Abstract

Through mutant selection on agar containing pyrazinoic acid (POA), the bioactive form of the prodrug pyrazinamide (PZA), we recently showed that missense mutations in the aspartate decarboxylase PanD and the unfoldase ClpC1, and loss-of-function mutation of polyketide synthases Mas and PpsA-E involved in phthiocerol dimycocerosate synthesis, cause resistance to POA and PZA in Mycobacterium tuberculosis. Here we first asked whether these in vitro-selected POA/PZA-resistant mutants are attenuated in vivo, to potentially explain the lack of evidence of these mutations among PZA-resistant clinical isolates. Infection of mice with panD, clpC1, and mas/ppsA-E mutants showed that whereas growth of clpC1 and mas/ppsA-E mutants was attenuated, the panD mutant grew as well as the wild-type. To determine whether these resistance mechanisms can emerge within the host, mice infected with wild-type M. tuberculosis were treated with POA, and POA-resistant colonies were confirmed for PZA and POA resistance. Genome sequencing revealed that 82 and 18% of the strains contained missense mutations in panD and clpC1, respectively. Consistent with their lower fitness and POA resistance level, independent mas/ppsA-E mutants were not found. In conclusion, we show that the POA/PZA resistance mechanisms due to panD and clpC1 missense mutations are recapitulated in vivo. Whereas the representative clpC1 mutant was attenuated for growth in the mouse infection model, providing a possible explanation for their absence among clinical isolates, the growth kinetics of the representative panD mutant was unaffected. Why POA/PZA resistance-conferring panD mutations are observed in POA-treated mice but not yet among clinical strains isolated from PZA-treated patients remains to be determined.
Keywords: tuberculosis, pyrazinamide, pyrazinoic acid, resistance, in vivo
Treatment of tuberculosis (TB) involves a combination of the four first-line drugs: isoniazid, rifampicin, ethambutol, and pyrazinamide (PZA). PZA plays a critical role in this regimen by sterilizing lesions and preventing disease relapse. Inclusion of this drug in the treatment regimen in the 1980s resulted in shortening the duration of therapy from 9 to 6 months.1 Since then, most new drug combinations in development include PZA.2 However, its mechanism of action remains controversial.3 Understanding how this critical drug works against TB may reveal new rational approaches for shortening TB treatment and preventing disease relapse.
PZA is a prodrug that requires conversion to its bioactive form, pyrazinoic acid (POA). Prodrug conversion is carried out by the bacterial pyrazinamidase PncA. The inactivation of PncA causes PZA resistance in vitro,4 in vivo,5 and in clinical isolates.6 Host enzymes also metabolize PZA to POA, which could contribute to its activity in vivo.7,8
Recently, we9,10 and others11 isolated spontaneous POA-resistant Mycobacterium tuberculosis mutants in vitro by plating bacteria on agar containing POA, thereby avoiding selection of mutations in pncA. Missense mutations in the aspartate decarboxylase panD(9,11,12) and in the unfoldase/ATPase clpC1(10) were found to cause resistance to PZA and POA. Loss-of-function mutations of the phthiocerol dimycocerosate (PDIM) virulence factor producing polyketide synthases Mas and PpsA-E caused a lower level of resistance to PZA and POA.9
PanD is required for coenzyme A biosynthesis, and metabolic pathway analyses suggested PanD may be a direct target for POA; that is, inhibition of coenzyme A biosynthesis appears to be a mechanism of action of PZA.9,11 Whether ClpC1, the unfoldase component of the caseinolytic protease complex, is a direct target of POA or whether missense mutations in this gene cause resistance by an indirect mechanism remains to be determined.10 It also remains to be determined if the Mas-PpsA-E-based resistance is direct or indirect.9
An unsolved puzzle associated with PZA is the lack of clear evidence that mutations in genes other than pncA are associated with PZA resistance in clinical isolates.12−15 Our finding that loss of PDIM virulence factor synthesis causes POA resistance abides with the hypothesis that spontaneous drug-resistant mutants that suffer a significant fitness cost in vivo may not survive or compete well with other drug-resistant mutants and hence cannot be identified in sputum isolates.16 Here we first asked whether the in vitro isolated POA/PZA-resistant panD, clpC1, and mas-ppsA-E mutants are attenuated in vivo, therefore providing a possible explanation for the observation that these mutations are not prominent in clinical isolates. Then we asked whether the POA/PZA resistance mechanisms identified in vitro, that is, via selection of mutants on POA-containing agar, represent in vitro artifacts or whether these mechanisms can be recapitulated in vivo, via selection of mutants in POA-treated mice.
Results
In Vitro Selected POA/PZA-Resistant M. tuberculosis Strains Harboring Mutations in clpC1 and mas/ppsA-E Show Attenuated Growth in Vivo
The current lack of evidence associating mutations in panD, clpC1, and mas/ppsA-E with PZA resistance in clinical isolates could be explained if such mutants suffer a fitness cost in vivo.16 To address this hypothesis, we selected three isogenic POA/PZA-resistant M. tuberculosis mutants isolated previously in vitro (POAR 1 [panD1], POAR 18 [clpC1-7], POAR 7 [ppsC1]), representing the three different POA/PZA resistance-conferring genotypes with mutations in panD, clpC1, and mas/ppsA-E, respectively,9,10 and carried out in vivo growth experiments in comparison with wild-type M. tuberculosis. Low-dose aerosol infection of BALB/c mice was performed, and colony-forming units (CFU) were quantified in lung and spleen homogenates obtained 2, 4, and 6 weeks postinfection. At 6 weeks postinfection, we confirmed that the POA-resistant strains POAR 1 [panD1] and POAR 18 [clpC1-7] had retained their relatively higher level resistance by plating lung homogenates on 7H11 agar containing 3 mM POA.
PDIMs, complex lipids residing in the cell wall of mycobacteria, have been well established as virulence factors, whereby loss of these lipids results in attenuation of M. tuberculosis in different animal models.17−19 As expected, POAR 7 [ppsC1] containing a frameshift mutation in ppsC:Ins2674C as determined by whole genome sequencing in,9 was significantly attenuated in both lungs and spleen as compared to the virulent wild-type strain (Figure 1), particularly during the early phase of infection in the lungs (Figure 1A). Interestingly, POAR 18 [clpC1-7] containing a mutation in clpC1:A625G/Lys209Glu, which has a higher level of POA resistance in vitro, mimicked the attenuation phenotype of POAR 7 [ppsC1] in both lungs and spleen (Figure 1), indicating that the mutation in clpC1 causes in vivo fitness loss similar to loss of PDIMs over the first 6 weeks postinfection. In contrast, POAR 1 [panD1] containing a C-terminal mutation in panD:Δ380A displayed no defect in in vivo growth (Figure 1). H&E staining of one lung from four mice in each infection group revealed similar pathology in those infected with either wild-type or panD mutant strains throughout the course of infection. At 2 weeks postinfection, similar and diffuse lymphocytic infiltration was seen across all infection groups (not shown). At 4 weeks postinfection, mice infected with the clpC1 and ppsC mutants had fewer visible granulomatous lesions and mostly perivascular lymphocytic infiltration, compared to the wild-type and panD mutant groups (Figure 2A). At 6 weeks postinfection, clpC1-infected mice had a lower number of granulomas ≥500 μm in diameter (Figure 2B). Ziehl–Neelsen staining showed clusters of acid-fast bacilli mostly in areas rich in foamy macrophages (Figure 2C–J). The abundance of bacilli in these clusters was clearly higher in mice infected with wild-type and panD mutant strains compared to those infected with ppsC and clpC1 mutants (Figure 2G–J), which has been quantified in the Supporting Information, Table S1.
Figure 1.
Growth in mice of wild-type M. tuberculosis H37Rv (ATCC 27294) and isogenic POA-resistant strains selected in vitro. Growth of POAR 1 [panD1], POAR 7 [ppsC1], and POAR 18 [clpC1-7] in the (A) lungs and (B) spleen of BALB/c mice is shown. Values represent the mean ± standard deviation of CFU counts obtained from four mice per group.
Figure 2.
Comparative lung pathology induced by infection with POA-resistant mutants versus wild-type M. tuberculosis H37Rv (ATCC 27294). (A, B) Hematoxylin and eosin (H&E) staining of one lung from groups of four mice infected with H37Rv wild-type, POAR 1 [panD1], POAR 7 [ppsC1], and POAR 18 [clpC1-7] at 4 (A) and 6 (B) weeks postinfection. Blue scale bars for each group represent 5 mm. (C–J) High-power magnification images of representative individual granulomas in the corresponding infection groups at 6 weeks, stained by H&E (C–F) and Ziehl–Neelsen (G–J). (C–F) Typical granulomas contained lymphocyte aggregates with interspersed epithelioid histocytes and foamy macrophages on the outer rim of the granulomatous lesion. Black scale bars on top right for each group represent 100 μm. (G–J) Clusters of acid-fast bacilli were found mostly in areas rich in foamy macrophages (insets).
These results show that the previously in vitro selected POA/PZA-resistant strains harboring mutations in clpC1 and mas/ppsA-E display attenuated growth in vivo, hence providing a possible explanation for their apparent absence in PZA-resistant clinical isolates. In contrast, the POA/PZA-resistant panD mutant displayed in vivo fitness indistinguishable from that of wild-type.
In Vivo Selected POA/PZA-Resistant M. tuberculosis Strains Harbor Missense Mutations in panD and clpC1
All reported panD, clpC1, and mas/ppsA-E POA/PZA resistance-conferring mutants have been selected on POA/PZA-containing agar,9−12 that is, under in vitro culture conditions on rich 7H10/7H11 medium, and might hence represent in vitro artifacts. We therefore asked whether these resistance mechanisms can be recapitulated in selection experiments performed in vivo. BALB/c and C3HeB/FeJ mice were infected with M. tuberculosis wild type and treated with a range of POA and PZA doses for 8 weeks, and 28 apparent POA-resistant colonies were isolated from lung homogenates on POA-containing agar as described previously.8 The details of the treatment and resistance mutation frequencies are listed in the Supporting Information, Table S2.8 To confirm POA resistance and carry out colony purification, the primary M. tuberculosis isolates were restreaked on agar containing 300 mg/L POA. As expected, all 28 suspected POA-resistant isolates grew, whereas the parental wild-type strain did not.
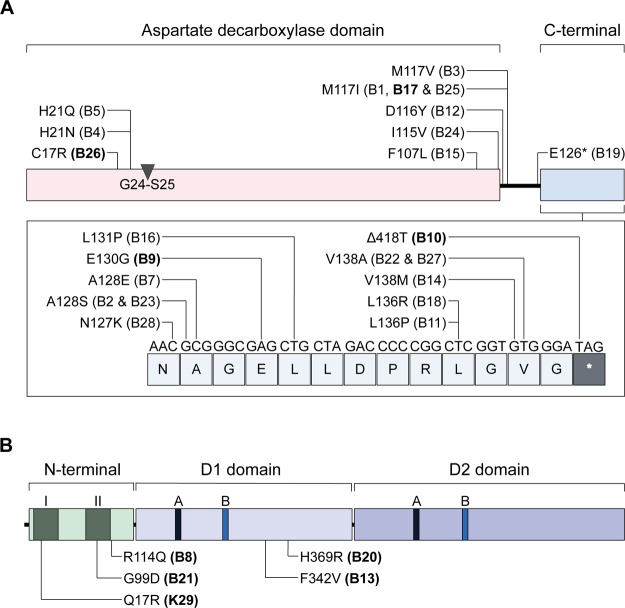
Because the in vivo-isolated strains displayed resistance to POA at a concentration higher than the MIC of in vitro-isolated low-level POA-resistant strains with loss of function mutations in mas/ppsA-E,9 we expected them to harbor a resistance mechanism causing a higher level of POA resistance, such as mutations in panD or clpC1 (with or without additional comutations in mas/ppsA-E).9,10 On the basis of our observation that POA resistance-conferring clpC1 mutations are attenuated in vivo, whereas panD mutations did not cause a growth defect in vivo, we hypothesized that panD mutations—if they do occur in vivo—may be found frequently in vivo. Targeted PCR sequencing of panD revealed that 23 (82%) of the 28 POA-resistant strains isolated after POA treatment of mice contained panD mutations observed in multiple selection experiments in vitro9,11,12 (Supporting Information, Table S2). The parental strain (as expected) and the five remaining POA-resistant M. tuberculosis strains exhibited wild-type panD genes (Supporting Information, Table S2). Similar to the previously in vitro-selected panD mutations, the in vivo-selected panD mutations mainly cluster in the C-terminal region of PanD (outside the aspartate decarboxylase domain), with the exception of three strains (B4, B5, and B26) containing mutations near the autocleavage site (Gly24-Ser259) and two strains (B15 and B24) with mutations at the C-terminal end of the aspartate decarboxylase domain (Figure 3A).9,20,21
Figure 3.
Location of amino acid sequence polymorphisms in (A) PanD and (B) ClpC1 of POA-resistant M. tuberculosis strains isolated from POA-treated mice. (A) Domain organization of PanD is shown as described in ref (20). The conserved autocleavage site between residues 24 (Gly) and 25 (Ser) in the aspartate decarboxylase domain is indicated. (B) Domain organization of ClpC1 is shown as described in ref (35). Within the N-terminal domain, two repeats are labeled I and II. A and B in the D1 and D2 domains indicate Walker A and Walker B motifs, respectively. The nine POA-resistant M. tuberculosis strains subjected to whole genome sequencing and described in Table 1 are labeled in bold.
Whole genome sequencing was carried out to identify the genetic basis of POA resistance in the five remaining in vivo-selected strains with wild-type panD genes (B8, B13, B20, B21, and K29). Whole genome sequencing was also performed for four strains containing various mutations in panD (B9, B10, B17, and B26) to detect potential common background mutations that may be contributing to POA resistance. As shown in Table 1, all five panD wild-type POA-resistant strains contained different missense mutations in clpC1. The mutations in clpC1 were confirmed by targeted PCR sequencing. Similar to the previously in vitro-selected clpC1 mutations, the in vivo-selected clpC1 mutations are located in the N-terminal domain and in the D1 domain, outside the Walker A and Walker B motifs (Figure 3B).10
Table 1. Sequence Polymorphisms and Susceptibility to POA and PZA of POA-Resistant M. tuberculosis Strains Subjected to Whole Genome Sequencing.
| mutations |
|||||
|---|---|---|---|---|---|
| M. tuberculosis H37Rv strain | panDa | clpC1a | other genes | POA broth MIC50b (mM) | PZA susceptibility result (S/R)c |
| H37Rv parent | 1.5 | S | |||
| POA B8 | G341A/Arg114Gln | 5.5 | R | ||
| POA B9 | A389G/Glu130Gly | Rv0980c (PE-PGRS 18): C129A/His43Gln | 6.0 | R | |
| Rv0980c (PE-PGRS 18): G217C/Glu73Gln | |||||
| Rv0980c (PE-PGRS 18): A1088C/Asn363Thr | |||||
| Rv2615c (PE-PGRS 45): G247A/Gly83Ser | |||||
| POA B10 | Δ418T | Rv1230c: T1078G/Cys360Gly | 6.0 | R | |
| POA B13 | T1024G/Phe342Val | ppsA: Ins2101C | 6.5 | R | |
| Rv1230c: T1078G/Cys360Gly | |||||
| trpA: A104G/Tyr35Cys | |||||
| cycA: A1297G/Thr433Ala | |||||
| secA2: G487T/Val163Leu | |||||
| POA B17 | G351T/Met117Ile | Rv0007: G451A/Ala151Thr | 6.0 | R | |
| Rv3645: G502C/Ala168Pro | |||||
| POA B20 | A1106G/His369Arg | Rv2061c: C39G/Tyr13* | 6.0 | R | |
| Rv2402: C1135A/Arg379Ser | |||||
| POA B21 | G296A/Gly99Asp | 6.0 | R | ||
| POA B26 | T49C/Cys17Arg | ltp1: C361A/Pro121Thr | 5.5 | R | |
| Rv3784: G965A/Gly322Asp | |||||
| POA K29 | A50G/Gln17Arg | pitA: Ins908G | 6.0 | R | |
| embA: A2695G/Thr899Ala | |||||
| POAR 1 [panD1]d | Δ380A | 6.0 | R | ||
| POAR 18 [clpC1-7]e | A625G/Lys209Glu | 6.0 | R | ||
Polymorphisms were identified by whole genome sequencing and verified by targeted sequencing as described in the text.
MIC50, POA concentration that inhibits 50% of growth compared to drug free control. Drug susceptibility tests were carried out three times independently, and mean values are shown.
BACTEC MGIT 960 test for susceptibility (S) or resistance (R) to 100 μg/mL PZA.
POAR 1 [panD1] was selected in vitro as described in ref (9).
POAR 18 [clpC1-7] was selected in vitro as described in ref (10).
To verify that these strains display resistance levels similar to our previously in vitro selected POA-resistant M. tuberculosis strains containing panD or clpC1 mutations, we performed MIC determinations for POA in broth as well as on agar in comparison with the susceptible parental H37Rv strain and representative in vitro isolated strains (POAR 1 [panD1] and POAR 18 [clpC1-7]). As shown in Table 1 and Supporting Information, Table S2 and Figure S1A, the strains selected in vivo displayed resistance to POA at levels similar to the in vitro-selected panD and clpC1 mutant strains, that is, about a 4-fold increase in broth MIC50 and at least a 4-fold increase in agar MIC values. It is interesting to note that one of the strains (POA B13) with a mutation in clpC1:T1024G/Phe342Val carries an additional frameshift mutation in ppsA:Ins2101C and that this double-mutant strain may display a marginally elevated level of POA resistance (Table 1).9 An additional polymorphism in Rv1230c (T1078G/Cys360Gly) was observed in two POA-resistant strains isolated from different mice. Whether this polymorphism is involved in POA resistance is questionable as it does not appear to result in an increase of resistance to POA. Moreover, this polymorphism has been observed occasionally in isolates obtained from mice treated with other drugs unrelated to PZA in independent unpublished experiments by some of the authors (R.T., J-P.L., and E.N.) who used the same parental strain. Therefore, we believe that the appearance of this polymorphism in the present experiment likely represents genetic heterogeneity in the parental strain. To confirm that POA resistance correlates with resistance to the PZA prodrug, we demonstrated that all 28 POA-resistant strains isolated from POA-treated mice were also resistant to PZA using the BACTEC MGIT 960 PZA susceptibility test as well as agar MIC determination (Table 1 and Supporting Information, Table S2). In contrast, MICs of two other first-line TB drugs, rifampicin and isoniazid, were similar in POA-resistant strains and the wild-type H37Rv strain (Supporting Information, Figure S1B,C), indicating that the described mutations in panD and clpC1 do not confer nonspecific antibiotic resistance.
Taken together, these results show that missense mutations in panD and clpC1 do occur under POA pressure in vivo and, hence, do not represent an in vitro artifact.
Selective Amplification of POA-Resistant Mutants in Vivo Occurs Only with High Systemic POA Exposures and Does Not Occur with a Standard Dose of PZA
PZA is converted to POA by the host as well as M. tuberculosis.7 Our previous mouse efficacy study showed that only POA doses producing systemic POA exposures higher than those observed in humans after administration of standard PZA doses produce bactericidal effects in mice and, even then, the magnitude of these effects is much lower than those observed with standard PZA doses.5,8 However, we did not previously examine the effect of POA dose on the selective amplification of POA-resistant mutants. Therefore, we reanalyzed the drug-susceptible and POA-resistant CFU counts from the previous experiment and found that only POA doses exhibiting bactericidal activity against the total, drug-susceptible population (i.e., daily doses of 450–900 mg/kg) resulted in selective amplification of POA-resistant mutants compared to untreated animals (mean proportion of POA-resistant CFU after 8 weeks of POA treatment: 1.2 × 10–6, 1.1 × 10–6, and 6.5 × 10–6 for mice receiving 0, 27.5–150, and 450–900 mg/kg of POA daily, respectively [p < 0.0001 for high-dose group vs other groups]) (Supporting Information, Figure S2). After 8 weeks of treatment, 27 (90%) of 30 BALB/c mice treated with a range of POA doses harbored POA-resistant mutants in the lungs, compared to 1 (20%) of 5 mice treated with PZA at 150 mg/kg and 4 (80%) of 5 untreated mice (p = 0.003) for POA versus PZA treatment. The low proportion of mice harboring POA-resistant bacteria after treatment with PZA at 150 mg/kg contrasts markedly with results of another previous study,5 in which selective amplification of PZA-resistant pncA mutants was observed after 8 weeks of treatment in five (100%) of five BALB/c mice receiving PZA at 100 mg/kg and in four (57%) of seven evaluable mice receiving 300–900 mg/kg/day. Thus, PZA treatment appears more likely to select for PZA-resistant pncA mutants than for panD and clpC1 mutants that are resistant to both PZA and POA, and POA treatment only selectively amplifies the latter mutants at systemic POA exposures (i.e., mean plasma AUC > 300 μg·h/mL) above those observed in humans after standard PZA doses.7,8
Discussion and Conclusion
PZA is a critical first-line TB drug owing to its treatment-shortening effects. Due to its remarkable properties, it is also a component of many novel drug combinations that are in development for treatment of both drug-susceptible and multidrug-resistant TB.2 This is accompanied, however, by an alarming prevalence of PZA resistance in >50% of MDR-TB isolates.22,23 The vast majority of PZA-resistant clinical isolates (70–97%) contain mutations in the pncA gene encoding the prodrug-activating pyrazinamidase,24 where a highly diverse and scattered range of mutations has been described.13−15 However, aside from evidence linking mutations in rpsA to PZA resistance,25 the basis for resistance in the minority of PZA-resistant clinical isolates with wild-type pncA sequence remains undefined.
Previous in vitro selection of M. tuberculosis mutants resistant to POA, the bioactive component of PZA, identified three different resistance mechanisms. A higher level of resistance to POA is caused by (i) missense mutations in panD,9,11 which encodes the aspartate decarboxylase involved in de novo pantothenate and coenzyme A biosynthesis, and (ii) missense mutations in clpC1, which encodes the unfoldase/ATPase of the caseinolytic protease complex.10 A lower level of POA resistance is caused by frameshift mutations in mas and ppsA-E, which encode polyketide synthases responsible for PDIM synthesis.9 However, to date, no significant association has been found between mutations in panD and PZA resistance in clinical isolates.26,27 Similarly, we did not find any clinical isolates in the Genome-wide M. tuberculosis Variation Database,28 containing the polymorphisms in clpC1 that we identified in POA-resistant M. tuberculosis strains selected in vitro.10 It has been speculated that acquisition of PZA resistance, including pncA mutations, results in a fitness cost that limits person-to-person transmission.16 We further hypothesized that the PZA/POA resistance mechanisms identified in vitro compromise the fitness of M. tuberculosis and limit the selective amplification of these mutants in vivo. If this is true, it would provide one explanation for the current lack of evidence for such mutations among PZA-resistant clinical isolates. To test this hypothesis, we compared the growth of representative in vitro-selected panD, clpC1, and ppsC mutants with that of the virulent wild-type parental strain in BALB/c mice.
The PDIM-deficient strain M. tuberculosis POAR 7 [ppsC1] displayed a similar pattern of growth attenuation in lungs and spleens of BALB/c mice infected via aerosol as described previously resulting from the loss of PDIMs.29,30 PDIMs are complex lipid virulence factors of M. tuberculosis and loss of these lipids on the bacterial cell surface has been known to cause attenuation in mice18,19 and guinea pigs.17 This is attributed to the critical role of PDIMs in multiplication of M. tuberculosis during the acute phase of infection,29 whereby the lack of these lipids makes M. tuberculosis susceptible to killing by early innate immune host response.30
Interestingly, despite having a somewhat higher level of PZA/POA resistance, the strain POAR 18 [clpC1-7] displays a similar attenuation phenotype, implying that the polymorphism in clpC1 affects the growth of M. tuberculosis during the early phase of infection. Our finding that a POA resistance mutation in clpC1 imposes a fitness cost on M. tuberculosis in vivo suggests that these mutations may be at a competitive disadvantage with regard to amplification within the host and person-to-person transmission. It is noted that complementation studies are required to rule out that unidentified non-ClpC1 polymorphisms elsewhere in the genome of the strain POAR 18 [clpC1-7] may contribute to the observed growth behavior. On the other hand, our representative POA-resistant strain with a mutation in the C-terminal of panD:POAR 1 [panD1] displayed no growth defect in vivo. This finding suggests that panD mutants are more likely than clpC1 or mas/ppsA-E mutants to be selectively amplified by POA treatment in vivo.
To test this hypothesis and to verify that the resistance mechanisms that we identified in vitro could be recapitulated in vivo, we characterized POA-resistant isolates previously obtained from M. tuberculosis wild-type infected mice treated with a range of POA doses.8 Indeed, as predicted, sequencing of 28 POA-resistant M. tuberculosis strains isolated from different mice revealed that the vast majority of the strains (82%) contained various mutations in panD. PanD is involved in de novo production of pantothenate, which comprises the first stage of the biosynthesis of the essential cofactor coenzyme A. The panD gene was shown to be essential in vitro for M. tuberculosis growth.31,32 Furthermore, an auxotrophic mutant of M. tuberculosis lacking panC and panD was highly attenuated in vivo,32 implying that this pathway is crucial for the survival and growth of M. tuberculosis in vivo. Recently, we demonstrated that treatment with POA depletes intracellular pantothenate and coenzyme A levels,9 later confirmed by ref (33). Resistance to POA/PZA and prevention of this POA-mediated coenzyme A depletion in vitro can be caused by mutations in panD(9,11,12) or by exogenous supplementation of pantothenate.9,11,34 Mutations in panD causing POA/PZA resistance both in vitro and in vivo are almost exclusively localized in the C-terminal of PanD, which lies outside the aspartate decarboxylase domain and comprises a 13 amino acid tail, which is specific to mycobacteria,20 indicating that this region of the protein may be involved in interaction with POA.9 Despite some evidence suggesting that POA inhibits PanD enzyme activity,11 whether POA directly interacts with PanD and how the different PanD mutations affect this interaction are currently under investigation.
In this study, we also confirmed the hypothesis that PZA/POA resistance causing mutations in clpC1 are less likely to be found in vivo by demonstrating that only 5 of 28 POA-resistant strains (18%) selected in mice contained mutations in clpC1, whereas in vitro they were identified at more than twice this frequency.10M. tuberculosis ClpC1 works together with the ClpP1 and ClpP2 proteins of the caseinolytic protease complex and displays unfoldase and ATPase activities.35,36 The Clp protease complex is crucial for viability of M. tuberculosis both in vitro and in vivo,37,38 whereas the clpC1 gene on its own has been demonstrated to be essential for growth in vitro31 and within macrophages.39 Thus, drugs targeting the Clp protease machinery are considered attractive.40,41 Our finding that the mechanism of an established treatment-shortening TB drug, PZA, involves ClpC1 and that resistance mutations in this gene cause attenuation in vivo is of particular significance as novel antimycobacterials discovered recently, such as cyclomarin A,42 lassomycin,43 and ecumicin,44 target ClpC1, and resistance against these compounds in vitro is attributed to mutations in clpC1. Curiously, one of the polymorphisms (Gln17Arg) identified in this work as causing PZA/POA resistance was previously found to cause resistance to lassomycin and lies in the highly acidic portion of the N-terminal repeat I. Gln17 was found to be the major interacting residue with lassomycin via formation of H-bonds.43 However, the exact role of clpC1 in the mechanisms of action and resistance involving PZA and POA remains to be established.
Our findings shed some light on why mutations in panD, clpC1, and mas/ppsA-E may be difficult to find among PZA-resistant clinical isolates. First, mutations in these three genes were associated with relatively small (between 2- and 8-fold) shifts in susceptibility to PZA and POA, whereas pncA mutations conferring PZA resistance are associated with much higher (≥20-fold) increases in PZA MIC.4,45 To investigate whether previous studies may have missed panD or clpC1 mutations in clinical isolates due to their relatively low-level PZA resistance, we analyzed 1849 publicly available genomes of M. tuberculosis clinical isolates from the Genome-wide M. tuberculosis Variation (GMTV) Database. None of the strains contained any of the polymorphisms in clpC1 that we find associated with POA/PZA resistance in the current or previous study.10 We, however, found eight isolates containing the C-terminal mutation G415T/Gly139Stop in panD. Seven of these isolates were categorized as PZA-susceptible, whereas one was described as PZA-resistant.13 This identical polymorphism in panD was also observed in a PZA-susceptible clinical isolate from Russia, and the authors suggested that these strains may have been wrongly classified as PZA-susceptible due to the limitations of the BACTEC MGIT 960 PZA susceptibility test (which detects PZA resistance only at a single concentration of 100 μg/mL) in measuring low-level PZA resistance.27 The greater level of PZA resistance conferred by pncA mutations is expected to give a greater survival advantage in the face of PZA treatment. This argument is reinforced by our data showing that PZA monotherapy in mice is more likely to select for pncA mutants than for panD or clpC1 mutants.5 Moreover, whereas pncA mutations do not appear to reduce M. tuberculosis fitness in vitro or in vivo,14 our finding that mutations in clpC1 and ppsC cause growth defects in vivo provides another selective advantage favoring amplification of pncA mutants under PZA treatment. Because PZA is converted to POA by the host in addition to the pathogen, even M. tuberculosis cells with pncA mutations are exposed to circulating concentrations of POA that might provide additional selective pressure for POA resistance mutations. For example, we previously showed comparable plasma POA AUCs whether mice were treated orally with PZA or POA at 150 mg/kg.7,8 Host-metabolized POA exposures after standard PZA doses are similar to these mouse exposures but are likely too low to exert significant selection pressure in vivo,7 because only POA doses of 450 mg/kg and higher produced bactericidal effects in mice and, even then, the magnitude of these effects is much lower than those observed with standard PZA doses.5,8 We reanalyzed the quantitative drug-susceptible and POA-resistant CFU data from the prior mouse experiment5,8 and found that only the POA doses that had bactericidal activity (i.e., daily doses of 450–900 mg/kg) resulted in selective amplification of POA-resistant mutants compared to untreated animals. Thus, the greater selective advantage of pncA mutations that prevent intrabacillary conversion of PZA to POA, the requirement for systemic POA exposures higher than those produced by host metabolism of PZA to POA to selectively amplify POA-resistant mutants in vivo, and the reduced fitness of clpC1 and mas/ppsA-E mutants likely all interact to limit the selection of POA-resistant mutants in vivo. Finally, it must be noted that, due to the relatively small shift in PZA susceptibility conferred by panD, clpC1, and mas/ppsA-E mutations, current breakpoints for identification of PZA resistance in clinical isolates also may not identify these mutants as being PZA-resistant.
In conclusion, we show that the POA/PZA resistance mechanisms due to panD and clpC1 missense mutations previously identified in vitro can be recapitulated in vivo. Strains harboring clpC1 mutations were attenuated in mouse infection models, indicating loss of in vivo fitness and hence providing a possible explanation for the absence of such mutations in clinical isolates. Given the unimpaired fitness of panD mutants, we offer hypotheses relating to in vivo POA exposure and the clinical diagnosis of PZA resistance to explain why panD mutations are not observed more commonly among the small minority of PZA-resistant clinical isolates that do not harbor pncA mutations.
Materials and Methods
Bacterial Strains, Culture Conditions, and Chemicals
The parental strain M. tuberculosis H37Rv (ATCC 27294) was used to obtain representative POA-resistant strains: POAR 1 [panD1], POAR 7 [ppsC1], and POAR 18 [clpC1-7] as described in refs (9 and 10). Additional POA-resistant strains were selected by 8 weeks of POA treatment in BALB/c or C3HeB/FeJ mice infected with the parental strain as described in ref (8) and were shipped to the biosafety level 3 facility at the National University of Singapore, where the phenotypic and genotypic characterization detailed in this study was carried out. All strains were maintained in complete Middlebrook 7H9 medium (BD Difco) supplemented with 0.05% (v/v) Tween 80 (Sigma-Aldrich), 0.5% (v/v) glycerol (Fisher Scientific), and 10% (v/v) Middlebrook albumin–dextrose–catalase (BD Difco) at 37 °C with agitation at 80 rpm. Pyrazinamide, pyrazinoic acid, isoniazid, and rifampicin were purchased from Sigma-Aldrich and were freshly dissolved in 90% DMSO (Merck) and sterilized using 0.2 μm PTFE membrane filters (Acrodisc PALL). POA-resistant strains selected in mice were first subjected to colony purification and resistance confirmation on Middlebrook 7H10 agar (BD Difco) supplemented with 0.5% (v/v) glycerol and 10% (v/v) oleic acid–albumin–dextrose–catalase (OADC) (BD Difco) containing 300 mg/L POA. Isolated colonies were expanded in 7H9 to an OD600 of 0.7–0.8 and stored in 25% glycerol in 1 mL aliquots at −80 °C.
BALB/c Mouse Infection with M. tuberculosis
Pathogen-free female BALB/c mice aged 8 weeks were purchased from Charles River Laboratories. Mice were group-housed in a biosafety level 3 animal facility and maintained with sterile bedding, water, and mouse chow. This study was performed under strict accordance with recommendations outlined in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health with protocol 140020D017 approved by Rutgers University’s Institutional Animal Care and Use Committee.
The M. tuberculosis wild-type strain (H37Rv) and mutant strains POAR 1 [panD1], POAR 7 [ppsC1], and POAR 18 [clpC1-7] were used for infection. BALB/c mice were infected in a Glas–Col inhalation exposure system. Titered frozen bacterial stocks were diluted in phosphate-buffered saline (PBS), and 3 × 106 CFU/mL was added to the nebulizer to achieve the implantation of 100–300 CFU in the lungs. Three mice were sacrificed via cervical dislocation 24 h later to determine the starting bacteria load.
BALB/c mice (n = 4 per group per time point) were sacrificed at 2, 4, and 6 weeks postinfection. Right lung and spleen were collected post-mortem and homogenized in 5 mL of PBS containing 0.05% Tween 80. CFU was determined by plating serial dilutions of homogenates onto Middlebrook 7H11 agar with OADC (GIBCO BRL). At week 6, lung homogenates were also plated on 7H11 agar containing 3 mM POA to determine resistance. Colonies were counted after at least 21 days of incubation at 37 °C.
At weeks 4 and 6 postinfection, formalin-fixed (Fisher Chemical) lung tissues of infected mice (one lung from four mice in each group) were paraffin-embedded and used for standard 5 μm sectioning. Tissue sections were stained with hematoxylin/eosin (H&E) for cellular composition or with Ziehl–Neelsen to reveal acid-fast mycobacteria. To quantify the acid-fast bacilli in lesions from mice infected with the different strains described above, five lesions were randomly picked from each group and acid-fast bacilli were counted at a magnification of 50× and field size 340 by 228 μm. Numbers are reported either as single bacilli or clusters, if there were too many bacilli per macrophage. Each cluster is representative of a single macrophage, and the range of numbers of bacilli within the respective clusters is reported.
Susceptibility Testing
MICs against POA, isoniazid, and rifampicin were performed by the broth dilution method. The strains were grown to mid log phase, spun down, resuspended in fresh 7H9 media, and adjusted to an OD600 = 0.1. One hundred microliters of cell suspension was added into wells containing 100 μL of 2-fold serially diluted compound in transparent flat-bottomed 96-well plates (Corning Costar), sealed with Breath-Easy membranes (Sigma-Aldrich). The plates were incubated for 7 days at 37 °C with shaking at 80 rpm. After incubation, the cultures were manually resuspended and OD600 was measured using a spectrophotometer (Tecan Infinite M200 Pro). Experiments were performed three times independently with technical replicates. MIC50 values represent the concentration of drug that inhibits bacterial growth by 50% as compared to the respective drug-free control.
Susceptibility to PZA was assessed using the BACTEC MGIT 960 PZA susceptibility test following the guidelines as described in ref (46).
PZA and POA agar MICs were defined as the concentration of drug that suppresses colony formation upon plating 104 CFU from mid log cultures on 7H10 agar plates in independent experiments and incubating them for 2 weeks at 37 °C as previously described.10,47
Whole Genome Sequencing
Genomic DNA was isolated from the different M. tuberculosis strains and subjected to whole genome sequencing on an Illumina MiSeq platform with AIT Biotech (Singapore). Sequencing and subsequent analysis was performed as described in ref (9).
panD and clpC1 Sequencing
The panD gene was amplified from the genomic DNA using the primers 5′-AGCTGCGCGATATCGGGCTT-3′ and 5′-TGCACGACCTTTGCGTGCTCTT-3′ and clpC1 with the primers 5′-ACATATGTTCGAACGATTTACCGACCGTGC-3′ and 5′-TGAATTCACCCATGTCAATCTGAATAAGCGC-3′ or 5′-GATGATGTCACCGCGGGTGTTG-3′ using Phusion High Fidelity DNA polymerase (Thermo Scientific) as per the manufacturer’s instructions. The purified PCR products were subjected to capillary sequencing via BigDyeTerminator chemistry by AIT Biotech (Singapore) and analyzed using BioEdit (North Carolina State University) and BLAST (NCBI).
Analysis of Selective Amplification of POA-Resistant Mutants in Vivo
CFU count data obtained from lung homogenates plated on 7H11 plates with or without POA from the POA dose-ranging mouse efficacy experiment,8 in which the POA-resistant mutants were selected, were reanalyzed to quantify and compare the selective amplification of resistant mutants during treatment of BALB/c mice with POA or PZA. Mice received a low-dose aerosol infection followed 4 weeks later by initiation of treatment with PZA 150 mg/kg alone or POA alone in one of the following doses (in mg/kg body weight): 37.5, 75, 150, or 450, once daily, or 75 or 450 twice daily. Total and POA-resistant CFU after 8 weeks of treatment were quantified by plating serial dilutions of lung homogenates on drug-free plates and plates containing POA 300 μg/mL (2–3× MIC for H37Rv parent), respectively. Mean frequencies of drug-resistant CFU were compared by one-way ANOVA with Dunnett’s post-test to control for multiple comparisons. Additionally, the proportions of PZA-treated mice and POA-treated mice harboring POA-resistant mutants after 8 weeks of treatment were compared by Fisher’s Exact test. Statistical analyses were performed using GraphPad Prism 6.
Acknowledgments
This research was supported by the Singapore Ministry of Health’s National Medical Research Council under its TCR Flagship Grant NMRC/TCR/011-NUHS/2014 and Centre Grant “MINE”, Research Core 4, NMRC/CG/013/2013 to T.D. and is part of the Singapore Programme of Research Investigating New Approaches to Treatment of Tuberculosis (SPRINT-TB; www.sprinttb.org) managed by Kristina Rutkute and led by Nick Paton. Additional support came from a supplement to the Johns Hopkins University Center for AIDS Research (NIH-NIAID-DAIDS Grant P30-AI094189 (to E.N.)). We thank Sabai Phyu and Martin Gengenbacher, Yong Loo Lin School of Medicine BSL3 Core facility, for support, and Meera Gurumurthy and Claire Naftalin for discussions. P.G. received a scholarship from the Yong Loo Lin School of Medicine.
Glossary
Abbreviations
- TB
tuberculosis
- PZA
pyrazinamide
- POA
pyrazinoic acid
- PDIM
phthiocerol dimycocerosate
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsinfecdis.7b00017.
Growth inhibition dose response curves for POA-resistant M. tuberculosis strains, selection of POA-resistant mutants in vivo as a function of POA dose, abundance of acid-fast bacilli within lesions after infection with POA-resistant mutants versus wild-type M. tuberculosis H37Rv, phenotypic and genotypic characteristics of POA-resistant M. tuberculosis with details for 28 strains isolated from mouse lungs (PDF)
Author Contributions
P.G., V.D., E.N., and T.D. designed the experiments and wrote the manuscript. P.G. and M.Y. characterized the POA-resistant strains isolated from POA-treated mice. J.-P.L., R.T., and E.N. carried out the POA treatment experiments in mice, isolated POA-resistant strains, and initially characterized their POA resistance. J.S., G.R., L.L., and V.D. carried out the mouse infection experiments with in vitro isolated POA/PZA-resistant mutants.
The authors declare no competing financial interest.
Supplementary Material
References
- Mitchison D. A. (1985) The action of antituberculosis drugs in short-course chemotherapy. Tubercle 66, 219–225. 10.1016/0041-3879(85)90040-6. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Shi W.; Zhang W.; Mitchison D. (2014) Mechanisms of Pyrazinamide Action and Resistance. Microbiol. Spectrum 2, 1–12. 10.1128/microbiolspec.MGM2-0023-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal P.; Dick T. (2014) Reactive dirty fragments: implications for tuberculosis drug discovery. Curr. Opin. Microbiol. 21, 7–12. 10.1016/j.mib.2014.06.015. [DOI] [PubMed] [Google Scholar]
- Scorpio A.; Zhang Y. (1996) Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat. Med. 2, 662–667. 10.1038/nm0696-662. [DOI] [PubMed] [Google Scholar]
- Lanoix J.-P.; Ioerger T.; Ormond A.; Kaya F.; Sacchettini J.; Dartois V.; Nuermberger E. (2016) Selective Inactivity of Pyrazinamide against Tuberculosis in C3HeB/FeJ Mice Is Best Explained by Neutral pH of Caseum. Antimicrob. Agents Chemother. 60, 735–743. 10.1128/AAC.01370-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorpio A.; Lindholm-Levy P.; Heifets L.; Gilman R.; Siddiqi S.; Cynamon M.; Zhang Y. (1997) Characterization of pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 41, 540–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Via L. E.; Savic R.; Weiner D. M.; Zimmerman M. D.; Prideaux B.; Irwin S. M.; Lyon E.; O’Brien P.; Gopal P.; Eum S.; Lee M.; Lanoix J.-P.; Dutta N. K.; Shim T.; Cho J. S.; Kim W.; Karakousis P. C.; Lenaerts A.; Nuermberger E.; Barry C. E.; Dartois V. (2015) Host-Mediated Bioactivation of Pyrazinamide: Implications for Efficacy, Resistance, and Therapeutic Alternatives. ACS Infect. Dis. 1, 203–214. 10.1021/id500028m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanoix J.-P.; Tasneen R.; O’Brien P.; Sarathy J.; Safi H.; Pinn M.; Alland D.; Dartois V.; Nuermberger E. (2016) High systemic exposure of pyrazinoic acid has limited anti-tuberculosis activity in murine and rabbit models of tuberculosis. Antimicrob. Agents Chemother. 60, 4197–4205. 10.1128/AAC.03085-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal P.; Yee M.; Sarathy J.; Low J. L.; Sarathy J. P.; Kaya F.; Dartois V.; Gengenbacher M.; Dick T. (2016) Pyrazinamide Resistance Is Caused by Two Distinct Mechanisms: Prevention of Coenzyme A Depletion and Loss of Virulence Factor Synthesis. ACS Infect. Dis. 2, 616–626. 10.1021/acsinfecdis.6b00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee M.; Gopal P.; Dick T. (2016) Missense mutations in the unfoldase ClpC1 of the caseinolytic protease complex are associated with pyrazinamide resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. AAC.02342-16. 10.1128/AAC.02342-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W.; Chen J.; Feng J.; Cui P.; Zhang S.; Weng X.; Zhang W.; Zhang Y. (2014) Aspartate decarboxylase (PanD) as a new target of pyrazinamide in Mycobacterium tuberculosis. Emerging Microbes Infect. 3, e58. 10.1038/emi.2014.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.; Chen J.; Shi W.; Liu W.; Zhang W.; Zhang Y. (2013) Mutations in panD encoding aspartate decarboxylase are associated with pyrazinamide resistance in Mycobacterium tuberculosis. Emerging Microbes Infect. 2, e34. 10.1038/emi.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casali N.; Nikolayevskyy V.; Balabanova Y.; Harris S. R.; Ignatyeva O.; Kontsevaya I.; Corander J.; Bryant J.; Parkhill J.; Nejentsev S.; Horstmann R. D.; Brown T.; Drobniewski F. (2014) Evolution and transmission of drug-resistant tuberculosis in a Russian population. Nat. Genet. 46, 279–286. 10.1038/ng.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoffels K.; Mathys V.; Fauville-Dufaux M.; Wintjens R.; Bifani P. (2012) Systematic Analysis of Pyrazinamide-Resistant Spontaneous Mutants and Clinical Isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 56, 5186–5193. 10.1128/AAC.05385-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto P.; Cabibbe A. M.; Feuerriegel S.; Casali N.; Drobniewski F.; Rodionova Y.; Bakonyte D.; Stakenas P.; Pimkina E.; Augustynowicz-Kopeć E.; Degano M.; Ambrosi A.; Hoffner S.; Mansjö M.; Werngren J.; Rüsch-Gerdes S.; Niemann S.; Cirillo D. M. (2014) Mycobacterium tuberculosis Pyrazinamide Resistance Determinants: a Multicenter Study. mBio 5, e01819. 10.1128/mBio.01819-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hertog A. L., Sengstake S., and Anthony R. M. (2015) Pyrazinamide resistance in Mycobacterium tuberculosis fails to bite? Pathog. Dis. 73, DOI: 10.1093/femspd/ftv037. [DOI] [PubMed] [Google Scholar]
- Goren M. B.; Brokl O.; Schaefer W. B. (1974) Lipids of Putative Relevance to Virulence in Mycobacterium tuberculosis: Phthiocerol Dimycocerosate and the Attenuation Indicator Lipid. Infect. Immun. 9, 150–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J. S.; Chen B.; McNeil M.; Jacobs W. R. (1999) Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature 402, 79–83. 10.1038/47042. [DOI] [PubMed] [Google Scholar]
- Camacho L. R.; Ensergueix D.; Perez E.; Gicquel B.; Guilhot C. (1999) Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol. Microbiol. 34, 257–267. 10.1046/j.1365-2958.1999.01593.x. [DOI] [PubMed] [Google Scholar]
- Gopalan G.; Chopra S.; Ranganathan A.; Swaminathan K. (2006) Crystal structure of uncleaved l-aspartate-α-decarboxylase from Mycobacterium tuberculosis. Proteins: Struct., Funct., Genet. 65, 796–802. 10.1002/prot.21126. [DOI] [PubMed] [Google Scholar]
- Chopra S.; Pai H.; Ranganathan A. (2002) Expression, purification, and biochemical characterization of Mycobacterium tuberculosis aspartate decarboxylase, PanD. Protein Expression Purif. 25, 533–540. 10.1016/S1046-5928(02)00039-6. [DOI] [PubMed] [Google Scholar]
- Pierre-Audigier C.; Surcouf C.; Cadet-Daniel V.; Namouchi A.; Heng S.; Murray A.; Guillard B.; Gicquel B. (2012) Fluoroquinolone and pyrazinamide resistance in multidrug-resistant tuberculosis. Int. J. Tuberc. Lung Dis. 16, 221–223. 10.5588/ijtld.11.0266. [DOI] [PubMed] [Google Scholar]
- Mphahlele M.; Syre H.; Valvatne H.; Stavrum R.; Mannsåker T.; Muthivhi T.; Weyer K.; Fourie P. B.; Grewal H. M. S. (2008) Pyrazinamide Resistance among South African Multidrug-Resistant Mycobacterium tuberculosis Isolates. J. Clin. Microbiol. 46, 3459–3464. 10.1128/JCM.00973-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njire M.; Tan Y.; Mugweru J.; Wang C.; Guo J.; Yew W.; Tan S.; Zhang T. (2016) Pyrazinamide resistance in Mycobacterium tuberculosis: Review and update. Adv. Med. Sci. 61, 63–71. 10.1016/j.advms.2015.09.007. [DOI] [PubMed] [Google Scholar]
- Shi W.; Zhang X.; Jiang X.; Yuan H.; Lee J. S.; Barry C. E.; Wang H.; Zhang W.; Zhang Y. (2011) Pyrazinamide Inhibits Trans-Translation in Mycobacterium tuberculosis. Science 333, 1630–1632. 10.1126/science.1208813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y.; Yu X.; Jiang G.; Wang X.; Ma Y.; Li Y.; Huang H. (2016) Pyrazinamide resistance among multidrug-resistant tuberculosis clinical isolates in a national referral center of China and its correlations with pncA, rpsA, and panD gene mutations. Diagn. Microbiol. Infect. Dis. 84, 207–211. 10.1016/j.diagmicrobio.2015.10.017. [DOI] [PubMed] [Google Scholar]
- Maslov D. A.; Zaĭchikova M. V.; Chernousova L. N.; Shur K. V.; Bekker O. B.; Smirnova T. G.; Larionova E. E.; Andreevskaya S. N.; Zhang Y.; Danilenko V. N. (2015) Resistance to pyrazinamide in Russian Mycobacterium tuberculosis isolates: pncA sequencing versus Bactec MGIT 960. Tuberculosis 95, 608–612. 10.1016/j.tube.2015.05.013. [DOI] [PubMed] [Google Scholar]
- Chernyaeva E. N.; Shulgina M. V.; Rotkevich M. S.; Dobrynin P. V.; Simonov S. A.; Shitikov E. A.; Ischenko D. S.; Karpova I. Y.; Kostryukova E. S.; Ilina E. N.; Govorun V. M.; Zhuravlev V. Y.; Manicheva O. A.; Yablonsky P. K.; Isaeva Y. D.; Nosova E. Y.; Mokrousov I. V.; Vyazovaya A. A.; Narvskaya O. V.; Lapidus A. L.; O’Brien S. J. (2014) Genome-wide Mycobacterium tuberculosis variation (GMTV) database: a new tool for integrating sequence variations and epidemiology. BMC Genomics 15, 308. 10.1186/1471-2164-15-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau C.; Winter N.; Pivert E.; Bordat Y.; Neyrolles O.; Avé P.; Huerre M.; Gicquel B.; Jackson M. (2004) Production of phthiocerol dimycocerosates protects Mycobacterium tuberculosis from the cidal activity of reactive nitrogen intermediates produced by macrophages and modulates the early immune response to infection. Cell. Microbiol. 6, 277–287. 10.1046/j.1462-5822.2004.00368.x. [DOI] [PubMed] [Google Scholar]
- Day T. A.; Mittler J. E.; Nixon M. R.; Thompson C.; Miner M. D.; Hickey M. J.; Liao R. P.; Pang J. M.; Shayakhmetov D. M.; Sherman D. R. (2014) Mycobacterium tuberculosis Strains Lacking Surface Lipid Phthiocerol Dimycocerosate Are Susceptible to Killing by an Early Innate Host Response. Infect. Immun. 82, 5214–5222. 10.1128/IAI.01340-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin J. E.; Gawronski J. D.; DeJesus M. A.; Ioerger T. R.; Akerley B. J.; Sassetti C. M. (2011) High-Resolution Phenotypic Profiling Defines Genes Essential for Mycobacterial Growth and Cholesterol Catabolism. PLoS Pathog. 7, e1002251. 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambandamurthy V. K.; Wang X.; Chen B.; Russell R. G.; Derrick S.; Collins F. M.; Morris S. L.; Jacobs W. R. (2002) A pantothenate auxotroph of Mycobacterium tuberculosis is highly attenuated and protects mice against tuberculosis. Nat. Med. 8, 1171–1174. 10.1038/nm765. [DOI] [PubMed] [Google Scholar]
- Rosen B. C.; Dillon N. A.; Peterson N. D.; Minato Y.; Baughn A. D. (2016) Long-Chain Fatty Acyl-CoA Ligase FadD2 Mediates Intrinsic Pyrazinamide Resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. AAC.02130-16. 10.1128/AAC.02130-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon N. A.; Peterson N. D.; Rosen B. C.; Baughn A. D. (2014) Pantothenate and Pantetheine Antagonize the Antitubercular Activity of Pyrazinamide. Antimicrob. Agents Chemother. 58, 7258–7263. 10.1128/AAC.04028-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar N. P.; Sikriwal D.; Rath P.; Choudhary R. K.; Batra J. K. (2008) Mycobacterium tuberculosis ClpC1. FEBS J. 275, 6149–6158. 10.1111/j.1742-4658.2008.06738.x. [DOI] [PubMed] [Google Scholar]
- Akopian T.; Kandror O.; Raju R. M.; UnniKrishnan M.; Rubin E. J.; Goldberg A. L. (2012) The active ClpP protease from M. tuberculosis is a complex composed of a heptameric ClpP1 and a ClpP2 ring. EMBO J. 31, 1529–1541. 10.1038/emboj.2012.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju R. M.; Jedrychowski M. P.; Wei J.-R.; Pinkham J. T.; Park A. S.; O’Brien K.; Rehren G.; Schnappinger D.; Gygi S. P.; Rubin E. J. (2014) Post-Translational Regulation via Clp Protease Is Critical for Survival of Mycobacterium tuberculosis. PLoS Pathog. 10, e1003994. 10.1371/journal.ppat.1003994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll P.; Faray-Kele M.-C.; Parish T. (2011) Identifying Vulnerable Pathways in Mycobacterium tuberculosis by Using a Knockdown Approach. Appl. Environ. Microbiol. 77, 5040–5043. 10.1128/AEM.02880-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengarajan J.; Bloom B. R.; Rubin E. J. (2005) Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc. Natl. Acad. Sci. U. S. A. 102, 8327–8332. 10.1073/pnas.0503272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju R. M.; Goldberg A. L.; Rubin E. J. (2012) Bacterial proteolytic complexes as therapeutic targets. Nat. Rev. Drug Discovery 11, 777–789. 10.1038/nrd3846. [DOI] [PubMed] [Google Scholar]
- Moreira W.; Ngan G. J. Y.; Low J. L.; Poulsen A.; Chia B. C. S.; Ang M. J. Y.; Yap A.; Fulwood J.; Lakshmanan U.; Lim J.; Khoo A. Y. T.; Flotow H.; Hill J.; Raju R. M.; Rubin E. J.; Dick T. (2015) Target Mechanism-Based Whole-Cell Screening Identifies Bortezomib as an Inhibitor of Caseinolytic Protease in Mycobacteria. mBio 6, e00253. 10.1128/mBio.00253-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan D.; Rao S. P. S.; Noble C. G. (2013) Structural Basis of Mycobacterial Inhibition by Cyclomarin A. J. Biol. Chem. 288, 30883–30891. 10.1074/jbc.M113.493767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrish E.; Sit C. S.; Cao S.; Kandror O.; Spoering A.; Peoples A.; Ling L.; Fetterman A.; Hughes D.; Bissell A.; Torrey H.; Akopian T.; Mueller A.; Epstein S.; Goldberg A.; Clardy J.; Lewis K. (2014) Lassomycin, a Ribosomally Synthesized Cyclic Peptide, Kills Mycobacterium tuberculosis by Targeting the ATP-Dependent Protease ClpC1P1P2. Chem. Biol. (Oxford, U. K.) 21, 509–518. 10.1016/j.chembiol.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W.; Kim J.-Y.; Anderson J. R.; Akopian T.; Hong S.; Jin Y.-Y.; Kandror O.; Kim J.-W.; Lee I.-A.; Lee S.-Y.; McAlpine J. B.; Mulugeta S.; Sunoqrot S.; Wang Y.; Yang S.-H.; Yoon T.-M.; Goldberg A. L.; Pauli G. F.; Suh J.-W.; Franzblau S. G.; Cho S. (2015) The Cyclic Peptide Ecumicin Targeting ClpC1 Is Active against Mycobacterium tuberculosis In Vivo. Antimicrob. Agents Chemother. 59, 880–889. 10.1128/AAC.04054-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemens S. P.; Sharpe C. A.; Cynamon M. H. (1996) Activity of pyrazinamide in a murine model against Mycobacterium tuberculosis isolates with various levels of in vitro susceptibility. Antimicrob. Agents Chemother. 40, 14–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqi S. H., and Rüsch-Gerdes S. (2006) MGIT Procedure Manual, Foundation for Innovative New Diagnostics, Geneva, Switzerland. [Google Scholar]
- Heifets L.; Sanchez T. (2000) New Agar Medium for Testing Susceptibility of Mycobacterium tuberculosis to Pyrazinamide. J. Clin. Microbiol. 38, 1498–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.