Abstract
The aim of this study was to systematically review articles that investigated the prognostic significance of different microRNAs in bladder cancer (BC). We systematically searched PubMed, Web of Science, and Embase to identify relevant studies until March 2016. After screening, 26 studies that involved 2753 patients were included. Results suggested that many miRs expression aberration may predict prognosis in patients with BC. There are six miRs (miR-21, miR-143, miR-155, miR-200, miR-214, and miR-222) were reported by at least two studies, and we performed meta-analysis in the corresponding studies. Accordingly, we found that high miR-21 expression was associated with poor overall survival [OS; hazard ratio (HR) = 3.94, 95% CI 2.08–7.44]. High miR-143 expression was associated with poor progression-free survival (PFS; HR = 3.78, 95% CI 1.61–8.89). High miR-155 expression was associated with poor PFS (HR = 8.10, 95% CI 2.92–22.48). High miR-222 expression was associated with poor OS (HR = 3.39, 95% CI 1.10–10.41). Meanwhile, low miR-214 expression was correlated with poor RFS(HR = 0.34, 95% CI 0.22–0.53). Our comprehensive systematic review concluded that microRNAs, particularly miR-21, miR-143, miR-155, miR-214, and miR-222, could serve as meticulous follow-up markers for early detection of progression or recurrence and even useful therapeutic targets for the treatment in patients with BC.
Introduction
Among urological cancers, bladder cancer (BC) is the leading cause of death, with an estimated 76,960 new cases and 16,390 deaths in the United States in 2016 alone1. BC is highly heterogeneous, and its two major subsets are non-muscle-invasive BC (NMIBC) and muscle-invasive BC (MIBC)2. Approximately 70% of BC patients have NMIBC at first diagnosis; however, about 50%–70% of them will relapse and roughly 10%–20% will progress to MIBC3. MIBC, which could rapidly progress and metastasize, is correlated with a high mortality despite the improved therapeutic strategies at the moment4. In this regard, prediction models identifying patients with unfavorable prognosis, who may benefit from early systematic therapy, are greatly needed. Based on clinicopathological parameters, the currently used system seems inferior in accurately predicting the prognosis of BC patients with diverse and complicated tumor backgrounds5. Therefore, novel biomarkers that can stratify patients with poor prognosis when used alone or in combination with other clinicopathological parameters must be identified to precisely guide clinical decisions.
The detecting technique of MicroRNA (miR) molecules is an available, novel approach to evaluate tumor prognosis; hence, miRs constitute an attractive biomarker source for cancer research6. MiRs are small non-coding RNAs (~22 nucleotide) transcribed from DNA into RNA hairpins. MiRs post-transcriptionally regulate gene expression by binding to the 3′-UTR of target mRNAs, resulting in target mRNA degradation or inhibition of their translation7. MiRs are involved in a variety of biological functions and in the majority of known hallmarks of cancer, including initiation, development, and metastasis8, 9. Moreover, many studies have suggested that miRs have a prognostic value in several human cancers, such as colorectal10, breast11, lung12, and ovarian13 cancers. Numerous miR studies that have focused on BC have recently been conducted in the fields of outcome prediction and potential therapeutic targets. Many studies have suggested the prognostic significance of miRs in patients with BC14–17. In this study, we perform systematic review and meta-analysis to further increase statistical power, improve clinical translation, and comprehensively investigate the prognostic value of different miRs in patients with BC.
Results
Search Results
A total of 1075 articles were retrieved from the primary literature search. A total of 225 duplicate reports were excluded. Accordingly, after screening the titles and abstracts, 799 articles were excluded because they were found to be non-human studies, genetic variation studies, letters, case reports, reviews, commentaries, and other obvious irrelevant studies. The remaining articles were viewed in full text. After a careful review of the potential articles, 26 articles were included in this study and used for data extraction (Table 1 and Fig. 1). Six prognostic miRs (miR-21, miR-143, miR-155, miR-200, miR-214, and miR-222) in BC repeatedly appeared in the included studies and triggered a meta-analysis. Only 12 articles evaluating the relationship between six specific miRs and BC prognosis satisfied the criteria for meta-analysis14, 16, 18–27. Figure 1 shows a flowchart of the study selection process.
Table 1.
The main characteristics of eligible studies.
| Study (year) | miR | Population | Study design | Stage | Case number | Gender (M/F) | Follow up (month) | Detecting method | Detected sample | Cut-off | Survival outcome | HR availability | Adjusted | Quality scorea |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Veerla 2009 | 452 | Sweden | R cohort study | Ta-T3 | 34 | NR | > 50 | ISH | Tissue | Upper tertile | OS | Report | NR | 6 |
| 452* | Sweden | R cohort study | Ta-T3 | 34 | NR | > 50 | ISH | Tissue | Upper tertile | OS | Report | NR | ||
| Dyrskjøt 2009 | 133b | Denmark | R cohort study | Ta-T4 | 106 | 81/25 | ~90 | qRT-PCR | Tissue | ROC curve | PFS | Report | Yes | 7 |
| 518c* | Denmark | R cohort study | Ta-T4 | 106 | 81/25 | ~90 | qRT-PCR | Tissue | ROC curve | PFS | Report | Yes | ||
| 129 | Denmark | R cohort study | Ta-T4 | 106 | 81/25 | ~90 | qRT-PCR | Tissue | ROC curve | PFS | Report | Yes | ||
| 29c | Denmark | R cohort study | Ta-T4 | 106 | 81/25 | ~90 | qRT-PCR | Tissue | ROC curve | PFS | Report | Yes | ||
| Wang 2012 | 100 | China | R cohort study | Ta-T4 | 126 | 87/39 | 36 | qRT-PCR | Tissue | Median | OS,PFS | Report | Yes | 7 |
| Yun 2012 | 200 | Korea | P cohort study | Ta-T1 | 138 | NR | 7–76 | qRT-PCR | Urine cell-free | ROC curve | RFS | Report | Yes | 8 |
| Zaravinos 2012 | 21 | Greece | R cohort study | Ta-T4 | 77 | 68/9 | ~50 | qRT-PCR | Tissue | Median | OS,RFS | Report | Yes | 8 |
| 210 | Greece | R cohort study | Ta-T4 | 77 | 68/9 | ~50 | qRT-PCR | Tissue | Median | OS | Report | Yes | ||
| 387 | Greece | R cohort study | Ta-T4 | 77 | 68/9 | ~50 | qRT-PCR | Tissue | Median | OS,RFS | Report | Yes | ||
| Puerta-Gil 2012 | 222 | Spain | P cohort study | Ta-T4 | 113 | 101/12 | 36 | qRT-PCR | Tissue | Median | OS,CSS, RFS,PFS | SC | NR | 6 |
| 143 | Spain | P cohort study | Ta-T4 | 113 | 101/12 | 36 | qRT-PCR | Tissue | Median | RFS,PFS | SC | NR | ||
| Kim 2013 | 214 | Korea | R cohort study | Ta-T1 | 138 | 110/28 | 16–82 | qRT-PCR | Urine cell-free | Median | RFS | Report | Yes | 7 |
| Wang 2013 | 31 | China | R cohort study | Ta-T4 | 126 | 87/39 | 36 | qRT-PCR | Tissue | Median | OS,PFS | Report | Yes | 8 |
| Rosenberg 2013 | 29c* | Israel | R cohort study | Ta-T1 | 75 | NR | 53 | ISH | Tissue | Upper tertile | PFS | SC | NR | 6 |
| Ratert 2013 | 141 | Germany | R cohort study | Ta-T4 | 40 | 32/8 | > 100 | qRT-PCR | Tissue | Median | OS | SC | NR | 6 |
| 205 | Germany | R cohort study | Ta-T4 | 40 | 32/8 | > 100 | qRT-PCR | Tissue | Median | OS | SC | NR | ||
| Pignot 2013 | 9 | France | R cohort study | T2-T4 | 72 | 58/15 | 15 | qRT-PCR | Tissue | Median | OS,RFS | SC | NR | 6 |
| 182 | France | R cohort study | T2-T4 | 72 | 58/15 | 15 | qRT-PCR | Tissue | Median | OS,RFS | SC | NR | ||
| 200 | France | R cohort study | T2-T4 | 72 | 58/15 | 15 | qRT-PCR | Tissue | Median | OS,RFS | SC | NR | ||
| Zhang 2014 | 101 | China | R cohort study | T1-T4 | 72 | 42/30 | ~80 | qRT-PCR | Tissue | T/N ratio > 0.45 | OS | Report | Yes | 8 |
| Zhang 2014 | 222 | China | R cohort study | Ta-T4 | 97 | 51/46 | ~60 | qRT-PCR | Tissue | Median | OS | Report | Yes | 8 |
| Lin 2014 | 26a | China | R cohort study | Ta-T4 | 126 | 80/46 | 40 | qRT-PCR | Tissue | Median | OS,DFS | Report | Yes | 9 |
| Drayton 2014 | 27a | UK | P cohort study | Ta-T4 | 139 | 100/39 | ~96 | qRT-PCR | Tissue | T/N ratio > 2 | RFS,PFS | DE | NR | 6 |
| 27b | UK | P cohort study | Ta-T4 | 139 | 100/39 | ~96 | qRT-PCR | Tissue | T/N ratio > 2 | RFS,PFS | DE | NR | 6 | |
| Zhang 2015 | 203 | China | R cohort study | T2-T4 | 108 | 83/25 | 51.5 | qRT-PCR | Tissue | ROC curve | OS,PFS | Report | Yes | 9 |
| Zhang 2015 | 21 | China | R cohort study | T1-T4 | 53 | 35/18 | ~60 | qRT-PCR | Tissue | T/N ratio > 6.7 | OS | Report | Yes | 8 |
| Wu 2015 | 424 | China | R cohort study | Ta-T4 | 124 | 77/47 | 94–144 | ISH | Tissue | X-tile algorithm | OS,DFS | SC/Report | NR/Yes | 6 |
| Wang 2015 | 141 | China | R cohort study | Ta-T4 | 114 | 86/28 | 43 | qRT-PCR | Tissue | Median | CSS,DFS | Report | Yes | 8 |
| Wang 2015 | 214 | China | P cohort study | T2-T4 | 129 | NR | 29 | qRT-PCR | Urine cell-free | Median | OS,RFS | Report | Yes | 8 |
| Wang 2015 | 155 | China | R cohort study | Ta-T4 | 102 | 61/41 | ~60 | qRT-PCR | Tissue | Median | PFS | Report | Yes | 8 |
| Jiang 2015 | 152 | China | R cohort study | Ta-T1 | 59 | NR | ~62 | qRT-PCR | Serum | Median | RFS | Report | Yes | 6 |
| 148b-3p | China | R cohort study | Ta-T1 | 59 | NR | ~62 | qRT-PCR | Serum | Median | RFS | Report | NR | ||
| 3187–3p | China | R cohort study | Ta-T1 | 59 | NR | ~62 | qRT-PCR | Serum | Median | RFS | Report | Yes | ||
| 15b-5p | China | R cohort study | Ta-T1 | 59 | NR | ~62 | qRT-PCR | Serum | Median | RFS | Report | NR | ||
| 27a-3p | China | R cohort study | Ta-T1 | 59 | NR | ~62 | qRT-PCR | Serum | Median | RFS | Report | NR | ||
| 30a-5p | China | R cohort study | Ta-T1 | 59 | NR | ~62 | qRT-PCR | Serum | Median | RFS | Report | NR | ||
| Avgeris 2015 | 143 | Greece | R cohort study | Ta-T4 | 133 | NR | ~48 | qRT-PCR | Tissue | X-tile algorithm | OS,PFS | Report | NR | 6 |
| 145 | Greece | R cohort study | Ta-T4 | 133 | NR | ~48 | qRT-PCR | Tissue | X-tile algorithm | OS,PFS | Report | NR | ||
| 224 | Greece | R cohort study | Ta-T4 | 133 | NR | ~48 | qRT-PCR | Tissue | X-tile algorithm | OS,PFS | Report | NR | ||
| Andrew 2015 | 34a | USA | R cohort study | Ta-T1 | 229 | 171/58 | 46 | ISH | Tissue | fluorescence scores 1+ | RFS | Report | Yes | 8 |
| Martínez-Fernández 2015 | 200 | Spain | R cohort study | Ta-T1 | 61 | NR | 28.8 | qRT-PCR | Serum | Median | RFS | SC | NR | 6 |
| Zhang 2016 | 155 | China | P cohort study | Ta-T1 | 162 | 126/36 | 51.5 | qRT-PCR | Urine cell-free | ROC curve | RFS,PFS | Report | Yes | 9 |
miR: microRNA; HR: hazard ratio; R: retrospective; P: prospective; qRT-PCR: quantities reverse transcription polymerase chain reaction; ISH: in situ hybridization; OS: overall survival; CSS: cancer-specific survival; RFS: recurrence-free survival; PFS: progression-free survival; DFS: disease-free survival; SC: survival curve; NR: not reported.
aThe quality of the included studies was evaluated using the Newcastle-Ottawa scale.
Figure 1.
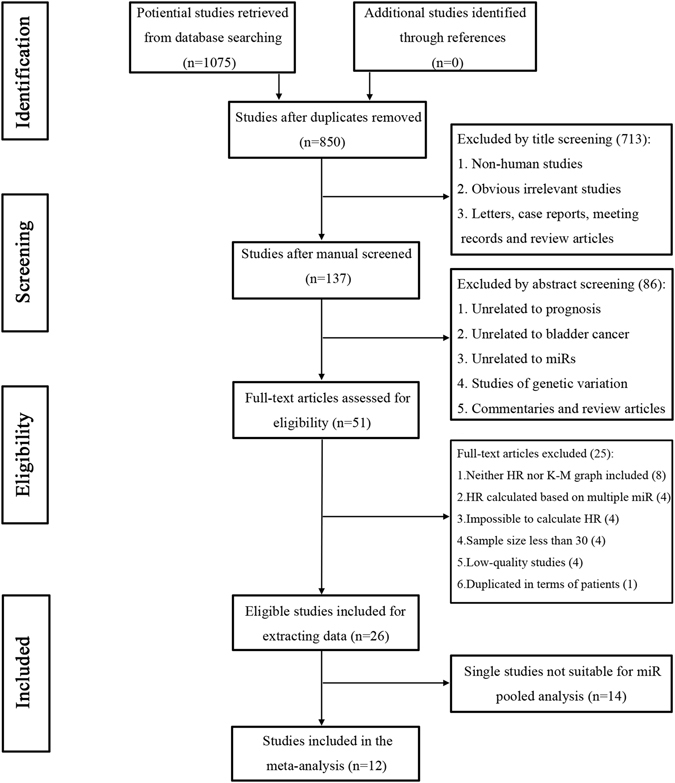
Flowchart of study selection. miR: microRNA; HR: hazard ratio.
Study Characteristics
All of the included studies were recently published (2009–2016). They reported the prognostic significance of 37 different miRs in BC patients with varying tumor stages. Thirteen studies originated from China, two from Korea, two from Greece, two from Spain, one from Sweden, one from Denmark, one from Israel, one from Germany, one from France, one from the United States, and one from the United Kingdom. Most studies applied the quantitative real-time polymerase chain reaction to measure the miR expression. Four studies applied in situ hybridization. The miR expressions were mainly detected in the tissue samples. However, two and four studies measured miRs in serum and urine cell-free, respectively. The HR was adjusted for corresponding covariates in 18 studies using the Cox regression multivariate analysis. Table 1 summarizes the detailed information of the twenty-six included studies.
MiRs and Prognosis
High miR-45228, miR-452*28, miR-2116, 25, miR-21025, miR-22222, 27, miR-923, miR-18223, miR-14321, 27, miR-133b29, miR-518c*29, miR-12929, miR-15514, 19, miR-14521, and miR-15230 expressions were associated with poor prognosis. Conversely, low miR-10031, miR-38725, miR-3132, miR-14133, miR-20533, miR-10134, miR-26a35, miR-20315, miR-42436, miR-21417, 24, miR-29c*37, miR-27a38, miR-27b38, miR-20315, and miR-34a39 expressions were correlated with unfavorable prognosis. The miR-20020, 23, 26, miR-22421, miR-29c29, miR-148b-3p30, miR-3187-3p30, miR-15b-5p30, miR-27a-3p30, and miR-30a-5p30 expression did not show any significant association with survival outcomes (Table 2 and Fig. 2). Six miRs (i.e., miR-21, miR-143, miR-155, miR-200, miR-214, and miR-222) were assessed by at least two studies. We performed a meta-analysis in the corresponding studies.
Table 2.
Summary of HR of miRNA expression in bladder cancer.
| Study | miR | Case number | OS | CSS | RFS | DFS/PFS | Expression associates with bad prognosis | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| High level | Low level | HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | |||
| Veerla 2009 | 452 | 11 | 23 | 8.6 (3.6–13.6) | < 0.025 | — | — | — | — | — | — | High |
| 452* | 11 | 23 | 8.2 (3.2–13.2) | < 0.025 | — | — | — | — | — | — | High | |
| Dyrskjøt 2009 | 133b | — | — | — | — | — | — | — | — | 3.5 (1.58–7.75)P | 0.002 | High |
| 518c* | — | — | — | — | — | — | — | — | 3.2 (1.49–6.89)P | 0.003 | High | |
| 129 | — | — | — | — | — | — | — | — | 3.0 (1.19–7.56)P | 0.02 | High | |
| 29c | — | — | — | — | — | — | — | — | 0.48 (0.23–1)P | 0.05 | High | |
| Wang 2012 | 100 | 48 | 78 | 0.10 (0.04–0.6) | 0.008 | — | — | — | — | 0.12 (0.04–0.77)P | 0.01 | Low |
| Yun 2012 | 200 | — | — | — | — | — | — | 0.449 (0.239–0.842) | 0.013 | — | — | Low |
| Zaravinos 2012 | 21 | 38 | 39 | 8.40 (1.90–37.04) | 0.005 | — | — | 4.88 (1.17–20.41) | 0.03 | — | — | High |
| 210 | 38 | 39 | 4.35 (1.13–16.67) | 0.033 | — | — | — | — | — | — | High | |
| 387 | 38 | 39 | 0.14 (0.03–0.65) | 0.012 | — | — | 0.17 (0.03–0.85) | 0.031 | — | — | Low | |
| Puerta-Gil 2012 | 222 | 56 | 57 | 1.96 (1.10–3.48) | 0.023 | 1.99 (1.05–3.76) | 0.034 | 2.08 (1.23–3.52) | 0.006 | 3.54 (1.54–8.18)P | 0.003 | High |
| 143 | 56 | 57 | — | — | — | — | 2.28 (1.21–4.31) | 0.011 | 3.01 (1.06–8.59)P | 0.039 | High | |
| Kim 2013 | 214 | 69 | 69 | — | — | — | — | 0.497 (0.254–0.974) | 0.041 | — | — | Low |
| Wang 2013 | 31 | 56 | 70 | 0.084 (0.033–0.833) | 0.008 | — | — | — | — | 0.114 (0.039–1)P | 0.01 | Low |
| Rosenbeg 2013 | 29c* | 18 | 57 | — | — | — | — | — | — | 0.2 (0.08–0.52)P | < 0.001 | Low |
| Ratert 2013 | 141 | 20 | 20 | 0.28 (0.09–0.82) | 0.02 | — | — | — | — | — | — | Low |
| 205 | 20 | 20 | 0.36 (0.13–0.95) | 0.04 | — | — | — | — | — | — | Low | |
| Pignot 2013 | 9 | 36 | 36 | 2.37 (1.36–4.15) | 0.003 | — | — | 1.86 (1.08–3.12) | 0.025 | — | — | High |
| 182 | 36 | 36 | 1.95 (1.09–3.47) | 0.024 | — | — | 1.95 (1.11–3.43) | 0.021 | — | — | High | |
| 200 | 36 | 36 | 1.86 (1.02–3.39) | 0.043 | — | — | 1.93 (1.09–3.39) | 0.023 | — | — | High | |
| Zhang 2014 | 101 | 46 | 26 | 0.451 (0.237–0.735) | 0.028 | — | — | — | — | — | — | Low |
| Zhang 2014 | 222 | 48 | 49 | 6.17 (2.33–10.39) | < 0.001 | — | — | — | — | — | — | High |
| Lin 2014 | 26a | 56 | 70 | 0.185 (0.088–0.762) | 0.01 | — | — | — | — | 0.192 (0.0891–0.745)D | 0.01 | Low |
| Drayton 2014 | 27a | 70 | 69 | — | — | — | — | 0.98 (0.49–1.96) | 0.96 | 0.44 (0.22–0.88)P | 0.02 | Low |
| 27b | 63 | 76 | — | — | — | — | 0.94 (0.47–1.90) | 0.87 | 0.38 (0.18–0.80)P | 0.01 | Low | |
| Zhang 2015 | 203 | 79 | 29 | 0.359 (0.209–0.616) | < 0.001 | — | — | — | — | 0.154 (0.082–0.288)P | < 0.001 | Low |
| Zhang 2015 | 21 | 28 | 25 | 3.32 (1.16–4.74) | 0.018 | — | — | — | — | — | — | High |
| Wu 2015 | 424 | 53 | 71 | 0.40 (0.24–0.69) | 0.001 | — | — | — | — | 0.152 (0.066–0.350)D | < 0.001 | Low |
| Wang 2015 | 141 | 54 | 60 | — | — | 0.314 (0.108–0.946) | 0.039 | — | — | 0.492 (0.254–0.954)D | 0.036 | Low |
| Wang 2015 | 214 | 64 | 65 | 0.282 (0.160–0.495) | < 0.001 | — | — | 0.264 (0.149–0.468) | < 0.001 | — | — | Low |
| Wang 2015 | 155 | 52 | 50 | — | — | — | — | — | — | 7.7 (1.4–14.7)P | 0.009 | High |
| Jiang 2015 | 152 | 29 | 30 | — | — | — | — | 2.324 (1.093–4.940) | 0.028 | — | — | High |
| 148b-3p | 29 | 30 | — | — | — | — | 0.872 (0.419–1.812) | 0.713 | — | — | Low | |
| 3187–3p | 29 | 30 | — | — | — | — | 0.483 (0.227–1.028) | 0.059 | — | — | Low | |
| 15b-5p | 29 | 30 | — | — | — | — | 1.023 (0.494–2.120) | 0.951 | — | — | High | |
| 27a-3p | 29 | 30 | — | — | — | — | 0.721 (0.344–1.510) | 0.386 | — | — | Low | |
| 30a-5p | 29 | 30 | — | — | — | — | 0.677 (0.323–1.419) | 0.302 | — | — | Low | |
| Avgeris 2015 | 143 | — | — | 3.329 (1.346–8.236) | 0.009 | — | — | — | — | 5.990 (1.351–26.55)P | 0.018 | High |
| 145 | — | — | 2.426 (0.985–5.976) | 0.054 | — | — | — | — | 4.164 (1.178–14.80)P | 0.027 | High | |
| 224 | — | — | 4.168 (0.557–31.183) | 0.164 | — | — | — | — | 2.654 (0.944–7.460)P | 0.064 | High | |
| Andrew 2015 | 34a | 63 | 166 | — | — | — | — | 0.57 (0.34–0.93) | 0.029 | — | — | Low |
| Martínez-Fernández 2015 | 200 | 31 | 30 | — | — | — | — | 0.49 (0.25–0.97) | 0.041 | — | — | Low |
| Zhang 2016 | 155 | 130 | 32 | — | — | — | — | 3.497 (1.722–7.099) | 0.001 | 9.466 (1.210–74.066)P | 0.032 | High |
miR: microRNA; HR: hazard ratio; CI: confidence interval; OS: overall survival; CSS: cancer-specific survival; RFS: recurrence-free survival; PFS: progression-free survival;
DFS: disease-free survival; —: not reported.
DDFS; PPFS.
Figure 2.
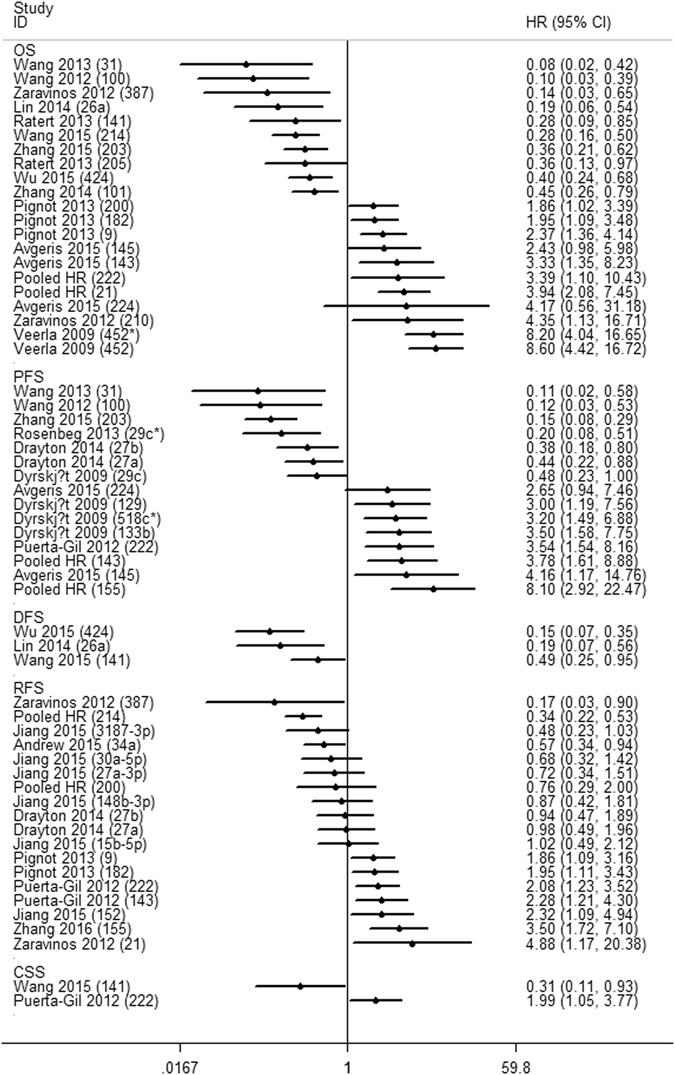
HR of miRs. The point estimate is bounded by a 95% CI (indicated by error bars), and the perpendicular line represents no increased risk for the outcome. HR: hazard ratio; CI: confidence interval; OS: overall survival; PFS: progression-free survival; RFS: recurrence-free survival; CSS: cancer-specific survival; DFS: disease-free survival. HR > 1 implied an unfavorable prognosis for the group with an elevated miR expression.
Two studies described the association of miR-21 with survival outcomes in BC, of which two reported overall survival (OS)16, 25 and one reported recurrence-free survival (RFS)25. Next, we conducted a meta-analysis on the miR-21 expression and OS relationship. The results showed that high miR-21 expression was correlated with poor OS [a fixed-effect model, hazard ratio (HR), 3.94; 95% CI: 2.08–7.44; p < 0.001; I 2 = 18.4%, p = 0.268]. Zaravinos et al.25 also reported shorter RFS in BC patients with an elevated level of miR-21 (HR, 4.88; 95% CI: 1.17–20.41; p = 0.03) (Fig. 3A).
Figure 3.

Forest plots of studies evaluating HR of six aberrant miRs expression. (A) miR-21, OS, RFS; (B) miR-143, RFS, PFS, OS; (C) miR-155, PFS, RFS; (D) miR-200, RFS, OS; (E) miR-214, RFS, OS; (F) miR-222, OS, CSS, RFS, PFS. HR: hazard ratio; OS: overall survival; PFS: progression-free survival; RFS: recurrence-free survival; CSS: cancer-specific survival. HR > 1 implied an unfavorable prognosis for the group with an elevated miR expression.
Two studies included survival outcomes for miR-143 in BC, of which two had progression-free survival (PFS)21, 27 data, one had OS21 data, and one contained RFS27 data. When we performed a meta-analysis on the relationship of miR-143 expression and PFS, the results showed that a higher miR-143 expression was predictive of shorter PFS (a fixed-effect model, HR, 3.78; 95% CI: 1.61–8.89; p = 0.002; I 2 = 0, p = 0.459). Avgeris et al.21 and Puerta-Gil et al.27 also reported shorter OS (HR, 3.33; 95% CI: 1.35–8.24; p = 0.009) and RFS (HR, 2.28; 95% CI: 1.21–4.31; p = 0.011) in BC patients with a higher miR-143 levels (Fig. 3B).
Two studies addressed the role of miR-155 in survival outcomes of BC patients, of which two reported PFS14, 19, and one reported RFS14. Then, we conducted a meta-analysis on the miR-155 expression and PFS relationship. This indicated that elevated miR-155 expression was correlated with poor PFS (a fixed-effect model, HR, 8.10; 95% CI: 2.92–22.48; p < 0.001; I 2 = 0, p = 0.864). Additionally, Zhang et al.14 also found shorter RFS in BC patients with an higher miR-155 level (HR, 3.50; 95% CI: 1.72–7.10; p = 0.001) (Fig. 3C).
Three studies investigated the association between miR-200 expression and the prognosis of BC patients, of which three focused on RFS20, 23, 26 and one focused on OS23. A meta-analysis was performed on the relationship of miR-200 expression and RFS. The results indicated that aberrant miR-200 expression was not related to RFS (a random-effect model, HR, 0.76; 95% CI: 0.29–2.00; p = 0.580; I 2 = 86.2%, p = 0.001). However, Pignot et al.23 reported that an increased miR-200 level was correlated with poor OS (HR, 1.86; 95% CI: 1.02–3.39; p = 0.043) (Fig. 3D).
Two studies addressed the relationship between miR-214 expression and survival outcomes in BC, of which two contained RFS18, 24 data, and one contained OS18 data. A meta-analysis was conducted on the miR-214 expression and RFS relationship. We found that low miR-214 expression was predicted poor RFS (a fixed-effect model, HR, 0.34; 95% CI: 0.22–0.53; p < 0.001; I 2 = 49.3%, p = 0.160). In addition, Wang et al.18 also found that a shorter OS in BC patients with an decreased level of miR-214 (HR, 0.28; 95% CI: 0.16–0.50; p < 0.001) (Fig. 3E).
For miR-222, two studies reported OS22, 27, one reported CSS27, one reported RFS27, and one reported PFS27. We performed a meta-analysis on the miR-222 expression and OS relationship. The results suggested that increased miR-222 expression tended to occur in patients with poor OS (a random-effect model, HR, 3.39; 95% CI: 1.10–10.41; p = 0.033; I 2 = 82.4, p = 0.017). Additionally, Puerta-Gil et al.27 found that a poor CSS (HR, 1.99; 95% CI: 1.05–3.77; p = 0.034), RFS (HR, 2.08; 95% CI: 1.23–3.52; p = 0.006), and PFS (HR, 3.54; 95% CI: 1.54–8.16; p = 0.003) in BC patients with an elevated miR-222 level (Fig. 3F).
Considering that few studies were included in this meta-analysis, the publication bias evaluated by Begg’s and Egger’s test was not considered necessary.
Discussion
We performed comprehensive systematic review and meta-analysis of the current literature on BC in response to the need for independent prognostic molecular markers that are readily assayable before, during, and/or after BC treatment. A total of 26 studies involving 2753 patients were analyzed to evaluate the relationship between the miRs and the BC prognosis in our study. Accordingly, 37 different miRs involved in the survival outcomes of patients with BC were compared. This study aimed to identify the miRs associated with BC prognosis, which could be further validated in future studies and eventually evaluated before the treatment, thereby improving BC management.
This study found that high miR-45228, miR-452*28, miR-2116, 25, miR-21025, miR-22222, 27, miR-923, miR-18223, miR-14321, 27, miR-133b29, miR-518c*29, miR-12929, miR-15514, 19, miR-14521, and miR-15230 expressions were correlated with poor prognosis. In comparison, low miR-10031, miR-38725, miR-3132, miR-14133, miR-20533, miR-10134, miR-26a35, miR-20315, miR-42436, miR-21417, 24, miR-29c*37, miR-27a38, miR-27b38, miR-20315, and miR-34a39 expressions were correlated with unfavorable prognosis (Table 2 and Fig. 2).
Our study systematically assessed the role of 37 different miRs in BC prognosis. However, most of the miRs were investigated only by a single study, and only six miRs (miR-21, miR-143, miR-155, miR-200, miR-214, and miR-222) were identified by at least two studies. Therefore, we conducted the meta-analysis on these six miRs to determine a pooled conclusion. We found that high miR-21 expression was associated with poor OS. High miR-143 expression was associated with poor PFS. High miR-155 expression was associated with poor PFS. Meanwhile, high miR-222 expression was associated with poor OS. Low miR-214 expression was correlated with poor RFS, and MiR-200 expression was not related to the RFS in the pooled analysis. Pignot et al.23 reported that an elevated miR-200 level was associated with a shorter OS. But it was important to note that the study was small (n = 72), had limited power, could lead to a premature result, and was not adjusted for other associated variables (covariates) that could also affect the survival outcome. Therefore, more multicenter prospective studies with large scale and long-term follow-up are needed to obtain a more persuasive conclusion.
MiR-21 is one of the most extensively studied cancer-related miRs; it is an abnormal expression in most cancers functioning as oncogene40–43. Zhou et al.44 published a meta-analysis evaluating the prognostic value of miR-21 in various cancers. They pooled 63 published studies and found that the increased miR-21 expression predicted worse OS in cancers. This finding is consistent with our results in BC. Some researchers explored the possible miR-21 mechanism in BC. Zhang et al.16 suggested that miR-21 through the maspin expression down-regulation up-regulated the VEGF-C expression, thereby increasing tumor growth, migration, and invasion in BC. Additionally, Zaravinos et al.25 demonstrated that miR-21 repressed the tumor suppressors, PTEN and PDCD4, to enhance angiogenesis, tumor cell proliferation, EMT, and metastasis activation in BC. MiR-143 is located in chromosome 5q33, which is a well-known fragile site in human genome and highly co-expressed with miR-145 in BC45, 46. Many studies have demonstrated the tumor suppressor role of miR-143 in BC. MiR-143 can suppress cell proliferation and migration and promote apoptosis in BC by inhibiting PI3K/Akt and MAPK signaling and targeting AKT47, KRAS48, ERK549, and PAI-145. Intriguingly, miR-143 has been found to be up-regulated in aggressive BC, and that the case of patients with evaluated miR-143 is associated with worse prognosis. This finding seemed to be contrary to the tumor suppressor functions of miR-143. Many studies have also observed this interesting outcome21, 23, 27, 29. However, the definite mechanism elucidating it remains unclear and needs further studies. A large number of studies confirmed the oncogene function of miR-155 in various cancers, including renal, thyroid, hematological, pancreatic, and bladder cancers50–54. High miR-155 expression has recently been correlated with BC recurrence and progression. Some studies might provide potential evidence linking the miR-155 expression and BC. Zhang et al.14 suggested that miR-155 overexpression promoted tumor cell growth via Wnt/β-catenin signaling activation, which is also a vital pathway in the BC tumorigenesis. In addition, Wang et al.19 reported that the oncogenic properties of miR-155 can be attributed to its antiapoptotic function through a blockade of caspase-3 activity or suppression of proapoptotic genes, such as TP53BP1, and promoted cell proliferation by down-regulating the SOCS1 gene, or activated PKB signaling via downregulation of tumor suppressors, including PTEN, PDCD4, and SHIP1. The role of miR-214 acting as an oncogene or a tumor suppressor is quite distinctive in different cancer types. MiR-214, serving as an oncogene, has been found in many human cancers, such as nasopharyngeal, gastric, and ovarian cancers as well as malignant melanomas55–58. Nevertheless, miR-214, serving as a tumor suppressor, has been found in several other cancers, such as cervical, breast, liver, esophagus, and bladder cancers17, 59–62. The functional discrepancies of miR-214 in different cancer types may be derived from its varied target genes or distinction among tissue types and cellular environments. Wang et al.17 showed that miR-214 decreased cellular proliferation, migration, and invasion and simultaneously increased apoptosis, suggesting that miR-214 functions a tumor suppressor in BC. The tumor-suppressive role of miR-214 might explain the unfavorable prognosis of BC patients with low miR-214 expression. High miR-222 expression has been observed in many human cancers, such as glioblastoma, prostate, colorectal, and pancreatic cancers63–66, suggesting that miR-222 might play a vital role in carcinogenesis. Puerta-Gil et al.27 recently found that high miR-222 expression is correlated with more advanced tumor stage and grade in BC, indicating its important role in tumorigenesis and metastasis. Moreover, Zhang et al.22 obtained the same conclusion in Asian patients with BC. Calderaro et al.67 explored the possible molecular mechanism of miR-222 in BC. They found that miR-222 decreased the tumor suppressor, PTEN, which is considered to enhance angiogenesis, tumor cell proliferation, EMT, and metastasis activation in BC. This finding might account for the poor prognosis of BC patients with low miR-222 expression.
Aside from the abovementioned miRs, the current study also systematically investigated the relationship between other miRs and BC prognosis. Figure 2 and Table 3 summarize the relation with prognosis and the possible role of other miRs in BC progression.
Table 3.
Summary of possible role and potential mechanism of miRs entered this study in bladder cancer.
| microRNA | Role | Mechanism | Reference |
|---|---|---|---|
| miR-155 | Proto-oncogene | Zhang: miR-155 overexpression promotes some tumor cell growth via Wnt/β-catenin signaling activation which is also a vital pathway in bladder cancer tumorigenesis. | 14, 19 |
| Wang: Oncogenic properties of miR-155 are attributed to its antiapoptotic function through a blockade of caspase-3 activity or suppression of proapoptotic genes such as TP53BP1 and promote cell proliferation by down-regulating the SOCS1 gene, or activate PKB signaling via downregulation of tumor suppressors, including PTEN, PDCD4, and SHIP1. | |||
| miR-203 | Tumor suppressor | miR-203 simultaneously suppressed antiapoptotic factors Bcl-w and Survivin. | 15 |
| miR-21 | Proto-oncogene | Zhang: miR-21 functions through regulation of maspin and VEGF-C, suggesting a miR-21/maspin/VEGF-C pathway in bladder cancer. | 16, 25 |
| Zaravinos: miR-21 represses the tumor suppressors PTEN deleted on chromosome 10, tropomyosin 1 and the PDCD4. | |||
| miR-424 | Tumor suppressor | miR-424 regulates multiple cellular biological behaviors, such as retarding growth, inducing apoptosis, and reducing invasion, by directly targeting EGFR in bladder cancer. | 36 |
| miR-214 | Tumor suppressor | Wang: miR-214 could exert tumor-suppressive effects in bladder cancer by directly down-regulating oncogene PDRG1. | 17, 24 |
| Kim: miR-214 could be related to the inhibition of angiogenesis, to cell proliferation, and to tumor recurrence. | |||
| miR-152 | Proto-oncogene | miR-152 acts through upregulation of DNA hypermethylation in bladder cancer. | 30 |
| miR-27a-3p | Tumor suppressor | miR-27a-3p as a target of mutant p53–273 H and uncovered a novel mutant p53–273 H/miR-27a-3p/EGFR pathway which played an important role in tumorigenesis. | 30 |
| miR-143 | Tumor suppressor | miR-143 can suppress cell proliferation and migration as well as promote apoptosis in bladder cancer by inhibiting PI3K/Akt and MAPK signaling. | 21 |
| miR-224 | Proto-oncogene | The upregulation of miR-224 levels has been observed to promote cell migration and tumor growth by targeting the tumor suppressors. | 21 |
| miR-34a | Tumor suppressor | miR-34a expression can inhibit cell migration and invasion by antagonizing Notch1 signaling. | 39 |
| miR-101 | Tumor suppressor | Abnormal down-regulation of miR-101 could frequently lead to the overexpression of EZH2 in cancer, which increased cell proliferation in bladder cancer cells and retarded transition of G phase to S phase. | 34 |
| miR-222 | Proto-oncogene | miR-222 decreased the tumor suppressor PTEN, which was considered to enhance angiogenesis, tumor cell proliferation, EMT and activation of metastasis in bladder cancer. | 67 |
| miR-26a | Tumor suppressor | miR-26a functions through regulation of HMGA1 in bladder cancer. | 35 |
| miR-29c | Tumor suppressor | miR-29c regulates the apoptotic protein MCL1 and thereby regulating apoptosis as well as DNA de novo methyltransferases DNMT3A and DNMT3B, key enzymes that are frequently up-regulated in cancer. | 29 |
| miR-210 | Proto-oncogene | miR-210 over expression activates VEGF and leads to the formation of capillary structures under hypoxic conditions during the early steps of tumor development. | 25 |
| miR-200 | Tumor suppressor | Higher levels of miR-200 might inhibit EMT and prevent non-muscle invasive bladder cancer recurrence through the silencing of various target genes. | 26 |
| miR-27a | Tumor suppressor | miR-27a functions through regulation of cystine/glutamate exchanger SLC7A11 in bladder cancer. | 38 |
| miR-129 | Proto-oncogene | miR-129 simultaneously repressed the tumor suppressors SOX4 and GALNT1 in bladder cancer. | 29 |
| miR-29c* | Tumor suppressor | miR-29c* acts through downregulation of DNA methyltransferases as well as upregulation of demethylating genes to keep the normal methylation pattern. | 37 |
| miR-9 | Proto-oncogene | miR-9 directly targeted the CDH-1 gene encoding E-cadherin, a regulator of EMT, considered to be an important initiating step for tumor metastasis. | 23 |
TP53BP1: tumor protein p53 binding protein 1; SOCS1: suppressor of cytokine signaling 1; PKB: protein kinase B; PTEN: phosphatase and tensin homolog; PDCD4: programmed cell death 4; VEGF-C: vascular endothelial growth factor-C; EGFR: epidermal growth factor receptor; PDRG1: p53 and DNA-damage regulated 1; EZH2: enhancer of zeste homologue 2; HMGA1: high mobility group AT-hook 1; MCL1: myeloid cell leukemia 1; DNMT3A: DNA-methyltransferase 3 alpha; DNMT3B: DNA-methyltransferase 3 beta; EMT: epithelial-to-mesenchymal transition; SLC7A11: solute carrier family 7 member 11; SOX4: SRY-box 4; GALNT1: polypeptide N-acetylgalactosaminyltransferase 1; CDH1: cadherin 1.
Accordingly, several limitations should be pointed out in the interpretation of the results of the current study. First, 26 involving 2753 patients were included in the systematic review. However, most of them investigated diverse miRs. Only six miRs (miR-21, miR-143, miR-155, miR-200, miR-214, and miR-222) were assessed by at least two studies that used different survival outcome assessments. Therefore, only two or three records were eligible in most of the meta-analyses in the current study. Furthermore, more multicenter prospective studies should be performed to verify our results and make a more mature conclusion. The publication bias was not evaluated because only few studies were included in this meta-analysis. The lack of analysis might have affected the interpretation of the results and make them less reliable. Second, error may have occurred because of inaccurate readings even if three independent reviewers extracted the data and the HR extrapolation method from a Kaplan–Meier graph was previously validated68. Therefore, the extrapolated HRs might be less dependable compared with the reported statistics. Third, the marked heterogeneity of studies was observed in some analyses. The heterogeneity of the pooled analysis might have been caused by several differences among the studies, including the baseline characteristics of the patients (i.e., study size, age, gender, ethnicity, and tumor stage), detecting methods, detecting sample, cut-off value, HR source, adjusted covariates, and follow-up duration. Finally, positive results were more likely to be published in most studies, whereas studies with negative results were often rejected or less assessable, which could compromise the validity of such analyses69. Urine markers have recently become more desirable than tissue or serum markers because urine is more convenient and less invasive to collect. Additionally, urine markers can be evaluated before, during, and/or after surgery and then monitored throughout a patient’s life. More well-conducted and appropriately designed studies are thus needed to establish the prognostic value of the miR urine levels. Some studies have already developed a combined expression signature of multiple miRs, which require a powerful validation strategy. More importantly, independent validation studies are needed to validate these results and evaluate the performance and prognostic power of the signature. Pignot et al.23 used three miRs to develop a molecular signature for BC with promising results. In the future, developing a new molecular signature using diverse miRs and then identifying their efficacy may also be useful.
In conclusion, our comprehensive systematic review and meta-analysis suggested that miRs, particularly miR-21, miR-143, miR-155, miR-214, and miR-222, could potentially serve as risk stratification markers and even therapeutic targets in BC despite the abovementioned limitations. However, more large scale, multicenter prospective studies with standardized methods and long-term follow up are needed to verify our results.
Methods
Search Strategy
This meta-analysis was conducted following the guidelines of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA)70.
A systematic literature search was performed in the electronic databases PubMed, Web of Science, and Embase on 15 March 2016 using the following search strategy: (microRNA or miRNA or miR) and (bladder cancer or bladder tumor or bladder carcinoma or bladder neoplasm or urothelial cancer or urinary tract cancer) and (prognosis or prognostic or survival or outcome or mortality). We also manually searched the reference lists of the relevant literature.
Selection Criteria
The studies were included based on the following criteria: (1) the association of miRs with the prognosis significance in BC should be described; (2) the studies defined the miR cut-off and clearly described the miR measurement; and (3) the studies correlated survival outcomes with a single miR expression. The exclusion criteria were as follows: (1) non-English papers; (2) case reports, letters, commentaries, meeting records, or review articles; (3) sample number fewer than 30 patients; (4) focused on animal models or cancer cells; (5) concerned genetic variation of an miR; (6) calculated HRs based on multiple miRs; and (7) the study lacked sufficient data for obtaining HR and 95% CI. All evaluations were independently performed by three individual researchers to ensure the accurate inclusion of studies. The discrepancies were resolved by discussion. We only retrieved the most informative and recently studied one for further analyses of duplicate studies.
This study was based on published literature. Therefore, ethical approval from ethics committees was not needed.
Data Extraction
Three investigators independently extracted data from eligible studies using a predefined form. The discrepancies in data extraction were resolved by discussion. The following data were extracted: surname of the first author, publication year, investigated miRs, origin of the studied population, study design, tumor stage, sample size, gender, follow-up time, detecting method, detected sample, cutoff value, and effect estimates, namely, HR of miR expression for OS, CSS, DFS, RFS or PFS, as well as their 95% CI (Table 1). We calculated HRs and their 95% CI based on the methods reported by Tierney et al.68 if the HR and 95% CI were not directly available.
Quality Assessment
The quality of the included studies was evaluated using the Newcastle–Ottawa scale, as recommended by the Cochrane Non-randomized Studies Methods Working Group71. Each study can be assessed by eight methodology items with a score ranging from 0 to 9. The high scores indicated high quality. We considered studies with scores of 6 or more as high quality for the meta-analysis. Only studies with high quality were included in the further analysis to assure the quality of this meta-analysis.
Statistical Analysis
Pooled HR with 95% CI was used to evaluate the association of the miR expression with the BC prognosis. An observed HR > 1 implied an unfavorable prognosis for the group with an elevated miR expression. Conversely, an observed HR < 1 implied a favorable prognosis for the group with an elevated miR expression. A heterogeneity test of the pooled HR was conducted using Cochran’s Q test and Higgins I-squared statistic72. A p value of less than 0.05 was considered significant. A random-effect model was used when between-study heterogeneity was observed (Cochran’s Q test, p < 0.05); otherwise, a fixed-effect model was used. All statistical analyses were performed using Stata 12.0 software (StatCorp, College Station, TX, USA), and p < 0.05 was considered statistically significant.
Author Contributions
Conceived and designed the experiments: Y.X, X.M. & X.Z.; Performed the experiments: Y.X. & L.C.; Analyzed the data: H.L., L.G., X.L., Y.G. &.Y.F.; Contributed analysis tools: Y.Z. & J.C.; Wrote the paper: Y.X. & X.Z.; Revised the manuscript: H.L. & L.C. All authors have reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Yongpeng Xie, Xin Ma and Luyao Chen contributed equally to this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Sanguedolce F, Bufo P, Carrieri G, Cormio L. Predictive markers in bladder cancer: do we have molecular markers ready for clinical use? Crit Rev Clin Lab Sci. 2014;51:291–304. doi: 10.3109/10408363.2014.930412. [DOI] [PubMed] [Google Scholar]
- 3.Kaufman DS, Shipley WU, Feldman AS. Bladder cancer. Lancet. 2009;374:239–249. doi: 10.1016/S0140-6736(09)60491-8. [DOI] [PubMed] [Google Scholar]
- 4.Chou R, et al. Treatment of muscle-invasive bladder cancer: A systematic review. Cancer. 2016;122:842–851. doi: 10.1002/cncr.29843. [DOI] [PubMed] [Google Scholar]
- 5.Luo Y, et al. High Ki-67 Immunohistochemical Reactivity Correlates With Poor Prognosis in Bladder Carcinoma: A Comprehensive Meta-Analysis with 13,053 Patients Involved. Medicine (Baltimore) 2016;95:e3337. doi: 10.1097/MD.0000000000003337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dietrich D, et al. Nucleic acid-based tissue biomarkers of urologic malignancies. Crit Rev Clin Lab Sci. 2014;51:173–199. doi: 10.3109/10408363.2014.906130. [DOI] [PubMed] [Google Scholar]
- 7.Shukla GC, Singh J, Barik S. MicroRNAs: Processing, Maturation, Target Recognition and Regulatory Functions. Mol Cell Pharmacol. 2011;3:83–92. [PMC free article] [PubMed] [Google Scholar]
- 8.Tilki D, et al. Urine markers for detection and surveillance of non-muscle-invasive bladder cancer. Eur Urol. 2011;60:484–492. doi: 10.1016/j.eururo.2011.05.053. [DOI] [PubMed] [Google Scholar]
- 9.Cortez MA, et al. MicroRNAs in body fluids–the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8:467–477. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma W, et al. miR-517a is an independent prognostic marker and contributes to cell migration and invasion in human colorectal cancer. Oncol Lett. 2016;11:2583–2589. doi: 10.3892/ol.2016.4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu JY, et al. miR-601 is a prognostic marker and suppresses cell growth and invasion by targeting PTP4A1 in breast cancer. Biomed Pharmacother. 2016;79:247–253. doi: 10.1016/j.biopha.2016.02.014. [DOI] [PubMed] [Google Scholar]
- 12.Jiang LP, Zhu ZT, He CY. Expression of miRNA-26b in the diagnosis and prognosis of patients with non-small-cell lung cancer. Future Oncol. 2016;12:1105–1115. doi: 10.2217/fon.16.21. [DOI] [PubMed] [Google Scholar]
- 13.Wang, F., Chang, J. T., Kao, C. J., Huang, R. S. High expression of miR-532-5p, a tumor suppressor, leads to better prognosis in ovarian cancer both in vivo and in vitro. Mol Cancer Ther (2016). [DOI] [PMC free article] [PubMed]
- 14.Zhang X, et al. Direct quantitative detection for cell-free miR-155 in urine: a potential role in diagnosis and prognosis for non-muscle invasive bladder cancer. Oncotarget. 2016;7:3255–3266. doi: 10.18632/oncotarget.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang, X. et al. MicroRNA-203 Is a Prognostic Indicator in Bladder Cancer and Enhances Chemosensitivity to Cisplatin via Apoptosis by Targeting Bcl-w and Survivin. Plos One10 (2015). [DOI] [PMC free article] [PubMed]
- 16.Zhang H-H, Qi F, Cao Y-H, Zu X-B, Chen M-F. Expression and clinical significance of microRNA-21, maspin and vascular endothelial growth factor-C in bladder cancer. Oncology Letters. 2015;10:2610–2616. doi: 10.3892/ol.2015.3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J, et al. MicroRNA-214 suppresses oncogenesis and exerts impact on prognosis by targeting PDRG1 in bladder cancer. PLoS One. 2015;10:e0118086. doi: 10.1371/journal.pone.0118086. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Wang J, et al. Downregulation of urinary cell-free microRNA-214 as a diagnostic and prognostic biomarker in bladder cancer. J Surg Oncol. 2015;111:992–999. doi: 10.1002/jso.23937. [DOI] [PubMed] [Google Scholar]
- 19.Wang H, Men C-P. Correlation of Increased Expression of MicroRNA-155 in Bladder Cancer and Prognosis. Labmedicine. 2015;46:118–122. doi: 10.1309/LMWR9CEA2K2XVSOX. [DOI] [PubMed] [Google Scholar]
- 20.Martínez-Fernández M, et al. A Polycomb-mir200 loop regulates clinical outcome in bladder cancer. Oncotarget. 2015;6:42258–42275. doi: 10.18632/oncotarget.5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Avgeris M, et al. Uncovering the clinical utility of miR-143, miR-145 and miR-224 for predicting the survival of bladder cancer patients following treatment. Carcinogenesis. 2015;36:528–537. doi: 10.1093/carcin/bgv024. [DOI] [PubMed] [Google Scholar]
- 22.Zhang DQ, Zhou CK, Jiang XW, Chen J, Shi BK. Increased expression of miR-222 is associated with poor prognosis in bladder cancer. World J Surg Oncol. 2014;12:241. doi: 10.1186/1477-7819-12-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pignot G, et al. microRNA expression profile in a large series of bladder tumors: identification of a 3-miRNA signature associated with aggressiveness of muscle-invasive bladder cancer. Int J Cancer. 2013;132:2479–2491. doi: 10.1002/ijc.27949. [DOI] [PubMed] [Google Scholar]
- 24.Kim SM, et al. Cell-Free microRNA-214 From Urine as a Biomarker for Non-Muscle-Invasive Bladder Cancer. Korean journal of urology. 2013;54:791–796. doi: 10.4111/kju.2013.54.11.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zaravinos A, et al. Expression of miRNAs involved in angiogenesis, tumor cell proliferation, tumor suppressor inhibition, epithelial-mesenchymal transition and activation of metastasis in bladder cancer. J Urol. 2012;188:615–623. doi: 10.1016/j.juro.2012.03.122. [DOI] [PubMed] [Google Scholar]
- 26.Yun SJ, et al. Cell-free microRNAs in urine as diagnostic and prognostic biomarkers of bladder cancer. Int J Oncol. 2012;41:1871–1878. doi: 10.3892/ijo.2012.1622. [DOI] [PubMed] [Google Scholar]
- 27.Puerta-Gil P, et al. miR-143, miR-222, and miR-452 are useful as tumor stratification and noninvasive diagnostic biomarkers for bladder cancer. Am J Pathol. 2012;180:1808–1815. doi: 10.1016/j.ajpath.2012.01.034. [DOI] [PubMed] [Google Scholar]
- 28.Veerla S, et al. MiRNA expression in urothelial carcinomas: important roles of miR-10a, miR-222, miR-125b, miR-7 and miR-452 for tumor stage and metastasis, and frequent homozygous losses of miR-31. Int J Cancer. 2009;124:2236–2242. doi: 10.1002/ijc.24183. [DOI] [PubMed] [Google Scholar]
- 29.Dyrskjot L, et al. Genomic profiling of microRNAs in bladder cancer: miR-129 is associated with poor outcome and promotes cell death in vitro. Cancer Res. 2009;69:4851–4860. doi: 10.1158/0008-5472.CAN-08-4043. [DOI] [PubMed] [Google Scholar]
- 30.Jiang X, et al. Serum microRNA expression signatures identified from genome-wide microRNA profiling serve as novel noninvasive biomarkers for diagnosis and recurrence of bladder cancer. Int J Cancer. 2015;136:854–862. doi: 10.1002/ijc.29041. [DOI] [PubMed] [Google Scholar]
- 31.Wang S, et al. Reduced expression of microRNA-100 confers unfavorable prognosis in patients with bladder cancer. Diagn Pathol. 2012;7:159. doi: 10.1186/1746-1596-7-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang S, et al. Decreased expression of microRNA-31 associates with aggressive tumor progression and poor prognosis in patients with bladder cancer. Clin Transl Oncol. 2013;15:849–854. doi: 10.1007/s12094-013-1014-4. [DOI] [PubMed] [Google Scholar]
- 33.Ratert N, et al. miRNA profiling identifies candidate mirnas for bladder cancer diagnosis and clinical outcome. J Mol Diagn. 2013;15:695–705. doi: 10.1016/j.jmoldx.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 34.Zhang H, Qi F, Cao Y, Chen M, Zu X. Down-regulated microRNA-101 in bladder transitional cell carcinoma is associated with poor prognosis. Med Sci Monit. 2014;20:812–817. doi: 10.12659/MSM.890300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin R, Shen W, Zhi Y, Zhou Z. Prognostic value of miR-26a and HMGA1 in urothelial bladder cancer. Biomed Pharmacother. 2014;68:929–934. doi: 10.1016/j.biopha.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 36.Wu C-T, et al. DNMT1-dependent suppression of microRNA424 regulates tumor progression in human bladder cancer. Oncotarget. 2015;6:24119–24131. doi: 10.18632/oncotarget.4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosenberg E, et al. Predicting progression of bladder urothelial carcinoma using microRNA expression. BJU Int. 2013;112:1027–1034. doi: 10.1111/j.1464-410X.2012.11748.x. [DOI] [PubMed] [Google Scholar]
- 38.Drayton RM, et al. Reduced Expression of miRNA-27a Modulates Cisplatin Resistance in Bladder Cancer by Targeting the Cystine/Glutamate Exchanger SLC7A11. Clinical Cancer Research. 2014;20:1990–2000. doi: 10.1158/1078-0432.CCR-13-2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andrew AS, et al. Expression of tumor suppressive microRNA-34a is associated with a reduced risk of bladder cancer recurrence. Int J Cancer. 2015;137:1158–1166. doi: 10.1002/ijc.29413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schee K, et al. Deep Sequencing the MicroRNA Transcriptome in Colorectal Cancer. PLoS One. 2013;8:e66165. doi: 10.1371/journal.pone.0066165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gu L, et al. MicroRNAs as prognostic molecular signatures in renal cell carcinoma: a systematic review and meta-analysis. Oncotarget. 2015;6:32545–32560. doi: 10.18632/oncotarget.5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jamali Z, et al. MicroRNAs as prognostic molecular signatures in human head and neck squamous cell carcinoma: a systematic review and meta-analysis. Oral Oncol. 2015;51:321–331. doi: 10.1016/j.oraloncology.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 43.Frampton AE, et al. microRNAs with prognostic significance in pancreatic ductal adenocarcinoma: A meta-analysis. Eur J Cancer. 2015;51:1389–1404. doi: 10.1016/j.ejca.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 44.Zhou X, et al. Prognostic value of miR-21 in various cancers: an updating meta-analysis. PLoS One. 2014;9:e102413. doi: 10.1371/journal.pone.0102413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Villadsen SB, et al. The miR-143/-145 cluster regulates plasminogen activator inhibitor-1 in bladder cancer. Br J Cancer. 2012;106:366–374. doi: 10.1038/bjc.2011.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kent OA, et al. Repression of the miR-143/145 cluster by oncogenic Ras initiates a tumor-promoting feed-forward pathway. Genes Dev. 2010;24:2754–2759. doi: 10.1101/gad.1950610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Noguchi S, et al. Replacement treatment with microRNA-143 and -145 induces synergistic inhibition of the growth of human bladder cancer cells by regulating PI3K/Akt and MAPK signaling pathways. Cancer Lett. 2013;328:353–361. doi: 10.1016/j.canlet.2012.10.017. [DOI] [PubMed] [Google Scholar]
- 48.Lin T, et al. MicroRNA-143 as a tumor suppressor for bladder cancer. J Urol. 2009;181:1372–1380. doi: 10.1016/j.juro.2008.10.149. [DOI] [PubMed] [Google Scholar]
- 49.Noguchi S, et al. MicroRNA-143 functions as a tumor suppressor in human bladder cancer T24 cells. Cancer Lett. 2011;307:211–220. doi: 10.1016/j.canlet.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 50.Gao Y. et al. miR-155 regulates the proliferation and invasion of clear cell renal cell carcinoma cells by targeting E2F2. Oncotarget (2016). [DOI] [PMC free article] [PubMed]
- 51.Zhang X, et al. Upregulated miR-155 in papillary thyroid carcinoma promotes tumor growth by targeting APC and activating Wnt/beta-catenin signaling. J Clin Endocrinol Metab. 2013;98:E1305–1313. doi: 10.1210/jc.2012-3602. [DOI] [PubMed] [Google Scholar]
- 52.Huang X, et al. Quantitative proteomics reveals that miR-155 regulates the PI3K-AKT pathway in diffuse large B-cell lymphoma. Am J Pathol. 2012;181:26–33. doi: 10.1016/j.ajpath.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu WJ, et al. MLH1 as a direct target of MiR-155 and a potential predictor of favorable prognosis in pancreatic cancer. J Gastrointest Surg. 2013;17:1399–1405. doi: 10.1007/s11605-013-2230-5. [DOI] [PubMed] [Google Scholar]
- 54.Peng Y, et al. MicroRNA-155 promotes bladder cancer growth by repressing the tumor suppressor DMTF1. Oncotarget. 2015;6:16043–16058. doi: 10.18632/oncotarget.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deng M, et al. miR-214 promotes tumorigenesis by targeting lactotransferrin in nasopharyngeal carcinoma. Tumour Biol. 2013;34:1793–1800. doi: 10.1007/s13277-013-0718-y. [DOI] [PubMed] [Google Scholar]
- 56.Ueda T, et al. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol. 2010;11:136–146. doi: 10.1016/S1470-2045(09)70343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu CX, et al. MicroRNA miR-214 regulates ovarian cancer cell stemness by targeting p53/Nanog. J Biol Chem. 2012;287:34970–34978. doi: 10.1074/jbc.M112.374611. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 58.Penna E, et al. miR-214 coordinates melanoma progression by upregulating ALCAM through TFAP2 and miR-148b downmodulation. Cancer Res. 2013;73:4098–4111. doi: 10.1158/0008-5472.CAN-12-3686. [DOI] [PubMed] [Google Scholar]
- 59.Peng RQ, et al. MicroRNA-214 suppresses growth and invasiveness of cervical cancer cells by targeting UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7. J Biol Chem. 2012;287:14301–14309. doi: 10.1074/jbc.M111.337642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Derfoul A, et al. Decreased microRNA-214 levels in breast cancer cells coincides with increased cell proliferation, invasion and accumulation of the Polycomb Ezh2 methyltransferase. Carcinogenesis. 2011;32:1607–1614. doi: 10.1093/carcin/bgr184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shih TC, et al. MicroRNA-214 downregulation contributes to tumor angiogenesis by inducing secretion of the hepatoma-derived growth factor in human hepatoma. J Hepatol. 2012;57:584–591. doi: 10.1016/j.jhep.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 62.Huang SD, et al. MicroRNA-98 and microRNA-214 post-transcriptionally regulate enhancer of zeste homolog 2 and inhibit migration and invasion in human esophageal squamous cell carcinoma. Mol Cancer. 2012;11:51. doi: 10.1186/1476-4598-11-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Quintavalle C, et al. miR-221/222 overexpession in human glioblastoma increases invasiveness by targeting the protein phosphate PTPmu. Oncogene. 2012;31:858–868. doi: 10.1038/onc.2011.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.He HC, et al. Global analysis of the differentially expressed miRNAs of prostate cancer in Chinese patients. BMC Genomics. 2013;14:757. doi: 10.1186/1471-2164-14-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu K, et al. MiR-222 modulates multidrug resistance in human colorectal carcinoma by down-regulating ADAM-17. Exp Cell Res. 2012;318:2168–2177. doi: 10.1016/j.yexcr.2012.04.014. [DOI] [PubMed] [Google Scholar]
- 66.Lee C, et al. Elevated expression of tumor miR-222 in pancreatic cancer is associated with Ki67 and poor prognosis. Med Oncol. 2013;30:700. doi: 10.1007/s12032-013-0700-y. [DOI] [PubMed] [Google Scholar]
- 67.Calderaro J, et al. PI3K/AKT pathway activation in bladder carcinogenesis. Int J Cancer. 2014;134:1776–1784. doi: 10.1002/ijc.28518. [DOI] [PubMed] [Google Scholar]
- 68.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sutton AJ, Song F, Gilbody SM, Abrams KR. Modelling publication bias in meta-analysis: a review. Stat Methods Med Res. 2000;9:421–445. doi: 10.1177/096228020000900503. [DOI] [PubMed] [Google Scholar]
- 70.Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6:e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25:603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 72.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]


