Abstract
Although large volumes of information are entered into our electronic health care records, radiation oncology information systems and treatment planning systems on a daily basis, the goal of extracting and using this big data has been slow to emerge. Development of strategies to meet this goal is aided by examining issues with a data farming instead of a data mining conceptualization. Using this model, a vision of key data elements, clinical process changes, technology issues and solutions, and role for professional societies is presented. With a better view of technology, process and standardization factors, definition and prioritization of efforts can be more effectively directed.
Summary.
Although large volumes of information are entered into our electronic health care records, radiation oncology information systems, and treatment planning systems on a daily basis, the goal of extracting and using these Big Data has been slow to emerge. Development of strategies to meet this goal is aided by examining issues with a data farming system instead of a data mining conceptualization. Using this model, a vision of key data elements, clinical process changes, technology issues and solutions, and role for professional societies is presented. With a better view of technology, process, and standardization factors, definition and prioritization of efforts can be more effectively directed.
Introduction
It should be common for clinics to have the ability to rapidly assemble datasets to address practice quality improvement (PQI), routine clinical translational research (CTR), and other arising questions to aid patients in our clinics today. We enter a wealth of information into electronic health records (EHR) and radiation oncology information systems (ROIS) on a daily basis. Shouldn’t it be the rule, rather than the exception, that clinics can seamlessly use this information to carry out tasks such as identifying the cohort of patients with a particular diagnosis and stage who were treated with specific technologies (eg, volumetric modulated arc therapy, breath hold) and examine the correlation of their survival and toxicities with dose delivered to target and organ-at-risk structures? We think it should be.
In addition, a wide range of analytics uses becomes viable, extensible, and automatable as availability of large, electronically gathered, comprehensive health care datasets emerge. Modern analytics approaches, such as machine learning, are poised to satisfy the promise of identifying and guiding response to factors affecting patient outcomes; however, these methods are more data-needy than ever. Moreover, broadening the scope of data elements to other departments within a single institution or to pooling data from multiple institutions is needed for development of realistic, comprehensive models of routine practice. Increased ability to participate in clinical trials and improved reporting and feedback mechanisms are crucial.
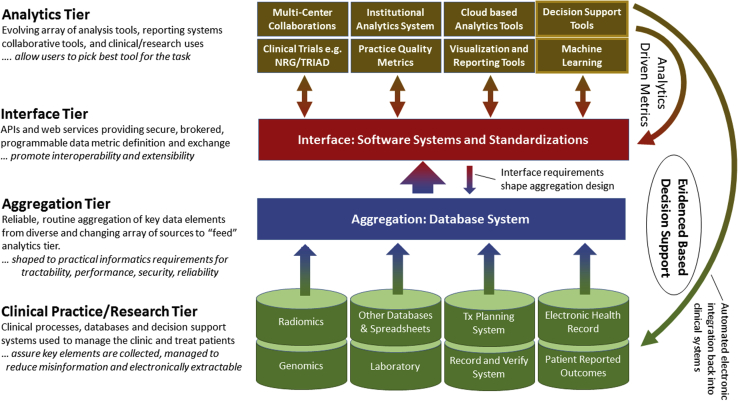
Reaching the goal of prospective automated, electronic incorporation of evidence-based decision support extracted from retrospective experience back into clinical and research efforts is a multi-level effort. Figure 1 illustrates 4 system tiers in constructing applications to support knowledge guided radiation therapy. Obtaining these analytics tier products depends on the ability to supply large volumes of useful data on a wide range of elements to their engines.
Figure 1.
The systems required for construction of a knowledge-guided radiation therapy system that supports machine learning, reporting, and participation in trials and other clinical efforts can be conceptualized in 4 tiers. The foundational clinical processes and aggregation tiers enable the benefits of the analytics tier. The integration tier promotes interoperability even when multiple technologies are used.
Reports focused on technologies or large-scale efforts highlighting the potential benefits of local and multi-institutional efforts are inspirational, but may make their realization seem distant and unapproachable for most clinics.1, 2, 3, 4, 5, 6, 7, 8, 9 Making local, routine use a reality and setting the stage to leverage machine learning and other analytics tier objectives require a multifront approach involving multiple data systems, changes to clinical processes, standardization, and database technologies to make more data available and accessible. Details on clinical experience with the foundational tiers could promote wider participation and more availability of multi-institutional datasets.
Recently, we have built on prior experience10, 11, 12, 13, 14, 15, 16, 17, 18 to construct a University of Michigan instance of a Radiation Oncology Analytics Resource (M-ROAR). It reduces information entropy by aggregating key multidisciplinary data elements from the clinical/research tier into a single system in the aggregation tier, encompassing an expanding range of key data elements; it currently contains data for ∼17,000 patients treated with radiation at the University of Michigan since 2002. By better framing our view of the current issues and tasks involved, our ability to leverage limited resources to develop solutions was improved. Our purpose in this manuscript is to share our vision of the issues, solutions, and key data elements that need to be addressed.
Data farming vs data mining
The standard conceptualization of data aggregation and analysis efforts is termed “data mining.” Unfortunately, this creates a misleading expectation that the data elements needed already exist in electronic systems and are just waiting to be “mined” (ie, found, extracted, and used in the analysis tier). Moreover, it assumes data are sufficiently curated to allow for accurate linkage to patients, identification of relationships among data elements, and extraction of reliable values. Embracing this conceptualization can lead to being overly receptive to promises for shiny new and eclectic technologies that are nominally able to pick through any “load” of the data in our ROIS or EHR to be able to meet all of our needs. Often, both the price tag and the level of dependence on these “one-of-a-kind” solutions are high. Moreover, detailed understanding of what key data elements are needed, how to accurately retrieve them from existing systems, and what clinical processes need to be addressed to fill in gaps in the data is frequently low.
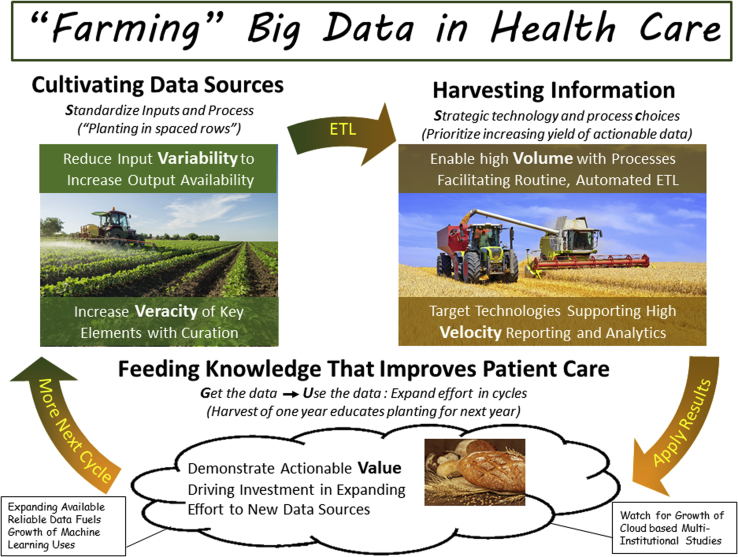
Data farming is a more realistic and functional conceptualization for shaping expectations of the type of work and commitment needed to construct reliable databases supporting practice quality improvement and clinical translational research (Fig 2). The objective is to harvest large volumes of data that we could use as raw materials for analyzing health care patterns and outcomes. Like the farmer who considers the implication of every part of the sowing, growing, and harvesting process on the yield of high-quality grain, we need to examine how best to use the tools available in our electronic systems to increase the volume of actionable data that are readily available. High-quality data sources rarely exist independent of our efforts, just waiting to be found, or mined. They result from intent and dedication of resources to grow these data sources and curate (weed out) misleading information.
Figure 2.
Farming is a useful metaphor for envisioning the issues in creating outcomes databases in health care.
A data farming conceptualization also helps highlight 5 of the Big Data “Vs” we have found to be prominent in technology and process discussions in radiation oncology.
-
•
Variability: Various given data types (eg, weight, laboratory values, dose-volume histogram [DVH] curves) may need to be aggregated from multiple sources based on criteria such as time range, stakeholder group, or vendor. Differences in location, access requirements, storage technology, nomenclature, formatting, units, and data quality contribute to complexity of extract, transform, and load (ETL) operations.
-
•
Veracity: Incorrect data values or missing data undermine the ability to draw accurate statistical conclusions about distributions of values and relationships between data elements. Many PQI and CTR efforts focused on data at the outer range or even in the tails of distributions where the “law of averages” cannot wash out errors.
-
•
Volume: Storage and processing requirements for data elements can drive technology decisions when very large (eg, >1 Pb). Thresholds for this classification evolve rapidly as technologies progress.
-
•
Velocity: Data input stream rates can drive technology decisions when very large (eg, >1 Tb/s). Processing speeds for the system of analytics, interface, and aggregation tiers drive tractability of incorporating analytics into clinical process flows. Thresholds for this classification evolve rapidly as technologies progress.
-
•
Value: Implementation of Big Data solutions has high costs: financial, technical, staffing resource allocation, process change, and political capital. Obtaining needed support depends on addressing cost vs benefit to PQI and clinical translational research efforts.
Blog postings for generalized Big Data discussions sometimes cite a “variety” of information/data types as an issue (eg, Facebook postings, Twitter feeds, image data, video data). Because key data elements are generally part of the EHR, ROIS, or treatment planning system (TPS), we have not found variety to be a driving issue for the specific case of radiation oncology.
Planting in spaced rows: Ensuring availability of key data elements
Big Data efforts in radiation oncology are challenged by high degree of variability in data types and sources, in both format and quality. Data elements are distributed among the ROIS and EHR across additional discipline specific (eg, pathology, chemotherapy, surgery, genetics) databases and in spreadsheets. The number of databases, versions, and quality caveats multiply as extractions reach further back into the historical record. Key data elements may not be routinely recorded as part of our clinical processes or are recorded in a format that makes eventual retrieval unlikely or very cumbersome. When information structuring is highly uncertain or only sparsely available, its value is compromised by the expense, complexity, and effort needed to accurately extract it.
In farming, the concept of a row as a process organizational principle was fundamental to improving effectiveness and enabling development of industrialized tools. The same principle applies to radiation oncology. Structuring routine practice processes to improve availability and accuracy of key data elements for automated, electronic extraction increases the volume of data available and reduces the cost of aggregation.
Where do we need to focus our efforts and what should we do? Table 1 lists key data element categories and characterizes challenges to aggregation of the information, ranking difficulty of ETL operations. Categories are ranked according to demand as elements of frequently requested PQI and CTR queries. Treatment details generated by the ROIS are typically available. In contrast, many highly ranked elements have multiple ETL challenges. For example, staging and outcomes data input are variable, and clinics frequently use free text entry in EHR notes instead of availing themselves of quantified fields in ROIS and tools to define linkages of metastatic to originating site diagnosis. We need to change multidisciplinary/provider processes to take advantage of data structuring tools already built in to the ROIS, TPS, or EHR to enable automated extraction with little expense. Better practice processes = better planting and thus a better harvest!
Table 1.
Categorization of key data element categories and summary of our experience of challenges to extract, transform, and load (ETL) of data from source systems to aggregation tier.
| Key element category | Demand ranking | ETL difficulty | Typical source systems | Access | Multiple source systems | Use or used free text entry | Missing data | Data accuracy | Lack of standardization | PHI constraints limit access | Legacy formats or systems | Require process changes | Extensive transformation | Other |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Demographics ● | 1 | L | EHR | × | E | |||||||||
| Health status factors | 2 | L | EHR | × | E | |||||||||
| Pathology ⊙ | 3 | M to H | EHR | × | × | × | × | × | ⊠ | E, X | ||||
| Surgery ⊙ | 2 | M to H | EHR | × | × | × | × | × | ⊠ | E, X | ||||
| Chemotherapy ● | 2 | M | EHR, ODB | × | E | |||||||||
| Encounter details ● Office, emergency room, hospitalization |
3 | L | EHR | ⊠ | × | R | ||||||||
Diagnosis ●, , ,
|
1 | M | EHR, ROIS | × | × | × | × | ⊠ | R, E | |||||
Staging ●, , ,
|
1 | H | EHR, ROIS | × | × | × | × | × | ⊠ | E | ||||
Prescription  ,♦ ,♦ |
1 | H | ROIS, ODB | ⊠ | × | E, X, R | ||||||||
| As-treated plan details ● | 1 | M | ROIS | × | ||||||||||
DVH ●, ,♦ ,♦ |
1 | M | TPS | × | × | × | ⊠ | × | ATPS | |||||
| Survival ● | 1 | M | EHR, XLS, ODB | × | ⊠ | UD, E | ||||||||
Recurrence  , ,
|
1 | H | EHR | × | × | × | × | × | ⊠ | E, X | ||||
Toxicity ●,
|
1 | H | EHR, ROIS | × | × | × | × | × | ⊠ | E, X | ||||
Patient-reported outcomes 
|
2 | H | EHR, P | × | × | × | × | ⊠ | E, X | |||||
| Laboratory values ● | 2 | M | EHR | ⊠ | × | × | E | |||||||
| Medications● | 2 | M | EHR | ⊠ | × | × | E | |||||||
| Height, weight, BMI● | 2 | M | EHR | ⊠ | × | × | E | |||||||
| Treatment imaging: Timeline details● | 3 | H | ROIS | × | R | |||||||||
| Diagnostic imaging details ⊙ |
3 | M | ODB | ⊠ | × | × | × | |||||||
| Radiomics ⊙,♦ | 3 | L | XLS | × | ⊠ | |||||||||
| Genomics ⊙ | 3 | L | XLS | × | ⊠ | |||||||||
| Charges ● | 3 | L | ROIS | |||||||||||
| Research datasets ⊙ | 4 | H | XLS | × | ⊠ | × | × | E | ||||||
| Registry data ⊙ | 4 | M | ODB | ⊠ | × | × | × | UD |
Demand ranking ranges from most (1) to least (4) frequently needed as part of queries. Range in ETL is specified when significant variation among institutions is anticipated; extensive transformation indicates need to construct sophisticated algorithms to process raw data from source systems to provide needed information.
APTS, special manual effort needed to construct as-treated plan sums; BMI, body mass index; DVH, dose-volume histogram; E, manual entry without process corrected curation are susceptible to random or system-related systematic errors; HER, electronic health records; ETL, extract, transform, and load; H, extensive process changes needed, data typically in unstructured free text fields; L, little modification required; M, changes to clinical processes required, interactions across different groups in the institution, significant computational processing; M-ROAR, Michigan Radiation Oncology Analytics Resource; NLP, natural language processing; ODB, other database systems; P, paper records; PHI, Patient Health Information; R, missing detail on key relationships to other data items; ROIS, radiation oncology information system; TPS, treatment planning system; UD, data values not being up to date; X, manual effort required to extract data; XLS, spreadsheet.
M-ROAR–specific ETL status for all patients: ●, current processes enable capture for all; ⊙, developing new extractions;  , exploring NLP-based process;
, exploring NLP-based process;  , piloting new clinical process; ♦, developing new software applications to improve availability or accuracy;
, piloting new clinical process; ♦, developing new software applications to improve availability or accuracy;  , developing extractions for legacy data with differing formats. The current database includes 17,956 patients treated since 2002. Records per patient vary with time period and key data element category.
, developing extractions for legacy data with differing formats. The current database includes 17,956 patients treated since 2002. Records per patient vary with time period and key data element category.
×, specific ETL challenges; ⊠, the primary issue for enabling automated extractions for multiple issues.
Missing data can be a problem. For example, recurrence and toxicity information are often entered into the EHR as free text notes because it is the fastest means of proceeding with the demands of a busy clinical day. The result of not using standardized inputs, including standardized “free text” formats (eg, smart lists in Epic EHR), is that accurately extracting information to define actionable statistics requires manual rather than electronic approaches. As a result, it is rarely done as part of routine practice for all patients.
Note that reliance on eventual emergence of natural language processing (NLP) methods as a catch-all, promising to eliminate need for any manual effort, leads to highly uncertain timelines for projecting when key data elements will be accurately extracted and available. Use of NLP as a filter, to augment manual efforts, is gradually gaining traction. Fully automated extractions, demonstrating high accuracy across a range of key elements, are areas of exploration. In the meantime, practice changes to use standardized, quantified entry of key data elements; enables gathering the data now; and will enhance the accuracy and reduce costs of NLP methods when they evolve in the future. For M-ROAR, our clinical practice committee reviewed the options and is overseeing the transition to the use of discrete fields in the ROIS for entry of staging data and use of quantified field objects (flow sheets in 1 EHR system) for toxicity data and recurrence.
Standard row spacing: Professional society-driven standardizations
Development and use of process standards promote ability to develop automated methods to aggregate key data elements for all patients. Standardizing definitions of key elements for treatment details, DVH metrics, toxicity, and patient-reported outcomes, segregated by disease site, that are recommended to be made available for automated extraction for all patients would aid in defining common practice.
Ideally, this standardization would be carried out with the combined effort of stakeholder societies (eg, American Association of Physicists in Medicine [AAPM], American Society for Radiation Oncology, European Society for Radiotherapy and Oncology, Southeast Asian Radiation Oncology Group) as part of data aggregation projects. Defining standards provides incentive for aggregating the information and for facilitating the ability of vendors to meet the defined need. For example, the AAPM Task Group 263 – Standardizing Nomenclature for Radiation Therapy has defined standards for naming of target and organ-at-risk structures and DVH metrics to facilitate the ability to automatically extract and analyze key data elements from DVHs.
Beginning with a small, core set and gradually expanding as use in multi-institutional collaborative data efforts demonstrates the value of adoption keeps the focus for success on volume of data harvested. Collaborative efforts to define standards that vendors can apply are important for developing common solutions that will ultimately increase pooling of federated datasets.
Cash crops: Defining key data elements in radiation oncology
Key data elements need to be quantified and prioritized to direct aggregation efforts. Arbitrarily increasing the number of data items gathered as part of clinical processes can have a high time cost for individual clinicians and other staff. Identifying subsets of data elements with high value to PQI and to CTR efforts, is an important starting point.
We used a combined approach to define key data elements and categories incorporated into M-ROAR.
-
•
Suggestions from clinicians and staff formed from their research and PQI experience.
-
•
Examination of recent queries from the ROIS and EHR systems to support PQI and research efforts.
-
•
Faculty surveys of questions for which they wished to use M-ROAR to address and deconstruction of responses to identify data elements and relationships required to meet those needs.
Identification of key data elements is not a fixed or one-time effort. It requires working with many stakeholders and recognition that these identifications may change or evolve over time.
Table 1 highlights categories of key elements that are common to a wide range of queries (eg, staging). The categories and elements continue to evolve as new capabilities lead to new queries and exploration of new data sources. The detailed list of specific elements is available upon request. They form the basis of radiation oncology translational research ontology. Standardizing radiation oncology translational research ontology as a joint effort of professional societies (eg, AAPM, American Society for Radiation Oncology, European Society for Radiotherapy and Oncology, Southeast Asian Radiation Oncology Group) using this and other existing ontologies6, 8 as a starting point would support long-term efforts to support multi-institutional research by defining a baseline of information and tools for information exchange. Ontologies will see wider utilization as part of vendor and institutional systems as they improve in detail, standardization, usability, and depth of information on data elements and interrelationships. Proactive engagement by professional societies will hasten this timeline. Awareness by task groups and working groups of implications of secondary effects from their efforts on facilitating Big Data aggregation is important to expand the range of information available (eg, AAPM TG174).
Careful consideration of the value of extracting and storing raw vs distilled information is needed and may be hotly debated. For example, recreating the functionality of picture archiving and communication systems to store pixel-based information for image series may not be as productive as developing automated access and processing capabilities to extract and store distilled features (eg, radiomics metric values, image access, characterization data). Raw genomics information, free text data, and dose arrays are similar examples encountered in these debates. Distilled data requires lower volume and may have higher value (cost/benefit) if provenance of the raw data is also recorded to preserve the ability to trace the raw source data for review.
To harvest you first have to plant: Ensuring key elements are present in the records
Much is known about the relatively small (∼5%) number of patients on clinical trials that systematically quantifies key data elements for participating patients. For the majority of patients, who are not treated on clinical trials, much less is known. Often, key data elements are simply not entered into the record as a part of routine practice because they are not required. Clinics should identify core key data items that are vital to their objectives that can be entered using existing tools in the EHR, ROIS, or TPS. Typically, these will include the following.
-
•
Basic disease details: diagnosis, staging, laterality, stratification factors.
-
•
Basic outcomes measures: survival, recurrence, toxicity.
-
•
Course composite dose data. This requires routine creation of As Treated Plan Sums showing composite doses of initial plan, boosts, and plan revisions. Automated extraction of DVH metrics, reflecting the full treatment course, is significantly undermined without creation of As Treated Plan Sums as part of clinical practice.
-
•
Prescriptions. A tabulated summary should reflect the fractionation groups (eg, initial plan, boosts, plan revisions), the gross tumor volume, clinical target volume, and planning target volume structures treated to differing dose levels as part of those fractionation groups, and sequential use of multimodality treatments (eg, external + brachytherapy).
-
•
Chemotherapy details. Agents used, because delivered infusion schedules.
Patient-Reported Outcomes (PRO) also fall into this area but require much more substantial changes in process, staffing, and development of technical resources to ensure routine collection of these data. In addition, because electronic PRO systems are deployed to reach patients outside of the clinic, additional coordination with information technology and compliance offices to protect patient health care information is required.
Weeding: Building data curation in practice processes
Reliability of manually entered data elements in the absence of proper curation incorporated into clinical processes is frequently a problem for the veracity of these datasets, requiring local expertise to assess and correct issues. The notion that noisy or inaccurate data (“dirty data”) values are acceptable because large volumes of correct data will wash out their effects undermines the ability to carry out cohort discovery for rare combinations of factors that might be most relevant. Unchecked, dirty data can lead to “garbage in - despair out” as confidence in the value of big data efforts erodes willingness to participate in practice changes.
For example, random errors compromise accurate characterization of integer counts of events when the incidence rate is low compared with the error rate. Manual entry errors or omissions for high-grade toxicities reduce ability to develop automated solutions to characterize distributions and correlate to contributing factors. Frequency of systematic errors and likelihood of missing data for core key data elements (eg, diagnosis, staging, laterality) undermine development of reliable, automated analytics. For example, systematic errors for patients treated for metastatic disease, by use of International Classification of Diseases codes for originating site (eg, prostate, breast, lung) instead of the correct International Classification of Diseases codes for the secondary site (eg, bone, lung, brain), or omitting connection between the two weakens accurate automated identification and characterization of treatment technologies used for these patients.
In farming, it is never possible to eradicate all weeds. Instead, applying sufficient effort so that the weeds do not overwhelm the grain is needed. In clinical processes, it is important not only to minimize noise, but also to have strong methods in place to identify major outliers and errors that could have a big impact on analysis. A practical approach to building curation into routine practice is needed to find a mean between requirements so burdensome to clinical processes that they reduce ability to obtain needed data and those that are so lax that they undermine ability to automate, accurate extraction, and analysis of data. Incorporating curation into clinical processes with a focus on high-priority data elements subject to manual entry errors (eg, recurrence type) or having low tolerance for random errors for values at the margins of distributions (eg, rare diseases, toxicities) is productive.
For example, peer review of diagnosis and staging as part of chart rounds or review of treatment plans enhances the accuracy of the data. Assuring compliance with nomenclature standards for target and organ-at-risk structures and the existence of “as treated” plan sums dramatically increases the reliability of automated processing of DVH data. Creation and review of monthly reports of toxicity values aid in weeding out incorrect values and minimizing missing values.
With loading of regularized data into data resources (Structured Query Language [SQL] or NoSQL) and inclusion of provenance information traceably linking to source systems, development of electronic algorithms to identify inaccuracies or missing data becomes plausible. Care must be taken with electronic fixing of data to avoid introducing bias or additional errors. This requires detailed understanding of clinical processes that produced the errors. For example, replacing missing toxicity values with grade 0 will skew comparisons of physician practices that systematically do not enter data for grade 0 vs occasionally neglecting to enter toxicity values for low grades.
Farming villages: Staffing resources and collaborations
Building an outcomes database is a community effort. Defining key data elements, gaining access to data sources outside the department, identifying and implementing optimal processes that align clinic flow with data objectives, and using the data in PQI and research require combined efforts of all staff member groups in the clinic. Physician, physics, dosimetry, therapist, nursing, administrative, and information technology staff groups all play multiple roles in the work. Providing encouragement, time, resources, and support for the members of the team motivated to build and apply a working system in the clinic increases likelihood of success in constructing a system that works for all. Gathering new data such as PROs requires investment in new staff to assist patients in setting up electronic portal accounts and initial navigation of electronic completion of surveys. Cross-departmental collaborations, leading to integration of aggregation processes and systems, are needed to form complete pictures of patient care (to make a meal, we need more than one crop).
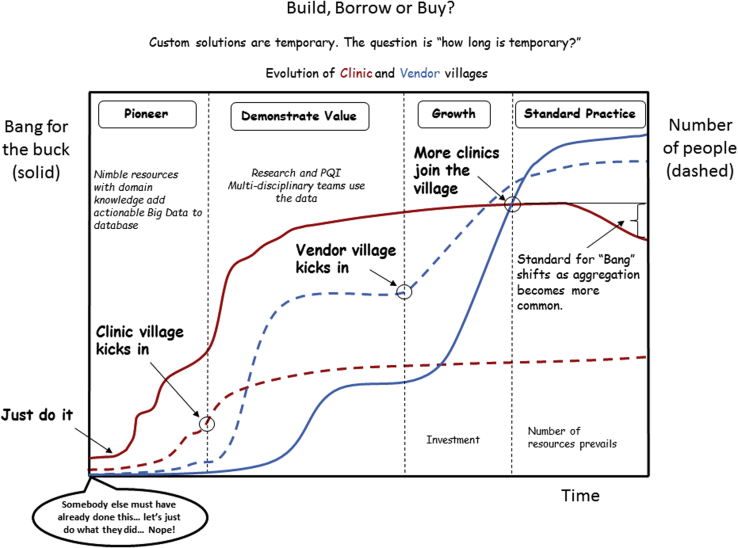
Figure 3 illustrates phases of development for creation of Big Data systems. Phases progress as the size and diversity of the village of contributors to the effort grows. Growth is fueled by demonstrations of value for research and PQI. Presently, most efforts are in the pioneering or demonstration of value phases. Transition to availability of viable, cost-effective, vended solutions is anticipated with demonstrations of value for use of Big Data in the clinic.
Figure 3.
Evolution of practical Big Data systems progresses from smaller highly skilled groups to large vendor-based systems as the multidisciplinary village of staked holders (physicians, physicists, administration, RTT, dosimetry, nursing) finds and demonstrates value for these systems. Value drives willingness to modify clinical practices to reduce data variability.
Multi-institutional collaborations, leveraging pooling of data to explore outcomes effects that are robust against practice variations, are important for lowering technical barriers and cost. They provide needed small-scale use cases for identification and proof of concept solutions for standardization, technology, practice, and policy issues that lead to viable large-scale approaches for health care. Integration of federated, multi-institutional data sources promotes better ability to develop evidence-based health care policy and analytics (to feed the world, we need a lot of farmland). These efforts provide collateral benefits to institutional objectives for improving quality and reducing cost. Health systems should be proactive in enabling these efforts through data use agreements, working with data compliance offices to standardizing secure server systems for federated exchange, and financial support.
Prioritizing demonstrations of value to PQI and research as new data elements are added builds community support and provides additional channels for feedback on key data elements and for curation. For example, the self-service dashboard illustrated in Figure 4 for patient cohort identification has the collateral benefit of highlighting issues with incorrect diagnosis codes.
Figure 4.
A self-service dashboard from M-ROAR illustrating high-velocity output from a large volume of data, value for supporting PQI, and research effort and means to improve veracity by bringing the consequences of “dirty data” directly into the view of end-users. PQI, practice quality improvement.
Farm machinery: Approaching technologies for radiation oncology big data
There are a large number of database classes and specific solutions in various stages of maturity to choose from for aggregation. Existing technologies destined for longevity evolve to adopt the best ideas of new technologies. Attempting to pick “the best” in this churning landscape may have uncertain outcome.19 Picking one that allows focus on progress in aggregating and analyzing the data with existing resources while also investigating strengths and weaknesses of alternative technologies provides a better mix for addressing both near-term needs and long-term vision.
Evaluation should include performance with realistic domain applicable datasets. Although high-velocity data input streams are typically not an issue, high speed in retrieval and analysis is for defining practical approaches to incorporate the data into clinical processes. Use realistic datasets to evaluate:
-
•
performance of query operations
-
•
ability to integrate into existing systems to carry out ETL operations
-
•
ability to integrate into development of clinical applications to use the data in practice
-
•
ability to interact with standard analytics or machine learning systems
-
•
implications for availability of staff required to implement the technology
-
•
longevity
-
•
cost (hardware, software, training, staff, time).
Our SQL data lake, which is used to stage multisource data for incorporation into M-ROAR, is currently 87 GB. The production version MS SQL database of M-ROAR that aggregates data for >17,000 patients treated since 2002 is currently 9 GB. The architecture is designed to allow the database to be refreshed periodically within a few hours. Reporting velocities are suitable for routine use. Benchmark queries combining multiple inner joins (intersections of datasets) and right outer joins (unions of datasets) over thousands of records in several tables (complex datasets) to stress the performance execute in less than 2 seconds. Examining the most recent 20 research query requests, each executed in less than 0.03 seconds and produced between 200 and 3500 records. Self-service reporting tools (Fig 4) allow users to sort over the full set of patients for cohort discovery in less than 1 second. So far, availability of time, resources, and access have been the rate-limiting factors for growth, not lack of use of visionary database technology.
Typically, the Big Data thresholds that challenge conventional technology paradigms are volume (eg, petabytes) or velocity (eg, terabytes/sec). For example, storage for genome sequences of ∼200,000 patients or sequence transmission rates of ∼200 patients/s reach these thresholds. Wide-scale availability of genomics data for all patients or use cases requiring storage of individual imaging or dose array pixels may eventually emerge to affect decisions on key data elements for routine aggregation. However, it is reasonable to anticipate emergence of a different landscape of database technologies before volume or input velocity thresholds become limiting factors that are more dominant for radiation oncology.
We anticipate that speed and maturity of query and analytics technologies (output velocity) will be important limiting factors as federated, multi-institutional databases emerge as part of routine research and clinical practice. Application of column/graphical store databases or use of high-capacity in-memory architectures to support faster queries will become more common as available data volumes catch up with potential.
Incorporating statistical analytics into database solutions to construct queries that are more sophisticated is emerging as a defining characteristic. For example, MS SQL 2016 will include incorporation of the open-source statistical platform R into the database server software. Similarly, availability analytics and reporting tools that function with a wide range of database technologies to improve end-user visual access (eg, Tableau) will be an increasingly important selection factor.
Differences in optimal characteristics of database technologies for aggregation vs analytics point away from one-size-fits-all solutions. For example, as aggregation systems emerge to allow rapid extraction of key elements from multiple source systems, their use to feed graphing databases (eg, Triple Store) in distributed analytics explorations of interactions among subsets of elements will have wider applicability for machine learning in both single- and multi-institutional efforts.6
Incorporation of high-performance encryption and watermarking technologies as part of routine practice to ensure security and data integrity for both institutional systems and for cloud-based systems is needed. Definition of data approaches that meet compliance and security standards and facilitate the ability to use cloud-based multi-institutional data pools containing key elements for longitudinal analysis are closely coupled to viability of these technologies.
Collaborating to find solutions to legal, policy, and security barriers to use of cloud-based systems to share database solutions with collaborators particularly as the volume of the data continues to grow is part of research in this area. As those solutions are developed, technologies that integrate well both with cloud-based architectures and enterprise source systems will have favorable cost (financial, staffing, space)/benefit ratios.
Summary
The vision for efforts required to make routine use of Big Data a part of clinical reality in radiation oncology is similar to the vision for creating a productive farm yielding large volumes of high-quality grain. We are both the consumers and the producers of the yield that serves to help us improve patient care. Farming cultures evolved their processes and technologies from sufficient for subsistence to enabling large-scale automation. An analogous evolution in radiation oncology data is within reach. It requires community effort leveraging the skills, insights, and data use needs of all clinical and information technology staffing groups as well as professional societies.
Cooperative development and adoption of standardizations by vendors and clinics to increase volume and availability of datasets created as part of routing processes is a vital part of that community effort. Engagement by government as part of these communities is needed to overcome barriers to combining these datasets so that the information learned through treating patients today can be used to improve treatments and health care policies for the patients of tomorrow.
Footnotes
Conflict of interest: None.
References
- 1.Palta J.R., Efstathiou J.A., Rose C.M. Developing a national radiation oncology registry: From acorns to oaks. Pract Radiat Oncol. 2012;2:10–17. doi: 10.1016/j.prro.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 2.Robertson S.P., Quon H., McNutt T.R. A data-mining framework for large scale analysis of dose-outcome relationships in a database of irradiated head and neck cancer patients. Med Phys. 2015;42:4329–4337. doi: 10.1118/1.4922686. [DOI] [PubMed] [Google Scholar]
- 3.Chen R.C., Gabiel P.E., Kavanagh B.D., McNutt T.R. How will big data impact clinical decision making and precision medicine in radiation therapy? Int J Oncol Radio Biol Phys. 2016;95:880–884. doi: 10.1016/j.ijrobp.2015.10.052. [DOI] [PubMed] [Google Scholar]
- 4.Skripcak T., Belka C., Bosch W., Baumann M. Creating a data exchange strategy for radiotherapy research: Towards federated databases and anonymised public datasets. Radiother Oncol. 2014;113:303–309. doi: 10.1016/j.radonc.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nyholm T., Olsson C., Montelius A. A national approach for automated collection of standardized and population-based radiation therapy data in Sweden. Radiother Oncol. 2016;119:344–350. doi: 10.1016/j.radonc.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 6.Roelofs E., Dekker A., Lambin P. International data-sharing for radiotherapy research: An open-source based infrastructure for multicentric clinical data mining. Radiother Oncol. 2014;110:370–374. doi: 10.1016/j.radonc.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kessel K.A., Combs S.E. Review of developments in electronic, clinical data collection, and documentation systems over the last decade - are we ready for big data in routine health care? Front Oncol. 2016;6:75. doi: 10.3389/fonc.2016.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalet A.M., Gennari J.H., Ford E.C., Phillips M.H. Bayesian network models for error detection in radiotherapy plans. Phys Med Biol. 2015;60:2735–2749. doi: 10.1088/0031-9155/60/7/2735. [DOI] [PubMed] [Google Scholar]
- 9.Meldolesi E., van Soest J., Valentini V. Standardized data collection to build prediction models in oncology: A prototype for rectal cancer. Future Oncol. 2016;12:119–136. doi: 10.2217/fon.15.295. [DOI] [PubMed] [Google Scholar]
- 10.Shumway D.A., Griffith K.A., Pierce L.J. Wide variation in the diffusion of a new technology: Practice-based trends in intensity-modulated radiation therapy (IMRT) use in the state of Michigan, with implications for IMRT use nationally. J Oncol Pract. 2015;11:e373–e379. doi: 10.1200/JOP.2014.002568. [DOI] [PubMed] [Google Scholar]
- 11.Jagsi R., Griffith K.A., Pierce L.J. Differences in the acute toxic effects of breast radiotherapy by fractionation schedule: Comparative analysis of physician-assessed and patient-reported outcomes in a large multicenter cohort. JAMA Oncol. 2015;1:918–930. doi: 10.1001/jamaoncol.2015.2590. [DOI] [PubMed] [Google Scholar]
- 12.Vallières M., Freeman C.R., Skamene S.R., El Naqa I. A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Phys Med Biol. 2015;60:5471–5496. doi: 10.1088/0031-9155/60/14/5471. [DOI] [PubMed] [Google Scholar]
- 13.Lee S., Ybarra N., El Naqa I. Bayesian network ensemble as a multivariate strategy to predict radiation pneumonitis risk. Med Phys. 2015;42:2421–2430. doi: 10.1118/1.4915284. [DOI] [PubMed] [Google Scholar]
- 14.El Naqa I., Ruijan L., Murphy M.J., editors. Machine Learning in Radiation Oncology Theory and Applications. Springer-Verlag; Switzerland: 2015. [Google Scholar]
- 15.Mayo C.S., Pisansky T.M., Petersen I.A. Establishment of practice standards in nomenclature and prescription to enable construction of software and databases for knowledge-based practice review. Pract Radiat Oncol. 2016;6:e117–e126. doi: 10.1016/j.prro.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 16.Mayo C., Conners S., Miller R. Demonstration of a software design and statistical analysis methodology with application to patient outcomes data sets. Med Phys. 2013;40:111718. doi: 10.1118/1.4824917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mayo C.S., Deasy J.O., Chera B.S. How can we effect culture change toward data driven medicine? Int J Radiat Oncol Biol Phys. 2016;95:916–921. doi: 10.1016/j.ijrobp.2015.12.355. [DOI] [PubMed] [Google Scholar]
- 18.Sloan J.A., Halyard M., El Naqa I., Naqa El, Mayo C. Lessons from large-scale collection of patient-reported outcomes: Implications for Big data aggregation and analytics. Int J Rad Oncol Biol Phys. 2016;95:922–929. doi: 10.1016/j.ijrobp.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bailis P, Hellerstein JM, Stonebraker M. Readings in Database Systems. 5th ed. Available at: http://www.redbook.io/pdf/redbook-5th-edition.pdf. Accessed October 13, 2016.