Abstract
Background
Accumulating evidence indicates that the long noncoding RNA, TINCR, plays a critical role in cancer progression and metastasis. However, the overall biological role and mechanisms of TINCR that were involved in human gastric cancer (GC) progression remain largely unknown.
Methods
TINCR expression was measured in 56 paired tumor and adjacent nontumor tissue samples by real-time polymerase chain reaction (PCR). Insights of the mechanism of competitive endogenous RNAs (ceRNAs) were gained from bioinformatic analysis, luciferase assays. The effects of TINCR and miR-375 on GC cell apoptosis and proliferation were studied by RNA interference approaches in vitro and in vivo. The correlation of TINCR and PDK1 was identified by real-time PCR and Western blot analysis.
Results
Our results showed that miR-375 level decreased and TINCR level increased in tumor tissues. In addition, TINCR was a target of miR-375 and inhibited its expression in GC cells. Furthermore, the low expression of TINCR increased cell apoptosis and inhibited the proliferation of GC cells, while the downregulation of miR-375 reversed the function. In particular, TINCR could negatively regulate the miR-375 expression and increased the PDK1 expression in GC cells. Finally, tumor growth suppression was retarded with miR-375 downregulated in TINCR knockdown of GC cell xenografts.
Conclusion
The long noncoding RNA TINCR functions as a competing endogenous RNA to regulate PDK1 expression by sponging miR-375 in GC. The ceRNA regulatory network of TINCR/miR-375/PDK1 allows us to better understand the pathogenesis of GC and facilitate the development of long noncoding RNA (lncRNA)-directed diagnostics in GC.
Keywords: lncRNA TINCR, ceRNA, PDK1, miR-375, gastric cancer
Introduction
Gastric cancer (GC) is the fourth most commonly diagnosed cancer and the second most cancer-related death worldwide.1 An estimated 420,000 patients were diagnosed with GC in 2011 and resulted in almost 300,000 deaths in China.2 An increasing number of studies have revealed that some lncRNAs exhibit multiple gene expression models and have significant roles during cellular development in multiple carcinomas.3–7
Long noncoding RNAs (lncRNAs) are functional RNAs more than 200 nucleotides in length.8 Increasing evidence has found that lncRNAs are important class of universal genes involved in carcinogenesis and metastasis and play multiple roles in regulating gene expression at the transcriptional, post-transcriptional, and epigenetic levels in GC.9 For example, Li et al10 found that the expression of lncRNA H19 was higher in GC and enhanced carcinogenesis and metastasis in GC. Zhang et al11 reported that lncRNA ANRIL indicated a poor prognosis of GC and promoted tumor growth by the epigenetically silencing of miR-99a/miR-449a. In addition, lncRNA SPRY4-IT1 plays a critical role in GC tumorigenesis and may represent a novel prognostic marker and potential therapeutic target in patients with GC.12
TINCR, a terminal differentiation-induced lncRNA producing a 3.7-kb transcript, is isolated from human well differentiated somatic tissues. TINCR interacts with a range of differentiation mRNAs through a specific motif. TINCR also binds to the stau1 protein and mediates the stabilization of differentiation mRNAs.13,14 Although Xu et al15 and Ma et al16 have reported that TINCR was strongly overexpressed in human GC and contributed to carcinogenesis and tumor progression, the specific mechanism that was involved in human GC progression still needed to be further studied.
To further understand the regulatory mechanisms of TINCR in GC progression, we analyzed the expression of TINCR and miR-375 in tumor and adjacent tissue samples from 56 patients with GC. TINCR expression was significantly decreased in GC tumor tissues; conversely, the expression of miR-375 was highly expressed in tumor tissues. Moreover, mechanistic analysis reveals that TINCR may function as a competitive endogenous RNA (ceRNA) to regulate the expression of PDK1 through competition for miR-375, thus playing an oncogenic role in gastric pathogenesis. Taken together, these data contribute to the understanding of the roles and molecular mechanisms of TINCR in GC progression.
Methods
Clinical ethics
Patient consent and Institutional Research Ethics Committee approval (Clinical Ethics Committee of Binzhou Central Hospital) were obtained prior to the use of these clinical materials for research purposes.
Cell lines and culture conditions
Human GC KATO III, NCI-N87, HGC-27, and SNU-1 cell lines were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum. All cell lines were cultured at 37°C in a water-saturated atmosphere with 5% CO2. These cell lines were obtained from the Chinese Academy of Sciences (Shanghai, China).
Tissue samples
Human primary GC tissue samples and paired adjacent nontu-mor tissue samples were obtained from 56 patients diagnosed at the Departments of Gastrointestinal Surgery and Pathology and immediately frozen in liquid nitrogen. Patient consent and Institutional Research Ethics Committee approval were obtained prior to the use of these clinical materials for research purposes.
Cell transfection
TINCR sequences were cloned into pcDNA.3.1 vector. We conducted the transfections by using a Lipofectamine 2000 kit (Thermo Fisher Scientific). Human GC HGC-27 and SNU-1 cell lines were transfected with miR-375 mimic, inhibitor, and negative control, which were purchased from GenePharma (Shanghai, China). miR-375 mimic, inhibitor, and negative control at a final concentration 50 nM were transfected using Lipofectamine® 2000 into cells seeded onto six-well plates. Twenty-four hours after transfection, cells were analysed as required. Small interfering RNAs (siRNAs) were purchased from Santa Cruz Biotechnology Inc. (Dallas, TX, USA). Transfection of siRNA duplexes into GC cells was carried out using Lipofectamine 2000 according to the manufacturer’s instructions. At 24 h posttransfection, cells were harvested using real-time PCR (polymerase chain reaction) or Western blot analysis.
Real-time PCR analysis
Total RNA was extracted from the frozen tissues or GC cells using TRIzol reagent according to the manufacturer’s instruction (TAKALA, Shiga, Japan). Reverse transcription of total RNA into cDNA was performed using reverse transcription reagents (TAKALA). The reaction was at 37°C for 15 min and 85°C for 5 s. And microRNA enrichment was conducted using an miRVana miRNA isolation kit (Ambion, Austin, TX, USA), according to the manufacturer’s instructions. microRNAs were reverse transcribed to cDNA using the TaqMan MicroRNA Reverse Transcription Kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. Real-time quantitative PCR was performed using the TaKaRa SYBRR1 Premix Ex TaqTM II PCR kit (TAKALA) in a Roche Lightcycler 480 instrument (Hoffman-La Roche Ltd., Basel, Switzerland). The expression level was normalized using GAPDH (mRNA) or U6 (microRNA) by the 2−ΔCt method.
Western blot analysis
Human GC HGC-27 and SNU-1 cell lines were harvested, and protein was extracted by using radio immunoprecipitation assay lysis buffer. Total protein (40 µg) was separated on sodium dodecyl sulfate polyacrylamide gel electrophoresis gels and electrophoretically transferred onto a polyvinylidene difluoride (PVDF) membrane (Bio-Rad Laboratories Inc., Hercules, CA, USA). The PVDF membrane was blocked with 5% nonfat dry milk for 30 min, and then, the PVDF membrane was incubated with PDK1 rabbit polyclonal (Abcam, Cambridge, MA, USA), and GAPDH mouse polyclonal (Cell Signalling Technology, Danvers, MA, USA) for overnight at 4°C. The PVDF membrane was washed with TBST for three times and incubated with horseradish peroxidase-linked secondary antibody for another 1 h. The PVDF membranes were washed, and the blots were visualized using ECL chemiluminescence (Bio-Rad Laboratories Inc.). Each experiment was performed in triplicate.
Cell proliferation assay
Cell proliferation was performed with Cell Counting Kit-8 (CCK-8) (Dojindo, Tokyo, Japan). According to the instructions, CCK-8 reagent was added at 0, 24, 48, and 72 h after seeding 3×103 cells/well into 96-well plates and transfected with miR-375 inhibitor or control, and incubated at 37°C for 2 h. The optical density (OD) 450 nm value was detected by using a microplate reader (Bio-Rad Laboratories Inc.).
Cell apoptosis analysis
Annexin V/propidium iodide (PI) staining was performed with the Apoptosis Detection Kit (BD Biosciences, San Jose, CA, USA) according to the manufacturer’s protocol. Cells were trypsinized (without EDTA) and washed twice with PBS. The cells were incubated in binding buffer containing Annexin V-FITC and PI. Stained cells were analyzed by flow cytometry using the CELLQUEST program (Becton Dickinson, Franklin Lakes, NJ, USA). Three independent experiments were performed.
Luciferase reporter assay
The sequence of TINCR containing the predicted miR-375 -binding site; the putative sequences of the binding site then cloned into a pmirGLO Dual-luciferase miRNA target expression vector to form the reporter vector pmirGLO-TINCR-wild type (TINCR-wt). To mutate the putative binding site of miR-375 in TINCR, the sequence of putative binding site was replaced as indicated and was named as pmirGLO-TINCR-mutated type (TINCR-mut). The pmirGLO-TINCR-wt or pmirGLO-TINCR-mut was cotransfected with miR-375 mimic or miR-375 NC into GC cells using Lipofectamine 2000. After 48 h transfection, the luciferase assay was used, a Dual-Luciferase Reporter Assay System (Promega Corporation, Fitchburg, WI, USA), according to the manufacturer’s protocol.
Nude mice model
All animal work was conducted under the institutional guidelines of Shandong Province and approved by the Use Committee for Animal Care (Animal Ethics Committee of Binzhou Central Hospital). Nude mice were purchased from the Vital River Laboratories (Beijing, China). 8×106 HGC-27 cells were transfected with miR-375 inhibitor, siTINCR, or control in replicate. Cells were cultured for 24 h before harvesting for transplantation into the animal. Before injection, cells were pooled and mixed with matrigel (BD Biosciences, San Jose, CA, USA). 3×106 cells were injected subcutaneously into the left flank of nude mice. Measurements were taken weekly, and tumor volumes were calculated using the formula V = length × width2/2. The animals were sacrificed 4 weeks after injection. Pictures were recorded using a Nikon d800 digital camera (Nikon Corporation, Tokyo, Japan).
Statistical analysis
Prism5 (GraphPad, USA) software was performed for statistical analysis, and data that were normally distributed were expressed as mean ± SD. P<0.05 was considered as statistically significant.
Results
The expression of miR-375 and TINCR in GC tissues and cell lines
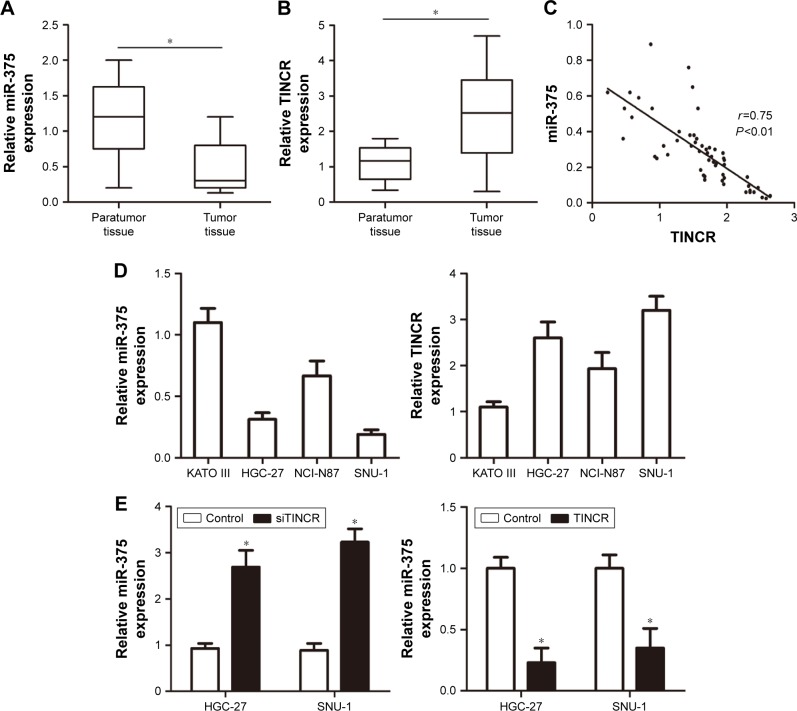
First, the real-time PCR analysis was performed to examine miR-375 and TINCR expressions in primary GC tissues compared with paired adjacent nontumor tissues. The U6 and GAPDH acted as the normalization control. Our results showed that miR-375 expression was downregulated in GC tissues compared with nontumor tissues (Figure 1A). On the contrary, the expression of TINCR was highly expressed in GC tissues compared with nontumor tissues (Figure 1B). Pearson correlation analysis suggested that TINCR expression levels were negatively correlated with the expression levels of miR-375 in GC tissues (Figure 1C). Therefore, we hypothesized that miR-375 was involved in the regulation of TINCR. To investigate the association between miR-375 and TINCR, we checked the expression levels of miR-375 and TINCR in human GC KATO III, NCI-N87, HGC-27, and SNU-1 cell lines by using real-time PCR. The results showed that miR-375 was downregulated and TINCR was upregulated in HGC-27 and SNU-1 cell lines compared with KATO III and NCI-N87 cell lines (Figure 1D). We transfected human GC cells HGC-27 and SNU-1 with TINCR-siRNA and pcDNA3.1-TINCR, and then we tested the expression levels of miR-375 by using real-time PCR. The results revealed that miR-375 was upregulated with TINCR-siRNA transfection and downregulated with pcDNA3.1-TINCR transfection compared to control (Figure 1E).
Figure 1.
The expression of miR-375 and TINCR in clinical GC tissues.
Notes: (A) miR-375 expression was examined by real-time PCR in 56 paired human GC tissues and adjacent noncancerous tissues; the normalized to U6 expression. (B) TINCR expression was examined by real-time PCR in 56 paired human GC tissues and adjacent noncancerous tissues; the normalized to GAPDH expression. (C) The correlation between miR-375 and TINCR expressions in GC cases was evaluated using Spearman’s correlation analysis; n=56; P<0.01. (D) miR-375 and TINCR expressions were examined by real-time PCR in human gastric cancer KATO III, NCI-N87, HGC-27, and SNU-1 cell lines. (E) miR-375 expression was examined by real-time PCR in HGC-27 and SNU-1 cells with TINCR-siRNA and pcDNA3.1-TINCR. Statistically significant differences are indicated: *P<0.05; Student’s t-test. The experiment was repeated at least three times.
Abbreviations: GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GC, gastric cancer; PCR, polymerase chain reaction.
TINCR is a target of miR-375 and inhibited its expression in GC cells
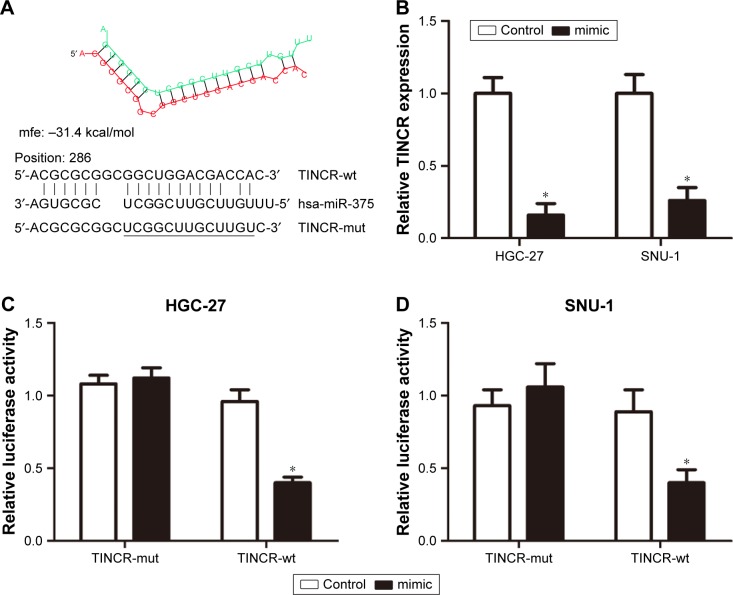
Based on the aforementioned results, we further investigate the correlation between TINCR and miR-375 in GC cells. We performed a search for miRNAs that have complementary base pairing with TINCR, using the online software program RNAhybrid (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid). The results demonstrated that miR-375 formed complementary base pairing with TINCR. According to the prediction results, there existed a putative miR-375-binding site in TINCR (Figure 2A). Moreover, overexpression of miR-375 by transfection with miR-375 mimic led to a substantial decreased expression of TINCR (Figure 2B). Furthermore, to confirm the direct target between TINCR and miR-375, we constructed luciferase reporter vectors. The luciferase activity was detected in HGC-27 and SNU-1 cells after cotransfecting with pmirGLO-TINCR-wt and miR-375 mimic or negative control and pmirGLO-TINCR-mut and miR-375 mimic or negative control. The results clearly indicated that miR-375 mimic significantly suppressed the activity of pmirGLO-TINCR-wt but has no effect on the activity of pmirGLO-TINCR-mut in both HGC-27 and SNU-1 cells (Figure 2C and D). Taken together, these results indicated that TINCR is a target of miR-375 and inhibited its expression in GC cells.
Figure 2.
TINCR was a target of miR-375 in GC cells.
Notes: (A) miR-375-binding sites on TINCR predicted by online soft RNAhybrid. (B) Relative expression of TINCR after transfecting miR-375 mimic into HGC-27 and SNU-1 cells was analyzed by using real-time PCR assays and normalized to GAPDH expression. (C and D) Luciferase activity of the indicated group in HGC-27 and SNU-1 cells by transfected miR-375 mimic + pmirGLO vector, miR-375 mimic + pmirGLO-TINCR-wt, or miR-375 mimic + pmirGLO-TINCR-mut. Statistically significant differences are indicated: *P<0.05; Student’s t-test. The experiment was repeated at least three times.
Abbreviations: GAPDH, glyceraldehyde-3-phosphate dehydrogenase; mut, mutant; GC, gastric cancer; PCR, polymerase chain reaction; wt, wild type.
TINCR mediates depression of PDK1 by blocking miR-375 in GC cells
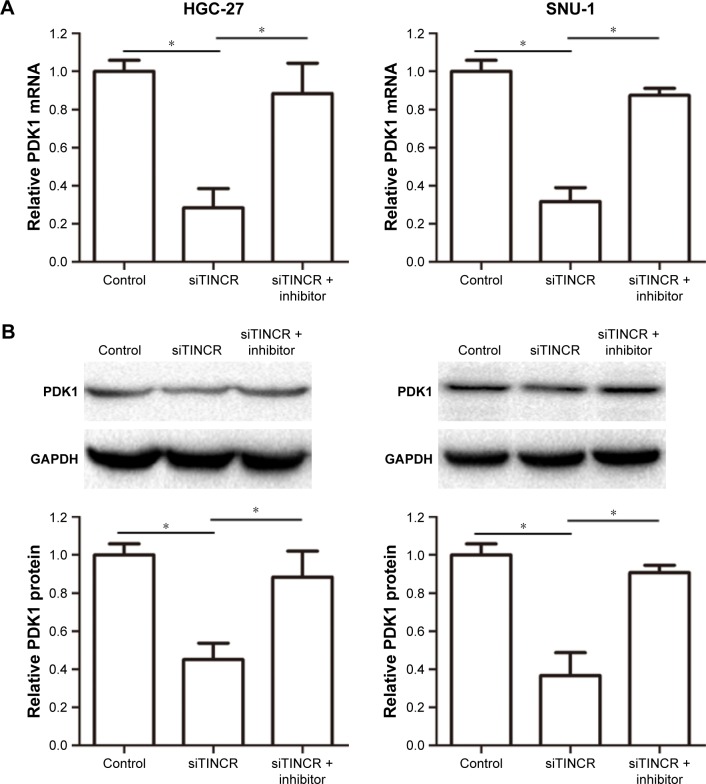
Tsukamoto et al reported that miR-375 overexpression inhibited apoptosis by suppressing caspase activity via targeting PDK1, so we designed experiments to verify whether TINCR could modulate PDK1 expression by targeting miR-375. Our results demonstrated that TINCR knockdown suppressed the PDK1 mRNA and protein expression levels in HGC-27 and SNU-1 cells but was abrogated by cotransfecting siTINCR and miR-375 inhibitor (Figure 3A and B). In general, our results indicated that TINCR could negatively regulate the miR-375 expression and increased the PDK1 expression in GC cells.
Figure 3.
TINCR promoted the PDK1 expression by sponging miR-375.
Notes: (A) Relative mRNA levels of PDK1 were analyzed by using real-time PCR analysis by transfecting negative control, siTINCR, and siTINCR + miR-375 inhibitor in HGC-27 and SNU-1 cells. (B) Relative protein expression levels of PDK1 were analyzed by Western blot analysis by transfecting negative control, siTINCR, and siTINCR + miR-375 inhibitor in HGC-27 and SNU-1 cells. Statistically significant differences are indicated: *P<0.05; Student’s t-test. The experiment was repeated at least three times.
Abbreviations: GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PCR, polymerase chain reaction.
TINCR inhibits cell apoptosis and promotes proliferation by sponging miR-375 in GC cells
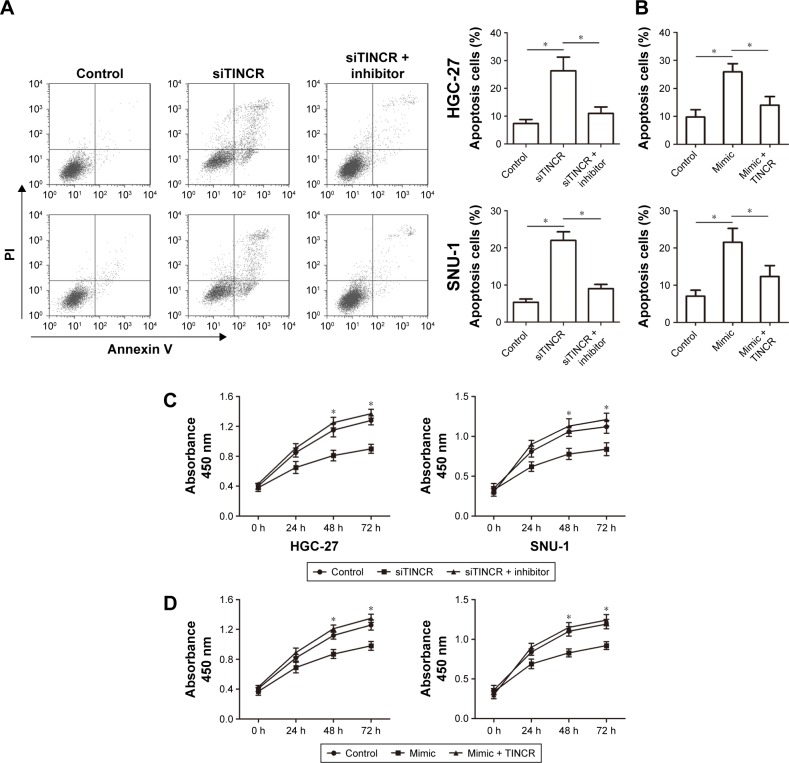
To determine whether TINCR has effects on malignant phenotypes of GC cells by targeting miR-375, we divided our experiments into the following three groups: transfection with siTINCR, transfection with siTINCR and miR-375 inhibitor, and transfection with both negative controls. As shown in Figure 4A, there was low expression of TINCR when transfecting with siTINCR increased cell apoptosis compared to negative control in HGC-27 and SNU-1 cells, whereas it failed to work when cotransfected with siTINCR and miR-375 inhibitor. In addition, the overexpression of miR-375 by transfecting with miR-375 mimic increased cell apoptosis compared to negative control in HGC-27 and SNU-1 cells, whereas it failed to work when cotransfected with mimic and pcDNA3.1-TINCR (Figure 4B). Moreover, the CCK-8 assay revealed that GC cells transiently transfected with siTINCR and had a significantly inhibited proliferation of GC cells but not transfected with negative control or siTINCR and miR-375 inhibitor together (Figure 4C). Additionally, GC cells transiently transfected with mimic and had a significantly inhibited proliferation of GC cells but not transfected with control or mimic and pcDNA3.1-TINCR together (Figure 4D). Thus, our findings verified that TINCR promoted the proliferation of GC cells and inhibited apoptosis by sponging miR-375.
Figure 4.
TINCR inhibited apoptosis and promoted cell proliferation by sponging miR-375.
Notes: (A) Flow cytometry analysis was performed using cells stained with Annexin V-FITC and PI after transfecting negative control, siTINCR, and siTINCR + miR-375 inhibitor in HGC-27 and SNU-1 cells. (B) Flow cytometry analysis was performed using cells stained with Annexin V-FITC and PI after transfecting negative control, mimic, and mimic + TINCR in HGC-27 and SNU-1 cells. (C) CCK-8 cell proliferation assays were analyzed by transfecting negative control, siTINCR, and siTINCR + miR-375 inhibitor in HGC-27 and SNU-1 cells. (D) CCK-8 cell proliferation assays were analyzed by transfecting negative control, mimic, and mimic + TINCR in HGC-27 and SNU-1 cells. Statistically significant differences are indicated: *P<0.05; Student’s t-test. The experiment was repeated at least three times.
Abbreviations: CCK-8, Cell Counting Kit-8; FITC, fluorescein isothiocyanate; PI, propidium iodide.
Tumor growth suppression was retarded with miR-375 downregulated in TINCR knockdown of GC cell xenografts
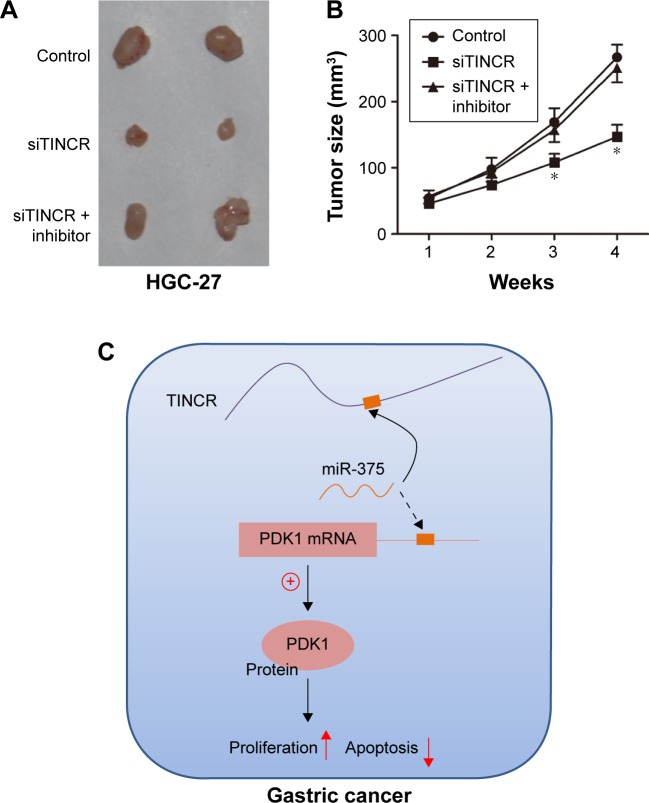
Moreover, we assessed tumor growth of xenografts derived from HGC-27 cells that were transfected with TINCR siRNA or control prior to subcutaneous injection into nude mice. As shown in Figure 5A, the knockdown of TINCR inhibited tumor growth of the HGC-27 xenograft. Furthermore, we next evaluated tumor growth of xenografts derived from HGC-27 cells that were cotransfecting with siTINCR and miR-375 inhibitor or control prior to subcutaneous injection into nude mice and found that tumor growth suppression was retarded with miR-375 downregulated in TINCR knockdown of GC cell xenografts. Moreover, tumor volumes of four time points are shown in Figure 5B. Together, these results supported the presence of a novel regulatory mechanism of miRNAs, lncRNAs, and mRNAs (Figure 5C).
Figure 5.
Tumor growth suppression was retarded with miR-375 downregulated in TINCR knockdown of gastric cancer cell xenografts.
Notes: (A) HGC-27 cells were transfected with negative control, siTINCR, or siTINCR + miR-375 inhibitor and injected subcutaneously into 10 nude mice per flank. Surgical resections of HGC-27 xenograft tumors on week 4 for animals were shown. (B) Measurements of tumor volumes were taken weekly, and tumor volumes were shown. Statistically significant differences are indicated: *P<0.05; Student’s t-test. (C) Schematic model of the TINCR functions as a competing endogenous RNA to regulate PDK1 expression by sponging miR-375 in gastric cancer.
Discussion
Accumulating evidence indicates that the abnormal expression of lncRNAs is a frequent molecular event in human malignances including GC.17–19 Although TINCR is strongly upregulated in GC and could promote cell growth,15,16 the specific mechanism that was involved in human GC progression still needed to be further studied.
LncRNAs and microRNAs represent two classes of important noncoding RNAs in eukaryotes, which have been implicated in various human tumor diseases. Recently, a new regulatory circuitry has been identified that lncRNAs could function as a “sponge” to titrate microRNAs.20 A study reported that MALAT1 upregulated the expression of miR-204 target gene SLUG through competitively “sponging” miR-204 in lung adenocarcinoma.21 Wang et al22 identified that lncRNA H19 contributes to gallbladder cancer cell proliferation by modulated miR-194-5p-targeting AKT2. In our study, we found that the expression of miR-375 decreased and the expression of TINCR increased in tumor compared to adjacent nontumor tissue samples from 56 patients with GC. Pearson correlation analysis also provided evidence that a negative association exists between the expression of miR-375 and TINCR in GC patients. Moreover, miR-375 was upregulated with TINCR-siRNA transfection and downregulated with pcDNA3.1-TINCR transfection. In addition, by the luciferase reporter assays, we showed that TINCR is a direct target of miR-375 in GC cells. Nevertheless, TINCR expression in GC tissue is not downregulated, which might be due to the overexpression of TINCR by other unknown mechanisms. However, RNA pull-down assay is still need to be performed to confirm the association of TINCR and miR-375.
Recent studies found that inhibition of Akt activity by direct targeting of PDK1 is involved in miR-375-induced apoptosis. By further study, they showed that knockdown of endogenous miR-375 increased the expression of PDK1 and was involved in the development of gastric carcinomas.23 Our results revealed that TINCR knockdown suppressed the PDK1 mRNA and protein expression levels in HGC-27 and SNU-1 cells, indicating that TINCR might modulate PDK1 by competing with miR-375. Furthermore, our experimental data showed that TINCR knockdown suppressed the PDK1 mRNA and protein expression levels but were abrogated by cotransfecting siTINCR and miR-375 inhibitor in GC cells, suggesting that TINCR could negatively regulate the miR-375 expression and promoted the PDK1 expression and targeting the TINCR/PDK1 interaction. Therefore, we proposed that the ceRNA activity of TINCR imparted a miRNA/lncRNA trans-regulatory function to regulate the expression of PDK1 in GCs. Moreover, knockdown of TINCR or overexpression of miR-375 inhibited cell proliferation and promoted cell apoptosis in GC cells. Besides, tumor growth suppression was retarded with miR-375 downregulated in TINCR knockdown of GC cell xenografts. Taken together, TINCR could involve in GC progression by sponging the miR-375. Apart from miRNA sponge, more functions of TINCR are still need to be revealed.
Conclusion
It is necessary to put forward that the lncRNA TINCR may sponge a handful of miRNAs at once, while one miRNA can also control a variety of genes. Therefore, the role of TINCR, which is dependent on targeting various targets simultaneously, may be multiple in GC. We have already verified that TINCR functioned as ceRNAs to regulate PDK1 expression by sponging miR-375 in GC, allowing us to better understand how TINCR function was involved in the progression of GC and ultimately provide a new target for lncRNA-based therapeutics in GC.
Acknowledgments
This work was supported by a grant from the Innovation Project of Education Department of Jiangsu Province (No 3231005701).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Baniak N, Senger JL, Ahmed S, Kanthan SC, Kanthan R. Gastric biomarkers: a global review. World J Surg Oncol. 2016;14(1):212. doi: 10.1186/s12957-016-0969-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun W, Yan L. Gastric cancer: current and evolving treatment landscape. Chin J Cancer. 2016;35(1):83. doi: 10.1186/s40880-016-0147-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pibouin L, Villaudy J, Ferbus D, et al. Cloning of the mRNA of overexpression in colon carcinoma-1: a sequence overexpressed in a subset of colon carcinomas. Cancer Genet Cytogenet. 2002;133(1):55–60. doi: 10.1016/s0165-4608(01)00634-3. [DOI] [PubMed] [Google Scholar]
- 5.Fu X, Ravindranath L, Tran N, Petrovics G, Srivastava S. Regulation of apoptosis by a prostate-specific and prostate cancer-associated noncoding gene, PCGEM1. DNA Cell Biol. 2006;25(3):135–141. doi: 10.1089/dna.2006.25.135. [DOI] [PubMed] [Google Scholar]
- 6.Calin GA, Liu CG, Ferracin M, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12(3):215–229. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 7.Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007;26(6):851–858. doi: 10.1038/sj.onc.1209846. [DOI] [PubMed] [Google Scholar]
- 8.Fang XY, Pan HF, Leng RX, Ye DQ. Long noncoding RNAs: novel insights into gastric cancer. Cancer Lett. 2015;356(2 pt B):357–366. doi: 10.1016/j.canlet.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 9.Zhao J, Liu Y, Huang G, Cui P, Zhang W, Zhang Y. Long non-coding RNAs in gastric cancer: versatile mechanisms and potential for clinical translation. Am J Cancer Res. 2015;5(3):907–927. [PMC free article] [PubMed] [Google Scholar]
- 10.Li H, Yu B, Li J, et al. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5(8):2318–2329. doi: 10.18632/oncotarget.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang EB, Kong R, Yin DD, et al. Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR-99a/miR-449a. Oncotarget. 2014;5(8):2276–2292. doi: 10.18632/oncotarget.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peng W, Wu G, Fan H, Wu J, Feng J. Long noncoding RNA SPRY4-IT1 predicts poor patient prognosis and promotes tumorigenesis in gastric cancer. Tumour Biol. 2015;36(9):6751–6758. doi: 10.1007/s13277-015-3376-4. [DOI] [PubMed] [Google Scholar]
- 13.Kretz M, Siprashvili Z, Chu C, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493(7431):231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kretz M. TINCR, staufen1, and cellular differentiation. RNA Biol. 2013;10(10):1597–1601. doi: 10.4161/rna.26249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu TP, Liu XX, Xia R, et al. SP1-induced upregulation of the long noncoding RNA TINCR regulates cell proliferation and apoptosis by affecting KLF2 mRNA stability in gastric cancer. Oncogene. 2015;34(45):5648–5661. doi: 10.1038/onc.2015.18. [DOI] [PubMed] [Google Scholar]
- 16.Ma X, Huang C, Luo D, et al. Tag SNPs of long non-coding RNA TINCR affect the genetic susceptibility to gastric cancer in a Chinese population. Oncotarget. 2016;7(52):87114–87123. doi: 10.18632/oncotarget.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Y, Shao Y, Zhu M, et al. Using gastric juice lncRNA-ABHD11-AS1 as a novel type of biomarker in the screening of gastric cancer. Tumour Biol. 2016;37(1):1183–1188. doi: 10.1007/s13277-015-3903-3. [DOI] [PubMed] [Google Scholar]
- 18.Lin XC, Zhu Y, Chen WB, et al. Integrated analysis of long non-coding RNAs and mRNA expression profiles reveals the potential role of lncRNAs in gastric cancer pathogenesis. Int J Oncol. 2014;45(2):619–628. doi: 10.3892/ijo.2014.2431. [DOI] [PubMed] [Google Scholar]
- 19.Zhao J, Liu Y, Zhang W, et al. Long non-coding RNA Linc00152 is involved in cell cycle arrest, apoptosis, epithelial to mesenchymal transition, cell migration and invasion in gastric cancer. Cell Cycle. 2015;14(19):3112–3123. doi: 10.1080/15384101.2015.1078034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cesana M, Cacchiarelli D, Legnini I, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Wang J, Chen Y, et al. LncRNA MALAT1 exerts oncogenic functions in lung adenocarcinoma by targeting miR-204. Am J Cancer Res. 2016;6(5):1099–1107. [PMC free article] [PubMed] [Google Scholar]
- 22.Wang SH, Wu XC, Zhang MD, Weng MZ, Zhou D, Quan ZW. Long noncoding RNA H19 contributes to gallbladder cancer cell proliferation by modulated miR-194-5p targeting AKT2. Tumour Biol. 2016;37(7):9721–9730. doi: 10.1007/s13277-016-4852-1. [DOI] [PubMed] [Google Scholar]
- 23.Zhou J, Song S, He S, et al. MicroRNA-375 targets PDK1 in pancreatic carcinoma and suppresses cell growth through the Akt signaling pathway. Int J Mol Med. 2014;33(4):950–956. doi: 10.3892/ijmm.2014.1638. [DOI] [PubMed] [Google Scholar]