Abstract
Recent studies indicate important roles for SMAD4 in SMCs proliferation, extracellular matrix maintenance, and blood vessel remodeling. However, the genetic effects of SMAD4 in the pathogenesis of thoracic aortic aneurysm and dissection (TAAD) are still largely unknown. Here we identified a functional variant of SMAD4 which might be involved in the pathological progression of TAAD. Five tagging SNPs of SMAD4 were genotyped in 202 TAAD cases and 400 controls using MALDI-TOF. rs12455792 CT or TT variant genotypes was associated with an significantly elevated TAAD risk (adjusted OR = 1.58, 95%CI = 1.09–2.30) under a dominant genetic model. It was located in the 5’UTR and predicted to influence transcription activity and RNA folding of SMAD4. In luciferase reporter assay, rs12455792 T allele markedly decreased luciferase activities. Accordingly, SMAD4 expression in tissues was lower in patients with CT or TT genotypes, compared with CC. Movat's pentachrome showed that rs12455792 T allele enhanced SMCs loss and fibers accumulation. With angiotensin II induction, rate of Apoptotic SMCs was significantly higher while SMAD4 silenced. Moreover, rs12455792 T allele also increased Versican degradation via ADAMTS-4. In conclusion, this variant might promote SMCs apoptosis and proteoglycans degradation, and further facilitate the progress of TAAD. Our findings identified rs12455792 as a predictor for progression of vascular media pathological changes related thoracic aortic disorders.
Abbreviations: TAAD, thoracic aortic aneurysm and dissection; SMCs, smooth muscle cells; AMD, aortic medial degeneration; TGF-β, transforming growth factor-β; MALDI-TOF MS, matrix-assisted laser desorption ionization time-of-flight mass spectrometry; SNP, single nucleotide polymorphism; 5′UTR, 5′ untranslated region
Keywords: SMAD4, Polymorphism, Thoracic aortic aneurysm and dissection, Proteoglycans degradation, SMCs, Apoptosis
Highlights
-
•
The variant on 5'UTR of SMAD4 gene significantly increased thoracic aortic aneurysm and dissection risk.
-
•
The variant rs12455792 reduced SMAD4 expression and influenced its effects on proteoglycans degradation, SMCs apoptosis and fiber accumulation.
-
•
rs1rs12455792 might be a potential therapeutic marker in vascular media pathological changes related thoracic aortic disorders.
Understanding the genetic features of thoracic aortic aneurysm and dissection can lead to precision surgery strategy. In this study, we demonstrated that SMAD4 rs12455792 is located in the transcription factor binding site and the allele change influences transcription activity and SMAD4 expression. Decrease of SMAD4 expression promotes proteoglycans degradation, vascular SMCs apoptosis and fiber accumulation, which are involved in pathological progression of TAAD. rs12455792 might be a potential therapeutic marker in vascular media pathological changes related thoracic aortic disorders.
1. Introduction
Aortic aneurysms and dissections are associated with high morbidity and mortality, accounting for over 152,000 deaths in the United States per annum (Benjamin et al., 2017). These are life-threatening diseases due to the predisposition for rupture. About 40% of patients with aortic dissection die immediately and have no enough time to reach a hospital. Aortic medial degeneration (AMD) is considered proceed thoracic aortic aneurysms and dissections (TAAD). AMD is histopathologically characterized by loss of smooth muscle cells (SMCs), and increased proteoglycans degradation.
SMAD4 plays a pivotal role in the pathological progression of vascular disorder (Zhang et al., 2016). SMAD4, a member of Smad family, is a central mediator in the canonical TGF-β signaling. It regulates biological processes important for the pathogenesis of TAAD, such as SMCs migration and proliferation, extracellular matrix degradation (Mao et al., 2012). SMAD4-deficient SMCs can trigger aortic wall inflammation by producing chemokines and recruiting macrophages (Zhang et al., 2016). Besides, SMAD4 also play essential roles in cardiogenesis, blood vessel remodeling, maturation and integrity (Jiao et al., 2003; Lan et al., 2007; Qi et al., 2007; Song et al., 2007).
The TGF-β signaling network consists of a complex of ligands, receptors, and transcriptional coregulators that have important effects in vascular remodeling and matrix degradation (Jones et al., 2009). Mutations of members (TGFBR1/2, FBN1) in this network are causative for disorders hallmarked by aortic aneurysm, e.g. Marfan syndrome (MFS) or Loeys–Dietz syndrome (LDS) (Loeys et al., 2005, 2006). Mutations in a TGF-β ligand-TGF-β2 gene have been recognized as a cause of TAAD with MFS (Boileau et al., 2012). SMAD4 encodes the only co-smad in TGF-β signaling (Mao et al., 2012). The mutations of SMAD4 gene are also important in progression of aortopathy. In Heald's retrospective study, the authors described a high prevalence (38%) of aortopathy in patients with juvenile polyposis (JPS) and hereditary hemorrhagic telangiectasia (HHT) is attributed to SMAD4 mutations (Heald et al., 2015). Gallione and Andrabi also reported that SMAD4 mutations could cause a combined JPS & HHT syndrome and vascular disorders (for example, aortic root dilation, multiple arteriovenous malformation) (Andrabi et al., 2011; Gallione et al., 2004). Thus, it is interesting to explore the association between variants of SMAD4 and the pathological progression of TAAD, which may shed light on the role of SMAD4 in pathogenesis of TAAD and provide a maker for disease diagnosis.
To investigate the genetic effect of SMAD4 on TAAD, five tagging SNPs were initially genotyped in 202 subjects and 400 healthy controls. The potential function of the significant SNP in the screening dataset was further analyzed by bioinformatic software. A series of experiments was conducted to investigate the potential molecular mechanism of the significant SNP.
2. Materials and Methods
2.1. Subjects
All the experiments involving human specimens were approved by Institutional Review Board (IRB) at the first affiliated hospital of Soochow University from January 2010 to December 2015. In the case-control study, the response rate for cases was 94% (n = 223) and for controls 92% (n = 437). Each participant has signed the written informed consent. After selecting, 202 patients with TAAD and 400 healthy controls during physical examination were enrolled in the study. All subjects were Han Chinese from eastern China. Patients with familial TAAD were exclude by the following criteria: (1) He/she had one first-degree relative with a documented TAAD history; (2) He/she had two second-degree relatives with TAAD. Controls were frequency matched by age and gender to cases.
The aortic status of subjects were evaluated by at least one type of imaging examination, such as echocardiography, angiography, CT, MRI. Each subject were interviewed in person and filled a structured questionnaire including demographic information, previous medical histories, diet, tobacco and alcohol use, weight, family history of aortopathy. Freshly frozen aortic tissues from 37 TAAD patients were obtained during the Bentall procedures or other large vessels replacement at Department of Cardiovascular Surgery, the first affiliated hospital of Soochow University. Normal aortic tissues were collected from patients who received aortic valve replacement.
2.2. Blood Sample Collection and Genomic DNA Extraction
Ethylenediamine tetraacetic acid (EDTA) anticoagulated peripheral blood samples were collected from patients before surgery and healthy controls. Genomic DNA was extracted using a RelaxGene Blood DNA System (TIANGEN biotech, Beijing, China) according to the manufacturer's instruction. The laboratory assistant was blinded to the samples about the case-control status.
2.3. SNP Selection and Genotyping
We selected 5 tagger SNPs by analyzing SMAD4-related Han Chinese data from 1000 Genome Project resources (http://www.1000genomes.org) using Run Tagger program in Haploview 4.2 (Broad Institute, Cambridge, MA, USA). These SNPs should meet the following criteria: (1) the minor allele frequency (MAF) > 0.05; (2) r2 > 0.80 for each paired SNPs (Fig. 1B). rs12455792 (in the 5′UTR, -650C > T) is located at a transcription factor binding site, with predicted proximal transcriptional regulatory potential (http://rsnp.psych.ac.cn/) (http://snpinfo.niehs.nih.gov/cgi-bin/snpinfo/snpfunc.cgi). rs61751987, rs12456284, rs3819122 and rs2282544 were also predicted as functional SNPs, which located in the enhancer, chromatin interactive region or other transcriptional regulatory region.
Fig. 1.
Representative MALDI-TOF MS spectra of rs12455792 and linkage disequilibrium (LD) analysis of the tagger SNPs in SMAD4 gene. (A) Genotypes of rs12455792 are determined by plotting peak intensity (y-axis) against mass (Da) (x-axis). (a) MALDI-TOF MS spectrum of a single peak at 8949.0 Da indicates homozygous genotype of CC. (b) MALDI-TOF MS spectrum of a single peak at 8932.5 Da indicates homozygous genotype of TT. (c) MALDI-TOF MS spectrum of 2 peaks at 8932.5 Da and 8949.0 Da represents heterozygous genotype of CT. (B) Pairwise LD among 5 tagger SNPs of SMAD4. The r2 value in each diamond indicates the pairwise correlation between SNPs. The color scale ranges from white to black reflects lower to higher r2 values.
SNPs were genotyped using matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) analysis on the MassARRAY platform (Sequenom, San Diego, CA, USA). The amplification and single-base extension primers applied in multiple PCR were synthesized by Benegene (Benegene Biotechnology, Shanghai, China) and listed in Supplement Table 1. The product of each sample was dispensed onto a 384-format SpectroCHIP with the MassARRAY Nanodispenser RS 1000. Then the MALDI-TOF MS assay was performed on a MassARRAY Compact Analyzer. Genotype calling was conducted in real time with MassARRAY RT software version 3.0. Data was analyzed with MassARRAY Typer software 4.0.3. Genotyping quality was assessed by Sanger sequencing of ~ 15% randomly selected samples, yielding a 100% concordance. The success rates of genotyping for these SNPs were > 99%.
2.4. Bioinformatic Prediction of Functional SNPs
Since the specific RNA structure determines certain function, we detected the potential change of rs12455792 on RNA folding structure. A 100 bp polymorphism-flanking region RNA was online analyzed using RNAfold (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi) and SNPfold (http://ribosnitch.bio.unc.edu/Downloads/SNPfold/). SNPs were predicted for putative transcription factor/miRNA binding site applying rSNP (http://rsnp.psych.ac.cn/), SNPinfo (http://snpinfo.niehs.nih.gov/cgi-bin/snpinfo/snpfunc.cgi) and JASPAR (http://jaspar.genereg.net/). The conservation analysis for transcription factor binding sites was performed using Seqlogo (http://www.bio.cam.ac.uk/seqlogo/logo.cgi).
2.5. Luciferase Reporter Assay
A 500 bp transcription regulatory sequence (TRS) centered at -650C > T of SMAD4 promoter was cloned into the pGL3 basic vector (Promega, Madison, WI, USA). This plasmid was named as pGL3-SMAD4 TRS reporter (Fig. 2B(b)). With C allele in the -650C > T site of TRS, the plasmid was pGL3-SMAD4 TRS-C allele. Otherwise, the plasmid was pGL3-SMAD4 TRS-T allele. Day 1, HASMCs and HAECs were seeded at 1 × 104 cells per well in 24-well plates. Day 2, cells were transfected with 800 ng pGL3 basic vector or pGL3-SMAD4 TRS reporter and 160 ng pRL-TK (Luciferase Assay System; Promega) using Lipofectamine 2000 (Invitrogen Life Technologies, Carlsbad, CA, USA). Day 3, cells were harvested to detect luciferase activity applying the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) on Synergy H1 microplate reader (BioTek Instruments, Winooski, VT, USA). Results were represented as relative luciferase activity to pRL-TK.
Fig. 2.
rs12455792 affected SMAD4 gene expression. (A) In silico prediction of rs12455792 impact on RNA folding structures. The structures corresponding to rs12455792C (a) or T allele (c). Mountain plots related to rs12455792C (b) or T allele (d). y-axis: minimum free energy or entropy. x-axis: sequence position. (B) Reporter plasmids construction. The schematic diagram of pGL3-basic vector (a) and pGL3-SMAD4-TRS reporter (b). (C) Representative graph of relative luciferase activity of cells transfected with pGL-3 basic vector or pGL3-SMAD4-TRS reporter. Six replicates for each group and the experiment was repeated at least three times. *P < 0.05. (D) Western blot analysis for SMAD4 expression in aorta tissues from patients with different genotypes (a) and corresponding quantitative analysis (b).
2.6. Western Blot
Protein was extracted from human aorta tissues of 12 TAAD patients or SMCs culture medium, and subjected to western blot with mouse monoclonal antibodies against SMAD4, rabbit polyclonal antibodies against Versican or Decorin (1:1000, Abcam, Cambridge, UK). β-actin was used as internal control. The grey scales of target proteins were normalized against β-actin using Gel-Pro analyzer 4.0 (Media Cybernetics, Silver Spring, MD, USA).
2.7. Movat's Pentachrome Staining
Human aortic tissues were fixed in 4% paraformaldehyde, dehydrated in gradient alcohol, transparented in xylene, embedded in paraffin. Histological cross-sections (5 μm) were cut and Movat's Pentachrome stained according to the Movat and Schmidt's protocols (Movat, 1955; Schmidt, 1996). In brief, sections were Bouin-fixed and incubated in Hypo solution. Then they were stained using 1% Alcian blue, 10% Weigert hematoxylin, Crocein Scarlet-Acid Fuchsin, phosphotungstic acid, alcoholic Saffron solution step by step.
Quantitative analysis of fibers (brownish-black) and cell counting of SMCs (purple-red) were performed using Image-Pro Plus 6.0 software (Media Cybernetics, Silver Spring, MD, USA). At 200 × magnification, 10 representative staining fields of each section were examined to generate the mean Integrate Optical Density (IOD). IOD represents the intensity of staining signals as detected positive pixels. The data was independently measured by 2 researchers (Y.W. and G.B.). They were blinded to the genetic features of patients. The average IOD difference between two groups (n = 5) was statistically analyzed using Student's t-test.
2.8. Flow Cytometry for Cell Apoptosis
After angiotensin II induction for 24 h, cells were digested by 0.25% trypsin (Beyotime, Shanghai, China) and stained by Annexin V-FITC and PI according to the manufacture's instruction (Invitrogen Life Technologies, Carlsbad, CA, USA). The fluorescence signals of apoptosis cells were detected on Guava EasyCyte HT Microcapillary flow cytometer (Milipore Corporation, Hayward, CA, USA). The percentages of cells in early-apoptosis or late-apoptosis stage were calculated applying Flowjo software (Tree Star Inc., Ashland, OR, USA).
2.9. Statistical Analysis
Differences of the demographic and clinical characteristics, and frequencies of genotypes in case-control study were tested by the Student's t-test (for continuous variables) or Chi-square test (for categorical variables). Hardy–Weinberg equilibrium (HWE) was calculated by online analytical tools (https://ihg.gsf.de/cgi-bin/hw/hwa1.pl). For the evaluation of main effect of SNPs, univariate and multivariate logistic regression models were conducted to calculate ORs and corresponding 95%CIs with adjustment for possible confounders. All statistical tests were two-sided and performed using Statistical Program for Social Sciences (SPSS 16.0, Chicago, IL, USA) and R (http://www.r-project.org/). P-value < 0.05 was considered as statistically significant.
3. Results
3.1. Characteristics of the Study Population
As shown in Table 1, the study included 202 TAAD patients and 400 healthy controls. The cases and controls were adequately frequency-matched for age and sex (P = 0.212 and 0.271, respectively). Compared with the cases, the controls were not significantly different in smoking status (P = 0.558). Thereafter, these variables were further adjusted for in multivariate logistic regression models.
Table 1.
Demographics and clinical features of the TAAD patients and healthy controls involved in this study.
| Items | Cases no. (%) n = 202 |
Controls no. (%) n = 400 |
P value |
|---|---|---|---|
| Age, years | 54.1 ± 14.2 | 52.4 ± 15.2 | 0.212 |
| Sex (male), n (%) | 152 (75.2) | 284 (71.0) | 0.271 |
| Smoking, n (%) | 105 (52.0) | 218 (54.5) | 0.558 |
| Diabetes, n (%) | 29 (14.4) | / | / |
| Hypertension, n (%) | 81 (40.1) | 96 (24.0) | 0.000 |
| Body mass index, kg/m2 | 23.93 ± 4.04 | 24.32 ± 4.22 | 0.124 |
| Total cholesterol, mmol/L | 4.16 ± 1.70 | 3.17 ± 1.46 | 0.033 |
| HDL-c, mmol/L | 1.15 ± 0.29 | 1.28 ± 0.31 | 0.000 |
| LDL-c, mmol/L | 2.45 ± 0.80 | 2.57 ± 0.97 | 0.141 |
The results were in bold, if the 95% CI excluded 1 and P < 0.05.
3.2. Associations of SMAD4 Genotypes and Hyplotypes With Risk of TAAD
The genotype distributions of the five tagger SNPs among the cases and controls were listed in Table 2. Representative graphs of MALDI-TOF MS spectra for rs12455792 were shown in Fig. 1A. The genotype frequencies in healthy controls were in consistent with the Hardy-Weinberg equilibrium (HWE) (all P > 0.05). Of the 5 SNPs, rs12455792 variant CT/TT genotypes were significantly associated with an increased risk of TAAD. Under the heterozygous model and dominant genetic model, the CT or CT + TT genotypes were significantly associated with higher risk of TAAD (Adjusted OR = 1.51, 1.58; 95%CI = 1.01–2.24, 1.09–2.30, respectively), compared with the wild-type homozygous CC genotype. The T allele of rs12455792 was significantly associated with higher risk of TAAD, compared with the C allele (Adjusted OR = 1.35, 95%CI = 1.07–1.72). However, no significant results were found of other 4 SNPs (rs61751987 G > A, rs12456284 A > G, rs3819122 A > C, rs2282544 G > C). In haplotype analysis, the frequency of inferred haplotypes of SMAD4 gene based on observed genotypes and their association with TAAD risk were shown in Table 3. Haplotypes CAACG (P = 0.000) and CAGCG (P = 0.015) was associated with TAAD risk significantly.
Table 2.
Logistic regression analysis of associations between the genotypes of SMAD4 and TAAD risk.
| Variants | Genotypes | Cases no. (%) n = 202 |
Controls no. (%) n = 400 |
Crude OR (95%CI) | Pa | Adjusted OR (95%CI) | Pb |
|---|---|---|---|---|---|---|---|
| rs12455792 | CC | 52 (25.7) | 142 (35.5) | 1.00 (Reference) | 1.00 (Reference) | ||
| (HWE = 0.936) | CT | 106 (52.5) | 192 (48.0) | 1.51 (1.01–2.24) | 0.026 | 1.51 (1.01–2.24) | 0.032 |
| TT | 44 (21.8) | 66 (16.5) | 1.82 (1.11–2.99) | 0.013 | 1.81 (1.11–2.99) | 0.016 | |
| CT + TT | 150 (74.3) | 258 (64.5) | 1.59 (1.09–2.31) | 0.009 | 1.58 (1.09–2.30) | 0.011 | |
| C allele | 210 (52.0) | 476 (59.5) | 1.00 (Reference) | 1.00 (Reference) | |||
| T allele | 194 (48.0) | 324 (40.5) | 1.36 (1.07–1.73) | 0.008 | 1.35 (1.07–1.72) | 0.010 | |
| rs61751987 | GG | 153 (75.7) | 299 (74.8) | 1.00 (Reference) | 1.00 (Reference) | ||
| (HWE = 0.098) | GA | 41 (20.3) | 89 (22.3) | 0.88 (0.54–1.42) | 0.624 | 0.85 (0.51–1.39) | 0.685 |
| AA | 8 (4.0) | 12 (3.0) | 1.33 (0.45–3.92) | 0.787 | 1.31 (0.42–3.81) | 0.829 | |
| GA + AA | 49 (24.3) | 101 (25.3) | 0.99 (0.63–1.55) | 1.000 | 0.97 (0.59–1.52) | 1.000 | |
| G allele | 347 (85.9) | 687 (85.9) | 1.00 (Reference) | 1.00 (Reference) | |||
| A allele | 57 (14.1) | 113 (14.1) | 0.99 (0.49–2.00) | 1.000 | 0.97 (0.43–1.91) | 1.000 | |
| rs12456284 | AA | 93 (46.0) | 195 (48.8) | 1.00 (Reference) | 1.00 (Reference) | ||
| (HWE = 0.065) | AG | 79 (39.1) | 157 (39.2) | 0.98 (0.66–1.47) | 1.000 | 0.96 (0.65–1.45) | 1.000 |
| GG | 30 (14.9) | 48 (12.0) | 1.19 (0.71–1.98) | 0.625 | 1.15 (0.69–1.89) | 0.676 | |
| AG + GG | 109 (54.0) | 205 (51.2) | 1.12 (0.79–1.57) | 0.546 | 1.11 (0.78–1.56) | 0.592 | |
| A allele | 265 (65.6) | 547 (68.4) | 1.00 (Reference) | 1.00 (Reference) | |||
| G allele | 139 (34.4) | 253 (31.6) | 1.10 (0.75–1.49) | 0.472 | 1.09 (0.74–1.48) | 0.535 | |
| rs3819122 | AA | 68 (33.7) | 122 (30.5) | 1.00 (Reference) | 1.00 (Reference) | ||
| (HWE = 0.297) | AC | 101 (50.0) | 207 (51.8) | 0.88 (0.60–1.28) | 0.497 | 0.87 (0.59–1.28) | 0.560 |
| CC | 33 (16.3) | 71 (17.7) | 0.83 (0.50–1.39) | 0.522 | 0.81 (0.48–1.37) | 0.583 | |
| AC + CC | 134 (66.3) | 278 (69.5) | 0.87 (0.60–1.24) | 0.458 | 0.84 (0.55–1.20) | 0.517 | |
| A allele | 237 (58.7) | 451 (56.4) | 1.00 (Reference) | 1.00 (Reference) | |||
| C allele | 167 (41.3) | 349 (43.6) | 0.84 (0.56–1.27) | 0.462 | 0.83 (0.55–1.26) | 0.507 | |
| rs2282544 | GG | 148 (73.3) | 282 (70.5) | 1.00 (Reference) | 1.00 (Reference) | ||
| (HWE = 0.086) | GC | 47 (23.2) | 102 (25.5) | 0.88 (0.59–1.31) | 0.548 | 0.86 (0.58–1.30) | 0.617 |
| CC | 7 (3.5) | 16 (4.0) | 0.83 (0.34–2.07) | 0.823 | 0.77 (0.29–1.94) | 0.865 | |
| GC + CC | 54 (26.7) | 118 (29.5) | 0.87 (0.60–1.27) | 0.505 | 0.86 (0.59–1.26) | 0.542 | |
| G allele | 343 (84.9) | 666 (83.2) | 1.00 (Reference) | 1.00 (Reference) | |||
| C allele | 61 (15.1) | 134 (16.8) | 0.85 (0.47–1.59) | 0.627 | 0.82 (0.45–1.58) | 0.679 | |
| Combined effect of risk genotypes | |||||||
| 0 | 0 (0) | 1 (0.2) | 1.00 (Reference) | 1.00 (Reference) | |||
| 1 | 3 (1.5) | 15 (3.7) | 0.39 (0.11–1.37) | 0.202 | 0.37 (0.09–1.31) | 0.217 | |
| 2 | 29 (14.3) | 65 (16.2) | 0.87 (0.54–1.41) | 0.635 | 0.84 (0.52–1.38) | 0.642 | |
| 3 | 86 (42.6) | 169 (42.3) | 1.01 (0.72–1.42) | 1.000 | 1.03 (0.73–1.46) | 0.981 | |
| 4 | 67 (33.2) | 123 (30.8) | 1.11 (0.77–1.60) | 0.577 | 1.13 (0.77–1.63) | 0.569 | |
| 5 | 17 (8.4) | 27 (6.8) | 1.28 (0.68–2.42) | 0.507 | 1.26 (0.66–2.41) | 0.496 | |
| Trend | |||||||
| 0–3 | 84 (41.6) | 150 (37.5) | 1.00 (Reference) | 1.00 (Reference) | |||
| 4–5 | 118 (58.4) | 250 (62.5) | 1.18 (0.84–1.67) | 0.374 | 1.19 (0.84–1.69) | 0.362 | |
CI, confidence interval; OR, odds ratio.
Chi-square test for genotype distributions between cases and controls.
Adjusted for age, sex, smoking status in logistic regress models.
Table 3.
The frequency of inferred haplotypes of SMAD4 gene based on observed genotypes and their associations with TAAD risk.
| Haplotypesa | Cases no. (%) n = 404 |
Controls no. (%) n = 800 |
Crude OR (95%CI) | P | Adjusted OR (95%CI) | Pb |
|---|---|---|---|---|---|---|
| CGAAG | 92 (22.8) | 168 (21.0) | 1.00 (Reference) | 1.00 (Reference) | ||
| CGACG | 64 (15.8) | 118 (14.8) | 1.08 (0.71–1.66) | 0.745 | 1.06 (0.67–1.68) | 0.769 |
| CGGAG | 38 (9.4) | 96 (12.0) | 0.71 (0.44–1.15) | 0.179 | 0.65 (0.37–1.09) | 0.152 |
| CGGCG | 25 (6.2) | 39 (4.9) | 1.18 (0.60–2.33) | 0.730 | 1.15 (0.53–2.41) | 0.795 |
| CAAAG | 17 (4.2) | 52 (6.5) | 0.58 (0.30–1.11) | 0.105 | 0.72 (0.28–1.42) | 0.179 |
| CGAAC | 30 (7.4) | 51 (6.4) | 1.23 (0.57–1.96) | 1.000 | 1.25 (0.61–2.12) | 1.000 |
| CGGAC | 12 (3.0) | 18 (2.3) | 0.73 (0.25–2.16) | 0.599 | 0.79 (0.21–2.35) | 0.623 |
| CGACC | 11 (2.7) | 18 (2.3) | 1.12 (0.42–2.96) | 1.000 | 1.23 (0.57–4.21) | 1.000 |
| CAACG | 6 (1.5) | 52 (6.5) | 0.21 (0.09–0.53) | 0.000 | 0.20 (0.08–0.51) | 0.000 |
| CAGCG | 0 (0) | 6 (0.8) | 0.49 (0.44–0.54) | 0.015 | 0.45 (0.41–0.52) | 0.023 |
| TAAAG | 39 (9.7) | 54 (6.8) | 0.53 (0.90–2.62) | 0.138 | 0.56 (0.90–2.69) | 0.159 |
| TGACG | 16 (4.0) | 26 (3.3) | 1.29 (0.57–2.89) | 0.680 | 1.26 (0.54–2.97) | 0.716 |
| TGGCG | 14 (3.5) | 24 (3.0) | 1.17 (0.53–2.59) | 0.840 | 1.15 (0.47–2.52) | 0.876 |
| TGAAG | 29 (7.2) | 44 (5.5) | 1.36 (0.75–2.45) | 0.369 | 1.32 (0.69–2.32) | 0.423 |
| TGACC | 5 (1.2) | 28 (3.5) | 0.32 (0.10–0.99) | 0.044 | 0.35 (0.11–1.05) | 0.076 |
| TGGAG | 6 (1.5) | 6 (0.8) | 2.01 (0.50–8.15) | 0.503 | 1.95 (0.45–7.59) | 0.582 |
The haplotypes order were rs12455792, rs61751987, rs12456284, rs3819122, and rs2282544.
Obtained in logistic regression models with adjustment for age, gender, and smoking status.
3.3. Association of rs12455792 Genotypes and SMAD4 Expression
rs12455792 is located in the 5′UTR of SMAD4 gene (-650) and it is predicted to affect the activity of transcription factor binding site (Supplement Fig. 1). While allele C changes to T, both the number of potential binding factors and affinity scores are decreased (Supplement Fig. 2A). Sequence logo graph indicated a high conservation of C allele in Sox6 binding site (Supplement Fig. 2B). In a word, this locus is important in transcriptional regulation. To evaluated the potential impact of rs12455792 on SMAD4 expression, we constructed two reporter plasmids containing either rs12455792 C or T allele and transfected them into HASMCs or HAECs. As shown in Fig. 2, the pGL3-SMAD4 TRS-C allele presented a significantly higher relative luciferase activity, compared with pGL3-SMAD4 TRS-T allele in HASMCs and HAECs. These data suggested that rs12455792 C → T change in 5′UTR might decrease the transcriptional activity of SMAD4. According to RNAfold prediction, local folding structures of rs12455792 flanking-region RNA in the 5′UTR of SMAD4 greatly changed with the C → T alteration, with the minimum free energy increasing from − 8.30 to − 7.80 kcal/mol (Fig. 2A). This change might affect RNA protein binding and SMAD4 expression. We further detected the SMAD4 expression in freshly frozen aorta tissues from patients with different genotype. The quantitative analysis of western blot indicated that SMAD4 expression was significant higher in samples with CC genotype than that in samples with variant CT or TT genotype (Fig. 2D). These findings were in consistent with the data of luciferase reporter assay.
3.4. The Variant rs12455792 Aggravated Versican Degradation via ADAMTS-4
Given the observed significant association between rs12455792 and TAAD risk, we further measured the levels of Versican and Decorin degradation products in aortic tissues from patients with CC, CT and TT genotypes. As shown in Fig. 3, the levels of Versican (fragment of 95 kDa and 70 kDa) and Decorin (fragment of 50 kDa) degradation products were significantly higher in tissues with variant CT or TT than in that with CC genotype. As ADAMTS-1 and ADAMTS-4 levels were strongly correlated with Versican degradation in aortic tissues (Ren et al., 2013), we hypothesized that SMAD4 might influence the expression of ADAMTS-1/4. The results of western blot assay suggested that ADAMTS-4 (not ADAMTS-1) expression was significantly higher in SMAD4-silenced HASMCs than that in negative control (NC) HASMCs (Fig. 3C). In cell medium, the Versican degradation was more serious after SMAD4-silenced, but rescued by ADAMTS-4 inhibitor (Fig. 4D). These evidences indicated that SMAD4 inhibited the ADAMTS-4 effects on Versican degradation.
Fig. 3.
SMAD4 inhibited Versican and Decorin degradation via ADAMTS-4. Western blot analysis and quantitative analysis for Versican (A) or Decorin (B) degradation products in aortic tissues of patients. *P < 0.05, ns: not significant. (C) Western blot analysis for ADAMTS-1/4 protein levels in SMAD4-silenced cells or not. (D) Versican degradation products detection in medium of cells with different treatment.
Fig. 4.
SMAD4 inhibited the apoptosis of HASMCs. (A) Movat's pentachrome staining of normal aortic tissues or tissues from TAAD patients with different genotype. In each group, Scale bar = 50 μm (left)/100 μm (right). (B) Representative scatterplots of flow cytometry for apoptosis analysis. (a) FSC/SSC scatterplot for cells gating. Annexin V-FITC/PI scatterplots of parental (b), siRNA-NC (c), siRNA-SMAD4 (d) group. Quantitative analysis of SMCs (C) and fibers (D) (n = 5). *P < 0.05. (E) Percentages of early or late-stage apoptotic cells in different groups.*P < 0.05. All experiments were performed in triplicates. Data was presented as mean ± SD.
3.5. The Variant rs12455792 Promoted the Apoptosis of HASMCs
It is known to all that loss of medial SMCs makes an important contribution to aortic aneurysm and dissection because the cell population is capable of directing connective tissue repair. To detect the alteration of SMCs and fibers, we performed Movat's pentachrome staining in aortic tissues from different group. Compared to normal aortic tissues, which were full of SMCs (purple-red) (Fig. 4A), the amount of SMCs was really significantly lower in tissues from TAAD patients. Quantitative analysis revealed that the numbers of SMCs were significantly higher in CC group than that in CT/TT group (Fig. 4C). However, the IOD of elastic and reticular fibers (brownish-black) in CC group was much lower, compared with that in CT or TT group (Fig. 4D). As described above, SMAD4 expression was associated with the variant of rs12455792. Thus we further detected the apoptosis of HASMCs transfected with siRNA-NC or siRNA-SMAD4 (Fig. 4B). After induction of angiotensin II for 24 h, both the rate of early and late apoptotic cells (%) in siRNA-SMAD4 group was notably higher, compared with that in siRNA-NC group. These phenomena suggested that the inhibition of SMAD4 expression increased angiotensin II-induced apoptosis of SMCs. Wild type C allele of rs12455792 might be a protect factor which prevented SMCs from apoptosis.
4. Discussion
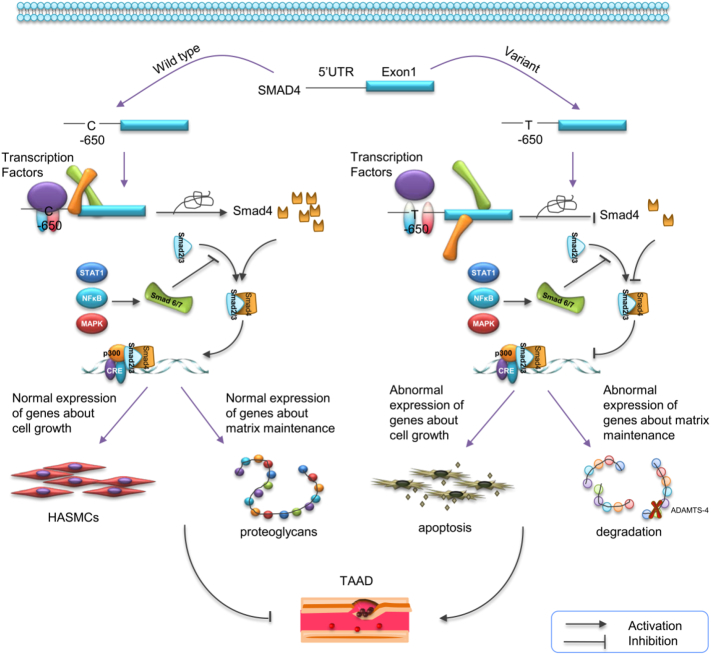
The TGF-β superfamily is composed of various secreted polypeptides which regulate cell proliferation, differentiation and tissue morphogenesis in many organisms (Xie et al., 2014). SMAD4 has been recently identified as the unique co-smad which mediates the responses to TGF-β and related factors. Multiple studies in animal models showed that the deficiency of SMAD4 contributed to the development of thoracic aortic aneurysm or other vascular disorder (Lan et al., 2007; Mao et al., 2012; Zhang et al., 2016). In the present study, we investigated the genetic effects of SMAD4 on the pathogenesis of TAAD. Five tagger SNPs were genotyped in 202 TAAD patients and 400 healthy controls. There was a significant association between individuals carrying at least one T allele of rs12455792 and an increased risk of TAAD. In functional analysis, we found that the variant reduced SMAD4 expression, thus promote proteoglycans degradation and SMCs apoptosis, which were important pathophysiological processes in TAAD (Fig. 5).
Fig. 5.
Schematic diagram for potential roles of the SMAD4 variant in pathological progression of TAAD. Variant T allele of rs12455792 might inhibit SMAD4 expression by affecting the affinity between transcription factors and 5′UTR of this gene, resulting in MSCs apoptosis and Versican degradation, and further facilitate the progress of TAAD.
rs12455792 is located in the 5′UTR of SMAD4. This locus is predicted as a transcription factors binding site, with proximal transcriptional regulatory potential. While allele C changes to T, both the number of potential binding factors and affinity scores are decreased. Sequence logo graph indicated a high conservation of C allele in Sox6 binding site (Supplemental Fig. 2). These analyses determined the significance of this locus in transcriptional regulation. In view of the evidences shown in Fig. 1, rs12455792 C → T change reduced the transcriptional activity, altered the RNA folding structure, and decreased SMAD4 expression. Except for the reduced affinity of transcription factor and -650 region DNA, the C to T substitution also changed RNA folding structure greatly. RNA hairpin structure might act like a hinge that open up DNA and bind the kinase domain. However, the variant of C to T might attenuated the function of RNA hairpin structure, thus down-regulated gene expression. To the best of our knowledge, rs12455792 is a functional SNP involved in the pathogenesis of TAAD. Although it has been reported in cancer metastasis (Li et al., 2012), the present study is the first to demonstrate potential function of this SNP.
Another intriguing finding was that rs12455792 might modulate the essential pathophysiological processes in TAAD-SMCs apoptosis, fibers accumulation and proteoglycans degradation, by fine tuning the SMAD4 expression. As we all known, the aorta is composed of 3 layers: tunica intima, tunica media and tunica adventitia. The tensile strength and elasticity of the aorta mainly depend on the tunica media, which is formed of concentrically arranged elastic fibers and SMCs (Milewicz et al., 2008). In response to pulsatile blood flow, SMCs regulate blood flow and pulse pressure via contracting and secreting vascular regulatory factors (Milewicz et al., 2008). Loss of vascular SMCs is a key event in progression of TAAD. Our study showed that the numbers of SMCs in tissues from CC group was significantly higher than variant group. The status of SMCs loss was more graveness in the latter. While SMAD4 expression was lower in variant group as described in Fig. 2, we hypothesized that SMCs loss might be related to decreased SMAD4 expression, since it played a pivotal role in SMCs proliferation and blood vessel remodeling. After induction of angiotensin II for 24 h, the percentage of both early and late-stage apoptotic cells were notably higher in siRNA-SMAD4 group. This suggested that SMAD4 was really important in maintenance of the SMCs number and normal physiological function of aorta. We also found that the elastic fibers and reticular fibers (brownish-black) were more disorganized and accumulated in tissues from patients with CT or TT genotype. This suggested that rs12455792 might be associated with fiber organization. Although there were no literatures about relationship between fibers accumulation and vascular disorders, we believed that the increase of components in connective tissues might destroy the structural and functional integrity of aortic wall, and reduce its reactive potency to changed blood pressure. SMAD4 might be involved in this process. These viewpoints needed more investigations to confirm.
We further demonstrated that the proteoglycans (e.g. Versican, Decorin) in matrix degraded more seriously in aorta tissues from CT or TT groups. Proteoglycans interact with hyaluronan to form large aggregates. They can retain water and create reversible compressive layer to protect vascular from lesions of pressure waves and shearing force (LeBaron et al., 1992). Thus, proteoglycans may play a pivotal role in maintaining the structural and functional integrity of the vascular wall. The degradation of proteoglycans may be responsible for vascular diseases (Evanko et al., 1999; Kenagy et al., 2006). In TAAD tissues, we observed that the degradation of Versican and Decorin was related to rs12455792 genotypes. Moreover, western blot data showed a significantly increased degradation of Versican in SMAD4-silenced SMCs, but it was rescued after the addition of ADAMTS-4 inhibitor. Our findings suggested that SMAD4 low expression might promote Versican degradation via ADAMTS-4. Versican was reported as main proteoglycan substrate of ADAMTS proteinases in the aorta (Sandy et al., 2001). With Movat's pentachrome (Fig. 4), we found that proteoglycans aggregates (aggregates without degradation were stained as bright-blue) were more in CC group than that in CT or TT group. This was consistent with the degradation status of relative groups which were reflected by western blot.
There were several limitations in the current study: (1) Since all the subjects were recruited from eastern Han Chinese population, the association between SMAD4 SNPs and risk of TAAD might not be generalized to other ethnic groups. (2) Large cohort, multi-center investigations should be performed to provide more potent statistical power. (3) TAAD is a complicated process. Inflammation and lymphocytes recruitment often accompany medial degeneration in the pathogenesis of TAAD (He et al., 2006). Activated T cells and macrophages may contribute to the apoptosis of SMCs and degradation of the matrix (He et al., 2006). Generous studies revealed that low SMAD4 expression might facilitate lymphocytes accumulation and infiltration (Huss et al., 2011; Inamoto et al., 2016; Sakata et al., 2017; Zhang et al., 2016). Therefore, the association between rs12455792 and inflammation or lymphocytes recruitment in the progression of TAAD is encouraging enough to warrant further investigations.
In summary, we demonstrated that SMAD4 rs12455792 CT, TT genotypes might increase the risk of TAAD by promoting proteoglycans degradation and SMCs apoptosis. To the best of our knowledge, this is the first study of relationship between functional polymorphisms of SMAD4 and pathogenic mechanism of TAAD. In a relatively complete investigation, the downstream impacts of SNPs on protein function and pathological progression should be clarified. Therefore the tissues from patients are very pivotal. The thoracic aorta tissues are difficult to obtain, for this reason studies in this area is infrequent. In the current study, SNP rs12455792 is located in the transcription factor binding site and the allele change influences transcription activity and SMAD4 expression. Decrease of SMAD4 expression promotes proteoglycans degradation, vascular SMCs apoptosis and fiber accumulation, which are involved in pathological progression of TAAD. rs12455792 might be a potential therapeutic marker in vascular media pathological changes related thoracic aortic disorders.
Funding Sources
This work is supported by National Clinical Key Specialty of Cardiovascular Surgery, Jiangsu Clinical Research Center for Cardiovascular Surgery (No. BL201451), and National Natural Science Foundation of China (No. 81401316 to F.H.). The findings had no role in study design, data collection, data analysis, interpretation, writing of the report.
Conflicts of Interest
The authors have declared that no competing interests exist.
Author Contributions
Y.W., H.H., G.B., Y.Y. and W.Y. designed the experiments and analyzed data. Y.W., G.B. and F.H. performed all the experiments. Z.S., Y.W. and Y.C. wrote the manuscript and prepared the figures. All authors reviewed and approved the final manuscript.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2017.06.022.
Appendix A. Supplementary data
Supplementary material
References
- Andrabi S., Bekheirnia M.R., Robbins-Furman P., Lewis R.A., Prior T.W., Potocki L. SMAD4 mutation segregating in a family with juvenile polyposis, aortopathy, and mitral valve dysfunction. Am. J. Med. Genet. A. 2011;155A:1165–1169. doi: 10.1002/ajmg.a.33968. [DOI] [PubMed] [Google Scholar]
- Benjamin E.J., Blaha M.J., Chiuve S.E., Cushman M., Das S.R., Deo R., de Ferranti S.D., Floyd J., Fornage M., Gillespie C. Heart disease and stroke statistics-2017 update: a report from the American Heart Association. Circulation. 2017;135:e146–e603. doi: 10.1161/CIR.0000000000000485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boileau C., Guo D.C., Hanna N., Regalado E.S., Detaint D., Gong L., Varret M., Prakash S.K., Li A.H., d'Indy H. TGFB2 mutations cause familial thoracic aortic aneurysms and dissections associated with mild systemic features of Marfan syndrome. Nat. Genet. 2012;44:916–921. doi: 10.1038/ng.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanko S.P., Angello J.C., Wight T.N. Formation of hyaluronan- and versican-rich pericellular matrix is required for proliferation and migration of vascular smooth muscle cells. Arterioscler. Thromb. Vasc. Biol. 1999;19:1004–1013. doi: 10.1161/01.atv.19.4.1004. [DOI] [PubMed] [Google Scholar]
- Gallione C.J., Repetto G.M., Legius E., Rustgi A.K., Schelley S.L., Tejpar S., Mitchell G., Drouin E., Westermann C.J., Marchuk D.A. A combined syndrome of juvenile polyposis and hereditary haemorrhagic telangiectasia associated with mutations in MADH4 (SMAD4) Lancet. 2004;363:852–859. doi: 10.1016/S0140-6736(04)15732-2. [DOI] [PubMed] [Google Scholar]
- He R., Guo D.C., Estrera A.L., Safi H.J., Huynh T.T., Yin Z., Cao S.N., Lin J., Kurian T., Buja L.M. Characterization of the inflammatory and apoptotic cells in the aortas of patients with ascending thoracic aortic aneurysms and dissections. J. Thorac. Cardiovasc. Surg. 2006;131:671–678. doi: 10.1016/j.jtcvs.2005.09.018. [DOI] [PubMed] [Google Scholar]
- Heald B., Rigelsky C., Moran R., LaGuardia L., O'Malley M., Burke C.A., Zahka K. Prevalence of thoracic aortopathy in patients with juvenile polyposis syndrome-hereditary hemorrhagic telangiectasia due to SMAD4. Am. J. Med. Genet. A. 2015;167A:1758–1762. doi: 10.1002/ajmg.a.37093. [DOI] [PubMed] [Google Scholar]
- Huss D.J., Winger R.C., Cox G.M., Guerau-de-Arellano M., Yang Y., Racke M.K., Lovett-Racke A.E. TGF-beta signaling via Smad4 drives IL-10 production in effector Th1 cells and reduces T-cell trafficking in EAE. Eur. J. Immunol. 2011;41:2987–2996. doi: 10.1002/eji.201141666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inamoto S., Itatani Y., Yamamoto T., Minamiguchi S., Hirai H., Iwamoto M., Hasegawa S., Taketo M.M., Sakai Y., Kawada K. Loss of SMAD4 promotes colorectal cancer progression by accumulation of myeloid-derived suppressor cells through the CCL15-CCR1 chemokine Axis. Clin. Cancer Res. 2016;22:492–501. doi: 10.1158/1078-0432.CCR-15-0726. [DOI] [PubMed] [Google Scholar]
- Jiao K., Kulessa H., Tompkins K., Zhou Y., Batts L., Baldwin H.S., Hogan B.L. An essential role of Bmp4 in the atrioventricular septation of the mouse heart. Genes Dev. 2003;17:2362–2367. doi: 10.1101/gad.1124803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.A., Spinale F.G., Ikonomidis J.S. Transforming growth factor-beta signaling in thoracic aortic aneurysm development: a paradox in pathogenesis. J. Vasc. Res. 2009;46:119–137. doi: 10.1159/000151766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenagy R.D., Plaas A.H., Wight T.N. Versican degradation and vascular disease. Trends Cardiovasc. Med. 2006;16:209–215. doi: 10.1016/j.tcm.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y., Liu B., Yao H., Li F., Weng T., Yang G., Li W., Cheng X., Mao N., Yang X. Essential role of endothelial Smad4 in vascular remodeling and integrity. Mol. Cell. Biol. 2007;27:7683–7692. doi: 10.1128/MCB.00577-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBaron R.G., Zimmermann D.R., Ruoslahti E. Hyaluronate binding properties of versican. J. Biol. Chem. 1992;267:10003–10010. [PubMed] [Google Scholar]
- Li Q., Wu H., Chen B., Hu G., Huang L., Qin K., Chen Y., Yuan X., Liao Z. SNPs in the TGF-beta signaling pathway are associated with increased risk of brain metastasis in patients with non-small-cell lung cancer. PLoS One. 2012;7:e51713. doi: 10.1371/journal.pone.0051713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeys B.L., Chen J., Neptune E.R., Judge D.P., Podowski M., Holm T., Meyers J., Leitch C.C., Katsanis N., Sharifi N. A syndrome of altered cardiovascular, craniofacial, neurocognitive and skeletal development caused by mutations in TGFBR1 or TGFBR2. Nat. Genet. 2005;37:275–281. doi: 10.1038/ng1511. [DOI] [PubMed] [Google Scholar]
- Loeys B.L., Schwarze U., Holm T., Callewaert B.L., Thomas G.H., Pannu H., De Backer J.F., Oswald G.L., Symoens S., Manouvrier S. Aneurysm syndromes caused by mutations in the TGF-beta receptor. N. Engl. J. Med. 2006;355:788–798. doi: 10.1056/NEJMoa055695. [DOI] [PubMed] [Google Scholar]
- Mao X., Debenedittis P., Sun Y., Chen J., Yuan K., Jiao K., Chen Y. Vascular smooth muscle cell Smad4 gene is important for mouse vascular development. Arterioscler. Thromb. Vasc. Biol. 2012;32:2171–2177. doi: 10.1161/ATVBAHA.112.253872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milewicz D.M., Guo D.C., Tran-Fadulu V., Lafont A.L., Papke C.L., Inamoto S., Kwartler C.S., Pannu H. Genetic basis of thoracic aortic aneurysms and dissections: focus on smooth muscle cell contractile dysfunction. Annu. Rev. Genomics Hum. Genet. 2008;9:283–302. doi: 10.1146/annurev.genom.8.080706.092303. [DOI] [PubMed] [Google Scholar]
- Movat H.Z. Demonstration of all connective tissue elements in a single section. Arch. Pathol. 1955;60:289–295. [PubMed] [Google Scholar]
- Qi X., Yang G., Yang L., Lan Y., Weng T., Wang J., Wu Z., Xu J., Gao X., Yang X. Essential role of Smad4 in maintaining cardiomyocyte proliferation during murine embryonic heart development. Dev. Biol. 2007;311:136–146. doi: 10.1016/j.ydbio.2007.08.022. [DOI] [PubMed] [Google Scholar]
- Ren P., Zhang L., Xu G., Palmero L.C., Albini P.T., Coselli J.S., Shen Y.H., LeMaire S.A. ADAMTS-1 and ADAMTS-4 levels are elevated in thoracic aortic aneurysms and dissections. Ann. Thorac. Surg. 2013;95:570–577. doi: 10.1016/j.athoracsur.2012.10.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata J., Yoshida R., Matsuoka Y., Nagata M., Hirosue A., Kawahara K., Nakamura T., Nakamoto M., Hirayama M., Takahashi N. Predictive value of the combination of SMAD4 expression and lymphocyte infiltration in malignant transformation of oral leukoplakia. Cancer Med. 2017;6:730–738. doi: 10.1002/cam4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandy J.D., Westling J., Kenagy R.D., Iruela-Arispe M.L., Verscharen C., Rodriguez-Mazaneque J.C., Zimmermann D.R., Lemire J.M., Fischer J.W., Wight T.N. Versican V1 proteolysis in human aorta in vivo occurs at the Glu441-Ala442 bond, a site that is cleaved by recombinant ADAMTS-1 and ADAMTS-4. J. Biol. Chem. 2001;276:13372–13378. doi: 10.1074/jbc.M009737200. [DOI] [PubMed] [Google Scholar]
- Schmidt R. Modification of movat pentachrome stain with improved reliability of elastin staining. J. Histotechnol. 1996;19:325–327. [Google Scholar]
- Song L., Yan W., Chen X., Deng C.X., Wang Q., Jiao K. Myocardial smad4 is essential for cardiogenesis in mouse embryos. Circ. Res. 2007;101:277–285. doi: 10.1161/CIRCRESAHA.107.155630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F., Zhang Z., van Dam H., Zhang L., Zhou F. Regulation of TGF-beta superfamily signaling by SMAD mono-ubiquitination. Cell. 2014;3:981–993. doi: 10.3390/cells3040981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Hou S., Chen J., Zhang J., Lin F., Ju R., Cheng X., Ma X., Song Y., Zhang Y. Smad4 deficiency in smooth muscle cells initiates the formation of Aortic aneurysm. Circ. Res. 2016;118:388–399. doi: 10.1161/CIRCRESAHA.115.308040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material