Abstract
A correctly functioning spinal cord is crucial for locomotion and communication between body and brain but there are fundamental gaps in our knowledge of how spinal neuronal circuitry is established and functions. To understand the genetic program that regulates specification and functions of this circuitry, we need to connect neuronal molecular phenotypes with physiological analyses. Studies using Xenopus laevis tadpoles have increased our understanding of spinal cord neuronal physiology and function, particularly in locomotor circuitry. However, the X. laevis tetraploid genome and long generation time make it difficult to investigate how neurons are specified. The opacity of X. laevis embryos also makes it hard to connect functional classes of neurons and the genes that they express. We demonstrate here that Tol2 transgenic constructs using zebrafish enhancers that drive expression in specific zebrafish spinal neurons label equivalent neurons in X. laevis and that the incorporation of a Gal4:UAS amplification cassette enables cells to be observed in live X. laevis tadpoles. This technique should enable the molecular phenotypes, morphologies and physiologies of distinct X. laevis spinal neurons to be examined together in vivo. We have used an islet1 enhancer to label Rohon-Beard sensory neurons and evx enhancers to identify V0v neurons, for the first time, in X. laevis spinal cord. Our work demonstrates the homology of spinal cord circuitry in zebrafish and X. laevis, suggesting that future work could combine their relative strengths to elucidate a more complete picture of how vertebrate spinal cord neurons are specified, and function to generate behavior.
Keywords: V0v, V0, Interneuron, Evx, Xhox3, Xvglut1, Glutamatergic, RB, Islet1, elavl3, slc17a7
Introduction
The spinal cord is a crucial part of the central nervous system, responsible for controlling movements as well as receiving and processing sensory information from the trunk and the limbs. Despite its relative simplicity when compared to the brain, there are still fundamental gaps in our knowledge of how spinal cord neuronal circuitry is established and functions. Traditionally, there have been two main approaches that have addressed different aspects of this question. Developmental studies have investigated neuronal specification (how cells are instructed to differentiate into neurons of particular types), while physiological studies have concentrated on identifying different functional types of neurons and determining their roles in particular behaviors. These different approaches usually identify specific populations of neurons using different criteria: gene expression in developmental studies and morphology and electrophysiological characteristics in physiological studies. They have also often exploited the strengths of different model systems. For example, Xenopus laevis has been an invaluable model for elucidating the components and functions of spinal cord circuitry, whereas zebrafish and mouse have contributed more to our understanding of how different spinal cord neurons are specified.
Spinal cord circuitry controlling vertebrate locomotor behavior is probably best understood in the X. laevis tadpole. X. laevis tadpoles are relatively easy to manipulate and have robust tissues and cells, which makes them ideal for electrophysiology. The morphologies, physiological properties, synaptic connections and activities of most classes of spinal interneuron during the two main X. laevis tadpole locomotion behaviors, swimming and struggling, have been established (Li et al., 2001; Li et al., 2003; Li et al., 2004a; Li et al., 2004c; Li et al., 2004d; Li et al., 2004b; Li et al., 2006; Li et al., 2007a; Li et al., 2007b; Sautois et al., 2007; Roberts et al., 2008; Li et al., 2009; Soffe et al., 2009; Roberts et al., 2010; Roberts et al., 2012). However, the tetraploid genome and long generation time of X. laevis makes it hard to make mutants and investigate how different populations of neurons are genetically specified using this animal. Furthermore, the opacity of X. laevis tadpoles, due to the yolk in each cell, makes it difficult to analyze double labeling experiments and determine which genes are expressed by functionally-defined spinal neurons. Consequently, to date, only one molecularly-identified spinal cord population, engrailed-expressing V1 cells, has been correlated with a physiologically/morphologically identified population of cells, aIN neurons, in X. laevis (Li et al., 2004a).
In contrast, zebrafish are a powerful model system for investigating how spinal cord neurons are specified because loss-of-function and gain-of-function experiments are more easy to perform using mutant lines and antisense reagents (e.g. Lewis and Eisen, 2001; Varga et al., 2001; Lewis and Eisen, 2003; Lewis and Eisen, 2004; Lewis et al., 2005; Gribble et al., 2007; Batista et al., 2008; Batista and Lewis, 2008; Bonner et al., 2008; Gribble et al., 2009; Yang et al., 2010; England et al., 2011; Hilinski et al., 2016; Juárez-Morales et al., 2016). Zebrafish embryos are also transparent making it relatively easy to identify genomic enhancers that drive expression in particular populations of zebrafish neurons (e.g. Higashijima et al., 2000; Bohm et al., 2016; Juárez-Morales et al., 2016) and use these to correlate gene expression with neuronal morphology (e.g. Kimura et al., 2006; Batista et al., 2008; Satou et al., 2012; Satou et al., 2013; Juárez-Morales et al., 2016). However, while it is possible to make electrophysiological recordings in zebrafish embryos (e.g. Higashijima et al., 2004; Kimura et al., 2006; Satou et al., 2009; Bohm et al., 2016) their small size and fragility makes these techniques very technically challenging, compared to X. laevis.
To really understand the genetic programs that regulate the specification and functions of neuronal circuitry, we need to be able to combine these different approaches so that the molecular phenotypes, morphologies and physiologies of spinal cord neurons can be examined together in vivo. Therefore, we decided to test if we could use genomic enhancers identified and validated in zebrafish spinal cord to drive expression of fluorescent proteins in equivalent X. laevis spinal cord neurons. We used the Tol2 method of transgenesis as this is widely used in zebrafish (Kawakami, 2004; Kwan et al., 2007; Villefranc et al., 2007; Asakawa et al., 2008), therefore maximizing the chances that, in the future, additional zebrafish constructs will be available for X. laevis researchers to use. We also exploited the gateway cloning system so that future constructs could be easily assembled using the same middle entry and destination plasmids (Kwan et al., 2007; Villefranc et al., 2007) (for a similar system in frogs see (Love et al., 2011)).
While several different methods of transgenesis have been used to label specific cells in X. laevis (e.g. Ogino et al., 2006; Amaya and Kroll, 2010; Haeri and Knox, 2012; Ishibashi et al., 2012; Zuber et al., 2012; Takagi et al., 2013; Tam et al., 2013; Wang and Szaro, 2015), Tol2 has only been used to test potential skeletal muscle enhancers (Loots et al., 2013). Therefore, before our study, it was unclear whether this method of transgenesis would be generally succesful in this animal. There have also been no reports where specific populations of transgenically-labeled spinal cord neurons have been observed in live tadpoles, which is what would be necessary to target specific molecularly-identified cells for electrophysiological analyses. Previous reports have usually used transgenic constructs to label and observe more superficially located cells in the skin, heart, eye, muscle or tail, where the opacity of X. laevis embryos presents less of a problem. (e.g. Moritz et al., 1999; Jansen et al., 2002; Lim et al., 2004; Smith et al., 2005; Scheenen et al., 2009; Yokoyama et al., 2011; Vivien et al., 2012; Haeri et al., 2013; Loots et al., 2013; Tam et al., 2013; Zhuo et al., 2013) (although also see (Love et al., 2011) and (L’Hostis-Guidet et al., 2009) as these authors used a few widely expressed neuronal promoters to test different types of transgenesis).
We tested different enhancers and promoters that label specific spinal cord neurons in zebrafish. The elavl3 (formerly HuC) enhancer drives expression in most post-mitotic spinal neurons (Park et al., 2000a; Park et al., 2000b; Sato et al., 2006), the islet1 enhancer drives expression primarily in skin sensory Rohon Beard cells (RBs) (Higashijima et al., 2000; Reyes et al., 2004) and the evx1 enhancer drives expression in a population of interneurons that form in the most dorsal part of the ventral spinal cord called V0v cells (e.g. Briscoe et al., 2000; Moran-Rivard et al., 2001; Pierani et al., 2001; Lanuza et al., 2004; Griener et al., 2015; Juárez-Morales et al., 2016) (also called V0e cells in zebrafish – see (Satou et al., 2012)) that develop into glutamatergic commissural interneurons in the zebrafish spinal cord (Juárez-Morales et al., 2016). We also identified a new evx2 enhancer region that drives expression in a similar manner to evx1. We found that while we could successfully label the expected spinal cord cells as assayed by immunohistochemistry, it was very hard to detect fluorescent spinal cord neurons in live tadpoles due to the location of the spinal cord deep inside the tadpole and the opacity of the tadpoles at these stages of development. However, when we incorporated a Gal4:UAS amplification cassette into our constructs we were able to observe EGFP in live spinal cords.
Interestingly, despite the extensive characterization of X. laevis spinal cord circuitry, and the similarity between zebrafish and X. laevis spinal cord neurons, V0v neurons have not yet been identified in X. laevis. Therefore, to confirm that the cells labeled by our evx transgenic constructs were indeed V0v cells we tested whether they also expressed X. laevis evx1 RNA and a marker of glutamatergic cells. We found that in all of the cases we examined, they did. This is important as it establishes that V0v neurons are present in the X. laevis spinal cord and further confirms the homology of spinal cord neurons and neuronal circuitry in fish, frogs and amniotes.
Methods
Xenopus laevis husbandry
X. laevis embryos were obtained following standard protocols from the Roberts lab colony at Bristol University, the Harris lab colony at the University of Cambridge and the Zuber lab colony at SUNY Upstate Medical University. Embryos were staged according to Nieuwkoop and Faber (Nieuwkoop and Faber, 1994). All experiments were approved by either the Syracuse University IACUC committee or the UK Home Office.
Construction of Transgenic Constructs
The elavl3 promoter was previously described (Park et al., 2000a; Park et al., 2000b). We PCR-amplified a 3.1Kb amplicon encompassing 2771bp upstream and 382bp downstream of the elavl3 coding sequence from the HuC cameleon 2.1 SV40poly(A) plasmid (Higashijima et al., 2003) with the following primers: Forward primer: ATTCACTAATTTGAATTTAA, Reverse primer: TCTTGACGTACAAAGATGAT. This PCR product was cloned into the pDONR™ P4-P1R vector from Invitrogen using Gateway technology (Sasaki et al., 2004; Suzuki et al., 2005). Two reporter constructs were generated using the resulting 3.1Kb elavl3 5’pDONR vector. One was assembled using the pME-EGFP plasmid and the pCSDest2 vector (Kwan et al., 2007; Villefranc et al., 2007) to generate the Tg(Tol2:3.1Kb5’zfish elavl3:EGFP:pA:Tol2) construct. The other was made using the Gal4VP16;UAS:EGFP middle entry construct (Koster and Fraser, 2001; Juárez-Morales et al., 2016) and the pCSDest2 vector to generate the Tg(Tol2:3.1Kb5’zfishelavl3:Gal4VP16;UAS:EGFP;pA:Tol2) construct.
The islet1 enhancer was also previously described (Uemura et al., 2005). We PCR-amplified this enhancer from zebrafish genomic DNA using the following primers: Forward primer: TGCAGCTTTAGACATTTAAA; Reverse primer: TCCAGCACCATAATTCACCA. The 750bp PCR product was cloned into the pDONR™ P4-P1R vector from Invitrogen using Gateway technology (Sasaki et al., 2004; Suzuki et al., 2005). Two reporter constructs were generated by assembling the 750bp islet1 5′ pDONR with either the pENTRbasegfp plasmid (which contains the βcarp minimal promoter) or the cfos minimal promoter:Gal4VP16;UAS:EGFP middle entry construct (Koster and Fraser, 2001; Juárez-Morales et al., 2016) and the pCSDest2 vector to generate either the Tg(Tol2:750bp3’zfish_islet1:βcarp minimal promoter:EGFP:pA:Tol2) or the Tg(Tol2:750bp3’zfish_isletl1:cfos minimal promoter:Gal4VP16;UAS:EGFP:pA:Tol2) construct respectively.
We identified the evx2 enhancer sequence through multispecies sequence comparisons using the global alignment program Shuffle-LAGAN (Brudno et al., 2003) and VISTA (Mayor et al., 2000) as described for evx1 in (Juárez-Morales et al., 2016) (Fig 1A). We identified three Conserved Non-coding Elements (CNEs) in the vicinity of evx2. The first is located 419bp upstream of zebrafish evx2 and extends for 97 bp. The other two CNEs are located downstream of evx2, one is 2052bp downstream of the stop codon and is 182bp long whereas the other is 2289bp downstream of the stop codon and extends for 700bp (Fig. 1A). We PCR-amplified one region encompassing the two 3′ CNEs and 3′ UTR. The forward primer was designed just after the stop codon. We used the BAC RP71-78H1 (BACPAC Resources Center) as a template with forward primer: GCGAGATGTAACGATGCTAT and reverse primer: CAAATGTGT TGAGGTGAGCA. The resulting amplicon was cloned into the pDONR™ P4-P1R vector from Invitrogen using Gateway technology (Sasaki et al., 2004; Suzuki et al., 2005). The final reporter construct was assembled using the pENTRbasegfp plasmid (which contains the βcarp minimal promoter) and the pCSDest2 vector (Villefranc., 2007). This produced the Tg(Tol2:3.9Kb3’zfish_evx2:βcarp_minimal promoter:EGFP:pA:Tol2) construct.
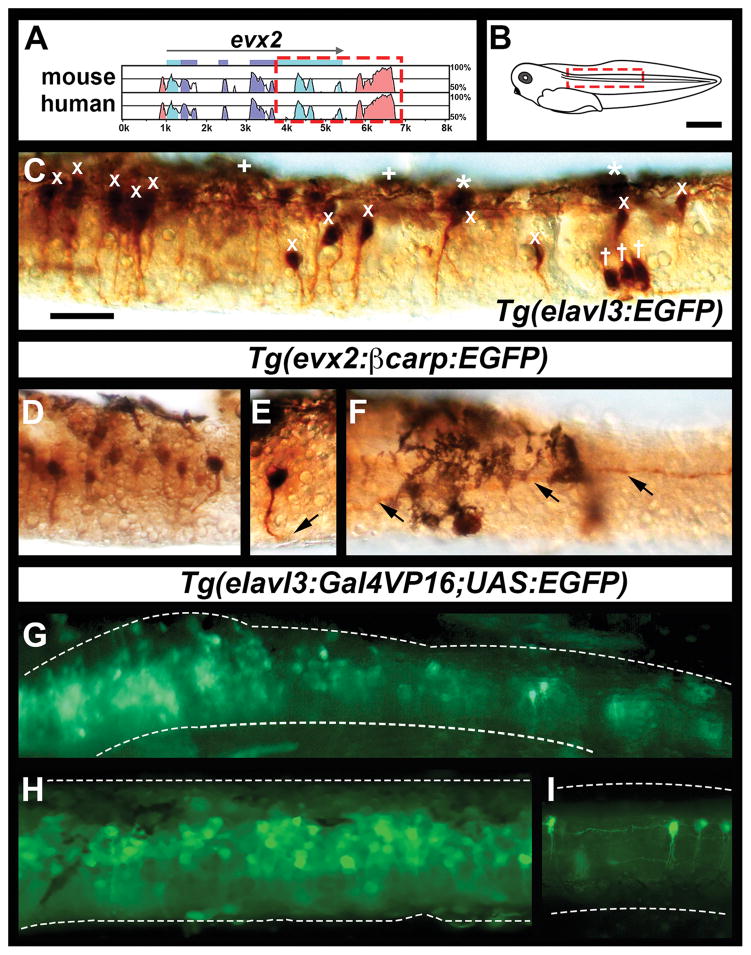
Figure 1. Zebrafish enhancers label appropriate neurons in Xenopus laevis spinal cord.
(A) Schematic of shuffle-LAGAN analysis of evx2 genomic region, with zebrafish evx2 used as the baseline and compared to orthologous genomic regions in mouse and human. Conserved coding sequences are indicated in purple and conserved UTR regions are indicated in light blue. Percentage of sequence conservation is indicated by peak heights (scale is provided on RHS), grey arrow indicates 5′-3′ gene orientation, CNEs are indicated in pink. Red dotted box indicates region amplified to create evx2 enhancer transgenic constructs. (B) Schematic of stage 41 X. laevis tadpole. Red box indicates the approximate spinal cord region shown in subsequent lateral views. (D–F and I) show only a small part of this region. (C – I) Lateral views of one side of stage 41 X. laevis spinal cord, dorsal top, rostral left. (C–F, H and I) show dissected spinal cords. The tissue shown is the full dorsal-ventral extent of the spinal cord and no other tissue is included except for a few pigment cells. (C) DAB immunohistochemistry (dark brown staining) for EGFP in transient transgenic Tg(elavl3:EGFP) spinal cord showing several different labeled post-mitotic neurons. For example, we have indicated a couple of RB neurons (*), a group of three motoneurons in the ventral spinal cord on the RHS of the panel (†) and some commissural cells (x). The dorsal black cells are pigment cells (+). (D–F) DAB immunohistochemistry for EGFP in transient transgenic Tg(evx2:βcarp:EGFP) spinal cords. (D) shows a region of rostral spinal cord with several labeled cells. The cells have pear shaped somata approximately 4.7 μm wide along the rostral-caudal axis and 6 μm tall in the dorsal-ventral axis, they are located in the dorsal 48–68% of the spinal cord, and they all have axons that project to the ventral spinal cord and then cross the midline to become commissural. (E & F) show two different focal planes of the same spinal cord in a region with just one labeled cell, black arrows indicate axon trajectory, black cells in F and dorsally in E are pigment cells. The cell soma is visible in (E) and its axon is visible on the contralateral side of the spinal cord in (F). (E) is slightly more rostral than (F). (G–I) Live expression of EGFP in transient transgenic Tg(elavl3:Gal4VP16;UAS:EGFP) spinal cords. (G) shows expression on one side of the mid-trunk spinal cord of an intact tadpole. The white dotted lines show the dorsal and ventral limits of the spinal cord. The expression appears weaker/more diffuse because we are looking through the skin and muscle overlying the spinal cord. (H & I) show expression in dissected live spinal cords where these other tissues have been removed. (H) shows an example where many cells are labeled. (I) shows an example of more sparse labeling where individual cells and their axons can be observed and identified. Scale bar in C = 20 μm (panels C–F) and 30 μm (panels G–I). Scale bar in B = 1mm.
The Tg(Tol2:1.3Kb 3’zfish evx1: βcarp minimal promoter:EGFP:Tol2) and the Tg(Tol2:1.3Kb 3’zfish evx1:cfos minimal promoter:Gal4VP16;UAS-EGFP:pA:Tol2) constructs were previously described (Juárez-Morales et al., 2016). We have previously demonstrated that these constructs specifically drive expression in evx1-expressing spinal cord cells in both transient transgenics and two stable transgenic lines in zebrafish (Juárez-Morales et al., 2016).
DNA and transposase mRNA preparation and microinjection
Plasmid DNA was prepared using QIAfilter plasmid purification kit (Qiagen, 12743) and transposase mRNA was prepared using the pCS2FA transposase plasmid (Kwan et al., 2007). After the transposase plamid was linearized and purified using the QIAquick PCR Purification Kit (Qiagen, 28104), mRNA in vitro transcription was conducted using the Ambion mMessage mMachine SP6 kit (Ambion, AM1340) and manufacturer’s protocols. Microinjection of DNA plus RNA was carried out using an air-pressure picospritzer II (General Valve Corporation). Glass microneedles were pulled in a P-2000 micropipette puller (Sutter Instruments Co.). Approximately 10 nl of a combination of Plasmid DNA [33-20 ng/μL] and transposase mRNA [30ng/μL] was injected into both blastomeres of 2-cell stage de-jellied X. laevis embryos.
Analysis of gene expression and transgenic labeling
X. laevis embryos were incubated at 180C until they reached stage 41. Just before fixation, tadpoles were analyzed for EGFP expression by examining the hindbrain to rostral spinal cord region on an Olympus SZX16 stereomicroscope. By stage 41 most of the yolk has been consumed making these internal structures more visible. In our experience, if we observe neurons expressing EGFP in these structures there is a high chance that there will also be EGFP-expressing neurons more caudally in the spinal cord.
For in situ hybridization and immunohistochemistry, embryos were fixed in 4% PFA for 1 hr at room temperature. Embryos were then washed in PBS + 0.01% Tween-20 three times for 5 minutes each, followed by 50% MeOH in PBS + 0.01% Tween-20 and then stored in 100% MeOH at −20°C. Whole mount in situ hybridizations were performed as previously described (Zuber et al., 2003; Viczian et al., 2006) with the following modifications: tadpoles were bleached for 11 mins with 0.5% SSC, 10% H2O2 and 5% formamide and proteinase K treatment [10μg/mL] was performed for 5 minutes at room temperature. RNA probes were detected with Anti-Digoxigenin-AP, Fab fragments (Sigma Aldrich 11093274910) and BM purple AP (Roche 11442074001).
RNA in situ hybridization probes were prepared using the following templates: evx1, (previously called Xhox3) was kindly provided by Jonathan Slack (Beck and Slack, 1998) and slc17a7 (previously called XVGlut1) was kindly provided by Margaret Saha (Gleason et al., 2003).
Primary antibodies used were rabbit anti-GFP (Invitrogen A6465, 1/500) or chicken anti-GFP (Abcam ab13970, 1/500) and secondary/tertiary antibodies were Alexa Fluor 488 goat anti-rabbit (Invitrogen A11034, 1/500) or Alexa Fluor 488 goat anti-chicken (Invitrogen A11039, 1/500) for fluorescent staining or goat anti-rabbit IgG (Covance SMI-5030C, 1/200) and rabbit PAP (Covance SMI-4010 L, 1/200) for DAB staining.
Tadpoles used for fluorescent immunohistochemistry were treated with Image-iT Signal Enhancer (Invitrogen, I36933) for 30 minutes, then incubated in block solution (2% goat serum, 1% BSA, 10% DMSO and 0.5% Triton) for 1 hour at room temperature followed by incubation in primary antibody in fresh block solution at 4°C overnight. Tadpoles were washed with PBT for 2 hours at room temperature and incubated with secondary antibody in block solution at 4°C overnight. Tadpoles were then washed with PBT for at least 2 hours at room temperature and stored in 2% DABCO (Acros Organics, AC11247-1000).
Tadpoles used for DAB immunohistochemistry were incubated at 4°C overnight with primary antibody and then incubated in fresh blocking solution with goat anti-rabbit IgG (Covance SMI-5030C, 1:200) at 4°C overnight. Embryos were then washed with PBT for 2 hours and incubated with rabbit PAP (Covance SMI-4010 L, 1:200) in block solution at 4°C overnight. Embryos were then washed in PBT for 2 hours. DAB staining was developed using SigmaFast™ 3,3′-diamino- benzidine tablets (Sigma, D4293-5set).
For in situ hybridization and immunohistochemistry on tissue sections, stage 42 tadpoles were fixed in 4% paraformaldehyde (PFA) for 1 hour at room temperature and cryoprotected in 30% sucrose in PBS at 4°C overnight. Tadpoles were embedded in Tissue Tek OCT (VWR, 25608-930) and cryostat sectioned at 12 μm. in situ hybridization was performed as previously described (Viczian et al., 2006; Martinez-De Luna et al., 2013) except that Proteinase K was used at a concentration of 10μg/ml and tissue sections were incubated in this solution for 1 min at room temperature. in situ hybridization was performed first and followed by immunohistochemistry as described above with the following modifications; tissue sections were fixed in 4% PFA for 30 min at room temperature, washed 3X 5 mins in PDT (1XPBS, 1% DMSO, 0.1% Triton X-100) then incubated in block solution (1XPBS, 1% DMSO, 0.1% Triton X-100, 2% goat serum, 1% BSA) for 1hour at room temperature, followed by incubation in primary antibody in fresh block solution at 4°C overnight. Sections were washed with PDT 3X 5 mins and then 4X 20 mins at room temperature and incubated with secondary antibody in block solution for 2 hours. After incubation, sections were washed with PDT 3X 5 mins and then 4X 20 mins at room temperature and stored in 2% DABCO (Acros Organics, AC11247-1000).
Image acquisition and processing
Whole-mount tadpoles were placed in a 1% agarose plate and covered in PBS for imaging using a Olympus SZX16 stereomicroscope and a Q-Imaging Micropublisher 5.0 RTV camera. Cross sections were mounted in 2% DABCO on microscope slides and brightfield pictures were taken using an AxioCam MRc5 camera mounted on a Zeiss Axio Imager M1 compound microscope. Fluorescent images were taken on a Zeiss LSM 710 confocal microscope. Images were processed using Adobe Photoshop software (Adobe, Inc) and Image J software (Abramoff et al., 2004).
Spinal cord dissections
After inmunohistochemistry, X. laevis tadpoles were placed in a dish which had a rotatable shaft with a sylgard platform on one side. The tadpoles were pinned down on this platform using two tungsten pins, one through the eye and the second through the muscle and notochord two thirds of the way down the body. Using a pair of dissecting pins, skin and muscle were removed from the tadpole to expose the spinal cord.
Results
To test whether we could use zebrafish enhancers and Tol2 transgenesis methods to label X. laevis spinal neurons we constructed three different Tol2 EGFP constructs with enhancers or promoters that had already been validated in zebrafish either by our lab or other groups and a fourth construct using a newly identified enhancer for evx2. In cases where we used enhancers, we combined these with either a cfos or βcarp basal promoter (Wang et al., 2000; Villefranc et al., 2007). In zebrafish, the elavl3 (previously called HuC) promoter drives expression in most post-mitotic neurons (Park et al., 2000a; Park et al., 2000b; Sato et al., 2006), the islet1 enhancer drives expression primarily in RB cells (Higashijima et al., 2000) and the evx1 enhancer drives expression in V0v neurons (Juárez-Morales et al., 2016). Given that evx1 and evx2 are expressed in the exact same cells in the zebrafish spinal cord we also investigated whether a similar enhancer to our previously identified evx1 enhancer (Juárez-Morales et al., 2016) existed for evx2. Our bioinformatic analyses (see methods) identified a similar region of high conservation downstream of evx2 (Fig. 1A and compare to Fig. 1D in (Juárez-Morales et al., 2016)) and, therefore, we also cloned and tested this enhancer. We injected these constructs into 1–2 cell stage X. laevis embryos. In most cases, we could not detect any obvious EGFP expression in the spinal cord of live tadpoles, but when we fixed the tadpoles and performed anti-GFP immunohistochemistry, detecting EGFP expression with a coloured substrate (3,3′-Diaminobenzidine; DAB), we could clearly observe expression in dissected spinal cords, in the types of spinal cord neurons that we would expect, based on the behavior of these enhancers/promoter in zebrafish (Fig. 1C–F and data not shown, for more detailed discussion of cell types observed see below). These immunohistochemistry results showed that the transgenesis method was working, suggesting that the EGFP was not bright enough to be detected in live spinal cords, probably due to the opacity of the X. laevis tadpoles at these stages of development.
To use transgenic constructs to label cells for electrophysiological experiments, we would need to visualize and identify individual cells in live tadpoles. To try and overcome the fact that we could not detect EGFP-labeled neurons in live X. laevis tadpoles using these initial constructs, we incorporated a Gal4:UAS amplification cassette (Koster and Fraser, 2001) into our Tol2 constructs. Using these new constructs we could observe EGFP-labeled neurons in live whole tadpoles (Fig. 1G). These labeled cells became even clearer after overlying skin and muscle was dissected away to view the spinal cords (Fig. 1H–I) and they had the same specificity as the cells labeled with the non-amplified constructs (e.g. Fig 1C and 1H). In each case, as expected for transient transgenesis assays, there was variability in the number of cells labeled in each tadpole, with some animals containing many labeled cells (e.g. Fig. 1C, D, G & H) and others fewer labeled cells (e.g. Fig. 1E & F).
As in fish larvae, the Tg(elavl3:EGFP) constructs labeled a wide range of different neurons in the X. laevis spinal cord (Fig. 1C, G & H). While in some tadpoles so many cells were labeled that it was hard to distinguish individual cell morphologies (e.g. Fig. 1G & H), in other cases there were stretches of spinal cord where individual cells could be easily identified (Fig. 1C–F & I).
Consistent with its activity in zebrafish, the islet1 enhancer drove expression mainly in RBs in the X. laevis spinal cord. We analyzed 7 tadpoles injected with Tg(islet1:cfos:Gal4VP16;UAS:EGFP) and identified 97 labeled cells. 72 of these (74%) were clearly RBs (Fig. 2A & B). Their cell bodies were located at the dorsal surface of the spinal cord and their axons extended in the dorsal tract. In more densely labeled animals, we observed labeled RBs along the whole spinal cord and into the caudal hindbrain. Interestingly, most of the non-RB cells labeled in these tadpoles had very similar morphologies to each other with oval somata and ventrally-directed axons (Fig 2C).
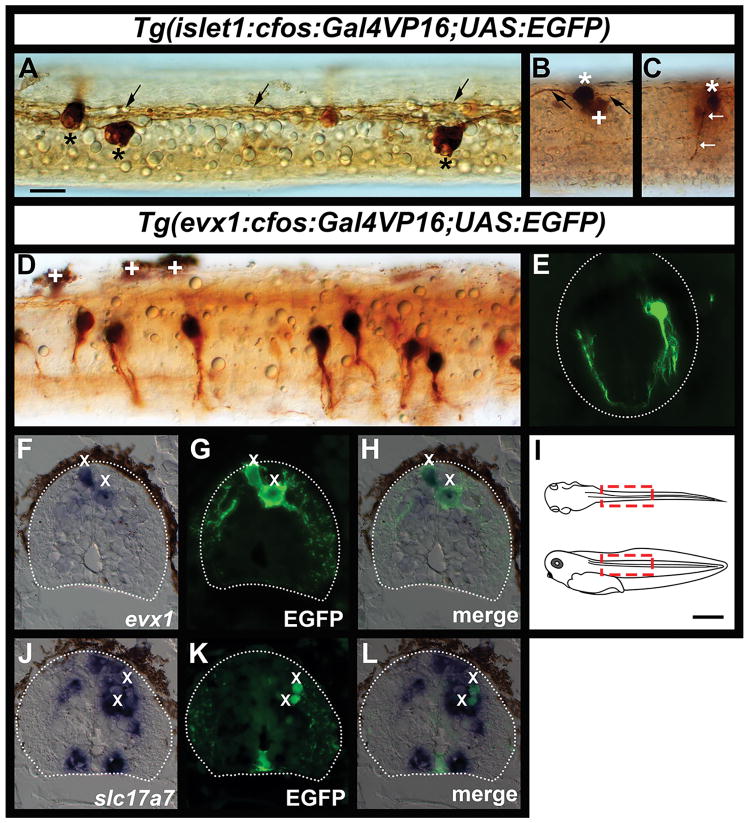
Figure 2. Zebrafish islet1 enhancer predominantly labels Rohon Beard neurons and evx1 enhancer labels V0v neurons in stage 41 Xenopus laevis spinal cord.
(A, B & C) DAB immunohistochemistry for EGFP in transient Tg(islet1:cfos:Gal4VP16;UAS:EGFP) transgenic spinal cord. (A) Dorsal view of spinal cord, rostral left showing labeled RB neuron soma (*) and longitudinal axons in the dorsal tract (black arrows). The smaller, more weakly labeled cells are probably not RB neurons. (B & C) lateral views, rostral left, dorsal top. (B) RB neuron with characteristic large round soma in dorsal spinal cord (*) and ascending and descending longitudinal axons (black arrows), + indicates a pigment cell located ventral and lateral to the RB neuron. (C) shows a slightly more ventrally located neuron in the same embryo as (B) with an oval soma and ventral axon (arrows). (D) Lateral view with rostral left and dorsal top showing DAB immunohistochemistry for EGFP in transient Tg(evx1:cfos:Gal4VP16;UAS;EGFP) transgenic spinal cord. Labeled cells have pear-shaped soma, axons that extend ventrally and then become commissural and are located in the dorsal 48–68% of the spinal cord. White crosses indicate dorsal pigment cells. (E) Fluorescent immunohistochemistry for EGFP in transient Tg(evx1:cfos:Gal4VP16;UAS:EGFP) spinal cord; cross section (dorsal up) showing a single labeled neuron soma and ventral commissural axon. White dotted lines show edge of spinal cord. (F–H & J–L) Double labels (in situ hybridization with BM purple plus fluorescent immunohistochemistry for EGFP) of Tg(evx1:cfos:Gal4VP16;UAS:EGFP) spinal cord cross sections. Spinal cord margins are delinated with dotted lines. (F) X. laevis evx1 RNA expression (purple). (G) EGFP expression (green) in same cross section as (F). (H) merged image of F and G. Two EGFP-labeled cells (X) co-express evx1 RNA. (J) X. laevis slc17a7 RNA expression (purple). (K) EGFP expression (green) in same cross section as (J). (L) merged image of J and K. Two EGFP-labeled cells (X) co-express slc17a7 RNA. (I) schematics showing approximate region of spinal cord shown in dorsal view in (A) and lateral views in (B–D). (B&C) show only part of this region. (A–L) are all stage 41. Scale bar in A = 20 μm (panels A–H & J–L) and scale bar in I = 1mm.
In addition, both the evx1 and evx2 enhancers drove expression mainly in X. laevis spinal cord neurons with pear shaped somata, located in a mid to dorsal dorsal-ventral position in the spinal cord. Several of these neurons had somata in a dorso-lateral position next to the dorsal tract of RB axons (e.g. Fig. 2D left 4 neurons). The labeled cells also had ventrally-directed axons that continued to the ventral commissure and then turned to ascend on the other side of the spinal cord where they also sometimes formed a caudally directed branch. For example, we analyzed 6 tadpoles injected with Tg(evx1:cfos:Gal4VP16;UAS:EGFP) and identified 291 labeled cells. Of these, 232 (~80%) had this morphology and dorso-ventral location (Fig 1D–F and Fig. 2D–E). The labeled neurons with different morphologies included 8 RBs in the dorsal spinal cord, 26 cells in the ventral spinal cord, 5 cells with ipsilateral axons in the mid-region of the spinal cord and 20 cells with a horizontal pear-shaped cell body that were also located in the mid-region of the spinal cord.
The dorso-ventral locations and commisural axon trajectories of the major class of labeled cells was highly reminiscent of zebrafish V0v neurons. However, while RB cells can be unambiguously identified by their morphology, this is not the case for V0v cells as they are not the only commissural neurons in the spinal cord and, their morphologies can differ (Satou et al., 2012). The characteristic that unambiguously identifies V0v cells in zebrafish and amniotes is their expression of evx1 and evx2 (they are the only cells in the spinal cord that express these genes). In addition, in all animals examined so far, V0v cells express glutamatergic markers (Moran-Rivard et al., 2001; Satou et al., 2012; Juárez-Morales et al., 2016). Therefore, to test whether cells labeled by our evx1 enhancer might be V0v cells, we performed double labeling experiments for EGFP and either X. laevis evx1 (previously called Xhox3) or slc17a7 (previously called vglut1 and Xvglut1), which labels glutamatergic cells. In the vast majority of the cases that we analyzed, spinal cord cells that expressed EGFP in Tg(evx1:cfos:Gal4VP16;UAS:EGFP) injected tadpoles also expressed these RNAs. 21/23 cells that expressed EGFP co-expressed evx1 RNA and in separate experiments 32/35 cells that expressed EGFP co-expressed the glutamatergic marker slc17a7 (Fig 2F–H, J–L). In both cases, rare EGFP single-positive cells were mainly located very ventrally in the spinal cord, suggesting that these are likely to be cells that were ectopically expressing EGFP (3 cells were ventral and 1 was very dorsal).
Discussion
All of the evidence so far, suggests that mechanisms of spinal cord patterning and resulting neuronal circuitry are highly conserved in vertebrates (e.g. Goulding and Pfaff, 2005; Lewis, 2006). In particular, comparisons between zebrafish, X. laevis and mouse suggest that the bony fish and tetrapod ancestor had a basic plan of spinal cord circuitry, where distinct classes of neurons with particular functions, were specified in the embryo by different transcription factors (e.g. Roberts, 2000; Higashijima et al., 2004; Li et al., 2004a; Goulding and Pfaff, 2005; Lewis, 2006). Data suggest that both the properties of these neurons and their method of specification have been conserved throughout the evolution of bony fish and tetrapods; with the proviso that in mammals and potentially other amniotes, some classes of neurons may have diversified during development, into specialized sub-classes of related neurons (e.g. Lewis and Eisen, 2001; Higashijima et al., 2004; Sapir et al., 2004; Alvarez et al., 2005; Goulding and Pfaff, 2005; Gosgnach et al., 2006; Kimura et al., 2006; Griener et al., 2015). Taken together, these observations argue strongly that we can use the simpler spinal cords of anamniote embryos such as zebrafish and X. laevis to determine fundamental aspects of vertebrate spinal cord neuronal specification and function and that findings from one vertebrate will usually apply more widely to the whole vertebrate family.
In this paper, we further demonstrate the homology of vertebrate spinal cord circuitry by showing that transgenic constructs using four different zebrafish genomic enhancers or promoters that label particular spinal cord neurons in zebrafish, are expressed by equivalent neurons in X. laevis tadpoles (see discussion below). Combined with data from previous reports that used mammalian (e.g. Beck and Slack, 1999; Lim et al., 2004; Suzuki et al., 2007; Yokoyama et al., 2011; Loots et al., 2013) or zebrafish (Concha et al., 2003; Love et al., 2011) enhancers to label specific cell types in frogs, this suggests that enhancers and promoters from other vertebrates can be used successfully for X. laevis transgenesis.
In addition, we identify a neuronal class, V0v interneurons, that has not been previously identified in X. laevis, but is present in both amniotes and zebrafish (Moran-Rivard et al., 2001; Suster et al., 2009; Satou et al., 2012; Juárez-Morales et al., 2016). As discussed in more detail below, we show that these cells have similar properties in frog to V0v cells in other vertebrates. As well as adding to our understanding of frog spinal circuitry, this again confirms that spinal cord neurons are highly conserved across vertebrates. This conservation is particularly strong for X. laevis and zebrafish. Both of these animals develop fast from eggs into free moving larvae in a similar way and their hatchling larvae have a relatively small number of different classes of spinal cord neurons and similar locomotor behaviours. Comparisons of neuronal morphology and function suggest that spinal cord circuitry in these vertebrates is also very similar, reflecting the basic common vertebrate plan for early spinal cord organisation (Roberts, 2000; Goulding and Pfaff, 2005). This suggests the intriguing possibility that X. laevis and zebrafish larvae could be used in a complementary way to study spinal cord neuronal circuits, enabling a more powerful and complete analysis than would be possible with either alone, using X. laevis to characterize the physiology of particular neurons and zebrafish to decipher how those same neurons are specified.
The transgenesis method that we test in this study enables us to identify EGFP-labeled spinal neurons in live X. laevis tadpoles. We used Gal4 and UAS components to amplify the expression of EGFP so that it can be observed even in the spinal cord of live tadpoles. We have also demonstrated that these constructs work well in injected F0 animals/transient transgenics. This is important as making stable lines in X. laevis is very time-consuming and laborious due to the long generation time of this animal. These new genetic tools should enable researchers to label specific neurons for electrophysiological studies and hence facilitate studies correlating the molecular, morphological and physiological properties of cells. In fact, mosaically-labeled transient transgenic larvae are arguably more useful for these sorts of studies than stable transgenic animals as labeling just a small subset of a particular population of neurons makes it easier to identify and characterize individual cells. These and other zebrafish enhancers could also potentially be used for optogenetics and/or to drive expression of constructs that alter neuronal behavior by silencing, activating or ablating specific neurons to study the links between neuronal circuitry and behaviour, although in some of these cases, stable transgenics may be needed for robust conclusions to be drawn.
We observed a reasonably high level of selectivity for both the islet1 and the evx1 enhancers in our transient transgenesis assays in X. laevis. For example, over 74% of cells labeled with the islet1 enhancer could be unambiguously identified as RB cells, by their unique cell morphology and dorsal spinal cord location. Interestingly, most of the remaining cells had an oval soma and ventrally-directed axon. While in zebrafish, the islet1 enhancer has only been reported to label RB cells (Higashijima et al., 2000; Reyes et al., 2004), islet1 is also expressed by motoneurons and interneurons in zebrafish (Inoue et al., 1994; Appel et al., 1995; Tokumoto et al., 1995; Lewis and Eisen, 2001; Tamme et al., 2002; Lewis and Eisen, 2004). The non-RB labeled cells did not have the morphology or ventral spinal cord position of motoneurons. However, it is possible that these cells are an islet1-expressing population of interneurons. Interneuron expression could have been missed in the previous zebrafish analyses particularly if the interneuron somas were located close to RBs. Even if these non-RB cells in X. laevis are examples of ectopic expression, which is very common with transient transgenesis/injected F0 animals, the frequency of correct labeling is high enough for this construct to be a very useful tool for identifying RB cells for physiological analyses in live tadpoles. As RBs have a unique characteristic morphology, labeled RBs can easily be distinguished from non-RB cells. Therefore, this construct should be invaluable for future studies, for example for examining synapse formation between RBs and dorsal sensory pathway neurons (Li et al., 2003, 2004c).
As mentioned above, we also found that our constructs using the evx1 enhancer that we identified in zebrafish (Juárez-Morales et al., 2016) labeled a population of interneurons with a unipolar pear-shaped cell body, a medial-dorsal dorsal-ventral spinal cord position and commissural ascending axons with no obvious dendrites. In all of these ways, they resemble zebrafish V0v cells which develop into excitatory Commissural Seconday Ascending (CoSA) neurons (Satou et al., 2012; Juárez-Morales et al., 2016). Approximately 80% of the labeled X. laevis cells had these characteristics. We have confirmed that these commissural cells do indeed express endogenous evx1 and that, as in zebrafish, they express the glutamatergic marker slc17a7, which strongly suggests that they are V0v neurons. In addition, we also demonstrated that a newly identified evx2 zebrafish enhancer sequence drives expression in cells with the same morphology (Fig. 1D–F), again consistent with these cells being evx1 and evx2-expressing V0v cells. This is the first time that V0v neurons have been identified in X. laevis. The only excitatory commissural cells that have been described so far in X. laevis spinal cord are dorsolateral commissural (dlc) (Li et al., 2003) and excitatory commissural (ecINs) sensory pathway interneurons (Li et al., 2007b). However, dlc neurons are multipolar with dorsal dendrites (Li et al., 2003) and at least most ecINs also have dendrites although these are variable (Li et al., 2007b). In contrast, our labeled cells have no obvious dendrites. Therefore, it is possible that at least some of our labeled cells are a different population of glutamatergic commissural cells, probably equivalent to excitatory CoSA neurons, the cell type labeled by these enhancers in zebrafish (Juárez-Morales et al., 2016). However, we can not rule out the possibility that dendrites may not have been clearly visible in our experiments, some ecIN neurons may be unipolar like zebrafish CoSA neurons (Li et al., 2007b) and at least some of our labeled cells are in a similar dorso-lateral position to dlc and ecIN neurons. Therefore, it is possible that at least some dlc and ecIN neurons correspond to evx-expressing V0v INs.
In conclusion, our data suggest that V0v interneurons exist in X. laevis tadpoles, further confirming the similarity of spinal cord circuitry and spinal neuron development across vertebrates. This is only the second molecularly-defined spinal cord cell type to be correlated with morphologically/physiologically defined neurons in X. laevis: the first was engrailed-expressing V1 cells that were shown to correspond to aIN neurons (Li et al., 2004a). In addition, we demonstrate that Tol2 transgenesis using zebrafish enhancers and a Gal4:UAS amplification cassette labels equivalent spinal neurons in X. laevis to those labeled in zebrafish and that EGFP expression driven by these constructs is visible in live X. laevis tadpole spinal cords. This should facilitate electrophysiological studies of specific interneurons in X. laevis tadpoles by reducing uncertainty in the selection of candidate neurons for recording. It should also enable additional connections between molecularly-distinct cells and functionally-distinct neurons to be made in this model system. This is important as X. laevis is arguably the animal in which we have the most complete understanding of spinal cord circuitry. Genetically modified mice and zebrafish have already been produced with fluorescent spinal neurons, which can be recorded electrically to study their physiology. These methods clearly work, but the mouse spinal cord is very complex and making recordings in zebrafish remains very difficult. Increasing the ease of electrophysiological analyses in X. laevis, while also confirming the homology of specific X. laevis neurons with equivalent neurons in zebrafish and amniotes, in which much more is known about neuronal specification, should help us to achieve our ultimate aim, which is a complete, integrated picture of how vertebrate spinal cord neurons and neuronal circuits are specified, function and generate behavior.
Acknowledgments
Funding
This work was funded by Leverhulme Trust project grant F/01 253/A to KEL and AR and NSF IOS-1257583 and NIH NINDS R21NS073979 to KEL.
We would like to thank the Harris lab and the Zuber/Viczian lab and in particular Karisa Rawlins for providing Xenopus laevis embryos. We would also like to thank Samantha England and Santanu Banerjee for helpful comments on a previous version of this manuscript.
Funding Information: Leverhulme Trust F/01 253/A, NSF IOS-1257583, NIH NINDS R21NS073979
Footnotes
Conflict of Interest/Competing Interests
The authors have no competing interests.
Author Contributions
J.M. performed all of the experiments and analyses in the paper, with the exception of some X. laevis embryo injections and X. laevis in situ hybridizations which were performed by R.M.D. A.R. and K.L designed and directed the study and provided expertise in frogs and zebrafish respectively. K.L and J.M wrote the paper with substantial input from A.R. All authors read, commented on and approved the final manuscript.
References
- Alvarez FJ, Jonas PC, Sapir T, Hartley R, Berrocal MC, Geiman EJ, Todd AJ, Goulding M. Postnatal phenotype and localization of spinal cord V1 derived interneurons. J Comp Neurol. 2005;493:177–192. doi: 10.1002/cne.20711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaya E, Kroll K. Production of transgenic Xenopus laevis by restriction enzyme mediated integration and nuclear transplantation. J Vis Exp. 2010 doi: 10.3791/2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appel B, Korzh V, Glasgow E, Thor S, Edlund T, Dawid IB, Eisen JS. Motoneuron fate specification revealed by patterned LIM homeobox gene expression in embryonic zebrafish. Development. 1995;121:4117–4125. doi: 10.1242/dev.121.12.4117. [DOI] [PubMed] [Google Scholar]
- Asakawa K, Suster ML, Mizusawa K, Nagayoshi S, Kotani T, Urasaki A, Kishimoto Y, Hibi M, Kawakami K. Genetic dissection of neural circuits by Tol2 transposon-mediated Gal4 gene and enhancer trapping in zebrafish. Proc Natl Acad Sci U S A. 2008;105:1255–1260. doi: 10.1073/pnas.0704963105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista MF, Jacobstein J, Lewis KE. Zebrafish V2 cells develop into excitatory CiD and Notch signalling dependent inhibitory VeLD interneurons. Dev Biol. 2008;322:263–275. doi: 10.1016/j.ydbio.2008.07.015. [DOI] [PubMed] [Google Scholar]
- Batista MF, Lewis KE. Pax2/8 act redundantly to specify glycinergic and GABAergic fates of multiple spinal interneurons. Dev Biol. 2008;323:88–97. doi: 10.1016/j.ydbio.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck CW, Slack JM. Analysis of the developing Xenopus tail bud reveals separate phases of gene expression during determination and outgrowth. Mech Dev. 1998;72:41–52. doi: 10.1016/s0925-4773(98)00015-x. [DOI] [PubMed] [Google Scholar]
- Beck CW, Slack JM. Gut specific expression using mammalian promoters in transgenic Xenopus laevis. Mech Dev. 1999;88:221–227. doi: 10.1016/s0925-4773(99)00217-8. [DOI] [PubMed] [Google Scholar]
- Bohm UL, Prendergast A, Djenoune L, Nunes Figueiredo S, Gomez J, Stokes C, Kaiser S, Suster M, Kawakami K, Charpentier M, Concordet JP, Rio JP, Del Bene F, Wyart C. CSF-contacting neurons regulate locomotion by relaying mechanical stimuli to spinal circuits. Nat Commun. 2016;7:10866. doi: 10.1038/ncomms10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner J, Gribble SL, Veien ES, Nikolaus OB, Weidinger G, Dorsky RI. Proliferation and patterning are mediated independently in the dorsal spinal cord downstream of canonical Wnt signaling. Dev Biol. 2008;313:398–407. doi: 10.1016/j.ydbio.2007.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J, Pierani A, Jessell TM, Ericson J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101:435–445. doi: 10.1016/s0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- Brudno M, Do CB, Cooper GM, Kim MF, Davydov E, Green ED, Sidow A, Batzoglou S. LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 2003;13:721–731. doi: 10.1101/gr.926603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concha ML, Russell C, Regan JC, Tawk M, Sidi S, Gilmour DT, Kapsimali M, Sumoy L, Goldstone K, Amaya E, Kimelman D, Nicolson T, Grunder S, Gomperts M, Clarke JD, Wilson SW. Local tissue interactions across the dorsal midline of the forebrain establish CNS laterality. Neuron. 2003;39:423–438. doi: 10.1016/s0896-6273(03)00437-9. [DOI] [PubMed] [Google Scholar]
- England S, Batista MF, Mich JK, Chen JK, Lewis KE. Roles of Hedgehog pathway components and retinoic acid signalling in specifying zebrafish ventral spinal cord neurons. Development. 2011;138:5121–5134. doi: 10.1242/dev.066159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason KK, Dondeti VR, Hsia HL, Cochran ER, Gumulak-Smith J, Saha MS. The vesicular glutamate transporter 1 (xVGlut1) is expressed in discrete regions of the developing Xenopus laevis nervous system. Gene Expr Patterns. 2003;3:503–507. doi: 10.1016/s1567-133x(03)00057-7. [DOI] [PubMed] [Google Scholar]
- Gosgnach S, Lanuza GM, Butt SJ, Saueressig H, Zhang Y, Velasquez T, Riethmacher D, Callaway EM, Kiehn O, Goulding M. V1 spinal neurons regulate the speed of vertebrate locomotor outputs. Nature. 2006;440:215–219. doi: 10.1038/nature04545. [DOI] [PubMed] [Google Scholar]
- Goulding M, Pfaff SL. Development of circuits that generate simple rhythmic behaviors in vertebrates. Curr Opin Neurobiol. 2005;15:14–20. doi: 10.1016/j.conb.2005.01.017. [DOI] [PubMed] [Google Scholar]
- Gribble SL, Kim HS, Bonner J, Wang X, Dorsky RI. Tcf3 inhibits spinal cord neurogenesis by regulating sox4a expression. Development. 2009;136:781–789. doi: 10.1242/dev.027995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribble SL, Nikolaus OB, Dorsky RI. Regulation and function of Dbx genes in the zebrafish spinal cord. Dev Dyn. 2007;236:3472–3483. doi: 10.1002/dvdy.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griener A, Zhang W, Kao H, Wagner C, Gosgnach S. Probing diversity within subpopulations of locomotor-related V0 interneurons. Dev Neurobiol. 2015 doi: 10.1002/dneu.22277. [DOI] [PubMed] [Google Scholar]
- Haeri M, Calvert PD, Solessio E, Pugh EN, Jr, Knox BE. Regulation of rhodopsin-eGFP distribution in transgenic xenopus rod outer segments by light. PLoS One. 2013;8:e80059. doi: 10.1371/journal.pone.0080059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeri M, Knox BE. Generation of transgenic Xenopus using restriction enzyme-mediated integration. Methods Mol Biol. 2012;884:17–39. doi: 10.1007/978-1-61779-848-1_2. [DOI] [PubMed] [Google Scholar]
- Higashijima S, Hotta Y, Okamoto H. Visualization of cranial motor neurons in live transgenic zebrafish expressing green fluorescent protein under the control of the islet-1 promoter/enhancer. J Neurosci. 2000;20:206–218. doi: 10.1523/JNEUROSCI.20-01-00206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashijima S, Masino M, Mandel G, Fetcho JR. Engrailed-1 Expression Marks a Primitive Class of Inhibitory Spinal Interneuron. J Neurosci. 2004;24:5827–5839. doi: 10.1523/JNEUROSCI.5342-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashijima S, Masino MA, Mandel G, Fetcho JR. Imaging neuronal activity during zebrafish behavior with a genetically encoded calcium indicator. J Neurophysiol. 2003;90:3986–3997. doi: 10.1152/jn.00576.2003. [DOI] [PubMed] [Google Scholar]
- Hilinski WC, Bostrom JR, England SJ, Juárez-Morales JL, de Jager S, Armant O, Legradi J, Strahle U, Link BA, Lewis KE. Lmx1b is required for the glutamatergic fates of a subset of spinal cord neurons. Neural Dev. 2016;11:16. doi: 10.1186/s13064-016-0070-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Takahashi M, Hatta K, Hotta Y, Okamoto H. Developmental regulation of islet-1 mRNA expression during neuronal differentiation in embryonic zebrafish. Dev Dyn. 1994;199:1–11. doi: 10.1002/aja.1001990102. [DOI] [PubMed] [Google Scholar]
- Ishibashi S, Love NR, Amaya E. A simple method of transgenesis using I-SceI meganuclease in Xenopus. Methods Mol Biol. 2012;917:205–218. doi: 10.1007/978-1-61779-992-1_12. [DOI] [PubMed] [Google Scholar]
- Jansen EJ, Holling TM, van Herp F, Martens GJ. Transgene-driven protein expression specific to the intermediate pituitary melanotrope cells of Xenopus laevis. FEBS Lett. 2002;516:201–207. doi: 10.1016/s0014-5793(02)02523-1. [DOI] [PubMed] [Google Scholar]
- Juárez-Morales JL, Schulte CJ, Pezoa SA, Vallejo GK, Hilinski WC, England SJ, de Jager S, Lewis KE. Evx1 and Evx2 specify excitatory neurotransmitter fates and suppress inhibitory fates through a Pax2-independent mechanism. Neural Dev. 2016;11:5. doi: 10.1186/s13064-016-0059-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K. Transgenesis and Gene Trap Methods in Zebrafish by Using the Tol2 Transposable Element. Methods in Cell Biology. 2004;77:201–222. doi: 10.1016/s0091-679x(04)77011-9. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Okamura Y, Higashijima S. alx, a zebrafish homolog of Chx10, marks ipsilateral descending excitatory interneurons that participate in the regulation of spinal locomotor circuits. J Neurosci. 2006;26:5684–5697. doi: 10.1523/JNEUROSCI.4993-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster RW, Fraser SE. Tracing transgene expression in living zebrafish embryos. Dev Biol. 2001;233:329–346. doi: 10.1006/dbio.2001.0242. [DOI] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien CB. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- L’Hostis-Guidet A, Recher G, Guillet B, Al-Mohammad A, Coumailleau P, Tiaho F, Boujard D, Madigou T. Generation of stable Xenopus laevis transgenic lines expressing a transgene controlled by weak promoters. Transgenic Res. 2009;18:815–827. doi: 10.1007/s11248-009-9273-0. [DOI] [PubMed] [Google Scholar]
- Lanuza G, Gosgnach S, Pierani A, Jessel T, Goulding M. Genetic Identification of Spinal Interneurons that Coordinate Left-Right Locomotor Activity Necessary for Walking Movements. Neuron. 2004;42:375–386. doi: 10.1016/s0896-6273(04)00249-1. [DOI] [PubMed] [Google Scholar]
- Lewis KE. How do genes regulate simple behaviours? Understanding how different neurons in the vertebrate spinal cord are genetically specified. Philos Trans R Soc Lond B Biol Sci. 2006;361:45–66. doi: 10.1098/rstb.2005.1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis KE, Bates J, Eisen JS. Regulation of iro3 expression in the zebrafish spinal cord. Dev Dyn. 2005;232:140–148. doi: 10.1002/dvdy.20215. [DOI] [PubMed] [Google Scholar]
- Lewis KE, Eisen JS. Hedgehog signaling is required for primary motoneuron induction in zebrafish. Development. 2001;128:3485–3495. doi: 10.1242/dev.128.18.3485. [DOI] [PubMed] [Google Scholar]
- Lewis KE, Eisen JS. From cells to circuits: development of the zebrafish spinal cord. Prog Neurobiol. 2003;69:419–449. doi: 10.1016/s0301-0082(03)00052-2. [DOI] [PubMed] [Google Scholar]
- Lewis KE, Eisen JS. Paraxial mesoderm specifies zebrafish primary motoneuron subtype identity. Development. 2004;131:891–902. doi: 10.1242/dev.00981. [DOI] [PubMed] [Google Scholar]
- Li WC, Cooke T, Sautois B, Soffe SR, Borisyuk R, Roberts A. Axon and dendrite geography predict the specificity of synaptic connections in a functioning spinal cord network. Neural Dev. 2007a;2:17. doi: 10.1186/1749-8104-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WC, Higashijima S, Parry DM, Roberts A, Soffe SR. Primitive roles for inhibitory interneurons in developing frog spinal cord. J Neurosci. 2004a;24:5840–5848. doi: 10.1523/JNEUROSCI.1633-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WC, Perrins R, Soffe SR, Yoshida M, Walford A, Roberts A. Defining classes of spinal interneuron and their axonal projections in hatchling Xenopus laevis tadpoles. J Comp Neurol. 2001;441:248–265. doi: 10.1002/cne.1410. [DOI] [PubMed] [Google Scholar]
- Li WC, Roberts A, Soffe SR. Locomotor rhythm maintenance: electrical coupling among premotor excitatory interneurons in the brainstem and spinal cord of young Xenopus tadpoles. J Physiol. 2009;587:1677–1693. doi: 10.1113/jphysiol.2008.166942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WC, Sautois B, Roberts A, Soffe SR. Reconfiguration of a vertebrate motor network: specific neuron recruitment and context-dependent synaptic plasticity. J Neurosci. 2007b;27:12267–12276. doi: 10.1523/JNEUROSCI.3694-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WC, Soffe SR, Roberts A. The spinal interneurons and properties of glutamatergic synapses in a primitive vertebrate cutaneous flexion reflex. J Neurosci. 2003;23:9068–9077. doi: 10.1523/JNEUROSCI.23-27-09068.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WC, Soffe SR, Roberts A. A direct comparison of whole cell patch and sharp electrodes by simultaneous recording from single spinal neurons in frog tadpoles. J Neurophysiol. 2004b;92:380–386. doi: 10.1152/jn.01238.2003. [DOI] [PubMed] [Google Scholar]
- Li WC, Soffe SR, Roberts A. Dorsal spinal interneurons forming a primitive, cutaneous sensory pathway. J Neurophysiol. 2004c;92:895–904. doi: 10.1152/jn.00024.2004. [DOI] [PubMed] [Google Scholar]
- Li WC, Soffe SR, Roberts A. Glutamate and acetylcholine corelease at developing synapses. Proc Natl Acad Sci U S A. 2004d;101:15488–15493. doi: 10.1073/pnas.0404864101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WC, Soffe SR, Wolf E, Roberts A. Persistent responses to brief stimuli: feedback excitation among brainstem neurons. J Neurosci. 2006;26:4026–4035. doi: 10.1523/JNEUROSCI.4727-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim W, Neff ES, Furlow JD. The mouse muscle creatine kinase promoter faithfully drives reporter gene expression in transgenic Xenopus laevis. Physiol Genomics. 2004;18:79–86. doi: 10.1152/physiolgenomics.00148.2003. [DOI] [PubMed] [Google Scholar]
- Loots GG, Bergmann A, Hum NR, Oldenburg CE, Wills AE, Hu N, Ovcharenko I, Harland RM. Interrogating transcriptional regulatory sequences in Tol2-mediated Xenopus transgenics. PLoS One. 2013;8:e68548. doi: 10.1371/journal.pone.0068548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love NR, Thuret R, Chen Y, Ishibashi S, Sabherwal N, Paredes R, Alves-Silva J, Dorey K, Noble AM, Guille MJ, Sasai Y, Papalopulu N, Amaya E. pTransgenesis: a cross-species, modular transgenesis resource. Development. 2011;138:5451–5458. doi: 10.1242/dev.066498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-De Luna RI, Ku RY, Lyou Y, Zuber ME. Maturin is a novel protein required for differentiation during primary neurogenesis. Dev Biol. 2013;384:26–40. doi: 10.1016/j.ydbio.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, Pachter LS, Dubchak I. VISTA : visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 2000;16:1046–1047. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
- Moran-Rivard L, Kagawa T, Saueressig H, Gross MK, Burrill J, Goulding M. Evx1 is a postmitotic determinant of V0 interneuron identity in the spinal cord. Neuron. 2001;29:385–399. doi: 10.1016/s0896-6273(01)00213-6. [DOI] [PubMed] [Google Scholar]
- Moritz OL, Tam BM, Knox BE, Papermaster DS. Fluorescent photoreceptors of transgenic Xenopus laevis imaged in vivo by two microscopy techniques. Invest Ophthalmol Vis Sci. 1999;40:3276–3280. [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal Table of Xenopus Laevis (Daudin): A Systematical & Chronological Survey of the Development from the Fertilized Egg till the end of Metamorphosis. New York: Garland Science; 1994. [Google Scholar]
- Ogino H, McConnell WB, Grainger RM. High-throughput transgenesis in Xenopus using I-SceI meganuclease. Nat Protoc. 2006;1:1703–1710. doi: 10.1038/nprot.2006.208. [DOI] [PubMed] [Google Scholar]
- Park HC, Hong SK, Kim HS, Kim SH, Yoon EJ, Kim CH, Miki N, Huh TL. Structural comparison of zebrafish Elav/Hu and their differential expressions during neurogenesis. Neurosci Lett. 2000a;279:81–84. doi: 10.1016/s0304-3940(99)00940-4. [DOI] [PubMed] [Google Scholar]
- Park HC, Kim CH, Bae YK, Yeo SY, Kim SH, Hong SK, Shin J, Yoo KW, Hibi M, Hirano T, Miki N, Chitnis AB, Huh TL. Analysis of upstream elements in the HuC promoter leads to the establishment of transgenic zebrafish with fluorescent neurons. Dev Biol. 2000b;227:279–293. doi: 10.1006/dbio.2000.9898. [DOI] [PubMed] [Google Scholar]
- Pierani A, Moran-Rivard L, Sunshine MJ, Littman DR, Goulding M, Jessell TM. Control of interneuron fate in the developing spinal cord by the progenitor homeodomain protein Dbx1. Neuron. 2001;29:367–384. doi: 10.1016/s0896-6273(01)00212-4. [DOI] [PubMed] [Google Scholar]
- Reyes R, Haendel M, Grant D, Melancon E, Eisen JS. Slow degeneration of zebrafish Rohon-Beard neurons during programmed cell death. Dev Dyn. 2004;229:30–41. doi: 10.1002/dvdy.10488. [DOI] [PubMed] [Google Scholar]
- Roberts A. Early functional organization of spinal neurons in developing lower vertebrates. Brain Res Bull. 2000;53:585–593. doi: 10.1016/s0361-9230(00)00392-0. [DOI] [PubMed] [Google Scholar]
- Roberts A, Li WC, Soffe SR. Roles for inhibition: studies on networks controlling swimming in young frog tadpoles. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2008;194:185–193. doi: 10.1007/s00359-007-0273-3. [DOI] [PubMed] [Google Scholar]
- Roberts A, Li WC, Soffe SR. How neurons generate behavior in a hatchling amphibian tadpole: an outline. Front Behav Neurosci. 2010;4:16. doi: 10.3389/fnbeh.2010.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Li WC, Soffe SR. A functional scaffold of CNS neurons for the vertebrates: the developing Xenopus laevis spinal cord. Dev Neurobiol. 2012;72:575–584. doi: 10.1002/dneu.20889. [DOI] [PubMed] [Google Scholar]
- Sapir T, Geiman EJ, Wang Z, Velasquez T, Mitsui S, Yoshihara Y, Frank E, Alvarez FJ, Goulding M. Pax6 and engrailed 1 regulate two distinct aspects of renshaw cell development. J Neurosci. 2004;24:1255–1264. doi: 10.1523/JNEUROSCI.3187-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Sone T, Yoshida S, Yahata K, Hotta J, Chesnut JD, Honda T, Imamoto F. Evidence for high specificity and efficiency of multiple recombination signals in mixed DNA cloning by the Multisite Gateway system. J Biotechnol. 2004;107:233–243. doi: 10.1016/j.jbiotec.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Sato T, Takahoko M, Okamoto H. HuC:Kaede, a useful tool to label neural morphologies in networks in vivo. Genesis. 2006;44:136–142. doi: 10.1002/gene.20196. [DOI] [PubMed] [Google Scholar]
- Satou C, Kimura Y, Higashijima S. Generation of multiple classes of V0 neurons in zebrafish spinal cord: progenitor heterogeneity and temporal control of neuronal diversity. J Neurosci. 2012;32:1771–1783. doi: 10.1523/JNEUROSCI.5500-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satou C, Kimura Y, Hirata H, Suster ML, Kawakami K, Higashijima S. Transgenic tools to characterize neuronal properties of discrete populations of zebrafish neurons. Development. 2013;140:3927–3931. doi: 10.1242/dev.099531. [DOI] [PubMed] [Google Scholar]
- Satou C, Kimura Y, Kohashi T, Horikawa K, Takeda H, Oda Y, Higashijima S. Functional role of a specialized class of spinal commissural inhibitory neurons during fast escapes in zebrafish. J Neurosci. 2009;29:6780–6793. doi: 10.1523/JNEUROSCI.0801-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sautois B, Soffe SR, Li WC, Roberts A. Role of type-specific neuron properties in a spinal cord motor network. J Comput Neurosci. 2007;23:59–77. doi: 10.1007/s10827-006-0019-1. [DOI] [PubMed] [Google Scholar]
- Scheenen WJ, Jansen EJ, Roubos EW, Martens GJ. Using transgenic animal models in neuroendocrine research: lessons from Xenopus laevis. Ann N Y Acad Sci. 2009;1163:296–307. doi: 10.1111/j.1749-6632.2008.03644.x. [DOI] [PubMed] [Google Scholar]
- Smith SJ, Ataliotis P, Kotecha S, Towers N, Sparrow DB, Mohun TJ. The MLC1v gene provides a transgenic marker of myocardium formation within developing chambers of the Xenopus heart. Dev Dyn. 2005;232:1003–1012. doi: 10.1002/dvdy.20274. [DOI] [PubMed] [Google Scholar]
- Soffe SR, Roberts A, Li WC. Defining the excitatory neurons that drive the locomotor rhythm in a simple vertebrate: insights into the origin of reticulospinal control. J Physiol. 2009;587:4829–4844. doi: 10.1113/jphysiol.2009.175208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suster ML, Kania A, Liao M, Asakawa K, Charron F, Kawakami K, Drapeau P. A novel conserved evx1 enhancer links spinal interneuron morphology and cis-regulation from fish to mammals. Dev Biol. 2009;325:422–433. doi: 10.1016/j.ydbio.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Satoh A, Ide H, Tamura K. Transgenic Xenopus with prx1 limb enhancer reveals crucial contribution of MEK/ERK and PI3K/AKT pathways in blastema formation during limb regeneration. Dev Biol. 2007;304:675–686. doi: 10.1016/j.ydbio.2007.01.019. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Kagawa N, Fujino T, Sumiya T, Andoh T, Ishikawa K, Kimura R, Kemmochi K, Ohta T, Tanaka S. A novel high-throughput (HTP) cloning strategy for site-directed designed chimeragenesis and mutation using the Gateway cloning system. Nucleic Acids Research. 2005;33:1–6. doi: 10.1093/nar/gni103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi C, Sakamaki K, Morita H, Hara Y, Suzuki M, Kinoshita N, Ueno N. Transgenic Xenopus laevis for live imaging in cell and developmental biology. Dev Growth Differ. 2013;55:422–433. doi: 10.1111/dgd.12042. [DOI] [PubMed] [Google Scholar]
- Tam BM, Lai CC, Zong Z, Moritz OL. Generation of transgenic X. laevis models of retinal degeneration. Methods Mol Biol. 2013;935:113–125. doi: 10.1007/978-1-62703-080-9_8. [DOI] [PubMed] [Google Scholar]
- Tamme R, Wells S, Conran JG, Lardelli M. The identity and distribution of neural cells expressing the mesodermal determinant spadetail. BMC Dev Biol. 2002;2:9. doi: 10.1186/1471-213X-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokumoto M, Gong Z, Tsubokawa T, Hew CL, Uyemura K, Hotta Y, Okamoto H. Molecular heterogeneity among primary motoneurons and within myotomes revealed by the differential mRNA expression of novel islet-1 homologs in embryonic zebrafish. Dev Biol. 1995;171:578–589. doi: 10.1006/dbio.1995.1306. [DOI] [PubMed] [Google Scholar]
- Uemura O, Okada Y, Ando H, Guedj M, Higashijima S, Shimazaki T, Chino N, Okano H, Okamoto H. Comparative functional genomics revealed conservation and diversification of three enhancers of the isl1 gene for motor and sensory neuron-specific expression. Dev Biol. 2005;278:587–606. doi: 10.1016/j.ydbio.2004.11.031. [DOI] [PubMed] [Google Scholar]
- Varga ZM, Amores A, Lewis KE, Yan YL, Postlethwait JH, Eisen JS, Westerfield M. Zebrafish smoothened functions in ventral neural tube specification and axon tract formation. Development. 2001;128:3497–3509. doi: 10.1242/dev.128.18.3497. [DOI] [PubMed] [Google Scholar]
- Viczian AS, Bang AG, Harris WA, Zuber ME. Expression of Xenopus laevis Lhx2 during eye development and evidence for divergent expression among vertebrates. Dev Dyn. 2006;235:1133–1141. doi: 10.1002/dvdy.20708. [DOI] [PubMed] [Google Scholar]
- Villefranc JA, Amigo J, Lawson ND. Gateway compatible vectors for analysis of gene function in the zebrafish. Dev Dyn. 2007;236:3077–3087. doi: 10.1002/dvdy.21354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivien C, Scerbo P, Girardot F, Le Blay K, Demeneix BA, Coen L. Non-viral expression of mouse Oct4, Sox2, and Klf4 transcription factors efficiently reprograms tadpole muscle fibers in vivo. J Biol Chem. 2012;287:7427–7435. doi: 10.1074/jbc.M111.324368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Szaro BG. A method for using direct injection of plasmid DNA to study cis-regulatory element activity in F0 Xenopus embryos and tadpoles. Dev Biol. 2015;398:11–23. doi: 10.1016/j.ydbio.2014.11.010. [DOI] [PubMed] [Google Scholar]
- Wang Y, Shen J, Arenzana N, Tirasophon W, Kaufman RJ, Prywes R. Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J Biol Chem. 2000;275:27013–27020. doi: 10.1074/jbc.M003322200. [DOI] [PubMed] [Google Scholar]
- Yang L, Rastegar S, Strahle U. Regulatory interactions specifying Kolmer-Agduhr interneurons. Development. 2010;137:2713–2722. doi: 10.1242/dev.048470. [DOI] [PubMed] [Google Scholar]
- Yokoyama H, Maruoka T, Aruga A, Amano T, Ohgo S, Shiroishi T, Tamura K. Prx-1 expression in Xenopus laevis scarless skin-wound healing and its resemblance to epimorphic regeneration. J Invest Dermatol. 2011;131:2477–2485. doi: 10.1038/jid.2011.223. [DOI] [PubMed] [Google Scholar]
- Zhuo X, Haeri M, Solessio E, Knox BE. An inducible expression system to measure rhodopsin transport in transgenic Xenopus rod outer segments. PLoS One. 2013;8:e82629. doi: 10.1371/journal.pone.0082629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber ME, Gestri G, Viczian AS, Barsacchi G, Harris WA. Specification of the vertebrate eye by a network of eye field transcription factors. Development. 2003;130:5155–5167. doi: 10.1242/dev.00723. [DOI] [PubMed] [Google Scholar]
- Zuber ME, Nihart HS, Zhuo X, Babu S, Knox BE. Site-specific transgenesis in Xenopus. Genesis. 2012;50:325–332. doi: 10.1002/dvg.22006. [DOI] [PMC free article] [PubMed] [Google Scholar]