Abstract
Strategies utilizing Toll-like receptor 4 (TLR4) agonists for treatment of cancer, infectious diseases, and other targets report promising results. Potent TLR4 antagonists are also gaining attention as therapeutic leads. Though some principles for TLR4 modulation by lipid A have been described, a thorough understanding of the structure-activity relationship (SAR) is lacking. Only through a complete definition of lipid A-TLR4 SAR is it possible to predict TLR4 signaling effects of discrete lipid A structures, rendering them more pharmacologically relevant. A limited ‘toolbox’ of lipid A-modifying enzymes has been defined and is largely composed of enzymes from mesophile human and zoonotic pathogens. Expansion of this ‘toolbox’ will result from extending the search into lipid A biosynthesis and modification by bacteria living at the extremes. Here, we review the fundamentals of lipid A structure, advances in lipid A uses in TLR4 modulation, and the search for novel lipid A-modifying systems in extremophile bacteria.
Keywords: Lipid A Structure, TLR4 Agonists, TLR4 Antagonists, TLR4 Immunomodulation, Lipid A Modifying Enzymes, Marine Bacteria
Introduction
Lipid A (also referred to as endotoxin) is the amphipathic lipid base structure of lipopolysaccharide (LPS). LPS is a unique bacterial lipid comprising the outer leaflet of the asymmetric outer membrane of most Gram-negative bacteria. LPS is composed of three distinct regions: O-antigen, core, and lipid A – descending from the outer bacterial surface to the membrane.[1,2] These structurally and functionally distinct regions have important roles in such diverse aspects as growth, virulence, stress adaptation, innate and adaptive immune avoidance, maintenance of membrane permeability, and resistance to antibiotics. O-antigen, the exterior polysaccharide of LPS, is a highly divergent structure and is one of the antigenic molecules forming the basis for bacterial serotyping. Diversity of O-antigen structure and length can vary widely, even within a single bacterial species. Structure, composition, regulation, antigenicity, and the functional consequences of compositional variation of O-antigen have been extensively reviewed.[3–5] Core oligosaccharide (‘core’ for the purposes of this review) links lipid A to O-antigen polysaccharide and is more structurally conserved in contrast. Two 3-deoxy-D-manno-octulosonic acid (KDO) sugars are attached to the non-reducing glucosamine of the lipid A backbone, typically followed by extension with heptose sugars. Though core is considerably less diverse than O-antigen, structural modifications are observed including phosphorylation and phosphoethanolamine addition.[6,7] Modifications to core have important consequences for virulence and resistance to cationic antimicrobial peptides (CAMPs) and represent an active field of study.[8,9] Relative to O-antigen and core the most conserved moiety of LPS is lipid A; however, substantial diversity in lipid A structure exists across the Gram-negative, LPS-bearing bacteria often with unique stimulatory properties based on the structurally-dependent interaction of lipid A with innate immune receptors. Potential therapeutic uses for lipid A are continually emerging as our understanding of the fundamental mechanisms of diseases and their links to both canonical and non-canonical innate immune components are gleaned. Discovery and characterization of novel lipid A structures in order to harness unique lipid A modifying systems is the goal of this review, specifically focusing on the structure-activity relationship (SAR) of lipid A and Toll-like receptor 4 (TLR4) to improve the effects of lipid A-based therapies through structural optimization.
Lipid A Innate Recognition and Signaling via TLR4
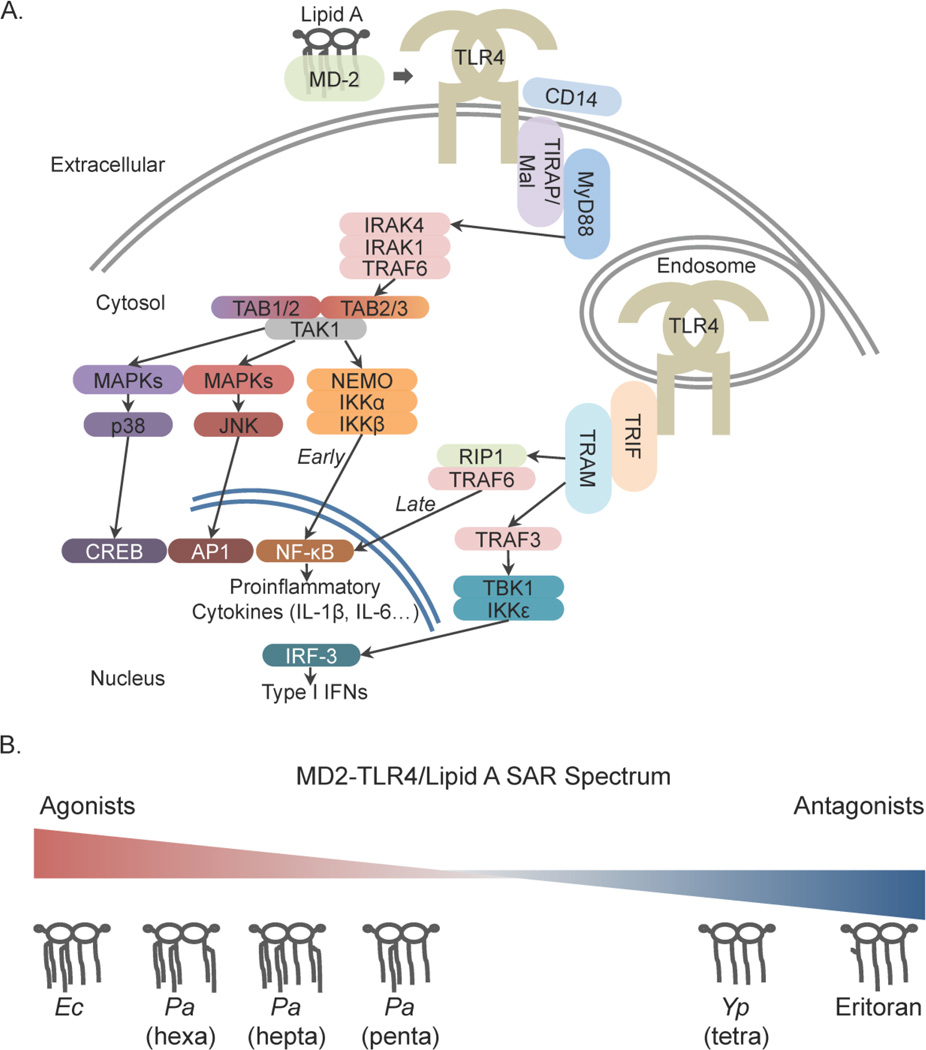
The endotoxic activity of LPS is derived from the interaction of lipid A and the cognate TLR4 receptor complex.[10–14] Agonism of TLR4 by extracellular, pro-inflammatory lipid A structures results in a strong nuclear factor kappa-B (NF-κB)-driven response (Figure 1A) via the MyD88-dependent pathway typified by release of inflammatory cytokines (IL-6, TNF-α, and IL-1β).[15–17] TLR4 signaling triggered from the endosomal compartment can signal through NF-κB via RIP1[18], but endosomal TLR4 signaling is typically associated with the IRF-3 axis via TRIF/TRAM resulting in IFN-β production.[19] Several studies have demonstrated differential activation of the NF-κB and IRF-3 axes as a result of lipid A structural modification.[19–21] In addition, mitogen-activated protein kinases (MAPKs) such as p38 and JNK are both directly activated (quickly through the MyD88 pathway, and slowly through the TRIF/TRAM pathway) and indirectly activated after cytokine and chemokine induction (through Ras GTPases). [22,23] Constitutive activation of these kinases is a hallmark of some cancer types, making modulation of their activities an attractive goal in oncological drug discovery. To date, few universal rules have been established delineating the contribution of discrete lipid A structural components to the resulting agonist/antagonist activity. In order to effectively exploit the therapeutic uses of lipid A, we must first improve the structure-activity relationship (SAR) definition (Figure 1B) and expand the current lipid A modifying enzyme ‘toolbox.’
Figure 1. Signaling pathways from TLR4 to early-/late-phase inflammation and the lipid A-MD-2-TLR4 (NF-κB-mediated) structure activity relationship (SAR) range.
(A) Surface and endosomal TLR4 signaling results in both divergent and convergent inflammatory processes. Convergent: surface TLR4 signaling via the MyD88 axis leads to early NF-κB activation and endosomal TLR4 signaling via TRIF/TRAM leads to late NF-κB activation, both resulting in proinflammatory cytokine production. Divergent: surface TLR4 activation of multiple MAP kinase cascades (involving p38 and JNK) results in the activation of CREB and AP1 transcription factors whereas endosomal TLR4 results in an IRF-3-mediated Type I interferon response via TRAF3. Selectively biasing these downstream signaling components is the goal of SAR refinement, resulting in a customized response. (B) Inflammatory activity of the lipid A-MD-2-TLR4 complex is dependent on lipid A structure. E. coli (Ec) hexa-acylated lipid A is a potent agonist of TLR4. Pseudomonas aeruginosa (Pa) hexa-, hepta-, and penta-acylated lipid A are associated with different human diseases and elicit diverse strong to weak TLR4 agonist responses. Two tetra-acylated molecules from Yersinia pestis (Yp) grown at 37°C (also lipid IVA) and Eritoran both result in TLR4 antagonism.
Lipid A stimulates the TLR4 receptor as a myeloid differentiation factor-2 (MD-2)-bound heterodimer complex in which two MD-2-lipid A complexes bind two TLR4 extracellular domains resulting in ligation of two TLR4 receptors (in the presence of CD14).[24,25] Species diversity among TLR4s is substantial and the TLR4 SAR interpretations and inferences here are limited to human (hu)TLR4. Reviews of TLR4 structure[26,27] and species diversity[28,29] are offered here. Our understanding of the basis for MD-2/TLR4 interaction with bound lipid A is expanding and the role of MD-2 in differential activity is coming into focus through analysis of binding with various lipid As.[30] Human MD-2 is a small (160 amino acid residues including the 16 amino acid secretion signal), secreted protein with two β sheets comprising an immunoglobulin fold.[31] The deep interior cavity is lined with hydrophobic residues conducive to acyl binding.[31,32] The F126 (referring to Phe126 residue) loop of MD-2 has been implicated in differential activity of the bound complex.[32–34] The structural change induced at the F126 loop of MD-2 following binding by pro-inflammatory hexa-acylated lipid A (example E. coli lipid A, Figure 1B) is a crucial component of TLR4 ligation.[35] In contrast, antagonistic tetra-acylated structures, such as the fundamentally minimal lipid A unit lipid IVA or Eritoran (example Eritoran[32], Figure 1B) bind MD-2 without displacement of the F126 loop resulting in a bound, but not ligated huTLR4 complex (evidenced by attenuated downstream NF-κB activity).[34] Regulated lipid A structures within a single species comprise a range of stimulatory potential illustrated by the diverse and conditional structures from Pseudomonas aeruginosa (Pa). Hexa-acylated Pa lipid A, associated with infections from patients with Cystic Fibrosis (CF), is a stronger agonist of MD-2-TLR4 than the bronchiectasis-associated penta-acylated Pa lipid A (Figure 1B).[36] Non-stimulatory lipid As, such as the penta-acylated structure derived from Rhodobacter sphaeroides (Rs, lipid A referred to as Rs LA) can also bind MD-2, forming a successful MD-2-lipid A complex capable of binding to huTLR4. However, in murine and human systems Rs LA is inhibitory due to flipped loading of the MD-2-RsLA complex.[34] Crystallographic structural studies of bound lipid A-MD-2-TLR4 complexes are few[25,32], but have been well-reviewed and the interpretations are supported by follow-up studies.[37] The time-consuming and fickle nature of ligand-bound crystallization precludes correlative studies of the full range of lipid A structures, but functional studies can be readily performed to evaluate the SAR using downstream reporters such as cytokine production. Accurately defining the SAR between discrete lipid A structures and MD-2/TLR4 is crucial to successful uses of engineered lipid A-based therapeutics.
Biosynthesis of Lipid A
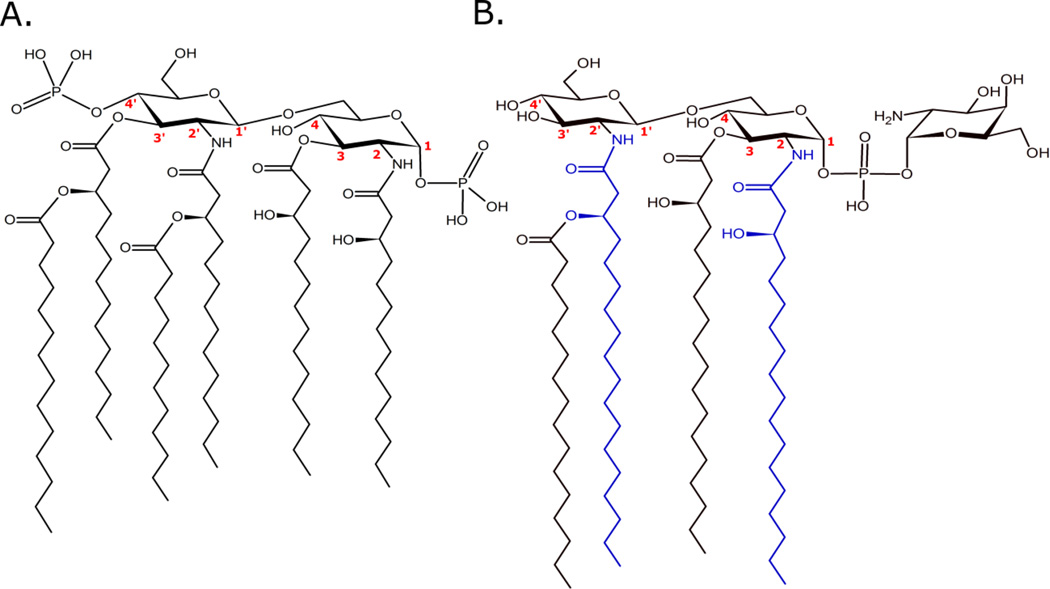
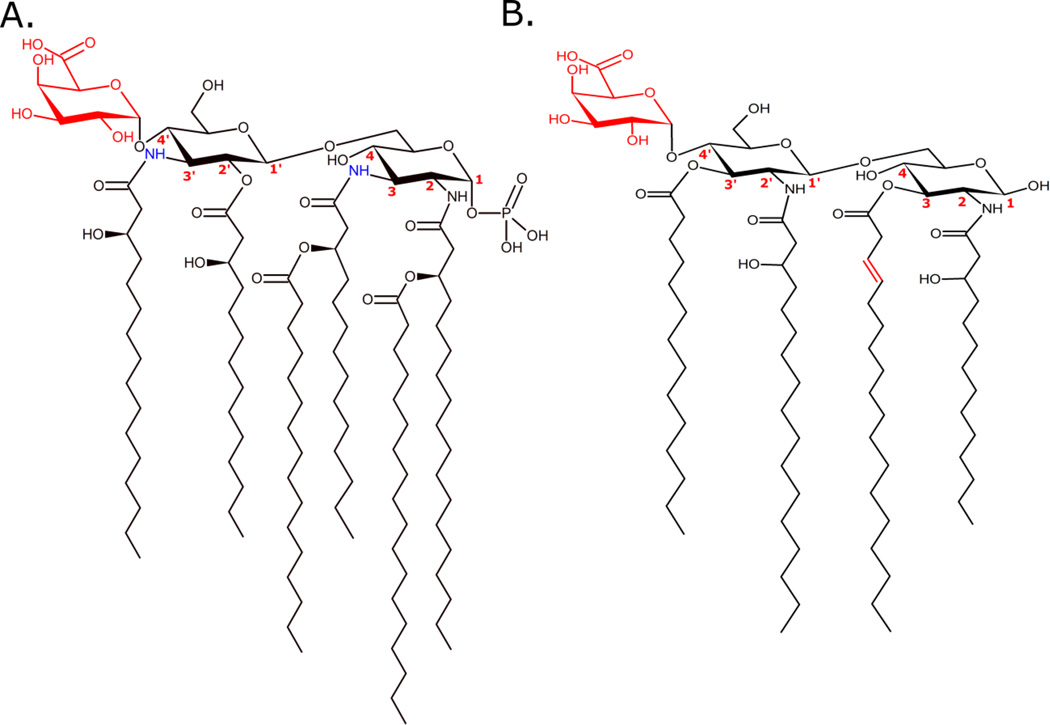
Lipid A produced by E. coli is the canonical pro-inflammatory lipid A structure shown in Figure 2A. E. coli lipid A is a hexa-acylated, bis-phosphorylated structure containing 3-hydroxylated 14-carbon primary acyl chains with one each of 14-carbon and 12-carbon secondary (acyl-oxo-acyl) chains.[38] In this configuration, E. coli lipid A results in a highly endotoxic structure with strong TLR4 agonism.[15,24,39] The fundamentals of lipid A synthesis, defined in E. coli, are well-established through the career-long work of Christian Raetz[40] and colleagues and is briefly described.[1,38,41]
Figure 2. Representative lipid A structures from E. coli (A) and F. novicida (B).
E coli lipid A is a hexa-acylated, bis-phosphorylated structure associated with highly proinflammatory properties. Carbon numbers are given for reference points in the text. F. novicida lipid A is a tetra-acylated, monophosphorylated structure with nonstimulatory activity through the MD-2/TLR4 complex. The blue acyl chains are C18 added by LpxD1 at warm temperatures. At cooler growth temperatures LpxD2 activity is higher resulting in one or both (2, 2’) N-linked positions being modified with a C16 addition.[41]
Lipid A synthesis begins with LpxA-mediated acylation of UDP-GlcNAc at the 3-position resulting in UDP-GlcNAc with an ester linked 14-carbon acyl chain (in E. coli). LpxC removes an acetate molecule from the 2-position leaving an exposed amino group and resulting in UDP-3-acyl-GlcN. Deacetylation of UDP-GlcNAc is an energetically unfavorable reaction and represents the thermodynamic commitment step toward lipid A biosynthesis, making LpxC a potential target for antibacterial strategies.[2,42] The aminotransferase LpxD further acylates UDP-3-acyl-GlcN at the 2-position, in E. coli with a 14-carbon acyl chain, resulting in UDP-2,3-diacyl-GlcN. The next enzyme in the series, LpxH relieves UMP from UDP-2,3-diacyl-GlcN resulting in lipid X, a 1-phosphorylated 2,3-diacyl-GlcN. To form the characteristic diglucosamine lipid A backbone, one lipid X molecule and one UDP-2,3-diacyl-GlcN are condensed by LpxB forming the β,1’-6 glycosidic bond. LpxK phosphorylates the 4’ position of the tetra-acylated 1-phosphorylated diglucosamine product of LpxB, thus completing biosynthesis of the base molecule lipid IVA. Lipid IVA is a tetra-acylated, 1,4’-bis-phosphorylated minimal lipid A unit and is a notable antagonist of huTLR4.
The remaining enzymes of lipid A synthesis, still in the inner membrane, prepare the final structure for export to the outer membrane. The process begins with KdtA-mediated attachment of two KDO sugars to the 6’ position of lipid IVA resulting in KDO2-lipid IVA. In Ec KDO2-lipid IVA is further processed by secondary acylations by the pair of acyltransferases, LpxL (adding a secondary 12-carbon acyl chain to the 3’ 3-hydroxymyristate) and LpxM (adding a secondary 14-carbon acyl chain to the 2’ 3-hydroxymyristate). The resulting product is complete E. coli KDO2-lipid A, a potent human and murine TLR4 agonist. The addition of core sugars follows and the entire, nascent E. coli lipooligosaccharide (LOS) is flipped to the periplasmic surface of the inner membrane via the ABC transporter, MsbA. In the periplasm O-antigen oligosaccharides are polymerized to complete the mature E. coli LPS molecule. Reviews of core attachment, LOS flipping, O-antigen polymerization, and final LPS flipping to the outer leaflet of the outer membrane are noted here as these topics are outside the scope of this review.[2–5]
The characteristic enzymes of the Raetz pathway of lipid A biosynthesis, defined in E. coli, often have variable or extended permissivity for alternative substrates in different LPS-bearing species. For example, LpxD in E. coli is permissive for both 3-OH C16 and 3-OH C14 with a substantial preference for the latter; alternative LpxD enzymes are discussed in detail vide infra. [43,44] Additionally, the substrate pool for lipid A biosynthetic enzymes often controls the final structure. LpxA from Leptospira interrogans (Li LpxA) is selective for the 3-aminated precursor UDP-GlcNAc3N; as such, Li LpxA cannot acylate UDP-GlcNAc and results in a four primary N-linked acyl chains in the mature Li lipid A.[45] While UDP-GlcNAc is abundant it is not used for lipid A synthesis by Li. UDP-GlcNAc3N is generated in Li as a result of two enzyme-directed modifications of UDP-GlcNAc by the enzymes GnnA and GnnB.[45] Complete primary amine-linked lipid A structures confer unique properties. The same use of UDP-GlcNAc3N in Campylobacter jejuni not only increased antimicrobial resistance but also reduced huTLR4 activation.[46] The use of alternative backbone sugars for lipid A synthesis may prove a useful tool for lipid A structural customization aimed to influence the lipid A/TLR4 SAR.
Lipid A Structural Modifications and Consequences
Natural structural modification of lipid A is achieved both through constitutive and regulated processes in response to alterations in growth condition (ex: temperature, nutrient, osmolarity), evasion of detection in vivo (ex: conversion of structures from agonist to antagonist), resistance to cationic antimicrobials (ex: modulation of surface exposed negative charge), and membrane-disrupting antimicrobials (ex: deacylases of the outer membrane), to list only a few. Through manipulation of the current lipid A modifying systems and biosynthetic enzymes, the basic fundamentals of the relationship between lipid A structure and TLR4 downstream activity has been established. In 2013, Needham and Trent published a thorough and still current review of lipid A modifying enzymes and the regulatory systems thereof.[47] Many of the lipid A modifying systems discussed herein are related directly to bacterial transmission (carriage and transmission via arthropod vectors) and/or bacterial pathogenesis (especially immunomodulation).[9,28] Pa isolates from patients with cystic fibrosis produce a highly proinflammatory lipid A structure, as compared to bronchiectasis patient isolates.[48,49] Yersinia pestis (Yp) modulates lipid A structure as a complex, regulated component of flea-to-mammalian host transmission using lipid A modifications.[50–52] A selection of lipid A modifications and the respective functional consequence follows to illustrate the purposeful use of lipid A modification in defining the SAR of lipid A/TLR4 to design appropriate lipid A-based therapeutics via exogenous or ectopic enzyme expression.
Early modifications of lipid A structure that alter TLR4 complex binding
To maintain membrane fluidity and accommodate growth in lower temperatures, Francisella novicida (Fn) incorporates shorter primary and secondary acyl chains during lipid A synthesis.[53,54] As above, the enzyme LpxD adds the amide-linked primary acyl chains to the 2 and 2’ sites of the diglucosamine backbone.[44,55] Reduced growth temperature was shown to result in lipid A acyl-shortening in the model species Fn, with longer chain fatty acids added at mammalian body temperature.[44,56] Two LpxD functional homologs, LpxD1 and LpxD2 were identified in Francisella with N-acyltransferase activity, though with different length hydrocarbon ruler specificites, optimal temperature ranges, and sharing only modest sequence identity (34%).[44] LpxD2 functions at lower temperature adding a 16-carbon acyl chain, whereas LpxD1 adds 18-carbon chains to the 2 and 2’ backbone sites at higher temperatures. Lipid A extracted from Fn wildtype (WT), ΔlpxD1, or ΔlpxD2 strains (Figure 2B) are astimulatory in human macrophages.[44,57] In Fn lipid A derived from these strains the 3’-position is empty, the result of the theorized Fn LpxR deacylase enzyme that has been reported in Salmonella[58] and Helicobacter[59] though a homolog has not been reported in any Francisella family member.[60] The empty 3’ position is one of the structural characteristics along with the longer primary acyl chains (C18) implicated in the astimulatory properties noted above. Several members of the Francisella family of mesophiles are culturable from niches ranging from cold, fresh water lakes in Scandinavia to arthropod vectors at ambient outdoor temperature in the mid-southern US and warm-blooded mammalian reservoirs; LpxD2 contributes to viability across this substantial temperature range.[61]
Late modifications of lipid A structure that alter TLR4 complex binding
Alteration in the overall number of fatty acids affects TLR4 stimulation.[62] Secondary or late acylation on lipid A structures are known to alter the ability of lipid A to bind to MD-2 and stimulate a strong innate immune response.[63] Late additions to lipid A at the inner membrane are made to the acyl chains at the 2’ and 3’ positions to form β-substituted acyl-oxo-acyl groups by the acyltransferases HtrB (LpxL) and MsbB (LpxM), respectively. In addition to position specificity, each enzyme has a substrate preference, laurate (C12:0) or myristate (C14:0) for LpxL or LpxM, respectively with LpxL acylation proceeding LpxM.[64,65]
Another widely recognized example of lipid A modification associated with a low temperature growth condition is the addition of a monounsaturated acyl chain by Yp. Yp is transmitted through the bite of infected fleas that have fed on a warm-blooded infected rodent.[52,66] This warm-cool-warm cycle of growth and transmission requires a responsive system to maintain membrane function. Yp lipid A at mammalian temperature is a tetra-acylated structure containing only primary 3-hydroxymyristates. Grown at the temperature of a flea (21–25°C) two secondary acyltransferases are active, Yp LpxP and Yp MsbB (LpxM homolog). Yp LpxP attaches a cis-9-palmitoleic acid (C16:1) to the hydroxyl group of the 2’-3-hydroxymyristate of Yp lipid A at flea temperatures.[67] Similarly, Yp MsbB attaches a lauroyl group to the 3’-3-hydroxymyristate at low temperature. While the activity of both contribute to maintenance of membrane fluidity, the presence of unsaturations in membranes correlates strongly with maintenance of membrane fluidity at lower temperatures.[68–70] In contrast to the Fn example above, Yp lipid A grown and extracted from low and high temperature are differentially active in TLR4 stimulations.[51,52] The hexa-acylated structure resulting from low temperature growth (21–25°C) is a TLR4 agonist, whereas the warm temperature (37°C) tetra-acylated structure is a TLR4 antagonist.[52,57] Overall acyl density, positioning, degree of unsaturation, and length all play a role in low temperature growth as described above and these substantial modifications have important consequences for TLR4 activity.
Subsequent to transport to the outer membrane by the Lpt system, lipid A can be further modified in the outer membrane by acyltransferases (PagP) or deacylases (PagL or LpxR). PagP is a palmitoyltransferase that transfers a palmitate (C16:0) to the hydroxyl group of the 3-hydroxymyristate chain at position 2 (as examples Salmonella and E. coli) or the 3’ position (Pa) resulting in a hepta- or hexa-acylated structure, respectively.[71,72] PagL[73] and LpxR[58] are 2 (as examples Salmonella and Pa) and 3’ (Hp, Salmonella, and Yersinia) position deacylases, respectively. PagP and PagL are regulated by the two component regulatory system, PhoP/PhoQ, whereas LpxR is a Ca2+-dependent enzyme in Salmonella and temperature regulated in Yersinia. Together, alterations in lipid A biosynthesis can lead to changes in membrane permeability that result in increased sensitivity of bacterial cells to environmental changes, susceptibility to alpha-helical antimicrobial peptides, a greatly reduced ability to stimulate NF-κB-mediated cytokines, and reduced virulence due to underacylated or less-toxic lipid A in mouse infection models.[74–77] These results indicate that targeting the “later” modifications could be a successful strategy for novel antimicrobial therapies.
In addition to acyl chain modifications, hydroxylation of the acyl-oxo-acyl fatty acids is observed at the 2-hydroxy position of the secondary acyl chains through the enzymatic activity of LpxO, an oxygen-dependent, aspartyl/asparaginyl β-hydroxylase homologue in a PhoP/PhoQ-dependent manner.[78] The 3-hydroxyl is commonly a site for secondary acyl additions; however, the addition of glycine and diglycine to the available hydroxyl group of the secondary 3’-3-hydroxylauroyl group was reported in Vibrio cholera (Vc) O1 El Tor.[79] Typically, these additions to the free hydroxyl are associated with membrane integrity and changes in antibiotic susceptibility patterns and in the case of glycine modification of lipid A little change was observed in TLR4 stimulation assays compared to unmodified Vc lipid A.[79] There are also important lipid A-TLR4 SAR implications from the study of branched acyl chain lipid A structures in the oral pathogen P. gingivalis (Pg) and the enteric commensal Bacterioides thetaiotaomicron (Bt). Pg lipid A structure is unique compared to that of the enteric pathogens and stimulates far lower pro-inflammatory cytokine production than E. coli lipid A.[80–82] Pg lipid A is a heterogeneous mixture of structures with several commonalities including methyl branches found on the primary N-acyl chain (methyl-C16), 1-phosphorylation, and a straight chain typically present at the 3-position, though 3-O-deacylated Pg lipid A is described.[83] Bt lipid A is characterized by the presence of a 4’-phosphorylation and a long branched 2’-N-acyl chain, yet Pg and Bt lipid As elicited opposite effects in TLR4 stimulation studies.[84] These results implicated the importance of phosphorylation position in potency of TLR4 stimulation, but the contribution of branched chains (and positioning) to lipid A-TLR4 SAR are still poorly defined.
Contribution of lipid A termini and modifications thereof to TLR4 complex activity
In studying the position of a mono-phosphorylation in Pg versus Bt and the outcomes in TLR4 stimulations, the Darveau group made a striking observation. Engineered E. coli mutants bearing either a 1- or 4’-monophosphorylation exhibited markedly different TLR4 activities with the 4’-monophosphorylated structure eliciting a lower NF-κB-driven response (E. coli wildtype structure given in Figure 2A for reference).[84] The same logic follows from the 4’-monophosphorylated structure of MPLA stimulating TLR4 at weak agonist levels compared to the parent 1- and 4’-phosphorylated Salmonella enterica LA molecule (commonly, diphosphoryl lipid A - DPLA) stimulating a robust agonist response. Mono- and bis- (or di-) phosphorylation status alone can drive differential activation, but these structural changes are often accompanied by other modifications and must be considered in the larger context for MD-2 binding and TLR4 ligation.
Finally, modifications to the terminal phosphate moieties, involved in modulating the bacterial surface charge and permeability have not been shown to play a role in recognition by the host innate immune system. However, such terminal phosphate modifications (including additions of the amino containing moieties, such as galactosamine, aminoarabinose, and phosphoethanolamine) can impart profoundly consequential antimicrobial resistance properties.[8,85] Addition of phosphoethanolamine and/or galactosamine to the lipid A terminal phosphate of Acinetobacter baumannii (Ab) renders the bacterium resistant to polymixin, an antibiotic of last resort for multiply drug resistant species.[86] The recent emergence of a phosphoethanolamine transferase gene (mcr-1) on a mobile element is a harbinger of extensive failure of the last effective antibiotics against MDR infections.[87] Though, with respect to MD2/TLR4 stimulation no alteration of the SAR has been noted. Specifically, modifications of lipid A by amino-containing sugars are predicted to reside outside the binding pocket of MD2 and therefore should not play a significant role in altering TLR4 activation.
Effective Strategies for Lipid A-Based Treatment Approaches Depend on Structure
Ribi et al. established the use of lipid A-based treatments for cancer immunotherapy in 1975.[88] In the intervening forty-one years, the suggested uses for both agonistic and detoxified lipid A as immune modulators have exploded.[89–91] Notably, the first new adjuvant approved by the FDA in decades is a lipid A derivative. Monophosphoryl lipid A (MPLA, commonly MPL) is chemically derived from a deep rough (Re)-LPS strain of Salmonella enterica (serovar Minnesota R595).[92] While MPLA has markedly reduced toxicity compared to Salmonella lipid A, it retains modest TLR4 stimulating properties.[93] Two of the major lipid A species in MPLA include 1-dephosphorylated lipid A and 1-dephosphorylated, 3-O-deacylated lipid A. MPLA is a success story in terms of productive uses for lipid A; however, chemical processing of lipid A (MPLA) or production of a similar molecule by synthetic processes (glucopyranosyl lipid A, GLA[94]) limits the structural possibilities, thus attenuating the potential for highly engineered or structurally customized lipid A. In contrast, several groups have demonstrated the use of structurally modified lipid A to modulate the TLR4 response, purposefully engineered through the expression or disruption of endogenous and exogenous lipid A modifying enzymes.[20,21,95] The formulary uses for MPLA-based (and similar – GLA and other synthetic TLR4 agonists) based protein-in-adjuvant vaccines are growing and mark a new era of the vaccine age. The approval of MPLA as an adjuvant served two important roles: one, new and efficacious vaccines and formulations are now available to help save lives and prevent disease including prophylactic cancer vaccines; two, establishing a path to human use for related, low toxicity lipid A-based TLR4 agonist and antagonist molecules. With these advances in mind novel sources of enzymes and regulatory systems must be discovered, characterized and understood to harness the potential uses for lipid A.
Similarly, novel prophylactic and therapeutic uses for MPLA and other lipid A structures are on the rise, outside of the vaccine context.[96,97] In 2013, Michaud and colleagues reported reduced Alzheimer’s-related pathology, including prevention of amyloid beta plaque accumulation in genetically disposed mice chronically dosed with MPLA.[98] Several attempts have been made to establish MPL or similar synthetic TLR4 agonists, such as Eritoran, as prophylactic treatments for sepsis with promising results, but limited success.[99–102] An interesting finding linking amyloidogenic blood clotting to LPS-precipitated fibrin net formation suggesting a role for low doses of circulating lipid A in hypercoagulation was reported in 2016.[103] The therapeutic and prophylactic uses of synthetic TLR4 antagonists deviating from the basic lipid A mimetic template are not discussed here, but have been tested for prevention of aortic aneurysms[104], neuropathic pain[105], and other TLR4-linked pathologies.[106] Together, these studies highlight the role of TLR4 in a wide variety of disease states and emphasize the need for improvements in TLR4 design with rational basis in SAR outcome. To achieve this goal we must have a diverse enzyme ‘toolbox’ for customization of therapeutic lipid As.
Finding Novel Sources of Lipid A Modifying Enzymes
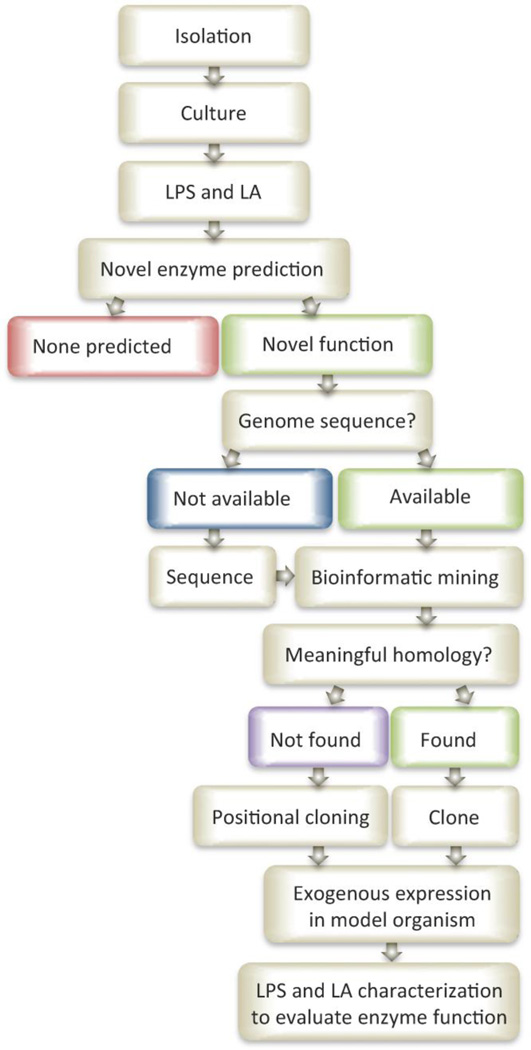
The lipid A modifying enzymes described to date are generally from mesophile species that grow best between 20° and 45°C. In order to accommodate growth in exotic, non-uniform conditions, unique mechanisms for maintaining membrane fluidity, resistance to detrimental solutes, and evasion of detection within their niche are required. Modification of lipid A structure is one such accommodation mechanism. The underlying molecular mechanisms for unique lipid A structures are of interest to cultivate new enzymatic resources for intentional lipid A modification aimed at customizing lipid A-based therapeutics. The following section is a review of the growing archive of novel lipid A structures from bacterial sources, description of molecular mechanism of unique structures (where possible), improved mass spectrometric approaches for structural elucidation of lipid A, and a rational approach (Figure 3) to identifying and cloning useful enzymes from sources sharing potentially low homology to known enzymes of the same function. By harnessing new lipid A modifying enzymes alongside the complete description of the lipid A-MD-2/TLR4 SAR, it will be possible to effectively design and test lipid A-based therapeutics to treat cancer, chronic inflammatory diseases of aging, acute endotoxemia, and other pathologies propagated by lipid A.
Figure 3. Pathway to systematic identification of novel lipid A biosynthetic and/or modifying enzymes.
Species from exotic source libraries (marine, thermal vents, etc.) must first be isolated and cultured for LPS and lipid A extraction. By comparing the known lipid A structures and their respective biosynthetic pathways to novel structures identified from exotic sources predictions can be made about new, unique, or alternative enzymatic functions. Where novel enzymes are predicted, homology searches of the available genome(s) can be performed using similar enzymes of known function and sequence. Some species may require genome sequencing and assembly prior to bioinformatic mining. Homology searches resulting in meaningful predictions can be cloned and exogenously expressed in a model organism and further evaluated at the lipid A structural modification level. Homology searches not yielding meaningful predictions may lead to gene identifications using a more laborious positional cloning approach for the most interesting potential modifying enzymes. Careful consideration of atypical growth conditions, unique inducing conditions, and unforeseen technical hurdles will be necessary.
Novel Lipid A Bacterial Sources from Extreme Environments
Marine
Marine bacteria can be found in myriad environments disparate from terrestrial ones. They are ubiquitous, inhabiting open and coastal waters at all depths, as well as in the sediment of the ocean floor and near hydrothermal vents. They can be planktonic, symbiotic, or pathogenic to other marine life. The observation of microbial life in these “extreme” environments has resulted in the generation of interesting hypotheses regarding differences in their intracellular machinery versus their terrestrial cousins. These, in turn, have led to the discovery of novel lipid A structures synthesized and utilized by marine bacteria as physical adaptations to their local environments. A literature review of endotoxin structures produced by Gram-negative marine bacteria was published by Leone et al.[107] and focused on diverse marine organisms, including several Gammaproteobacteria genera, Flavobacteria, Cellulophaga, Arenibacter, and Chryseobacteria. Currently, there is a resurgence in the discovery of lipid A structural variants from marine bacterial sources owing to their generally low inflammatory potentials and consequent toxicities.
One particularly interesting source for new bacterial species, and consequently new lipid A structures and modifying enzymes, are the marine sponges of the phylum Porifera. Sponges contain diverse and abundant microbiomes comprising, in part, prey organisms, pathogens, and symbionts. Sponges have also been shown to possess a TLR-mediated innate immune response and can respond to LPS from E. coli.[108] Interestingly, most of the structures discovered, when tested against human innate immune cells, have exhibited mild or no inflammatory potential. These observations could be due to four factors: i) the degree of acylation on marine bacterial lipid As trends toward hypo-acylation (primarily penta-acylated) when compared to E. coli, ii) three-dimensional conformational changes caused by unsaturated fatty acyl chain incorporation, iii) hypo-phosphorylation relative to E. coli lipid A, and iv) fatty acyl chains incorporated into marine bacterial lipid As tend to be shorter in carbon chain length than their highly inflammatory counterparts. Many of the marine bacteria with reported lipid A structures in the literature can be divided into several groups, including cyanobacteria, psychrophiles and thermophiles, and halophiles.[109] These classifications have extensive overlap because they do not fall on the same continuum.
Lone species
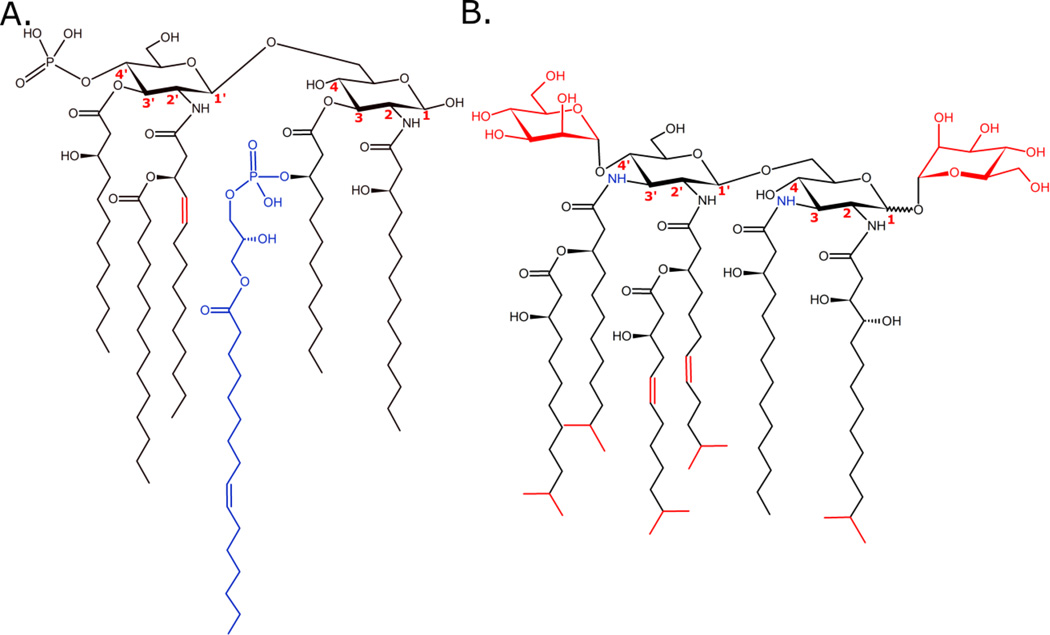
Three bacteria, for which lipid A structures have been reported, that do not fall into any of the following categories are Vibrio fischeri[110], Bdellovibrio bacteriovorus[111], and Gemmata obscuriglobus.[112] Representative lipid A structures for V. fischeri and B. bacteriovorus are shown in Figure 4. In both cases, the structures represented are for the base peak in the single stage scan mass spectrum. V. fischeri is a symbiont of the Hawaiian bobtail squid, Euprymna scolopes, and lends its bioluminescence to the squid’s light organ.[110] Several interesting discoveries have been made with regard to its lipid A structure, as shown in Figure 4A. First, a β-hydroxyl-C14:1 fatty acyl chain appears to be incorporated through amide linkage at the 2’ position. This is, so far, a unique finding in Gram-negative bacteria whose biosynthesis pathway is largely conserved. Other unique substitutions have been observed at the C-3 secondary position, namely phosphoglycerol, lysophosphatidic acyl groups, and phosphatidic acyl groups.[110] The mechanism by which these modifications are made to V. fischeri lipid A is unknown, and no modifying enzymes have been identified thus far.
Figure 4. Representative lipid A structures from V. fischeri (A) and B. bacteriovorus (B).
V. fischeri produces mono-phosphorylated lipid A with unusual modifications, including secondary phosphoglycerol, lysophosphatidic acyl groups, and phosphatidic acyl groups. The amide-linked, β-hydroxyl-substituted C14:1 fatty acyl group at the C-2’ position is, so far, unique among Gram-negative bacteria.[110] B. bacteriovorus produces a unique, neutral lipid A with α-D-mannopyranose residue substitutions at positions C-1 and C-4’. It also incorporates unsaturated and branched chain fatty acyl groups in the lipophilic domain.[111] (Red and blue colors were used for contrast only and do not denote any physicochemical properties. Bond positioning of terminal attachments are arbitrary.)
B. bacteriovorus is a bacterium that survives by predation on other Gram-negative bacteria. It is known to have an “attack” phase, wherein it invades the periplasm of its prey, presumably by means of its inherent motility. Once inside the prey bacterium, it grows, replicates, and ultimately lyses its victim.[111] It has been inferred that Bdellovibrio species are at least partially responsible for bacterial count reduction in the environment and within mammalian gut microbiomes.[113,114] B. bacteriovorus was the first Gram-negative bacterium identified that synthesizes a neutral lipid A (depicted in Figure 4B) for incorporation into its membrane. Unlike most other Gram-negative bacteria, which produce negatively charged lipid A with phosphate moieties at the 1 and 4’ position, B. bacteriovorus adds α-D-mannopyranose residues at those same positions.[111] So far, this is a unique finding in prokaryotes. These neutral modifications are believed to drive the ~1000-fold decrease in inflammatory response from exposed human mononuclear cells as compared to Ec lipid A. Additionally, B. bacteriovorus decorates its lipid A with at least two unsaturated fatty acyl chains and branched fatty acids, producing a large proportion of hexa-acylated structures. The degree of unsaturation and neutral sugar modifications are thought to contribute to a more fluid outer membrane than that found in other mesophilic bacteria.
G. obscuriglobus belongs to the phylum Planctomycetes. Planctomycetes are unique prokaryotes that contain both genotypic and phenotypic characteristics of eukaryotic cells. G. obscuriglobus is the first organism of its phylum to have a putative lipid A structure reported[112], as can be seen in Figure 5A. Most notably, the lipid A structure is mono-phosphorylated at C-1, with a GalA residue modification at the C-4’ position. This lipid A also contains relatively long chain fatty acyl groups, amide linkages at C-3 and C-3’, and an ester linkage at C-2’. These are all unusual modifications with potentially unique enzymes responsible for their existence. It should, however, be noted that more descriptive structural work is required to confirm the structure proposed in this study.
Figure 5. Representative lipid A structures from G. obscuriglobus (A) and Synechococcus sp. CC9311 (B).
G. obscuriglobus produces mono-phosphorylated lipid A with unusual modifications, including an acidic GalA substitution at the C-4’ position and amide-linked fatty acyl chains at the C-3 and C-3’ positions. The ester-linked, β-hydroxyl-substituted myristate group at the C-2’ position is a unique finding among Gram-negative bacteria.[112] Synechococcus sp. CC9311 produces a rudimentary, tetra-acylated lipid A with no phosphate substitutions. It also incorporates at least one unsaturated fatty acyl group in the lipophilic domain and an acidic GalA substitution at the C-4’ position, as in G. obscuriglobus.[116] (Red and blue colors were used for contrast only and do not denote any physicochemical properties. Bond positioning of terminal attachments are arbitrary.)
Cyanobacteria
Cyanobacteria are ubiquitous, heterotrophic, photosynthetic Gram-negative organisms. Many species are found in marine environments and they are the most abundant phytoplankton phylum on earth. Endotoxin structures from cyanobacteria were reviewed in 2015[115], and the authors refer the reader to that publication for more information about specific components and physiology. However, very few complete structures have been elucidated. Interestingly, the LPS structure reported for Synechococcus strains contains mostly glucose as the saccharide component of core, with no heptose or KDO sugars detected. Additionally, no phosphate modifications were detected. This is supported by the absence of a 4’ kinase.[116] The authors propose that all of these observations are indications of earlier inception of LPS synthesis than in enteric bacteria.[116] It is certain that cyanobacteria have been identified in earlier strata than their enteric cousins. These findings are in stark contrast to the aforementioned G. obscuriglobus, and a comparison is presented in Figure 5. Marine cyanobacterial LPS structures (or structural components) have been identified in Synechococcus[116], Microcystis[117], Anacystis[118], Agmenellum[119], Shizothrix[120], Anabaena[121], Spirulina[122], and Oscillatoria[123] species. All of the species reported to date make unique endotoxins, and presumably have unique endotoxin synthesis and modifying enzymes. Some of these structures include long fatty acyl chains with varying degrees of unsaturation and all lacking phosphate modifications as in Synechococcus. Most of the structures reported have not been directly supported in the literature, thus far, with genomic, transcriptomic, or proteomic data.
Psychrophiles and Thermophiles
Lipid A structures have been reported for five cold water-adapted (psychrophilic) bacteria and one hot water-adapted (thermophilic) bacterium[124] including two strains of the Antarctic isolate psychrophile Pseudoalteromonas haloplanktis.[125,126] In both strains, mass spectra of lipid A extracts produced base peaks due to a bis-phosphorylated, penta-acylated structure containing saturated, C12:0 acyl chains, including three β-hydroxyl substituted chains. However, the location of the only secondary acyl chain differed by occurring as part of an acyloxyamide in the TAB 23[126] strain and as an acyloxyacyl in the TAC 125[125] strain. One interesting observation in P. haloplanktis is that its overall acyl chain incorporation, when taking into account all structures available, tends to be highly heterogeneous. This can be inferred from the many ions at differing m/z values observed in its MALDI-TOF mass spectrum as well as the differences in structures between strains of the same species. However, this is not due to an abnormal number of acyltransferase domains in the genome, which indicates that its acyltransferases are more promiscuous than most and/or they have multiple proteoforms not coded for in the genome. One advantage of using promiscuous acyltransferases in a lipid A-by-design biosynthesis (in an organism that makes many different fatty acids) would be an increased number of compounds synthesized per unit time.
Lipid As from the obligate psychrophile Psychromonas marina and the psychrotolerant Psychrobacter cryohalolentis were characterized in 2014.[127] In both of these species and the aforementioned P. haloplanktis, only unusual acyl chain incorporations were observed due to the analytical experimental design. In P. marina, a very unusual C14:2 fatty acid methyl ester was observed in the gas chromatograph/mass spectrometer (GC/MS) analysis of hydrolyzed fatty acyl chains. It is unclear whether this is an adaptation to low temperatures. In P. cryohalolentis lipid A extracts, as in P. haloplanktis, a high acyl variability was observed including shorter chain fatty acyls and odd chain fatty acyls. The diversity of acyl forms may allow this bacterium to adapt to a wide range of temperatures, while the short chain lengths and odd chains may allow it to maintain a fluid membrane at low temperatures. In fact, lipid A acyl structure was shown to be temperature dependent in P. cryohalolentis.[128] The P. cryohalolentis genome does not contain an unusual number of Lpx-type acyltransferases, so it is likely that the heterogeneity and unusual fatty acyl chains incorporated in its lipid A are a result of enzyme promiscuity. The highest abundance lipid A form observed in P. cryohalolentis is a bis-phosphorylated, hexa-acylated form with C10:0 and C12:0 fatty acyl chains, including β-hydroxyl substituted chains.
In the thermophile Thermomonas hydrothermalis galacturonic acid (GalA) residues were reported as modifications to the backbone phosphates.[124] These GalA residues probably contribute to the observed low immunostimulatory activity in human MD2/TLR4 models, as compared to E. coli. The acylation profile, inferred from mass spectra of lipid A extracts and fatty acid methyl ester analysis, resulted in a putative hexa-acylated structure including only undecylic acid (C11:0) residues. The C11:0 chains were covalently bound through both ester and amide linkages on the commonly observed diglucosamine lipid A backbone, and some were β-hydroxyl substituted. These shorter fatty acyl substitutions provide another possibility for differential inflammatory response, as compared to E. coli. Additionally, one of the postulated mechanisms by which T. hydrothermalis is able to survive temperatures up to 50°C is a highly negatively charged LOS structure that enables it to form ionic bonds with divalent cations common on the membrane surface. The additional charge at physiological pH is due to the two GalA residues attached to the lipid A moiety as well as an additional phosphate residue and acidic sugar moiety in the core region.
Halophiles
Lipid A structures for four halophilic bacteria have been published for Pseudoalteromonas issachenkonii[129], Halomonas magadiensis[130], Salinivibrio sharmensis[131], and Halomonas pantelleriensis.[132] All of these species synthesize lipid As that are, on average, hypo-acylated and/or comprise shorter fatty acyl chains when compared with lipid A from E. coli. However, it should be noted that, for both Halomonas species, an acid hydrolysis step was used to liberate lipid A from full length LPS. So, it is impossible to know whether the net hypo-acylation is an artifact of ester hydrolysis or a true depiction of lipid A distribution in the cell membrane. The lipid As (after acid hydrolysis of LPS) from both Halomonas species also produced a weaker inflammatory response, measured by TLR4 induced cytokine production, when compared to LPS from E. coli. All of the halophilic bacteria studied to date produce bis-phosphorylated lipid A.
Exotic Lipid A Modifying Enzymes
Non-terrestrial Gram-negative bacteria have genes that code for the enzymes described in the canonical lipid A synthesis pathway.[1] However, none of these enzymes were found with sufficient description in the literature for non-terrestrial organisms whose lipid A structures have been elucidated to date. P. haloplanktis has been fully sequenced.[133] However genes coding for lipid A synthesis or modification were not specifically reported. The fact that odd-chain fatty acids and/or short-chain fatty acids, with varying degrees of unsaturation, and unusual decorations at the O-1 and O-4’ positions are incorporated into some lipid As from non-terrestrial bacteria is cause for further exploration of their parent organisms’ genomes, transcriptomes, and proteomes. It is a relatively safe assumption that new homologs of the lipid A synthesis genes will be discovered (Figure 3) that will be useful in the lipid A-by-design toolbox.
Structural Characterization
Historically, lipid A has been structurally assigned after liberation from full length LPS using a combined analytical approach. At first, lipid A structures were postulated from mass-to-charge ratio (m/z) obtained from matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectra, and fatty acyl components were liberated from the GlcN backbone and separately analyzed by a GC/MS or gas chromatograph-flame ionization detector (GC/FID) to strengthen structural inferences. The analytical methodologies utilized have gradually become more sophisticated due to both advancements in instrumentation and greater adoption of state-of-the-art techniques. The most widely used method begins with the extraction of LPS followed by a separate hydrolysis step to liberate lipid A and electrospray ionization tandem mass spectrometry (ESI-MS/MS) to obtain product ion spectra. Often, ion trap instruments have been used for these experiments because of their tandem-in-time configuration. This allows the analyst to perform multiple, sequential tandem events (MSn) using the same original pool of ions. Ultimately, this achieves a relatively simple to interpret, data rich set of tandem mass spectra that can then be used to infer primary chemical structure of the original precursor ion isolated. Several different dissociation methods, including collision-induced dissociation (CID)[134], ultraviolet photodissociation (UVPD)[135], higher-energy collisional dissociation (HCD), and combinations thereof, have been used in an attempt to disambiguate tandem mass spectra and increase probability of correct assignment.
Recently, efforts have been directed toward top-down analysis of full length rough-type LPS (rLPS, or LOS).[136] Advantages to these methods include reduced sample preparation time, increased species and strain specificity, greater clarity with regard to which chemical structures (rather than components of structures) actually exist in the membrane, and a greater overall understanding of how a bacterium survives in a given environment. Regarding the latter, one example is that many marine bacteria have greater net negative charge associated with their membranes. It is thought that this charge stabilizes the membrane in harsh environments by allowing more ionic interactions between divalent cations and anionic modifications to the membrane glycolipid. In general, both quantity and quality of data are increased by the analysis of intact biological molecules when it is feasible to do so.
To date, lipid As have been presumed too difficult to separate by liquid chromatography coupled to mass spectrometry (LC/MS), so most published research in this area has been conducted either by MALDI-TOF or direct infusion ESI-MS/MS. These are not ideal approaches to structural assignment because lipid A extracts are always complex mixtures. Therefore, tandem mass spectra acquired are never produced by a single precursor ion. Recently, more thorough attempts have been made at chromatographic separation of both lipid A[137] and rLPS[136] with great success. Every extract analyzed thus far has been quite heterogeneous. This is an important finding because proper structure-activity relationships cannot be established through the administration of mixtures. It should also change a common perception in the field that a single characterized lipid A structure from a bacterium is sufficient for drawing structure-driven biological conclusions. It is, in fact, impossible to know which molecules elicit which biological response without administering them one at a time. However, it is often possible and sometimes proper to derive species-driven biological conclusions about activity using the widely adopted mixture administration workflow. As is the case for all biological sciences research, along with advances in analytical technology and methods have come great advances in general understanding about biological processes on the molecular level.
Conclusions/Perspectives
The emerging uses for TLR4-binding therapeutics are growing rapidly as both the role of TLR4- mediated inflammation in pathogenesis and customized control of downstream signaling events become more apparent. Through customization of lipid A structure, TLR4-modulating therapies can be engineered to alter disease course. Here we have covered lipid A biosynthesis and modifying enzymes and unique potential sources for their discovery outside the human and zoonotic pathogens previously curated for lipid A-based drugs. By expanding the structural diversity of lipid A therapeutic candidates the potential applications increase.
Lipid A structural elucidation is accessible through current methods as discussed herein. However, isolation of pure product for structural characterization and drug discovery is paramount. For instance, characterizing the SAR of lipid A with the MD-2/TLR4 complex demands individual structure. Much of our understanding of the biological activity caused by lipid A administration is based on stimulations with mixed lipid A products (biological extracts). While this may achieve acceptable results for regulatory acceptance criteria, rational drug design efforts would be greatly improved through establishment of fundamental rules about structure-function. In addition, we propose that administering a single active ingredient would reduce off-target effects. Batch-to-batch reproducibility would be easier to achieve in the scale-up manufacturing process as well. With the continual advancements in separation science and analytical capability these goals should be increasingly achievable.
Highlights.
Biosynthesis of lipid A is reviewed in the context of structure modification
Contributions of lipid A moieties to TLR4 activity are reviewed for immunomodulatory potential
Novel and inferred modification enzymes are reviewed in the context of drug discovery and design
Advancements in separation and structural characterization of endotoxin is reviewed
Acknowledgments
The authors are grateful to Dr. Erin M. Harberts for generous critical review of this work. This work was supported by the National Institutes of Health [grant numbers R21AI101685, R01GM111066, and R01AI23820-01].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Raetz CRH, Whitfield C. Lipopolysaccharide endotoxins. Annual Review of Biochemistry. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whitfield C, Trent MS. Biosynthesis and export of bacterial lipopolysaccharides. Annual Review of Biochemistry. 2014;83:99–128. doi: 10.1146/annurev-biochem-060713-035600. [DOI] [PubMed] [Google Scholar]
- 3.Whitfield C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annual Review of Biochemistry. 2006;75:39–68. doi: 10.1146/annurev.biochem.75.103004.142545. [DOI] [PubMed] [Google Scholar]
- 4.Sperandeo P, Dehò G, Polissi A. The lipopolysaccharide transport system of Gram-negative bacteria. Biochim. Biophys. Acta. 2009;1791:594–602. doi: 10.1016/j.bbalip.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 5.Greenfield LK, Whitfield C. Synthesis of lipopolysaccharide O-antigens by ABC transporter-dependent pathways. Carbohydrate Research. 2012;356:12–24. doi: 10.1016/j.carres.2012.02.027. [DOI] [PubMed] [Google Scholar]
- 6.White KA, Lin S, Cotter RJ, Raetz CR. A Haemophilus influenzae gene that encodes a membrane bound 3-deoxy-D-manno-octulosonic acid (Kdo) kinase. Possible involvement of kdo phosphorylation in bacterial virulence. The Journal of Biological Chemistry. 1999;274:31391–31400. doi: 10.1074/jbc.274.44.31391. [DOI] [PubMed] [Google Scholar]
- 7.Reynolds CM, Kalb SR, Cotter RJ, Raetz CRH. A phosphoethanolamine transferase specific for the outer 3-deoxy-D-manno-octulosonic acid residue of Escherichia coli lipopolysaccharide. Identification of the eptB gene and Ca2+ hypersensitivity of an eptB deletion mutant. The Journal of Biological Chemistry. 2005;280:21202–21211. doi: 10.1074/jbc.M500964200. [DOI] [PubMed] [Google Scholar]
- 8.Gunn JS. Bacterial modification of LPS and resistance to antimicrobial peptides. J. Endotoxin Res. 2001;7:57–62. [PubMed] [Google Scholar]
- 9.Trent MS, Stead CM, Tran AX, Hankins JV. Diversity of endotoxin and its impact on pathogenesis. J. Endotoxin Res. 2006;12:205–223. doi: 10.1179/096805106X118825. [DOI] [PubMed] [Google Scholar]
- 10.Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 11.Poltorak A, Ricciardi-Castagnoli P, Citterio S, Beutler B. Physical contact between lipopolysaccharide and Toll-like receptor 4 revealed by genetic complementation. Proceedings of the National Academy of Sciences. 2000;97:2163–2167. doi: 10.1073/pnas.040565397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beutler B, Du X, Poltorak A. Identification of Toll-like receptor 4 (Tlr4) as the sole conduit for LPS signal transduction: genetic and evolutionary studies. 2001 [PubMed] [Google Scholar]
- 13.Hoshino K, Takeuchi O, Kawai T, Sanjo H, Ogawa T, Takeda Y, et al. Cutting Edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: Evidence for TLR4 as the Lps gene product. Journal of Immunology. 1999 [PubMed] [Google Scholar]
- 14.Mackay IR, Rosen FS, Medzhitov R, Janeway C., Jr Innate immunity. N. Engl. J. Med. 2000;343:338–344. doi: 10.1056/NEJM200008033430506. [DOI] [PubMed] [Google Scholar]
- 15.Raetz CRH, Ulevitch RJ, Wright SD, Sibley CH, Ding A, Nathan CF. Gram-negative endotoxin: an extraordinary lipid with profound effects on eukaryotic signal transduction1. FASEB Journal. 1991;5:2652–2660. doi: 10.1096/fasebj.5.12.1916089. [DOI] [PubMed] [Google Scholar]
- 16.Akira S, Takeda K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 17.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 18.Cusson-Hermance N, Khurana S, Lee TH, Fitzgerald KA, Kelliher MA. Rip1 mediates the Trif-dependent toll-like receptor 3- and 4-induced NF-{kappa}B activation but does not contribute to interferon regulatory factor 3 activation. The Journal of Biological Chemistry. 2005;280:36560–36566. doi: 10.1074/jbc.M506831200. [DOI] [PubMed] [Google Scholar]
- 19.Mata-Haro V, Cekic C, Martin M, Chilton PM, Casella CR, Mitchell TC. The vaccine adjuvant monophosphoryl lipid A as a TRIF-biased agonist of TLR4. Science. 2007;316:1628–1632. doi: 10.1126/science.1138963. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Wang Z, Chen J, Ernst R, Wang X. Influence of lipid A acylation pattern on membrane permeability and innate immune stimulation. Marine Drugs. 2013;11:3197–3208. doi: 10.3390/md11093197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Needham BD, Carroll SM, Giles DK, Georgiou G, Whiteley M, Trent MS. Modulating the innate immune response by combinatorial engineering of endotoxin. Proceedings of the National Academy of Sciences. 2013;110:1464–1469. doi: 10.1073/pnas.1218080110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawai T, Adachi O, Ogawa T, Takeda K, Akira S. Unresponsiveness of MyD88-deficient mice to endotoxin. Immunity. 1999;11:115–122. doi: 10.1016/s1074-7613(00)80086-2. [DOI] [PubMed] [Google Scholar]
- 23.David MD, Cochrane CL, Duncan SK, Schrader JW. Pure Lipopolysaccharide or Synthetic Lipid A Induces Activation of p21Ras in Primary Macrophages through a Pathway Dependent on Src Family Kinases and PI3K. Journal of Immunology (Baltimore, Md. : 1950) 2005;175:8236–8241. doi: 10.4049/jimmunol.175.12.8236. [DOI] [PubMed] [Google Scholar]
- 24.Dobrovolskaia MA, Vogel SN. Toll receptors, CD14, and macrophage activation and deactivation by LPS. Microbes and Infection. 2002;4:903–914. doi: 10.1016/s1286-4579(02)01613-1. [DOI] [PubMed] [Google Scholar]
- 25.Park BS, Song DH, Kim HM, Choi B-S, Lee H, Lee J-O. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458:1191–1195. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 26.Jin MS, Lee JO. Structures of the Toll-like receptor family and its ligand complexes. Immunity. 2008 doi: 10.1016/j.immuni.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 27.Kang JY, Lee J-O. Structural biology of the Toll-like receptor family. Annual Review of Biochemistry. 2011;80:917–941. doi: 10.1146/annurev-biochem-052909-141507. [DOI] [PubMed] [Google Scholar]
- 28.Miller SI, Ernst RK, Bader MW. LPS, TLR4 and infectious disease diversity. Nature Reviews Microbiology. 2005;3:36–46. doi: 10.1038/nrmicro1068. [DOI] [PubMed] [Google Scholar]
- 29.Hajjar AM, Ernst RK, Fortuno ES, III, Brasfield AS, Yam CS, Newlon LA, et al. Humanized TLR4/MD-2 mice reveal LPS recognition differentially impacts susceptibility to Yersinia pestis and Salmonella enterica. 2012;8:e1002963. doi: 10.1371/journal.ppat.1002963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kovach NL. Lipid IVA inhibits synthesis and release of tumor necrosis factor induced by lipopolysaccharide in human whole blood ex vivo. Journal of Experimental Medicine. 1990;172:77–84. doi: 10.1084/jem.172.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohto U, Fukase K, Miyake K, Satow Y. Crystal structures of human MD-2 and its complex with antiendotoxic lipid IVa. Science. 2007;316:1632–1634. doi: 10.1126/science.1139111. [DOI] [PubMed] [Google Scholar]
- 32.Kim HM, Park BS, Kim JI, Kim SE, Lee J, Oh SC. Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist Eritoran. Cell. 2007;130:906–917. doi: 10.1016/j.cell.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 33.Kobayashi M, Saitoh SI, Tanimura N, Takahashi K, Kawasaki K, Nishijima M, et al. Regulatory roles for MD-2 and TLR4 in ligand-induced receptor clustering. The Journal of Immunology. 2006;176:6211–6218. doi: 10.4049/jimmunol.176.10.6211. [DOI] [PubMed] [Google Scholar]
- 34.Anwar MA, Panneerselvam S, Shah M, Choi S. Insights into the species-specific TLR4 signaling mechanism in response to Rhodobacter sphaeroides lipid A detection. Sci. Rep. 2015;5:7657. doi: 10.1038/srep07657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu L, Phillips RL, Zhang D, Teghanemt A, Weiss JP, Gioannini TL. NMR studies of hexaacylated endotoxin bound to wild-type and F126A mutant MD-2 and MD-2{middle dot}TLR4 ectodomain complexes. Journal of Biological Chemistry. 2012;287:16346–16355. doi: 10.1074/jbc.M112.343467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moskowitz SM, Ernst RK. The role of Pseudomonas lipopolysaccharide in cystic fibrosis airway infection. Subcell. Biochem. 2010;53:241–253. doi: 10.1007/978-90-481-9078-2_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Billod J-M, Lacetera A, Guzmán-Caldentey J, Martín-Santamaría S. Computational Approaches to Toll-Like Receptor 4 Modulation. Molecules. 2016;21 doi: 10.3390/molecules21080994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raetz CRH. Biochemistry of endotoxins. Annual Review of Biochemistry. 1990;59:129–170. doi: 10.1146/annurev.bi.59.070190.001021. [DOI] [PubMed] [Google Scholar]
- 39.Raetz CRH. Bacterial endotoxins: Extraordinary lipids that activate eucaryotic signal transduction. Journal of Bacteriology. 1993;175:5745–5753. doi: 10.1128/jb.175.18.5745-5753.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dowhan W. The Raetz Pathway for Lipid A Biosynthesis: Christian Rudolf Hubert Raetz, MD PhD, 1946–2011. The Journal of Lipid Research. 2011;52:1857–1860. doi: 10.1194/jlr.E020701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raetz CRH, Reynolds CM, Trent MS, Bishop RE. Lipid A modification systems in gram-negative bacteria. Annual Review of Biochemistry. 2007;76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jackman JE, Fierke CA, Tumey LN, Pirrung M, Uchiyama T, Tahir SH, et al. Antibacterial agents that target lipid A biosynthesis in Gram-negative bacteria. The Journal of Biological Chemistry. 2000;275:11002–11009. doi: 10.1074/jbc.275.15.11002. [DOI] [PubMed] [Google Scholar]
- 43.Bartling CM, Raetz CRH. Crystal structure and acyl chain selectivity of Escherichia coli LpxD, the N-acyltransferase of lipid A biosynthesis. Biochemistry. 2009;48:8672–8683. doi: 10.1021/bi901025v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y, Powell DA, Shaffer SA, Rasko DA, Pelletier MR, Leszyk JD, et al. LPS remodeling is an evolved survival strategy for bacteria. Proc. Natl. Acad. Sci. U.S.a. 2012;109:8716–8721. doi: 10.1073/pnas.1202908109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sweet CR, H WA, Karbarz MJ, Werts C, Kalb SR, Cotter RJ, et al. Enzymatic synthesis of lipid A molecules with four amide-linked acyl chains. Journal of Biological Chemistry. 2004;279:25411–25419. doi: 10.1074/jbc.M400597200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Mourik A, Steeghs L, van Laar J, Meiring HD, Hamstra HJ, van Putten JPM, et al. Altered linkage of hydroxyacyl chains in lipid A of Campylobacter jejuni reduces TLR4 activation and antimicrobial resistance. Journal of Biological Chemistry. 2010;285:15828–15836. doi: 10.1074/jbc.M110.102061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Needham BD, Trent MS. Fortifying the barrier: the impact of lipid A remodelling on bacterial pathogenesis. Nature Reviews Microbiology. 2013;11:467–481. doi: 10.1038/nrmicro3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ernst RK, Yi EC, Guo L, Lim KB, Burns JL, Hackett M, et al. Specific lipopolysaccharide found in cystic fibrosis airway Pseudomonas aeruginosa. Science. 1999;286:1561–1565. doi: 10.1126/science.286.5444.1561. [DOI] [PubMed] [Google Scholar]
- 49.Hajjar AM, Ernst RK, Tsai JH, Wilson CB, Miller SI. Human Toll-like receptor 4 recognizes host-specific LPS modifications. Nat Immunol. 2002;3:354–359. doi: 10.1038/ni777. [DOI] [PubMed] [Google Scholar]
- 50.Aussel L, Thérisod H, Karibian D, Perry MB, Bruneteau M, Caroff M. Novel variation of lipid A structures in strains of different Yersinia species. FEBS Lett. 2000;465:87–92. doi: 10.1016/s0014-5793(99)01722-6. [DOI] [PubMed] [Google Scholar]
- 51.Kawahara K, Tsukano H, Watanabe H, Lindner B, Matsuura M. Modification of the Structure and Activity of Lipid A in Yersinia pestis Lipopolysaccharide by Growth Temperature. Infection and Immunity. 2002;70:4092–4098. doi: 10.1128/IAI.70.8.4092-4098.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rebeil R, Ernst RK, Gowen BB, Miller SI, Hinnebusch BJ. Variation in lipid A structure in the pathogenic yersiniae. Mol. Microbiol. 2004;52:1363–1373. doi: 10.1111/j.1365-2958.2004.04059.x. [DOI] [PubMed] [Google Scholar]
- 53.Phillips NJ, Schilling B, McLendon MK, Apicella MA, Gibson BW. Novel modification of lipid A of Francisella tularensis. Infection and Immunity. 2004;72:5340–5348. doi: 10.1128/IAI.72.9.5340-5348.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schilling B, McLendon MK, Phillips NJ, Apicella MA, Gibson BW. Characterization of lipid A acylation patterns in Francisella tularensis, Francisella novicida, and Francisella philomiragia using multiple-stage mass spectrometry and matrix-assisted laser desorption/ionization on an intermediate vacuum source linear ion trap. Anal. Chem. 2007;79:1034–1042. doi: 10.1021/ac061654e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shaffer SA, Harvey MD, Goodlett DR, Ernst RK. Structural heterogeneity and environmentally regulated remodeling of Francisella tularensis subspecies novicida lipid A characterized by tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 2007;18:1080–1092. doi: 10.1016/j.jasms.2007.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li Y, Wang X, Ernst RK. A rapid one-step method for the characterization of membrane lipid remodeling in Francisella using matrix-assisted laser desorption ionization time-of-flight tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2011;25:2641–2648. doi: 10.1002/rcm.5168. [DOI] [PubMed] [Google Scholar]
- 57.Hajjar AM, Harvey MD, Shaffer SA, Goodlett DR, Sjöstedt A, Edebro H, et al. Lack of in vitro and in vivo recognition of Francisella tularensis subspecies lipopolysaccharide by Toll-like receptors. Infection and Immunity. 2006;74:6730–6738. doi: 10.1128/IAI.00934-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reynolds CM, Ribeiro AA, McGrath SC, Cotter RJ, Raetz CRH, Trent MS. An outer membrane enzyme encoded by Salmonella typhimurium lpxR that removes the 3'-acyloxyacyl moiety of lipid A. The Journal of Biological Chemistry. 2006;281:21974–21987. doi: 10.1074/jbc.M603527200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stead CM, Beasley A, Cotter RJ, Trent MS. Deciphering the unusual acylation pattern of Helicobacter pylori lipid A. Journal of Bacteriology. 2008;190:7012–7021. doi: 10.1128/JB.00667-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang X, Ribeiro AA, Guan Z, McGrath SC, Cotter RJ, Raetz CRH. Structure and biosynthesis of free lipid A molecules that replace lipopolysaccharide in Francisella tularensis subsp. novicida. Biochemistry. 2006;45:14427–14440. doi: 10.1021/bi061767s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sjöstedt A. Tularemia: History, epidemiology, pathogen physiology, and clinical manifestations. Annals of the New York Academy of Sciences. 2007;1105:1–29. doi: 10.1196/annals.1409.009. [DOI] [PubMed] [Google Scholar]
- 62.Hone DM, Powell J, Crowley RW, Maneval D, Lewis GK. Lipopolysaccharide from an Escherichia coli htrB msbB mutant induces high levels of MIP-1 alpha and MIP-1 beta secretion without inducing TNF-alpha and IL-1 beta. J. Hum. Virol. 1998;1:251–256. [PubMed] [Google Scholar]
- 63.Somerville JE, Jr, Cassiano L, Bainbridge B, Cunningham MD, Darveau RP. A novel Escherichia coli lipid A mutant that produces an antiinflammatory lipopolysaccharide. J. Clin. Invest. 1996;97:359. doi: 10.1172/JCI118423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brozek KA, Raetz CR. Biosynthesis of lipid A in Escherichia coli. Acyl carrier protein-dependent incorporation of laurate and myristate. 1990 [PubMed] [Google Scholar]
- 65.Clementz T, Zhou Z, Raetz CR. Function of the Escherichia coli msbB gene, a multicopy suppressor of htrB knockouts, in the acylation of lipid A. Acylation by MsbB follows laurate incorporation by HtrB. The Journal of Biological Chemistry. 1997;272:10353–10360. doi: 10.1074/jbc.272.16.10353. [DOI] [PubMed] [Google Scholar]
- 66.Perry RD, Fetherston JD. Yersinia pestis--etiologic agent of plague. Clinical Microbiology Reviews. 1997 doi: 10.1128/cmr.10.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rebeil R, Ernst RK, Jarrett CO, Adams KN, Miller SI, Hinnebusch BJ. Characterization of late acyltransferase genes of Yersinia pestis and their role in temperature-dependent lipid A variation. Journal of Bacteriology. 2006;188:1381–1388. doi: 10.1128/JB.188.4.1381-1388.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang Y-M, Rock CO. Membrane lipid homeostasis in bacteria. Nature Reviews Microbiology. 2008;6:222–233. doi: 10.1038/nrmicro1839. [DOI] [PubMed] [Google Scholar]
- 69.Mykytczuk NCS, Trevors JT, Leduc LG, Ferroni GD. Fluorescence polarization in studies of bacterial cytoplasmic membrane fluidity under environmental stress. Progress in Biophysics and Molecular Biology. 2007;95:60–82. doi: 10.1016/j.pbiomolbio.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 70.Russell NJ. Mechanisms of thermal adaptation in bacteria: blueprints for survival. Trends in Biochemical Sciences. 1984;9:108–112. [Google Scholar]
- 71.Bishop RE, Gibbons HS, Guina T, Trent MS, Miller SI, Raetz CR. Transfer of palmitate from phospholipids to lipid A in outer membranes of gram-negative bacteria. Embo J. 2000;19:5071–5080. doi: 10.1093/emboj/cdd507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thaipisuttikul I, Hittle LE, Chandra R, Zangari D, Dixon CL, Garrett TA, et al. A divergent Pseudomonas aeruginosa palmitoyltransferase essential for cystic fibrosis-specific lipid A. Mol. Microbiol. 2014;91:158–174. doi: 10.1111/mmi.12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Geurtsen J, Steeghs L, Hove JT, van der Ley P, Tommassen J. Dissemination of lipid A deacylases (pagL) among gram-negative bacteria: identification of active-site histidine and serine residues. The Journal of Biological Chemistry. 2005;280:8248–8259. doi: 10.1074/jbc.M414235200. [DOI] [PubMed] [Google Scholar]
- 74.Vaara M, Nurminen M. Outer membrane permeability barrier in Escherichia coli mutants that are defective in the late acyltransferases of lipid A biosynthesis. Antimicrobial Agents and Chemotherapy. 1999;43:1459–1462. doi: 10.1128/aac.43.6.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Swords WE, Chance DL, Cohn LA, Shao J, Apicella MA, Smith AL. Acylation of the lipooligosaccharide of Haemophilus influenzae and colonization: an htrB mutation diminishes the colonization of human airway epithelial cells. Infection and Immunity. 2002;70:4661–4668. doi: 10.1128/IAI.70.8.4661-4668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bainbridge BW, Coats SR, Pham T-TT, Reife RA, Darveau RP. Expression of a Porphyromonas gingivalis lipid A palmitylacyltransferase in Escherichia coli yields a chimeric lipid A with altered ability to stimulate interleukin-8 secretion. Cellular Microbiology. 2006;8:120–129. doi: 10.1111/j.1462-5822.2005.00605.x. [DOI] [PubMed] [Google Scholar]
- 77.Ranallo RT, Kaminski RW, George T, Kordis AA, Chen Q, Szabo K, et al. Virulence, inflammatory potential, and adaptive immunity induced by Shigella flexneri msbB mutants. Infection and Immunity. 2010;78:400–412. doi: 10.1128/IAI.00533-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gibbons HS, Lin S, Cotter RJ, Raetz CRH. Oxygen requirement for the biosynthesis of the S-2-hydroxymyristate moiety in Salmonella typhimurium Lipid A function of LpxO, a new Fe2+/α-ketoglutarate-dependent dioxygenase homologue. Journal of Biological Chemistry. 2000;275:32940–32949. doi: 10.1074/jbc.M005779200. [DOI] [PubMed] [Google Scholar]
- 79.Hankins JV, Madsen JA, Giles DK, Brodbelt JS, Trent MS. Amino acid addition to Vibrio cholerae LPS establishes a link between surface remodeling in gram-positive and gram-negative bacteria. Proceedings of the National Academy of Sciences. 2012;109:8722–8727. doi: 10.1073/pnas.1201313109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Reife RA, Shapiro RA, Bamber BA, Berry KK, Mick GE, Darveau RP. Porphyromonas gingivalis lipopolysaccharide is poorly recognized by molecular components of innate host defense in a mouse model of early inflammation. Infection and Immunity. 1995;63:4686–4694. doi: 10.1128/iai.63.12.4686-4694.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Martin M, Katz J, Vogel SN, Michalek SM. Differential induction of endotoxin tolerance by lipopolysaccharides derived from Porphyromonas gingivalis and Escherichia coli. Journal of Immunology (Baltimore, Md. : 1950) 2001;167:5278–5285. doi: 10.4049/jimmunol.167.9.5278. [DOI] [PubMed] [Google Scholar]
- 82.Coats SR, Reife RA, Bainbridge BW, Pham TT-T, Darveau RP. Porphyromonas gingivalis lipopolysaccharide antagonizes Escherichia coli lipopolysaccharide at toll-like receptor 4 in human endothelial cells. Infection and Immunity. 2003;71:6799–6807. doi: 10.1128/IAI.71.12.6799-6807.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reife RA, Coats SR, Al Qutub M, Dixon DM, Braham PA, Billharz RJ, et al. Porphyromonas gingivalis lipopolysaccharide lipid A heterogeneity: differential activities of tetra- and penta-acylated lipid A structures on E-selectin expression and TLR4 recognition. Cellular Microbiology. 2006;8:857–868. doi: 10.1111/j.1462-5822.2005.00672.x. [DOI] [PubMed] [Google Scholar]
- 84.Coats SR, Berezow AB, To TT, Jain S, Bainbridge BW, Banani KP, et al. The lipid A phosphate position determines differential host Toll-like receptor 4 responses to phylogenetically related symbiotic and pathogenic bacteria. Infection and Immunity. 2010;79:203–210. doi: 10.1128/IAI.00937-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ernst RK, Guina T, Miller SI. Salmonella typhimurium outer membrane remodeling: role in resistance to host innate immunity. Microbes and Infection. 2001;3:1327–1334. doi: 10.1016/s1286-4579(01)01494-0. [DOI] [PubMed] [Google Scholar]
- 86.Pelletier MR, Casella LG, Jones JW, Adams MD, Zurawski DV, Hazlett KRO, et al. Unique structural modifications are present in the Lipopolysaccharide from Colistin-resistant strains of Acinetobacter baumannii. Antimicrobial Agents and Chemotherapy. 2013;57:4831–4840. doi: 10.1128/AAC.00865-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. The Lancet Infectious Diseases. 2016;16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 88.Ribi EE, Granger DL, Milner KC, Strain SM. Tumor regression caused by endotoxins and mycobacterial fractions. J. Natl. Cancer Inst. 1975;55:1253–1257. doi: 10.1093/jnci/55.5.1253. [DOI] [PubMed] [Google Scholar]
- 89.Ribi E, Cantrell JL, Takayama K, Qureshi N, Peterson J, Ribi HO. Lipid A and immunotherapy. Clinical Infectious Diseases. 1984;6:567–572. doi: 10.1093/clinids/6.4.567. [DOI] [PubMed] [Google Scholar]
- 90.Rockwell CE, Morrison DC, Qureshi N. Lipid A-mediated tolerance and cancer therapy. Adv. Exp. Med. Biol. 2010;667:81–99. doi: 10.1007/978-1-4419-1603-7_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Awasthi S. Toll-like receptor-4 modulation for cancer immunotherapy. Frontiers in Immunology. 2014;5 doi: 10.3389/fimmu.2014.00328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Qureshi N, Takayama K, Ribi E. Purification and structural determination of nontoxic lipid A obtained from the lipopolysaccharide of Salmonella typhimurium. Journal of Biological Chemistry. 1982 [PubMed] [Google Scholar]
- 93.Steinhagen F, Kinjo T, Bode C, Klinman DM. TLR-based immune adjuvants. Vaccine. 2011;29:3341–3355. doi: 10.1016/j.vaccine.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Coler RN, Bertholet S, Moutaftsi M, Guderian JA, Windish HP, Baldwin SL, et al. Development and characterization of synthetic glucopyranosyl lipid adjuvant system as a vaccine adjuvant. PLoS ONE. 2011;6:e16333. doi: 10.1371/journal.pone.0016333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dixon DR, Darveau RP. Lipopolysaccharide heterogeneity: Innate host responses to bacterial modification of lipid A structure. Journal of Dental Research. 2005;84:584–595. doi: 10.1177/154405910508400702. [DOI] [PubMed] [Google Scholar]
- 96.Molteni M, Gemma S, Rossetti C. The role of Toll-like receptor 4 in infectious and noninfectious inflammation. Mediators Inflamm. 2016;2016:6978936. doi: 10.1155/2016/6978936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rossignol DP, Lynn M. TLR4 antagonists for endotoxemia and beyond. Curr Opin Investig Drugs. 2005;6:496–502. [PubMed] [Google Scholar]
- 98.Michaud J-P, Hallé M, Lampron A, Thériault P, Préfontaine P, Filali M, et al. Toll-like receptor 4 stimulation with the detoxified ligand monophosphoryl lipid A improves Alzheimer's disease-related pathology. Proceedings of the National Academy of Sciences. 2013;110:1941–1946. doi: 10.1073/pnas.1215165110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Salkowski CA, Detore G, Franks A, Falk MC, Vogel SN. Pulmonary and hepatic gene expression following cecal ligation and puncture: Monophosphoryl lipid A prophylaxis attenuates sepsis-induced cytokine and chemokine expression and neutrophil infiltration. Infection and Immunity. 1998;66:3569–3578. doi: 10.1128/iai.66.8.3569-3578.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lynn M, Rossignol DP, Wheeler JL, Kao RJ, Perdomo CA, Noveck R, et al. Blocking of responses to endotoxin by E5564 in healthy volunteers with experimental endotoxemia. J. Infect. Dis. 2003;187:631–639. doi: 10.1086/367990. [DOI] [PubMed] [Google Scholar]
- 101.Bennett-Guerrero E, Grocott HP, Levy JH. A Phase II, Double-Blind, Placebo-Controlled, Ascending-Dose. Anesthesia & Analgesia. 2007;104:378–383. doi: 10.1213/01.ane.0000253501.07183.2a. [DOI] [PubMed] [Google Scholar]
- 102.Tse MT. Trial watch: Sepsis study failure highlights need for trial design rethink. Nat Rev Drug Discov. 2013;12:334–334. doi: 10.1038/nrd4016. [DOI] [PubMed] [Google Scholar]
- 103.Pretorius E, Mbotwe S, Bester J, Robinson CJ, Kell DB. Acute induction of anomalous and amyloidogenic blood clotting by molecular amplification of highly substoichiometric levels of bacterial lipopolysaccharide. J R Soc Interface. 2016;13 doi: 10.1098/rsif.2016.0539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Huggins C, Pearce S, Peri F, Neumann F, Cockerill G, Pirianov G. A novel small molecule TLR4 antagonist (IAXO-102) negatively regulates non-hematopoietic toll like receptor 4 signalling and inhibits aortic aneurysms development. Atherosclerosis. 2015;242:563–570. doi: 10.1016/j.atherosclerosis.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 105.Bettoni I, Comelli F, Rossini C, Granucci F, Giagnoni G, Peri F, et al. Glial TLR4 receptor as new target to treat neuropathic pain: efficacy of a new receptor antagonist in a model of peripheral nerve injury in mice. Glia. 2008;56:1312–1319. doi: 10.1002/glia.20699. [DOI] [PubMed] [Google Scholar]
- 106.Peri F, Calabrese V. Toll-like receptor 4 (TLR4) modulation by synthetic and natural compounds: an update. J. Med. Chem. 2014;57:3612–3622. doi: 10.1021/jm401006s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Leone S, Silipo A, Nazarenko EL, Lanzetta R, Parrilli M, Molinaro A. Molecular structure of endotoxins from Gram-negative marine bacteria: an update. Marine Drugs. 2007;5:85–112. doi: 10.3390/md503085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wiens M, Korzhev M, Perovic-Ottstadt S, Luthringer B, Brandt D, Klein S, et al. Toll-like receptors are part of the innate immune defense system of sponges (demospongiae: Porifera) Mol. Biol. Evol. 2007;24:792–804. doi: 10.1093/molbev/msl208. [DOI] [PubMed] [Google Scholar]
- 109.Anwar MA, Choi S. Gram-negative marine bacteria: structural features of lipopolysaccharides and their relevance for economically important diseases. Marine Drugs. 2014;12:2485–2514. doi: 10.3390/md12052485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Phillips NJ, Adin DM, Stabb EV, McFall-Ngai MJ, Apicella MA, Gibson BW. The Lipid A from Vibrio fischeri Lipopolysaccharide: A unique structure bearing a phosphoglycerol moeity. Journal of Biological Chemistry. 2011;286:21203–21219. doi: 10.1074/jbc.M111.239475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schwudke D, Linscheid M, Strauch E, Appel B, Zahringer U, Moll H, et al. The obligate predatory Bdellovibrio bacteriovorus possesses a neutral lipid A containing alpha-D-Mannoses that replace phosphate residues: similarities and differences between the lipid As and the lipopolysaccharides of the wild type strain B. bacteriovorus HD100 and its host-independent derivative HI100. Journal of Biological Chemistry. 2003;278:27502–27512. doi: 10.1074/jbc.M303012200. [DOI] [PubMed] [Google Scholar]
- 112.Mahat R, Seebart C, Basile F, Ward NL. Global and targeted lipid analysis of Gemmata obscuriglobus reveals the presence of Lipopolysaccharide, a signature of the classical Gram- negative outer membrane. J. Bacteriol. 2015;198:221–236. doi: 10.1128/JB.00517-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lambina VA, Ledova LA, Churkina LG. Importance of Bdellovibrio in regulating microbial cenoses and self-purification processes in domestic sewage. Mikrobiologiia. 1987;56:860–864. [PubMed] [Google Scholar]
- 114.Schwudke D, Strauch E, Krueger M, Appel B. Taxonomic studies of predatory Bdellovibrios based on 16S rRNA analysis, ribotyping and the hit locus and characterization of isolates from the gut of animals. Systematic and Applied Microbiology. 2001;24:385–394. doi: 10.1078/0723-2020-00042. [DOI] [PubMed] [Google Scholar]
- 115.Durai P, Batool M, Choi S. Structure and Effects of Cyanobacterial Lipopolysaccharides. Marine Drugs. 2015;13:4217–4230. doi: 10.3390/md13074217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Snyder DS, Brahamsha B, Azadi P, Palenik B. Structure of Compositionally Simple Lipopolysaccharide from Marine Synechococcus. J. Bacteriol. 2009;191:5499–5509. doi: 10.1128/JB.00121-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Raziuddin S, Siegelman HW, Tornabene TG. Lipopolysaccharides of the cyanobacterium Microcystis aeruginosa. Eur. J. Biochem. 1983;137:333–336. doi: 10.1111/j.1432-1033.1983.tb07833.x. [DOI] [PubMed] [Google Scholar]
- 118.Weise G, Drews G, Jann B, Jann K. Identification and analysis of a lipopolysaccharide in cell walls of the blue-green alga Anacystis nidulans. Arch Mikrobiol. 1970;71:89–98. doi: 10.1007/BF00412238. [DOI] [PubMed] [Google Scholar]
- 119.Buttke TM, Ingram LO. Comparison of lipopolysaccharides from Agmenellum quadruplicatum to Escherichia coli and Salmonella typhimurium by using thin-layer chromatography. J. Bacteriol. 1975;124:1566–1573. doi: 10.1128/jb.124.3.1566-1573.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Keleti G, Sykora JL, Lippy EC, Shapiro MA. Composition and biological properties of lipopolysaccharides isolated from Schizothrix calcicola (Ag.) Gomont (Cyanobacteria) Appl. Environ. Microbiol. 1979;38:471–477. doi: 10.1128/aem.38.3.471-477.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Keleti G, Sykora JL. Production and properties of cyanobacterial endotoxins. Appl. Environ. Microbiol. 1982;43:104–109. doi: 10.1128/aem.43.1.104-109.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tornabene TG, Bourne TF, Raziuddin S, Ben-Amotz A. Lipid and lipopolysaccharide constituents of cyanobacterium Spirulina platensis (Cyanophyceae, Nostocales) Marine Ecology. Progress Series. 1985;22:121–125. [Google Scholar]
- 123.Carillo S, Pieretti G, Bedini E, Parrilli M, Lanzetta R, Corsaro MM. Structural investigation of the antagonist LPS from the cyanobacterium Oscillatoria planktothrix FP1. Carbohydrate Research. 2014;388:73–80. doi: 10.1016/j.carres.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 124.Di Lorenzo F, Paciello I, Fazio LL, Albuquerque L, Sturiale L, da Costa MS, et al. Thermophiles as potential source of novel endotoxin antagonists: the full structure and bioactivity of the lipo-oligosaccharide from Thermomonas hydrothermalis. ChemBioChem. 2014;15:2146–2155. doi: 10.1002/cbic.201402233. [DOI] [PubMed] [Google Scholar]
- 125.Corsaro MM, Piaz FD, Lanzetta R, Parrilli M. Lipid A structure of Pseudoalteromonas haloplanktis TAC 125: use of electrospray ionization tandem mass spectrometry for the determination of fatty acid distribution. J. Mass. Spectrom. 2002;37:481–488. doi: 10.1002/jms.304. [DOI] [PubMed] [Google Scholar]
- 126.Carillo S, Pieretti G, Parrilli E, Tutino ML, Gemma S, Molteni M, et al. Structural investigation and biological activity of the lipooligosaccharide from the psychrophilic bacterium Pseudoalteromonas haloplanktis TAB 23. Chemistry. 2011;17:7053–7060. doi: 10.1002/chem.201100579. [DOI] [PubMed] [Google Scholar]
- 127.Sweet C, Alpuche G, Landis C, Sandman B. Endotoxin Structures in the Psychrophiles Psychromonas marina and Psychrobacter cryohalolentis Contain Distinctive Acyl Features. Marine Drugs. 2014;12:4126–4147. doi: 10.3390/md12074126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sweet C, Watson R, Landis C, Smith J. Temperature-Dependence of Lipid A Acyl Structure in Psychrobacter cryohalolentis and Arctic Isolates of Colwellia hornerae and Colwellia piezophila. Marine Drugs. 2015;13:4701–4720. doi: 10.3390/md13084701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Silipo A, Leone S, Lanzetta R, Parrilli M, Sturiale L, Garozzo D, et al. The complete structure of the lipooligosaccharide from the halophilic bacterium Pseudoalteromonas issachenkonii KMM 3549T. Carbohydrate Research. 2004;339:1985–1993. doi: 10.1016/j.carres.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 130.Ialenti A, Di Meglio P, Grassia G, Maffia P, Di Rosa M, Lanzetta R, et al. A novel lipid A from Halomonas magadiensis inhibits enteric LPS-induced human monocyte activation. Eur. J. Immunol. 2006;36:354–360. doi: 10.1002/eji.200535305. [DOI] [PubMed] [Google Scholar]
- 131.Carillo S, Pieretti G, Lindner B, Romano I, Nicolaus B, Lanzetta R, et al. The Lipid A from the haloalkaliphilic bacterium Salinivibrio sharmensis strain BAG(T) Marine Drugs. 2013;11:184–193. doi: 10.3390/md11010184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Carillo S, Pieretti G, Casillo A, Lindner B, Romano I, Nicolaus B, et al. Structural characterization of the lipid A from the LPS of the haloalkaliphilic bacterium Halomonas pantelleriensis. Extremophiles. 2016:1–8. doi: 10.1007/s00792-016-0858-2. [DOI] [PubMed] [Google Scholar]
- 133.Médigue C, Krin E, Pascal G, Barbe V, Bernsel A, Bertin PN, et al. Coping with cold: the genome of the versatile marine Antarctica bacterium Pseudoalteromonas haloplanktis TAC125. Genome Res. 2005;15:1325–1335. doi: 10.1101/gr.4126905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Murphy RC, Raetz CRH, Reynolds CM, Barkley RM. Mass spectrometry advances in lipidomica: collision-induced decomposition of Kdo2–lipid A. Prostaglandins & Other Lipid Mediators. 2005;77:131–140. doi: 10.1016/j.prostaglandins.2004.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.O'Brien JP, Needham BD, Henderson JC, Nowicki EM, Trent MS, Brodbelt JS. 193 nm ultraviolet photodissociation mass spectrometry for the structural elucidation of lipid A compounds in complex mixtures. Anal. Chem. 2014;86:2138–2145. doi: 10.1021/ac403796n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.O'Brien JP, Needham BD, Brown DB, Trent MS, Brodbelt JS. Top-Down Strategies for the Structural Elucidation of Intact Gram-negative Bacterial Endotoxins. Chem Sci. 2014;5:4291–4301. doi: 10.1039/C4SC01034E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sándor V, Kilár A, Kilár F, Kocsis B, Dörnyei Á. Characterization of complex, heterogeneous lipid A samples using HPLC-MS/MS technique II. Structural elucidation of non-phosphorylated lipid A by negative-ion mode tandem mass spectrometry. J. Mass. Spectrom. 2016;51:615–628. doi: 10.1002/jms.3786. [DOI] [PubMed] [Google Scholar]