Abstract
Background
Recently, it was demonstrated that proteins can be translated from alternative open reading frames (altORFs), increasing the size of the actual proteome. Top-down mass spectrometry-based proteomics allows the identification of intact proteins containing post-translational modifications (PTMs) as well as truncated forms translated from reference ORFs or altORFs.
Methods
Top-down tissue microproteomics was applied on benign, tumor and necrotic-fibrotic regions of serous ovarian cancer biopsies, identifying proteins exhibiting region-specific cellular localization and PTMs. The regions of interest (ROIs) were determined by MALDI mass spectrometry imaging and spatial segmentation.
Findings
Analysis with a customized protein sequence database containing reference and alternative proteins (altprots) identified 15 altprots, including alternative G protein nucleolar 1 (AltGNL1) found in the tumor, and translated from an altORF nested within the GNL1 canonical coding sequence. Co-expression of GNL1 and altGNL1 was validated by transfection in HEK293 and HeLa cells with an expression plasmid containing a GNL1-FLAG(V5) construct. Western blot and immunofluorescence experiments confirmed constitutive co-expression of altGNL1-V5 with GNL1-FLAG.
Conclusions
Taken together, our approach provides means to evaluate protein changes in the case of serous ovarian cancer, allowing the detection of potential markers that have never been considered.
Keywords: Top-down, Microproteomics, Hidden proteome, Biomarkers, Ovarian cancer, Alternative proteins
Highlights
-
•
Top-down spatially-resolved microproteomics associated to MALDI-Mass spectrometry imaging
-
•
Identification of proteoform markers of ovarian cancer
-
•
Identification of proteins translated from alternative open reading frames
Mass spectrometry (MS) imaging is a molecular imaging tool which can be used to map distribution of molecules and distinguish between normal and diseased states based on molecular content.
In this work, MALDI MS imaging was combined with intact protein identification by top-down microproteomics analysis. Using ovarian cancer tissues, tumor and benign zones can be distinguished from each other with great correlation and in accord with standard pathological examination. We then extracted proteins in each of the regions and identified several potential ovarian cancer biomarkers as well as non-canonical proteins translated from alternative open reading frames. In summary, our approach allows molecular classification of the tissue and identification of protein biomarkers including proteins issued from alternative open reading frames.
1. Introduction
With the recent advances in mass spectrometry (MS) based-proteomics, the application of top-down MS-based proteomic strategies now allows the analysis of complex protein mixtures in their intact state without the need for enzymatic digestion (Tran et al., 2011). In a study by Ye et al., top-down MS-based proteomics coupled to Matrix Assisted Laser Desorption Ionization (MALDI) MS imaging (MALDI-MSI) of a rat brain post-treated with the NMDA receptor antagonist MK801 revealed 34 proteins with their specific post-translational modifications (PTMs) (Ye et al., 2014). Recently, we performed MALDI-MSI coupled to top-down tissue microproteomics on 3 rat brain regions and demonstrated the possibility to identify specific proteoforms linked to the physiology of the tissue region; several unique markers were identified showing different proteoforms of brain-specific proteins (data not shown). In this work, we investigated the pathological heterogeneity in ovarian serous cancer tumor microenvironment utilizing a top-down microproteomics approach. Specifically, we investigated proteome microenvironment alterations aiming to delineate and characterize specific protein profiles in benign, tumor and necrotic/fibrotic tumor regions by taking into account their PTMs and assessing their cleaved forms.
Our assessment also takes into account the identification of alternative proteins (AltProts) (Vanderperre et al., 2013). AltProts are translated from alternative open reading frames (AltORFs). AltORFs can have different localizations: they can overlap annotated protein-coding sequences in a different reading frame, or can be present within untranslated regions (UTRs) of mature mRNAs (Mouilleron et al., 2016, Vanderperre et al., 2013). Thus, alternative proteins are completely different from annotated or reference proteins (Mouilleron et al., 2016, Vanderperre et al., 2013). AltORFs may also be present in transcripts annotated as non-coding RNAs (ncRNAs). Indeed, proteins translated from non-annotated AltORFs were detected in our previous studies by MS. Some of these alternative translation products have also been validated biologically and assessed for their biological activity. For example, we have shown that AltMRVI1 is translated from an AltORF overlapping the MRVI1 coding sequence in a different reading frame and interacts with BRCA1 (Vanderperre et al., 2013). Translation of AltORFs in addition to annotated coding sequences opens the door to proteins that cannot be detected using conventional protein databases. Thus, due to their intriguing role, we aimed at investigating the profiles of the “hidden proteome” and assess their contribution in serous ovarian cancer. Additionally, these AltProts are mainly small proteins and the top-down proteomics strategy seems to be a better alternative rather than the shotgun proteomics for their detection. This is so because, even if the shotgun approach remains the most efficient strategy for high throughput proteomics, the identification of small proteins in this approach can be hampered due to the low amount of generated tryptic peptides and the generally fewer presence of enzyme cleavage sites. Therefore, top-down proteomics offers a good alternative to identify small proteins or truncated forms as well as some PTMs from the reference or the hidden proteome. Overall, our aim is to identify and characterize reference and altprots as potential markers for serous ovarian cancer pathology.
2. Experimental Procedures
2.1. Tissue Collection
The ovarian biopsies were obtained from patients of the Centre Oscar Lambret (Lille, France) and from the CHRU de Lille Pathology Department. All experiments were approved by the local Ethics Committee (CPP Nord Ouest IV 12/10) in accordance with the French and European legislation on this topic. Methods of collection for human ovaries were performed in accordance with procedures that were approved by the Ethics Committee of the CHRU Lille. The study adhered to the principles of the Declaration of Helsinki and the Guidelines for Good Clinical Practice. All patients gave written informed consent before enrolment. The flash-frozen biopsies were stored at − 80 °C until use.
2.2. Experimental Design and Statistical Rationale
We first performed MS imaging of lipids in order to perform spatial segmentation analysis to identify regions of interest (ROIs), which were then subjected to liquid micro-junction (LMJ) (Quanico et al., 2013, Wisztorski et al., 2016) or parafilm-assisted manual microdissection (PAM) (Franck et al., 2013, Quanico et al., 2014) methods of microextraction. LMJ and PAM were followed by top-down proteomics for protein identification from necrotic/fibrotic tumor, tumor and benign (B) regions (technical triplicate). Reference and alternative proteins were then identified and localized in the 3 tissue regions of the ovarian serous cancer biopsies.
2.3. Chemicals
MS grade water (H2O), acetonitrile (ACN), methanol (MeOH), ethanol (EtOH), and chloroform were purchased from Biosolve (Valkensvaard, Netherlands). The cleavable detergent ProteaseMAX was purchased from Promega (Charbonnieres, France). Parafilm M was purchased from Pechiney Plastic Packaging (Chicago, Illinois). 2,5-dihydroxybenzoic acid (DHB), sodium dodecyl sulphate (SDS), dl-dithiothreitol (DTT) trifluoroacetic acid (TFA) and formic acid (FA) were purchased from Sigma (Saint-Quentin-Fallavier, France).
2.4. Tissue Section Preparation
For MALDI-MSI experiments, tissues were cut in 10-μm slices using a cryostat (Leica Microsystems, Nanterre, France) and mounted on Indium Tin Oxide (ITO)-coated glass slides (LaserBio Labs, Sophia-Antipolis, France) by finger-thawing. For LMJ and PAM, consecutive tissue slices were also obtained but with a 30-μm thickness. For LMJ, the tissues were mounted on a polylysine glass slide. For PAM, on the other hand, the tissue sections were mounted on a parafilm M-covered glass slide (Franck et al., 2013, Quanico et al., 2014). After tissue section preparation, the sections were immediately dehydrated under vacuum at room temperature for 20 min. The slides were then scanned using a Nikon scanner and stored at − 80 °C until use.
2.5. MALDI-MSI
DHB matrix (50 mg/mL) dissolved in 6:4 (v/v) MeOH/0.1% TFA in water was manually sprayed at a flow rate of 300 μL/h using a syringe pump connected to an electrospray nebulizer. The nebulizer was connected to a nitrogen line operated at 1 bar. The nebulizer was moved uniformly throughout the tissue until crystallization was sufficient to ensure optimal lipid detection. The tissue was then analyzed using an UltraFlex II MALDI-TOF/TOF (Time Of Flight) mass spectrometer equipped with a Smartbeam Nd-YAG laser (355 nm) and controlled by FlexControl software (Bruker Daltonics, Bremen, Germany). Lipid image acquisition was performed in positive reflector mode within an m/z range of 50 to 900 at a 300 μm resolution, and the obtained spectra were averaged from 300 laser shots per pixel. Peak detection and spatial segmentation analysis were then performed on the acquired images using SCiLS Software 2015b (SCiLS Lab GmbH, Bremen, Germany). For spatial segmentation, the Bisecting k-Means approach with Correlation as the distance metric was used. Spectra were subjected to median normalization and medium denoising prior to peak picking. After analysis, the ROIs were determined by selecting segments where the correlation distance metric is significantly distant from the other.
2.6. Intact Protein Extraction Buffer
To ensure minimal protein hydrolysis by endogenous proteases, every step from buffer preparation to nanoflow Liquid Chromatography (nanoLC)-MS/MS analysis was carried out within the same day with on-ice conservation in between sample processing steps. A 0.1% (v/v) aliquot of temperature- and acid-cleavable commercial detergent (ProteaseMAX) was prepared from a 1% (v/v) stock solution prepared in 50 μM DTT and immediately stored at − 20 °C until use according to manufacturer's recommendations. Aliquots were processed within the day of sample extraction to ensure minimal degradation of the detergent over time, and remaining solutions were discarded.
2.7. LMJ Experiments
To ensure optimal protein extraction, lipids were removed from the tissue section by immersing the glass slide consecutively for 1 min each in 70% EtOH and in 95% EtOH then 30 s in chloroform with complete solvent evaporation under reduced pressure at room temperature between each washing step. The slide was then scanned again as washing steps improve structure visibility. The slide used for LMJ microextraction was placed inside a TriVersa NanoMate (Advion, Ithaca, NY, USA) instrument. Proteins were then extracted from every ROI by completing six cycles of extraction composed of the following steps: 1) aspirate 1.5 μL of detergent solution, 2) dispense 0.8 μL of the extraction buffer on the surface of the selected ROI with 10 iterations of up-and-down pipetting, 3) aspirate 2.5 μL volume, and 4) expel 4 μL from the pipette tip into a clean tube to ensure complete retrieval of the initial 1.5 μL volume. Per ROI, the final collected volume was 9 μL. Each extract was immediately stored on-ice until further processing.
2.8. PAM Experiments
ROIs generated from spatial segmentation of MS images were cut using a scalpel. The pieces of parafilm M containing the tissue were then placed in a tube containing 10 μL of the extraction buffer and stored on-ice until further processing.
2.9. nanoLC-MS/MS
The extracts obtained with the LMJ or PAM strategy were sonicated for 5 min and the proteins were denatured at 55 °C for 15 min. The tubes were then quickly centrifuged to collect the extracts at the bottom of the tube. For extracts obtained using the PAM strategy, the parafilm pieces were carefully removed from the tubes using a pipette tip and the extracts were incubated at 95 °C for 10 min to ensure complete detergent dissociation. The tubes were then quickly centrifuged and stored on ice. 11 μL of 10% ACN in 0.4% FA in water were added to each tube so that the final ACN concentration is equal to the concentration of ACN at the beginning of the LC gradient. Samples were subjected to nanoLC-MS/MS analysis on the same day of sample preparation and were kept in the autosampler with the thermostat set at 4 °C.
5 μL of the sample was loaded onto a 2 cm * 150 μm internal diameter IntegraFrit sample trap-column (New Objective, Woburn, Massachusetts, USA) at a maximum pressure 280 bar using a Proxeon EASY nLC-II (Proxeon, Thermo Scientific, Bremen, Germany). Proteins were separated on a 15 cm * 100 μm internal diameter PLRP-S column (Varian, Palo Alto, California, USA) with a linear gradient of ACN from 5 to 55% for 110 min and 55% to 90% for 25 min and a flow rate of 300 nL/min.
Data were acquired on a Q-Exactive mass spectrometer (Thermo Scientific, Bremen, Germany) equipped with a nanoESI (Electrospray Ionization) source (Proxeon, Thermo Scientific, Bremen, Germany) and a PicoTip nanospray emitter (New Objective, Woburn, Massachusetts, USA). Data were acquired in data-dependent mode using a top 3 strategy. Full scans were acquired by averaging 4 microscans at 70,000 resolution within a m/z mass range of 800–2000 with an AGC target of 1 * 106 and a maximum accumulation time of 200 ms. The three most abundant ions with charge superior than 3 or unassigned were selected for fragmentation. Precursors were selected within a 15 m/z selection window by the quadrupole and fragmented with a Normalized Collision Energy (NCE) of 25; the (Automatic Gain Control) AGC target was set to 1 * 106 with a maximum accumulation time of 500 ms. For each MS/MS spectrum, two microscans at a resolution of 70,000 at m/z 400 were acquired and averaged. Dynamic exclusion was set to 20 s.
2.10. Data Analysis
RAW files were processed with ProSight PC 3.0 (Thermo Fisher Scientific, Bremen, Germany). Spectral data were deisotoped using the cRAWler algorithm and searched against the complex Homo sapiens ProSightPC database version 2014_07 containing every canonical protein and its known PTMs. Files were searched with the “absolute mass” then “biomarker” search modes (Kellie et al., 2010) in ProsightPC considering every PTM available in the complex database. A second search was performed to detect altORF products with a concatenated database composed of the H. sapiens UniProt Reference proteome (canonical and isoforms) of 01.16.2015 and an in-silico translated database of the H. sapiens of the transcripts from GenBank containing every ORF with potential protein product that had at least 29 amino acids with the same search strategy. Identification was considered positive when one of the two strategies gave an expected score (E-value) that was lower than 10− 4.
Raw files were also processed with ProSightPC 3.0 (Thermo Scientific) and Proteome Discoverer 2.1 (Thermo Scientific) utilizing the ProSightPD 1.0 node. Spectra were then searched using a three-tiered search tree. The first search was an Absolute Mass search with MS1 tolerance of 100 Da and MS2 tolerance of 10 ppm, against the complex Homo sapiens ProSightPC database version 2014_07 containing every canonical protein and its known PTMs. The second search was a ProSight Biomarker search with MS1 tolerance of 10 ppm, MS2 tolerance of 10 ppm, against the same database. Lastly, a second Absolute Mass search was performed with MS1 tolerance of 1000 Da, MS2 tolerance of 10 ppm, using Delta M mode, against the same database.
False discovery rates (FDR) were estimated as described previously (Kellie et al., 2012). Briefly, data were searched using scrambled protein sequences as decoys using identical strategies as above (absolute and biomarker modes). Logarithmic P-score distributions of decoy protein hits were analyzed for each search mode (absolute and biomarker) separately. Area under score distributions were calculated to reach 5% of total distribution starting from the best score (highest –log P), thus giving P-score cutoffs at 5% FDR for each search strategy. Proteins that had greater P-scores were removed from identification files.
UniProt accession numbers from each ovarian tissue technical replicate were combined and exported to UniProt “Retrieve/ID mapping” tool to recover files with accession numbers, Gene names and protein names (Supplementary Data 1). Venn diagrams were then generated by entering the UniProt combined accession number of each region into the University of Gent Venn diagram Webtool. The mass spectrometry top-down proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Vizcaíno et al., 2016) partner repository with the dataset identifier PXD005420.
2.11. Subnetwork Enrichment Pathway Analyses and Statistical Testing
The gene names of identified proteins were used as input to retrieve a network from STRING (Szklarczyk et al., 2015). The Elsevier's Pathway Studio version 10.0 (Ariadne Genomics, Elsevier) was used to deduce relationships among differentially expressed proteomics protein candidates using the Ariadne ResNet database (Bonnet et al., 2009, Yuryev et al., 2009). “Subnetwork Enrichment Analysis” (SNEA) algorithm was selected to extract statistically significant altered biological and functional pathways pertaining to each identified set of protein hits among the different groups. SNEA utilizes Fisher's statistical test set to determine if there are nonrandom associations between two categorical variables organized by specific relationship. Integrated Venn diagram analysis was performed using “the InteractiVenn”: a web-based tool for the analysis of complex data sets (Heberle et al., 2015).
2.12. Alternative Protein Validation
To validate the altprot product identified by top-down proteomics, one of the identified altprots was selected and cloned in the context of its reference protein. The plasmid contained the canonical G protein nucleolar 1 (GNL1) coding sequence with a C-terminal FLAG tag and the AltGNL1 coding sequence nested within the GNL1 coding sequence, but in a frameshifted ORF with a C-terminal V5 tag. The DNA sequence was built using Gblocks which were assembled using the Gibson assembly (Gibson et al., 2009) protocol using the NEBuilder HiFi DNA Assembly Cloning Kit (New England BioLabs, Ipswich, Massachusetts, USA) according to manufacturer's recommendation into a pcDNA 3.1-expression vector (Invitrogen, Carlsbad, California, USA). GNL1 and AltGNL1 co-expression was validated by western blot in HEK 293 cells transfected with polyethylenimine (PEI, Sigma) as transfection reagent (Hsu and Uludağ, 2012). Briefly, cells were grown in complete Dulbecco's Modified Eagle's Medium (DMEM, Wisent, St-Bruno, Québec, Canada) into a 6-well plate until 70–80% confluent. 1.6 μg plasmidic DNA was mixed into 80 μL of serum-free DMEM and 8 μL of 0.1% (w/v) PEI. The mixture was then gently mixed and let stand at room temperature for 10 min. 480 μL of complete media was then added to the mixture immediately and dropwise onto cells without media renewal. After 24 h of transfection, the cells were washed twice with phosphate-buffered saline (PBS) and lysed using 4% SDS. The lysate was sonicated and centrifuged and the protein content was estimated using Bicinchoninic Acid (BCA) assay (Thermo Scientific). 100 μg of protein was denatured and loaded onto a 15% SDS-PAGE gel. After migration, the proteins were transferred onto a polyvinyldiene difluoride membrane. The membrane was then blocked using 2.5% milk–supplemented, Tris-buffered saline with tween 20 (TBST, 1/1000 v/v). The membrane was rinsed with PBS and probed with anti-FLAG M2 mouse antibody (F1804, Sigma) and anti-V5 mouse antibody (V8012, Sigma) overnight. The membrane was then washed three times with TBST and rinsed with PBS and probed with anti-mouse horseradish peroxidase antibody (7076S, Cell signaling technology) and visualized.
GNL1 and AltGNL1 co-expression was also validated at the cellular level by immunofluorescence. HeLa cells were transfected with GeneCellIn transfection reagent (BioCellChallenge, Toulon, France). Briefly, 25,000 HeLa cells per well were seeded into a 24-well plate and let to grow in complete DMEM media for 24 h. The cells were then transfected by adding 250 ng of plasmidic DNA into 100 μL of serum-free DMEM media and 1.5 μL of GeneCellIn transfection reagent. The mixture was let at room temperature for 15 min and then added dropwise to cells without media renewal. After 24 h of transfection, the cells were rinsed twice with PBS and fixed with 4% paraformaldehyde for 20 min and rinsed twice again. The cell membranes were permeabilized using 0.15% Triton X-100 for 5 min, then rinsed twice with PBS and then twice with Normal Goat Serum (NGS) blocking buffer for 20 min. Anti-FLAG rabbit antibody (F7425, Sigma) and anti-V5 mouse antibody were then added and cells were incubated overnight at 4 °C. The cells were then rinsed twice with NGS and probed with anti-Mouse 488 antibody (A11017, Invitrogen) and anti-rabbit 568 antibody (A21069, Invitrogen) for an hour. The cells were rinsed twice with PBS and incubated with 4′,6′-diamidino-2-phenylindole (DAPI) for 30 min. The cells were rinsed twice with PBS, mounted on a microscope slide with SlowFade (Thermo Scientific) and sealed. The slides were stored in the dark at 4 °C until observation via confocal microscopy.
3. Results
3.1. Tumor Proteome Microenvironment Investigation
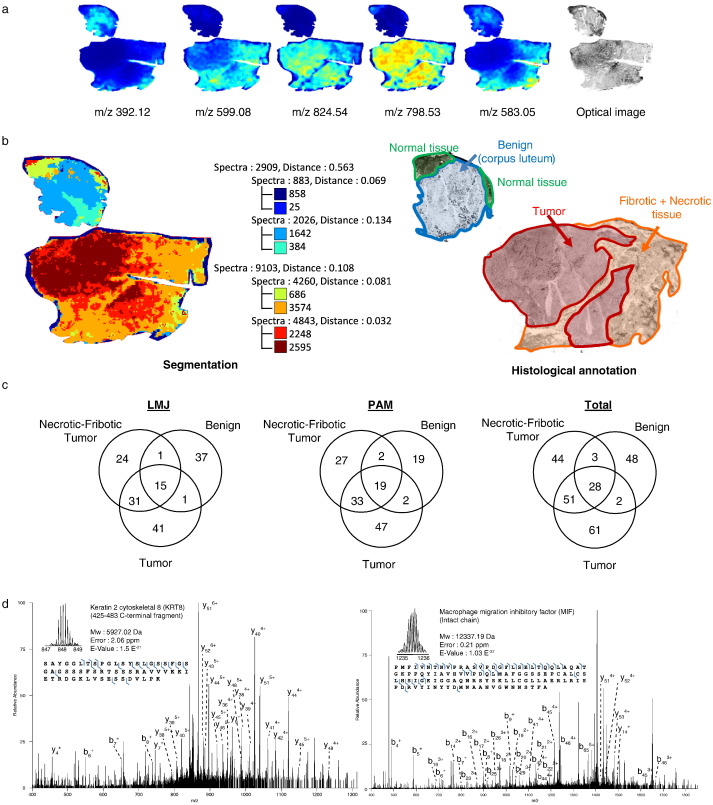
MALDI-MSI was performed on ovarian high grade serous carcinoma sections in order to perform non-supervised spatial segmentation analysis and identification of ROIs (Fig. 1a). The tissue was thus successfully classified by MALDI-MSI with three main clusters (Fig. 1b). One was associated with the benign region (blue-cyan), whereas the two others matched the tumor and necrotic/fibrotic tumor regions (brown-red and orange-yellow, respectively). The presence of the three regions was confirmed by a pathologist (Fig. 1b). A total of 18 samples from the three clustered regions were extracted and analyzed in triplicate employing the two extraction strategies (Fig. 1c). This resulted in the identification of 150 proteins in LMJ and 149 in PAM (Fig. 1c) at an estimated FDR of 5% for reference and “reference plus AltProts” concatenated protein databases (Supplementary Fig. 1a and b, respectively). The distribution using LMJ or PAM is as follows: 41 vs. 47 specific proteins in tumor regions, 24 vs. 27 in necrotic/fibrotic tumor regions, and 37 vs. 19 in benign regions (Fig. 1c). Overall, 61 proteins are specific to the tumor region, 44 to the necrotic/fibrotic tumor region and 48 to the benign region. Thus, 237 proteins were identified by combining the data (see Supplementary Data 1) which is, to our knowledge, the highest number of identified proteins for tissue top-down microproteomics. From the list of identified proteins, some were already known to be involved in ovarian cancer (Table 1) but the ones we identified are mainly fragments of proteins e.g. a fragment of 58 amino acid residues derived from KRT8 (Fig. 1d). Among the identified proteins some are particularly interesting due to the fact that they are found in both tumor and necrotic-fibrotic tumor regions e.g. gamma-synuclein, Lupus la protein (SSB), Nucleophosmin (NPM1), Nuclease-sensitive element-binding protein 1 (YBX1), Probable ATP-dependent RNA helicase DDX17 (DDX17), and Hematological and neurological expressed 1-like protein (JPT2). Others are found specifically in benign and necrotic/fibrotic tumor regions, such as salivary acidic proline-rich phosphoprotein ½ (PRH1). G antigen 7 (GAGE7), High mobility group protein B1 (HMGB1), Glycogen synthase (GYS1), G antigen 2B/2C (GAGE2B), and Cilia- and flagella-associated protein 44 (CFAP44) are only found in the necrotic/fibrotic tumor region.
Fig. 1.
Association of MALDI-MSI and top-down microproteomics. (a) MALDI-MSI of lipids and optical image, (b) histological annotation and segmentation analysis using the Bisecting k-Means and Correlation Distance approach (left). (c) Venn diagram of the top-down gene reference products identified in the ovarian cancer tissue by the LMJ approach, the PAM approach and total, (d) Precursor and HCD fragmentation scan of Keratin 2 cytoskeletal 8 fragment 425–483 and Macrophage migration inhibitory factor (MIF).
Table 1.
List of proteins and potential biomarkers identified within the necrotic/fibrotic tumor and tumor regions with referenced pathological involvement. Here, the identification of the intact or fragmented form is emphasized (see also Supplementary Data 1).
| Uniprot accession number | Protein name | Region | Ref. |
|---|---|---|---|
| P05787 | Cytokeratin 8 (fragment) | Tumor, necrotic/fibrotic tumor | (Wang et al., 2012) |
| P08729 | Cytokeratin 7 (fragment) | Tumor | (Waldemarson et al., 2012) |
| P14174 | Macrophage migration inhibition factor | Tumor, necrotic/fibrotic tumor | (Hagemann et al., 2005) |
| P23528 | Cofilin-1 (fragment) | Tumor | (Li et al., 2013) |
| P31949 | Protein S100-A11 | Tumor, necrotic/fibrotic tumor | (Liu et al., 2015) |
| P46939 | Utrophin (fragment) | Tumor | (Lomnytska et al., 2006) |
| P53985 | Monocarboxylate transporter 1 (fragment) | Necrotic/fibrotic tumor | (Chen et al., 2010) |
| Q12906 | Interleukin enhancer-binding factor 3 (fragment) | Necrotic/fibrotic tumor | (Guo et al., 2012) |
| Q14247 | Src substrate contactin (fragment) | Tumor, necrotic/fibrotic tumor | (Bourguignon et al., 2001) |
| Q86Z02 | Homeodomain-interacting protein kinase 1 | Necrotic/fibrotic tumor | (Kondo et al., 2003) |
| Q8NC51 | Plasminogen activator inhibitor 1 RNA-binding protein (fragment) | Tumor, necrotic/fibrotic tumor | (Koensgen et al., 2007) |
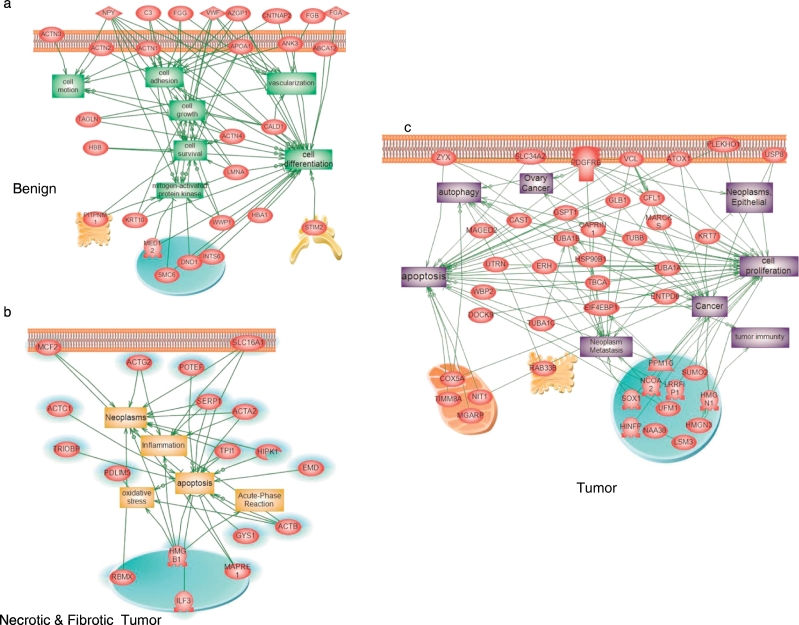
STRING protein analysis of the tumor region associated with GO term analyses led to the identification of two major pathways (RNA binding (GO: 0003727), and poly(A) RNA binding (GO: 0044822). Cellular component GO overrepresentation analysis revealed that the proteins identified in the tumor are mainly found in exosomes - extracellular vesicles (42.3%) and in the nucleus (57.6%). In the necrotic/fibrotic tumor region, 50% of the proteins are found in the extracellular exosomes and in vesicles, 16% are in the nucleus and 34% in various organelles. In benign regions, 50% of the proteins are involved in cellular traffic, 12.3% are cytoplasmic and 27.7% are membrane-bound proteins. Global subnetwork analyses in both tumor, necrotic/fibrotic tumor and benign regions clearly showed differences in protein pathway involvement (Fig. 2). In the benign region, protein pathways are mainly implicated in cell survival, growth, motion, adhesion, differentiation and vascularization (Fig. 2a), whereas in the necrotic/fibrotic tumor region the proteins are mainly implicated in apoptosis, inflammation, neoplasm, acute phase reaction and oxidative stress (Fig. 2b). Tumor subnetwork global analysis showed pathways in neoplasm, autophagy, apoptosis, cell proliferation and tumor immunity (Fig. 2c).
Fig. 2.
Systems biology analysis. Global network identification of the proteins present in benign (a), necrotic-fibrotic tumor (b) and tumor regions (c).
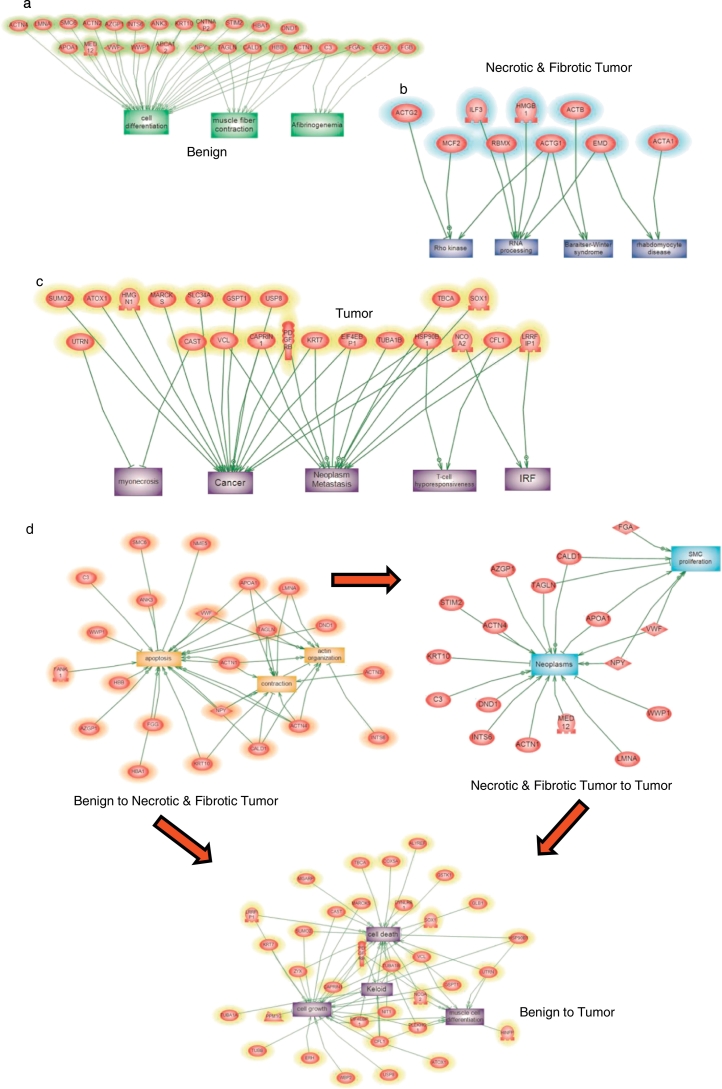
Subnetwork enrichment analysis confirmed the global analysis (Fig. 3). In the benign region, the subnetworks revealed implication in muscle contraction and cell differentiation (Fig. 3a). For necrotic/fibrotic tumor, the subnetworks are involved in Rho Kinase, RNA processing and Rhabdomyocyte disease pathways (Fig. 3b). Tumor subnetworks revealed implication in necrosis, interferon regulatory factor signaling pathway, myonecrosis, and cancer and T cell hypo responsiveness (Fig. 3c). Global network analysis between benign and necrotic/fibrotic tumor regions (Fig. 3d) revealed proteins involved in apoptosis, contraction and actin organization pathways. The same analysis between tumor and necrotic/fibrotic tumor revealed proteins involved in neoplasm and Smooth Muscle Cell (SMC) proliferation pathways. Comparison of proteins from tumor and benign regions showed proteins involved in cell death, cell growth, keloid and muscle cell differentiation.
Fig. 3.
GO Enrichment analysis of benign (a), necrotic/fibrotic tumor (b) and tumor region (c). (d) Global network analysis between benign and necrotic-fibrotic tumor region, between tumor and necrotic-fibrotic tumor, and between benign and tumor showed proteins involved in cell death, cell growth, keloid and muscle cell differentiation.
3.2. Hidden Proteome: Alternative Proteins
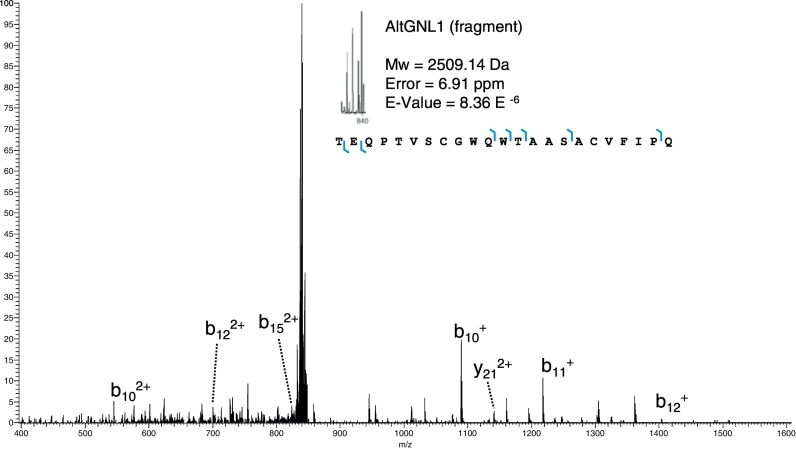
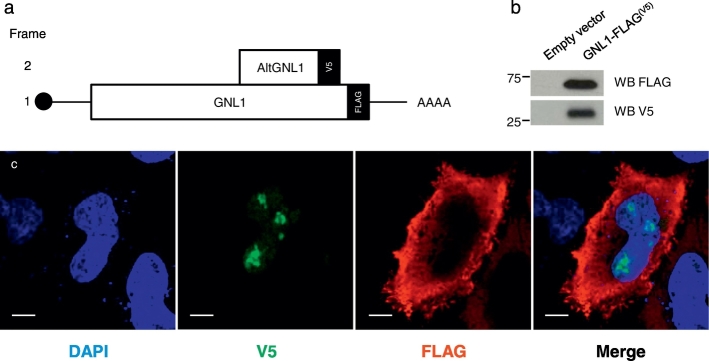
We previously identified 6 AltProts using the shot-gun proteomic approach i.e. AltADCY1, AltCCDC152, AltKART34, AltMOBKL2B, AltPALLD, AltSMCHD1 (Vanderperre et al., 2013). With the top-down microproteomics approach, 15 unknown proteins were identified in patient biopsies including: AltApol6, AltCMBL, AltTLR5, AltPKHD1L1, AltLARS2-AS1, AtltSERPINE1, AltCSNK1A1L, AltGPC5, AltLTB4R, AltTMP1, AltGRAMD4, AltMTHFR, AltAGAP1, AltGNL1 and AltRP11-576E20.1 (Table 2). Six altprots were identified in the benign region (AltTLR5, AltPKHD1L1, AtltSERPINE1, AltGPC5, AltGRAMD4, AltAGAP1), 5 in the necrotic/fibrotic tumor region (AltApol6, AltLARS2-AS1, AltLTB4R, AltTMP1, AltMTHFR) and 4 in the tumor (AltCMBL, AltGNL1, AltRP11-576E20.1, AltCSNK1A1L). The function of these proteins remains unknown. AltGNL1 was selected for further analysis (Fig. 4) based on immunofluorescence data provided by the Human Protein Atlas confirming the presence of its reference protein GNL1 in ovarian cancer tissue. In order to validate the co-expression of GNL1 and non-annotated AltGNL1 proteins from the same gene, we transfected cells with an expression plasmid containing a GNL1-FLAG(V5) construct in HEK 293 cells (Fig. 5). In this construct, the Flag and V5 tags are in-frame with GNL1 and AltGNL1, respectively. Both GNL1FLAG and AltGNL1V5 are expressed and detected with anti-FLAG and anti-V5 antibodies, respectively (Fig. 5a & b). Co-expression at single cell level was confirmed by immunofluorescence (Fig. 5c). AltGNL1 displays a nuclear localization whereas GNL1 is present in the cytosol.
Table 2.
Alternative protein products from ovarian cancer biopsies identified by tissue top-down microproteomics (see also Supplementary Data 1). AltORF localization are either “CDS” for AltORFs nested within the canonical CDS but in a frameshifted ORF, “3′” for AltORF located in the 3′UTR region, CDS-3′ for AltORF overlapped in the CDS and 3′UTR region or “ncRNA” for AltORFs found in putative non-coding transcripts.
Alternative protein products from ovarian cancer biopsies identified by tissue top-down microproteomics. AltORF localization are either “CDS” for AltORFs nested within the canonical CDS but in a frameshifted ORF, “3′” for AltORF located in the 3′UTR region, CDS-3′ for AltORF overlapped in the CDS and 3′UTR region or “ncRNA” for AltORFs found in putative non-coding transcripts.
| E-value (P-score) | Theoretical mass | Gene | Transcript | Tissue | AltORF localization |
|---|---|---|---|---|---|
| 9.34E-06 (3.5E-11) | 3864.85 | APOL6 | NM_030641.3 | LMJ-necrotic-fibrotic tumor | 3′ |
| 5.88E-05 (2.2E-10) | 3802.9 | CMBL | NM_138809.3 | PAM-tumor | 3′ |
| 2.15E-05 (8.04E-11) | 4332.03 | TLR5 | NM_003268.5 | LMJ-benign | CDS |
| 3.91E-06 (1.46E-11) | 2814.3 | PKHD1L1 | NM_177531.4 | PAM-benign | 3′ |
| 8.36E-06 (3.13E-11) | 2509.14 | GNL1 | NM_005275.3 | PAM-tumor | CDS |
| 6.67E-07 (1.25E-15) | 4977.49 | LARS2-AS1 | NR_048543.1 | LMJ-necrotic-fibrotic tumor | ncRNA |
| 7.63E-06 (1.43E-11) | 3564.83 | RP11-576E20.1 | XR_241690.1 | PAM-tumor | ncRNA |
| 4.58E-06 (8.59E-12) | 3943.89 | SERPINE1 | NM_000602.4 | LMJ benign | CDS |
| 1.75E-06 (3.28E-12) | 5429.66 | CSNK1A1L | NM_145203.5 | LMJ tumor | 3′ |
| 6.23E-06 (1.17E-11) | 4832.21 | GPC5 | NM_00446.5.1 | LMJ benign | CDS |
| 6.12E-07 (1.15E-12) | 4873.55 | LTB4R | NM_001143919.2.1 | LMJ necrotic-fibrotic tumor | 3′ |
| 2.95E-06 (5.54E-12) | 3875.90 | TMP1 | XM_005254648.1.1 | LMJ necrotic-fibrotic tumor | 3′ |
| 1.76E-06 (3.30E-12) | 4911.42 | GRAMD4 | XM_005261398.1.1 | LMJ benign | 3′ |
| 3.39E-06 (6.36E-12) | 4852.45 | MTHFR | NM_005957.4 | LMJ necrotic-fibrotic tumor | CDS-3′ |
| 8.57E-06 (1.61E-11) | 4908.42 | AGAP1 | XM_006712240.1 | LMJ benign | 3′ |
Fig. 4.
Precursor and HCD fragmentation scan of Alternative Guanine Nucleotide-binding Protein-like 1 (AltGNL1).
Fig. 5.
Validation of co-expression of reference protein GNL1 and its alternative protein AltGNL1. (a) Schematic representation of the mRNA product from GNL1 (AltGNL1) plasmid used for validation. (b) Western blot showing co-expression of GNL1 (FLAG tagged) and AltGNL1 (V5 tagged) and (c) immunofluorescence assay showing co-expression at cell level with nucleus staining (DAPI, blue) AltGNL1-V5 (green), GNL1 (red) and merge panel. White bars represent 10 μm.
4. Discussion
This work involves the use of tissue microproteomics to characterize the local proteome in three regions (necrotic/fibrotic tumor, tumor and benign region) of human ovarian cancer. These regions were analyzed by MALDI-MSI and discerned by spatial segmentation analysis (Alexandrov et al., 2011, Bonnel et al., 2011, Bruand et al., 2011), and the proteins were microextracted utilizing LMJ and PAM approaches (Franck et al., 2013, Quanico et al., 2014, Quanico et al., 2013, Wisztorski et al., 2016). A total of 237 gene products within the three regions were identified. 61 proteins were specific to the tumor region, 44 to the necrotic/fibrotic tumor region, and 48 to the benign region. The extracted protein profiles from the 3 regions are clearly different and subnetwork analysis revealed a possible progression in the nature of the protein pathways involved in the 3 regions. These results suggest a mechanism in cancer progression from benign to tumor and necrotic/fibrotic tumor regions by a progressive switch in the cell phenotype because we detected proteins common to these regions e.g. SSB, NPM1, YBX1, DDX17, HN1L or PHR1, HMGB1, GYS1, GAGE2B, CFAP44. Utilizing a systems biology approach, pathways implicated in muscle proliferation, cell differentiation, actin, cytoskeleton disorganization, apoptosis, neoplasia, and necrosis with Rho kinase activation are enriched and are likely to be involved in the switch in cell phenotype. In addition, T cell response is observed to be inhibited, leading a tolerant immune response towards the tumor. These results are consistent with spatial segmentation analysis showing that the tumor and necrotic-fibrotic tumor regions had a close histological molecular profile distinct from that of benign regions (see cluster tree, Fig. 1a).
Tissue top-down microproteomics gives insight on the tumor microenvironment with the identification of proteins involved in cancer processes, diagnosis and/or progression (Table 1). For example, the C-terminal fragment (aa425–483) of Cytokeratin-8 (KRT8) has been detected in our experiments in the necrotic/fibrotic tumor and tumor regions (Fig. 1d). KRT8 was previously referenced as a potential biomarker for ovarian cancer (Wang et al., 2012). We demonstrate here that in cancer regions, it is not the complete protein that is present but a C-terminal fragment of 58 amino acid residues. We previously obtained similar results for the C-terminal fragment of the immunoproteasome 11S, PA28 or Reg alpha, a marker for Grade III-IV serous ovarian cancer (Lemaire et al., 2007), as well as for Grade I and tumor relapse (Longuespée et al., 2012). Similarly, a fragment (aa55–72) of Cytokeratin-7 (KRT7) was detected in the tumor region. KRT7 is already a marker for ovarian adenocarcinoma (Chu et al., 2000, Waldemarson et al., 2012), but here we demonstrate that, in fact, the fragment composed of 17 amino acid residues is potentially the actual marker in ovarian tumor. KRT8 and 7 were also reported to be highly expressed in ovarian cancer cell lines (Chu et al., 2000). Protein S100-A11 was detected in the tumor and necrotic/fibrotic tumor regions and was also observed as being particularly highly expressed in ovarian cancer (Liu et al., 2015). The pro-inflammatory cytokine Macrophage migration inhibitory factor (MIF) was detected in the necrotic/fibrotic tumor and tumor regions (Fig. 3d). MIF is already a potential biomarker for ovarian cancer and is associated with tumor growth, metastasis and poor prognosis (Simpson et al., 2012). This protein is also a serum biomarker that distinguishes benign from malignant ovarian tumors in combination with other biomarkers (Agarwal et al., 2007, Krockenberger et al., 2008), and is associated with loss of p53 suppressor activity (Hudson et al., 1999), inhibiting apoptosis and DNA damage repair. Several other proteins already linked to cancer were also identified, including nitrilase-1(Nit1), melanoma antigen family D 2 (MAGED2), Zyxin (ZYX), and ATX1 antioxidant protein 1 homolog (ATOX1). Nit1 is a negative regulator in primary T cells and is classified as a tumor suppressor in association with the fragile histidine-triad protein Fhit (Semba et al., 2006) over-produced in non-small cell lung cancer (NSCLC) and may be a therapeutic target in ovarian cancer (Croce et al., 1999). MAGED2 is also over-expressed in NSCLC (Sienel et al., 2004). Zyxin, a Smad3-mediated TGF-β1 signaling target, regulates cancer cell motility and epithelial-mesenchymal transition during lung cancer development and progression (Beaino et al., 2014, Mise et al., 2012). Interestingly, some proteins identified in the present work have not yet been identified by the Cancer Network Galaxy (TCNG) e.g. Dolichyl-diphosphooligosaccharide-protein glycosyltransferase subunit 4 (OST4), Signal recognition particle receptor subunit alpha (SRPRA), and U6 snRNA-associated Sm-like protein LSm8 (LSM8).
In addition to reference proteins, we identified altprots by top-down microproteomics. 6 altprots were detected in the benign region, 5 in the necrotic/fibrotic tumor region, and 4 in the tumor region. None of these 15 altprots were previously identified. Genes coding for these altprots are annotated as genes coding for receptors (TLR5, LTB4R, AGAP1R), enzymes (CMBL, SERPINE1, MTHF, CSNK1A1L, TMP1), or cytoplasmic or nuclear proteins (Apol6, GRAMD4, GNL1, PKHD1L1). AltLARS2-AS1 and AltRP11-576E20.1 are expressed from genes annotated as non-coding genes, and thus should be re-annotated. We focused our interest on AltProts detected in the cancer region, specifically AltGNL1. Indeed, the reference GNL1 protein was previously detected in ovarian cancer according to the Protein Atlas. None of the other 13 reference proteins were previously identified in proteomic or genomic large-scale studies on ovarian cancer. We validated the co-expression of the reference GNL1 protein with its AltProt AltGNL1 (Fig. 5b–c). Our results clearly demonstrate that both proteins are co-expressed from a single mRNA expressed from a cDNA construct. Immunofluorescence experiments showed that AltGNL1 displays nuclear localization whereas GNL1 is present in the cytosol (Fig. 5c). Our results confirm the presence of a hidden proteome which can constitute a reservoir of potential biomarkers and therapeutic targets.
Taken together, our results show that top-down microproteomics coupled with MALDI MSI can be used to detect proteins expressed from altORFs. These proteins can be used as putative diagnostic biomarkers that may have been missed in conventional proteomics approaches utilizing reference protein databases only. Our approach will be useful to determine the function of altprots in health and disease.
The following are the supplementary data related to this article.
FDR estimation via interrogation of scrambled databases. Area of densities of highest negative logarithm of P-score distributions gave P-score cutoffs for absolute and biomarker search modes at 5% FDR. Results were generated using “reference” (a) and concatenated “reference and AltORFs” protein databases (b). P-score cutoffs were reported on density plots.
List of identified proteins by top-down microproteomics using LMJ and PAM approaches.
Funding Sources
Supported by grants from Région Nord Pas-de-Calais (CM/YB N°2015.2097/12) and PROTEO (FRQNT-RS-188158) (V. Delcourt), University Lille 1 (BQR to Dr. Julien Franck, 2015), Canadian Institutes for Health Research (MOP-136962) and Canada Research Chairs in Functional Proteomics and Discovery of New Protein (Prof. X. Roucou), PRISM (Prof. M. Salzet), Ministère de l'Enseignement Supérieur et de la Recherche via Institut Universitaire de France (ESRS 0900500E) (Prof. I. Fournier), SIRIC ONCOLille (Prof. I. Founier), and Grant INCa-DGOS-Inserm 6041.
Conflict of Interest
The authors declare no competing financial interests in this work.
Author Contributions
MS IF and JF designed the study.
VD, JF, MW and JQ performed all experiments.
JPG and JFJ provided the technical support.
FK performed the subnetwork analyses.
XR has provided the human altORF database.
EL, FN, YMR are clinicians that have provided the biopsies, performed the pathological analyses and the correlation with clinical data.
IF, MS, VD and JF wrote the manuscript.
IF, MS, XR provided funding for this work.
All authors have read and corrected the manuscript.
Contributor Information
Michel Salzet, Email: Michel.salzet@univ-lille1.fr.
Isabelle Fournier, Email: isabelle.fournier@univ-lille1.fr.
References
- Agarwal R., Whang D.H., Alvero A.B., Visintin I., Lai Y., Segal E.A., Schwartz P., Ward D., Rutherford T., Mor G. Macrophage migration inhibitory factor expression in ovarian cancer. Am. J. Obstet. Gynecol. 2007;196:348.e1–348.e5. doi: 10.1016/j.ajog.2006.12.030. [DOI] [PubMed] [Google Scholar]
- Alexandrov T., Meding S., Trede D., Kobarg J.H., Balluff B., Walch A., Thiele H., Maass P. Super-resolution segmentation of imaging mass spectrometry data: solving the issue of low lateral resolution. J. Proteome. 2011;75:237–245. doi: 10.1016/j.jprot.2011.08.002. [DOI] [PubMed] [Google Scholar]
- Beaino W., Guo Y., Chang A.J., Anderson C.J. Roles of Atox1 and p53 in the trafficking of copper-64 to tumor cell nuclei: implications for cancer therapy. J. Biol. Inorg. Chem. 2014;19:427–438. doi: 10.1007/s00775-013-1087-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnel D., Longuespee R., Franck J., Roudbaraki M., Gosset P., Day R., Salzet M., Fournier I. Multivariate analyses for biomarkers hunting and validation through on-tissue bottom-up or in-source decay in MALDI-MSI: application to prostate cancer. Anal. Bioanal. Chem. 2011;401:149–165. doi: 10.1007/s00216-011-5020-5. [DOI] [PubMed] [Google Scholar]
- Bonnet A., Lagarrigue S., Liaubet L., Robert-Granié C., Sancristobal M., Tosser-Klopp G. Pathway results from the chicken data set using GOTM, pathway studio and ingenuity softwares. BMC Proc. 2009;3(Suppl. 4):S11. doi: 10.1186/1753-6561-3-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourguignon L.Y., Zhu H., Shao L., Chen Y.W. CD44 interaction with c-Src kinase promotes cortactin-mediated cytoskeleton function and hyaluronic acid-dependent ovarian tumor cell migration. J. Biol. Chem. 2001;276:7327–7336. doi: 10.1074/jbc.M006498200. [DOI] [PubMed] [Google Scholar]
- Bruand J., Alexandrov T., Sistla S., Wisztorski M., Meriaux C., Becker M., Salzet M., Fournier I., MacAgno E., Bafna V. AMASS: algorithm for MSI analysis by semi-supervised segmentation. J. Proteome Res. 2011;10:4734–4743. doi: 10.1021/pr2005378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Wang L., Beretov J., Hao J., Xiao W., Li Y. Co-expression of CD147/EMMPRIN with monocarboxylate transporters and multiple drug resistance proteins is associated with epithelial ovarian cancer progression. Clin. Exp. Metastasis. 2010;27:557–569. doi: 10.1007/s10585-010-9345-9. [DOI] [PubMed] [Google Scholar]
- Chu P., Wu E., Weiss L.M. Cytokeratin 7 and cytokeratin 20 expression in epithelial neoplasms: a survey of 435 cases. Mod. Pathol. 2000;13:962–972. doi: 10.1038/modpathol.3880175. [DOI] [PubMed] [Google Scholar]
- Croce C.M., Sozzi G., Huebner K. Role of FHIT in human cancer. J. Clin. Oncol. 1999;17:1618–1624. doi: 10.1200/JCO.1999.17.5.1618. [DOI] [PubMed] [Google Scholar]
- Franck J., Quanico J., Wisztorski M., Day R., Salzet M., Fournier I. Quantification-based mass spectrometry imaging of proteins by parafilm assisted microdissection. Anal. Chem. 2013;85:8127–8134. doi: 10.1021/ac4009397. [DOI] [PubMed] [Google Scholar]
- Gibson D.G., Young L., Chuang R.-Y., Venter J.C., Hutchison C. a, Smith H.O., Iii C.A.H., America N. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Guo Y., Fu P., Zhu H., Reed E., Remick S.C., Petros W., Mueller M.D., Yu J.J. Correlations among ERCC1, XPB, UBE2I, EGF, TAL2 and ILF3 revealed by gene signatures of histological subtypes of patients with epithelial ovarian cancer. Oncol. Rep. 2012;27:286–292. doi: 10.3892/or.2011.1483. [DOI] [PubMed] [Google Scholar]
- Hagemann T., Wilson J., Kulbe H., Li N.F., Leinster D.A., Charles K., Klemm F., Pukrop T., Binder C., Balkwill F.R. Macrophages induce invasiveness of epithelial cancer cells via NF-kappa B and JNK. J. Immunol. 2005;175:1197–1205. doi: 10.4049/jimmunol.175.2.1197. [DOI] [PubMed] [Google Scholar]
- Heberle H., Meirelles G.V., da Silva F.R., Telles G.P., Minghim R. InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics. 2015;16:169. doi: 10.1186/s12859-015-0611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C.Y.M., Uludağ H. A simple and rapid nonviral approach to efficiently transfect primary tissue-derived cells using polyethylenimine. Nat. Protoc. 2012;7:935–945. doi: 10.1038/nprot.2012.038. [DOI] [PubMed] [Google Scholar]
- Hudson J.D., Shoaibi M.A., Maestro R., Carnero A., Hannon G.J., Beach D.H. A proinflammatory cytokine inhibits p53 tumor suppressor activity. J. Exp. Med. 1999;190:1375–1382. doi: 10.1084/jem.190.10.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellie J.F., Tran J.C., Lee J.E., Ahlf D.R., Thomas H.M., Ntai I., Catherman A.D., Durbin K.R., Zamdborg L., Vellaichamy A., Thomas P.M., Kelleher N.L. The emerging process of top down mass spectrometry for protein analysis: biomarkers, protein-therapeutics, and achieving high throughput. Mol. BioSyst. 2010;6:1532–1539. doi: 10.1039/c000896f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellie J.F., Catherman A.D., Durbin K.R., Tran J.C., Tipton J.D., Norris J.L., Witkowski C.E., Thomas P.M., Kelleher N.L. Robust analysis of the yeast proteome under 50 kDa by molecular-mass-based fractionation and top-down mass spectrometry. Anal. Chem. 2012;84:209–215. doi: 10.1021/ac202384v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koensgen D., Mustea A., Klaman I., Sun P., Zafrakas M., Lichtenegger W., Denkert C., Dahl E., Sehouli J. Expression analysis and RNA localization of PAI-RBP1 (SERBP1) in epithelial ovarian cancer: association with tumor progression. Gynecol. Oncol. 2007;107:266–273. doi: 10.1016/j.ygyno.2007.06.023. [DOI] [PubMed] [Google Scholar]
- Kondo S., Lu Y., Debbas M., Lin A.W., Sarosi I., Itie A., Wakeham A., Tuan J., Saris C., Elliott G., Ma W., Benchimol S., Lowe S.W., Mak T.W., Thukral S.K. Characterization of cells and gene-targeted mice deficient for the p53-binding kinase homeodomain-interacting protein kinase 1 (HIPK1) Proc. Natl. Acad. Sci. U. S. A. 2003;100:5431–5436. doi: 10.1073/pnas.0530308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krockenberger M., Dombrowski Y., Weidler C., Ossadnik M., Hönig A., Häusler S., Voigt H., Becker J.C., Leng L., Steinle A., Weller M., Bucala R., Dietl J., Wischhusen J. Macrophage migration inhibitory factor contributes to the immune escape of ovarian cancer by down-regulating NKG2D. J. Immunol. 2008;180:7338–7348. doi: 10.4049/jimmunol.180.11.7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire R., Menguellet S.A., Stauber J., Marchaudon V., Lucot J.P., Collinet P., Farine M.O., Vinatier D., Day R., Ducoroy P., Salzet M., Fournier I. Specific MALDI imaging and profiling for biomarker hunting and validation: fragment of the 11S proteasome activator complex, reg alpha fragment, is a new potential ovary cancer biomarker. J. Proteome Res. 2007;6:4127–4134. doi: 10.1021/pr0702722. [DOI] [PubMed] [Google Scholar]
- Li M., Yin J., Mao N., Pan L. Upregulation of phosphorylated cofilin 1 correlates with taxol resistance in human ovarian cancer in vitro and in vivo. Oncol. Rep. 2013;29:58–66. doi: 10.3892/or.2012.2078. [DOI] [PubMed] [Google Scholar]
- Liu Y., Han X., Gao B. Knockdown of S100A11 expression suppresses ovarian cancer cell growth and invasion. Exp. Ther. Med. 2015;9:1460–1464. doi: 10.3892/etm.2015.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomnytska M., Dubrovska A., Hellman U., Volodko N., Souchelnytskyi S. Increased expression of cSHMT, Tbx3 and utrophin in plasma of ovarian and breast cancer patients. Int. J. Cancer. 2006;118:412–421. doi: 10.1002/ijc.21332. [DOI] [PubMed] [Google Scholar]
- Longuespée R., Boyon C., Castellier C., Jacquet A., Desmons A., Kerdraon O., Vinatier D., Fournier I., Day R., Salzet M. The C-terminal fragment of the immunoproteasome PA28S (Reg alpha) as an early diagnosis and tumor-relapse biomarker: evidence from mass spectrometry profiling. Histochem. Cell Biol. 2012;138:141–154. doi: 10.1007/s00418-012-0953-0. [DOI] [PubMed] [Google Scholar]
- Mise N., Savai R., Yu H., Schwarz J., Kaminski N., Eickelberg O. Zyxin is a transforming growth factor-β (TGF-β)/Smad3 target gene that regulates lung cancer cell motility via integrin α5β1. J. Biol. Chem. 2012;287:31393–31405. doi: 10.1074/jbc.M112.357624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouilleron H., Delcourt V., Roucou X. Death of a dogma: eukaryotic mRNAs can code for more than one protein. Nucleic Acids Res. 2016;44:14–23. doi: 10.1093/nar/gkv1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quanico J., Franck J., Dauly C., Strupat K., Dupuy J., Day R., Salzet M., Fournier I., Wisztorski M. Development of liquid microjunction extraction strategy for improving protein identification from tissue sections. J. Proteome. 2013;79:200–218. doi: 10.1016/j.jprot.2012.11.025. [DOI] [PubMed] [Google Scholar]
- Quanico J., Franck J., Gimeno J.P., Sabbagh R., Salzet M., Day R., Fournier I. Parafilm-assisted microdissection: a sampling method for mass spectrometry-based identification of differentially expressed prostate cancer protein biomarkers. Chem. Commun. (Camb.) 2014;51:4564–4567. doi: 10.1039/c4cc08331h. [DOI] [PubMed] [Google Scholar]
- Semba S., Han S.-Y., Qin H.R., McCorkell K.A., Iliopoulos D., Pekarsky Y., Druck T., Trapasso F., Croce C.M., Huebner K. Biological functions of mammalian Nit1, the counterpart of the invertebrate NitFhit Rosetta stone protein, a possible tumor suppressor. J. Biol. Chem. 2006;281:28244–28253. doi: 10.1074/jbc.M603590200. [DOI] [PubMed] [Google Scholar]
- Sienel W., Varwerk C., Linder A., Kaiser D., Teschner M., Delire M., Stamatis G., Passlick B. Melanoma associated antigen (MAGE)-A3 expression in stages I and II non-small cell lung cancer: results of a multi-center study. Eur. J. Cardiothorac. Surg. 2004;25:131–134. doi: 10.1016/j.ejcts.2003.09.015. [DOI] [PubMed] [Google Scholar]
- Simpson K.D., Templeton D.J., Cross J.V. Macrophage migration inhibitory factor promotes tumor growth and metastasis by inducing myeloid-derived suppressor cells in the tumor microenvironment. J. Immunol. 2012;189:5533–5540. doi: 10.4049/jimmunol.1201161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P., Kuhn M., Bork P., Jensen L.J., von Mering C. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran J.C., Zamdborg L., Ahlf D.R., Lee J.E., Catherman A.D., Durbin K.R., Tipton J.D., Vellaichamy A., Kellie J.F., Li M., Wu C., Sweet S.M.M., Early B.P., Siuti N., LeDuc R.D., Compton P.D., Thomas P.M., Kelleher N.L. Mapping intact protein isoforms in discovery mode using top-down proteomics. Nature. 2011;480:254–258. doi: 10.1038/nature10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderperre B., Lucier J.-F., Bissonnette C., Motard J., Tremblay G., Vanderperre S., Wisztorski M., Salzet M., Boisvert F.-M., Roucou X. Direct detection of alternative open reading frames translation products in human significantly expands the proteome. PLoS One. 2013;8:e70698. doi: 10.1371/journal.pone.0070698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno J.A., Csordas A., del-Toro N., Dianes J.A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q.-W., Wang R., Hermjakob H. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44 doi: 10.1093/nar/gkw880. 11033-11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldemarson S., Krogh M., Alaiya A., Kirik U., Schedvins K., Auer G., Hansson K.M., Ossola R., Aebersold R., Lee H., Malmström J., James P. Protein expression changes in ovarian cancer during the transition from benign to malignant. J. Proteome Res. 2012;11:2876–2889. doi: 10.1021/pr201258q. [DOI] [PubMed] [Google Scholar]
- Wang L.N., Tong S.W., Hu H.D., Ye F., Li S.L., Ren H., Zhang D.Z., Xiang R., Yang Y.X. Quantitative proteome analysis of ovarian cancer tissues using a iTRAQ approach. J. Cell. Biochem. 2012;113:3762–3772. doi: 10.1002/jcb.24250. [DOI] [PubMed] [Google Scholar]
- Wisztorski M., Desmons A., Quanico J., Fatou B., Gimeno J.P., Franck J., Salzet M., Fournier I. Spatially-resolved protein surface microsampling from tissue sections using liquid extraction surface analysis. Proteomics. 2016;16:1622–1632. doi: 10.1002/pmic.201500508. [DOI] [PubMed] [Google Scholar]
- Ye H., Mandal R., Catherman A., Thomas P.M., Kelleher N.L., Ikonomidou C., Li L. Top-down proteomics with mass spectrometry imaging: a pilot study towards discovery of biomarkers for neurodevelopmental disorders. PLoS One. 2014;9 doi: 10.1371/journal.pone.0092831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuryev A., Kotelnikova E., Daraselia N. Ariadne's ChemEffect and pathway studio knowledge base. Expert Opin. Drug Discovery. 2009;4:1307–1318. doi: 10.1517/17460440903413488. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FDR estimation via interrogation of scrambled databases. Area of densities of highest negative logarithm of P-score distributions gave P-score cutoffs for absolute and biomarker search modes at 5% FDR. Results were generated using “reference” (a) and concatenated “reference and AltORFs” protein databases (b). P-score cutoffs were reported on density plots.
List of identified proteins by top-down microproteomics using LMJ and PAM approaches.