Abstract
Epigenetic mechanisms play a key role in gastrointestinal cancer (GIC) development and progression, and most studies have been focused on aberrant DNA methylation and histone modifying enzymes. However, the histone H3–H4 chaperone ASF1A is an important factor regulating chromatin assembling and gene transcription, while it is currently unclear whether ASF1A is involved in cancer pathogenesis. The present study is thus designed to address this issue. Here we showed that ASF1A expression was widespread in GIC-derived cell lines and up-regulated in primary GIC. Higher levels of ASF1A expression predicted significantly shorter patient overall survival in colorectal cancer (P = 0.0012). The further analyses of the GEO dataset validate higher ASF1A expression as a prognostic factor for CRC patients. Mechanistically, ASF1A interacted with β-catenin and promoted the transcription of β-catenin target genes including c-MYC, cyclin D1, ZEB1 and LGR5, thereby stimulating proliferation, stemness and migration/invasion of GIC cells. β-Catenin inhibition abolished these effects of ASF1A. Moreover, the ASF1A-β-catenin-ZEB1 axis down-regulated E-Cadherin expression, thereby contributing to enhanced migration/invasion of GIC cells. ASF1A over-expression and depletion facilitated and inhibited in vivo tumor growth and/or metastasis in mouse xenograft models, respectively. Taken together, ASF1A is aberrantly over-expressed in GIC tumors and plays key roles in GIC development and progression by stimulating the transcription of β-catenin target genes. ASF1A may thus be a novel target for GIC therapy and a potential prognostic marker.
Abbreviations: ChIP, chromatin immunoprecipitation; CRC, colorectal cancer; GC, gastric cancer; GIC, gastrointestinal cancer; IHC, immunohistochemistry; IP, immunoprecipitation; qPCR, quantitative PCR
Keywords: ASF1A, β-Catenin, Gastrointestinal cancer, Histone modification, ZEB1
Graphical Abstract
Highlights
-
•
ASF1A is over-expressed in GIC and predicts poor patient outcomes.
-
•
ASF1A interacts with β-catenin, facilitates the expression of its targets and promotes stemness and invasion of GIC cells.
-
•
Targeting ASF1A could be a novel epigenetic strategy against GIC.
Gastrointestinal cancer (GIC) is the leading cause of cancer-related death worldwide and invasion or distant metastasis account for the majority of mortalities due to the limited treatment choices. We determined the effect of ASF1A, the histone H3–H4 chaperone, on GIC pathogenesis. We found that ASF1A expression was up-regulated in primary GIC and higher ASF1A levels predicted significantly shorter patient overall survival in colorectal cancer. ASF1A interacted with β-catenin and promoted the transcription of β-catenin target genes, thereby stimulating proliferation, stemness and migration/invasion of GIC cells. ASF1A may thus be a novel target for GIC therapy and a useful prognostic marker.
1. Introduction
Gastrointestinal cancer (GIC) is one of the most common malignancies and the leading cause of cancer-related death worldwide. An estimated 1,157,000 new cases of GIC with 798,500 deaths occurred in China in 2015 (Chen et al., 2016), whereas in the United States, although gastric cancer (GC) incidence is relatively low, colorectal cancer (CRC) is ranked among the top three, with 132,700 new cases and 49,700 deaths in 2015 (Schreuders et al., 2015). Early diagnosis and intervention is the key to patient cure or long-term survival. However, most GIC patients are diagnosed at an advanced stage, and invasion or distant metastasis account for the majority of mortalities due to the limited treatment choices (Schreuders et al., 2015). Therefore, better defining of GIC pathogenesis, elucidating the mechanism(s) underlying metastasis, identifying reliable diagnostic and prognostic biomarkers and exploring novel therapeutic targets are urgently demanding tasks.
Recent genomic landscape analyses have identified a spectrum of recurrent somatic alterations driving GIC formation and progression, and significantly broadened our understanding of the disease (Vogelstein et al., 2013). However, the pathogenesis of GIC involves not only the accumulation of genetic alterations, but also the disruption of epigenetic modifications. Especially in CRC, epigenetic aberrations are even much more frequent than genetic alterations (Okugawa et al., 2015, Vogelstein et al., 2013). These epigenetic alterations include mutations or dysregulation of epigenetic modifiers that are responsible for DNA methylation or post-translational histone modifications such as methylations or acetylations (Okugawa et al., 2015, Vogelstein et al., 2013). The contribution of aberrant DNA methylation to GIC development has long been established (Okugawa et al., 2015), whereas recent studies have also shed light on the important role of dysregulated histone modifying enzymes in GIC biology. For instance, up-regulation of histone deacetylases while down-regulation of histone acetyltransferases is widespread in GIC (Kramer, 2009, Ozdag et al., 2006, Song et al., 2005); the histone demethylase RBP2 is frequently over-expressed in GC and promotes cancer cell proliferation and invasion (Zeng et al., 2010). In addition, altered expression and activities of JMJD2B, EZH2 and other histone modifying enzymes have been observed in GIC tumors, too (Crea et al., 2012, Kim and Roberts, 2016, Li et al., 2011, Zhao et al., 2014). Consistent with these alterations, there exist widespread aberrations of histone modification profiles in GIC, which has been shown to be closely associated with disease prognosis and treatment efficacy (Benard et al., 2015, Goossens-Beumer et al., 2014, Paul et al., 2015, Saldanha et al., 2014, Tamagawa et al., 2013, Wei et al., 2016).
Despite extensive studies of histone modifying enzymes dysregulated in GIC, little has been known about the role of histone variants and their chaperones in GIC, while recent studies indicate that they are emerging as key regulatory molecules in cancer (Vardabasso et al., 2014, Zuber et al., 2011). ASF1 is the histone H3–H4 chaperone that regulates chromatin assembly in both replication-dependent and replication-independent manners, and encompasses the isoforms ASF1A and 1B (Mousson et al., 2007, Richet et al., 2015). Both ASF1A and 1B are involved in nucleosome assembling, DNA replication, DNA damage repair and gene transcription (Das et al., 2009, Mousson et al., 2007), while they also have different functions. ASF1A, but not ASF1B, is required for histone H3-K56 acetylation (Das et al., 2009, Das et al., 2014), whereas the H3-K56 acetylation state plays a key role in enabling rapid transcriptional changes. ASF1B was previously shown to stimulate breast cancer proliferation and predict shorter patient survival (Corpet et al., 2011), however, little is known about the relationship between ASF1A and oncogenesis. To address this issue, we determined the potential role of ASF1A in GIC tumors. Our present results revealed multiple effects of ASF1A on GIC by stimulating the transcription of β-catenin target genes, demonstrating its potential value in predicting GIC prognosis and identifying new treatment targets.
2. Materials and Methods
2.1. Cell Culture
Gastric cancer cell lines (AGS, BGC-823 and HGC-27) and colorectal cancer cell lines (HCT116, SW480, Caco2 and HT29) were cultured in RPMI-1640 medium (Thermo Fisher Scientific, Waltham, MA) supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher Scientific), 100 U/ml penicillin, 100 μg/ml streptomycin and 4 mM l-glutamine.
2.2. Patients
Two hundreds and eighty-six of patients with GIC (106 with gastric cancer, 90 with colon cancer and 90 with rectal cancer) who underwent surgical operation at Qilu Hospital and Second Hospital of Shandong University between 2007 and 2008 were included in the study. The tumor specimens were collected after surgery and paraffin-embedded. The study was approved by the Shandong University Second Hospital ethics committees.
2.3. ASF1A mRNA Expression in Progression and Prognosis of CRC and Correlation With c-MYC, ZEB1 and Cyclin D1 Expression Derived From GEO Datasets
The value of ASF1A mRNA expression in the prognosis of CRC patient survival was determined using GEO datasets (http://www.prognoscan.org/). The data were from GSE17536 and GSE17537 and the cut-offs of ASF mRNA levels (higher and lower) defined by the database were 0.86 and 0.76, respectively.
The data set for 32 colorectal adenoma and 32 normal mucosa samples were obtained from GEO (GSE8671) and analyzed online through GEO2R. The level of ASF1A mRNA was downloaded and the graph was constructed accordingly. With respect to progressive colorectal diseases, from adenoma (17 samples) to carcinoma (17 samples) and carcinoma with metastasis (11 samples), the data were obtained from GEO (GSE77953) and ASF1A mRNA levels were downloaded and analyzed. For the correlation analysis of gene expression, the data were obtained from GEO (GSE75315) including 211 primary CRC samples. GEO2R was used to analyze the data and mRNA data of ASF1A, c-MYC and CCND1 in those 211 CRC tumors were downloaded.
2.4. SiRNA Transfection
ASF1A, β-catenin and ZEB1 Stealth Interference RNAs (Thermo Fisher Scientific) were transfected into cells with Lipofactamine2000 (Thermo Fisher Scientific) according to the protocol provided. Sequences for these siRNAs are listed in Table 1.
Table 1.
Oligo sequences used in the present study.
| Primers for qRT-PCR |
| ASF1A |
| Forward: 5′-CAGATGCAGATGCAGTAGGC-3′ |
| Reverse: 5′-CCTGGGATTAGATGCCAAAA-3′ |
| E-Cadherin |
| Forward: 5′-TTCCTCCCAATACATCTCCC-3′ |
| Reverse: 5′-TTGATTTTGTAGTCACCCACC-3 |
| ZEB1 |
| Forward: 5′-AGCAGTGAAAGAGAAGGGAATGC-3′ |
| Reverse: 5′-GGTCCTCTTCAGGTGCCTCAG-3′ |
| MYC |
| Forward: 5′-5′-TACCCTCTCAACGACAGCAGCTCGCCCAACTCCT-3′ |
| Reverse: 5′-TCTTGACATTCTCCTCGGTGTCCGAGGACCT-3′ |
| CCND1 |
| Forward: 5′-GCC GAG AAG CTG TGC ATC T-3′ |
| Reverse: 5′-CTC CTC CGC CTC TGG CAT T-3′ |
| β2-M |
| Forward: 5′-GAA TTG CTA TGT GTC TGG GT-3′ |
| Reverse: 5′-CAT CTT CAA ACC TCC ATG ATG-3′ |
| Primers for ChIP |
| ZEB1 |
| Forward: 5′-TGGAAGGGAAGGGAAGGGAGTC-3′ |
| Reverse: 5′-AGGCAGGGCTACCATCAGTC-3′ |
| MYC |
| Forward: 5′-CAA CTT TGA ACA ATG AGC AC-3′ |
| Reverse: 5′-CCA GAA ACA TGA GTC TTT T-3′ |
| CCND1 |
| Forward: 5′-CGC TCC CAT TCT CTG CCG G-3′ |
| Reverse: 5′-GGG GCT CTT CCT GGG CAG C-3′ |
| GAPDH |
| Forward: 5′-AAA GGG CCC TGA CAA CTC TT 3′ |
| Reverse: 5′-GGT GGT CCA GGG GTC TTA CT 3′ |
| siRNAs |
| ASF1A1 (A1): 5′-UGA CUG UAG AUU UGG AUU ACU GCU C-3′ |
| ASF1A1 (A2): 5′-UGA ACA AUG UAG UGG UGC UGG AUA-3′ |
| ZEB1: 5′-GCU GAG AAG CCU GAG UCC UCU GUU U-3′ |
| β-Catenin: 5′-CCA CAG CUC CUU CUC UGA GUG GUA A-3′ |
| Control: 5′-CCU ACA UCC CGA UCG AUG AUG UUG A-3′ |
2.5. Lenti-Viral Infection
HEK293T cells were cultured at 90% cell confluence in 75 cm3 flasks and transfected with 15 μg viral vector, 10.5 μg psPAX2 vector and 6 μg pMD2.G plasmid using polyethylenimine method. Supernatant was collected 48–72 h post-infection and filtered through 45-mm pore size filters. The virus was concentrated with PEG-it (System Biosciences, Palo Alto, CA). 5 ml of viral supernatant were then added to infect 25,000 cells in the presence of 4 μg/ml polybrene. HT29 cells expressing ectopic ASF1A were isolated using FACS sorting, while stable ASF1A knocked down HCT116 cells were selected using 5 μg/ml puromycine.
2.6. RNA Extraction, Reverse Transcription and Quantitative PCR (qPCR)
Total RNA was extracted with Trizol-Reagent (Thermo Fisher Scientific). RNAs were reversely transcribed using the kit from (Thermo Fisher Scientific). qPCR was carried out in TaqmMan ABI PRISM 7900 HT using SYBR Green kit (Thermo Fisher Scientific). Levels of target mRNA were calculated based on the ΔCT values and normalization of human β2-M expression. Primers used in this study are in Table 1.
2.7. Clonogenic Assay
Cells with different treatments (1000 cells/well) were seeded in 6-well plates for 2 week incubation. Cells were then fixed and stained with Giemsa. Colonies (> 50 cells) were counted.
2.8. Mono-Spheroid Formation Assay
Cells (2000/well) were cultured in ultra-low-attachment 96-well plates (Corning Life Sciences) with 100 μl RPMI-1640/10 mM HEPES serum-free medium supplemented with cocktails of following growth factors: 10 ng/ml bFGF (PeproTech Nordic, Stockholm, Sweden) and 20 ng/ml EGF (PeproTech Nordic). Fresh medium was supplemented every 3 days. Fifteen days later, the spheroid colonies were examined under light microscopy and counted.
2.9. Flow Cytometry
For cell cycle analyses, cells were fixed with 70% ethanol at + 4 °C overnight and stained with RNAse A (0.5 μg)-containing Propidium Iodide (50 μg/ml). Cell cycle distribution was determined using flow cytometry with ModFit (BD Biosciences, Franklin Lakes, NJ). For Lrg5 staining, cells were washed with PBS followed by blocking buffer treatment. Cells were incubated with primary antibodies against LGR5 (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany, Cat# 130-100-856) for 1 h at room temperature, then with secondary antibodies for half an hour at room temperature, and analyzed.
2.10. Migration and Invasion Assays
Cells (5 × 104 for HGC-27, HCT116 and SW480 cells and 1 × 105 for HT29 cells) were seeded into the upper chamber. The low chamber contained RPMI-1640 medium with 20% FBS. Forty-eight hours later, the migrated cells were stained with crystal violet, counted and photographed. For invasion assay, 50 μl matrigel (Corning Life Sciences, Flintshire, UK) was first loaded into the bottom of the upper chamber followed by the identical method as described above.
2.11. Western Blot
Proteins were extracted from cells with RIPA Lysis Buffer (Thermo Fisher Scientific). Samples were run on SDS-PAGE and transferred to the PVDF membrane (Bio-Rad, Hercules, California). The membrane was blocked with 5% non-fat milk for 2 h at room temperature. Primary antibodies of ASF1A (Cell Signaling Technology, Danvers, MA, Cat# 2990), β-actin (Santa Cruz Biotechnology, Santa Cruz, CA, Cat# sc-1615), ZEB1 (Novus Biologicals, USA), β-catenin (Cell Signaling Technology, Cat# 8480), E-cadherin (Cell Signaling Technology, Cat# 3195 s), c-Myc (Santa Cruz Biotechnology, Cat# sc-789) and cyclin D1 (Cell Signaling Technology, Cat# 92G2) were incubated with the membrane at 4 °C overnight. Secondary antibodies were incubated for 1 h at room temperature. Antibody binding was detected using chemiluminescence method (Thermo Fisher Scientific).
2.12. Immunoprecipitation (IP)
Pierce™ Crosslink Magnetic IP/Co-IP Kit (Thermo Fisher Scientific) was used. In brief, IP antibodies (3.0 μg) were incubated with 50 μg protein extracts overnight at + 4 °C, and then bound to Protein A/G Magnetic Beads and covalently crosslinked to the beads using disuccinmidyl suberate. The antibody-crosslinked beads were then incubated with cell lysate that contained the protein of interest. Non-bound materials were removed by washing the beads. A low pH elution buffer was used to remove bound antigen from the antibody-crosslinked beads followed by neutralization buffer washing to prevent precipitation of the isolated antigen. The precipitated material was finally re-suspended in Lane Marker Sample Buffer for SDS-PAGE analysis.
2.13. Immunofluorescence (IF)
Cells grown on coverslips were washed with PBS and non-specific antigen binding blocked with blocking buffer. Triton-100 was used to increase the permeability of cell and nuclear membrane. Primary antibodies of ASF1A (Cell Signaling cyclin D1 (Cell Signaling Technology, Cat# 2990), F-actin (Thermo Fisher Scientific, Cat# A12381), ZEB1 (Novus Biologicals, Littleton, CO, Cat# NBP1-05987), H3K56Ac (Active Motif, Carlsbad, CA, Cat# 39281), and E-cadherin (Cell Signaling cyclin D1 (Cell Signaling Technology, Cat# 3195s) were added to bind the antigens of interest followed by secondary antibody incubation and DAPI mounting.
2.14. Chromatin Immunoprecipitation (ChIP)
SimpleChIP® Enzymatic Chromatin IP Kit (Magnetic Beads) #9003 (Cell Signaling Technology) was used according to the protocol provided. In brief, HCT116 cells transfected with ASF1A siRNA were harvested at 48 h and then crosslinked with formaldehyde. Chromatin digestion was performed with Micrococcal Nuclease and analyzed by agarose gel. DNA concentration was assessed by Nanodrop (Thermo Fisher Scientific). The antibodies against ASF1A (Abcam, Cambridge, UK, Cat# AB190747) and β-catenin (Cell Signaling Technology, Cat# 8480, negative (IgG) and positive control (histone H3) antibodies were added into the digested samples and incubated overnight at 4 °C with rotation. Protein G Magnetic beads were used to precipitate the DNA-antigen-antibody complex followed by the elution of chromatin from Antibody/Protein G Magnetic Beads and reversal of cross-links. Spin columns were used to purify DNA and the collected DNA was amplified using PCR with specific primers (Table 1).
2.15. Assessment of TCF/LEF Reporter and E-Cadherin Promoter Activity
TCF/LEF reporter activity assay kit was purchased from QIAGEN (QIAGEN, Hilden, Germany). HCT116 cells were transfected with β-catenin and/or ASF1A expression vectors and incubated for 24 h followed by TCF/LEF reporter transfection. Cells were then harvested and TCF/LEF-driven Luciferase activity was determined using a dual luciferase reporter assay system (Promega, Madison, WI). The target promoter-driven firefly luciferase activity was normalized to the renilla activity included in the kit.
WT E-cadherin promoter and E-box-mutated (E-boxes 1 and 3) E-cadherin promoter were purchased from addgene (Middlesex, UK). HCT116 cells were transfected with ASF1A siRNA and incubated for 48 h followed by co-transfection of E-cadherin promoter constructs and pRL-TK (reference). Luciferase activity was detected as described above.
2.16. Immunohistochemistry (IHC)
Paraffin embedded slides were deparaffinized and rehydrated followed by antigen-retrieval using citric acid buffer. Endogenous peroxidase was deactivated by H2O2. Slides were blocked using 10% goat serum and incubated with the corresponding primary antibodies overnight at 4 °C. After incubation with secondary antibodies for 45 mins at room temperature, DAB staining (Thermo Fisher Scientific) was used to detect the antigen-antibody binding. The primary antibodies used were: ASF1A (Cell Signaling Technology, Cat# 2990), PCNA (Santa Cruz Biotechnology, Cat# sc-7907), and E-cadherin (Cell Signaling Technology, Cat# 3195s). The slides were examined by two of the co-authors (XL and DX) and mean values of ASF1A positive cells were presented based on the results from two observers. For each slide, a total of 200 cells in two fields were analyzed.
2.17. Subcutaneous Tumor Formation and Metastasis of CRC Cells in the Murine Xenograft Model
The study was approved by the Shandong University Second Hospital ethics committees. Six weeks old male nude mice were bought from Huafukang Company (Beijing, China). HT29 and HCT116 cells with and without ASF1A manipulations were injected subcutaneously (2 × 106 cells/mouse, 5 mice/group). The size of tumors was measured each week. Mice were sacrificed 6 weeks later and tumors were collected for further analyses including IHC staining.
For metastasis assays, six weeks old male NOD-SCID mice were used (Huafukang Company, Beijing, China). Control and HT29-ASF1A cells were injected mice (6 mice/group) via the tail vein (3 × 106 cells/mouse). After 7 weeks, mice were sacrificed and the lungs were collected. Half of mouse lungs were stored in formalin to make paraffin-embedded slides and the other half were stored at − 80 °C until use for additional analyses.
2.18. Statistical Analyses
Mann-Whitney U test or Student's t-test was used for analyses of differences between experiment groups. Overall survival was illustrated by Kaplan-Meier plots, and significance was calculated by log-rank test. All the tests were two-tailed and computed using SigmaStat3.1® software (Systat Software, Inc., Richmond, CA). P values of < 0.05 were considered as statistically significant.
3. Results
3.1. ASF1A Expression in Gastrointestinal Cancer-Derived Cell Lines
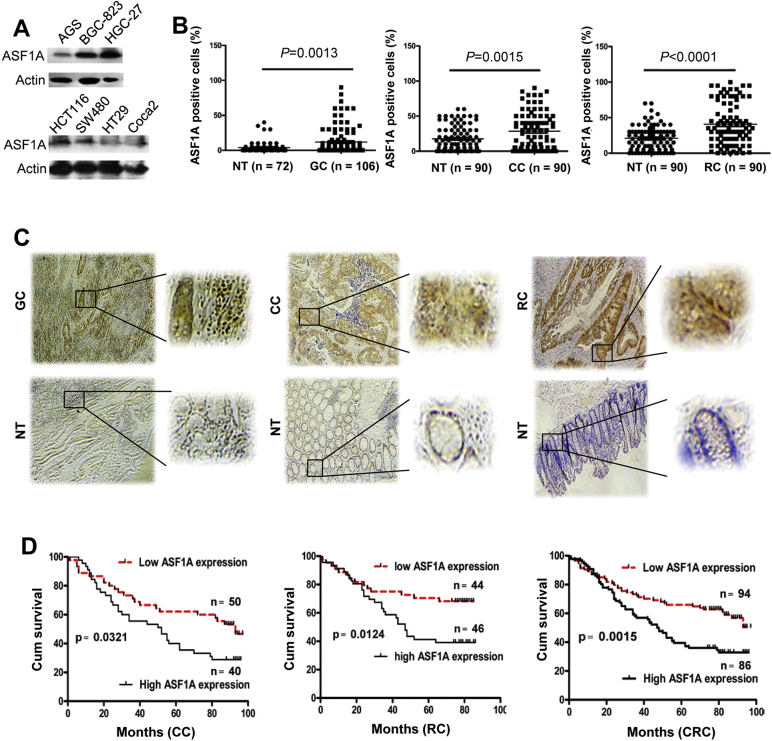
Little is known about ASF1A expression in human malignancies. We thus first determined whether GIC cells expressed ASF1A. A panel of gastric (AGS, BGC-823 and HGC-27) and colorectal cancer (HCT116, SW480, HT29 and Caco2)-derived cell lines were analyzed using immuno-blotting. As shown in Fig. 1A, all these cell lines expressed ASF1A protein at different levels. Of note, ASF1A was most abundant in poorly differentiated or aggressive cell lines HGC-27 and HCT116 (Fig. 1A).
Fig. 1.
ASF1A over-expression in gastrointestinal cancer (GIC) and its association with patient survival. (A) ASF1A expression in GIC-derived cell lines. Cells were analyzed for ASF1A protein levels using immunoblotting. Two independent experiments were performed. (B and C) Immunohistochemical (IHC) staining of ASF1A expression in primary GIC tumors and their adjacent normal tissues (NT). Primary tumors derived from 106 patients with gastric cancer (GC) and 180 patients with colorectal cancer (CRC) [90 colon cancer (CC) and 90 rectal cancer (RC)] were analyzed for ASF1A expression using IHC. (B) shows percentages of ASF1A-positive cells in tumors and adjacent tissues and representative IHC images are shown in (C). The right panel was enlarged in the rectangle area of the left ones. (D) Higher ASF1A expression as a predictor for shorter overall survival in CRC patients. The median level of 35% was used as a cut-off to categorize patients into low and high ASF1A groups. Overall survival of patients with colon and rectal cancer either separately or combined together was analyzed and presented.
3.2. Over-Expression of ASF1A as a Predictor of Poor Outcomes in Primary Gastrointestinal Cancer
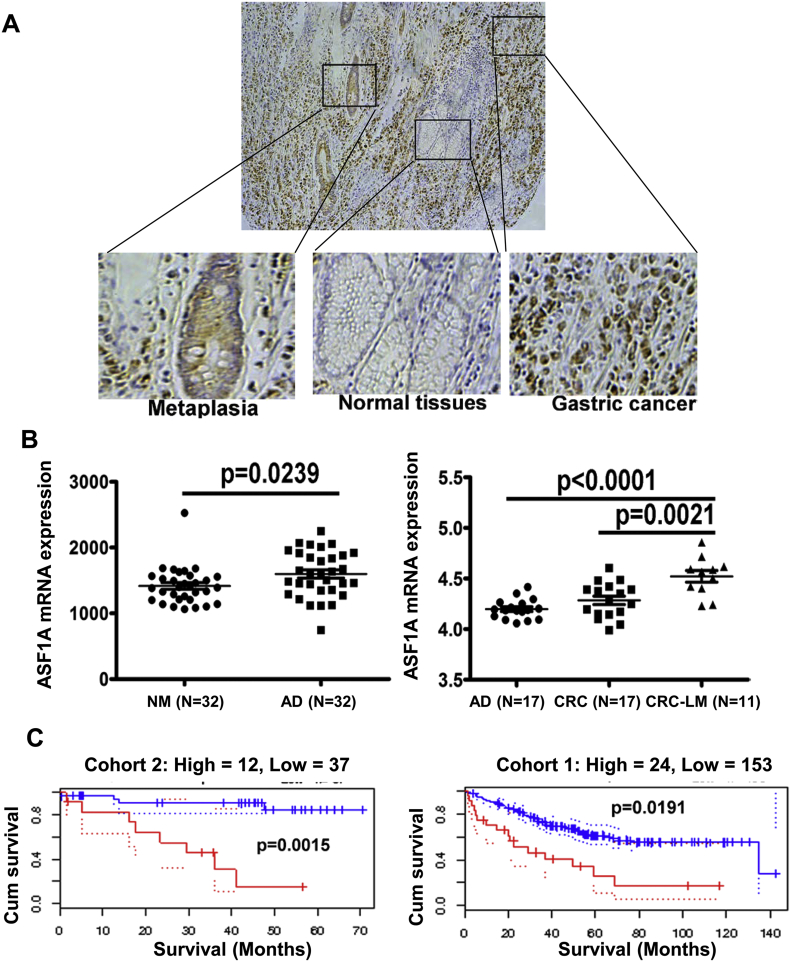
We then sought to examine ASF1A expression in primary GIC tumors. IHC staining of ASF1A was performed on both primary tumors and their adjacent normal tissues derived from a total of 286 GIC patients (106 gastric and 180 colorectal cancer patients). Most adjacent non-cancerous gastric tissues in general exhibited negative or weak ASF1 staining, while their colorectal non-cancerous counterparts had slightly stronger ASF1A expression (Fig. 1B and C). Compared to those adjacent non-cancerous tissues, GIC tumors expressed significantly higher levels of ASF1A, evidenced by the presence of increased percentages of positive cells and much more intensive staining [Fig. 1B and C, adjacent tissues vs tumors for gastric, colon and rectal (mean ± SD), 4% ± 8% vs 12% ± 18%, P = 0.0013, 18% ± 18% vs 28% ± 26%, P = 0.0015, and 21% ± 16% vs 41% ± 30%, P < 0.0001, respectively, Mann-Whitney U test]. Interestingly, in gastric sections containing normal, precancerous neoplasia and cancerous tissues, expression of ASF1A increased progressively (Fig. 2A), indicating that ASF1A over-expression is associated with acquisition of a malignant phenotype.
Fig. 2.
The progressive upregulation of ASF1A expression in evolution from premalignant lesions to gastrointestinal cancer and its association with patient survival derived from GEO datasets. (A) Immunohistochemical staining was performed on gastric cancer samples and representatives are shown. In normal areas, ASF1A expression is almost undetectable, and a positive staining is seen in areas with metaplasia while cancerous tissue areas exhibit highest ASF1A expression. (B) Relative levels of ASF1A mRNA expression in normal intestinal mucosa (NM), adenoma (AD), colorectal cancer (CRC) tissues and CRC with liver metastasis (CRC-LM). (Derived from GEO database, https://www.ncbi.nlm.nih.gov/geo/.) (C) Higher ASF1A mRNA expression as a predictor for shorter overall survival in CRC patients. The data were derived from PrognoScan website (http://www.prognoscan.org/). Blue and red curves: Low and high ASF1A expression, respectively.
ASF1A expression was then examined for its prognostic value in patients with colorectal cancer from whom survival information was available. The median percentage of ASF1A-positive tumor cells 35% was used as a cutoff to define low (< 35%) and high (≥ 35%) expression of ASF1A. The Kaplan-Meier analysis revealed that a high ASF1A expression was significantly associated with shorter patient overall survival in both colon and rectal cancer (Fig. 1E, P = 0.0321 and 0.0124 for colon and rectal cancer, respectively, and for both patient groups, P = 0.0015) (Fig. 1D).
We further validated the up-regulation of ASF1A in CRC tumors and its prognostic value by analyzing GEO data (http://www.prognoscan.org/). ASF1A mRNA expression in adenomas was much higher than that in normal intestinal mucosa (Fig. 2B left). Moreover, in 17 patients, the disease evolved from adenoma to carcinoma with or without liver metastasis and the correlation between higher expression of ASF1A and disease progression was already observed (Fig. 2B right). Fig. 2C further showed that higher levels of ASF1A mRNA expression were significantly associated with shorter overall survival in two cohorts of CRC patients.
3.3. ASF1A Stimulation of Cell Cycle Progression, Clonogenicity, Stemness and Migration of GIC Cells
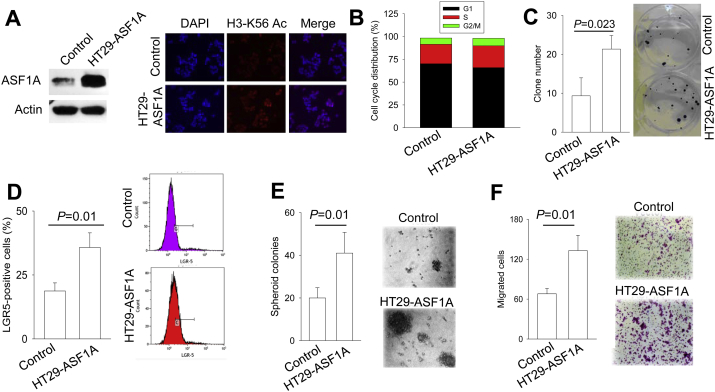
Given the findings above, we sought to determine functional activities of ASF1A in GIC cells. HT29 cells, the CRC line expressing a low level of ASF1A (Fig. 1A), were infected with lenti-viral GFP-ASF1A expression vectors to generate ASF1A over-expressing sublines (HT29-ASF1A). Higher ASF1A expression was verified using immunoblotting and IF staining, and as expected, increased histone H3-K56 acetylation was observed in those ASF1A over-expressing HT29 cells (Fig. 3A). The cell cycle analysis revealed that HT29-ASF1A cells exhibited moderate, but significant declines in G1 phase coupled with increased S phases, compared to their control cells [Fig. 3B, and HT29 control vs HT29 ASF1A (the mean values ± SD from 4 independent experiments): G1, 68.4 ± 3.8% vs 63.9 ± 4.0%, P = 0.008; S, 23.9 ± 5.3% vs 27.4 ± 7.1%, P = 0.038; G2/M, 6.2 ± 2.4% vs 7.0 ± 2.7%, P = 0.246, Student's t-test]. Consistently, ASF1A over-expression significantly stimulated clonogenic potentials of HT29 cells and > 2-fold increase in clones was observed in HT29-ASF1A cells (Fig. 3C) (clones 21.33 ± 3.51 vs 9.33 ± 4.62, P = 0.023).
Fig. 3.
Ectopic ASF1A expression promotes proliferation, clonogenic potential, stemness and migration of CRC-derived HT29 cells. Three or more independent experiments were performed. (A) Ectopic expression of ASF1A in HT29 cells verified using immunoblotting. HT29 cells were infected with control- and ASF1A-lentiviral vectors and over-expression of ASF1A was revealed by immunoblotting (Left panel). Immuno-fluorescence (IF) staining shows enhanced histone H3-K56 acetylation in HT29 cells expressing ectopic ASF1A (Right panel). (B) ASF1A over-expression in HT29 cells stimulates cell cycle progression. Cells with and without ASF1A over-expression were stained with Propidium Iodide (PI) and cell cycle analyses then performed using Flow cytometry. Shown is the result from one representative experiment. (C) Increased clonogenic ability of HT29 cells with ASF1A over-expression. Cells were seeded into 6-well plates with 1000 cells/well for 2 week incubation and foci then counted. (D) Up-regulation of the CRC stem cell marker LGR5 in HT29 cells by ASF1A over-expression, as shown by flow cytometry analysis. (E) Augmented formation of spheroid colonies by ASF1A over-expression. HT29 cells with and without ASF1A over-expression were seeded in ultra-low attachment 96-well plates (2000/well) and incubated in serum-free medium containing bFGF and EGF for 15 days. Spheroid colonies were then counted. Left panel: The spheroid colony numbers from three independent experiments. Right panel: Shown are representative. (F) ASF1A-mediated enhancement of cellular migration. Control- and ASF1A-HT29 cells were loaded into the upper chamber at 1 × 105 and migrated cells into the bottom chamber were stained with crystal violet and counted. Left panel: The migrated cell numbers from three independent experiments. Right panel: Shown are representative.
We further evaluated the effect of ASF1A over-expression on cellular stemness and migration/invasion. HT29-ASF1A cells expressed significantly higher levels of CRC stem cell marker Lgr5 than did their control counterparts (35.8 ± 5.7% vs 18.7 ± 3.2% for GFP + vs −, P = 0.011). In accordance with this, control HT29 cells had low abilities to form CRC-spheres, while ASF1A over-expression led to the highly increased number of CRC-spheres (Fig. 3D, spheres in control vs HT29-ASF1A cells, 20 ± 5 vs 41 ± 10, P = 0.009). Fig. 3E shows the difference in cellular migration between control and ASF1A over-expressing cells: HT29-ASF1A cells exhibited a significantly enhanced migration potential than did their control counterparts (133 ± 23 vs 68 ± 8, P = 0.009). The invasion potential of these cells could not be assessed due to their poor ability to pass through Matrigel.
3.4. Diminished Migration and Invasion of GIC Cells Caused by ASF1A Depletion
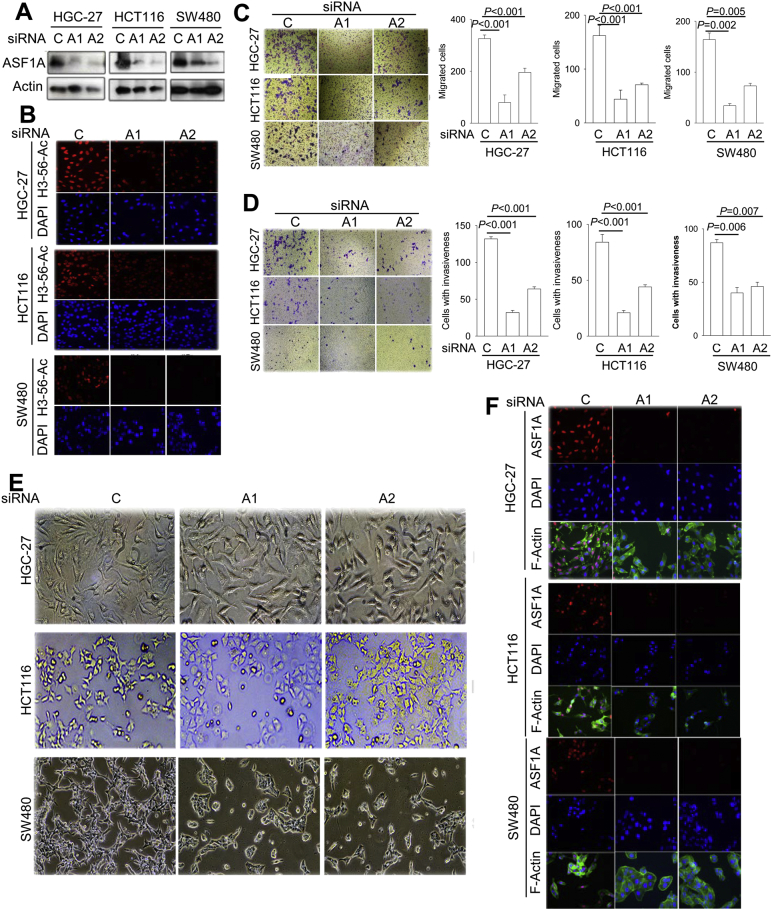
To further verify the role of ASF1A in regulating migration/invasion of GIC cells, we knocked-down ASF1A expression in GIC cells expressing high levels of ASF1A (HGC-27, HCT116 and SW480). The efficient ASF1A depletion using two different siRNAs was confirmed by immunoblotting and IF staining (Fig. 4A). In accordance with diminished ASF1A expression, H3-K56 acetylation was inhibited in these same cells (Fig. 4B). ASF1A depletion significantly inhibited migration in all three cell lines (Fig. 4C), which was in sharp contrast to that seen in ASF1A over-expressing HT29 cells. Moreover, the cells passing through Matrigel were substantially diminished after ASF1A knocking-down (Fig. 4D), suggesting highly impaired abilities of cellular invasion.
Fig. 4.
Impaired migration and invasion of GIC cells by ASF1A inhibition. Two ASF1A-specific siRNAs (A1 and A2) were used to knock down its expression in GIC cell lines including HGC-27, HCT116 and SW480 and cells then analyzed for migration/invasion, morphological alterations and F-actin re-arrangement. C: Control siRNA. Three or more independent experiments were performed. (A) Efficient ASF1A depletion was verified using two different ASF1A specific siRNAs. (B) IF staining shows diminished histone H3-K56 acetylation, consistent with reduced ASF1A expression. (C) Migration and (D) invasion inhibition by ASF1A depletion in GIC cells. (E) Diminished elongated/spindle-like cellular shapes or cell cluster formation upon ASF1A knock-down in GIC cells. (F) F-Actin staining showed reduced dendrites/branching or a switch from a pattern of central stress fibers to a predominant peripheral rearrangement in ASF1A-depleted cells.
Because epithelial-mesenchymal transition (EMT) plays a key role in cancer cell migration/invasion (Liu et al., 2013, Puisieux et al., 2014), we further examined morphological changes in ASF1A-depleted cells. As documented in Fig. 4E, the ASF1A-depleted cells underwent less elongated/spindle-like shapes or grew into cell clusters. F-Actin staining showed similar changes in cellular morphology and reduced dendrites or branching (Fig. 4F). A switch of F-actin from a pattern of central stress fibers to a predominant peripheral rearrangement was also observed in ASF1A-depleted HGC-27 and SW480 cells (Fig. 4F).
3.5. ASF1A Interaction with β-Catenin to Potentiate the β-Catenin Activity
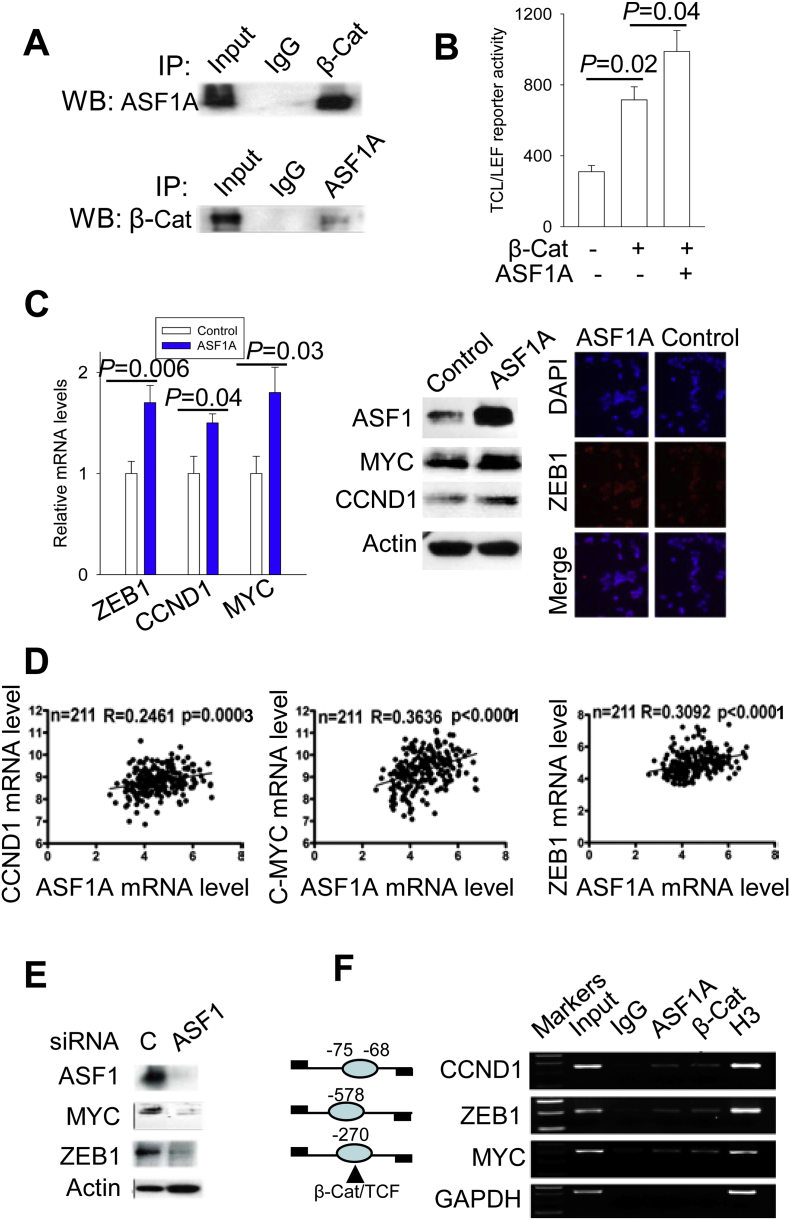
How ASF1A exerts its oncogenic activities as observed above was then addressed. Because the aberrant Wnt/β-catenin pathway plays a key part in the GIC pathogenesis (Basu et al., 2016, Zhao et al., 2014, Zhao et al., 2017), we asked whether there are potential links between ASF1A and β-catenin. Toward this end, we first performed IP to determine ASF1A interaction with β-catenin. Reciprocal IP experiments demonstrated a physical association of ASF1A with β-catenin (Fig. 5A). To evaluate the functional consequence of the observed interaction, we measured the effect of ASF1A on the TCF/LEF reporter activity, an assay for the transcription activity of β-catenin (Liu et al., 2013). As expected, the luciferase activity driven by the TCF/LEF transcriptional response elements increased significantly when HCT116 cells were transfected with β-catenin expression vectors (Fig. 5B). Furthermore, the TCF/LEF transactivation mediated by β-catenin was further enhanced in the presence of ASF1A expression vectors (Fig. 5B). Taken together, ASF1A interacts with β-catenin, thereby promoting TCF/LEF transactivation.
Fig. 5.
ASF1A interaction with β-catenin to promote β-catenin target transcription in CRC cells. β-Cat: β-catenin. Three or more independent experiments were performed. (A) The physical association between ASF1A and β-catenin as demonstrated using reciprocal immunoprecipitation (IP). (B) Regulation of TCF/LEF reporter activity by ASF1A. HCT116 cells were transfected with TCF/LEF reporters together with β-catenin vectors alone or β-catenin plus ASF1A expression vectors and luciferase activity was assessed 48 h post-transfection. (C) The expression of β-catenin target genes ZEB1, CCND1 and MYC was up-regulated by ASF1A over-expression in HT29 cells. Right panel: qRT-PCR quantification of ZEB1, CCND1 and MYC mRNA levels in control and ASF1A-overexpressing HT29 cells. Middle and left panels: Immunoblotting and immunofluorescence analyses of ZEB1, CCND1 and MYC protein expression in those same sets of cells. (D) The positive correlation of ASF1A mRNA expression with CCND1, MYC and ZEB1 mRNA levels in tumors from patients with colorectal cancer (CRC). The data were derived from GEO database (https://www.ncbi.nlm.nih.gov/geo/). A total of 211 CRC patients were analyzed. (E) Down-regulated expression of β-catenin target genes by ASF1A-depletion. HCT116 cells were transfected with control and ASF1A siRNAs, respectively, and protein expression was then determined using immunoblotting. (F) ChIP assay for ASF1A and β-catenin occupancy on the promoters of β-catenin targets CCND1, ZEB1 and MYC. ChIP was performed with chromatin derived from HCT116 cells as described in Materials and Methods. Histone H3 antibody was used as a positive control. Left: Schematic expression of β-catenin/TCF sites and ChIP primer positions at the promoters of CCND1, ZEB1 and MYC.
3.6. ASF1A Stimulation of β-Catenin Target Expression in CRC Cells
Having observed the ASF1A effect on GIC cell growth and invasion, and its functional interaction with β-catenin, we sought to probe whether it up-regulated the expression of β-catenin target genes. It has been well documented that β-catenin targets, critical for cancer cell proliferation/progression, include LGR5, cyclin D1, c-MYC and ZEB1 (Basu et al., 2016, He et al., 1998, Sanchez-Tillo et al., 2011, Tetsu and McCormick, 1999). We thus determined their expression in HT29 control and HT29-ASF1A cells. The enhanced LGR5 expression in HT29-ASF1A cells was already documented above (Fig. 3C), and expression of three other targets was shown in Fig. 5C. HT29-ASF1A cells expressed significantly higher levels of cyclin D1, c-MYC and ZEB1 mRNA coupled with increased protein abundances compared these of control cells (cyclin D1: 1.43 ± 0.09 vs 1.0 ± 0.17, P = 0.042; c-MYC: 1.74 ± 0.27 vs 1.10 ± 0.18, P = 0.025; ZEB1: 1.71 ± 0.17 vs 1.07 ± 0.12, P = 0.005) (Fig. 4C). We also examined the effect of ASF1A depletion. HCT116 cells exhibited down-regulated expression of c-MYC and ZEB1 after ASF1A inhibition (Fig. 5E).
To determine whether the correlation between ASF1A expression and β-catenin targets was also present in primary CRC tumors, we performed the analyses using GEO datasets. As shown in Fig. 5D, ASF1A mRNA expression was positively correlated with CCND1, MYC and ZEB1 mRNA levels in tumors derived from 210 patients with CRC.
3.7. ASF1A Association with ZEB1, Cyclin D1 and c-MYC Promoters in CRC Cells
Given the observations above, we then wanted to determine whether ASF1A occupied these β-catenin target promoters. ChIP was performed on HCT116 cells where endogenous ASF1A expression and β-catenin activity were high, by using the primers spanning the β-catenin binding region of each promoter. As shown in Fig. 5F, both ASF1A and β-catenin were enriched on the promoters of ZEB1, cyclin D1 and c-MYC, while control IgG did not give rise to detectable signals.
3.8. Inhibition of E-Cadherin Expression by the ASF1A/β-Catenin-ZEB1 Axis in GIC Cells
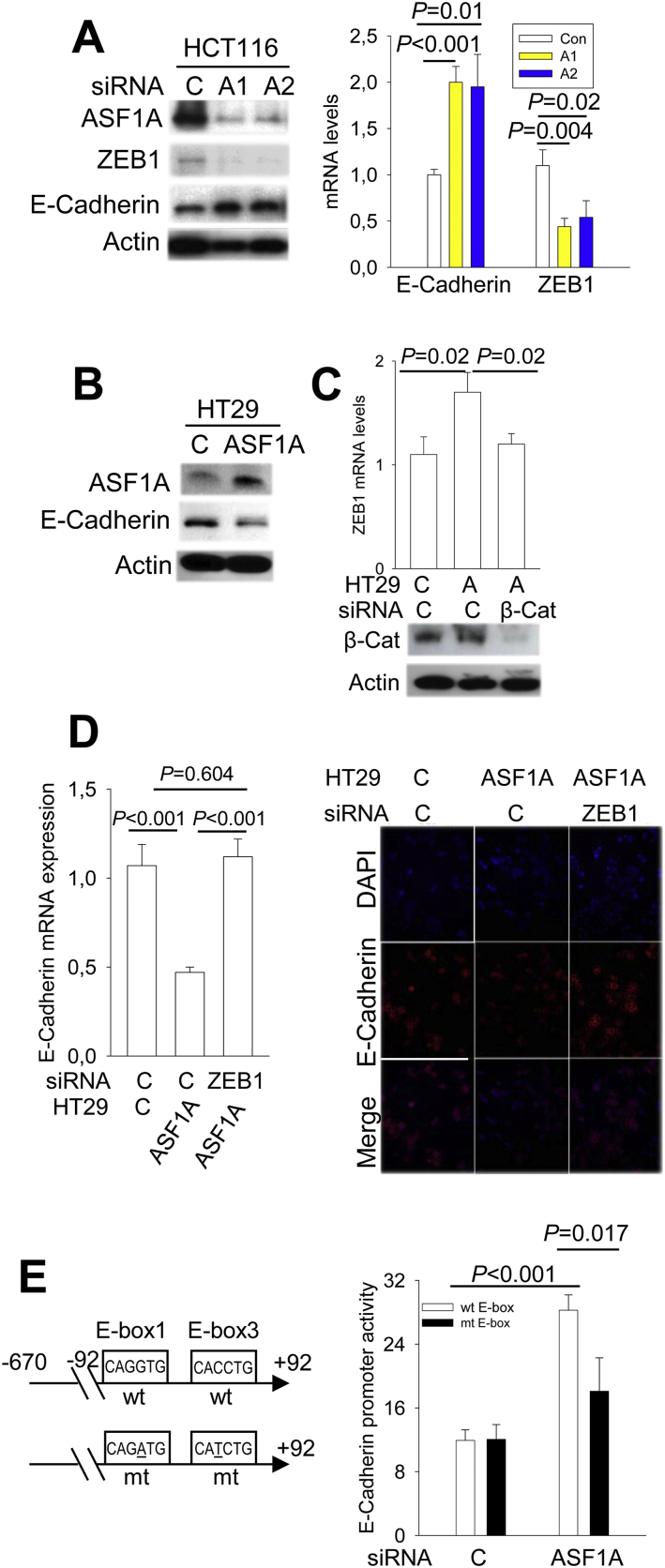
The loss of the epithelial marker E-Cadherin is known as a driving-force promoting cellular mobility, while ZEB1 inhibits E-Cadherin expression, thereby facilitating cancer invasion (Gheldof et al., 2012). We thus asked whether the ASF1A/β-catenin-ZEB1 axis contributes to invasion of GIC cells by regulating E-Cadherin expression. To this end, we first determined E-Cadherin expression and its relationship with ZEB1 in GIC cells where ASF1A expression was manipulated. HCT116 cells were transfected with ASF1A siRNA as above, and then analyzed for E-Cadherin and ZEB1 expression. ASF1A depletion in HCT116 cells significantly up-regulated E-Cadherin expression coupled with the diminished abundance of ZEB1 at both transcriptional and protein levels (Fig. 6A). On the other hand, the inhibition of E-Cadherin expression coupled with enhanced ZEB1 levels were observed in HT29 cells over-expressing ASF1A (Fig. 6B). To address whether β-catenin is required for ASF1A-mediated ZEB1 up-regulation, we knocked down β-catenin in HT29-ASF1A cells and as shown in Fig. 6, this ASF1A effect was totally abolished by β-catenin inhibition, which was coupled with recovery of E-Cadherin expression (Fig. 6C). To further ascertain a causal relationship between E-Cadherin and ZEB1 expression, we blocked ZEB1 up-regulation mediated by ASF1A over-expression in HT29 cells using ZEB1 siRNA. As documented above, HT-29-ASF1A cells expressed higher levels of ZEB1 accompanied by decreased E-Cadherin expression, however, ZEB1 depletion led to the recovery of E-Cadherin expression, comparable with that observed in control cells (Fig. 6D).
Fig. 6.
E-Cadherin expression regulated by the ASF1A- β-catenin-ZEB1 axis in CRC cells. Three or more independent experiments were performed. (A) Diminished ZEB1 expression coupled with E-Cadherin expression in ASF1A-depleted HTC116 cells. Right and left panels: Protein and mRNA expression of ZEB1 and E-Cadherin as determined using immunoblotting and qPCR, respectively. (B) Down-regulated of E-Cadherin expression mediated by ASF1A. HT29 cells with and without ASF1A over-expression were analyzed for E-Cadherin expression using immunoblotting. (C) The abolishment of ASF1A-induced ZEB1 up-regulation by β-catenin inhibition. HT29 cells with ASF1A over-expression were transfected with β-catenin siRNA (Bottom panel) and ZEB1 mRNA levels then determined using qPCR (Top panel). A: ASF1A. (D) The abolishment of ASF1A-induced E-Cadherin down-regulation by ZEB1 inhibition. HT29 cells with ASF1A over-expression were transfected with ZEB1 siRNA, and ZEB1 and E-Cadherin mRNA and protein levels then determined using qPCT (left) and immunofluorescence (right), respectively. (E) Significantly attenuated E-Cadherin promoter activity by the mutation of ZEB1 binding sites in ASF1A-depleted HCT116 cells. Left panel: Schematics of the E-Cadherin promoter (+ 92 - -670) with wt and mutant E-boxes. Right panel: The wt E-Cadherin promoter reporter and its variant carrying mutant E-boxes (for ZEB1 binding) were transfected into HCT116 cells with and without ASF1A knocking-down, and luciferase activity was then assessed and expressed as arbitrary units.
It is known that ZEB1 binds to the E-boxes in the E-Cadherin core promoter through which E-Cadherin transcription is inhibited (Gheldof et al., 2012). We thus further evaluated the E-Cadherin promoter activity in ASF1A-depleted cells. The E-Cadherin promoter activity was significantly up-regulated upon ASF1A knocking down in HCT116 cells, which, however, was substantially attenuated by the E-box mutation (Fig. 6E).
3.9. The ASF1A Effect on In Vivo Growth and Metastasis of CRC Cells in the Murine Xenograft Model
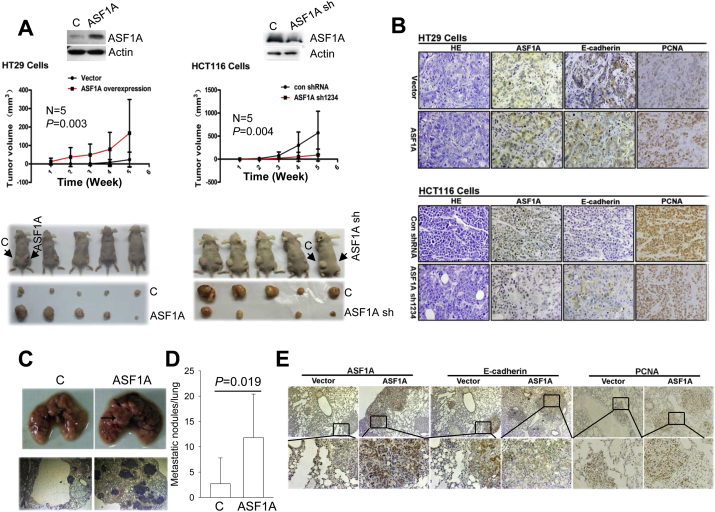
Finally, we determined whether the in vitro ASF1A effects on GIC, as observed above, could be recapitulated under in vivo conditions. For this purpose, HT29-ASF1A cells and HCT116 cells with stable ASF1A depletion (HCT116/ASF1A-sh), together with their corresponding control counterparts, were injected subcutaneously into nude mice, and mice were killed 6 weeks post-injection. During the 6 week period, tumor growth was monitored weekly. As shown in Fig. 7A, ASF1A over-expression in HT29 cells and depletion in HCT116 cells significantly promoted and retarded in vivo tumor growth, respectively: Tumors grew much faster and were significantly bigger in mice receiving HT29-ASF1A cells than those injected with control HT29 cells (Fig. 7A, left), whereas ASF1A-depleted HCT116 cells gave rise to tumors significantly smaller than did their control counterparts (Fig. 7A, right). IHC analyses showed that ASF1A expression in these tumors was consistent with that in the injected original cells and negatively correlated with E-Cadherin expression (Fig. 7B). In addition, higher levels of PCNA expression were observed in HT29-ASF1A tumors, which indicate their enhanced proliferation ability, while weaker PCNA staining was seen in HCT116-ASF1A-depleted tumors (Fig. 7B).
Fig. 7.
ASF1A stimulation of in vivo growth and metastasis of CRC cells in the murine xenograft model. C: Control cells. The mouse experiments were performed once. (A) ASF1A-mediated tumor formation in nude mice (5 mice/group). HT29 cells with and without ASF1A over-expression were subcutaneously injected into nude mice. Tumor growth was monitored weekly, and mice were killed 6 week latter. The identical experiments were also performed on HCT116 cells with (HCT116-ASF1A sh) and without ASF1A depletion. (B) Immunohistochemical (IHC) analyses of ASF1A, E-Cadherin and PCNA expression in tumors grown in nude mice. (C–E) Lung metastases of HT29 cells with and without ASF1A over-expression in NOD-SCID mice (6 mice/group). Cells were injected into mice via the tail vein and mice were killed 7 weeks after injection. Metastatic nodules in the mouse lungs were counted and analyzed for ASF1A, E-Cadherin and PCNA expression using IHC.
To assess the in vivo effect of ASF1A on tumor metastasis, we further injected control HT29 and HT29-ASF1A cells into NOD-SCID mice via the tail vein. Mice were killed 7 weeks post-injection and tumors seeded in the lungs were counted. Lung metastasis occurred in 2/7 (29%) and 6/8 (75%) mice receiving control and ASF1A-overexpressed HT29 cells, respectively. There were a total of 19 tumors in the lungs from mice injected with control HT29 cells, and 93 from mice with ASF1A-HT29 cells (P = 0.019). Moreover, bigger tumor colonies were more frequently seen in the latter group (Fig. 7C and D). IHC staining of those metastatic tumors revealed similar ASF1A and E-Cadherin expression patterns (Fig. 7E), as observed in tumors grown subcutaneously (Fig. 7B).
4. Discussion
Given the epigenetic aberration as one of the key oncogenic mechanisms in GIC (Vogelstein et al., 2013), and important functions of the histone chaperone ASF1A in nucleosome assembling/histone modifications (Mousson et al., 2007), it is highly reasonable to explore ASF1 effects in GIC pathogenesis. From the result presented herein, we uncover crucial oncogenic activities of ASF1A. First, a widespread over-expression of ASF1A was observed in GIC cell lines and primary tumors. During evolution from precursor lesions to fully transformed gastric cancer, ASF1A expression increased significantly. Second, ASF1A interacts with β-catenin and potentiates the transcription of β-catenin target genes crucial for GIC cell proliferation, migration or invasion and stemness. Third, ASF1A over-expression and depletion promotes and inhibits CRC tumor growth and metastasis in mouse xenograft models. Finally, higher levels of ASF1A expression predict shorter patient survival in patients with CRC. Taken together, our findings suggest that ASF1A plays significant roles in GIC pathogenesis and may serve as prognostic factor and a potentially therapeutic target for GIC.
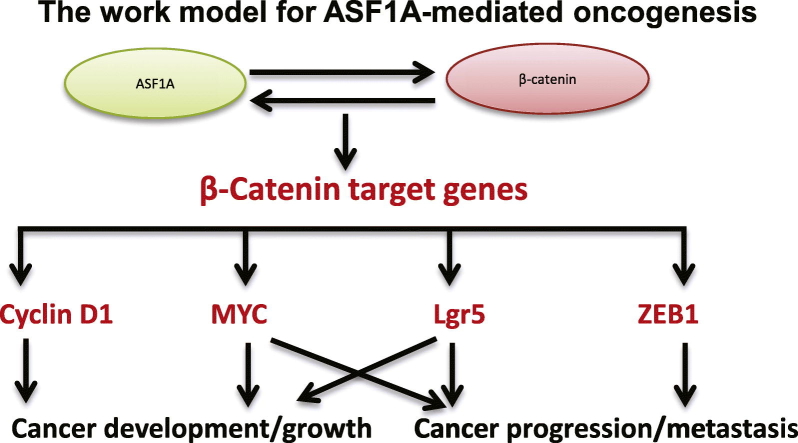
In a limited number of studies, researchers have shown the dysregulation of ASF1 in human malignancies (Corpet et al., 2011, Das et al., 2009, Wang et al., 2015), however, it is largely unclear whether and how ASF1 contributes to cancer development or progression. Human ASF1 exists as two isoforms, ASF1A and ASF1B, and they share certain common functions, but also have different activities (Mousson et al., 2007). More recently, Gonzalez-Munoz et al. found that ASF1A promoted iPS cell induction by interacting with OCT4 and stimulating the transcription of stem cell factors including NANOG and SOX2 (Gonzalez-Munoz et al., 2014); and moreover, ASF1A and OCT4 over-expression was sufficient to convert human normal fibroblasts into pluripotent stem cells (Gonzalez-Munoz et al., 2014). These findings promoted us to examine the regulatory effect of ASF1A on gene transcription. Because the Wnt/β-catenin axis is frequently altered in CRC (Basu et al., 2016), we sought to determine the relationship between ASF1A and β-catenin. Indeed, the present results provide convincing evidence that ASF1A acts as a co-factor for the transcription of β-catenin target genes. (a) ASF1A and β-catenin interacts with each other; (b) ASF1A and β-catenin synergistically stimulate the TCF/LEF reporter promoter activity; (c) ASF1A over-expression up-regulates whereas its inhibition down-regulates expression of β-catenin target genes including ZEB1, cyclin D1, c-MYC and LGR5, respectively; (d) ASF1A is associated with ZEB1, cyclin D1 and c-MYC promoters in the TCF binding regions. Based on all these findings, we suggest a model for the oncogenic activity of ASF1A as presented in Fig. 8.
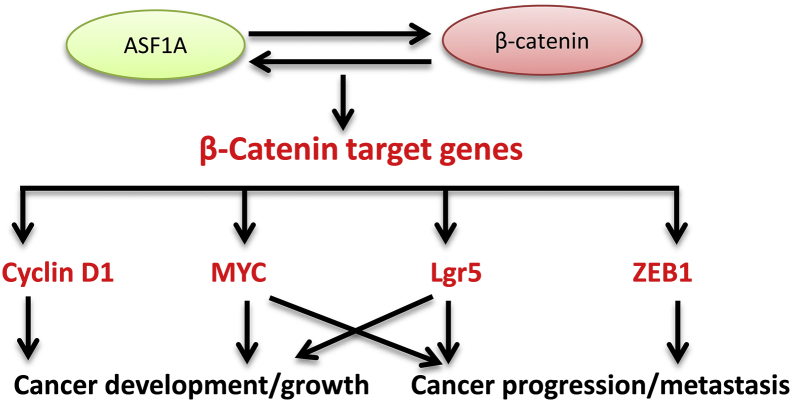
Fig. 8.
The work model for ASF1A-mediated oncogenesis. ASF1A interacts with β-catenin, thereby acting as a co-factor to stimulate the expression of β-catenin target genes including cyclin D1, c-MYC, ZEB1 and LGR5. The up-regulated expression of these β-catenin targets consequently promotes proliferation, stemness, invasion and metastasis of malignant cells. All these ASF1A effects eventually lead to cancer development and progression.
Dysregulation or mutations of β-catenin are widespread in GIC, and altered β-catenin plays fundamental roles in GIC initiation and progression (Basu et al., 2016, Oguma et al., 2008, Vogelstein et al., 2013). By interacting with β-catenin, ASF1A thus exhibits multiple oncogenic activities, as documented in the present study. It promoted proliferation of CRC cells by stimulating c-MYC and cyclin D1 expression, while inhibiting E-Cadherin expression via up-regulation of ZEB1, thereby augmenting cellular migration or invasion. Moreover, the CRC stem cell marker LGR5 increased coupled with the enhanced self-renewal potential in ASF1A-over-expressed CRC cells, suggesting its requirement for maintaining CRC stem cell phenotype. Thus, the ASF1A-β-catenin axis is functionally essential to GIC tumors and disruption of their interaction may have therapeutic implications.
In addition to OCT4 and β-catenin, ASF1A has been shown to interact with other transcriptional regulators such as the histone acetyltransferase Creb-binding protein (CBP)/P300 and the transcription factor Mef2 (Das et al., 2009, Das et al., 2014). Furthermore, ASF1A is required for histone H3-K56 acetylation, while the H3-K56 acetylation state plays a key role in enabling rapid transcriptional changes (Das et al., 2009, Das et al., 2014). These data collectively indicate that ASF1A may be an important co-factor participating in transcriptional regulation by interacting and cooperating with transcription factors or their co-factors, and via modification of H3-K56. Likely, many more genes are transcriptionally controlled by ASF1A, and the elucidation of ASF1A target genes will gain profound insights into biological and oncogenic activities of ASF1A.
Corpet et al. previously reported that ASF1B was required for proliferation of breast cancer cells, and higher ASF1B expression predicted poor patient outcomes, whereas ASF1A expression did not increase and contributed no prognostic information to breast cancer (Corpet et al., 2011). This observation is in contrast to what we found in GIC tumors. Thus it is likely that tumors derived from different tissues/organs utilize different isoforms of ASF1 for their malignant phenotype maintenance.
The bromodomain and extra terminal domain (BET) family proteins including BRD3 and BRD4. They are another type of epigenetic modulators that read acetylated lysine residues and transfer cellular signals through which the transcription of oncogenes is augmented (Zuber et al., 2011). Small-molecule BET bromodomain inhibitors, for instance JQ1, have been developed to competitively bind to acetyl–lysine recognition pockets, and disrupt BET bromodomain protein interaction with chromatin, and subsequently reduce the expression of oncogenes, thereby leading to cancer cell growth inhibition and/or apoptosis (Zuber et al., 2011). Intriguingly, JQ1 was recently shown to inhibit H3-K56 acetylation (Das et al., 2014). H3-K56 acetylation is known to drive chromatin toward the disassembled state during transcriptional activation whereas loss of histone H3-K56 acetylation drives the chromatin toward the assembled state. It is thus necessary to elucidate whether JQ1-mediated blockade of H3-K56 acetylation is associated with its anti-cancer activity and whether there exists a cross-talk between BRDs and ASF1A.
In summary, our present results show that ASF1A is over-expressed in GIC tumors and could serve as a prognostic factor in CRC. Mechanistically, ASF1A interacts with β-catenin and stimulates its target transcription, thereby facilitating proliferation, stemness, and migration/invasion of GIC cells. Collectively, ASF1A plays an important part in GIC pathogenesis and progression, and thus targeting ASF1A may be a novel strategy against GIC.
Grant Support
This work was supported by grants from The National Basic Research Program of China (grant no. 2012CB911202), Swedish Cancer Society (CAN 2016/427), the Swedish Research Council (#K2013-67X-14201-12-4), Cancer Society in Stockholm (141373), the National Natural Science Foundation of China (81070224, 81071721, 81000868 and 81171536), and Shandong Provincial Natural Science Foundation (2016ZDJS07A09). The funding sources have no roles in the writing of the manuscript or the decision to submit it for publication, data collection, analysis, or interpretation or patient recruitment. None of the authors have been paid to write this article by a pharmaceutical company or other agency.
Conflict of Interest
The authors declare no potential conflicts of interest.
Author Contributions
XL and DX designed the study. XY, JY, YW, KL, CS, SL, LS, and FK performed experiments. XL, XY, JY, JJ and DX analyzed data. JJ and DX obtained funding; XL and DX prepared the figures. XL, XY, MB and DX wrote the manuscript. MB, JJ and DX supervised the study.
Acknowledgements
We thank professor Cibelli JB (Centro Andaluz de Nanomedicina y Biotecnología Andalucía) for ASF1A shRNA and its control plasmids.
Contributor Information
Xiuming Liang, Email: liangxm@sdu.edu.cn.
Dawei Xu, Email: Dawei.Xu@ki.se.
References
- Basu S., Haase G., Ben-Ze'ev A. Wnt signaling in cancer stem cells and colon cancer metastasis. F1000Res. 2016;5 doi: 10.12688/f1000research.7579.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benard A., Goossens-Beumer I.J., van Hoesel A.Q., Horati H., de Graaf W., Putter H., Zeestraten E.C., Liefers G.J., van de Velde C.J., Kuppen P.J. Nuclear expression of histone deacetylases and their histone modifications predicts clinical outcome in colorectal cancer. Histopathology. 2015;66:270–282. doi: 10.1111/his.12534. [DOI] [PubMed] [Google Scholar]
- Chen W., Zheng R., Baade P.D., Zhang S., Zeng H., Bray F., Jemal A., Yu X.Q., He J. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- Corpet A., De Koning L., Toedling J., Savignoni A., Berger F., Lemaitre C., O'Sullivan R.J., Karlseder J., Barillot E., Asselain B., Sastre-Garau X., Almouzni G. Asf1b, the necessary Asf1 isoform for proliferation, is predictive of outcome in breast cancer. EMBO J. 2011;30:480–493. doi: 10.1038/emboj.2010.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crea F., Fornaro L., Paolicchi E., Masi G., Frumento P., Loupakis F., Salvatore L., Cremolini C., Schirripa M., Graziano F., Ronzoni M., Ricci V., Farrar W.L., Falcone A., Danesi R. An EZH2 polymorphism is associated with clinical outcome in metastatic colorectal cancer patients. Ann. Oncol. 2012;23:1207–1213. doi: 10.1093/annonc/mdr387. [DOI] [PubMed] [Google Scholar]
- Das C., Lucia M.S., Hansen K.C., Tyler J.K. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459:113–117. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das C., Roy S., Namjoshi S., Malarkey C.S., Jones D.N., Kutateladze T.G., Churchill M.E., Tyler J.K. Binding of the histone chaperone ASF1 to the CBP bromodomain promotes histone acetylation. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E1072–E1081. doi: 10.1073/pnas.1319122111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheldof A., Hulpiau P., van Roy F., De Craene B., Berx G. Evolutionary functional analysis and molecular regulation of the ZEB transcription factors. Cell. Mol. Life Sci. 2012;69:2527–2541. doi: 10.1007/s00018-012-0935-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Munoz E., Arboleda-Estudillo Y., Otu H.H., Cibelli J.B. Cell reprogramming. Histone chaperone ASF1A is required for maintenance of pluripotency and cellular reprogramming. Science. 2014;345:822–825. doi: 10.1126/science.1254745. [DOI] [PubMed] [Google Scholar]
- Goossens-Beumer I.J., Zeestraten E.C., Benard A., Christen T., Reimers M.S., Keijzer R., Sier C.F., Liefers G.J., Morreau H., Putter H., Vahrmeijer A.L., van de Velde C.J., Kuppen P.J. Clinical prognostic value of combined analysis of Aldh1, Survivin, and EpCAM expression in colorectal cancer. Br. J. Cancer. 2014;110:2935–2944. doi: 10.1038/bjc.2014.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He T.C., Sparks A.B., Rago C., Hermeking H., Zawel L., da Costa L.T., Morin P.J., Vogelstein B., Kinzler K.W. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- Kim K.H., Roberts C.W. Targeting EZH2 in cancer. Nat. Med. 2016;22:128–134. doi: 10.1038/nm.4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer O.H. HDAC2: a critical factor in health and disease. Trends Pharmacol. Sci. 2009;30:647–655. doi: 10.1016/j.tips.2009.09.007. [DOI] [PubMed] [Google Scholar]
- Li W., Zhao L., Zang W., Liu Z., Chen L., Liu T., Xu D., Jia J. Histone demethylase JMJD2B is required for tumor cell proliferation and survival and is overexpressed in gastric cancer. Biochem. Biophys. Res. Commun. 2011;416:372–378. doi: 10.1016/j.bbrc.2011.11.045. [DOI] [PubMed] [Google Scholar]
- Liu Z., Li Q., Li K., Chen L., Li W., Hou M., Liu T., Yang J., Lindvall C., Bjorkholm M., Jia J., Xu D. Telomerase reverse transcriptase promotes epithelial-mesenchymal transition and stem cell-like traits in cancer cells. Oncogene. 2013;32:4203–4213. doi: 10.1038/onc.2012.441. [DOI] [PubMed] [Google Scholar]
- Mousson F., Ochsenbein F., Mann C. The histone chaperone Asf1 at the crossroads of chromatin and DNA checkpoint pathways. Chromosoma. 2007;116:79–93. doi: 10.1007/s00412-006-0087-z. [DOI] [PubMed] [Google Scholar]
- Oguma K., Oshima H., Aoki M., Uchio R., Naka K., Nakamura S., Hirao A., Saya H., Taketo M.M., Oshima M. Activated macrophages promote Wnt signalling through tumour necrosis factor-alpha in gastric tumour cells. EMBO J. 2008;27:1671–1681. doi: 10.1038/emboj.2008.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okugawa Y., Grady W.M., Goel A. Epigenetic alterations in colorectal cancer: emerging biomarkers. Gastroenterology. 2015;149(1204–1225):e1212. doi: 10.1053/j.gastro.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdag H., Teschendorff A.E., Ahmed A.A., Hyland S.J., Blenkiron C., Bobrow L., Veerakumarasivam A., Burtt G., Subkhankulova T., Arends M.J., Collins V.P., Bowtell D., Kouzarides T., Brenton J.D., Caldas C. Differential expression of selected histone modifier genes in human solid cancers. BMC Genomics. 2006;7:90. doi: 10.1186/1471-2164-7-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul B., Barnes S., Demark-Wahnefried W., Morrow C., Salvador C., Skibola C., Tollefsbol T.O. Influences of diet and the gut microbiome on epigenetic modulation in cancer and other diseases. Clin. Epigenetics. 2015;7:112. doi: 10.1186/s13148-015-0144-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puisieux A., Brabletz T., Caramel J. Oncogenic roles of EMT-inducing transcription factors. Nat. Cell Biol. 2014;16:488–494. doi: 10.1038/ncb2976. [DOI] [PubMed] [Google Scholar]
- Richet N., Liu D., Legrand P., Velours C., Corpet A., Gaubert A., Bakail M., Moal-Raisin G., Guerois R., Compper C., Besle A., Guichard B., Almouzni G., Ochsenbein F. Structural insight into how the human helicase subunit MCM2 may act as a histone chaperone together with ASF1 at the replication fork. Nucleic Acids Res. 2015;43:1905–1917. doi: 10.1093/nar/gkv021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha S.N., Kala R., Tollefsbol T.O. Molecular mechanisms for inhibition of colon cancer cells by combined epigenetic-modulating epigallocatechin gallate and sodium butyrate. Exp. Cell Res. 2014;324:40–53. doi: 10.1016/j.yexcr.2014.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Tillo E., de Barrios O., Siles L., Cuatrecasas M., Castells A., Postigo A. Beta-catenin/TCF4 complex induces the epithelial-to-mesenchymal transition (EMT)-activator ZEB1 to regulate tumor invasiveness. Proc. Natl. Acad. Sci. U. S. A. 2011;108:19204–19209. doi: 10.1073/pnas.1108977108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreuders E.H., Ruco A., Rabeneck L., Schoen R.E., Sung J.J., Young G.P., Kuipers E.J. Colorectal cancer screening: a global overview of existing programmes. Gut. 2015;64:1637–1649. doi: 10.1136/gutjnl-2014-309086. [DOI] [PubMed] [Google Scholar]
- Song J., Noh J.H., Lee J.H., Eun J.W., Ahn Y.M., Kim S.Y., Lee S.H., Park W.S., Yoo N.J., Lee J.Y., Nam S.W. Increased expression of histone deacetylase 2 is found in human gastric cancer. APMIS. 2005;113:264–268. doi: 10.1111/j.1600-0463.2005.apm_04.x. [DOI] [PubMed] [Google Scholar]
- Tamagawa H., Oshima T., Numata M., Yamamoto N., Shiozawa M., Morinaga S., Nakamura Y., Yoshihara M., Sakuma Y., Kameda Y., Akaike M., Yukawa N., Rino Y., Masuda M., Miyagi Y. Global histone modification of H3K27 correlates with the outcomes in patients with metachronous liver metastasis of colorectal cancer. Eur. J. Surg. Oncol. 2013;39:655–661. doi: 10.1016/j.ejso.2013.02.023. [DOI] [PubMed] [Google Scholar]
- Tetsu O., McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- Vardabasso C., Hasson D., Ratnakumar K., Chung C.Y., Duarte L.F., Bernstein E. Histone variants: emerging players in cancer biology. Cell. Mol. Life Sci. 2014;71:379–404. doi: 10.1007/s00018-013-1343-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B., Papadopoulos N., Velculescu V.E., Zhou S., Diaz L.A., Jr., Kinzler K.W. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Chang J.F., Yan H., Wang D.L., Liu Y., Jing Y., Zhang M., Men Y.L., Lu D., Yang X.M., Chen S., Sun F.L. A conserved RAD6-MDM2 ubiquitin ligase machinery targets histone chaperone ASF1A in tumorigenesis. Oncotarget. 2015;6:29599–29613. doi: 10.18632/oncotarget.5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei T.T., Lin Y.T., Chen W.S., Luo P., Lin Y.C., Shun C.T., Lin Y.H., Chen J.B., Chen N.W., Fang J.M. Dual targeting of 3-hydroxy-3-methylglutaryl coenzyme A reductase and histone deacetylase as a therapy for colorectal cancer. EBioMed. 2016;10:124–136. doi: 10.1016/j.ebiom.2016.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J., Ge Z., Wang L., Li Q., Wang N., Bjorkholm M., Jia J., Xu D. The histone demethylase RBP2 is overexpressed in gastric cancer and its inhibition triggers senescence of cancer cells. Gastroenterology. 2010;138:981–992. doi: 10.1053/j.gastro.2009.10.004. [DOI] [PubMed] [Google Scholar]
- Zhao C.M., Hayakawa Y., Kodama Y., Muthupalani S., Westphalen C.B., Andersen G.T., Flatberg A., Johannessen H., Friedman R.A., Renz B.W. Denervation suppresses gastric tumorigenesis. Sci. Transl. Med. 2014;6:250ra115. doi: 10.1126/scitranslmed.3009569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Li W., Zang W., Liu Z., Xu X., Yu H., Yang Q., Jia J. JMJD2B promotes epithelial-mesenchymal transition by cooperating with beta-catenin and enhances gastric cancer metastasis. Clin. Cancer Res. 2014;19:6419–6429. doi: 10.1158/1078-0432.CCR-13-0254. [DOI] [PubMed] [Google Scholar]
- Zhao L., Liu Y., Tong D., Qin Y., Yang J., Xue M., Du N., Liu L., Guo B., Hou N. MeCP2 promotes gastric cancer progression through regulating FOXF1/Wnt5a/beta-catenin and MYOD1/caspase-3 signaling pathways. EBioMed. 2017;16:87–100. doi: 10.1016/j.ebiom.2017.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J., Shi J., Wang E., Rappaport A.R., Herrmann H., Sison E.A., Magoon D., Qi J., Blatt K., Wunderlich M., Taylor M.J., Johns C., Chicas A., Mulloy J.C., Kogan S.C., Brown P., Valent P., Bradner J.E., Lowe S.W., Vakoc C.R. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]