Abstract
SB 9200 is a novel, first-in-class oral modulator of innate immunity that is believed to act via the activation of the RIG-I and NOD2 pathways. SB 9200 has broad-spectrum antiviral activity against RNA viruses including hepatitis C virus (HCV), norovirus, respiratory syncytial virus and influenza and has demonstrated activity against hepatitis B virus (HBV) in vitro and in vivo. In phase I clinical trials in chronically infected HCV patients, SB 9200 has been shown to reduce HCV RNA by up to 1.9 log10. Here, we demonstrate the antiviral activity of SB 9200 against a HCV replicon system and patient derived virus. Using the HCV capture-fusion assay, we show that SB 9200 is active against diverse HCV genotypes and is also effective against HCV derived from patients who relapse following direct-acting antiviral treatment, including viruses containing known NS5A resistance-associated sequences. These data confirm the broad antiviral activity of SB 9200 and indicate that it may have clinical utility in HCV patients who have failed to respond to current antiviral regimens.
Introduction
New direct-acting antiviral (DAAs) agents for chronic hepatitis C virus (HCV) infection have substantially increased the rates of sustained virological response (SVR). Pan-genotypic regimens, with response rates in excess of 95%, are being marketed [Chahine et al., 2016; Feld et al., 2015; Foster et al., 2015], leading to the hope that viral elimination may now be possible. However, despite the effectiveness of the new drugs, a small proportion of patients do not respond to first-line therapy and, although re-treatment with extended duration may benefit some patients [Lawitz et al., 2016], current therapies may not lead to virological cure for all. A concern is the relatively limited targets for current antivirals – all the licensed drugs inhibit either the viral NS3/4 protease, the NS5A protein or NS5B, the RNA-dependent RNA-polymerase, leading to the possibility of cross-resistant virus emerging after repeated cycles of therapy; mutations within the NS5A protein are replication competent and persist long-term [Sarrazin et al., 2016]. To successfully treat the small number of patients who develop resistance to current agents, novel antivirals with new modes of action may be required.
Mammalian cells have evolved a wide range of restriction factors to inhibit viral replication and protect the host from infection. Some of these factors are specific to viral families, such as TRIM5α [Stremlau et al., 2004] and REAF [Marno et al., 2014] which are active against retroviruses. Others target large numbers of viruses, such as the IFN or the APOBEC systems [Schneider et al., 2014] [Harris and Dudley, 2015]. These host antiviral mechanisms are an attractive target for novel antiviral therapeutics. We have previously described a group of compounds that augment antiviral defences by targeting the Retinoic Acid Inducible Gene (RIG-I) and Nucleotide Oligomerization Domain protein 2 (NOD2) signalling cascades [Iyer et al., 2010]. SB 9000 has previously been shown to bind RIG-I in a dose-dependent manner and induce rapid translocation (or repetitive shuttling) towards viral double-stranded RNA (dsRNA). When transfected into A549 or HEK cells, IRF3 and NF-KB reporter constructs revealed that SB 9000/9200 is capable of activating the IFN signaling cascade, whereas siRNA-knockdown of RIG-I and NOD2 expression abrogated IFN signalling in SB 9200-treated cells [Sreerupa Challa, 2016] (conference paper). Other studies have also reported antiviral effects of SB 9200 against RNA viruses including, Flaviviridae, (HCV), Paramyxoviridae (human respiratory syncytial virus, hRSV), Caliciviridae (norovirus) and Orthomyxoviridae (influenza) [Iyer RP, 2014a; Iyer RP, 2014b] (conference papers).
An orally bioavailable prodrug, SB 9200, of the active SB 9000 has been advanced to preclinical and clinical studies. Studies in tissue culture and animal models have shown the safety and efficacy of this compound for the treatment of chronic HBV infection [Korolowicz et al., 2016], In chronically woodchuck hepatitis virus (WHV)-infected woodchucks, the antiviral effects of SB 9200 were associated with dose-dependent and long-lasting induction of IFN-α, IFN-β and ISGs in liver tissues. SB 9200-treatment also increased liver expression levels of RIG-I, NOD2, STING, and IRF3 compared with pre-treatment levels [Korolowicz et al., 2016].
A SAD and MAD seven-day Phase I clinical study in chronic HCV patients showed that SB 9200 was safe, well tolerated and also reduced viral replication during short term dosing [Thompson AJ, 2015] (conference paper). Herein, we report the activity of SB 9200 against several HCV viral genotypes and virus containing resistant motifs. In addition, we have analysed the effects of SB 9200 against a wide range of HCV clinical isolates. We used the ‘capture-fusion’ replication system for HCV [Cunningham et al., 2015] and show that SB 9200 is a potent inhibitor of HCV replication and, importantly, retains activity against known viral variants that confer resistance to other drug classes.
Materials and methods
Cells, reagents and clinical material
Antiviral activity against HCV was assessed in a using the stably-expressing HCV replicon cell line, AVA5 (sub-genomic (CON1), genotype 1b [Blight et al., 2000] and a replicon cell line containing H/FL-Neo (genotype 1a (H77), full length construct) [Blight et al., 2003]. For the capture fusion assay, all Huh7 derivative cells (Lunet and ICP+1, kind gift from Gilead, Foster City, CA, USA, Huh7.5, kind gift form C. Rice, The Rockefeller University, New York, NY, USA) were propagated in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum. THP1 cells were propagated in RPMI supplemented with 10% fetal calf serum. HCV sera were obtained from patients with chronic HCV infection, with informed consent. Cytokines used were phorbol 12-myristate 13-acetate (PMA, Sigma-Aldrich, Dorset, UK) and interferon γ (IFNγ; Invitrogen, Paisley, UK).
SB 9200, an oral prodrug of SB 9000, was manufactured by Spring Bank Pharmaceuticals, Inc. and the structure and antiviral characteristics of SB 9200/SB 9000 have previously been described [Iyer et al., 2005]. Other antiviral reagents were kindly supplied by Janssen Virology (telaprevir), Novartis Pharmaceuticals (alisporivir), Gilead (sofosbuvir). All antiviral drugs were reconstituted in DMSO. Interferon α-2a (IFNα-2a) was purchased from Cambridge Bioscience (Cambridge, UK) and recombinant interferon alpha 2b was from PBL Laboratories, England).
Ethical approval for the study was given by London – City Research Ethics Committee and informed consent was obtained from all patients for the use of their samples in laboratory research. The study was conducted in accordance with the Declaration of Helsinki.
Replicon cell culture based antiviral assays
Antiviral activity was determined by blot hybridization analysis of intracellular HCV RNA (normalized to the level of cellular B-actin RNA in each culture sample) as previously described [Okuse et al., 2005]. Cytotoxicity was assessed by neutral red dye uptake in parallel cultures. EC50, EC90 and CC50 values were calculated by linear regression analysis (MS EXCEL®, QuattroPro®) using data combined from all treated cultures. Standard deviations for EC50 and EC90 values were calculated from the standard errors generated by the regression analyses. EC50 and EC90 are drug concentrations at which a 2-fold, or a 10-fold depression of intracellular HCV RNA (relative to the average levels in untreated cultures), respectively, was observed. CC50 is the drug concentration at which a 2-fold lower level of neutral red dye uptake (relative to the average levels in untreated cultures) was observed. The Selectivity index (S.I.) was calculated as CC50/EC50. Recombinant human interferon alpha 2b and 2’C-methyl Cytidine (Moravek Laboratories) were used as positive antiviral assay controls. For combination treatment studies, compounds were mixed at ratios centred around achieving equipotent concentrations (based on EC50 observed in monotherapies) and this molar ratio was maintained during serial dilution [Korba, 1996] [Iyer et al., 2004]. Corresponding monotherapies were included in parallel for each experiment. Treatment and analysis methods of antiviral and toxicity were as described above. Evaluation of drug interactions in the combination treatments was conducted against the corresponding monotherapies in the same experiments using the Combostat® (Biosoft, Inc.) analysis software. For combination treatments, EC50, EC90, CC50 and S.I. are presented for the first compound listed. The molar ratio of the compounds in each combination is also indicated.
Capture fusion assay
The capture fusion was performed as previously described with minor modifications [Cunningham et al., 2015]. THP-1 cells were seeded into 6 well plates (106 cells/mL) and maintained for 18 hours with IFNγ [10ng/ml] and PMA [200ng/ml]. The cells were washed thrice with PBS and the medium replaced with RPMI/2% FCS and patient serum (1 HCV IU/cell). Following overnight incubation at 37°C for 18–24 hours, the supernatant was removed and cells were washed with PBS. Adherent cells were removed by scraping and centrifuged with the Huh7 derivatives (1:1 ratio). The cell pellet was re-suspended in polyethylene glycol (PEG) 1500 (Roche Diagnostics, Burgess Hill, UK). After incubation at 37°C for 2 minutes, pre-warmed medium (DMEM) was added dropwise and the cells washed by centrifugation. The fused cells were seeded into 6-well plates (5×105 cells/mL) and maintained at 37°C in the presence or absence of antiviral drugs for 5 days.
HCV RNA quantification by reverse-transcription PCR
Total RNA was extracted with TRIzol reagent (Invitrogen) and quantified using the RiboGreen reagent (Invitrogen), according to the manufacturer’s instructions. One-step reverse transcription PCR (RT-qPCR) with the QuantiTect Virus Kit (Qiagen, Crawley, UK) and TaqMan Gene Expression Assay HCV primer and probe (Applied Biosystems, Paisley, UK) was used for HCV quantification. PCR reactions were performed in triplicate. Absolute quantification of HCV RNA was performed by inclusion of serial dilutions of an RNA standard in each PCR run, and expressed relative to total sample RNA [Bustin et al., 2009]
Statistical analysis
Statistical analyses were performed using Student t-test or Mann-Whitney U-test in Prism 4.0, with p <0.05 considered significant.
Results
SB 9200 is an effective inhibitor of HCV replication in cell culture
The antiviral activity of SB 9200 against HCV was assessed using genotype 1 HCV replicon systems in duplicate experiments using 4 drug concentrations. Results are expressed as EC50 and EC90, as a 2-fold, or a 10-fold reduction of intracellular HCV RNA relative to that of the untreated controls [Dot blot hybridization assay normalized to β–actin RNA]. SB 9200 was shown to inhibit HCV replication and the range of inhibition was comparable between genotypes 1a and 1b (Table 1). Combination studies of SB 9200 with other anti-HCV agents were carried out using the HCV genotype 1b CON1 sub-genomic replicons. Prior to the combination studies, the anti-HCV activity of individual compounds was determined to calculate ratios based on relative EC50 values. Synergy studies were initiated by combining SB 9200 with each agent in different ratios based on individual EC50 values to achieve equipotent anti-HCV activity of each combination (ratios remained constant throughout the dilution series). Combination and monotherapy studies were conducted side-by-side with using 8, 3-fold dilutions to assess antiviral activity and drug interactions. In combination treatments with interferon alpha, ribavirin, or two different classes of anti-HCV direct-acting antiviral agents (nucleoside analogue, protease inhibitor), SB 9200 displayed predominantly synergistic interactions (Table 2).
Table 1.
Anti-HCV activity of SB 9200 in replicons.
| Compound | HCV genotype | EC50 (μM) | EC90 (μM) | CC50 (μM) | S.I. (CC50/EC50) |
|---|---|---|---|---|---|
| SB 9200 | 1A* | 2.2 ± 0.3 | 8.0 ± 0.8 | >100 | > 45 |
| 1B** | 1.0 ± 0.2 | 6.0 ± 0.7 | >100 | >100 | |
| Interferon αIFNB2 IU/mL | 1A | 1.8 ± 0.2 | 8.0 ± 0.6 | >10,000 | >5556 |
| NM 283 | 1B | 1.4 ± 0.1 | 5.3 ± 0.6 | >300 | >214 |
| 1A | 1.6 ± 0.2 | 6.0 ± 0.5 | >300 | >188 |
Replicon cell line containing H/FL-Neo (genotype 1a (H77), full length construct) (Blight, et al., 2003, J. Virol. 77:3181);
Replicon cell line AVA5 (sub-genomic (CON1), genotype 1b; (Blight, et al., 2000, Science 290:1972). EC50 and EC90 are drug concentrations that result in a 2-fold, or a 10-fold depression of intracellular HCV RNA relative to that of the untreated controls [Dot blot hybridization assay normalized to β–actin RNA]. S.I. corresponds to selectivity index.
Table 2. Combination studies of SB 9200 with anti-HCV agents using HCV replicon assays.
Combination studies of SB 9200 with other anti-HCV agents were carried out using the HCV genotype 1b CON1 sub-genomic replicons. All compounds were dissolved in DMSO to get 100mM stock solution. Before initiating combination studies, the anti-HCV activity of individual compounds was determined using protocols described in the primary assays. Synergy studies were initiated by combining SB 9200 with each agent in different ratios based on individual EC50 values to achieve equipotent anti-HCV activity of each combination.
| SB 9200 | EC50 (μM) (individual agent) | Ratio | EC50 (μM) | EC90 (μM) | CC50 (μM) | S.I. (CC50/EC50) | Type of Interaction |
|---|---|---|---|---|---|---|---|
|
| |||||||
| SB 9200 | 1.0 ± 0.2 | N/A | 1.0 ± 0.2 | 6.0 | >100 | >100 | N/A |
|
| |||||||
| + IFN α | 2.1 ± 0.2 | 3:1 | 0.662 ± 0.072 | 2.8 | >100 | >151 | Synergistic |
| 1:1 | 0.643 ± 0.060 | 2.9 | >100 | >156 | Synergistic | ||
| 1:3 | 0.658 ± 0.055 | 8.8 | >100 | >152 | Synergistic | ||
|
| |||||||
| + Ribavirin | >30 | 1:30 | 2.3 ± 0.3 | >100 | >43 | Synergistic | |
|
| |||||||
| + Ribavirin (30μM) + IFN α | N/A | 3:1 | 0.777 ± 0.087 | 2.7 | >100 | >129 | Synergistic |
| 1:1 | 0.629 ± 0.082 | 2.6 | >100 | >159 | Synergistic | ||
| 1:3 | 0.686 ± 0.099 | 2.5 | >100 | >146 | Synergistic | ||
|
| |||||||
| + 2CMeCyt (NM 283) | 1.4 ± 0.1 | 3:1 | 0.522 ± 0.046 | 1.3 | >100 | >192 | Synergistic |
| 1:1 | 0.494 ± 0.050 | 1.8 | >100 | >202 | Synergistic | ||
| 1:3 | 0.462 ± 0.061 | 1.5 | >100 | >216 | Additive | ||
|
| |||||||
| + VX-950 (telaprevir) | 0.250 ± 0.024 | 100:1 | 0.108 ± 0.028 | 0.95 | >100 | >926 | Synergistic |
| 30:1 | 0.126 ± 0.026 | 1.0 | >100 | >794 | Synergistic | ||
| 10:1 | 0.118 ± 0.023 | 0.93 | >100 | >847 | Synergistic | ||
Synergy analysis using CalcuSynTM program (Biosoft, Inc.) (B. E Korba (1996) Antiviral Res. 29:49). N/A not applicable.
SB 9200 demonstrates pan-genotypic antiviral activity against HCV
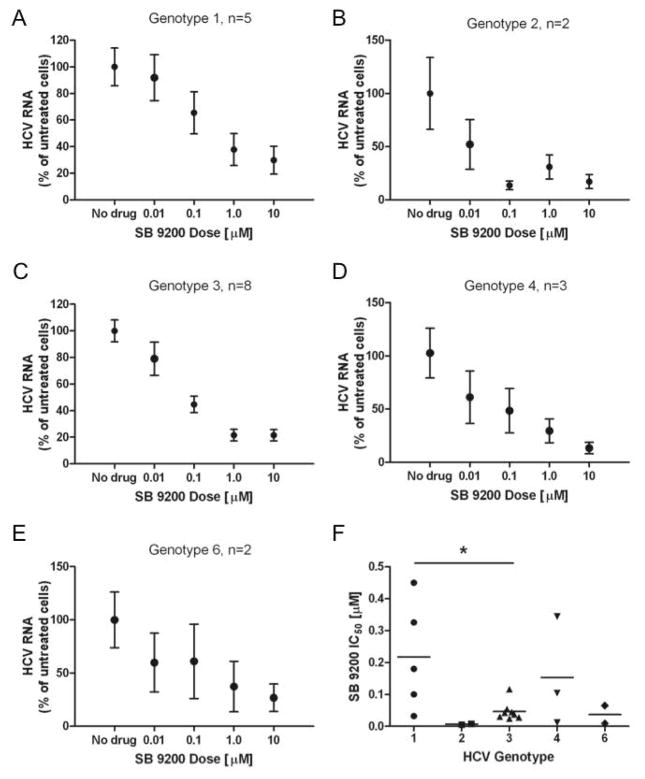
The capture fusion assay allows in vitro determination of the sensitivity of patient-derived HCV to antiviral drugs. The effect of SB 9200 on viral samples from a number of patients was determined (Patient details - supplementary Table 1). Cell viability in the capture fusion assay was not affected by the presence of SB 9200 (data not shown). Figure 1A–E shows the pooled 50% inhibitory concentration (IC50) data for Genotypes 1,2,3,4, and 6. SB 9200 inhibited replication of all the genotypes tested in a dose-dependent manner. G3 isolates showed significantly greater sensitivity to SB 9200 than G1 strains (IC50 0.035 ± 0.013 versus 0.22 ± 0.076, p = 0.048).
Figure 1. SB 9200 demonstrates pan-genotypic antiviral activity against HCV.
The sensitivity of patient derived HCV to SB 9200 was assessed in the phenotypic capture fusion assay. HCV-containing fused cells were treated with varying concentrations of SB 9200 and HCV RNA was quantified. Pooled results, normalised to untreated cells to account for variability in HCV RNA yield between samples from different donors are shown for each genotype (A–E) values are mean ± s.e.m. Comparison of the individual IC50 for SB 9200 across the HCV genotypes tested (F) p value (*=p<0.05) was calculated using Mann Whitney U test
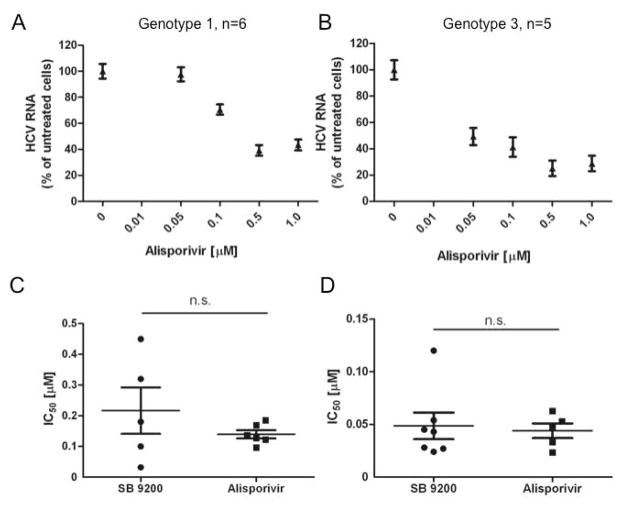
Sensitivity of both G1 and G3 HCV to SB 9200 compares favourably to Alisporivir
The host-targeting agent, alisporivir, has been shown to be a potent in vitro inhibitor of HCV replication [Paeshuyse et al., 2006]. Using the capture fusion assay we assessed the antiviral activity of alisporivir against genotype 1 and 3 patient derived samples (figure 2A–B) (G1; IC50 = 0.099μM, G3; IC50 = 0.036μM). No significant difference was seen between the effect of alisporivir and SB 9200 on either G1 or G3 patient derived HCV (figure 2C–D).
Figure 2. Sensitivity of both G1 and G3 HCV to SB 9200 compares favourably to Alisporivir.
Sensitivity of patient-derived G1 (A) or G3 (B) HCV to Alisporivir was assessed in the capture-fusion assay and a dose response calculated. Values are mean ± s.e.m. The individual IC50 for SB 9200 and Alisporivir are plotted for G1 (C) and G3 (D).
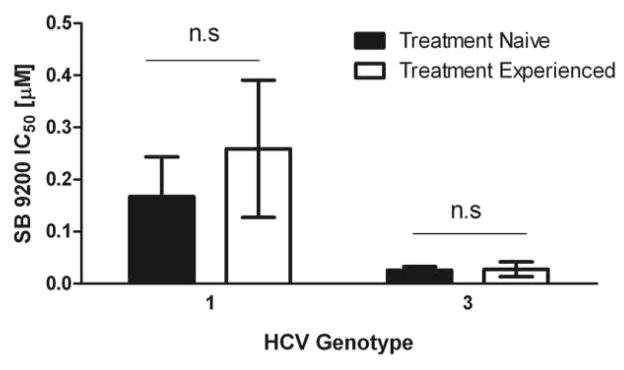
SB 9200 shows activity against HCV variants derived from patients who have relapsed after IFN based therapy
Prior to the introduction of all oral antiviral therapy, interferon was widely used to treat HCV infection and patient response was dependent on both viral genotype and host factors [Suppiah et al., 2009] [Tanaka et al., 2009]. We examined the activity of SB 9200 against viral samples from patients who had failed to respond to interferon-based therapy. No significant difference in SB 9200 efficacy was found between treatment naïve and treatment experienced patients in either G1 or G3 (figure 3).
Figure 3. SB 9200 shows activity against HCV derived from patients who have relapsed after IFN based therapy.
Capture fusion assay was performed with sera from patients who had relapsed after IFN based therapy. Comparison of the SB 9200 IC50 between treatment naïve (g1 n=5 and g3 n=7) and treatment experienced (g1 n=3 and g3 n=4 ) revealed no significant difference between the groups.
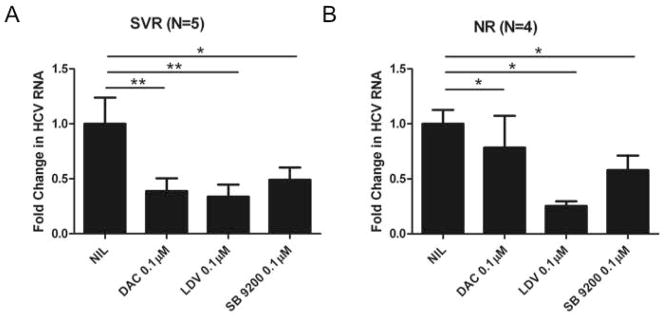
SB 9200 shows activity against HCV derived from patients who have relapsed after DAA therapy
HCV treatment has now progressed to highly effective IFN-free regimens. To evaluate if SB 9200 had a differential effect on viruses derived from patients who had failed all oral regimens, we studied nine G3 patients from the English early access programme who had been treated with sofosbuvir plus an NS5A inhibitor-containing regime (patient details are shown in Supplementary Table 2). Serum from these patients with low levels of viraemia was limited. Therefore we assessed drug sensitivity with a single concentration of the antiviral agent and determined sensitivity by calculating fold change in HCV RNA from the non-treated controls. SB 9200 demonstrated antiviral activity against both the SVR and NR samples (figure 4). The fold change in the SVR samples was similar to the tested NS5A inhibitors. In the NR samples, counter-intuitively, although blunted, we showed antiviral activity of the NS5A inhibitors, suggesting that resistance to the NS5A inhibitors had not developed.
Figure 4. SB 9200 shows activity against HCV derived from patients who have relapsed after DAA therapy.
Capture fusion assay was performed with sera from G3 patients who had responded (A) or failed to respond (B) to DAA treatment. A single dose [0.1μM] in quadruplicate of either Daclatasvir, Ledipasvir or SB 9200 was used and the fold change for each drug ± s.e.m plotted.
SB 9200 is active against DAA-resistant HCV variants
To expand our analysis of the activity of SB 9200 against NS5A, we selectively examined the effect of SB 9200 in samples with known NS5A resistance associated substitutions (RAS). Samples and sequencing data from Gilead’s sofosbuvir clinical trial programme were kindly supplied by Gilead Sciences (Foster City, CA). Table 3 shows the sensitivity of these viruses to SB 9200. In these G1a samples with known NS5A RAS, SB 9200 was generally potent although one sample (with the L31M RAS) did have a reduced sensitivity to the drug.
Table 3. SB 9200 is active against DAA resistant HCV variants.
The activity of SB 9200 against serum from g1a patients who subsequently failed sofosbuvir/ledipasvir treatment was evaluated. Sequencing analysis revealed the presence of several HCV variants associated with poor response to current NS5A inhibitors. SB 9200 demonstrated antiviral activity against the known NS5A RAVs L31M and Q30H.
| Sub type | HCV RNA (IU/ml) | Change From Ref NS5A | Consensus Sequencing: NS5A RAS | SB 9200 IC50 [uM] |
|---|---|---|---|---|
| 1a | 12955297 | S1A L31M R78K I90V K107T S131T I144V R176K E181D A213T M226E A245T N246K D285E V296I A310G R311P V315I V326L R348Q L368V T370N A400S G403D V410A Y413C T442A | L31M | 0.0026 |
| 1a | 4850952 | A25S Q30H V37L T64A R78K T99V K107T S131T T135A I144V E171D E181D M226V A245T D248E V296I R308K A310S R311P V315I V326L P347S R348R/Q S349P L368V N392E P399S A400G G403A P405L V410A G439E T442A | Q30H | 0.2143 |
| 1a | 3019319 | A25S Q30H V37L T64A R78K T99V K107T S131T T135A I144V E171D E181D M226V A245T D248E V296I R308K A310S R311P V315I V326L P347S S349P L368V N392E P399S A400G G403A P405L V410A G439E T442A | Q30H | 0.046 |
| 1a | 1420666 | S1A L31M R78K K107T S131T I144V E171D E181D G215R M226E K240R A245T I280V D285E E293D A310G R311P V315I V326L R348Q T367S L368V T395A G403V P405L V410A Y413C D441E | L31M | >10 |
| 1a | 5933317 | I8I/V F36L V37V/M S85S/N I121I/V S131T I144V E171D E181D A197T L199V A213T G215K M226L A241G D285E V288V/M V296I V298T A310T R311P V315I R348K/Q T367S L368V I388I/V G390S T393M T395A S397P P401S V410A G439E D441G | None | 0.02043 |
Activity of SB 9200 against pre-, and post-relapse HCV variants in a patient with emerging NS5A resistance
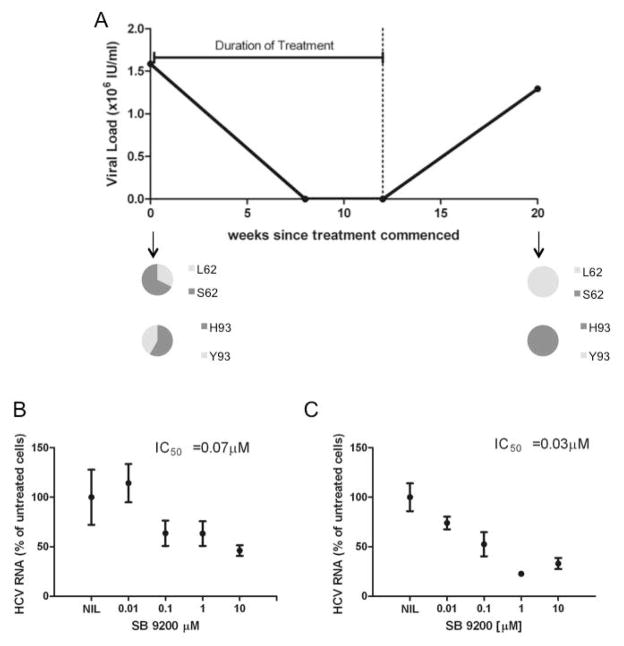
Pre-, and post-treatment-failure serum was available from a patient who failed to respond to 12 weeks SOF/DAC/RBV treatment. The patient’s viral load initially fell on treatment but virological breakthrough and relapse occurred after week 8 on treatment. Deep sequencing was performed on the serum pre-treatment and at week 20 after start of treatment. Analysis of the NS5A sequence showed that RASs, including those known to inhibit the activity of current NS5A inhibitors - S62L and Y93H - were present in the pre-treatment samples and these were enriched following unsuccessful treatment (figure 5A). No significant difference in the activity of SB 9200 against the pre-, (figure 5B) and post treatment (figure 5C) quasi-species was seen when evaluated by capture fusion, suggesting that SB 9200 retains activity against treatment emergent NS5A substitutions.
Figure 5. Activity of SB 9200 against pre and post-relapse HCV variants in a g3 DAA failure.
A decline in viral load (A) was observed during treatment with SOF/DAC/RBV but the viral load rebounded 8 weeks after the end of the DAA therapy. Pre-treatment and post-relapse samples were subjected to high throughput viral genome sequencing. Despite enrichment for known NS5A RAS (L62 and H93) in the post-relapse sample no significant difference was seen in out phenotypic capture fusion assay in the IC50 between pre-treatment (B) and post-relapse (C) for SB 9200.
Discussion
All oral DAA therapy has transformed the management of patients with chronic HCV infection. Despite the marked improvement in outcome, a minority of patients remain refractory to current therapy and, as treatment programmes expand, increasing numbers of such patients will emerge. It is unclear how to manage such patients but concerns have been expressed that multi-drug resistant HCV may emerge. Given the probability of treatment failure with the current, rather restricted antiviral regimens, new drugs that target different host targets will be required. SB 9200 is a novel antiviral agent with a unique mechanism of action. The drug targets intracellular innate viral inhibitors including RIGI and NOD-2. Previous work has shown activity against HCV in patients as well as activity against HBV in animal and tissue culture models [Korolowicz et al., 2016]. Here we have evaluated the activity of SB 9200 in samples from patients with potentially difficult to cure HCV.
To confirm the activity of SB 9200 against HCV in established assays we used the well-described HCV replicon system. The EC50 values obtained for the individual compounds we tested compared favourably to those previously reported in the literature [Horscroft et al., 2005] and confirmed that SB 9200 does have potent antiviral activity in this model. However the replicon assay does not allow an evaluation of patient derived viral samples and is sub-optimal for phenotyping studies. The ‘capture-fusion’ model is a recently described assay that allows patient derived virus to be analysed [Cunningham et al., 2015]. Although the assay is cumbersome and can only be applied to samples with a high concentration of virus (usually >105 IU/ml) it has the advantage of allowing studies on patient derived, drug resistant virus. In this study, we have deployed the assay to analyse SB 9200 in a wide range of samples and potent antiviral activity is seen against nearly all the isolates studied. Although SB 9200 had pan-genotypic antiviral activity, we noted increased sensitivity among G3 isolates with a minority of G1 samples being relatively insensitive to the drug. Of importance, isolates from patients who had failed to respond to current treatment regimens were inhibited by SB 9200, suggesting that this drug may be a valuable option in patients requiring ‘rescue’ therapy. One g1a sample with a L31M RAS in the NS5a appeared to be refractory to the effects of SB 9200, no other known RAS were identified in the NS5A protein, however we cannot discount the presence of other mutations within this sample. Further work will be required to identify the mechanisms underlying these differences.
The observed anti-HCV activity of SB 9200 is supported by the mechanism of action (MOA) studies of SB 9200/SB 9000. The activation of both RIG-I and NOD2, and the induction of the IFN signalling cascade could result in the expression of IFNs, ISGs and antiviral cytokines all of which may play an important role in the antiviral activity of SB 9200/SB 9000.
It is pertinent to mention that in the classical HCV replicon cells, RIG-I signalling is reported to be impaired. The HCV NS3/4A protease has been shown to inhibit phosphorylation of IRF-3, which is a key transcriptional regulator of the IFN response, thereby abrogating the antiviral response to infection [Foy et al., 2005; Sumpter et al., 2005]. The Huh7.5 cells used in this study [Blight et al., 2002] contain a mutation in the RIG-I gene which disrupts IRF-3 activation. Nevertheless, the observed antiviral activity of SB 9200 against the HCV replicon may be partly due to SB 9200-mediated rapid shuttling of RIG-I on dsRNA that displaces viral polymerase from dsRNA template thereby inhibiting viral replication [Jankowsky et al., 2001]. Fusion of Huh7 derived cell lines with the RIG-I competent monocytes is thought to re-constitute RIG-I activity, and in these hybrid cells SB 9200 was found to be a potent inhibitor of HCV replication.
In summary, we find that the broad-spectrum antiviral agent SB 9200 shows activity against a wide range of HCV isolates, including those that are refractory to current antiviral agents. These data indicate that trials of SB 9200 in patients with resistant chronic HCV infection are warranted and this drug may play an important role in the global elimination of HCV by increasing treatment options for patients who have failed to respond to first-line therapies.
Supplementary Material
Acknowledgments
The authors wish to thank Prof John McLauchlan and Dr Ana Da Silva Filipe, MRC-University of Glasgow, Centre for Virus Research for the HCV sequencing and data analysis from the EAP sample.
HCV replicon studies were conducted under and supported by NIAID contract HHSN2722011000008C to the Georgetown University Medical Center.
Partial support of this work from NIAID through R01 grant AI094469 (RPI, PI) to Spring Bank Pharmaceuticals, Inc. is gratefully acknowledged.
Abbreviations
- DAA
direct acting antiviral
- SVR
sustained virological response
- G
genotype
- SOF
sofosbuvir
- DAC
daclatasvir
- RBV
ribavirin
- RAS
resistance associated substitutions
- SAD
single ascending dose
- MAD
multiple ascending dose
References
- Blight KJ, Kolykhalov AA, Rice CM. Efficient initiation of HCV RNA replication in cell culture. Science. 2000;290(5498):1972–1974. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- Blight KJ, McKeating JA, Marcotrigiano J, Rice CM. Efficient replication of hepatitis C virus genotype 1a RNAs in cell culture. J Virol. 2003;77(5):3181–3190. doi: 10.1128/JVI.77.5.3181-3190.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol. 2002;76(24):13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Chahine EB, Sucher AJ, Hemstreet BA. Sofosbuvir/Velpatasvir: The First Pangenotypic Direct-Acting Antiviral Combination for Hepatitis C. Ann Pharmacother. 2016 doi: 10.1177/1060028016668897. [DOI] [PubMed] [Google Scholar]
- Cunningham ME, Javaid A, Waters J, Davidson-Wright J, Wong JL, Jones M, Foster GR. Development and validation of a “capture-fusion” model to study drug sensitivity of patient-derived hepatitis C. Hepatology. 2015;61(4):1192–1204. doi: 10.1002/hep.27570. [DOI] [PubMed] [Google Scholar]
- Feld JJ, Jacobson IM, Hezode C, Asselah T, Ruane PJ, Gruener N, Abergel A, Mangia A, Lai CL, Chan HL, Mazzotta F, Moreno C, Yoshida E, Shafran SD, Towner WJ, Tran TT, McNally J, Osinusi A, Svarovskaia E, Zhu Y, Brainard DM, McHutchison JG, Agarwal K, Zeuzem S Investigators A. Sofosbuvir and Velpatasvir for HCV Genotype 1, 2, 4, 5, and 6 Infection. N Engl J Med. 2015;373(27):2599–2607. doi: 10.1056/NEJMoa1512610. [DOI] [PubMed] [Google Scholar]
- Foster GR, Afdhal N, Roberts SK, Brau N, Gane EJ, Pianko S, Lawitz E, Thompson A, Shiffman ML, Cooper C, Towner WJ, Conway B, Ruane P, Bourliere M, Asselah T, Berg T, Zeuzem S, Rosenberg W, Agarwal K, Stedman CA, Mo H, Dvory-Sobol H, Han L, Wang J, McNally J, Osinusi A, Brainard DM, McHutchison JG, Mazzotta F, Tran TT, Gordon SC, Patel K, Reau N, Mangia A, Sulkowski M Investigators A, Investigators A. Sofosbuvir and Velpatasvir for HCV Genotype 2 and 3 Infection. N Engl J Med. 2015;373(27):2608–2617. doi: 10.1056/NEJMoa1512612. [DOI] [PubMed] [Google Scholar]
- Foy E, Li K, Sumpter R, Jr, Loo YM, Johnson CL, Wang C, Fish PM, Yoneyama M, Fujita T, Lemon SM, Gale M., Jr Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc Natl Acad Sci U S A. 2005;102(8):2986–2991. doi: 10.1073/pnas.0408707102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS, Dudley JP. APOBECs and virus restriction. Virology. 2015;479–480:131–145. doi: 10.1016/j.virol.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horscroft N, Lai VC, Cheney W, Yao N, Wu JZ, Hong Z, Zhong W. Replicon cell culture system as a valuable tool in antiviral drug discovery against hepatitis C virus. Antivir Chem Chemother. 2005;16(1):1–12. doi: 10.1177/095632020501600101. [DOI] [PubMed] [Google Scholar]
- Iyer R, Coughlin J, Padmanabhan S, Korba B, Myong S. Activation of Retinoic Acid Inducible Gene (RIG-I) by Nucleotide Analogs: A Potential Novel Mechanism for Antiviral Discovery. Antiviral Research. 2010;86(1):A35. [Google Scholar]
- Iyer RP, Jin Y, Roland A, Morrey JD, Mounir S, Korba B. Phosphorothioate di- and trinucleotides as a novel class of anti-hepatitis B virus agents. Antimicrob Agents Chemother. 2004;48(6):2199–2205. doi: 10.1128/AAC.48.6.2199-2205.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer RP, Padmanabhan S, Zhang GR, Morrey JD, Korba BE. Nucleotide analogs as novel anti-hepatitis B virus agents. Curr Opin Pharmacol. 2005;5(5):520–528. doi: 10.1016/j.coph.2005.04.019. [DOI] [PubMed] [Google Scholar]
- Iyer RPSA, Pandey RK, Padmanabhan S, Korba BE, Bose S, et al. Activation of intracellular viral sensors by the anti-hepatitis Agent SB 9200–Implications for broad-spectrum antiviral activity. Abstracts of the 27th International Conference on Antiviral Research; Raleigh, NC. 2014a. [Google Scholar]
- Iyer RPSA, Pandey RK, Padmanabhan S, Korba BE, Bose S, et al. Activation of intracellular viral sensors by the anti-hepatitis Agent SB 9200–Implications for broad-spectrum antiviral activity. International Society for Antiviral Research; Washington, DC: 2014b. [Google Scholar]
- Jankowsky E, Gross CH, Shuman S, Pyle AM. Active disruption of an RNA-protein interaction by a DExH/D RNA helicase. Science. 2001;291(5501):121–125. doi: 10.1126/science.291.5501.121. [DOI] [PubMed] [Google Scholar]
- Korba BE. In vitro evaluation of combination therapies against hepatitis B virus replication. Antiviral Res. 1996;29(1):49–51. doi: 10.1016/0166-3542(95)00915-9. [DOI] [PubMed] [Google Scholar]
- Korolowicz KE, Iyer RP, Czerwinski S, Suresh M, Yang J, Padmanabhan S, Sheri A, Pandey RK, Skell J, Marquis JK, Kallakury BV, Tucker RD, Menne S. Antiviral Efficacy and Host Innate Immunity Associated with SB 9200 Treatment in the Woodchuck Model of Chronic Hepatitis B. PLoS One. 2016;11(8):e0161313. doi: 10.1371/journal.pone.0161313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawitz E, Poordad F, Hyland RH, Wang J, Liu L, Dvory-Sobol H, Brainard DM, McHutchison JG, Gutierrez JA. Ledipasvir/sofosbuvir-based treatment of patients with chronic genotype-1 HCV infection and cirrhosis: results from two Phase II studies. Antivir Ther. 2016 doi: 10.3851/IMP3062. [DOI] [PubMed] [Google Scholar]
- Marno KM, Ogunkolade BW, Pade C, Oliveira NM, O’Sullivan E, McKnight A. Novel restriction factor RNA-associated early-stage anti-viral factor (REAF) inhibits human and simian immunodeficiency viruses. Retrovirology. 2014;11:3. doi: 10.1186/1742-4690-11-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuse C, Rinaudo JA, Farrar K, Wells F, Korba BE. Enhancement of antiviral activity against hepatitis C virus in vitro by interferon combination therapy. Antiviral Res. 2005;65(1):23–34. doi: 10.1016/j.antiviral.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Paeshuyse J, Kaul A, De Clercq E, Rosenwirth B, Dumont JM, Scalfaro P, Bartenschlager R, Neyts J. The non-immunosuppressive cyclosporin DEBIO-025 is a potent inhibitor of hepatitis C virus replication in vitro. Hepatology. 2006;43(4):761–770. doi: 10.1002/hep.21102. [DOI] [PubMed] [Google Scholar]
- Sarrazin C, Dvory-Sobol H, Svarovskaia ES, Doehle BP, Pang PS, Chuang SM, Ma J, Ding X, Afdhal NH, Kowdley KV, Gane EJ, Lawitz E, Brainard DM, McHutchison JG, Miller MD, Mo H. Prevalence of Resistance-Associated Substitutions in HCV NS5A, NS5B, or NS3 and Outcomes of Treatment With Ledipasvir and Sofosbuvir. Gastroenterology. 2016;151(3):501–512. e501. doi: 10.1053/j.gastro.2016.06.002. [DOI] [PubMed] [Google Scholar]
- Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreerupa Challa AS, Padmanabhan Seetharamaiyer, Korba Brent E, Chang Te-Hung, Shil Niraj K, Bose Santanu, Iyer Radhakrishnan P. Prophylactic and therapeutic anti-RSV activity of SB 9200 – A Novel Agent that activates RIG-I and NOD2. 29th International Conference on Antiviral Research; la Jolla, CA. 2016. [Google Scholar]
- Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427(6977):848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- Sumpter R, Jr, Loo YM, Foy E, Li K, Yoneyama M, Fujita T, Lemon SM, Gale M., Jr Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol. 2005;79(5):2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, Bassendine M, Spengler U, Dore GJ, Powell E, Riordan S, Sheridan D, Smedile A, Fragomeli V, Muller T, Bahlo M, Stewart GJ, Booth DR, George J. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41(10):1100–1104. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, Nakagawa M, Korenaga M, Hino K, Hige S, Ito Y, Mita E, Tanaka E, Mochida S, Murawaki Y, Honda M, Sakai A, Hiasa Y, Nishiguchi S, Koike A, Sakaida I, Imamura M, Ito K, Yano K, Masaki N, Sugauchi F, Izumi N, Tokunaga K, Mizokami M. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41(10):1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- Thompson AJRS, Cheng W, Angus P, Visvanathan K, Iyer RP, Barclay M. SB 9200, a novel immunomodulator for patients with viral hepatitis: Phase I MAD study in patients with hepatitis C Virus (HCV) infection. The European Association for the Study of the Liver (EASL); Vienna: 2015. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.