Abstract
The numerous secondary metabolites in Streptomyces spp. are crucial for various applications. For example, cephamycin C is used as an antibiotic, and avermectin is used as an insecticide. Specifically, antibiotic yield is closely related to many factors, such as the external environment, nutrition (including nitrogen and carbon sources), biosynthetic efficiency and the regulatory mechanisms in producing strains. There are various types of regulatory genes that work in different ways, such as pleiotropic (or global) regulatory genes, cluster-situated regulators, which are also called pathway-specific regulatory genes, and many other regulators. The study of regulatory genes that influence antibiotic biosynthesis in Streptomyces spp. not only provides a theoretical basis for antibiotic biosynthesis in Streptomyces but also helps to increase the yield of antibiotics via molecular manipulation of these regulatory genes. Currently, more and more emphasis is being placed on the regulatory genes of antibiotic biosynthetic gene clusters in Streptomyces spp., and many studies on these genes have been performed to improve the yield of antibiotics in Streptomyces. This paper lists many antibiotic biosynthesis regulatory genes in Streptomyces spp. and focuses on frequently investigated regulatory genes that are involved in pathway-specific regulation and pleiotropic regulation and their applications in genetic engineering.
Keywords: Antibiotic biosynthetic gene cluster, Regulatory gene, Regulatory mechanism, Streptomyces, Secondary metabolites
Introduction
Secondary metabolites of Streptomyces, including antibiotics, immunomodulators, enzyme inhibitors and other bioactive substances, often have significant medicinal value. However, wild strains usually produce low levels of antibiotics. There is a large and complicated regulatory network in many Streptomyces strains, and the biosynthesis of one antibiotic in one strain may be controlled by more than one regulatory mechanism. For example, the production of actinorhodin (ACT) in Streptomyces coelicolor is regulated by both the cluster-situated regulator (CSR) actII-ORF4 (Fernández-Moreno et al. 1991) and the pleiotropic regulatory gene cprB (Onaka et al. 1998). Further, there is also more than one regulatory gene that affects the biosynthesis of a single antibiotic. For example, dnrI, dnrO and dnrN (Kitani et al. 2008; Parajuli et al. 2005) all regulate the production of daunorubicin (DNR) in Streptomyces peucetius.
To better and more comprehensively understand the mechanisms of antibiotic biosynthesis regulation, in this paper, we classify the antibiotic biosynthetic regulators in various Streptomyces strains that are associated with antibiotic production into three types. This work will provide a theoretical basis for the molecular perturbation of regulatory genes and will help with manipulating the antibiotic biosynthetic pathways and accordingly improving antibiotic production.
Regulatory genes involved in pathway-specific regulation
The CSRs, located within the antibiotic biosynthetic clusters, can modulate the antibiotic biosynthetic genes of the clusters in which they are included (Martín and Liras 2010; Rodríguez et al. 2013). The CSRs can only affect the biosynthetic pathway of a single, specific antibiotic and act as a master switch for biosynthesis of that individual antibiotic; this regulation is called pathway-specific regulation (Bibb 1996; Novakova et al. 2011). Some CSRs encode proteins that belong to a family known as the Streptomyces antibiotic regulatory proteins (SARPs). SARPs contain two characteristic structural domains: an OmpR DNA-binding domain and a bacterial transcription activation domain (BTAD) (Tanaka et al. 2007). The SARPs include the positive regulators ActII-ORF4 (Fernández-Moreno et al. 1991) and RedD (Wilson et al. 2001) in S. coelicolor and DnrI (Parajuli et al. 2005) in S. peucetius. In addition, some CSRs encode proteins belonging to another family, called the LuxR family, which is often found in Gram-negative bacteria (Lei et al. 2007). LuxR-type regulators contain a nucleotide triphosphate (NTP) binding motif at the N-terminus and a helix-turn-helix (HTH) motif at the C-terminus; examples include the positive regulators PimR (Antón et al. 2004) in Streptomyces natalensis and PikD (Xue et al. 1998) in Streptomyces venezuelae. There are many additional families of regulatory proteins, including the LysR and TetR families. The CSRs are shown in Table 1. Huang et al. (2005) reported that some CSRs can also control the expression of pleiotropic genes, and some pleiotropic regulators can affect the expression of CSRs. In this paper, we will not describe the cross-regulation between various types of regulators.
Table 1.
Regulatory genes involved in pathway-specific regulation
| Regulatory gene(s) | Gene(s) from | Product(s) of regulation | Function | Notes | References |
|---|---|---|---|---|---|
| actII-ORF4 | S. coelicolor | ACT | + | Encodes a SARP | Fernández-Moreno et al. (1991) |
| mmyR | S. coelicolor | Methylenomycin | − | A gene adjacent to mmyR provides positive regulation | Arias et al. (1999) |
| cdaR | S. coelicolor | Mmy and CDA | + | Encodes a SARP | Bibb (2005) |
| redD and redX | S. coelicolor | RED | + | Encode SARPs, and the transcription of redX regulates the transcription of redD | Romero-Rodríguez et al. (2015), Takano et al. (1992) and Wilson et al. (2001) |
| gdmRI and gdmRII | S. hygroscopicus 179977 | Geldanamycin | + | Regulate the transcription of pks, gdmF and gdnA, which are involved in biosynthesis of geldanamycin | (He et al. 2008) |
| tylR, tylS, tylP, tylQ tylT and tylU | S. fradiae | Tylosin |
tylR, tylS, tyl T and tylU: + tylQ: − |
tylT and tylS encode SARPs, and TylP is similar to γ-butyrolactone receptor proteins | Bate et al. (1999, 2006) and Stratigopoulos and Cundliffe (2002) |
| aveR, aveR1, aveR2 and aveT | S. avermitilis | Avermectin | + | aveR contains a HTH motif; AveT belongs to the TetR family and activates transcription of aveR | Ikeda et al. (2003) |
| alpT, alpU, alpV, alpW and alpZ | S. ambofaciens | Alpomycin | alpT, alpU, and alpV: +; alpW: − | alpT, alpU and alpV encode SARPs, alpW encodes a transcriptional repressor protein, and alpZ encodes a γ-butyrolactone receptor protein | Aigle et al. (2005) |
| ccaR | S. clavuligerus | Cephamycin C and clavulanic acid | + | Encodes a SARP | Pérez-Llarena et al. (1997) |
| pimR | S. natalensis | Pimaricin | + | Encodes a LuxR family protein, does not regulate its own transcription | Antón et al. (2004) |
| pikD | S. venezuelae | Pikromycin | + | Encodes a LuxR family protein | Xue et al. (1998) |
| monH, monRI and monRII | S. cinnamonensis | Monensin | + | MonH is similar to PikD, and monRI encodes a SARP | Oliynyk et al. (2003) |
| spbR | S. pristinaespiralis | Pristinamycin | + | SpbR is a γ-butyrolactone receptor | Mast et al. (2015) |
| papR1-R5 | S. pristinaespiralis | Pritinamycin | papR1, papR2 and papR4: +; papR3 and papR5: − | PapR1, PapR2, and PapR4 are SARPs, and PapR3 and PapR5 belong to the TetR family | Mast et al. (2015) |
| jadR* and jadR3 | S. venezuelae | Jadomycin B | − | JadR* is a TetR-like protein, and JadR3 represses jadR2 and jadR3 but activates jadR1 | Yang et al. (2001), Zhang et al. (2013) and Zou et al. (2014) |
| nysRI-RIII | S. noursei ATCC | Nystatin | + | Deletion of nysRI abolishes the transcription of nysRII-III | Sekurova et al. (2004) |
| amph RI- RIII | S. nodosus | Amphotericin | + | All contain a HTH motif in C-terminal | Carmody et al. (2004) |
| fscRI-RIII | S. pp. FR008 | Candicidin | + | Encodes a LuxR family protein | Chen et al. (2003) |
| dnrI, dnrO and dnrN | S. peucetius | Daunorubicin | dnrI: +; dnrO: −; dnrN: + | DnrN is a RR belonging to the Uhp-LuxR superfamily and activates the transcription of dnrI, and DnrO positively controls dnrN | Otten et al. (2000) and Parajuli and Moon (2002) |
| strR | S. griseus | Streptomycin | + | Regulates streptomycin by activating the expression of strA and strB | Distler et al. (1987) |
| rapH, rapG and rapY | S. hygroscopicus | Rapamycin |
rapH, rapG: + rapY: − |
RapG and RapY each contain a HTH motif, and RapH contains a DNA-binding motif and an ATP-binding site | Yoo et al. (2015) |
| srrX, srrY, srrZ and srrB | S. rochei | Lankamycin and lancadicin |
srrX and ssrY: + for both ssrZ: + for lankamycin srrB: − for both |
srrY and srrZ encode SARPs, and ssrY positively regulates ssrZ | Arakawa et al. (2007) and Suzuki et al. (2010) |
| scbR and scbR2 | S. coelicolor | ACT, RED, CDA and yCPK | ACT, CDA and RED: + yCPK: − |
ScbR is a γ-butyrolactone receptor protein, and ScbR2 is an antibiotic receptor protein | Li et al. (2015a) |
| polR and polY | S. cacaoi | Polyoxin | + | Both encode SARPs, and the transcription of polR is positively regulated by polY | Hwang et al. (2003) |
| aur1PR3 and aur1PR4 | S. aureofaciens | Auricin | + | Both encode SARPs, aur1PR3 is controlled by Aur1R, and Aur1P directly regulates the expression of aur1PR4 | Rehakova et al. (2013) |
| barA, barB and varR, | S. virginiae | Virginiamycin | − | BarA, BarB and VarR are TetR-like regulators, and the transcription of barB is tightly repressed by BarA | Matsuno et al. (2003), Nakano et al. (2000) and Namwat et al. (2001) |
| vmsS, vmsT and vmsR | S. virginiae | Virginiamycin M and virginiamycin S | vmsS and vmsR: + for virginiamycin M and virginiamycin S; vmsT: − for virginiamycin M | VmsS and VmsR are SARPs, and VmsT is a RR of a TCS | Pulsawat et al. (2009) |
| aur1R | S. aureofaciens | Auricin | − | aur1R encodes a homolog of the TetR family, and Aur1R represses the expression of aur1P | Novakova et al. (2010) |
| fdmR1 | S. griseus | Fredericamycin | + | Encodes a homologue of SARPs | Chen et al. (2008) |
| thnI | S. cattleya | Thienamycin | + | ThnI resembles LysR-type transcriptional activators and contains a HTH motif | Rodríguez et al. (2008) |
| thnU | S. cattleya | Cephamycin C | + | Encodes a SARP | Rodríguez et al. (2008) |
| asuR1, asuR2, asuR4 and asuR6 | S. nodosus | Asukamycin | + | AsuR1 and AsuR6 belong to the LuxR family, AsuR2 belongs to the TetR family, and asuR5 encodes a SARP | Xie et al. (2012) |
| farR3 and farR4 | S. lavendulae FRI-5 | Indigoidine |
farR3: + farR4: − |
Both encode SARPs, FarR3 positively controls the biosynthesis of indigoidine, and FarR4 negatively controls the expression of farX, farA, farR1 and farR2 | Kitani et al. (2008) and Kurniawan et al. (2014) |
| papR6 | S. pristinaespiralis | Pritinamycin II | + | PapR6 is an orphan RR | Dun et al. (2015) and Mast et al. (2015) |
| redZ | S. coelicolor | RED | + | Encodes a NarL-type RR, and the transcription of redD depends on redZ and the translation of redZ depends on bldA | Guthrie et al. (1998) and Wang et al. (2009) |
| ssaA | Streptomyces sp. strain SS | Sansanmycin | + | SsaA has a N-terminal fork head-associated (FHA) domain and a C-terminal LuxR-type HTH motif | Li et al. (2013) |
| vlmI | S. viridifaciens | Valanimycin | + | vlmI encodes a SARP and can complement a redD mutation; | Garg and Parry (2010) |
| nanR1, nanR2 and nanR4 | S. nanchangensis | Nanchangmycin |
nanR1, nanR2: + nanR4: − |
nanR1 and nanR2 encode SARPs, and nanR4 is an AraC-family transcriptional regulator and represses the transcription of nanR1 and nanR2 | Yu et al. (2012) |
+ Represents positive regulation, − represents negative regulation
tylR, tylP, tylQ, tylS and tylT
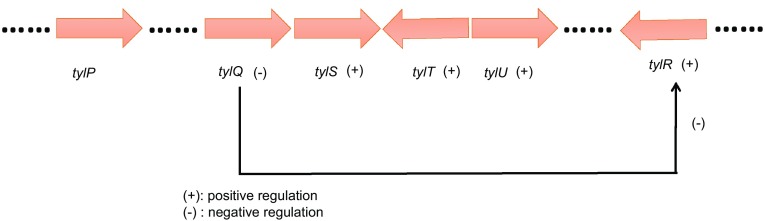
Eli Lilly and Company obtained the tylosin biosynthetic gene cluster from Streptomyces fradiae. Five regulators of this cluster are all CSRs. Sequence analysis identified TylP as a γ-butyrolactone (GBL) signal receptor (Bate et al. 1999; Wilson et al. 2001), and there were several indications that TylP is an effector-binding regulator and a regulator of tylosin biosynthesis (Bignell et al. 2007). The protein TylQ was reported to be a transcriptional repressor that blocked tylosin biosynthesis by controlling the expression of tylR (Stratigopoulos and Cundliffe 2002). Studies revealed that tylT and tylS encode proteins belonging to the SARPs family (Bate et al. 1999). Bate et al. (2006) identified a new regulatory gene, tylU, in the tylosin biosynthetic gene cluster. Targeted disruption of tylU decreased tylosin yield by approximately 80%, demonstrating that tylU is a positive regulatory gene for tylosin biosynthesis. TylR, a CSR, was able to regulate the core polyketide genes, but it primarily affected tylosin biosynthesis (Bate et al. 1999). The tylosin biosynthetic gene cluster is shown in Fig. 1.
Fig. 1.
Tylosin biosynthetic gene cluster of S. fradiae. Five regulatory genes are shown: TylP is a GBL signal receptor. TylQ is a transcriptional repressor and blocks tylosin biosynthesis by controlling the expression of tylR. tylT and tylS encode cluster-situated regulatory proteins of the SARPs family. tylU is a positive regulatory gene of tylosin biosynthesis. TylR is also a CSR
dnrI, dnrO and dnrN
A previous report (Parajuli and Moon 2002) has demonstrated that the DNR producer S. peucetius possesses two DNA segments, dnrR1 and dnrR2. Sequence analysis of dnrR1 and the subsequent inactivation of dnrI, which is contained within dnrR1, suggested the involvement of dnrI in the transcription of the biosynthetic genes of DNR. The disruption of dnrI resulted in the absence of DNR production (Madduri and Hutchinson 1995), and the overexpression of dnrI under the control of the strong ermE* promoter increased the production of DNR (Malla et al. 2010). These results indicated that dnrI positively regulates DNR biosynthesis. dnrN encodes a response regulator (RR) of UhpA-LuxR superfamily of regulatory proteins and has a motif that is highly similar to the HTH DNA-binding motif. An earlier study showed that reintroduction of dnrN into a dnrI:aphII mutant failed to restore DNR production. This suggested that DnrN activates the transcription of dnrI in the regulatory cascade; dnrI, in turn, positively triggers the transcription of DNR biosynthetic genes (Otten et al. 1995). dnrO, which is a negative regulatory gene, encodes a DNA-binding protein. DnrO has been shown to regulate antibiotic yield in S. peucetius by positively controlling dnrN (Otten et al. 2000; Parajuli and Moon 2002).
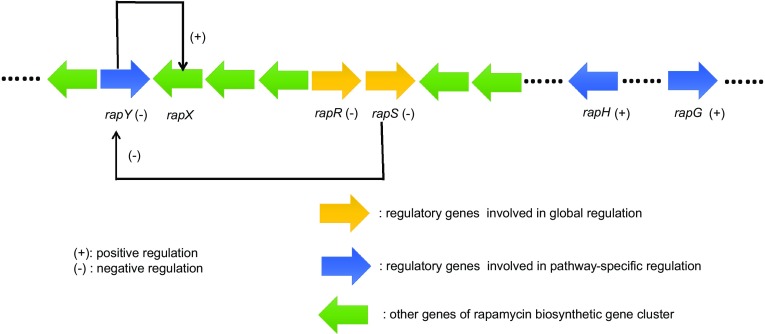
rapH, rapG and rapY
DNA sequence analysis of rapH and rapG in Streptomyces hygroscopicus revealed that RapH and RapG share significant similarity with two positive transcriptional families, the LAL and AraC families, respectively (Kuščer et al. 2007). RapH contained a DNA-binding motif and an ATP-binding site, while RapG contained a HTH DNA-binding motif (Yoo et al. 2015). In one study, antibiotic production was increased by 50% only if copies of both rapH and rapG under the control of their native promoter regions were introduced. Further, the complementation of rapH and rapG deletion mutants under the control of their native promoters led to a restoration of rapamycin production to parental levels (Yoo et al. 2015). The overexpression of both genes led to an abundant rapamycin synthesis, while the deletion of rapG and rapH caused a total shutdown of antibiotic production, suggesting that rapG and rapH play active roles in antibiotic biosynthesis. Furthermore, these genes cannot exert regulatory effects on the rapamycin biosynthetic gene cluster independent of each other. Yoo et al. (2015) found that the overexpression of rapY caused a drastic reduction in antibiotic production, while deletion of rapY increased antibiotic production by approximately fivefold. RapY, which contains an HTH motif near its N-terminus, plays a negative role in rapamycin production. The antibiotic regulatory genes in the biosynthetic gene cluster of rapamycin are shown in Fig. 2.
Fig. 2.
The location of regulatory genes in rapamycin biosynthetic gene cluster of S. hygroscopicus. RapY inhibits the transcription of rapX which is an ABC-transporter gene. Together, rapS and rapR negatively regulate most of the rapamycin biosynthetic genes. RapS also represses the expression of rapY. RapH and RapG positively regulate rapamycin biosynthesis
polY and polR
Li et al. (2009) found that the deletion of polR completely blocked polyoxin (POL) biosynthesis, which was complemented by introducing a copy of polR into the mutant. Similarly, the yield of POL was found to increase with the presence of an additional copy of polR in the mutant. PolR is necessary for the transcription of many structural genes in the POL biosynthetic gene cluster. Another CSR, polY in S. cacaoi, positively controls the production of this antibiotic. Both polR and polY encode proteins belonging to the SARPs family, and the expression of polR depends on the activity of PolY (Kitani et al. 2001).
aur1R, aur1P, aur1PR3 and aur1PR4
Some studies (Novakova et al. 2010, 2011; Rehakova et al. 2013) described four genes regulating auricin production in Streptomyces aureofaciens. aur1R encodes a homologue similar to the members of the TetR family. Antibiotic production was much higher in the disrupted strain than in the parental strain, which suggested a negative regulatory effect of aur1R on auricin production (Novakova et al. 2010). Aur1R can specifically negatively control the expression of aur1P, and this repression is released by auricin or its intermediates. In another experiment (Rehakova et al. 2013), aur1P was deleted from the chromosome, and no auricin was produced in the mutant, which suggested that aur1P is critical for the biosynthesis of auricin and exerts a positive effect on the expression of the biosynthetic genes of auricin. Aur1P belongs to the OmpR subfamily, which is similar to the RRs of two-component systems (TCSs). aur1PR3 and aur1PR4 encode proteins that are highly similar to those belonging to SARP family. The disruption of aur1PR3, an activator of auricin, resulted in a dramatic decrease in antibiotic production compared to the wild-type parent. Additionally, the expression of aur1PR3 was controlled by Aur1R (Novakova et al. 2011). Aur1P directly regulated the expression of aur1PR4 because its promoter was dependent on Aur1P (Rehakova et al. 2013).
thnI and thnU
There are two regulators, thnI and thnU, located in the thienamycin gene cluster of Streptomyces cattleya. ThnI is similar to the members of LysR family, and they all have a highly conserved HTH DNA-binding domain. A deletion mutant constructed by gene replacement failed to produce thienamycin, thereby revealing the importance of thnI in thienamycin synthesis. Gene expression analysis of the thienamycin gene cluster demonstrated that ThnI is a positive regulator and that it can control the expression of several genes involved in the assembly and export of thienamycin (Rodríguez et al. 2008). thnU encodes a positive regulator that belongs to the SARPs family. HPLC–MS analysis of a thnU mutant constructed using the same method that was used for thnI indicated that inactivation of thnU resulted in a loss of the production of Cephamycin C, whereas thienamycin synthesis was not affected. These results revealed the positive role of ThnI in thienamycin biosynthesis and the relevance of ThnU in cephamycin C biosynthesis (Rodríguez et al. 2008).
Regulatory genes involved in pleiotropic regulation
Many regulatory genes, which are mostly located outside of biosynthetic gene clusters, have pleiotropic (or global) effects on the production of multiple secondary metabolites or on both secondary metabolites and morphological development. In Streptomyces, the most abundant pleiotropic regulators belong to the TCSs, which are the predominant signal transduction systems in bacteria (Stock et al. 2000; Hakenbeck and Stock 1996). Other pleiotropic regulators can regulate antibiotic production in association with many small molecules called γ-butyrolactones (GBLs). This paper divides pleiotropic regulators into two subtypes. One type includes TCSs, orphan response regulation [e.g., farR1 (Kitani et al. 2008)], orphan histidine kinase regulation [e.g., ohkA (Lu and Jiang 2013)] and other special TCSs [e.g., abrC1/C2/C3 (Yepes et al. 2011)]. This subtype of pleiotropic regulatory genes is shown in Table 2a. The other includes the pleiotropic regulators closely associated with GBLs. To date, approximately 13 GBLs have been discovered, including A-factor from S. griseus (Ohnishi et al. 1999), IM-2 from S. lavendulae (Sato et al. 1989), SCB1, SCB2 and SCB3 (Hsiao et al. 2009; Takano et al. 2005), VBs from S. virginiae (Yamada et al. 1987) and factor 1 from S. viridochromogenes (Sato et al. 1989). Moreover, a few pleiotropic regulators contain a TPR structural domain [such as nsdA (Yu et al. 2006)], a protein repeat sequence consisting of 34 amino acids, which encodes an HTH secondary structural fragment. This type of pleiotropic genes is shown in Table 2b.
Table 2.
Regulatory genes involved in pleiotropic regulation
| Regulatory gene(s) | Gene(s) from | Product(s) of regulation | Function | Notes | References |
|---|---|---|---|---|---|
| a) | |||||
| afsQ1/afsQ2 | S. lividans | ACT | + | Ishizuka et al. (1992) | |
| afsK/afsR | S. coelicolor and S. griseus | ACT | + | AfsR is the transcriptional activator of afsS, which can activate actII-ORF4 |
Atsushi et al. (1994) |
| afsR-p/afsS | S. peucetius | Adriamycin | + | Parajuli et al. (2005) | |
| cutR/cutS | S. coelicolor and S. lividans | ACT | − | Chang et al. (1996) | |
| ecrA1/ecrA2 and ecrE2/ecrE1 | S. coelicolor | RED | + | Coordinates with the expression of redD | Huang et al. (2001) |
| orfX/orf41 | S. avermitilis | Avermectin | + | orfX exerts regulation by itself or by the collaboration of orf41 with orfX | Hwang et al. (2003) |
| phoR/phoP | S. coelicolor and S. lividans | ACT and RED | − | PhoP belongs to the Ormp family | Sola-Landa et al. (2003) |
| valP/valQ | S. hygroscopicus 5008 | Validamycin | Bai et al. (2006) | ||
| absA1/absA2 | S. coelicolor | CDA, yCPK and albaflavenone | − | Sheeler et al. (2005) | |
| rapR/rapS | S. hygroscopicus | Rapamycin | − | RapS represses the expression of rapY | Yoo et al. (2015) |
| rapA1/rapA2 | S. coelicolor | ACT and yCPK | + | The regulation of RapA1/A2 depends on ActII-ORF4 and KasO | Lu et al. (2007) |
| draK/R | S. coelicolor | ACT, yCPK and RED | ACT: + RED and yCPK: − |
DraR binds to the promoter regions of actII-ORF4 and cpkO | Rodríguez et al. (2013) |
| abrA1/A2 | S. coelicolor | ACT, CDA and RED | − | Yepes et al. (2011) | |
| SCO0203/0204 | S. coelicolor | ACT | − | Wang et al. (2009b) | |
| ohkA | S. coelicolor | ACT, CDA and RED | − | No identified RR matches OhkA | Lu et al. (2011) |
| aur1P | S. aureofaciens | Auricin | + | aur1P encodes a protein similar to the RRs | Novakova et al. (2005) |
| farR1 | S. lavendulae FRI-5 | Nucleoside antibiotics and indigoidine | FarR1 is an orphan RR | Kitani et al. (2008) | |
| glnR | S. coelicolor | RED and ACT | + | GlnR belongs to the OmpR family and indirectly regulates the production of antibiotics in response to changes in nitrogen availability | Pullan et al. (2011) |
| SCO3818 | S. coelicolor | ACT | − | SCO0203 can phosphosphorylate SCO0204 and SCO3818, and there is a functional correlation between SCO0203 and SCO3818 | Wang et al. (2009b) |
| jadR1/jadR2 | S. venezuelae | Jadomycin B |
jadR1: + jadR2: − |
The jadR1 and jadR2 genes represent a novel TCS linking antibiotic synthesis to stress; jadR1 encodes a RR; jadR2 encodes a TetR-like protein, and JadR2 is a pseudo γ-butyrolactone receptor | Yang et al. (2001) |
| abrC1/C2/C3 | S. coelicolor | ACT, CDA and RED | + | AbrC1 and AbrC2 are HKs, and AbrC3 is a RR | Yepes et al. (2011) |
| b) | |||||
| nsdA and nsdB | S. coelicolor | ACT and CDA | − | Each encodes a protein containing a TPR structure | Yu et al. (2006) |
| tcrA | S. coelicolor | All secondary metabolites | − | Liu and Yang (2006) | |
| afsR2 | S. lividans and S. coelicolor | ACT and RED | + | Lee et al. (2000) | |
| afsB | S. lividans and S. coelicolor | ACT, methylenomycin, CDA and RED | + | Horinouchi et al. (1989) | |
| barX | S. virginiae | Virginiamycin | BarX is an AfsA-like protein | Bate et al. (1999) and Pulsawat et al. (2007) | |
| farA | S. lavendulae | Nucleoside antibiotics | − | IM-2 binds to the FarA receptor to regulate the signal transduction of secondary metabolism | Kitani et al. (2001) |
| arpA | S. griseus | Streptomycin | − | ArpA is an A-factor receptor protein | Hong et al. (2007) and Kato et al. (2004) |
| adpA | S. griseus | Streptomycin | + | Encodes an Arac/XylS family protein and has two HTH motifs at the C-terminal | Higo et al. (2012), Ohnishi et al. (1999, 2005) and Zhu et al. (2005) |
| bldD | S. coelicolor | ACT, indigoidine, CDA and methylenomycin | + | BldD has a C-terminal domain of unknown function and an N-terminal domain that mediates DNA binding and dimerization | Den Hengst et al. (2010) |
| bldA | S. coelicolor | ACT | + | BldA regulates the production of antibiotics by controlling the activator ActII-ORF4 | Fernández-Moreno et al. (1991) |
| crp | S. coelicolor | ACT, RED and CDA | + | Crp is a member of the cAMP receptor protein/fumarate-nitrate-reductase family of regulators | Gao et al. (2012) |
| wblA | S. coelicolor | ACT, RED and CDA | − | WblA is a protein of the WhiB family | Kang et al. (2007) |
| atrA | S. coelicolor | ACT | + | AtrA is a TetR-like protein, AtrA positively controls the transcription of actII-ORF4 | Li et al. (2015b) |
| rrdA | S. coelicolor | RED and ACT | RED: − ACT: + |
RrdA belongs to the TetR family, and RrdA negatively regulates RED by controlling the abundance of RedD mRNA | Ou et al. (2009) |
| avaR3 | S. avermitilis | Avermectin and filipin | Avermectin: +; filipin: − | AvaR3 is a γ-butyrolactone autoregulator receptor homologue | Miyamoto et al. (2011) |
| cprA | S. coelicolor | ACT and RED | + | Encodes an AprA analogue | Onaka et al. (1998) |
| cprB | S. coelicolor | ACT | − | CprB shows high sequence similarity to CprA | Onaka et al. (1998) |
+ Represents positive regulation, − represents negative regulation
For TCSs, there are two types of kinase super-families in Streptomyces that regulate secondary metabolites. One is the histidine kinase family. This family consists of two proteins, a phosphate donor—HK (histidine kinase) and a phosphate receptor—RR. Most of the HKs that belong to membrane-spanning proteins have three functional structural domains: a sensing domain, a transmitter domain and an ATPase domain. RRs contain two domains: a receiver domain and an effector domain. A HK activates itself by first phosphorylating its conserved histidine residues and then transferring the phosphate groups to the conserved asparagine acid residues of the RR. The phosphorylated RR then regulates the expression of other genes (Lu and Jiang 2013). This signal transduction process is shown in Fig. 3. Genes belonging to this family include afsQ1/afsQ2 (Ishizuka et al. 1992), cutR/cutS (Tanaka et al. 2007) and ecrA1/ecrA2 (Huang et al. 2001). The other type is the serine/threonine and tyrosine kinase family, whose members transmit signals to regulate secondary metabolism via a series of cascade reactions, such as phosphorylation or dephosphorylation of proteins triggered by external environmental changes. AfsR/AfsS (Tanaka et al. 2007) belongs to the serine/threonine and tyrosine kinase family. The genomic sequence analysis of S. coelicolor which is a model strain was performed in 2002. Bioinformatic analysis revealed that S. coelicolor contains many TCSs, encompassing 84 HKs and 80 RRs. Of these, 67 HKs have been matched with 67 RRs and are adjacent to genes encoding RRs, while the remaining TCSs are orphan HKs and orphan RRs (Hutchings et al. 2004).
Fig. 3.
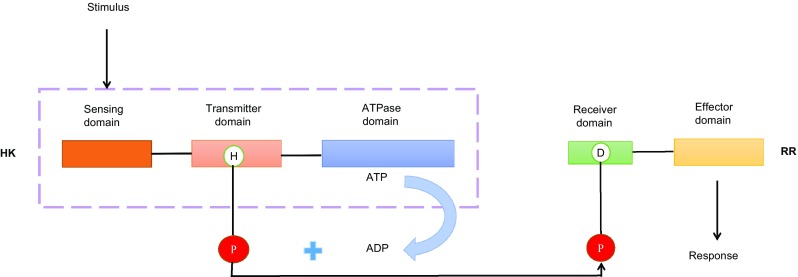
The process of signal transduction in TCSs
nsdA and nsdB
The pleiotropic negative regulatory gene nsdA can be found in many Streptomyces spp. The nsdA genes from various Streptomyces spp. share 77–100% similarity with each other, and nsdA homologous genes are also present in many Streptomyces strains (Yu et al. 2006). NsdA, which contains a TPR structural domain, plays a negative role in sporulation, morphological differentiation and antibiotic synthesis. The overproduction of actinorhodin (ACT), calcium-dependent antibiotic (CDA) and methylenomycin was detected in a nsdA mutant, and the deletion of the nsdA in Streptomyces lividans also resulted in the expression of silent ACT biosynthetic genes. These results indicated that NsdA may silence the ACT biosynthetic gene cluster by repressing the expression of CSRs (Yu et al. 2006). There is also a TPR-like structural domain in the nsdB gene product whose disruption gives rise to ACT production, but NsdB does not affect morphological differentiation. The deletion of nsdB, which is a negative regulator of CDA, resulted in an increase in CDA production. nsdB has been shown to control antibiotic biosynthesis along with nsdA but has no influence on the expression of nsdA at the RNA level (Yu et al. 2006).
afsR2 and afsB
The protein encoded by afsR2 is 63 amino acids long (Lee et al. 2000), and AfsR2 is a positive regulator of ACT and undecylprodigiosin (RED). Lee et al. (2000) cloned a regulatory gene from S. avermitilis that was homologous to afsR2 from S. lividans and S. coelicolor. The integration of multiple copies of afsR2 into the wild-type strain resulted in approximately twofold overproduction of avermectin. ACT and RED can be regulated by pleiotropic regulatory genes in addition to synthetic gene clusters. afsB encodes a DNA-binding protein and is a pleiotropic regulator that is essential for ACT biosynthesis. An increase in copies of afsB significantly improved the production of ACT and RED. The introduction of afsB into S. lividans triggered the transcription of ACT biosynthetic genes that were otherwise silent (Horinouchi et al. 1989). afsB provides positive regulation by stimulating its target genes and can trigger RED biosynthesis in S. lividans.
cprA and cprB
In S. coelicolor, the gene products of cprA and cprB are highly similar to each other. Onaka et al. (1998) found that the deletion of cprA led to a sharp reduction of ACT and RED. The introduction of cprA into the parental strain resulted in increased antibiotic production. Those results demonstrated that cprA positively regulated the biosynthesis of ACT and RED (Onaka et al. 1998). The disruption of cprB led to a precocious overproduction of ACT, but no change was detected in the production of RED. These results revealed that cprB only negatively regulated the biosynthesis of ACT (Onaka et al. 1998). Another study showed that cprA and cprB activated antibiotic biosynthesis in S. coelicolor via the GBL quorum-sensing pathway (Bhukya et al. 2014).
adpA and arpA
adpA, which is a pleiotropic regulator, encodes a 405-amino-acid protein containing a HTH DNA-binding motif in its central region. AdpA showed high sequence similarity to the transcriptional regulators of the AraC/XylS family (Gallegos et al. 1997). To determine the function of adpA in S. griseus, a disruption strain was generated using in-frame deletion. No streptomycin was detected in the disrupted strain, but streptomycin production was recovered when adpA was introduced, showing that adpA exerted a positive effect on streptomycin biosynthesis (Higo et al. 2012; Ohnishi et al. 1999). ArpA contains a region resembling the HTH motif that is present in many transcriptional families. ArpA is an A-factor receptor protein that negatively controls the production of streptomycin in S. griseus. ArpA behaved as a repressor-type regulator for streptomycin production (Onaka et al. 1995). adpA, arpA and A-factor form a widespread regulatory cascade in Streptomyces, termed the A-factor-adpA-arpA regulatory cascade. There is a model for the A-factor regulatory cascade that leads to streptomycin biosynthesis (Ohnishi et al. 1999): A-factor gradually accumulates, and when the concentration of A-factor reaches a certain point, it binds ArpA and causes ArpA to dissociate from the promoter, thus leading to the transcription and translation of adpA. AdpA then activates the transcription of strR. Therefore, the induction of StrR positively regulates the transcription of most of the streptomycin biosynthetic genes (Retzlaff and Distler 1995).
afsQ1/afsQ2 and cutR/cutS
afsQ1/afsQ2 is representative of the TCSs, with afsQ1 encoding an aspartic acid RR and AfsQ2 belonging to the sensing kinases. AfsQ2, located in the membrane, is autophosphorylated at His-294 upon sensing an environmental signal, and the signal is then transferred to the Asp-52 residue of the AfsQl protein in the cytoplasm (Ishizuka et al. 1992). afsQ1/afsQ2 plays a pleiotropic role in the secondary metabolism of antibiotics. cutR/cutS is the second TCS found in Streptomyces that represses secondary metabolism, and cutR/cutS also negatively regulates antibiotic production. The time taken to produce ACT is 20 h shorter in the cutR/cutS mutant than in the parental strain. The inactivation of cutR/cutS triggers an increase in ACT production, and the introduction of cutR reverses this change (Chang et al. 1996).
afsR-p/afsS
Parajuli et al. (2005) isolated the afsR-p gene from S. peucetius ATCC 27952, and it had greater than 50% sequence similarity to afsR from S. coelicolor. The overproduction of doxorubicin, γ-actinorhodin, clavulanic acid and streptomycin (slight), respectively, was detected in strains of S. peucetius, S. lividans, S. clavuligerus and S. griseus strains that carried the afsR-p gene (Parajuli et al. 2005). This demonstrated that AfsR-p may activate various CSRs in antibiotic biosynthetic gene clusters in different ways, leading to the speculation that phosphorylated AfsR-p binds to the promoter region of afsS and that the latter then triggers other regulators to induce the production of certain secondary metabolites (Parajuli et al. 2005).
orfX/orf41 and phoR/phoP
Disruption of the orfX gene resulted in a significant but incomplete loss of the production of avermectin in S. avermitilis (Hwang et al. 2003). The increase in avermectin may result from the role of orfX itself or from the collaboration of orf41 with orfX. Recently, a new TCS, phoR-phoP, was found in S. lividans and S. coelicolor, and PhoP was indeed a member of the OmpR family. The PhoR-PhoP system may activate the formation of ACT and RED via a specific repressor protein with phosphate-controlled promoters, acting via a cascade mechanism (Sola-Landa et al. 2003).
valP/valQ and rapR/rapS
valP and valQ, which can regulate validamycin in S. hygroscopicus 5008, may encode a TCS regulatory protein consisting of a HK and a Sigma B PP2C-like phosphatase (Bai et al. 2006). RapR and RapS in Streptomyces rapamycinicus ATCC 29253 share high sequence identities with the RRs and HKs, respectively, of TCSs. Gene expression analysis demonstrated that most of the rapamycin biosynthetic genes were negatively controlled by rapS (probably in collaboration with rapR) and rapY (a CSR for the biosynthesis of rapamycin) (Yoo et al. 2015). In addition, RapS represses the expression of rapY gene and RapR/S is a repressor of rapamycin biosynthesis.
rapA1/A2, abrA1/A2 and abrC1/C2/C3
The rapA1 in S. coelicolor encodes a protein that belongs to the OmpR family, while the sequence of RapA2 shows characteristics that are typical of HKs (Lu et al. 2007). Lu et al. (2007) found that RapA1/A2 is an activator of ACT and a yellow cryptic polyketide (yCPK). The effect exerted by rapA1/A2 on the biosynthesis of these antibiotics may also depend on two CSRs, actII-ORF4 and kasO. The disruption of rapA1/A2 and subsequent experiments with the disrupted strain indicated that rapA1/A2 was a positive TCS regulator of ACT [which is encoded by a type II polyketide synthase (PKS)] and yCPK [which is encoded by a type I PKS in S. coelicolor (Lu et al. 2007)]. abrA1/A2 is also a TCS regulator that negatively regulates ACT, RED and CDA in S. coelicolor. However, abrC1/C2/C3 is composed of two HKs and one RR that positively regulate the abovementioned antibiotics. AbrC1 and AbrC2 are HKs, while AbrC3 is a RR (Yepes et al. 2011).
ohkA
OhkA was reported to be an orphan HK in S. coelicolor that repressed the biosynthesis of five known secondary metabolites: ACT, RED, CDA, yCPK and albaflavenone (Lu and Jiang 2013). The deletion of ohkA in S. coelicolor caused a drastic increase in the biosynthesis of antibiotics, especially of ACT and CDA (Räty et al. 2002). OhkA negatively regulates secondary metabolites by repressing CSRs in S. coelicolor.
Other regulatory genes
In addition to CSRs and pleiotropic regulators, there are many other regulatory genes that deserve attention. These include barZ in Streptomyces virginiae, which regulates virginiamycin (Pulsawat et al. 2007); ppk from S. lividans (Ghorbel et al. 2006 ); hyg1 and hyg3 (Palaniappan et al. 2006) from S. hygroscopicus NRRL 2388; and some regulatory genes whose functions remain unknown, such as claR, which is related to cephamycin C and clavulanic acid. Regulatory genes of this type are shown in Table 3.
Table 3.
Other regulatory genes
| Regulatory gene(s) | Gene(s) from | Product(s) of regulation | Function | Notes | References |
|---|---|---|---|---|---|
| hyg1 and hyg3 | S. hygroscopicus | Hygromycin A | Hyg1 and Hyg3 show sequence similarities to AfsR and StrR, respectively | Palaniappan et al. (2006) | |
| ppk | S. lincolnensis | ACT | − | Positively controlled by phoR/phoP | Ghorbel et al. (2006) |
| barZ | S. virginiae | Virginiamycin | Bate et al. (1999 and Pulsawat et al. (2007) | ||
| farR2 | S. lavendulae FRI-5 | Nucleoside antibiotics and indigoidine | FarR2 is a GBL receptor | Kitani et al. (2008) | |
| claR | S. clavuligerus | Cephamycin C and clavulanic acid | Cephamycin C: − Clavulanic acid: + |
ClaR contains two HTH motifs and is homologous with the LysR family | Pérez-Redondo et al. (1998) |
| stgR | S. coelicolor | ACT and RED | − | StgR belongs to the LysR family of transcriptional regulators and regulates antibiotics by indirectly repressing redD and actII-ORF4 | Mao et al. (2013) |
| lndYR | S. globisporus | Landomycin | + | Encodes a GntR-like regulator of the YtrA subfamily | Ostash et al. (2011) |
+ Represents positive regulation, − represents negative regulation
Discussion
The regulation of secondary metabolite biosynthetic gene clusters in Streptomyces spp. is currently receiving substantial attention. Many transcription units are apparently regulated by several metabolite regulatory genes via transcriptional activation or repression, and biosynthetic gene clusters often collaborate with transcriptional regulators. Furthermore, it is reported that many Streptomyces strains can produce antibiotics. Analyses of the genomic sequences of many Streptomyces spp., such as S. coelicolor and S. avermitilis, have also been completed. These analyses provide a basis for post-genomic projects involving Streptomyces and will effectively promote studies of the structure, function and expression regulation of biosynthetic genes. In addition, many regulatory genes can also be expressed in other strains and can influence antibiotic production by heterologous expression combined with other methods. For example, the integration of aveR, orfX, or afsR in the chromosomes of S. hygroscopicus promoted rapamycin production by approximately 3.8-fold, 1.2-fold or slightly, respectively (Huang et al. 2011). Currently, we can effectively influence antibiotic production by manipulating regulatory genes involved in pathway-specific regulation or pleiotropic regulation in Streptomyces. For example, as introduced in this paper, the aveR mutants in S. avermitilis lost the ability to synthesize avermectin, whereas the overexpression of this gene increased the yield (Ikeda et al. 2003). The overproduction of ACT, CDA and methylenomycin was detected in the nsdA mutant of S. coelicolor (Yu et al. 2006). Hence, studying the regulatory mechanisms of secondary metabolite biosynthesis, inactivating the transcriptional repressors and overexpressing the transcriptional activators in natural producing strains may allow the optimization of antibiotic production. Further, it will provide a crucial theoretical basis for improving antibiotic production and using regulators to activate silent gene clusters, thereby leading to the discovery of new drugs.
Acknowledgements
This work is supported by the Open Research Program of Key Laboratory of Synthetic Biology, the Chinese Academy of Sciences (SYN201612) and the Key Program of Sichuan Science and Technology Project (2017GZ0430). The authors thank all the supporting institutions.
Compliance with ethical standards
Funding
This study was funded by the Open Research Program of Key Laboratory of Synthetic Biology, the Chinese Academy of Sciences (SYN201612) and the Key Program of Sichuan Science and Technology Project (2017GZ0430).
Conflict of interest
All of the authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
References
- Aigle B, Pang X, Decaris B, Leblond P. Involvement of AlpV, a new member of the Streptomyces antibiotic regulatory protein family, in regulation of the duplicated type II polyketide synthase alp gene cluster in Streptomyces ambofaciens. J Bacteriol. 2005;187:2491–2500. doi: 10.1128/JB.187.7.2491-2500.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antón N, Mendes MV, Martín JF, Aparicio JF. Identification of PimR as a positive regulator of pimaricin biosynthesis in Streptomyces natalensis. J Bacteriol. 2004;186:2567–2575. doi: 10.1128/JB.186.9.2567-2575.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakawa K, Mochizuki S, Yamada K, Noma T, Kinashi H. γ-Butyrolactone autoregulator-receptor system involved in lankacidin and lankamycin production and morphological differentiation in Streptomyces rochei. Microbiology. 2007;153:1817–1827. doi: 10.1099/mic.0.2006/002170-0. [DOI] [PubMed] [Google Scholar]
- Arias P, Fernández-Moreno MA, Malpartida F. Characterization of the pathway-specific positive transcriptional regulator for actinorhodin biosynthesis in Streptomyces coelicolor A3(2) as a DNA-binding protein. J Bacteriol. 1999;181:6958–6968. doi: 10.1128/jb.181.22.6958-6968.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsushi M, Soon-Kwang H, Hiroshi I, Sueharu H, Teruhiko B. Phosphorylation of the AfsR protein involved in secondary metabolism in Streptomyces species by a eukary otic-type protein kinase. Gene. 1994;146:47–56. doi: 10.1016/0378-1119(94)90832-X. [DOI] [PubMed] [Google Scholar]
- Bai L, et al. Functional analysis of the validamycin biosynthetic gene cluster and engineered production of validoxylamine A. Chem Biol. 2006;13:387–397. doi: 10.1016/j.chembiol.2006.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bate N, Butler AR, Gandecha AR, Cundliffe E. Multiple regulatory genes in the tylosin biosynthetic c luster of Streptomyces fradiae. Chem Biol. 1999;6:617–624. doi: 10.1016/S1074-5521(99)80113-6. [DOI] [PubMed] [Google Scholar]
- Bate N, Bignell DRD, Cundliffe E. Regulation of tylosin biosynthesis involving ‘SARP-helper’ activity. Mol Microbiol. 2006;62:148–156. doi: 10.1111/j.1365-2958.2006.05338.x. [DOI] [PubMed] [Google Scholar]
- Bhukya H, Bhujbalrao R, Bitra A, Anand R. Structural and functional basis of transcriptional regulation by TetR family protein CprB from S. coelicolor A3(2) Nucleic Acids Res. 2014 doi: 10.1093/nar/gku587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb M. The regulation of antibiotic production in Streptomyces coelicolor A3(2) Microbiology. 1996;142:1335–1344. doi: 10.1099/13500872-142-6-1335. [DOI] [PubMed] [Google Scholar]
- Bibb MJ. Regulation of secondary metabolism in streptomycetes. Curr Opin Microbiol. 2005;8:208–215. doi: 10.1016/j.mib.2005.02.016. [DOI] [PubMed] [Google Scholar]
- Bignell DRD, Bate N, Cundliffe E. Regulation of tylosin production: role of a TylP-interactive ligand. Mol Microbiol. 2007;63:838–847. doi: 10.1111/j.1365-2958.2006.05541.x. [DOI] [PubMed] [Google Scholar]
- Carmody M, et al. Analysis and manipulation of amphotericin biosynthetic genes by means of modified phage KC515 transduction techniques. Gene. 2004;343:107–115. doi: 10.1016/j.gene.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Chang H, Chen MY, Shieh YT, Bibb MJ, Chen CW. The cutRS signal transduction system of Streptomyces lividans represses the biosynthesis of the polyketide antibiotic actinorhodin. Mol Microbiol. 1996;21:1075–1085. [PubMed] [Google Scholar]
- Chen S, et al. Organizational and mutational analysis of a complete FR-008/Candicidin gene cluster encoding a structurally related polyene complex. Chem Biol. 2003;10:1065–1076. doi: 10.1016/j.chembiol.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Chen Y, Wendt-Pienkowski E, Shen B. Identification and utility of FdmR1 as a Streptomyces antibiotic regulatory protein activator for fredericamycin production in Streptomyces griseus ATCC 49344 and heterologous hosts. J Bacteriol. 2008;190:5587–5596. doi: 10.1128/JB.00592-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Hengst CD, Tran NT, Bibb MJ, Chandra G, Leskiw BK, Buttner MJ. Genes essential for morphological development and antibiotic production in Streptomyces coelicolor are targets of BldD during vegetative growth. Mol Microbiol. 2010;78:361–379. doi: 10.1111/j.1365-2958.2010.07338.x. [DOI] [PubMed] [Google Scholar]
- Distler J, Ebert A, Mansouri K, Pissowotzki K, Stockmann M, Piepersberg W. Gene cluster for streptomycin biosynthesis in Streptomyces griseus: nucleotide sequence of three genes and analysis of transcriptional activity. Nucleic Acids Res. 1987;15:8041–8056. doi: 10.1093/nar/15.19.8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dun J, et al. PapR6, a putative atypical response regulator. Functions as a pathway-specific activator of pristinamycin II biosynthesis in Streptomyces pristinaespiralis. J Bacteriol. 2015;197:441–450. doi: 10.1128/JB.02312-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Moreno MA, Caballero J, Hopwood DA, Malpartida F. The act cluster contains regulatory and antibiotic export genes, direct targets for translational control by the bldA tRNA gene of streptomyces. Cell. 1991;66:769–780. doi: 10.1016/0092-8674(91)90120-N. [DOI] [PubMed] [Google Scholar]
- Gallegos MT, Schleif R, Bairoch A, Hofmann K, Ramos JL. Arac/XylS family of transcriptional regulators. Microbiol Mol Biol Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Hindra, Mulder D, Yin C, Elliot MA. Crp is a global regulator of antibiotic production in Streptomyces. MBio. 2012;3:1–12. doi: 10.1128/mBio.00407-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg RP, Parry RJ. Regulation of valanimycin biosynthesis in Streptomyces viridifaciens: characterization of VlmI as a Streptomyces antibiotic regulatory protein (SARP) Microbiology. 2010;156:472–483. doi: 10.1099/mic.0.033167-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghorbel S, Smirnov A, Chouayekh H, Sperandio B, Esnault C, Kormanec J, Virolle M-J. Regulation of ppk expression and in vivo function of Ppk in Streptomyces lividans TK24. J Bacteriol. 2006;188:6269–6276. doi: 10.1128/JB.00202-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie EP, Flaxman CS, White J, Hodgson DA, Bibb MJ, Chater KF. A response-regulator-like activator of antibiotic synthesis from Streptomyces coelicolor A3(2) with an amino-terminal domain that lacks a phosphorylation pocket. Microbiology. 1998;144(3):727–738. doi: 10.1099/00221287-144-3-727. [DOI] [PubMed] [Google Scholar]
- Hakenbeck R, Stock JB. Analysis of two-component signal transduction systems involved in transcriptional regulation. Methods Enzymol. 1996;273:281–300. doi: 10.1016/S0076-6879(96)73026-4. [DOI] [PubMed] [Google Scholar]
- He W, Lei J, Liu Y, Wang Y. Regulatory genes of geldanamycin biosynthesis. Chin J Biotechnol. 2008;24:717–722. doi: 10.1016/S1872-2075(08)60036-9. [DOI] [PubMed] [Google Scholar]
- Higo A, Hara H, Horinouchi S, Ohnishi Y. Genome-wide distribution of AdpA, a global regulator for secondary metabolism and morphological differentiation in Streptomyces. Revealed the extent and complexity of the AdpA regulatory network. DNA Res. 2012;19:259–274. doi: 10.1093/dnares/dss010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong B, Phornphisutthimas S, Tilley E, Baumberg S, McDowall KJ. Streptomycin production by Streptomyces griseus can be modulated by a mechanism not associated with change in the adpA component of the A-factor cascade. Biotechnol Lett. 2007;29:57–64. doi: 10.1007/s10529-006-9216-2. [DOI] [PubMed] [Google Scholar]
- Horinouchi S, Malpartida F, Hopwood D, Beppu T. afsB stimulates transcription of the actinorhodin biosynthetic pathway in Streptomyces coelicolor A3(2) and Streptomyces lividans. Mol Gen Genet. 1989;215:355–357. doi: 10.1007/BF00339742. [DOI] [PubMed] [Google Scholar]
- Hsiao N-H, et al. Analysis of two additional signaling molecules in Streptomyces coelicolor and the development of a butyrolactone-specific reporter system. Chem Biol. 2009;16:951–960. doi: 10.1016/j.chembiol.2009.08.010. [DOI] [PubMed] [Google Scholar]
- Huang J, Lih CJ, Pan KH, Cohen SN. Global analysis of growth phase responsive gene expression and regulation of antibiotic biosynthetic pathways in Streptomyces coelicolor using DNA microarrays. Genes Dev. 2001;15:3183–3192. doi: 10.1101/gad.943401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, et al. Cross-regulation among disparate antibiotic biosynthetic pathways of Streptomyces coelicolor. Mol Microbiol. 2005;58:1276–1287. doi: 10.1111/j.1365-2958.2005.04879.x. [DOI] [PubMed] [Google Scholar]
- Huang M, Li M, Feng Z, Liu Y, Chu Y, Tian Y. Enhanced rapamycin production in Streptomyces hygroscopicus by integrative expression of aveR, a LAL family transcriptional regulator. World J Microbiol Biotechnol. 2011;27:2103–2109. doi: 10.1007/s11274-011-0673-y. [DOI] [Google Scholar]
- Hutchings MI, Hoskisson PA, Chandra G, Buttner MJ. Sensing and responding to diverse extracellular signals? Analysis of the sensor kinases and response regulators of Streptomyces coelicolor A3(2) Microbiology. 2004;150:2795–2806. doi: 10.1099/mic.0.27181-0. [DOI] [PubMed] [Google Scholar]
- Hwang YS, Kim ES, Biró S, Choi CY. Cloning and analysis of a DNA fragment stimulating avermectin production in various Streptomyces avermitilis strains. Appl Environ Microbiol. 2003;69:1263–1269. doi: 10.1128/AEM.69.2.1263-1269.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda H et al. (2003) Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat Biotech 21:526–531. http://www.nature.com/nbt/journal/v21/n5/suppinfo/nbt820_S1.html [DOI] [PubMed]
- Ishizuka H, Horinouchi S, Kieser HM, Hopwood DA, Beppu T. A putative two-component regulatory system involved in secondary metabolism in Streptomyces spp. J Bacteriol. 1992;174:7585–7594. doi: 10.1128/jb.174.23.7585-7594.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S-H, Huang J, Lee HN, Hur YA, Cohen SN, Kim KS. Interspecies DNA microarray analysis identifies WblA as a pleiotropic down-regulator of antibiotic biosynthesis in Streptomyces. J Bacteriol. 2007;189:4315–4319. doi: 10.1128/JB.01789-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato J-Y, Miyahisa I, Mashiko M, Ohnishi Y, Horinouchi S. A single target is sufficient to account for the biological effects of the a-factor receptor protein of Streptomyces griseus. J Bacteriol. 2004;186:2206–2211. doi: 10.1128/JB.186.7.2206-2211.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitani S, Yamada Y, Nihira T. Gene replacement analysis of the butyrolactone autoregulator receptor (FarA) reveals that FarA acts as a novel regulator in secondary metabolism of Streptomyces lavendulae FRI-5. J Bacteriol. 2001;183:4357–4363. doi: 10.1128/JB.183.14.4357-4363.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitani S, Iida A, Izumi T, Maeda A, Yamada Y, Nihira T. Identification of genes involved in the butyrolactone autoregulator cascade that modulates secondary metabolism in Streptomyces lavendulae FRI-5. Gene. 2008;425:9–16. doi: 10.1016/j.gene.2008.07.043. [DOI] [PubMed] [Google Scholar]
- Kurniawan Y, Kitani S, Maeda A, Nihira T. Differential contributions of two SARP family regulatory genes to indigoidine biosynthesis in Streptomyces lavendulae FRI-5. Appl Microbiol Biotechnol. 2014;98:9713–9721. doi: 10.1007/s00253-014-5988-9. [DOI] [PubMed] [Google Scholar]
- Kuščer E, Coates N, Challis I, Gregory M, Wilkinson B, Sheridan R, Petković H. Roles of rapH and rapG in positive regulation of rapamycin biosynthesis in Streptomyces hygroscopicus. J Bacteriol. 2007;189:4756–4763. doi: 10.1128/JB.00129-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Hwang YS, Kim SS, Kim ES, Choi CY. Effect of a global regulatory gene, afsR2, from Streptomyces lividans on avermectin production in Streptomyces avermitilis. J Biosci Bioeng. 2000;89:606–608. doi: 10.1016/S1389-1723(00)80065-1. [DOI] [PubMed] [Google Scholar]
- Lei J, He W, Wang Y. Regulatory mechanism of morphological differentiation and secondary metabolism in Streptomyces. Pharm Biotechnol. 2007;14:225–229. [Google Scholar]
- Li R, et al. polR, a pathway-specific transcriptional regulatory gene, positively controls polyoxin biosynthesis in Streptomyces cacaoi subsp. asoensis. Microbiology. 2009;155:1819–1831. doi: 10.1099/mic.0.028639-0. [DOI] [PubMed] [Google Scholar]
- Li Q, Wang L, Xie Y, Wang S, Chen R, Hong B. SsaA, a member of a novel class of transcriptional regulators, controls sansanmycin production in Streptomyces sp. Strain SS through a feedback mechanism. J Bacteriol. 2013;195:2232–2243. doi: 10.1128/JB.00054-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wang J, Li SS, Ji JJ, Wang WS, Yang KQ. ScbR- and ScbR2-mediated signal transduction networks coordinate complex physiological responses in Streptomyces coelicolor. Sci Rep. 2015;5:14831. doi: 10.1038/srep14831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, et al. Binding of a biosynthetic intermediate to AtrA modulates the production of lidamycin by Streptomyces globisporus. Mol Microbiol. 2015;96:1257–1271. doi: 10.1111/mmi.13004. [DOI] [PubMed] [Google Scholar]
- Liu J, Yang K. Functional analyses of TcrA-a TPR-containing regulatory protein in Streptomyces coelicolor A3(2) Wei Sheng Wu Xve Bao. 2006;46:33–37. [PubMed] [Google Scholar]
- Lu Y, Jiang W. Perspectives on two-component systems involved in regulation of secondary metabolism in Streptomyces. Microbiol China. 2013;40:1847–1859. [Google Scholar]
- Lu Y, et al. Characterization of a novel two-component regulatory system involved in the regulation of both actinorhodin and a type I polyketide in Streptomyces coelicolor. Appl microbiol biotechnol. 2007;77:625–635. doi: 10.1007/s00253-007-1184-5. [DOI] [PubMed] [Google Scholar]
- Lu Y, et al. An orphan histidine kinase, OhkA, regulates both secondary metabolism and morphological differentiation in Streptomyces coelicolor. J Bacteriol. 2011;193:3020–3032. doi: 10.1128/JB.00017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madduri K, Hutchinson CR. Functional characterization and transcriptional analysis of the dnrR1 locus, which controls daunorubicin biosynthesis in Streptomyces peucetius. J Bacteriol. 1995;177:1208–1215. doi: 10.1128/jb.177.5.1208-1215.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malla S, Niraula NP, Liou K, Sohng JK. Improvement in doxorubicin productivity by overexpression of regulatory genes in Streptomyces peucetius. Res Microbiol. 2010;161:109–117. doi: 10.1016/j.resmic.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Mao X, Sun Z, Liang B, Wang Z, Feng W, Huang F, Li Y. Positive feedback regulation of stgR expression for secondary metabolism in Streptomyces coelicolor. J Bacteriol. 2013;195:2072–2078. doi: 10.1128/JB.00040-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín J-F, Liras P. Engineering of regulatory cascades and networks controlling antibiotic biosynthesis in Streptomyces. Curr Opin Microbiol. 2010;13:263–273. doi: 10.1016/j.mib.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Mast Y, Guezguez J, Handel F, Schinko E. A complex signaling cascade governs pristinamycin biosynthesis in Streptomyces pristinaespiralis. Appl Env Microbiol. 2015;81:6621–6636. doi: 10.1128/AEM.00728-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuno K, Yamada Y, Lee CK, Nihira T. Identification by gene deletion analysis of barB as a negative regulator controlling an early process of virginiamycin biosynthesis in Streptomyces virginiae. Arch Microbiol. 2003;181:52–59. doi: 10.1007/s00203-003-0625-5. [DOI] [PubMed] [Google Scholar]
- Miyamoto KT, Kitani S, Komatsu M, Ikeda H, Nihira T. The autoregulator receptor homologue AvaR3 plays a regulatory role in antibiotic production, mycelial aggregation and colony development of Streptomyces avermitilis. Microbiology. 2011;157:2266–2275. doi: 10.1099/mic.0.048371-0. [DOI] [PubMed] [Google Scholar]
- Nakano H, Lee C-K, Nihira T, Yamada Y. A null mutant of the Streptomyces virginiae barA gene encoding a butyrolactone autoregulator receptor and its phenotypic and transcriptional analysis. J Biosci Bioeng. 2000;90:204–207. doi: 10.1016/S1389-1723(00)80111-5. [DOI] [PubMed] [Google Scholar]
- Namwat W, Lee CK, Kinoshita H, Yamada Y, Nihira T. Identification of the varR gene as a transcriptional regulator of virginiamycin S resistance in Streptomyces virginiae. J Bacteriol. 2001;183:2025–2031. doi: 10.1128/JB.183.6.2025-2031.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novakova R, Homerova D, Feckova L, Kormanec J. Characterization of a regulatory gene essential for the production of the angucycline-like polyketide antibiotic auricin in Streptomyces aureofaciens CCM 3239. Microbiology. 2005;151:2693–2706. doi: 10.1099/mic.0.28019-0. [DOI] [PubMed] [Google Scholar]
- Novakova R, Kutas P, Feckova L, Kormanec J. The role of the TetR-family transcriptional regulator Aur1R in negative regulation of the auricin gene cluster in Streptomyces aureofaciens CCM 3239. Microbiology. 2010;156:2374–2383. doi: 10.1099/mic.0.037895-0. [DOI] [PubMed] [Google Scholar]
- Novakova R, Rehakova A, Kutas P, Feckova L, Kormanec J. The role of two SARP family transcriptional regulators in regulation of the auricin gene cluster in Streptomyces aureofaciens CCM 3239. Microbiology. 2011;157:1629–1639. doi: 10.1099/mic.0.047795-0. [DOI] [PubMed] [Google Scholar]
- Ohnishi Y, Kameyama S, Onaka H, Horinouchi S. The A-factor regulatory cascade leading to streptomycin biosynthesis in Streptomyces griseus: identification of a target gene of the A-factor receptor. Mol Microbiol. 1999;34:102–111. doi: 10.1046/j.1365-2958.1999.01579.x. [DOI] [PubMed] [Google Scholar]
- Ohnishi Y, Yamazaki H, Kato J, Tomono A, Horinouchi S. AdpA, a central transcriptional regulator in the A-factor regulatory cascade that leads to morphological development and secondary metabolism in Streptomyces griseus. Biosci Biotechnol Biochem. 2005;69:431–439. doi: 10.1271/bbb.69.431. [DOI] [PubMed] [Google Scholar]
- Oliynyk M, et al. Analysis of the biosynthetic gene cluster for the polyether antibiotic monensin in Streptomyces cinnamonensis and evidence for the role of monB and monC genes in oxidative cyclization. Mol Microbiol. 2003;49:1179–1190. doi: 10.1046/j.1365-2958.2003.03571.x. [DOI] [PubMed] [Google Scholar]
- Onaka H, Ando N, Nihira T, Yamada Y, Beppu T, Horinouchi S. Cloning and characterization of the A-factor receptor gene from Streptomyces griseus. J Bacteriol. 1995;177:6083–6092. doi: 10.1128/jb.177.21.6083-6092.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onaka H, Nakagawa T, Horinouchi S. Involvement of two A-factor receptor homologues in Streptomyces coelicolor A3(2) in the regulation of secondary metabolism and morphogenesis. Mol Microbiol. 1998;28:743–753. doi: 10.1046/j.1365-2958.1998.00832.x. [DOI] [PubMed] [Google Scholar]
- Ostash B, et al. Identification and characterization of the Streptomyces globisporus 1912 regulatory gene lndYR that affects sporulation and antibiotic production. Microbiology. 2011;157:1240–1249. doi: 10.1099/mic.0.045088-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otten SL, Ferguson J, Hutchinson CR. Regulation of daunorubicin production in Streptomyces peucetius by the dnrR2 locus. J Bacteriol. 1995;177:1216–1224. doi: 10.1128/jb.177.5.1216-1224.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otten SL, Olano C, Hutchinson CR. The dnrO gene encodes a DNA-binding protein that regulates daunorubicin production in Streptomyces peucetius by controlling expression of the dnrN pseudo response regulator gene. Microbiology. 2000;146:1457–1468. doi: 10.1099/00221287-146-6-1457. [DOI] [PubMed] [Google Scholar]
- Ou X, Zhang B, Zhang L, Zhao G, Ding X. Characterization of rrdA, a TetR family protein gene involved in the regulation of secondary metabolism in Streptomyces coelicolor. Appl Environ Microbiol. 2009;75:2158–2165. doi: 10.1128/AEM.02209-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaniappan N, Ayers S, Gupta S, Habib ES, Reynolds KA. Production of hygromycin A Analogs in Streptomyces hygroscopicus NRRL 2388 through identification and manipulation of the biosynthetic gene cluster. Chem Biol. 2006;13:753–764. doi: 10.1016/j.chembiol.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Parajuli N, Moon YH. Analysis of doxorubicin biosynthetic gene cluster and intensive study of regulatory system of Streptomyces peucetious ATCC 27952. Theor Appl Chem Eng. 2002;8:3909–3912. [Google Scholar]
- Parajuli N, Viet HT, Ishida K, Tong HT, Lee HC, Liou K, Sohng JK. Identification and characterization of the afsR homologue regulatory gene from Streptomyces peucetius ATCC 27952. Res Microbiol. 2005;156:707–712. doi: 10.1016/j.resmic.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Pérez-Llarena FJ, Liras P, Rodríguez-García A, Martín JF. A regulatory gene (ccaR) required for cephamycin and clavulanic acid production in Streptomyces clavuligerus: amplification results in overproduction of both beta-lactam compounds. J Bacteriol. 1997;179:2053–2059. doi: 10.1128/jb.179.6.2053-2059.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Redondo R, Rodríguez-García A, Martín JF, Liras P. The claR gene of Streptomyces clavuligerus, encoding a LysR-type regulatory protein controlling clavulanic acid biosynthesis, is linked to the clavulanate-9-aldehyde reductase (car) gene. Gene. 1998;211:311–321. doi: 10.1016/S0378-1119(98)00106-1. [DOI] [PubMed] [Google Scholar]
- Pullan ST, Chandra G, Bibb MJ, Merrick M. Genome-wide analysis of the role of GlnR in Streptomyces venezuelae provides new insights into global nitrogen regulation in actinomycetes. BMC Genom. 2011;12:1–14. doi: 10.1186/1471-2164-12-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulsawat N, Kitani S, Nihira T. Characterization of biosynthetic gene cluster for the production of virginiamycin M, a streptogramin type A antibiotic, in Streptomyces virginiae. Gene. 2007;393:31–42. doi: 10.1016/j.gene.2006.12.035. [DOI] [PubMed] [Google Scholar]
- Pulsawat N, Kitani S, Fukushima E, Nihira T. Hierarchical control of virginiamycin production in Streptomyces virginiae by three pathway-specific regulators: VmsS, VmsT and VmsR. Microbiology. 2009;155:1250–1259. doi: 10.1099/mic.0.022467-0. [DOI] [PubMed] [Google Scholar]
- Räty K, Kantola J, Hautala A, Hakala J, Ylihonko K, Mäntsälä P. Cloning and characterization of Streptomyces galilaeus aclacinomycins polyketide synthase (PKS) cluster. Gene. 2002;293:115–122. doi: 10.1016/S0378-1119(02)00699-6. [DOI] [PubMed] [Google Scholar]
- Rehakova A, Novakova R, Feckova L, Mingyar E, Kormanec J. A gene determining a new member of the SARP family contributes to transcription of genes for the synthesis of the angucycline polyketide auricin in Streptomyces aureofaciens CCM 3239. FEMS Microbiol Lett. 2013;346:45–55. doi: 10.1111/1574-6968.12200. [DOI] [PubMed] [Google Scholar]
- Retzlaff L, Distler J. The regulator of streptomycin gene expression, StrR, of Streptomyces griseus is a DNA binding activator protein with multiple recognition sites. Mol Microbiol. 1995;18:151–162. doi: 10.1111/j.1365-2958.1995.mmi_18010151.x. [DOI] [PubMed] [Google Scholar]
- Rodríguez M, Núñez LE, Braña AF, Méndez C, Salas JA, Blanco G. Identification of transcriptional activators for thienamycin and cephamycin C biosynthetic genes within the thienamycin gene cluster from Streptomyces cattleya. Mol Microbiol. 2008;69:633–645. doi: 10.1111/j.1365-2958.2008.06312.x. [DOI] [PubMed] [Google Scholar]
- Rodríguez H, Rico S, Díaz M, Santamaría RI. Two-component systems in Streptomyces: key regulators of antibiotic complex pathways. Microb Cell Fact. 2013;12:127. doi: 10.1186/1475-2859-12-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Rodríguez A, Robledo-Casados I, Sánchez S. An overview on transcriptional regulators in Streptomyces. Biochem Biophys Acta. 2015;1849:1017–1039. doi: 10.1016/j.bbagrm.2015.06.007. [DOI] [PubMed] [Google Scholar]
- Sato K, Nihira T, Sakuda S, Yanagimoto M, Yamada Y. Isolation and structure of a new butyrolactone autoregulator from Streptomyces sp. FRI-5. J Ferment Bioeng. 1989;68:170–173. doi: 10.1016/0922-338X(89)90131-1. [DOI] [Google Scholar]
- Sekurova ON, et al. In vivo analysis of the regulatory genes in the nystatin biosynthetic gene cluster of Streptomyces noursei ATCC 11455 reveals their differential control over antibiotic biosynthesis. J Bacteriol. 2004;186:1345–1354. doi: 10.1128/JB.186.5.1345-1354.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeler NL, MacMillan SV, Nodwell JR. Biochemical activities of the absA two-component system of Streptomyces coelicolor. J Bacteriol. 2005;187:687–696. doi: 10.1128/JB.187.2.687-696.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola-Landa A, Moura RS, Martín JF. The two-component PhoR-PhoP system controls both primary metabolism and secondary metabolite biosynthesis in Streptomyces lividans. Proc Natl Acad Sci USA. 2003;A100:6133–6138. doi: 10.1073/pnas.0931429100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- Stratigopoulos G, Cundliffe E. Expression analysis of the tylosin-biosynthetic gene cluster: pivotal regulatory role of the tylQ product. Chem Biol. 2002;9:71–78. doi: 10.1016/S1074-5521(01)00095-3. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Mochizuki S, Yamamoto S, Arakawa K, Kinashi H. Regulation of lankamycin biosynthesis in Streptomyces rochei by two SARP genes, srrY and srrZ. Biosci Biotechnol Biochem. 2010;74:819–827. doi: 10.1271/bbb.90927. [DOI] [PubMed] [Google Scholar]
- Takano E, Gramajo HC, Strauch E, Andres N, White J, Bibb MJ. Transcriptional regulation of the redD transcriptional activator gene accounts for growth-phase-dependent production of the antibiotic undecylprodigiosin in Streptomyces coelicolor A3(2) Mol Microbiol. 1992;6:2797–2804. doi: 10.1111/j.1365-2958.1992.tb01459.x. [DOI] [PubMed] [Google Scholar]
- Takano E, et al. A bacterial hormone (the SCB1) directly controls the expression of a pathway-specific regulatory gene in the cryptic type I polyketide biosynthetic gene cluster of Streptomyces coelicolor. Mol Microbiol. 2005;56:465–479. doi: 10.1111/j.1365-2958.2005.04543.x. [DOI] [PubMed] [Google Scholar]
- Tanaka A, Takano Y, Ohnishi Y, Horinouchi S. AfsR recruits RNA polymerase to the afsS promoter: a model for transcriptional activation by SARPs. J Mol Biol. 2007;369:322–333. doi: 10.1016/j.jmb.2007.02.096. [DOI] [PubMed] [Google Scholar]
- Wang L, et al. Autoregulation of antibiotic biosynthesis by binding of the end product to an atypical response regulator. Proc Natl Acad Sci USA. 2009;106:8617–8622. doi: 10.1073/pnas.0900592106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Shu D, Chen L, Jiang W, Lu Y. Cross-talk between an orphan response regulator and a noncognate histidine kinase in Streptomyces coelicolor. FEMS Microbiol Lett. 2009;294:150–156. doi: 10.1111/j.1574-6968.2009.01563.x. [DOI] [PubMed] [Google Scholar]
- Wilson DJ, Xue Y, Reynolds KA, Sherman DH. Characterization and analysis of the PikD regulatory factor in the pikromycin biosynthetic pathway of Streptomyces venezuelae. J Bacteriol. 2001;183:3468–3475. doi: 10.1128/JB.183.11.3468-3475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie PF, Sheng Y, Ito T, Mahmud T. Transcriptional regulation and increased production of asukamycin in engineered Streptomyces nodosus subsp. asukaensis strains. Appl Microbiol Biotechnol. 2012;96:451–460. doi: 10.1007/s00253-012-4084-2. [DOI] [PubMed] [Google Scholar]
- Xue Y, Zhao L, Liu HW, Sherman DH. A gene cluster for macrolide antibiotic biosynthesis in Streptomyces venezuelae: architecture of metabolic diversity. Proc Natl Acad Sci USA. 1998;95:12111–12116. doi: 10.1073/pnas.95.21.12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y, Sugamura K, Kondo K, Yanagimoto M, Okada H. The structure of inducing factors for virginiamycin production in Streptomyces virginiae. J Antibiot. 1987;40:496–504. doi: 10.7164/antibiotics.40.496. [DOI] [PubMed] [Google Scholar]
- Yang K, Han L, He J, Wang L, Vining LC. A repressor-response regulator gene pair controlling jadomycin B production in Streptomyces venezuelae ISP5230. Gene. 2001;279:165–173. doi: 10.1016/S0378-1119(01)00723-5. [DOI] [PubMed] [Google Scholar]
- Yepes A, Rico S, Rodríguez-García A, Santamaría RI, Díaz M. Novel two-component systems implied in antibiotic production in Streptomyces coelicolor. PLoS ONE. 2011;6:e19980. doi: 10.1371/journal.pone.0019980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo YJ, Hwang JY, Shin HL. Characterization of negative regulatory genes for the biosynthesis of rapamycin in Streptomyces rapamycinicus and its application for improved production. Microbiol Biotechnol. 2015;42:125–135. doi: 10.1007/s10295-014-1546-9. [DOI] [PubMed] [Google Scholar]
- Yu Z, Wang Q, Deng Z, Tao M. Activation of silent antibiotic synthesis in Streptomyces lividans by disruption of a negative regulator nsdA, a gene conserved in Streptomyces Chinese. J Biotechnol. 2006;22:757–762. [PubMed] [Google Scholar]
- Yu Q, Du A, Liu T, Deng Z, He X. The biosynthesis of the polyether antibiotic nanchangmycin is controlled by two pathway-specific transcriptional activators. Arch Microbiol. 2012;194:415–426. doi: 10.1007/s00203-011-0768-8. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zou Z, Niu G, Tan H. jadR* and jadR2 act synergistically to repress jadomycin biosynthesis. Sci China Life Sci. 2013;56:584–590. doi: 10.1007/s11427-013-4508-y. [DOI] [PubMed] [Google Scholar]
- Zhu D, He X, Zhou X, Deng Z. Expression of the melC operon in several streptomyces strains is positively regulated by AdpA, an AraC family transcriptional regulator involved in morphological development in Streptomyces coelicolor. J Bacteriol. 2005;187:3180–3187. doi: 10.1128/JB.187.9.3180-3187.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z, Du D, Zhang Y, Zhang J, Niu G, Tan H. A γ-butyrolactone-sensing activator/repressor, JadR3, controls a regulatory mini-network for jadomycin biosynthesis. Mol Microbiol. 2014;94:490–505. doi: 10.1111/mmi.12752. [DOI] [PubMed] [Google Scholar]