Recent studies provided evidence for a role of var intron-derived antisense long noncoding RNA (lncRNA) in var gene activation and monoallelic expression (3, 17, 18). However, microarray and RNA sequencing data from studies of endogenous var genes demonstrated that transcription originating from the var intron in either the sense or antisense direction showed no overall correlation with var gene activation or silencing (19, 20). Thus, it remains unclear if var intron-derived sterile transcripts are involved in var gene regulation (reviewed in reference 21).
An alternative, but perhaps related, mechanism of noncoding RNA-mediated var gene regulation is the specific binding of the intron to regulatory proteins. An 18-bp binding element present in the introns of a subset of var genes was shown to bind to the putative AP2 transcription factor and to actin (22). This intron element was able to localize episomes to the nuclear periphery, which is often associated with heterochromatin and transcriptional repression (5, 23, 24). Moreover, induction of actin polymerization led to the disruption of var gene localization and monoallelic expression (22). Thus, the var intron may mediate var gene transcription via binding to regulatory proteins and/or nuclear positioning.
The conflicting data supporting a role for the var intron in regulation of var gene transcription may be a consequence of the almost exclusive use of episomes, reporter genes, and selectable drug markers for the study of this genetic element. Because var gene transcription is regulated epigenetically, it is essential to study the underlying regulatory genetic elements in a natural, native chromatin context (25). To address the transcriptional role of the var intron, we used the recently developed clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 system (26) to directly delete the var2csa intron, producing an intronless var gene in its native locus without the introduction of a drug-selectable marker. We show that deletion of the var2csa intron leads to increased transcription of the var2csa gene (Pf3D7_1200600) in ring-stage parasites and can, in a minority of clones, lead to activation of transcription of other var genes. The intronless var2csa gene can be silenced in late-stage parasites and via long-term culture or panning with the CD36 receptor, although this silencing can be reversed via panning with chondroitin sulfate A (CSA). Our data suggest that, while the var intron is not required for silencing or activation of the var2csa gene, it may play a role in the transcriptional regulation and switching of the members of the var gene family.
RESULTS
Generation of the intronless var gene mutant.
To gain insight into the role of the var intron in var gene transcription regulation, we targeted the intron of the upsE var2csa gene in the P. falciparum 3D7 strain. Among the approximately 60 var genes found in the genomes of various P. falciparum isolates, var2csa is well conserved and is present even in the genome of the non-human primate parasite P. reichenowi (27). The expression of this particular var gene has been strongly implicated in the pathogenesis of placental malaria, as the resultant PfEMP1 protein binds strongly to CSA expressed on placental syncytiotrophoblasts (28–30). As the var2csa gene is unique and clinically relevant, it was ideal for our study because it was easily targeted by CRISPR/Cas9 and several useful reagents are available for studying its expression. However, the findings of our study might not necessarily apply to all subtypes of the var genes.
The var2csa gene is located in the subtelomeric region of chromosome 12 and consists of an 8,004-bp exon I, an 847-bp intron, and a 1,167-bp exon II (Fig. 1A, green, orange, and blue, respectively). We completely deleted the endogenous var2csa intron using the CRISPR/Cas9 genome-editing system recently described in reference 26. Cells were cotransfected with the pUF1_Cas9 and pL7_var2csa_IntKO plasmids (Fig. 1A). The Cas9 endonuclease was directed to the extreme 3′ end of exon I by a highly specific single guide RNA (sgRNA) designed with the newly developed Protospacer software (31). The resultant DNA double-strand break was repaired via homologous recombination with the intron knockout DNA construct provided on the pL7 plasmid. This construct consists of 500 bp of the 3′ end of exon I immediately upstream of the intron (Fig. 1A, homology region 1 in green) fused in frame to 500 bp of the 5′ end of exon II immediately downstream of the intron (Fig. 1A, homology region 2 in blue). The protospacer adjacent motif (PAM) sequence targeted by the sgRNA/Cas9 was mutated in the knockout construct to prevent further Cas9 endonuclease activity while conserving amino acid sequence (Fig. 1A, red star).
The result of the CRISPR/Cas9 process was an intronless var2csa gene: exon I fused in frame to exon II without the use of drug-selectable markers at the endogenous locus (Fig. 1A). After subcloning of the transfectants was performed, genomic DNA was prepared and the absence of the var2csa intron was confirmed with PCR and next-generation sequencing (Fig. 1A and B, respectively). Curiously, the vast majority of clones contained an unintended mutation in which endogenous DNA between the end of homology region 2 and the telomere was lost, leading to truncation of exon II (Fig. 1B, red). The sequencing data also revealed that in these telomere mutants, the Cas9-induced double-strand break resulted in chromosome breakage and repair with telomere repeats, a repair mechanism that has also been reported for spontaneous chromosome breakage in in vitro culture (32). While unexpected, the intronless var2csa telomere mutant clone served as a useful control for experiments discussed below. In the end, through genomic sequencing, we identified six clones from two independent transfections with correctly edited genomes. Representative genomic sequencing data are shown for clone A (Fig. 1B, green).
The var intron has been implicated in both silencing and activation of var gene transcription (reviewed in reference 7). To determine if the var2csa gene is transcribed in the absence of the intron, RNA was harvested from a highly synchronous 3D7 wild-type (WT) bulk culture, three var2csa intronless clones, and one var2csa intronless telomere mutant clone at 12 h post infection (hpi), a time when var genes are highly transcribed. Stranded RNA sequencing was performed using poly(A)-enriched RNA, confirming that mRNA containing exon I and exon II, but not the intron, is produced in WT and var2csa intronless mutant clones A, B, and C (Fig. 1C, WT in black and intron knockout [KO] mutant clone A in green). The intronless telomere mutant clone also transcribes var2csa but lacks all endogenous RNA sequence downstream of homology region II (Fig. 1C, in red).
var2csa intron deletion results in upregulation of transcription in ring-stage parasites.
To determine if the var gene transcriptional profile is altered in the absence of the var2csa intron, we analyzed RNA levels of the ~60 var genes in the var2csa intronless mutants. A CSA-panned FCR3 culture, the 3D7 WT parent strain of the var2csa intronless mutants (bulk culture), one var2csa intronless telomere mutant clone, and six clones of the var2csa intron KO were tightly synchronized, and total RNA was harvested at 12 hpi (ring stage). Reverse transcription-quantitative PCR (RT-qPCR) was performed with primers specific to each of the ~60 var genes (from reference 28).
As the FCR3 culture had been enriched for parasites expressing VAR2CSA, it predominantly transcribed the var2csa gene, or Pf3D7_1200600 (Fig. 2A, top panel in yellow). The 3D7 WT bulk parent culture was not clonal; thus, multiple different var genes, especially upsC types, were transcribed to various degrees in ring-stage parasites (Fig. 2A, second panel in black). The var2csa gene was not highly transcribed in the 3D7 culture; however, higher levels of var2csa transcription were seen in all intronless mutant clones (Fig. 2A, green/gray, purple, blue), including the telomere mutant (Fig. 2A, red).
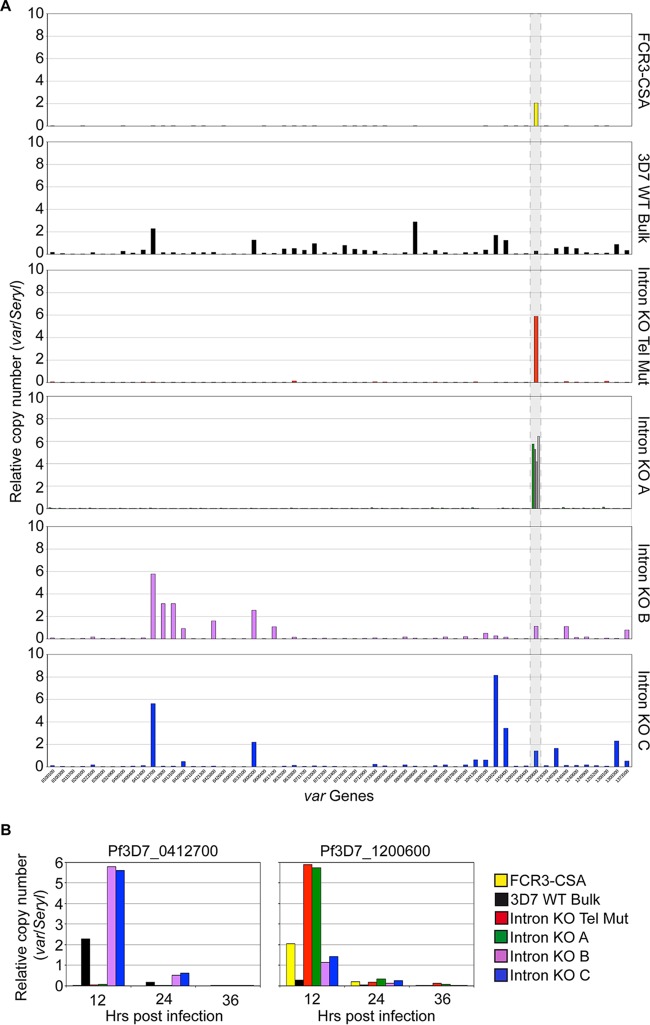
FIG 2 .
var2csa intron deletion results in upregulation of transcription in ring-stage parasites. (A) RNA was isolated from highly synchronized ring-stage parasites (12 hpi) from a CSA-panned FCR3 culture (top panel, yellow), a 3D7 WT bulk culture (second panel, black), an intron KO telomere mutant clone (third panel, red), and six var2csa intron KO mutant clones (fourth, fifth, and sixth panels). RT-qPCR analysis of all var genes was performed, and cDNA levels were normalized to those of serine-tRNA ligase. Intron KO clone A is shown in the fourth panel in green along with three other clones (in gray) showing similar monoallelic transcriptional var profiles. Intron KO clones B (fifth panel in purple) and C (sixth panel in blue) transcribed multiple var genes. A gray dashed box indicates the var2csa gene, Pf3D7_1200600. (B) RNA was isolated from highly synchronized parasites from a CSA-panned FCR3 culture (yellow); a 3D7 WT bulk culture (black); intron KO mutant clones A (green), B (purple), and C (blue); and an intron KO telomere mutant clone (red) at 12 (ring), 24 (trophozoite), and 36 (schizont) hpi. RT-qPCR analysis of Pf3D7_0412700 (upsC var gene) and Pf3D7_1200600 (var2csa) was performed, and var gene cDNA levels were normalized to those of serine-tRNA ligase.
In a clonal population of parasites, usually only a single var gene is transcribed (2). Indeed, transcription of a single var gene—the intronless var2csa gene—was observed in four different intron KO clones (Fig. 2A, fourth panel: intron KO clone A in green and three additional clones in gray) and the intron KO telomere mutant (Fig. 2A, third panel in red). However, intron KO clones B (purple) and C (blue) transcribed distinct subsets of var genes, both subtelomeric and central, in addition to var2csa (Fig. 2A, fifth and sixth panels). The latter pattern of transcription suggests loss of monoallelic expression or an increase in var gene switching within the clonal population. These data were supported by the stranded RNA sequencing analysis performed for intron KO clones A, B, and C and the telomere mutant clone.
RT-qPCR analysis was repeated with RNA harvested at 24 and 36 hpi, when var gene transcription is normally repressed, using primers against the Pf3D7_0412700 central var gene and the Pf3D7_1200600 var gene. In 3D7 WT bulk culture parasites and var2csa intron KO clones B and C, both the Pf3D7_0412700 and Pf3D7_1200600 var genes were transcribed at 12 hpi but at background levels at 24 and 36 hpi (Fig. 2B, left and right panels, respectively). Similarly, the Pf3D7_1200600 var gene was transcribed in FCR3-CSA; var2csa intron KO clones A, B, and C; and the var2csa intronless telomere mutant clone at 12 hpi but was transcriptionally silenced in later-stage parasites (Fig. 2B, right panel). Together, these data suggest that the intron is not required for var2csa gene activation in rings or silencing in trophozoites and schizonts but may be involved in switching of the var gene family.
A var2csa intronless mutant produces VAR2CSA PfEMP1 protein.
The VAR2CSA PfEMP1 binds to the proteoglycan CSA, which is highly expressed on the surface of placental syncytiotrophoblasts (29, 30). It has been previously reported that VAR2CSA expression is regulated at the translational level, as var2csa mRNA is sometimes expressed in infected individuals and cultured parasites without detectable VAR2CSA PfEMP1 expression (33–35). This translational regulation is believed to be controlled by an open reading frame upstream of the actual var2csa open reading frame (36, 37). To determine if the var2csa intron knockout strains produce PfEMP1, we performed Western blot analysis and immunofluorescence assay (IFA).
Western blot analysis of 30-hpi iRBC membrane fractions with an antibody against the VAR2CSA PfEMP1 revealed that var2csa intron knockout clone A produces VAR2CSA at levels that are similar to, if not higher than, those of VAR2CSA produced by a CSA-panned FCR3 bulk culture (Fig. 3A). VAR2CSA expression is undetectable in the bulk 3D7 parent strain of the var2csa intron knockout, although expression of other PfEMP1 proteins can be detected with an antibody against the acidic transmembrane segment (ATS), which is highly similar among all PfEMP1 proteins (Fig. 3A).
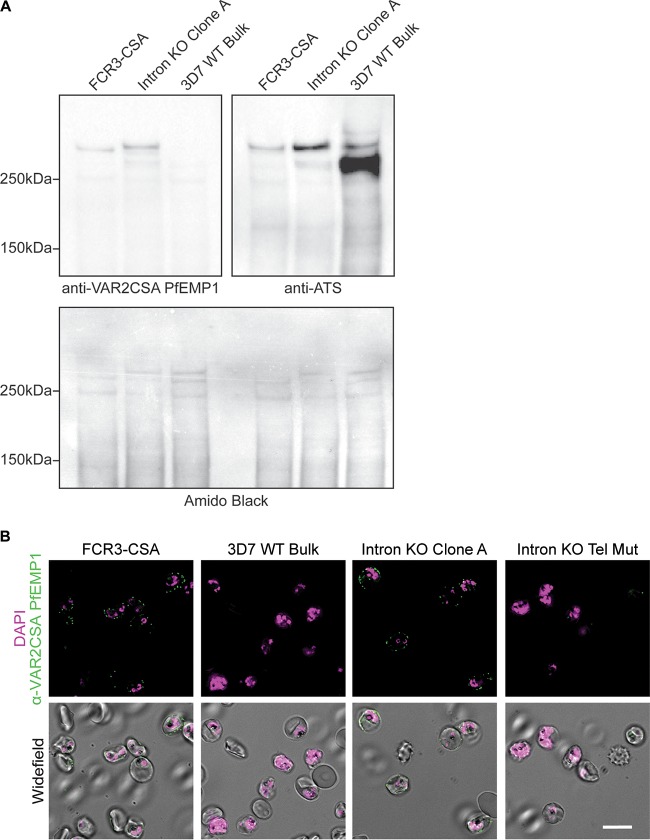
FIG 3 .
var2csa intronless mutant produces VAR2CSA PfEMP1 protein. (A) Western blot analysis of 30-hpi iRBC membrane extracts performed using antibodies against VAR2CSA PfEMP1 (top left panel) or the PfEMP1 ATS region (top right panel). Extracts were prepared from a CSA-panned FCR3 culture, intron KO clone A, and a 3D7 WT bulk culture. An amido black staining of the Western blot is shown as a loading control. Molecular masses are indicated at the left. (B) Immunofluorescence analysis of live 30-hpi iRBCs containing CSA-panned FCR3, 3D7 WT bulk, intron KO clone A, or intron KO telomere mutant clone parasites. DNA was stained with DAPI (magenta), and iRBCs were surface labeled with specific antibodies against VAR2CSA PfEMP1 (green). Fluorescent images are shown in the top panel, and wide-field merged images are shown in the bottom panel. Scale bar, 10 μm.
A similar pattern of VAR2CSA expression was seen with surface IFA of live 30-hpi iRBCs using DAPI (4′,6-diamidino-2-phenylindole) and the antibody against VAR2CSA PfEMP1. The CSA-panned FCR3 culture showed expression of VAR2CSA at the membrane of approximately 90% iRBCs, while var2csa intron KO clone A showed expression of VAR2CSA at the membrane of approximately 80% iRBCs (Fig. 2B). VAR2CSA was present in a small percentage of 3D7 WT iRBCs but was undetectable by IFA in the var2csa intron KO telomere mutant clone (Fig. 2B) and also in var2csa intron KO clone C. These data suggest that the intron is not needed for proper mRNA processing or translation but support previous studies demonstrating VAR2CSA PfEMP1 expression control at the translational level (36, 37).
An intronless var2csa gene can be silenced via long-term culture or CD36 panning, then reactivated with CSA panning.
To determine if the intron is required for var gene silencing, we attempted to induce the var2csa intronless mutant parasites to switch var transcription in a natural manner that did not involve the use of episomes. It has been shown that var gene switching occurs spontaneously when a parasite population is kept in culture for long periods of time, especially in the case of subtelomeric var genes (38). Switching can also be induced by selective binding, or panning, of iRBCs to various host receptors. As many as 84% of PfEMP1 proteins—excluding VAR2CSA—are believed to bind to human endothelial receptor CD36 (1). Thus, we attempted to induce var gene switching via long-term culture and panning with CD36. A 3D7 bulk culture and var2csa intron KO clone A were tightly synchronized, and RT-qPCR was performed on RNA harvested at 12 hpi using primers against all ~60 var genes. These cultures were then split and kept in continuous culture for 2 months or were used to perform three rounds of CD36 panning, after which we confirmed maintenance of the intron knockout mutation with PCR and sequencing. RNA analysis was then repeated with synchronized parasites.
The transcript levels of several different var genes changed noticeably between the initial time point (Fig. 4, top panel) and 2 months in culture (Fig. 4, second panel) or CD36 panning (Fig. 4, third panel) in the 3D7 bulk culture and the var2csa intron KO clone (Fig. 4, black and gray bars, respectively). Notably, var2csa transcription was reduced substantially in the var2csa intron KO clone after 2 months of continuous culture and was barely detectable after CD36 panning.
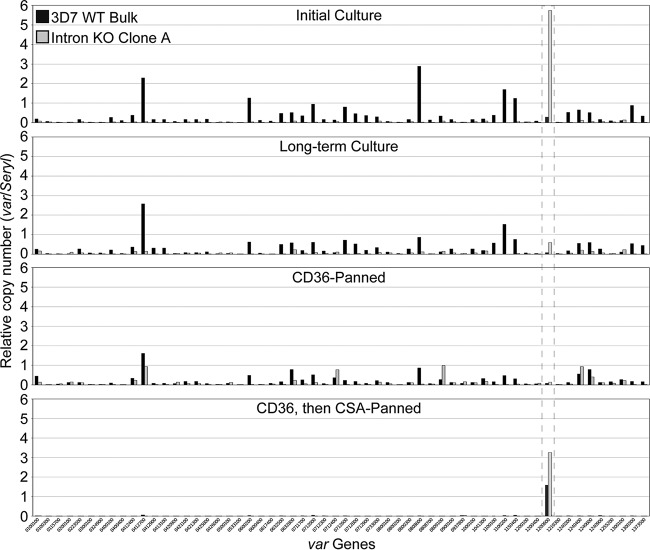
FIG 4 .
An intronless var2csa gene can be silenced via long-term culture or CD36 panning, then reactivated with CSA panning. RNA was isolated from highly synchronized ring-stage parasites (12 hpi) from a 3D7 WT bulk culture (black) and var2csa intron KO mutant clone A (gray). RT-qPCR analysis of all var genes was performed before (“Initial Culture,” top panel) and after (“Long-term Culture,” second panel) 2 months of continuous culture. The initial cultures were also subjected to three rounds of CD36 panning (third panel), followed by three rounds of CSA panning (bottom panel). var gene cDNA levels are normalized to those of serine-tRNA ligase. A gray dotted box indicates the var2csa gene, Pf3D7_1200600.
To determine if the intronless var2csa gene could be transcriptionally reactivated once silenced, we performed three rounds of CSA panning with the CD36-panned cultures. While three rounds of CSA panning were required to observe adherence of the 3D7 bulk culture to CSA, the var2csa intron KO culture adhered well after just one round of CSA panning. These panned cultures were synchronized, and var transcriptional analysis was carried out at 12 hpi. Indeed, the transcriptional profile demonstrates almost exclusive transcription of the var2csa gene in both strains, although it was transcribed to a higher degree by the var2csa intron KO culture (Fig. 4, bottom panel). These data further demonstrate that the intron is not required for var2csa transcriptional silencing, activation, or switching and that the lack of the intron perhaps allows easier transcriptional activation and switching of the var2csa gene.
DISCUSSION
In P. falciparum, var gene transcription is under tight control. Although the 5′ upstream promoter and intron have been implicated in this process, the true role of these genetic elements has been difficult to elucidate, as most previous studies have relied on episomes and/or transgenes. Using the CRISPR/Cas9 system, we directly deleted a var intron without insertion of a drug-selectable marker at the endogenous locus. Although previous studies of var gene transcription have primarily used genetic elements of central var genes, we focused our study on the subtelomeric Pf3D7_1200600 (var2csa) gene, as this var gene is well conserved across many P. falciparum strains and has been strongly implicated in the pathogenesis of pregnancy-associated malaria (reviewed in reference 39).
Our data show that deletion of the var2csa intron leads to transcriptional upregulation of the targeted var2csa gene in ring-stage parasites (Fig. 1C and 2A). These data might suggest that the intron is required for var2csa transcriptional silencing, but we show that transcription of the intronless var2csa gene could be downregulated in late-stage parasites and in response to long-term culture or CD36 panning (Fig. 2B and 4). It is possible that the intron is needed for var2csa gene activation and that the initial increase in transcription of the intronless var2csa gene was the result of transcriptionally activating epigenetic changes taking place in response to the Cas9-induced DNA damage and repair rather than to the loss of an intron-binding silencing factor. However, we show that transcription of the intronless var2csa gene can be easily reactivated by CSA panning after being silenced by CD36 panning (Fig. 4).
Thus, in the case of var2csa, the intron is not required for transcriptional silencing or activation. Our data do not support earlier studies that used episomes and transgenes to demonstrate the requirement of the var intron in silencing the var 5′ UTR promoter (8–10). Conflicting reports have provided evidence that antisense and/or sense sterile transcripts originating from the intron play a role in both var gene silencing and activation (3, 14, 17–19). However, our RT-qPCR and RNA sequencing data demonstrate that the var intron and any sense or antisense sterile transcripts that originate from the intron are not strictly required for var2csa gene activation or silencing (Fig. 1C, 2A, and 4). It is possible, however, that these intron-based mechanisms may modulate var gene activation or silencing for other var subtypes and maybe even switching or counting for var2csa. Given that functional differences between different var introns were previously observed (22), it is possible that some introns have distinct roles in the control of mutually exclusive var gene transcription.
Our data do support previous studies showing that PfEMP1 protein expression is not required for monoallelic expression, as multiple clones of the intron KO telomere mutant transcribed only the targeted var2csa gene in the absence of functional VAR2CSA PfEMP1 (Fig. 2A, red) (40). Previous studies have shown that VAR2CSA PfEMP1 expression is subject to posttranscriptional regulation (35–37). Our data support these studies, as we saw a lack of VAR2CSA PfEMP1 expression via IFA in intron KO clone C, which clearly transcribed var2csa (Fig. 2A [blue] and data not shown). It is unclear, however, which of the many var genes transcribed in these intron KO clones are expressed at the protein level, as specific antibodies are currently unavailable.
This report provides new insight into the plastic nature of intron-mediated var gene switching and substantiates the complexity of var gene transcriptional regulation in the chromosomal context. In our study, different transcriptional phenotypes were seen in different genetically identical mutant clones, as two of six intron KO clones transcribed distinct sets of multiple var genes (Fig. 2A, purple and blue). Considering that transcription of a single var gene is usually stable over the course of cloning and synchronization in a WT clonal population of parasites, we postulate that our mutant clones have an increased rate of var gene switching. It has been shown that P. falciparum cultures tend to switch preferentially from transcription of subtelomeric var genes to transcription of central var genes and that var2csa may play a unique coordinating role in var gene switching, serving as a default switching node (38, 41). Interestingly, half of the var genes that were transcribed in addition to the intronless var2csa in our two mutant clones were subtelomeric var genes, and none were adjacent to the var2csa gene (Fig. 2A, purple and blue). This observation might hint at an intron-based telomeric clustering mechanism used to silence var genes and/or to coordinate var gene switching in trans, as has been suggested previously (5). It is possible that putative repressive var gene clusters are controlled by a delicate balance of intron-binding proteins or associated ncRNAs, and it will be critical to identify these potential regulators in the future.
The var2csa gene has been shown to be a unique case among var genes, being subject to transcriptional and posttranscriptional control. Thus, it remains to be seen if intron deletion in other var subtypes would result in a phenotype similar to that of var2csa. We attempted to create several different intron knockouts in upsA, upsB, and upsC var genes but were unsuccessful due to chromosomal aberrations resulting from Cas9-induced DNA damage. Each time we attempted to delete the intron from a different subtelomeric var gene, close to 100% of the transfected parasite population acquired the same telomeric mutation that we saw to a lesser extent in the var2csa intron KO mutants. Moreover, induced DNA damage occurring with any type of var gene may lead to interchromosomal recombination events, a mechanism that P. falciparum is known to use to survive in vivo. Thus, the CRISPR/Cas9 system may be challenging to use in the context of the subtelomeric or even central var genes, and we recommend caution and genomic sequencing for any mutant created with CRISPR/Cas9.
MATERIALS AND METHODS
Parasite culture.
Asexual blood-stage P. falciparum parasites were cultured as previously described in reference 5. Parasites were synchronized by sorbitol lysis at the ring stage, by Plasmagel enrichment 24 h later, and by an additional sorbitol lysis 6 h after Plasmagel enrichment. Parasite development was monitored by Giemsa staining. Parasites were harvested at 4% hematocrit and approximately 5% parasitemia.
Strain creation.
var intron knockout parasites were created from the 3D7 wild-type strain using the CRISPR/Cas9 system as previously described in reference 26. For var2csa (Pf3D7_1200600), a 20-nucleotide sgRNA (GGTTTTTTGCAGAATGTCAC) was designed using Protospacer software (31) and inserted into the pL6 sgRNA expression plasmid. To facilitate knockout via homologous recombination, homology regions were ordered from GenScript, PCR amplified, and inserted into the same pL6 plasmid. Homology regions were designed by fusion of a 500-bp sequence of exon I directly upstream of the intron to a 500-bp sequence of exon II directly downstream of the intron. Silent shield mutations were introduced into and near the protospacer adjacent motif sequence of the homology region (GGTTTTTTGCAGAATGTCGCTCG [the mutations are underlined]). All cloning was performed using HiFi DNA polymerase (Kapa Biosystems), an In-Fusion HD cloning kit (Clontech), and XL10-Gold ultracompetent Escherichia coli (Agilent Technologies). Ring-stage parasites were transfected with pUF1-Cas9 and pL7 plasmids as previously described in reference 26. Cloning of parasites was carried out by limiting dilution.
Genomic DNA preparation, PCR, and sequencing.
Genomic DNA was isolated with a DNeasy Blood and Tissue kit (Qiagen) from late-stage parasites isolated with manual cell separation (MACS) columns (Miltenyi Biotec) or Plasmagel. For sequencing, purified genomic DNA was sheared with a Bioruptor standard ultrasonicator (Diagenode). Libraries were prepared for sequencing using MicroPlex Library Preparation kit v2 (Diagenode). Libraries were sequenced on a NextSeq 500 or HiSeq platform (Illumina). Sequenced reads were mapped to the P. falciparum genome (42) (plasmoDB.org, version 3, release 29) using “bwa mem” (43) and allowing a read to align only once to the reference genome (option “-c 1”). Alignments were subsequently filtered for duplicates and a mapping quality value of ≥30 using samtools (44) and visualized in the Integrative Genomics Viewer (45).
RNA extraction and qPCR analysis.
Total RNA was isolated from synchronized parasite cultures by saponin lysis followed by purification performed with an miRNeasy kit (Qiagen). RNA was subjected to DNase (Qiagen) treatment, and reverse transcription was performed using random hexamer primers and SuperScript VILO reverse transcriptase (Thermo Fisher Scientific). Quantitative PCR was performed with the resultant cDNA in triplicate on a CFX real-time PCR system (BioRad) using Power SYBR green (Life Technologies, Inc.) and primers from a previous study (28). Transcript levels were determined using the quantity mean determined for each triplicate as calculated from a standard curve based on a serial dilution of genomic DNA. var gene transcription was normalized to that of a housekeeping gene, the serine-tRNA ligase gene (PF3D7_0717700).
Stranded RNA sequencing and analysis.
Total RNA was extracted as described above. RNA samples were depleted of rRNA by the use of a Dynabeads mRNA Direct kit (Ambion 61012), and libraries were prepared with a TruSeq Stranded mRNA LT Sample Prep kit (Illumina). Libraries were sequenced on a NextSeq 500 platform (Illumina). Sequenced RNA reads were mapped, aligned, and visualized as described above for the genomic DNA.
Western blot analysis.
iRBCs containing synchronous parasites were isolated at 30 hpi by Plasmagel enrichment. iRBCs were washed once with phosphate-buffered saline (PBS), and soluble proteins were extracted with NETT buffer (50 mM Tris [pH 8], 150 mM NaCl, 5 mM EDTA, 1% Triton X-100) containing protease inhibitors (Roche 11836170001). This extract was centrifuged at 16,000 relative centrifugal force (rcf), and the membrane fraction was extracted with Tris-saline buffer (50 mM Tris [pH 8], 150 mM NaCl, 2% SDS) containing protease inhibitors (Roche 11836170001). The extract was sonicated for 5 min (30 s on, 30 s off) with a Diagenode Bioruptor and then centrifuged at 16,000 rcf. Proteins in the supernatant were separated on a 3% to 8% Tris-acetate NuPage gel and transferred to a polyvinylidene difluoride (PVDF) membrane. The membrane was blocked for an hour with 5% milk–TBST (50 mM Tris, 150 mM NaCl, 0.1% Tween 20). PfEMP1 proteins were detected with guinea pig anti-PfEMP1 acidic transmembrane segment (ATS) (46) and rabbit anti-VAR2CSA PfEMP1 (47) primary antibodies, followed by goat anti-guinea pig horseradish peroxidase (HRP) (Abcam, Inc.; ab6908) and donkey anti-rabbit HRP (GE Healthcare Life Sciences; NA934V) secondary antibodies. HRP signal was developed with SuperSignal West Pico chemiluminescent substrate (Thermo Fisher Scientific; 34080) and imaged with a ChemiDoc XRS+ system (Bio-Rad).
Immunofluorescence assay.
Prior to labeling, polyclonal rabbit anti-VAR2CSA antibody was diluted 1:500 and subjected to preadsorption with 10 µl uninfected red blood cells for 1 h in 4% bovine serum albumin (BSA)–PBS. A Plasmagel flotation assay was performed on parasite culture to enrich for late-stage iRBCs. After washing 10 µl of iRBCs with PBS, cells were blocked with 4% BSA–PBS for 30 min, followed by incubation with 50 µl preadsorped primary antibody dilution for 1 h, three washes with PBS, and detection with Alexa Fluor 488-conjugated anti-rabbit IgG (Invitrogen) diluted 1:1,000 in 4% BSA–PBS. After three final washes in PBS, cells were mounted in Vectashield containing DAPI for nuclear staining. Images were captured using a Nikon Eclipse 80i microscope with a CoolSnap HQ2 camera (Photometrics). NIS elements 3.0 software (Nikon) was used for acquisition and Fiji software (http://fiji.sc/) for analysis.
Receptor panning.
Plastic cell culture dishes were coated with CD36 receptor (Life Technologies, Inc.; 10752-H08H) (4 µg/ml PBS) or CSA (Sigma; C9819) (1 mg/ml PBS) overnight at 4°C. The dishes were blocked with 1% BSA (Sigma, A4503)–PBS for 1 h at 37°C. iRBCs containing trophozoites and schizonts were isolated by Plasmagel enrichment and resuspended in 8 ml binding medium (RPMI 1640 with 25 mM HEPES [pH 6.8 for CD36 and pH 7.2 for CSA]) at a concentration of 5 × 107 iRBCs/ml. After the dish was washed once with binding medium, iRBCs were added and allowed to adhere for 1 h with gentle agitation at 37°C. The dish was washed with binding medium to eliminate all unbound cells, and the bound cells were recovered with normal medium. The culture was allowed to recover, and panning was repeated twice more. Binding was assessed at each panning by microscope.
Data availability.
Genomic DNA (3D7 WT clone, intron KO clone A, and intron KO telomere mutant clone) and RNA (3D7 WT bulk, intron KO clone A, and intron KO telomere mutant clone) sequencing fastq files are available from the National Centre for Biotechnology Information’s Sequence Read Archive (SRA) under project accession number PRJNA385099.
ACKNOWLEDGMENTS
We thank Benoit Gamain for providing important reagents and advice and Anna Barcons-Simon for critical reading of the manuscript.
This work was supported by a European Research Council Advanced grant (PlasmoSilencing 670301) and grants from the Agence Nationale de la Recherche (ANR-11-LABEX-0024-01 ParaFrap, ANR-13-ISV3-0003-01 NSFC MalVir) to A.S. J.M.B. was supported by a European Molecular Biology Organization long-term postdoctoral fellowship (EMBO ALTF 180-2015) and the Institut Pasteur Roux-Cantarini postdoctoral fellowship. J.G. was supported by a Human Frontier Science Program fellowship (LT000620/2012-L). The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
We all declare that we have no conflicts of interest.
Footnotes
Citation Bryant JM, Regnault C, Scheidig-Benatar C, Baumgarten S, Guizetti J, Scherf A. 2017. CRISPR/Cas9 genome editing reveals that the intron is not essential for var2csa gene activation or silencing in Plasmodium falciparum. mBio 8:e00729-17. https://doi.org/10.1128/mBio.00729-17.
REFERENCES
- 1.Smith JD, Rowe JA, Higgins MK, Lavstsen T. 2013. Malaria’s deadly grip: cytoadhesion of Plasmodium falciparum-infected erythrocytes. Cell Microbiol 15:1976–1983. doi: 10.1111/cmi.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scherf A, Lopez-Rubio JJ, Riviere L. 2008. Antigenic variation in Plasmodium falciparum. Annu Rev Microbiol 62:445–470. doi: 10.1146/annurev.micro.61.080706.093134. [DOI] [PubMed] [Google Scholar]
- 3.Jiang L, Mu J, Zhang Q, Ni T, Srinivasan P, Rayavara K, Yang W, Turner L, Lavstsen T, Theander TG, Peng W, Wei G, Jing Q, Wakabayashi Y, Bansal A, Luo Y, Ribeiro JM, Scherf A, Aravind L, Zhu J, Zhao K, Miller LH. 2013. PfSETvs methylation of histone H3K36 represses virulence genes in Plasmodium falciparum. Nature 499:223–227. doi: 10.1038/nature12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lopez-Rubio JJ, Gontijo AM, Nunes MC, Issar N, Hernandez Rivas R, Scherf A. 2007. 5’ flanking region of var genes nucleate histone modification patterns linked to phenotypic inheritance of virulence traits in malaria parasites. Mol Microbiol 66:1296–1305. doi: 10.1111/j.1365-2958.2007.06009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lopez-Rubio JJ, Mancio-Silva L, Scherf A. 2009. Genome-wide analysis of heterochromatin associates clonally variant gene regulation with perinuclear repressive centers in malaria parasites. Cell Host Microbe 5:179–190. doi: 10.1016/j.chom.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 6.Dzikowski R, Deitsch KW. 2009. Genetics of antigenic variation in Plasmodium falciparum. Curr Genet 55:103–110. doi: 10.1007/s00294-009-0233-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guizetti J, Scherf A. 2013. Silence, activate, poise and switch! Mechanisms of antigenic variation in Plasmodium falciparum. Cell Microbiol 15:718–726. doi: 10.1111/cmi.12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deitsch KW, Calderwood MS, Wellems TE. 2001. Malaria. Cooperative silencing elements in var genes. Nature 412:875–876. doi: 10.1038/35091146. [DOI] [PubMed] [Google Scholar]
- 9.Frank M, Dzikowski R, Costantini D, Amulic B, Berdougo E, Deitsch K. 2006. Strict pairing of var promoters and introns is required for var gene silencing in the malaria parasite Plasmodium falciparum. J Biol Chem 281:9942–9952. doi: 10.1074/jbc.M513067200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gannoun-Zaki L, Jost A, Mu J, Deitsch KW, Wellems TE. 2005. A silenced Plasmodium falciparum var promoter can be activated in vivo through spontaneous deletion of a silencing element in the intron. Eukaryot Cell 4:490–492. doi: 10.1128/EC.4.2.490-492.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calderwood MS, Gannoun-Zaki L, Wellems TE, Deitsch KW. 2003. Plasmodium falciparum var genes are regulated by two regions with separate promoters, one upstream of the coding region and a second within the intron. J Biol Chem 278:34125–34132. doi: 10.1074/jbc.M213065200. [DOI] [PubMed] [Google Scholar]
- 12.Dzikowski R, Li F, Amulic B, Eisberg A, Frank M, Patel S, Wellems TE, Deitsch KW. 2007. Mechanisms underlying mutually exclusive expression of virulence genes by malaria parasites. EMBO Rep 8:959–965. doi: 10.1038/sj.embor.7401063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voss TS, Healer J, Marty AJ, Duffy MF, Thompson JK, Beeson JG, Reeder JC, Crabb BS, Cowman AF. 2006. A var gene promoter controls allelic exclusion of virulence genes in Plasmodium falciparum malaria. Nature 439:1004–1008. doi: 10.1038/nature04407. [DOI] [PubMed] [Google Scholar]
- 14.Epp C, Li F, Howitt CA, Chookajorn T, Deitsch KW. 2009. Chromatin associated sense and antisense noncoding RNAs are transcribed from the var gene family of virulence genes of the malaria parasite Plasmodium falciparum. RNA 15:116–127. doi: 10.1261/rna.1080109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kyes SA, Christodoulou Z, Raza A, Horrocks P, Pinches R, Rowe JA, Newbold CI. 2003. A well-conserved Plasmodium falciparum var gene shows an unusual stage-specific transcript pattern. Mol Microbiol 48:1339–1348. doi: 10.1046/j.1365-2958.2003.03505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Su XZ, Heatwole VM, Wertheimer SP, Guinet F, Herrfeldt JA, Peterson DS, Ravetch JA, Wellems TE. 1995. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell 82:89–100. doi: 10.1016/0092-8674(95)90055-1. [DOI] [PubMed] [Google Scholar]
- 17.Amit-Avraham I, Pozner G, Eshar S, Fastman Y, Kolevzon N, Yavin E, Dzikowski R. 2015. Antisense long noncoding RNAs regulate var gene activation in the malaria parasite Plasmodium falciparum. Proc Natl Acad Sci U S A 112:E982–E991. doi: 10.1073/pnas.1420855112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Q, Siegel TN, Martins RM, Wang F, Cao J, Gao Q, Cheng X, Jiang L, Hon CC, Scheidig-Benatar C, Sakamoto H, Turner L, Jensen AT, Claes A, Guizetti J, Malmquist NA, Scherf A. 2014. Exonuclease-mediated degradation of nascent RNA silences genes linked to severe malaria. Nature 513:431–435. doi: 10.1038/nature13468. [DOI] [PubMed] [Google Scholar]
- 19.Ralph SA, Bischoff E, Mattei D, Sismeiro O, Dillies MA, Guigon G, Coppee JY, David PH, Scherf A. 2005. Transcriptome analysis of antigenic variation in Plasmodium falciparum—var silencing is not dependent on antisense RNA. Genome Biol 6:R93. doi: 10.1186/gb-2005-6-11-r93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siegel TN, Hon CC, Zhang Q, Lopez-Rubio JJ, Scheidig-Benatar C, Martins RM, Sismeiro O, Coppée JY, Scherf A. 2014. Strand-specific RNA-Seq reveals widespread and developmentally regulated transcription of natural antisense transcripts in Plasmodium falciparum. BMC Genomics 15:150. doi: 10.1186/1471-2164-15-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vembar SS, Scherf A, Siegel TN. 2014. Noncoding RNAs as emerging regulators of Plasmodium falciparum virulence gene expression. Curr Opin Microbiol 20:153–161. doi: 10.1016/j.mib.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Q, Huang Y, Zhang Y, Fang X, Claes A, Duchateau M, Namane A, Lopez-Rubio JJ, Pan W, Scherf A. 2011. A critical role of perinuclear filamentous actin in spatial repositioning and mutually exclusive expression of virulence genes in malaria parasites. Cell Host Microbe 10:451–463. doi: 10.1016/j.chom.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freitas-Junior LH, Hernandez-Rivas R, Ralph SA, Montiel-Condado D, Ruvalcaba-Salazar OK, Rojas-Meza AP, Mâncio-Silva L, Leal-Silvestre RJ, Gontijo AM, Shorte S, Scherf A. 2005. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell 121:25–36. doi: 10.1016/j.cell.2005.01.037. [DOI] [PubMed] [Google Scholar]
- 24.Ralph SA, Scheidig-Benatar C, Scherf A. 2005. Antigenic variation in Plasmodium falciparum is associated with movement of var loci between subnuclear locations. Proc Natl Acad Sci U S A 102:5414–5419. doi: 10.1073/pnas.0408883102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duffy MF, Selvarajah SA, Josling GA, Petter M. 2014. Epigenetic regulation of the Plasmodium falciparum genome. Brief Funct Genomics 13:203–216. doi: 10.1093/bfgp/elt047. [DOI] [PubMed] [Google Scholar]
- 26.Ghorbal M, Gorman M, Macpherson CR, Martins RM, Scherf A, Lopez-Rubio JJ. 2014. Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nat Biotechnol 32:819–821. doi: 10.1038/nbt.2925. [DOI] [PubMed] [Google Scholar]
- 27.Trimnell AR, Kraemer SM, Mukherjee S, Phippard DJ, Janes JH, Flamoe E, Su XZ, Awadalla P, Smith JD. 2006. Global genetic diversity and evolution of var genes associated with placental and severe childhood malaria. Mol Biochem Parasitol 148:169–180. doi: 10.1016/j.molbiopara.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 28.Salanti A, Staalsoe T, Lavstsen T, Jensen AT, Sowa MP, Arnot DE, Hviid L, Theander TG. 2003. Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Mol Microbiol 49:179–191. doi: 10.1046/j.1365-2958.2003.03570.x. [DOI] [PubMed] [Google Scholar]
- 29.Salanti A, Dahlbäck M, Turner L, Nielsen MA, Barfod L, Magistrado P, Jensen AT, Lavstsen T, Ofori MF, Marsh K, Hviid L, Theander TG. 2004. Evidence for the involvement of VAR2CSA in pregnancy-associated malaria. J Exp Med 200:1197–1203. doi: 10.1084/jem.20041579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Viebig NK, Gamain B, Scheidig C, Lépolard C, Przyborski J, Lanzer M, Gysin J, Scherf A. 2005. A single member of the Plasmodium falciparum var multigene family determines cytoadhesion to the placental receptor chondroitin sulphate A. EMBO Rep 6:775–781. doi: 10.1038/sj.embor.7400466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacPherson CR, Scherf A. 2015. Flexible guide-RNA design for CRISPR applications using Protospacer Workbench. Nat Biotechnol 33:805–806. doi: 10.1038/nbt.3291. [DOI] [PubMed] [Google Scholar]
- 32.Bottius E, Bakhsis N, Scherf A. 1998. Plasmodium falciparum telomerase: de novo telomere addition to telomeric and nontelomeric sequences and role in chromosome healing. Mol Cell Biol 18:919–925. doi: 10.1128/MCB.18.2.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duffy MF, Caragounis A, Noviyanti R, Kyriacou HM, Choong EK, Boysen K, Healer J, Rowe JA, Molyneux ME, Brown GV, Rogerson SJ. 2006. Transcribed var genes associated with placental malaria in Malawian women. Infect Immun 74:4875–4883. doi: 10.1128/IAI.01978-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lavstsen T, Magistrado P, Hermsen CC, Salanti A, Jensen AT, Sauerwein R, Hviid L, Theander TG, Staalsoe T. 2005. Expression of Plasmodium falciparum erythrocyte membrane protein 1 in experimentally infected humans. Malar J 4:21. doi: 10.1186/1475-2875-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mok BW, Ribacke U, Rasti N, Kironde F, Chen Q, Nilsson P, Wahlgren M. 2008. Default pathway of var2csa switching and translational repression in Plasmodium falciparum. PLoS One 3:e1982. doi: 10.1371/journal.pone.0001982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amulic B, Salanti A, Lavstsen T, Nielsen MA, Deitsch KW. 2009. An upstream open reading frame controls translation of var2csa, a gene implicated in placental malaria. PLoS Pathog 5:e1000256. doi: 10.1371/journal.ppat.1000256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bancells C, Deitsch KW. 2013. A molecular switch in the efficiency of translation reinitiation controls expression of var2csa, a gene implicated in pregnancy-associated malaria. Mol Microbiol 90:472–488. doi: 10.1111/mmi.12379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frank M, Dzikowski R, Amulic B, Deitsch K. 2007. Variable switching rates of malaria virulence genes are associated with chromosomal position. Mol Microbiol 64:1486–1498. doi: 10.1111/j.1365-2958.2007.05736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rogerson SJ, Hviid L, Duffy PE, Leke RF, Taylor DW. 2007. Malaria in pregnancy: pathogenesis and immunity. Lancet Infect Dis 7:105–117. doi: 10.1016/S1473-3099(07)70022-1. [DOI] [PubMed] [Google Scholar]
- 40.Dzikowski R, Frank M, Deitsch K. 2006. Mutually exclusive expression of virulence genes by malaria parasites is regulated independently of antigen production. PLoS Pathog 2:e22. doi: 10.1371/journal.ppat.0020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ukaegbu UE, Zhang X, Heinberg AR, Wele M, Chen Q, Deitsch KW. 2015. A unique virulence gene occupies a principal position in immune evasion by the malaria parasite Plasmodium falciparum. PLoS Genet 11:e1005234. doi: 10.1371/journal.pgen.1005234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, Paulsen IT, James K, Eisen JA, Rutherford K, Salzberg SL, Craig A, Kyes S, Chan MS, Nene V, Shallom SJ, Suh B, Peterson J, Angiuoli S, Pertea M, Allen J, Selengut J, Haft D, Mather MW, Vaidya AB, Martin DM, Fairlamb AH, Fraunholz MJ, Roos DS, Ralph SA, McFadden GI, Cummings LM, Subramanian GM, Mungall C, Venter JC, Carucci DJ, Hoffman SL, Newbold C, Davis RW, Fraser CM, Barrell B. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 GPDPS . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wickert H, Wissing F, Andrews KT, Stich A, Krohne G, Lanzer M. 2003. Evidence for trafficking of PfEMP1 to the surface of P. falciparum-infected erythrocytes via a complex membrane network. Eur J Cell Biol 82:271–284. doi: 10.1078/0171-9335-00319. [DOI] [PubMed] [Google Scholar]
- 47.Avril M, Hathaway MJ, Srivastava A, Dechavanne S, Hommel M, Beeson JG, Smith JD, Gamain B. 2011. Antibodies to a full-length VAR2CSA immunogen are broadly strain-transcendent but do not cross-inhibit different placental-type parasite isolates. PLoS One 6:e16622. doi: 10.1371/journal.pone.0016622. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Genomic DNA (3D7 WT clone, intron KO clone A, and intron KO telomere mutant clone) and RNA (3D7 WT bulk, intron KO clone A, and intron KO telomere mutant clone) sequencing fastq files are available from the National Centre for Biotechnology Information’s Sequence Read Archive (SRA) under project accession number PRJNA385099.