Abstract
The use of non-Saccharomyces yeast for wine making is becoming a common trend in many innovative wineries. The application is normally aimed at increasing aromas, glycerol, reducing acidity, and other improvements. This manuscript focuses on the reproduction of the native microbiota from the vineyard in the inoculum. Thus, native selected yeasts (Hanseniaspora uvarum, Metschnikowia pulcherrima, Torulaspora delbrueckii, Starmerella bacillaris species and three different strains of Saccharomyces cerevisiae) were inoculated sequentially, or only S. cerevisiae (three native strains together or one commercial) was used. Inoculations were performed both in laboratory conditions with synthetic must (400 mL) as well as in industrial conditions (2000 kg of grapes) in red winemaking in two different varieties, Grenache and Carignan. The results showed that all the inoculated S. cerevisiae strains were found at the end of the vinifications, and when non-Saccharomyces yeasts were inoculated, they were found in appreciable populations at mid-fermentation. The final wines produced could be clearly differentiated by sensory analysis and were of similar quality, in terms of sensory analysis panelists’ appreciation.
Keywords: indigenous yeast, Torulaspora, Metschnikowia, Hanseniaspora, Starmerella, Priorat, wine
Introduction
Due to the increasingly competitive global market, there is a trend for local wine producers to attempt to link their products with geographical identity (Harvey et al., 2014), which has been identified as the terroir, including soil, climate, grape varieties and microbial population (Bokulich et al., 2013). Native microorganisms, particularly yeasts, have been highlighted as key factors for preserving typicality, characteristic flavors and the high quality of wines (Tofalo et al., 2014), which could be considered the microbial fingerprint. However, this microbial fingerprint is not probably static and can change along the time and climatic conditions of the harvest as can be seen by comparing in the Priorat region results form our group (Torija et al., 2001; Padilla et al., 2016). Also new results obtained in a 3 years study (Vigentini et al., 2015) are opening the debate if the microbial population permanently remains in vineyards.
The Priorat Qualified Appellation of Origin (DOQ in Catalan language) is a traditional area for wine production located in the south of Catalonia, Spain, where Carignan and Grenache are the typical and characteristic red grape varieties. Although limited data are available concerning the use of locally selected yeast for must inoculation in Catalonia, several studies developed in different wine-producing areas have noted the use of native yeasts as an innovative approach to obtain wines reflecting terroir (Vilanova and Massneuf-Pomarède, 2005; Carrascosa et al., 2012; Scacco et al., 2012).
The use of locally selected yeast is normally based on a study on natural biodiversity. Yeast biodiversity during the spontaneous fermentation of grape juice includes the presence of different species. It has been widely reported that non-Saccharomyces species dominate the first phase of alcoholic fermentation, and some of these yeasts can also be present at advanced stages, even while the species Saccharomyces cerevisiae dominates the process (Fleet, 1993). This extensive yeast biodiversity is the reason supporting the design and implementation of yeast starter cultures that are not pure or single-species. The defense of the wine typicality should actually include a combination of non-Saccharomyces and S. cerevisiae strains with the aim to obtain wines exhibiting complexity but avoiding the risks related to natural fermentations (Comitini et al., 2011; Tristezza et al., 2011; Suzzi et al., 2012; Gobbi et al., 2013; Medina et al., 2013).
Thus, the proper design of an autochthonous starter culture is essential to reproduce the local sensory properties, including the incorporation of a mixture of different non-Saccharomyces species and different strains of S. cerevisiae to mimic spontaneous alcoholic fermentations. Among non-Saccharomyces species, Hanseniaspora uvarum, Starmerella bacillaris (previously known as Candida zemplinina), Torulaspora delbrueckii and Metschnikowia pulcherrima have been isolated in different wines (Lopandic et al., 2008; Kraková et al., 2012; Albertin et al., 2014) and have been described as characteristic of the Priorat (Torija et al., 2001; Wang et al., 2015; Padilla et al., 2016; Portillo and Mas, 2016). However, the combination of several non-Saccharomyces and strains of S. cerevisiae can be challenged by the winemaking conditions (i.e., SO2 dosage, temperature, etc…) as well as the initial yeast population in grapes (Constantí et al., 1998; Vigentini et al., 2014). Thus, special care in the winery has to be taken for this kind of procedures.
This work aims to test the industrial use of locally selected yeast strains reproducing the vineyard for wine production in the Priorat DOQ. For this purpose, a specific multistarter culture consisting of different strains of S. cerevisiae and non-Saccharomyces species mimicking Priorat natural musts has been developed. This study was done using synthetic must in order to have all the conditions of incubation and sterility under control as well as natural Grenache and Carignan grape juices at industrial scale. The mix of different species was used to inoculate the four non-Saccharomyces species and sequentially (24 h later) the mix of three different S. cerevisiae strains. Additionally, control fermentations containing only the three native S. cerevisiae strains or a S. cerevisiae commercial strain have been performed to evaluate the contribution of non-Saccharomyces and native inoculum to fermentation kinetics, yeast dynamics, and different oenological parameters as well as the production of major volatile compounds. Additionally, a sensory evaluation based on triangle tests was performed.
Materials and Methods
Strains
Four non-Saccharomyces yeast strains: H. uvarum CECT 13130, S. bacillaris CECT 13129, T. delbrueckii CECT 13135 and M. pulcherrima CECT 13131; and three S. cerevisiae strains: CECT 13132, CECT 13133 and CECT 13134, were used in this work. All strains were previously isolated from DOQ Priorat spontaneous fermentations (Padilla et al., 2016) and deposited in the Spanish Type Culture Collection (CECT). The non-Saccharomyces species were selected by the absence of off-odor production (especially acetic acid), prevalence in musts during fermentations and ester production. Instead, resistance to high sugar concentration was the main criteria for selection of S. cerevisiae strains, which is one of the main characteristics of Priorat musts, but also competitiveness in front of other Saccharomyces strains and the production of esters and acetates in single fermentations (Torija et al., 2001). Additionally, commercial S. cerevisiae wine strains GR in Grenache (provided by AB Mauri, Sydney, NSW, Australia), CA in Carignan or QA23 in Synthetic must (both from Lallemand Inc., Montreal, QC, Canada) were included in this study as a control. Yeasts were maintained in glycerol stocks at -80°C.
Biomass Production
Native yeasts were grown in plates with 25 cm of diameter containing YPD medium (10 g/L yeast extract, 20 g/L peptone, 20 g/L glucose, 17 g/L agar) at 28°C before use. Plates were washed with saline solution (NaCl 0.9% w/v) for yeast collecting, and cells were then quantified and used as inocula for laboratory and industrial vinifications. Commercial S. cerevisiae strains were purchased as active dry yeast and rehydrated following the manufacturer’s instructions.
Fermentation and Sampling
The laboratory scale fermentations were conduct using synthetic must (as reported in Andorrà et al., 2012) with nitrogen content of 300 mg/L and 200 g/L of total sugar and pH adjusted to 3.3. Fermentations were performed in triplicate in continuous shaking at 120 rpm at 25°C in 500 mL glass bottles filled each one with 400 mL of synthetic must and covered with cotton caps. The inoculation process is described in Table 1, monitoring and sampling was done as in the industrial scale.
Table 1.
Yeast composition of starter cultures employed in different fermentation modalities (cells/mL).
| Grenache | Carignan and Synthetic must | |||||
|---|---|---|---|---|---|---|
| Yeast strains | A | B | C | A | B | C |
| H. uvarum CECT 13130 | 1.2×105 | 1.2×106 | ||||
| S. bacillaris CECT 13129 | 6×104 | 6×105 | ||||
| T. delbruecki CECT 13135 | 104 | 105 | ||||
| M. pulcherrima CECT 13131 | 104 | 105 | ||||
| S. cerevisiae CECT 13132 | 7×104 | 7×104 | 7×105 | 7×105 | ||
| S. cerevisiae CECT 13133 | 7×104 | 7×104 | 7×105 | 7×105 | ||
| S. cerevisiae CECT 13134 | 7×104 | 7×104 | 7×105 | 7×105 | ||
| S. cerevisiae GR | 2×106 | |||||
| S. cerevisiae CA | 2×106 | |||||
| S. cerevisiae QA 23 | 2×106 | |||||
On the other hand, six industrial fermentations were conducted in stainless steel tanks filled with 2000 kg of crushed grapes, rendering 1050 L of Grenache (GR) or Carignan (CA) wine in a cellar from DOQ Priorat. Due to the specific characteristics of the vineyards in Priorat, this volume is very common in the area and is the volume routinely used in this cellar. Before inoculation, musts were chemically analyzed. The musts had a density around 1100 g/L, pH between 3.19 and 3.29 with total acidity of 4.6 and 5.2 g/L, which are typical values from the area. Due to the low levels of yeast assimilable nitrogen (66 and 80 mg/L), juices were gradually supplemented throughout the alcoholic fermentation with a total of 50 mg inorganic nitrogen/L (as Diammonium Phosphate) and 15 mg organic nitrogen/L (as yeast lysates). For each must variety, three vinifications containing different yeast strain combinations were performed and monitored (Table 1). The A fermentations (A-SM = in Synthetic must, A-GR = in Grenache must and A-CA = in Carignan must) contained a combination of the seven native strains, which were sequentially inoculated. At time 0, non-Saccharomyces strains were added into the must, mimicking the natural yeast composition found in previous studies, and the mixture of S. cerevisiae was incorporated 24 h later. In contrast, the B fermentations (B-SM, B-GR and B-CA) contained only the mixture of the three S. cerevisiae autochthonous strains. Experiments with commercial S. cerevisiae strains (C-SM, C-GR and C-CA) were conducted as a control for each type of must.
From each bottle and tank, daily samples were taken to monitor sugar concentration by measuring must density using an electronic densitometer (Mettler-Toledo S.A.E., Barcelona, Spain). Additionally, samples of the grape juice before inoculation (day 0), 1 day after inoculation with non-Saccharomyces in the case of mixed fermentations (24 h; day 1), 1 day after inoculation with S. cerevisiae (24 h Sc; day 1 or 2), at a mid-fermentation point (M; day 4–5) and at the end of the fermentation (F; day 8–9) were also aseptically withdrawn for yeast counting and molecular identification. Moreover during industrial fermentations, cells from 1 mL at each sampling point were collected after centrifugation (Spectrafuge, Labnet, United States) at 9200 g for 5 min for quantitative PCR (qPCR) analysis.
The synthetic wines were analyzed after the alcoholic fermentation. The final industrial wines were stabilized for 30 days at 4°C, and then 30 ppm of sulfur dioxide was added as potassium metabisulfite, and the final product was bottled. These conditions were maintained for 2 months until the sensory evaluation took place.
Yeast Content and Isolation
Yeast counts were conducted in duplicate on solid YPD and agar-Lysine (LYS) plates (Oxoid, United Kingdom, prepared according to manufacturer’s instructions) after serial decimal dilution with distilled sterile water of the samples. Plates were incubated at 28°C for 3 days. For yeast isolation and identification, from plates containing 30–300 colonies approximately, 25 colonies from each medium and each sampling point were picked randomly.
Yeast Identification: RFLPs of 5.8S-ITS rRNA Region and Sequencing of D1/D2 of 26S rRNA Gene
Yeast isolates were identified by PCR-RFLP analysis of 5.8S-ITS rDNA according to Esteve-Zarzoso et al. (1999), using primers ITS1 and ITS4 (White et al., 1990). PCR products were digested, without further purification, with the restriction enzymes CfoI, HaeIII, DdeI, and HinfI. The PCR products and their restriction fragments were separated by gel electrophoresis on 1.5 and 3% (w/v) agarose gels, respectively. The sizes of the DNA fragments were estimated by comparison against a DNA ladder (100 bp Roche Diagnostics GmBh, Mannheim, Germany). The obtained restriction profiles were compared with the profiles reported in Esteve-Zarzoso et al. (1999) and in the Yeast-id database1.
Sequencing of the D1/D2 domains of 26S rRNA gene was conducted to confirm yeast identification using primers NL1 and NL4 (Kurtzman and Robnett, 1998). The PCR product was purified and sequenced by Macrogen Inc. (Seoul, South Korea) using an ABI3730XL automated capillary DNA sequencer. The sequences were compared with the ones in GenBank using the BLASTN tool (NCBI) and deposited in GenBank database with the accession numbers described in Padilla et al. (2016).
Yeast Typing
The isolates from the dominant yeast species were genetically characterized. S. cerevisiae strains were typified by the analysis of inter-delta regions, as described by Legras and Karst (2003) using the primers delta12 and delta21. DNA was extracted from yeast cultures grown in YPD broth for 24 h at 28°C (Querol et al., 1992). Interdelta PCR products were separated by electrophoresis on 2% (w/v) agarose gels. The sizes of the DNA fragments were estimated by comparison against a DNA ladder (100 bp Roche Diagnostics GmBh, Mannheim, Germany).
Quantitative PCR
Yeast DNA was extracted from 1 ml pelleted cells using the DNeasy PLANT kit (Qiagen, United States). Quantitative PCR (qPCR) was performed in a 7300 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, United States). Power SybrGreen master mix was used according to the manufacturer’s instructions (Applied Biosystems, Foster City, CA, United States). An AB 0–600 96-well optical plate (Thermo Scientific, Waltham, MA, United States) was used for the reaction. The primers used for each species were those described by Hierro et al. (2007) (Saccharomyces and Hanseniaspora), Andorrà et al. (2010) (S. bacillaris), Zott et al. (2010) (T. delbrueckii), and Díaz et al. (2013) (M. pulcherrima). The cycle threshold (CT) was automatically determined. All samples were analyzed in duplicate, and cell concentrations were quantified by CT measurement using the calibration curves of a relevant concentration series of yeast cells for each species (see calibration curves for each species in Supplementary Table S1).
Chemical Analysis of Musts and Wines
Density, pH, total acidity and ethanol were determined according to the methods in the Compendium of International Methods of Analysis of Musts and Wines (OIV, 2015). Yeast assimilable nitrogen was measured according to the formol method (Gump et al., 2000). Sugars (glucose and fructose), acetic acid and glycerol were quantified using the Miura one enzymatic autoanalyzer (BioGamma I.S.E. S.r.L., Rome, Italy) with the corresponding enzymatic kits (BioSystems S.A., Barcelona, Spain).
Determination of Volatile Compound Production
The six final wines obtained using industrial conditions were analyzed for major volatile compounds by gas chromatographic–flame ionization detection (GC-FID) by an external analytical service (L.A.A.E., Zaragoza, Spain) according to Ortega et al. (2001). In summary, 3 mL of each wine were diluted with 7 mL of water, salted with 4.5 g of ammonium sulfate and extracted with 0.2 mL of dichloromethane. The extract was injected in the split mode into a Varian CP-3800 GC system (Palo Alto, CA, United States), separated on a DB-WAX polyethylene glycol column (30 m × 0.32 mm and 0.5 μm film thickness) from J&W Scientific (Folsom, CA, United States), and detected by FID.
Sensory Analysis of the Industrial Wines Obtained
The panel for wine sensory evaluation consisted of two groups of tasters. Group A consisted of eleven judges (six females and five males) recruited from the Faculty of Oenology of the University Rovira i Virgili. Group B consisted of six oenologists from cellars belonging to the Priorat DOQ (four females and two males). Panelists were placed in individual sensory booths. Fifty milliliters of each wine was served at room temperature, and the order of presentation was randomized. For each grape variety, two different discriminating triangle tests were presented, one containing samples from fermentations A and B and the other from B and C.
Statistical Analysis
Significant differences in sensory analysis were defined using the critical number of correct answers for the triangle test (Roessler et al., 1948).
Results
Yeasts in Natural Musts
A specific characteristic of these juices was the very healthy status of the grapes, which reached concentrations of 2 × 103 (GR) to 4 × 104 (CA) CFU/mL when plated. Yeast populations in the grapes of the area are generally higher, approximately 105 cells/g grapes or mL of must. A total of 153 colonies were identified from these musts. This low yeast population, especially in GR juices, led to the isolation of many different yeast species, the most abundant being Rhodotorula mucilaginosa (30%), Debaryomyces hansenii (21%), and M. pulcherrima (19%). Additionally, other species such as Wickerhamomyces anomalus and Zygoascus hellenicus were isolated in minor numbers (less than 10%). Instead, in the case of CA musts, the more common H. uvarum (74%) and S. bacillaris (25%) were present, reaching 99% of the isolates, and only one additional isolate of M. pulcherrima was found.
Fermentation Kinetics and Yeast Population
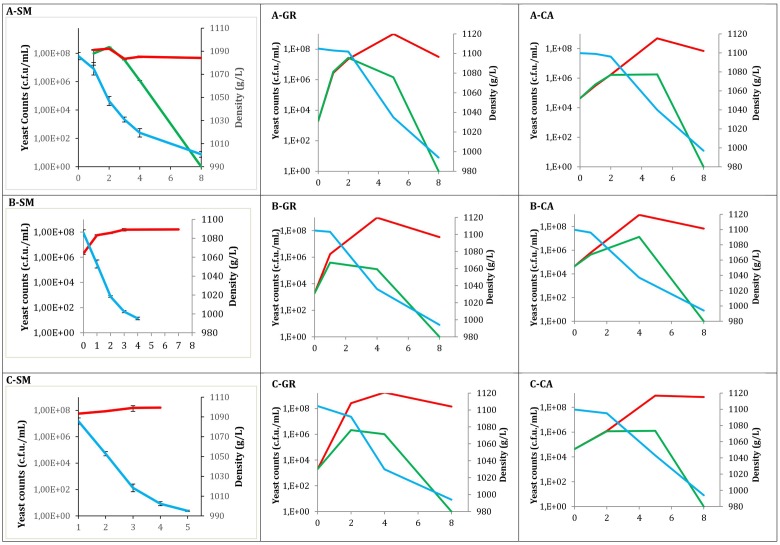
Total yeast counts (YPD), non-Saccharomyces yeast counts (LYS) and must density throughout all fermentations are shown in Figure 1. Values at time 0 correspond to must samples before inoculation in the case of industrial fermentations, while for synthetic must fermentations correspond to the inoculated population. In all cases, the typical growth kinetic was observed, exhibiting high total yeast viability until the end of the fermentations, with values of approximately 108 CFU/mL. In contrast, there was no growth of non-Saccharomyces species at this point, with counts at the middle fermentation point ranging between 105 and 107 CFU/mL. When non-Saccharomyces yeasts were inoculated (Figure 1A), the population recovered in LYS plates reached concentrations of 106 (A-CA) to 108 (A-SM) CFU/mL. These non-Saccharomyces populations decreased when Saccharomyces was inoculated to synthetic must fermentations; however, in natural musts this high population size was maintained until the middle of fermentation, declining afterward. Additionally, three strains of S. cerevisiae were sequentially inoculated after 24 h. When the three S. cerevisiae native strains were inoculated (Figure 1A), according to the type of must used, a similar pattern was observed. In all fermentations the non-Saccharomyces population was able to increase during the 1st days to decrease afterward. However, in synthetic must, the decreased appears shortly after S. cerevisiae inoculation, whereas in natural musts these decreases were later.
FIGURE 1.
Evolution of different fermentations modalities. Total yeast ( ) counts obtained from YPD; Non-Saccharomyces yeast (
) counts obtained from YPD; Non-Saccharomyces yeast ( ) counts on LYS; and Density (
) counts on LYS; and Density ( ). (A) Mixed fermentation (four species of non-Saccharomyces and the three strains of Saccharomyces), (B) Fermentation performed using the three native strains of Saccharomyces, (C) Fermentations conducted by industrial yeast starter belonging to Saccharomyces. Fermentations were performed using different musts. SM, Synthetic must; GR, Grenache must; CA, Carignan must.
). (A) Mixed fermentation (four species of non-Saccharomyces and the three strains of Saccharomyces), (B) Fermentation performed using the three native strains of Saccharomyces, (C) Fermentations conducted by industrial yeast starter belonging to Saccharomyces. Fermentations were performed using different musts. SM, Synthetic must; GR, Grenache must; CA, Carignan must.
Mixed fermentation in synthetic must revealed the maximum yeast diversity on YPD plates at mid fermentation. During the initial sampling points, a high presence of H. uvarum was detected; however, these non-Saccharomyces yeast species were not identified at the last sampling point, in which all of the colonies were identified as S. cerevisiae.
When the three selected S. cerevisiae strains were used (Figure 1B) the non-Saccharomyces populations had a similar pattern, with Grenache must samples reaching slightly lower populations. When the commercial yeast strains were used (Figure 1C) the pattern was also very similar to the inoculation of the three S. cerevisiae strains.
The fermentation kinetics observed by density monitoring showed that the fermentations finished within 8 days when a mixture of Saccharomyces and non-Saccharomyces are present, independently of the origin (natural or synthetic) of the must. However, for fermentation using Saccharomyces inoculation this time is reduced to 4 days (synthetic must). This fact can be explained for the presence of lag phase when non-Saccharomyces yeasts are present. In the case of GR mixed fermentation, the sugar consumption was slightly slower compared with pure S. cerevisiae, although the process finished at the same time. For the three CA experiments, a similar fermentative pattern was observed.
Yeast Population Dynamics
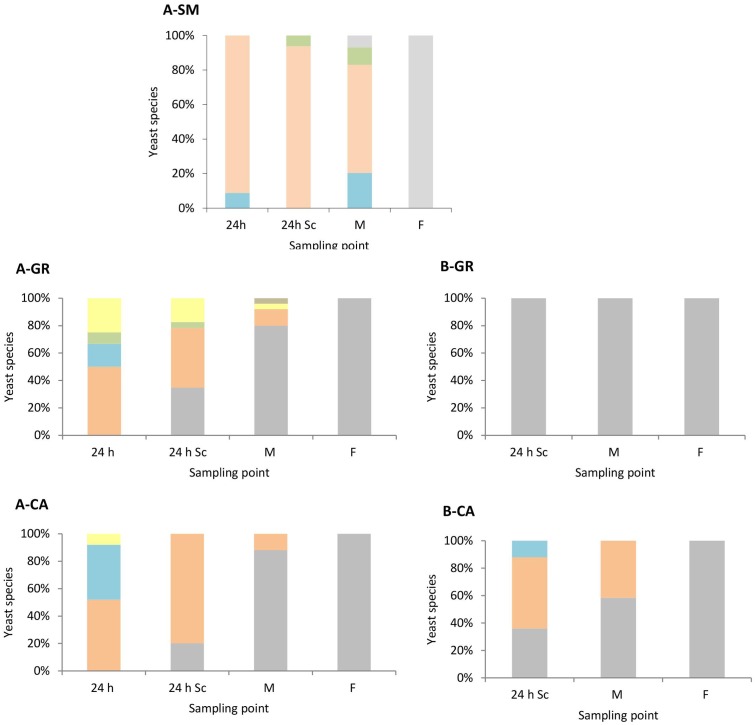
As expected, the colonies recovered on YPD medium (Figure 2), at later time sampling points were identified as S. cerevisiae (100%). Only S. cerevisiae was recovered from the colonies isolated in synthetic must fermentations inoculated with this species (data not shown). However, in natural must fermentations, a clear difference was observed between mixed (Figure 2A) and pure S. cerevisiae vinifications (Figure 2B). In the natural must fermentations inoculated with a mixture of non-Saccharomyces, a higher biodiversity during the first stages of the process was observed compared with those inoculated with S. cerevisiae. In the case of A-GR, all inoculated non-Saccharomyces species, H. uvarum (50%), S. bacillaris (17%), T. delbrueckii (8%), and M. pulcherrima (25%), were recovered after 1 day of inoculation (24 h); however, in A-CA, all inoculated yeasts except T. delbrueckii were found, with values of 52% for H. uvarum, 40% for S. bacillaris and 8% for M. pulcherrima. Additionally, H. guilliermondii was also recovered at the middle point in A-GR. This high biodiversity present in mixed inocula fermentations decreased as the fermentation proceeded. The percentages of non-Saccharomyces species found were between 65 and 80% 24 h after inoculation with S. cerevisiae and 12 and 20% at the mid-fermentation point.
FIGURE 2.
Yeast species population dynamics established by RFLP-ITS-PCR of YPD-cultured isolates. (A) Mixed fermentation (four species of non-Saccharomyces and the three strains of Saccharomyces), (B) Fermentation performed using the three native strains of Saccharomyces. SM, Synthetic must; GR, Grenache must; CA, Carignan must. The species identified were Hanseniaspora guilliermondii
 , Metschnikowia pulcherrima
, Metschnikowia pulcherrima
 , Torulaspora delbrueckii
, Torulaspora delbrueckii
 , Starmerella bacillaris
, Starmerella bacillaris
 , Hanseniaspora uvarum
, Hanseniaspora uvarum
 , Saccharomyces cerevisiae
, Saccharomyces cerevisiae
 . Columns are indicated as: 24 h (24 h after non-Saccharomyces inoculation), 24 h Sc (24 h after S. cerevisiae inoculation), M (middle fermentation), F (end of fermentation).
. Columns are indicated as: 24 h (24 h after non-Saccharomyces inoculation), 24 h Sc (24 h after S. cerevisiae inoculation), M (middle fermentation), F (end of fermentation).
Once S. cerevisiae was incorporated into the must, it was possible to isolate it after 24 h (24 h Sc). In A-GR as well as in A-CA, S. cerevisiae gradually dominated the process. In the fermentation with the native strains of S. cerevisiae B-GR, only the yeast S. cerevisiae was isolated at the three sampling points. In contrast, in B-CA, the imposition occurred gradually, as non-Saccharomyces species were also recovered up to the mid-fermentation point (42%).
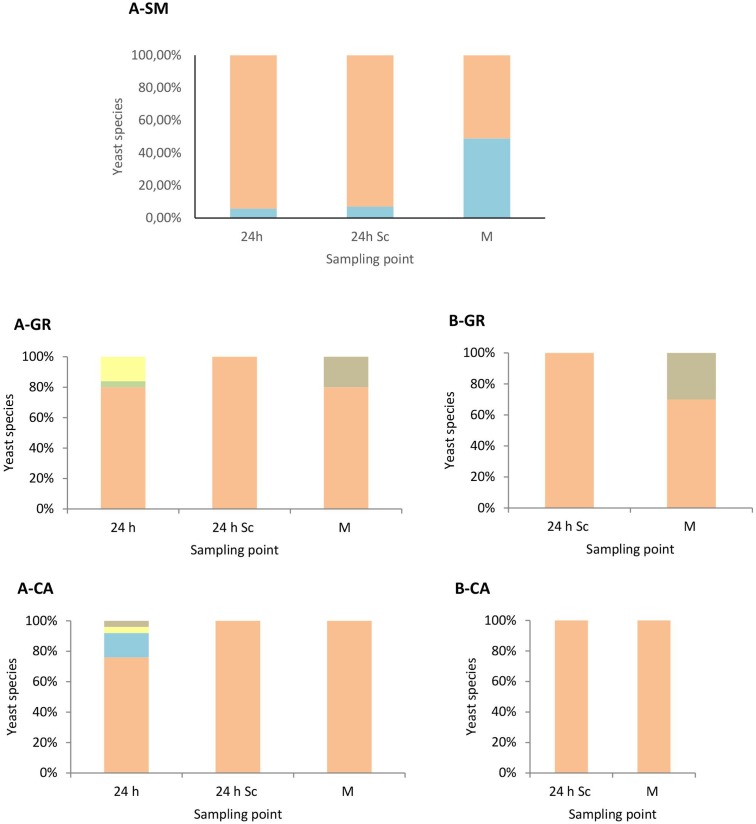
These results were confirmed when the yeast population dynamics in LYS media were analyzed, where non-Saccharomyces species were isolated until the mid-fermentation point but undetectable at later stages (Figure 3). In synthetic must fermentations only H. uvarum and S. bacillaris were detected, showing an increase of the S. bacillaris presence as the fermentation proceeded (Figure 3A). In natural must fermentations inoculated with non-Saccharomyces yeast (Figure 3A) showed some species diversity at the beginning of the fermentation compared with pure S. cerevisiae fermentations (Figure 3B), although the most abundant yeast in all cases was H. uvarum, with proportions of approximately 86% and 95% in the GR and CA varieties, respectively. Among non-Saccharomyces species, this yeast dominated throughout fermentation, with just one non-inoculated species, H. guilliermondii, detected in small percentages at mid-fermentation in the case of GR wines.
FIGURE 3.
Yeast species population dynamics established by RFLP-ITS-PCR of LYS-cultured isolates. (A) Mixed fermentation (four species of non-Saccharomyces and the three strains of Saccharomyces), (B) Fermentation performed using the three native strains of Saccharomyces. SM, Synthetic must; GR, Grenache must; CA, Carignan must. The species identified were Hanseniaspora guilliermondii
 , Metschnikowia pulcherrima
, Metschnikowia pulcherrima
 , Torulaspora delbrueckii
, Torulaspora delbrueckii
 , Starmerella bacillaris
, Starmerella bacillaris
 , Hanseniaspora uvarum
, Hanseniaspora uvarum
 , Saccharomyces cerevisiae
, Saccharomyces cerevisiae
 . Columns are indicated as: 24 h (24 h after non-Saccharomyces inoculation), 24 h Sc (24 h after S. cerevisiae inoculation), M (middle fermentation), F (end of fermentation).
. Columns are indicated as: 24 h (24 h after non-Saccharomyces inoculation), 24 h Sc (24 h after S. cerevisiae inoculation), M (middle fermentation), F (end of fermentation).
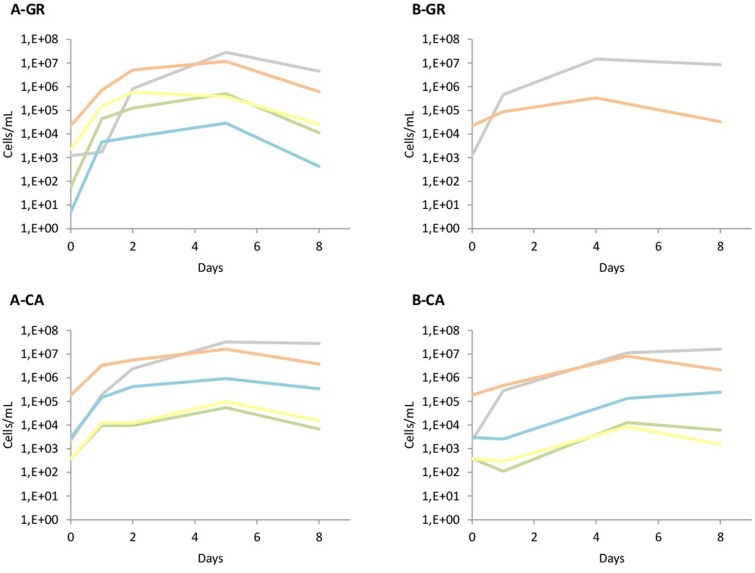
The yeast dynamics for natural must fermentations were also analyzed by culture-independent techniques, specifically by qPCR (Figure 4). The data obtained from qPCR analysis overall agree with the plating results, with some particularities. First, the quantification of yeast in both musts in GR showed the presence of Hanseniaspora species at higher levels (1 × 104 cells/mL) than the colonies recovered on plates (2 × 103 CFU/mL). Furthermore, S. cerevisiae initial counts detected by qPCR were at concentrations that had to be detected in plates, although no S. cerevisiae isolates were identified using the RFLPs of 5.8S ITS rDNA. However, when S. bacillaris was recovered on plates, its quantification by qPCR was very low. In contrast, in the CA musts, the massive presence of Hanseniaspora cells agreed with the observations on plates, as well as the numbers of S. bacillaris, although in this case the qPCR counts were slightly lower than expected.
FIGURE 4.
Population dynamics of different yeast species established by qPCR. The species tested were Saccharomyces spp. ( ); Hanseniaspora spp. (
); Hanseniaspora spp. ( ); S. bacillaris (
); S. bacillaris ( ); T. delbrueckii (
); T. delbrueckii ( ); M. pulcherrima (
); M. pulcherrima ( ). (A) Mixed fermentation (four species of non-Saccharomyces and the three strains of Saccharomyces), (B) Fermentation performed using the three native strains of Saccharomyces. GR, Grenache must, CA, Carignan must.
). (A) Mixed fermentation (four species of non-Saccharomyces and the three strains of Saccharomyces), (B) Fermentation performed using the three native strains of Saccharomyces. GR, Grenache must, CA, Carignan must.
The increase of cell concentration due to inoculation was observable in all cases when the addition of the starter culture was performed. When the non-Saccharomyces species were inoculated, the observed increase agreed with the inoculated populations, except for S. bacillaris in GR, likely due to its low presence in the grape juice (Figure 4A). In the musts inoculated with autochthonous S. cerevisiae (Figure 4B), the increase was also seen in the quantification with qPCR. Finally, the levels of non-Saccharomyces during all these fermentations were very similar to the levels detected on LYS plates, with a clear dominance of H. uvarum. The presence of non-Saccharomyces throughout B-CA fermentation is remarkable, likely due to the presence of higher populations of these yeasts in the CA must.
Yeast Typing
To test the dominance of the major inoculated species, S. cerevisiae strains isolated during the fermentations were typified at strain level. In the case of S. cerevisiae, the analysis was performed using the colonies at the end of fermentations. In fermentations A and B, the interdelta fingerprint of S. cerevisiae colonies isolated at the end of the process corresponded with the three native strains inoculated, CECT 13132, CECT 13133 and CECT 13134. Although the three profiles were present at the end of the fermentations, the main profile recovered was that of the strain CECT 12132, followed by CECT 12134 and CECT 12133 (Table 2). In the fermentations with the commercial strain, only the inoculated strain was recovered (data not shown).
Table 2.
Percentages of the inoculated S. cerevisiae strains recovered at the end of different fermentations.
| CECT 13132 | CECT 13133 | CECT 13134 | |
|---|---|---|---|
| A-SM | 54 | 18 | 27 |
| B-SM | 56 | 25 | 18 |
| A-GR | 48 | 8 | 44 |
| B-GR | 63 | 8 | 29 |
| A-CA | 68 | 14 | 18 |
| B-CA | 64 | 12 | 24 |
A, Mixed fermentation (four species of non-Saccharomyces and the three strains of Saccharomyces); B, Fermentation performed using the three native strains of Saccharomyces. Fermentations were performed using different musts: SM, Synthetic must; GR, Grenache; CA, Carignan.
Chemical Analysis of Wines
The main oenological parameters of the wines obtained are shown in Table 3. All wines contained less than 2 g/L of residual sugars. Additionally, all wines presented an alcohol level expected according to the sugar content of the musts. Small variations were observed in the synthetic and CA wines, with alcohol content from 12.43 to 12.83% for synthetic must or from 13.9 to 14.3% for Carignan wines. Within each wine, the natural must fermentations performed with commercial S. cerevisiae strains contained higher levels of glycerol and acetic acid than fermentations performed with autochthonous strains, however, when synthetic must was used, the highest values were exhibited by the mixed fermentation (Table 3).
Table 3.
Analytical parameters of final wines.
| Glucose+Fructose (g/L) | Glycerol (g/L) | Acetic acid (g/L) | Alcohol (% v/v) | pH | |
|---|---|---|---|---|---|
| A-SM | 3.79 ± 1.50 | 11.06 ± 0.21 | 0.84 ± 0.01 | 12.83 ± 0.11 | 3.23 ± 0.01 |
| B-SM | 0.11 ± 0.15 | 8.58 ± 0.10 | 0.44 ± 0.03 | 12.43 ± 0.11 | 3.21 ± 0.01 |
| C-SM | 0.17 ± 0.10 | 9.80 ± 1.12 | 0.64 ± 0.03 | 12.50 ± 0.34 | 3.19 ± 0.02 |
| A-GR | 1.09 | 6.01 | 0.27 | 14.9 | 3.20 |
| B-GR | 0.25 | 5.41 | 0.31 | 14.9 | 3.20 |
| C-GR | 0.17 | 7.10 | 0.42 | 14.9 | 3.13 |
| A-CA | 0.18 | 7.92 | 0.45 | 14.3 | 3.20 |
| B-CA | 0.45 | 8.13 | 0.44 | 13.9 | 3.18 |
| C-CA | 0.15 | 8.97 | 0.56 | 13.9 | 3.16 |
Abbreviations as in Table 2. Standard deviations were indicated only when triplicates has been used (Synthetic Must fermentations).
Volatile Compound Production in Industrial Fermentations
The volatile profiles of the six final wines were evaluated (Supplementary Table S2). A total of twenty-nine volatile compounds were quantified and classified into esters (10), alcohols (8), acids (7), carbonyl compounds (3) and lactones (1). Among esters, the most abundant in all fermentations was ethyl acetate, followed by ethyl lactate. However, ethyl and isoamyl acetates, ethyl hexanoate and ethyl butyrate were detected above the odor threshold only in GR wines. In the case of alcohols, isoamyl alcohol, isobutanol and ß-phenylethanol were the main alcohols detected in all wines. All of them and methionol were present above the odor threshold, except for isobutanol in GR-C wine. Acetic acid was by far the most abundant acid in both wine varieties. All except isobutyric acid and decanoic acid were present above the odor threshold. Additionally, the major carbonyl compounds acetaldehyde and butyrolactone were present in all fermentations in a similar range, but the latter was detected below the odor threshold.
Sensory Analysis of the Industrial Wines
The wines obtained at industrial scale for the three different treatments underwent sensory evaluation by triangle tests. Table 4 presents the results obtained for the two different varieties. Statistically significant differences among the wines were found in three of the four tests performed, as more than 10 of 17 panelists were able to differentiate wines produced with different inocula. In the case of GR wines, native S. cerevisiae fermentations were different from the fermentation produced with the commercial strain of S. cerevisiae. When the CA variety was tasted, the wines presented in both sensory tests were perceived as different.
Table 4.
Triangle test evaluation of final industrial wines.
| Triangle test | Correct answers (Total) |
|---|---|
| A-GR against B-GR | 7 (17) |
| B-GR against C-GR | 12∗∗ (17) |
| A-CA against B-CA | 12∗∗ (17) |
| B-CA against C-CA | 10∗ (17) |
Abrreviations as in Table 2. ∗∗Significant difference p-value < 0.01. ∗Significant difference p-value < 0.05.
Discussion
In this work, the effects of native multi-starter yeast inocula on industrial and laboratory alcoholic fermentations have been studied. Concerning fermentation kinetics and total yeast population, similar results were obtained for the fermentations, and thus similar behavior was found between commercial and native yeast inocula. Additionally, the data obtained followed the typical growth pattern, with values of total yeasts at the end of the alcoholic fermentation close to 108 CFU/mL. This value is consistent with results obtained from pure S. cerevisiae fermentations as well as from combined S. cerevisiae and non-Saccharomyces vinifications (Beltran et al., 2002; Gobbi et al., 2013; Belda et al., 2015). However, minor differences have been observed when natural and synthetic must were compared. Natural must was the best medium to grow the Saccharomyces yeast, because the recovery on YPD plates was more than 60% at mid fermentation, while in synthetic must the presence of Saccharomyces was reduced to 5%. However, S. cerevisiae was the only isolated at the end of all fermentations. Although synthetic must tries to mimic natural musts, these are more complex and most likely will be richer in nutrients, which could be a determining factor.
The low yeast population size, such as the observed in GR musts, is normally related to low recovery of the main non-Saccharomyces species (H. uvarum and S. bacillaris), which allows minor species to be easily detected (Beltran et al., 2002). Alternatively, CA must exhibits the typical Priorat microbial fingerprint consisting mainly of H. uvarum and S. bacillaris (Padilla et al., 2016). It is important to highlight that all non-Saccharomyces species isolated at this point have been previously reported on grapes or wine fermentations from the Priorat region (Torija et al., 2001) and are fairly universal, as reviewed by Jolly et al. (2014).
When comparing the populations obtained using culture-dependent and culture-independent techniques in fresh must samples, total yeast plate counts were approximately 1-log lower than qPCR data when yeast populations were low. The qPCR of the Saccharomyces spp. population in both varieties found approximately 103 cells/mL, but no isolates from these species were recovered from the fresh juice. Similar qPCR results had been reported during the characterization of Merlot musts, but in that case, the culturing of S. cerevisiae was directly excluded due to the choice of a non-Saccharomyces growth media (Zott et al., 2010). However, the qPCR determination of H. uvarum population overestimated it at this initial point, as previously reported, and therefore our data support the suggestion that qPCR is a more sensitive method concerning detection of this species (Zott et al., 2010). In contrast, S. bacillaris was slightly underestimated. The reasons for these differences could be different: on one side the differential growth of different species on plates, and on the other side due to limited specificity of the qPCR probes and the method efficiency.
Yeast counts and population dynamics after the incorporation of native yeast were also monitored. The initial growth of the non-Saccharomyces yeasts was only observed clearly using synthetic must, while the use of natural must seems to be more restrictive to the growth of this type of yeasts. However, this fact is not so clear in all fermentations, because the increase of non-Saccharomyces populations by plating has been detected in the GR fermentations inoculated with commercial starter and CA fermentations inoculated with native starter, while this increase is no so evident in other fermentations. The overall detection and quantification of yeast during different fermentation strategies by both culture-dependent and independent methods were very similar, as also reported by Zott et al. (2010), likely due to the high yeast population levels and small number of dominant species. However, small differences need to be further described. One of the main differences is that the non-Saccharomyces yeasts were detected up to mid fermentation by plating but until the end of the fermentation by qPCR analysis, as previously reported (Hierro et al., 2006). In these fermentations, Hanseniaspora spp. values ranged from 3 × 104 to 3 × 106 cells/mL at the end of the different industrial fermentations, while S. bacillaris counts were approximately 3 × 105 cells/mL in the final CA wines, which was in agreement with previous studies (Hierro et al., 2007; Andorrà et al., 2010; Zott et al., 2010). Additionally, T. delbrueckii was detected and quantified by qPCR in fermentations A-GR, A-CA and B-CA, but it was only isolated from experiment A-GR at 24 and 48 h after non-Saccharomyces inoculation. At these points, qPCR detected T. delbrueckii populations at 4 × 104 and 1 × 105 cells/mL, values above the cell concentrations found in CA fermentations. Disagreements in the detection of this species in plates and qPCR were also reported by Zott et al. (2010).
In addition to yeast identification, isolates from the main species were typified to assess the dominance of the starter culture. In experiments where a mixture of three native S. cerevisiae strains was inoculated, 100% of S. cerevisiae isolates exhibited the electrophoretic pattern of the inoculated strains. This result indicates that the three native S. cerevisiae strains included in the yeast consortium coexisted throughout the alcoholic fermentation and dominated the process, being in all types of fermentations (synthetic, Grenache and Carignan) a clear predominance of CECT13132, independently of the composition of the must or the presence of other native yeasts. Similarly, in a study conducted in Albariño white wines where three native strains were singly inoculated, all strains were recovered, and the percentage of imposition was between 90 and 100% in the different stages of fermentation (Carrascosa et al., 2012). In contrast, other authors (Esteve-Zarzoso et al., 2000; Sun et al., 2015) have reported that not all commercial yeast starters can dominate the fermentations in comparison with natural yeast present or isolated from their area, showing that the native microbiota prevailed over the commercial starter culture used, mainly isolated from other oenological region. This result supports the idea that autochthonous yeasts are well adapted to particular fermentation conditions, and thus their incorporation in a mixed inoculum is highly recommendable.
Wines produced with commercial strains rendered higher levels of glycerol and acetic acid, but in all cases, the final content was acceptable. These commercial strains are among the most used in the region and are able to perform the alcoholic fermentation of high sugar content to dryness. In the case of GR, all fermentations produced the same final alcohol content. However, for CA wines, there were some differences in the alcohol production. This result could be due to the heterogeneity of the starting must, which could include slight differences in the sugar content.
The volatile profiles of the different wines were also studied. The total acid content was higher in fermentations conducted using commercial S. cerevisiae strains, in agreement with results obtained in the general chemical characterization. However, the concentrations of the other analyzed volatile compounds were very similar among different treatments. The contributions of H. uvarum, C. zemplinina, T. delbrueckii, and M. pulcherrima to wine aroma have been studied (Comitini et al., 2011; Andorrà et al., 2012; González-Royo et al., 2014; Loira et al., 2014; Renault et al., 2015). Most studies concluded that the incorporation of these species exhibited a positive impact on aroma development. Nevertheless, most articles focused on evaluating the effects of single strains or mixed starters composed of one S. cerevisiae strain and one non-Saccharomyces species. Therefore, the interactions among different non-Saccharomyces wine yeast species need to be further elucidated. The results obtained in this paper highlight that complex interactions among yeast strains are likely to occur during the industrial fermentation of grape juice, and thus it is difficult to identify clear trends among different inoculation strategies. Still, the sensory evaluation concluded that most of the wines produced could be identified as different from the organoleptic point of view. However, high ethanol content and the full body characterize the Priorat wines, which is the consequence of its high complexity. Thus, although tasters could differentiate all the produced wines, there was not a significant preference: in all the cases the preferences were close to 50% of the tasters that identified the differences.
Author Contributions
BP, LZ, AF, RP, and BE-Z performed the experiments; BP, BE-Z, GB, and AM designed the experiments, analyzed and interpreted the results; wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Wildwine EU Project (grant agreement 315065). Authors would like to thank Ferrer Bobet winery for their assistance in the experimental work.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01293/full#supplementary-material
References
- Albertin W., Miot-Sertier C., Bely M., Marullo P., Coulon J., Moine V., et al. (2014). Oenological prefermentation practices strongly impact yeast population dynamics and alcoholic fermentation kinetics in Chardonnay grape must. Int. J. Food Microbiol. 178 87–97. 10.1016/j.ijfoodmicro.2014.03.009 [DOI] [PubMed] [Google Scholar]
- Andorrà I., Berradre M., Mas A., Esteve-Zarzoso B., Guillamón J. M. (2012). Effect of mixed culture fermentations on yeast populations and aroma profile. LWT Food Sci. Technol. 491 8–13. 10.1016/j.lwt.2012.04.008 [DOI] [Google Scholar]
- Andorrà I., Landi S., Mas A., Esteve-Zarzoso B., Guillamón J. M. (2010). Effect of fermentation temperature on microbial population evolution using culture-independent and dependent techniques. Food Res. Int. 43 773–779. 10.1016/j.foodres.2009.11.014 [DOI] [Google Scholar]
- Belda I., Navascués E., Marquina D., Santos A., Calderon F., Benito S. (2015). Dynamic analysis of physiological properties of Torulaspora delbrueckii in wine fermentations and its incidence on wine quality. Appl. Microbiol. Biotechnol. 99 1911–1922. 10.1007/s00253-014-6197-2 [DOI] [PubMed] [Google Scholar]
- Beltran G., Torija M. J., Novo M., Ferrer N., Poblet M., Guillamón J. M., et al. (2002). Analysis of yeast populations during alcoholic fermentation: a six year follow-up study. Syst. Appl. Microbiol. 25 287–293. 10.1078/0723-2020-00097 [DOI] [PubMed] [Google Scholar]
- Bokulich N. A., Thorngate J. H., Richardson P. M., Mills D. A. (2013). Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc. Natl. Acad. Sci. U.S.A. 111 139–148. 10.1073/pnas.1317377110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrascosa A. V., Bartolome B., Robredo S., León A., Cebollero E., Juega M. (2012). Influence of locally-selected yeast on the chemical and sensorial properties of Albariño white wines. LWT Food Sci. Technol. 46 319–325. 10.1016/j.lwt.2011.09.01 [DOI] [Google Scholar]
- Comitini F., Gobbi M., Domizio P., Romani C., Lencioni L., Mannazzu I., et al. (2011). Selected non-Saccharomyces wine yeasts in controlled multistarter fermentations with Saccharomyces cerevisiae. Food Microbiol. 28 873–882. 10.1016/j.fm.2010.12.001 [DOI] [PubMed] [Google Scholar]
- Constantí M., Reguant C., Poblet M., Zamora F., Mas A., Guillamon J. M. (1998). Molecular analysis of yeast population dynamics: effect of sulphur dioxide and the inoculum in must fermentation. Int. J. Food Microbiol. 41 169–175. 10.1016/S0168-1605(98)00041-5 [DOI] [PubMed] [Google Scholar]
- Díaz C., Molina A. M., Nähring J., Fischer R. (2013). Characterization and dynamic behavior of wild yeast during spontaneous wine fermentation in steel tanks and amphorae. BioMed Res. Int. 2013:540465 10.1155/2013/540465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteve-Zarzoso B., Belloch C., Uruburu F., Querol A. (1999). Identification of yeasts by RFLP analysis of the 5.8S rRNA gene and the two ribosomal internal transcribed spacers. Int. J. Syst. Bacteriol. 49 329–337. 10.1099/00207713-49-1-329 [DOI] [PubMed] [Google Scholar]
- Esteve-Zarzoso B., Gostíncar A., Bobet R., Uruburu F., Querol A. (2000). Selection and molecular characterization of wine yeasts isolated from the “El Penedès” area (Spain). Food Microbiol. 17 553–562. 10.1006/fmic.2000.0347 [DOI] [Google Scholar]
- Fleet G. H. (1993). “The microorganisms of winemaking-isolation enumeration and identification,” in Wine Microbiology and Biotechnology, ed. Fleet G. H. (Chur: Harwood Academic Publishers; ), 1–27. [Google Scholar]
- Gobbi M., Comitini F., Domizio P., Romani C., Lencioni L., Mannazzu I., et al. (2013). Lachancea thermotolerans and Saccharomyces cerevisiae in simultaneous and sequential co-fermentation: a strategy to enhance acidity and improve the overall quality of wine. Food Microbiol. 33 271–281. 10.1016/j.fm.2012.10.004 [DOI] [PubMed] [Google Scholar]
- González-Royo E., Pascual O., Kontoudakis N., Esteruelas M., Esteve-Zarzoso B., Mas A., et al. (2014). Oenological consequences of sequential inoculation with non-Saccharomyces yeasts (Torulaspora delbrueckii or Metschnikowia pulcherrima) and Saccharomyces cerevisiae in base wine for sparkling wine production. Eur. Food Res. Technol. 240 999–1012. 10.1007/s00217-014-2404-8 [DOI] [Google Scholar]
- Gump B. H., Zoecklein B. W., Fugelsang K. C. (2000). “Prediction of prefermentation nutritional status of grape juice-The formol method,” in Food Microbiology Protocols Vol. 14 eds Spencer J. F. T., Ragout de Spencer A. L. (Totowa, NJ: Humana Press, Inc; ), 283–296. [Google Scholar]
- Harvey M., White L., Frost W. (2014). “Exploring wine and identity,” in Wine and Identity: Branding, Heritage, Terroir, eds Harvey M., White L., Frost W. (Oxon: Routledge; ), 1–13. [Google Scholar]
- Hierro N., Esteve-Zarzoso B., González A., Mas A., Guillamón J. M. (2006). Real-time quantitative PCR (QPCR) and reverse transcription-QPCR (RT- QPCR) for the detection and enumeration of total yeasts in wine. Appl. Environ. Microbiol. 72 7148–7155. 10.1128/AEM.00388-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hierro N., Esteve-Zarzoso B., Mas A., Guillamón J. M. (2007). Monitoring of Saccharomyces and Hanseniaspora populations during alcoholic fermentation by real-time quantitative PCR. FEMS Yeast Res. 7 1340–1349. 10.1111/j.1567-1364.2007.00304.x [DOI] [PubMed] [Google Scholar]
- Jolly N. P., Varela C., Pretorius I. S. (2014). Not your ordinary yeast: non-Saccharomyces yeasts in wine production uncovered. FEMS Yeast Res. 14 215–237. 10.1111/1567-1364.12111 [DOI] [PubMed] [Google Scholar]
- Kraková L., Chovanová K., Ženišová K., Turcovská V., Brežná B., Kuchta T., et al. (2012). Yeast diversity investigation of wine-related samples from two different Slovakian wine-producing areas through a multistep procedure. LWT Food Sci. Technol. 46 406–411. 10.1016/j.lwt.2011.12.010 [DOI] [Google Scholar]
- Kurtzman C. P., Robnett C. J. (1998). Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek 73 331–371. 10.1023/A:1001761008817 [DOI] [PubMed] [Google Scholar]
- Legras J. L., Karst F. (2003). Optimisation of interdelta analysis for Saccharomyces cerevisiae strain characterisation. FEMS Microbiol. Lett. 221 249–255. 10.1016/S0378-1097(03)00205-2 [DOI] [PubMed] [Google Scholar]
- Loira I., Vejarano R., Bañuelos M. A., Morata A., Tesfaye W., Uthurry C., et al. (2014). Influence of sequential fermentation with Torulaspora delbrueckii and Saccharomyces cerevisiae on wine quality. LWT Food Sci. Technol. 59 915–922. 10.1016/j.lwt.2014.06.019 [DOI] [Google Scholar]
- Lopandic K., Tiefenbrunner W., Gangl H., Mandl K., Berger S., Leitner G., et al. (2008). Molecular profiling of yeasts isolated during spontaneous fermentations of Austrian wines. FEMS Yeast Res. 8 1063–1075. 10.1111/j.1567-1364.2008.00385.x [DOI] [PubMed] [Google Scholar]
- Medina K., Boido E., Fariña L., Gioia O., Gómez M. E., Barquet M., et al. (2013). Increased flavour diversity of Chardonnay wines by spontaneous fermentation and co-fermentation with Hanseniaspora vineae. Food Chem. 141 2513–2521. 10.1016/j.foodchem.2013.04.056 [DOI] [PubMed] [Google Scholar]
- OIV (2015). Compendium of International Methods of Wine and Must Analysis. Paris: International Organisation of Vine and Wine. [Google Scholar]
- Ortega C., López R., Cacho J., Ferreira V. (2001). Fast analysis of important wine volatile compounds Development and validation of a new method based on gas chromatographic – flame ionisation detection analysis of dichloromethane microextracts. J. Chromatogr. A 923 205–214. 10.1016/S0021-9673(01)00972-4 [DOI] [PubMed] [Google Scholar]
- Padilla B., García-Fernández D., González B., Izidoro I., Esteve-Zarzoso B., Beltran G., et al. (2016). Yeast biodiversity from DOQ Priorat uninoculated fermentations. Front. Microbiol. 7:930 10.3389/fmicb.2016.00930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portillo M. C., Mas A. (2016). Analysis of microbial diversity and dynamics during wine fermentation of Grenache grape variety by high-throughput barcoding sequencing. LWT Food Sci. Technol. 72 317–321. 10.1016/j.lwt.2016.05.009 [DOI] [Google Scholar]
- Querol A., Barrio E., Ramón D. (1992). A comparative study of different methods of yeast strain characterization. Syst. Appl. Microbiol. 15 439–446. 10.1016/S0723-2020(11)80219-5 [DOI] [Google Scholar]
- Renault P., Coulon J., de Revel G., Barbe J. C., Bely M. (2015). Increase of fruity aroma during mixed T. delbrueckii/S. cerevisiae wine fermentation is linked to specific esters enhancement. Int. J. Food Microbiol. 207 40–48. 10.1016/j.ijfoodmicro.2015.04.037 [DOI] [PubMed] [Google Scholar]
- Roessler E. B., Warren J., Guymon J. F. (1948). Significance in triangular taste tests. J. Food Sci. 13 503–505. 10.1111/j.1365-2621.1948.tb16650.x [DOI] [PubMed] [Google Scholar]
- Scacco A., Oliva D., Di Maio S., Polizzotto G., Genna G., Tripodi G., et al. (2012). Indigenous Saccharomyces cerevisiae strains and their influence on the quality of Catarratto, Inzolia and Grillo white wines. Food Res. Int. 46 1–9. 10.1016/j.foodres.2011.10.038 [DOI] [Google Scholar]
- Sun Y., Li E., Qi X., Liu Y. (2015). Changes of diversity and population of yeasts during the fermentations by pure and mixed inoculation of Saccharomyces cerevisiae strains. Ann. Microbiol. 65 911–919. 10.1007/s13213-014-0934-8 [DOI] [Google Scholar]
- Suzzi G., Schirone M., Sergi M., Marianella R. M., Fasoli G., Aguzzi I., et al. (2012). Multistarter from organic viticulture for red wine Montepulciano d’Abruzzo production. Front. Microbiol. 3:135 10.3389/fmicb.2012.00135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tofalo R., Perpetuini G., Fasoli G., Schirone M., Corsetti A., Suzzi G. (2014). Biodiversity study of wine yeasts belonging to the “terroir” of Montepulciano d’Abruzzo “Colline Teramane” revealed Saccharomyces cerevisiae strains exhibiting atypical and unique 5.8S-ITS restriction patterns. Food Microbiol. 39 7–12. 10.1016/j.fm.2013.10.001 [DOI] [PubMed] [Google Scholar]
- Torija M. J., Rozés N., Poblet M., Guillamón M., Mas A. (2001). Yeast population dynamics in spontaneous fermentations?: comparison between two different wine-producing areas over a period of three years. Antonie Van Leeuwenhoek 79 345–352. 10.1023/A:1012027718701 [DOI] [PubMed] [Google Scholar]
- Tristezza M., Vetrano C., Bleve G., Grieco F., Tufariello M., Quarta A., et al. (2011). Autochthonous fermentation starters for the industrial production of Negroamaro wines. J. Ind. Microbiol. 39 81–92. 10.1007/s10295-011-1002-z [DOI] [PubMed] [Google Scholar]
- Vigentini I., De Lorenzis G., Fabrizio V., Valdetara F., Faccincani M., Panont C. A., et al. (2015). The vintage effect overcomes the terroir effect: a three year survey on the wine yeast biodiversity in Franciacorta and Oltrepo‘ Pavese, two northern Italian vine-growing areas. Microbiology 161 362–373. 10.1099/mic.0.000004 [DOI] [PubMed] [Google Scholar]
- Vigentini I., Fabrizio V., Faccincani M., Picozzi C., Comasio A., Foschino R. (2014). Dynamics of Saccharomyces cerevisiae populations in controlled and spontaneous fermentations for Franciacorta D.O.C.G. base wine production. Ann. Microbiol 64 639–651. 10.1007/s13213-013-0697-7 [DOI] [Google Scholar]
- Vilanova M., Massneuf-Pomarède I. (2005). Characterization of yeast strains from Rías Baixas (NW Spain) and their contribution to the fermentation of Albariño wine. Ann. Microbiol. 55 23–26. [Google Scholar]
- Wang C., García-Fernández D., Mas A., Esteve-Zarzoso B. (2015). Fungal diversity in grape must and wine fermentation assessed by massive sequencing, quantitative PCR and DGGE. Front. Microbiol. 6:1156 10.3389/fmicb.2015.01156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T. J., Bruns S., Lee S., Taylor J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide to Methods and Applications, ed. Innis M. (Orlando, FL: Academic Press; ), 315–322. [Google Scholar]
- Zott K., Claisse O., Lucas P., Coulon J., Lonvaud-Funel A., Masneuf-Pomarede I. (2010). Characterization of the yeast ecosystem in grape must and wine using real-time PCR. Food Microbiol. 27 559–567. 10.1016/j.fm.2010.01.006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.