Abstract
Purpose
Prospective detection of patients with advanced rectal cancer (LARC) who have a higher probability of responding to preoperative chemoradiotherapy (CRT) may provide individualized therapy. Lipidomics is an emerging science dedicated to the characterization of lipid fingerprint involved in different pato-physiological conditions. The purpose of this study is to highlight a typical lipid signature able to predict the tumor response to CRT.
Experimental Design
A prospective global analysis of lipids in 54 sera from 18 LARC patients treated with preoperative CRT was performed. Samples were collected at 3 time points: before (T0), at 14th day and at 28th day of CRT. An open LC-MS/MS analysis was performed to characterize lipid expression at T0. Differential lipids were validated by an independent approach and studied during treatment.
Results
From 65 differential lipids highlighted between responder (RP) vs not responder (NRP) patients, five lipids were validated to predict response at T0: SM(d18:2/18:1), LysoPC (16:0/0:0), LysoPC (15:1(9z)/0:0), Lyso PE (22:5/0:0) and m/z= 842.90 corresponding to a PC containing 2 fatty acids of 40 carbons totally. The levels of these lipids were lower in NRP before treatment. The ROC curve obtained by combining these five lipid signals showed an AUC of 0.95, evidence of good sensitivity and specificity in discriminating groups.
Conclusion
Our results are in agreement with previous evidences about the role of lipids in determining the tumor response to therapy and suggest that the study of serum lipid could represent a useful tool in prediction of CRT response and in personalizing treatment.
Summary.
This study aims to highlight a typical lipid signature able to predict the tumor response to preoperative chemoradiation therapy in advanced rectal cancer by using a lipidomics approach. Five lipids were validated as biomarkers able to predict response before treatment, resulting in a receiver operating characteristic curve characterized by an area under the curve of 0.95. Results suggest serum lipids could represent a useful tool in prediction of chemoradiation therapy response, toward a personalized treatment.
Introduction
Colorectal cancer (CRC) is the third most frequently occurring cancer globally.1 Preoperative fluoropyrimidine-based chemoradiation therapy (CRT) or short-course radiation therapy followed by total mesorectal excision are the standard treatments for CRC.2, 3, 4 In the effort to personalize treatments, there is increasing interest in predicting which patients will respond to neoadjuvant CRT,5 especially via investigating easily accessible biological fluids,6 and in improving response rate and survival outcomes. Several biomarkers have been investigated for their ability to predict outcome in locally advanced rectal cancer (LARC) treated with CRT, but few works have investigated lipids.7, 8, 9 Bioactive lipids are fundamental mediators of a number of biological processes,10, 11, 12 and the implication of lipids in cancer growth and diffusion have already been demonstrated.13 In this work, we aimed to study serum polar lipids in a prospective cohort of LARC patients before CRT (t0 group), including patients naïve to chemotherapy and radiation therapy. Samples were also collected during CRT (t14 and t28 days), in the effort to correlate the global lipid signature to response to treatment.
Methods
See Appendix E1, available as supplementary material online only at www.practicalradon.org.
Results
Lipidomics biomarker discovery
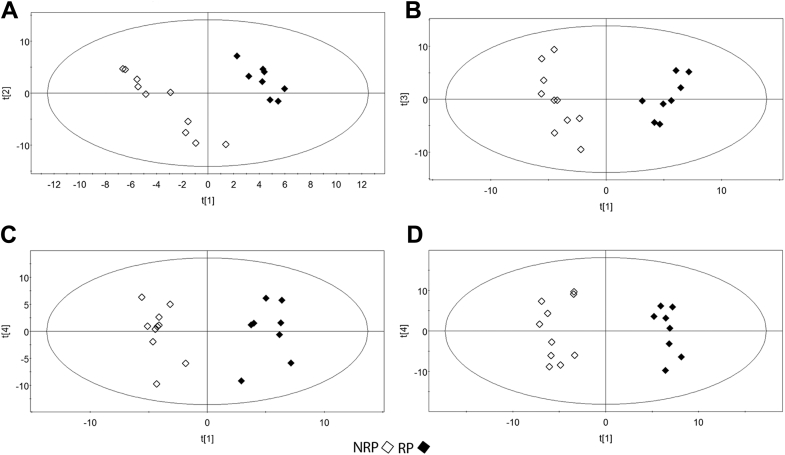
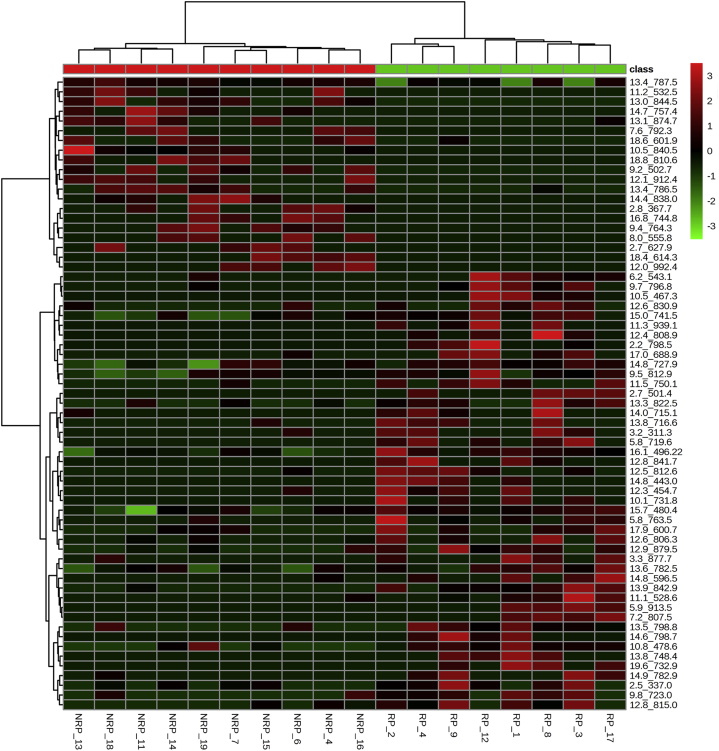
The serum from 18 patients with LARC (7 women, 11 men)—8 of whom were classified as responders (RPs) and 10 as not responders (NRPs) according to Mandard's tumor regression grading—treated with preoperative CRT was analyzed by liquid chromatography electrospray ionization tandem mass spectometry. Data were converted into a matrix containing m/z signals coupled with retention time as variables and the patient codes as observations. This dataset was reduced by considering only variables present in at least 50% of patients. Figure 1 shows the lipid classes (including lyso forms) screened. The studied lipids were sphingomyelins (SMs) and phosphatidylcholines (PCs; Fig 1A), phosphatidylethanolamine (Fig 1B), phosphatidylglycerols (Fig 1C), and phosphatidylserines (Fig 1D). Each lipid class screened was reported. In Fig 1A, the score plot of phosphatidylcholine/SM phospholipids is shown, whereas Fig 1B shows the score plot of the phosphatidylethanolamine class; Fig 1C shows the phosphatidylglycerol lipids; and the phosphatidylserine class is reported in Fig1D. The resulting PLS-DA models are reported as score scatter plots in Fig 1, showing clear separation between RP and NRP before treatment. The lipids identified as variable important for the projection (VIP >1) were confirmed through a univariate test. At t0, 65 lipids were identified as significant, with the criteria of VIP >1.5 and P < .05 in the univariate test, depicted in Fig 2 as a heat map. The heat map provides an overview of the different lipid signals (reported as a combination of the retention time and mass/charge [m/z]) and their relative intensity, in terms of overexpression (in red) or underexpression (in green), in RP versus NRP sera. These results help highlight the differential lipid patterns between RP and NRP sera and are summarized in Table 1.
Figure 1.
Partial least squares discriminant analysis score plots based on the lipidomics data. Responders (RPs) (represented as full diamonds) and not responders (NRPs; represented as open diamonds) before treatment (t0). The panels show partial least squares discriminant analysis score plots for the analyzed lipids, in particular the phosphatidylcholine/sphingomyelin class (A), phosphatidylethanolamine class (B), phosphatidylglycerol class (C), and phosphatidylserine class (D).
Figure 2.
Heat map showing the relative intensity of the 65 differential serum lipids (listed on the right) of each sample (listed at the bottom) before treatment (t0). Samples are divided in 2 groups: RP and NRP. Lipid levels are indicated by a color code: high (red) and low (green). See Fig 1 for abbreviations.
Table 1.
Significant lipids obtained from statistical analysis (VIP >1.5; P < .05) in RPs and NRPs at the t0 time point
| RT_m/z | VIP | NRPs |
RPs |
t test value | |||
|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | ||||
| PCs/SMs | 14.79_727.86 | 1.86 | 18.03 | 6.28 | 23.84 | 3.91 | 0.037 |
| 16.14_495.7 | 2.59 | 9.88 | 1.53 | 13.49 | 2.42 | 0.001 | |
| 15.72_480.42 | 2.27 | 1.46 | 0.62 | 2.24 | 0.39 | 0.008 | |
| 13.44_787.52 | 2.13 | 333.95 | 56.02 | 183.28 | 155.44 | 0.011 | |
| 13.51_798.84 | 1.96 | 2.21 | 4.71 | 8.47 | 6.02 | 0.025 | |
| 13.86_842.90 | 1.71 | 0.08 | 0.25 | 1.88 | 1.26 | 0.0001 | |
| 12.57_830.92 | 1.70 | 0.22 | 0.48 | 1.01 | 0.93 | 0.034 | |
| 12.58_806.35 | 1.87 | 0.25 | 0.43 | 1.53 | 1.78 | 0.042 | |
| 12.51_812.58 | 2.21 | 0.02 | 0.07 | 0.29 | 0.26 | 0.008 | |
| 14.75_757.37 | 1.83 | 1.72 | 2.14 | 0.00 | 0.00 | 0.038 | |
| 14.91_782.88 | 1.93 | 0.11 | 0.36 | 1.53 | 1.78 | 0.025 | |
| 14.04_715.12 | 1.89 | 0.04 | 0.14 | 0.40 | 0.50 | 0.047 | |
| PEs | 9.53_812.96 | 1.92 | 22.90 | 15.93 | 41.24 | 20.47 | 0.048 |
| 10.80_478.63 | 1.78 | 4.40 | 8.72 | 15.31 | 7.55 | 0.013 | |
| 9.83_723.00 | 1.78 | 3.15 | 6.77 | 12.80 | 9.44 | 0.023 | |
| 11.08_528.61 | 2.08 | 1.49 | 4.71 | 20.81 | 25.01 | 0.028 | |
| 11.54_750.09 | 2.46 | 0.63 | 2.01 | 7.78 | 7.04 | 0.007 | |
| 9.67_796.81 | 2.23 | 0.93 | 2.96 | 17.07 | 19.23 | 0.018 | |
| 9.23_502.71 | 2.39 | 7.47 | 7.17 | 0.00 | 0.00 | 0.010 | |
| 11.18_532.48 | 2.05 | 7.55 | 9.03 | 0.00 | 0.00 | 0.032 | |
| 12.35_454.71 | 1.96 | 0.73 | 2.31 | 6.06 | 7.27 | 0.043 | |
| 9.43_764.26 | 2.15 | 6.60 | 7.41 | 0.00 | 0.00 | 0.024 | |
| 8.01_555.83 | 1.92 | 3.24 | 4.22 | 0.00 | 0.00 | 0.046 | |
| 10.10_731.77 | 2.21 | 0.00 | 0.00 | 7.10 | 8.62 | 0.018 | |
| 7.60_792.30 | 1.91 | 3.81 | 4.98 | 0.00 | 0.00 | 0.047 | |
| 11.27_939.11 | 2.16 | 0.00 | 0.00 | 3.70 | 4.67 | 0.023 | |
| 12.36_808.91 | 1.98 | 0.00 | 0.00 | 3.24 | 4.61 | 0.039 | |
| PGs | 2.55_337.05 | 2.11 | 7.66 | 10.13 | 34.40 | 31.77 | 0.023 |
| 6.22_543.15 | 2.07 | 1.60 | 5.07 | 15.31 | 16.87 | 0.026 | |
| 5.77_763.48 | 1.86 | 2.73 | 8.65 | 22.98 | 28.48 | 0.048 | |
| 3.16_311.30 | 2.01 | 1.15 | 3.66 | 9.54 | 10.76 | 0.034 | |
| 12.14_912.39 | 2.14 | 9.69 | 10.56 | 0.00 | 0.00 | 0.020 | |
| 2.79_367.71 | 2.04 | 9.12 | 10.56 | 0.00 | 0.00 | 0.027 | |
| 5.83_719.64 | 2.54 | 0.00 | 0.00 | 13.05 | 12.30 | 0.004 | |
| 3.34_877.71 | 1.92 | 1.93 | 6.11 | 14.62 | 16.73 | 0.040 | |
| 5.96_913.52 | 2.33 | 0.00 | 0.00 | 9.74 | 10.60 | 0.010 | |
| 2.73_627.94 | 1.70 | 7.51 | 9.94 | 0.00 | 0.00 | 0.050 | |
| 2.25_798.51 | 2.02 | 0.00 | 0.00 | 20.04 | 28.36 | 0.039 | |
| 2.71_501.43 | 2.34 | 0.00 | 0.00 | 8.52 | 9.19 | 0.009 | |
| 7.16_807.46 | 2.32 | 0.00 | 0.00 | 6.22 | 6.78 | 0.010 | |
| PSs | 13.60_782.52 | 2.35 | 48.23 | 37.83 | 104.02 | 52.04 | 0.018 |
| 15.05_741.50 | 2.44 | 11.11 | 9.65 | 24.79 | 11.20 | 0.013 | |
| 12.92_879.50 | 2.45 | 3.65 | 7.03 | 18.01 | 14.25 | 0.013 | |
| 12.84_815.03 | 2.49 | 2.95 | 5.18 | 18.46 | 16.20 | 0.011 | |
| 13.26_822.49 | 2.43 | 4.19 | 7.38 | 20.42 | 16.66 | 0.014 | |
| 17.99_600.69 | 2.13 | 2.40 | 3.97 | 10.04 | 9.65 | 0.036 | |
| 10.48_840.46 | 2.08 | 7.75 | 9.91 | 0.00 | 0.00 | 0.043 | |
| 13.03_844.46 | 2.78 | 12.36 | 10.11 | 0.00 | 0.00 | 0.003 | |
| 13.45_786.54 | 2.50 | 11.73 | 10.59 | 0.71 | 2.03 | 0.011 | |
| 18.56_601.86 | 2.11 | 4.04 | 4.40 | 0.39 | 1.12 | 0.038 | |
| 13.09_874.69 | 2.04 | 7.73 | 8.80 | 0.76 | 2.16 | 0.045 | |
| 14.40_838.03 | 2.04 | 10.66 | 13.79 | 0.00 | 0.00 | 0.045 | |
| 12.85_841.74 | 2.12 | 0.81 | 2.56 | 8.56 | 10.51 | 0.038 | |
| 17.02_688.96 | 2.23 | 0.56 | 1.79 | 7.07 | 8.23 | 0.026 | |
| 14.85_596.55 | 2.26 | 0.55 | 1.74 | 9.20 | 10.95 | 0.025 | |
| 13.81_716.64 | 2.22 | 0.78 | 2.46 | 6.97 | 7.66 | 0.028 | |
| 16.79_744.77 | 2.01 | 2.48 | 3.29 | 0.00 | 0.00 | 0.050 | |
| 13.76_748.40 | 2.47 | 0.00 | 0.00 | 5.36 | 5.99 | 0.012 | |
| 10.50_467.35 | 2.23 | 0.00 | 0.00 | 4.43 | 5.76 | 0.026 | |
| 14.58_798.73 | 2.29 | 0.00 | 0.00 | 3.66 | 4.59 | 0.022 | |
| 18.36_614.29 | 2.08 | 2.34 | 3.07 | 0.00 | 0.00 | 0.048 | |
| 18.81_810.59 | 2.03 | 2.71 | 3.58 | 0.00 | 0.00 | 0.049 | |
| 14.83_443.05 | 2.45 | 0.00 | 0.00 | 2.89 | 3.28 | 0.013 | |
| 19.61_732.99 | 2.43 | 0.00 | 0.00 | 3.20 | 3.68 | 0.014 | |
| 12.03_992.42 | 2.03 | 2.41 | 3.17 | 0.00 | 0.00 | 0.048 | |
Bold type indicates confirmed biomarkers. Lipids are reported as a combination of RT_m/z.
NRP, not responder; PC, phosphatidylcholine; RP, responder; RT_m/z, retention time and mass/charge; SM, sphingomyelin; SD, standard deviation; PE, phatidylethanolamine; PG, phosphatidylglycerol; PS, phosphatidylserine; VIP, variable important for the projection.
Biomarker confirmation
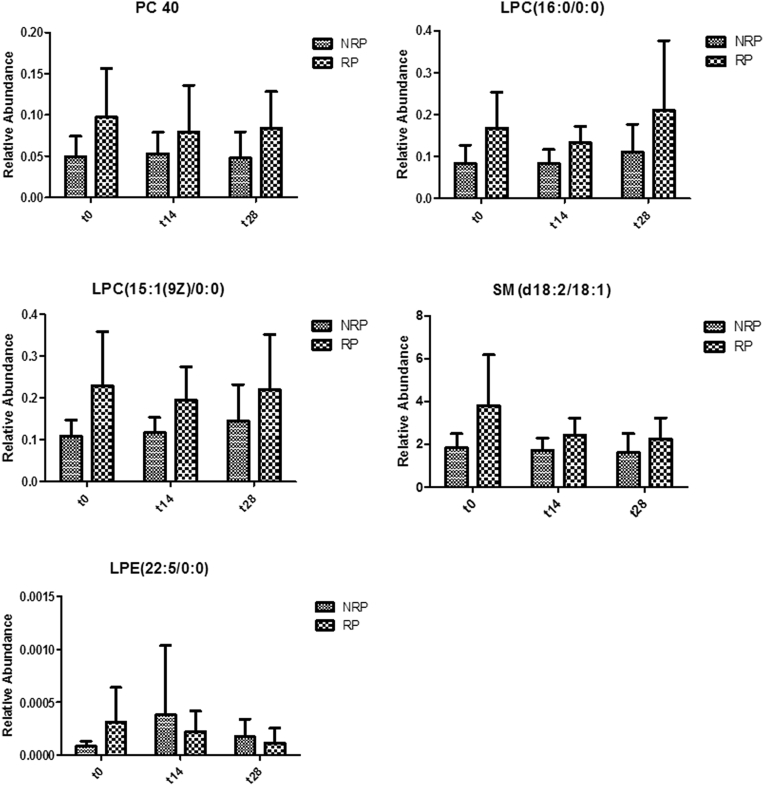
To further validate the reliability of the highlighted biomarkers, an independent validation analysis was performed through targeted liquid chromatograph tandem mass spectometry. Results confirmed the lower levels in NRP of 5 differentially expressed lipids (P < .05) that were identified as follows: SM (d18:2/18:1) at m/z = 727.86; lysophosphatidylcholine (LPC;16:0/0:0) at m/z = 496.22; LPC (15:1(9z)/0:0) at m/z = 480.42; lysophosphatidylethanolamine (LPE;22:5/0:0) at m/z = 528.6; and PC (40:2) at m/z = 842.90. These 5 lipids were regarded as the more reliable predictive biomarkers and quantified at 14 and 28 days to evaluate their prognostic value. As shown in Fig 3, PC (40:2), the 2 LPCs, and SM confirmed their lower levels in NRP with respect to RP during the entire therapy (P < .05). Conversely, the levels of LPE varied during CRT. No significant difference between males and females was found in the highlighted biomarkers (data not shown).
Figure 3.
Histograms reporting the relative abundance of potential biomarkers in RPs and NRPs during chemoradiation therapy (CRT). Relative abundances of phosphatidylcholine (PC; 40:2), lysophosphatidylcholine (LPC;16:0/0:0), LPC (15:1 (9Z)/0:0), sphingomyelin (SM; d18:2/18:1), and lysophosphatidylethanolamine (LPE;22:5/0:0) are shown, respectively, before treatment (t0), during CRT (t14), and at the last therapy day (t28). See Fig 1 for abbreviations.
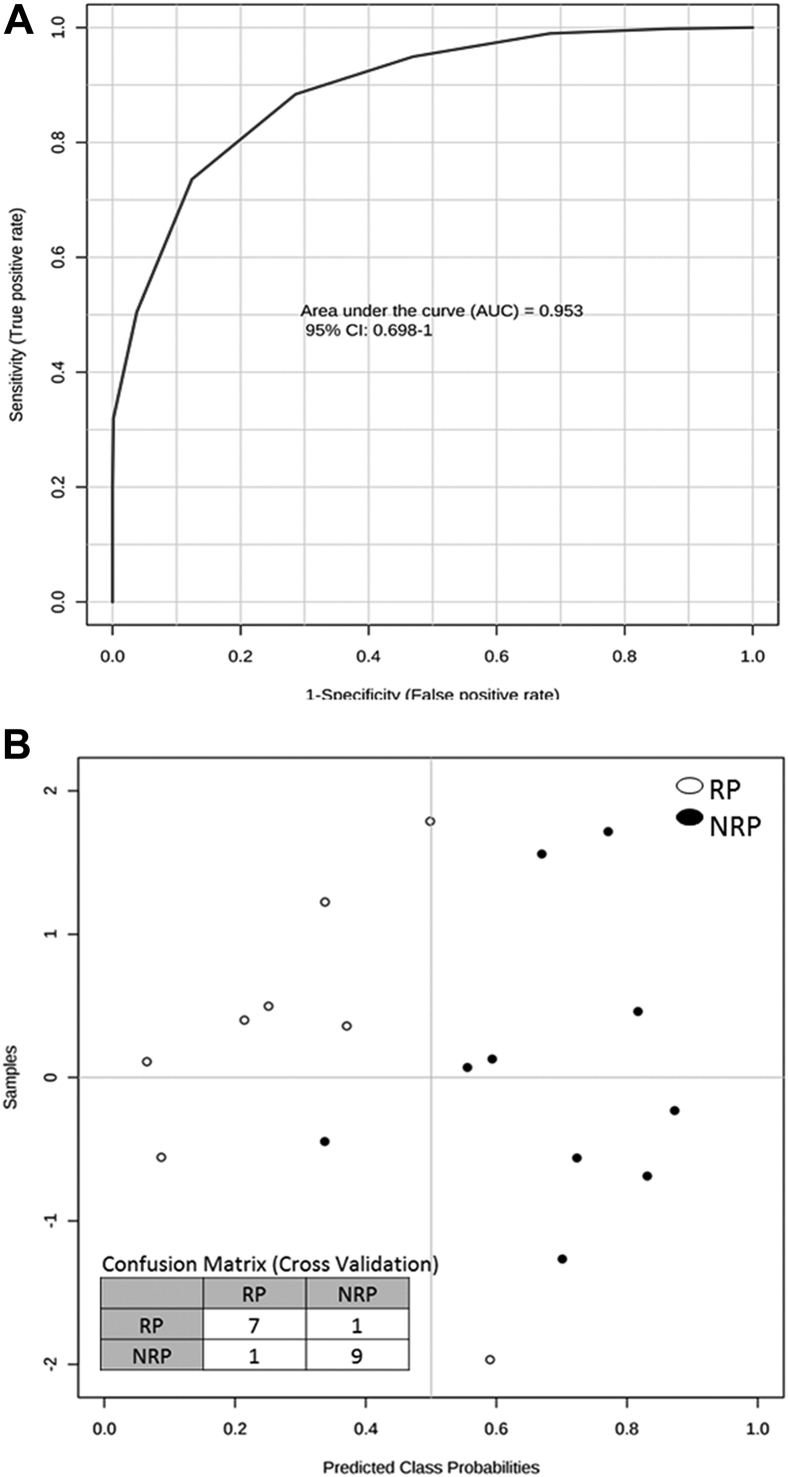
Predictive power of lipid biomarkers
Figure 4A shows the receiver operating characteristic curve generated combining the 5 validated lipids. The area under the curve is 0.95, showing good sensitivity and specificity in discriminating between RP and NRP. The 100 cross-validations performed show the predicted class probabilities of each sample, as reported in Fig 4B, underlying the good predictivity of the proposed model (P = .03) in suggesting patients who may better respond to therapy.
Figure 4.
Predictive power of 5 validated lipids at the t0 time point. (A) Receiver operating characteristic curve generated combining the 5 validated lipids; (B) predicted class probabilities (RP or NRP) of each sample across the 100 cross-validations and the related confusion matrix generated. See Fig 1 for abbreviations.
Discussion
Predictive response biomarkers to neoadjuvant CRT in LARC could personalize treatment strategy to improve response rate and survival outcomes. In this study, we focus on serum lipids to define a discriminatory profile able to predict CRT response in LARC. Despite the small sample size analyzed, our results indicate 5 lipids that drive the separation of RP and NRP. We found that LPE (22:5/0:0), SM (d18:2/18:1), LPC (16:0/0:0), LPC (15:1(9z)/0:0), and PC (40:2) are significantly lower in NRP at t0, whereas the LPE level significantly increases in NRP during CRT. The involvement of these lipids in radioresistance may be supported by the known correlation between human phosphatidylethanolamine-binding protein 4 (hPEBP4) and inhibition of apoptosis.14, 15, 16 Qiu et al have already demonstrated that hPEBP4 is a predictive marker of radioresistance in rectal cancer by activating Akt in a reactive oxygen species–dependent manner.17, 18
PC (40:2) is lower in NRP compared with RP before and during treatment, probably resulting from dysregulation of choline metabolism, a known metabolic hallmark associated with oncogenesis and cancer progression.19 Moreover, we highlighted low levels of LPCs in NRP, which is consistent with several studies that correlate higher blood LPC levels with reduced risk of cancer,18 thus suggesting that LPCs may represent a useful circulating biomarker for early detection of CRC.20 The low levels of SM in NRP may be due to the high activity of SM, resulting in high levels of ceramide. Even if ceramide is involved in cell-cycle arrest, apoptosis, and senescence in CRC cells,21, 22 its degradation product, sphingosine1P, induces cell proliferation and angiogenesis and triggers cell motility.23 Bearing in mind the limitations of this pilot study, these results provide novel insights regarding lipid metabolism in the modulation of CRT response in LARC patients. If confirmed in a more extensive clinical cohort, these biomarkers could represent a useful tool for predicting outcome as part of efforts to personalize therapy.
Footnotes
Conflicts of interest: None.
Supplementary material for this article (http://dx.doi.org/10.1016/j.adro.2016.12.005) can be found at www.practicalradonc.org.
Supplementary data
References
- 1.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Edge S.B., Compton C.C. The American Joint Committee on Cancer: The 7th edition of the AJCC Cancer Staging Manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 3.Glimelius B., Tiret E., Cervantes A., Arnold D., Group E.G.W. Rectal cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2013;24(Suppl 6):vi81–vi88. doi: 10.1093/annonc/mdt240. [DOI] [PubMed] [Google Scholar]
- 4.Valentini V., Glimelius B., Haustermans K. EURECCA consensus conference highlights about rectal cancer clinical management: The radiation oncologist's expert review. Radiother Oncol. 2014;110:195–198. doi: 10.1016/j.radonc.2013.10.024. [DOI] [PubMed] [Google Scholar]
- 5.Conde-Muino R., Cuadros M., Zambudio N., Segura-Jimenez I., Cano C., Palma P. Predictive biomarkers to chemoradiation in locally advanced rectal cancer. BioMed Res Int. 2015;2015:921435. doi: 10.1155/2015/921435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pieragostino D., Agnifili L., Fasanella V. Shotgun proteomics reveals specific modulated protein patterns in tears of patients with primary open angle glaucoma naive to therapy. Molecular bioSystems. 2013;9:1108–1116. doi: 10.1039/c3mb25463a. [DOI] [PubMed] [Google Scholar]
- 7.Kuremsky J.G., Tepper J.E., McLeod H.L. Biomarkers for response to neoadjuvant chemoradiation for rectal cancer. Int J Radiat Oncol Biol Phys. 2009;74:673–688. doi: 10.1016/j.ijrobp.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 8.Bathen T.F., Engan T., Krane J., Axelson D. Analysis and classification of proton NMR spectra of lipoprotein fractions from healthy volunteers and patients with cancer or CHD. Anticancer Res. 2000;20:2393–2408. [PubMed] [Google Scholar]
- 9.Perrotti F., Rosa C., Cicalini I. Advances in lipidomics for cancer biomarkers discovery. Int J Mol Sci. 2016;17:1992. doi: 10.3390/ijms17121992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brizuela L., Martin C., Jeannot P. Osteoblast-derived sphingosine 1-phosphate to induce proliferation and confer resistance to therapeutics to bone metastasis-derived prostate cancer cells. Mole Oncol. 2014;8:1181–1195. doi: 10.1016/j.molonc.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schneider G., Bryndza E., Abdel-Latif A. Bioactive lipids s1p and c1p are prometastatic factors in human rhabdomyosarcoma, and their tissue levels increase in response to radio/chemotherapy. Mole Cancer Res. 2013;11:793–807. doi: 10.1158/1541-7786.MCR-12-0600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schneider G., Sellers Z.P., Abdel-Latif A., Morris A.J., Ratajczak M.Z. Bioactive lipids, LPC and LPA, are novel prometastatic factors and their tissue levels increase in response to radio/chemotherapy. Mole Cancer Res. 2014;12:1560–1573. doi: 10.1158/1541-7786.MCR-14-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Jaarsveld M.T.M., Houthuijzen J.M., Voest E.E. Molecular mechanisms of target recognition by lipid gpcrs: Relevance for cancer. Oncogene. 2014;12:1560–1573. doi: 10.1038/onc.2015.467. [DOI] [PubMed] [Google Scholar]
- 14.Wang X., Li N., Li H. Silencing of human phosphatidylethanolamine-binding protein 4 sensitizes breast cancer cells to tumor necrosis factor-alpha-induced apoptosis and cell growth arrest. Clin Cancer Res. 2005;11:7545–7553. doi: 10.1158/1078-0432.CCR-05-0879. [DOI] [PubMed] [Google Scholar]
- 15.Li H., Wang X., Li N., Qiu J., Zhang Y., Cao X. Hpebp4 resists trail-induced apoptosis of human prostate cancer cells by activating akt and deactivating erk1/2 pathways. TJ Biol Chemi. 2007;282:4943–4950. doi: 10.1074/jbc.M609494200. [DOI] [PubMed] [Google Scholar]
- 16.Li P., Wang X., Li N. Anti-apoptotic hpebp4 silencing promotes trail-induced apoptosis of human ovarian cancer cells by activating ERK and JNK pathways. Int J Mole Med. 2006;18:505–510. [PubMed] [Google Scholar]
- 17.Qiu J., Yang G., Lin A., Shen Z., Wang D., Ding L. Human phosphatidylethanolamine-binding protein 4 promoted the radioresistance of human rectal cancer by activating akt in an ROS-dependent way. PloS One. 2014;9:e90062. doi: 10.1371/journal.pone.0090062. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Qiu J., Yang G., Shen Z., Xie Y., Wang L. Hpebp4 as a predictive marker for the pathological response of rectal cancer to preoperative radiotherapy. Int J Colorect Ds. 2013;28:241–246. doi: 10.1007/s00384-012-1534-3. [DOI] [PubMed] [Google Scholar]
- 19.Glunde K., Bhujwalla Z.M., Ronen S.M. Choline metabolism in malignant transformation. Nat Rev Cancer. 2011;11:835–848. doi: 10.1038/nrc3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Z., Xiao Y., Elson P. Plasma lysophosphatidylcholine levels: Potential biomarkers for colorectal cancer. J Clini Oncol. 2007;25:2696–2701. doi: 10.1200/JCO.2006.08.5571. [DOI] [PubMed] [Google Scholar]
- 21.Adada M., Luberto C., Canals D. Inhibitors of the sphingomyelin cycle: Sphingomyelin synthases and sphingomyelinases. Chem Phys Lipids. 2016;197:49–59. doi: 10.1016/j.chemphyslip.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 22.Ogretmen B., Hannun Y.A. Biologically active sphingolipids in cancer pathogenesis and treatment. Nat Rev Cancer. 2004;4:604–616. doi: 10.1038/nrc1411. [DOI] [PubMed] [Google Scholar]
- 23.Hannun Y.A., Obeid L.M. Many ceramides. J Biol Chem. 2011;286:27855–27862. doi: 10.1074/jbc.R111.254359. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.