Abstract
Glioblastoma (GBM) is the most aggressive primary brain tumor in adults. The mechanisms that confer GBM cells their invasive behavior are poorly understood. The electroneutral Na+-K+-2Cl− co-transporter 1 (NKCC1) is an important cell volume regulator that participates in cell migration. We have shown that inhibition of NKCC1 in GBM cells leads to decreased cell migration, in vitro and in vivo. We now report on the role of NKCC1 on cytoskeletal dynamics. We show that GBM cells display a significant decrease in F-actin content upon NKCC1 knockdown (NKCC1-KD). To determine the potential actin-regulatory mechanisms affected by NKCC1 inhibition, we studied NKCC1 protein interactions. We found that NKCC1 interacts with the actin-regulating protein Cofilin-1 and can regulate its membrane localization. Finally, we analyzed whether NKCC1 could regulate the activity of the small Rho-GTPases RhoA and Rac1. We observed that the active forms of RhoA and Rac1 were decreased in NKCC1-KD cells. In summary, we report that NKCC1 regulates GBM cell migration by modulating the cytoskeleton through multiple targets including F-actin regulation through Cofilin-1 and RhoGTPase activity. Due to its essential role in cell migration NKCC1 may serve as a specific therapeutic target to decrease cell invasion in patients with primary brain cancer.
Keywords: Glioblastoma, NKCC1, Actin cytoskeleton, Cell migration
Highlights
-
•
NKCC1 ion co-transporter regulates actin cytoskeleton dynamics in glioblastoma, which plays a key role in cell migration.
-
•
NKCC1 interacts with actin-regulating protein Cofilin, potentially regulating its activity.
-
•
The NKCC1-Cofilin interaction brings the opportunity of finding new targets to treat patients with brain cancer.
Glioblastoma is a devastating brain tumor with prominent cell migration and invasion capabilities. Its invasiveness makes surgical resection ineffective, resulting in a high recurrence rate. Glioblastoma cells utilize multiple mechanisms to travel through the brain. One of such mechanisms is cell volume modulation, regulated by ion transporters like NKCC1. In our study, we observed that NKCC1 also regulates glioblastoma cell migration in a volume-independent way by controlling the cytoskeleton. Specifically, by interacting with the actin-regulating protein Cofilin. Our observations identify a significant target to inhibit glioblastoma cell dispersal, which is crucial given that current therapies only target cell proliferation.
1. Introduction
Glioblastoma (GBM) is the most common and aggressive primary brain tumor of the Central Nervous System (Siegel et al., 2013, Chaichana et al., 2011, Chaichana et al., 2014). GBM has cellular heterogeneity and displays key features of invasion and infiltration of healthy brain tissue (Quinones-Hinojosa and Chaichana, 2007, Giese et al., 2003). Despite multimodal therapy including surgery, radiation, and chemotherapy, the median survival is 14.6 months, and the prognosis remains dismal due to tumor recurrence (Stupp et al., 2005, Chaichana et al., 2014). This high rate of recurrence can be attributed to the high migration capacity of GBM cells as well as brain tumor initiating cells that are resistant to treatment (Guerrero-Cazares et al., 2012, Lathia et al., 2015). For these reasons, targeting proteins that promote cell migration may potentially result in better therapeutic strategies.
During cell migration, GBM cells require modifying their cellular volume to go through narrow spaces; these changes in volume are regulated through ionic transport (Watkins and Sontheimer, 2011). Ion co-transporters, such as the Sodium Potassium Chloride co-transporter (NKCC1) 1, regulate intracellular volume and Cl2− accumulation, allowing the movement of Na+, K+ and Cl− ions across the plasma membrane using the energy generated by the Na+/K+ ATPase.
Previously, we and others have determined that NKCC1 inhibition decreases GBM cell migration and invasion in vitro and in vivo (Garzon-Muvdi et al., 2012, Haas and Sontheimer, 2010). In addition, we found that NKCC1 expression levels are increased in GBM tissues (with respect to normal cortex) and that NKCC1 modulates glioma cell invasion through the regulation of cell contractility and focal adhesion dynamics (Garzon-Muvdi et al., 2012). Furthermore, we found that EGF stimulation increases the presence of active (i.e. phosphorylated) NKCC1 (Garzon-Muvdi et al., 2012). However, the intracellular mechanisms utilized by NKCC1 to regulate cell migration and adhesion changes have not been elucidated. Cytoskeleton dynamics have the potential of being the converging phenomenon that links NKCC1 activity, cell migration, and cell adhesion.
Cell movement is driven by the assembling of actin filaments at the leading edge of the cell, providing a major force to drive cell protrusions, changes in shape, migration and invasion (Insall and Machesky, 2009, Pollard and Borisy, 2003). One of the key regulators of actin assembly is Cofilin 1, which is involved in determining the direction of the protrusion and promotes lamellipodium extension and cell migration (Chen et al., 2001). Cofilin 1 severs actin filaments to produce free actin barbed ends, required for new actin polymerization (Desmarais et al., 2005, Chan et al., 2000). Actin dynamics regulated by Cofilin 1 are coupled with the activation of Rho and Rac1 family of GTPases, which are key intermediates in signal transduction driving cytoskeleton organization (Lauffenburger and Horwitz, 1996, Lang et al., 1998, Fortin Ensign et al., 2013, Nakada et al., 2007, Kwiatkowska and Symons, 2013). Interestingly, Cofilin 1 has been implicated in promoting metastasis and invasion in breast and prostate cancer, allowing the formation of filopodia and enhancing migration activity (Bravo-Cordero et al., 2013, Sidani et al., 2007).
Here, we report that NKCC1 regulates the actin cytoskeleton in primary patient-derived GBM cells serving as a protein scaffold to Cofilin, thus facilitating its localization at the plasma membrane. Upon NKCC1 knockdown, there is a decreased expression of Cofilin1 at the plasma membrane coupled with a decrease of RhoA and Rac1 activity. These events lead to a reduction in the formation of filamentous actin, delayed cell spreading, and reduced migration. Our data shows NKCC1 as a potential component of the actin cytoskeleton machinery of primary-derived GBM cells. Our results suggest that targeting NKCC1 in GBM will decrease cell dispersal by disrupting cytoskeleton dynamics.
2. Materials and Methods
2.1. Cell Lines
Patient samples of glioma tissues were obtained at the Johns Hopkins Hospital under the approval of the Institutional Review Board (IRB). All human brain tumor cell lines were derived from intraoperative tissue samples from patients treated surgically for newly diagnosed glioblastoma without prior treatment. Clinical data for primary GBM cell lines 318, 612 and 965 is described in detail in Table S1. Additionally, we used Human Embryonic Kidney 293 (HEK293) and MCF10A (mammary gland/breast derived cells) cells which were obtained from ATCC (American Type Culture Collection, Manassas, VA, USA) and cultured according to manufacturer instructions. Primary GBM cell lines 318, 612 and 965 have been analyzed by our group previously (Garzon-Muvdi et al., 2012, Yang et al., 2017, Smith et al., 2016) and were cultured using Dulbecco's Modified Eagle Medium: Nutrient Mixture F-12, B27 serum free supplement (Gibco), 20 ng/mL epidermal growth factor (EGF), and 20 ng/mL fibroblast-derived growth factor (FGF). HEK293 and MCF10A were cultured according to manufacturer instructions.
2.2. Viral Transduction
We used a human clone set (Sigma Aldrich Mission) of sequence verified lentiviral particles (pLKO.1, TRC0000296498) that target human NKCC1 (SLC12A2) and TRC2-pLKO-puro empty vector control (ref: SHC201) to generate NKCC1-shRNA stably expressing cell lines. Seventy-two hours after transduction, cells were cultured in the presence of puromycin to select cells with successful transduction. Knock-down of NKCC1 was confirmed by immunoblot before each experiment. In addition, the F-actin biosensor Lifeact (Riedl et al., 2008) cloned into lentiviral particles (kindly provided by Peter Devreotes laboratory) was used to transduce GBM cells. Throughout the text empty vector control cells are referred as EV and NKCC1-shRNA cells are referred as NKCC1-KD.
2.3. Cloning of Full-length Human NKCC1 and Generation of EGFP Fusion Protein
As previously described by our group (Garzon-Muvdi et al., 2012) we have cloned the human NKCC1 protein into a PCDNA3-EGFP expression vector (NKCC1-GFP plasmids).
2.4. Immunoblotting
Cells were plated on a 25 cm2 flask or 6 well plates and exposed to the different experimental conditions. Cell were harvested using RIPA lysis buffer (150 mM NaCl, 10 mM Tris, pH 7.5, 1% NP40, 1% deoxycholate, 0.1% SDS, protease inhibitor cocktail (Roche)), and HaltTM Phosphatase inhibitor cocktail (Thermo). Proteins from whole cell lysates were resolved using the NuPAGE 4–12% Bis-Tris gradient gel (Invitrogen). The Subcellular Protein Fractionation Kit was used for membrane and cytosol protein extraction (Thermo Scientific). Proteins were transferred to PVDF membrane, blocked in 5% non-fat milk or 2% bovine serum albumin in TBS-Tween-20, and probed with the antibodies for NKCC1 (ref: 8351, Cell signaling), MLC2 (ref: 3672, Cell signaling), pMLC2 (ref: 3671, Cell signaling), Cofilin1 (ref:5175, Cell signaling), pCofilin (ref: 3311, Cell signaling, Ser3); GFP (ref:632459, Clontech), Rac1 (ref; sc-95, Santa cruz), RhoA (ref: sc-418, Santa Cruz), GAPDH (ref: sc-32233, Santa Cruz), EGFR (ref: 06-847, Millipore), pEGFR (ref: 2234, Cell Signaling) and Actin (ref: ab8227, AbCAM).
MCF10A cells were used to test pMLC2 and tMLC2 phosphorylation due to a high signal-to-noise ratio using the GBM primary-derived cells. Detection was performed with the appropriate horseradish-peroxidase conjugated secondary antibodies and using enhanced chemiluminescence reagent (GE Healthcare Life Science). Densitometry analysis was performed using the Gel Analysis tool from ImageJ.
2.5. Assessment of Migration on Nano-patterned Groove Surface
Migration of glioma cells was quantified using a directional migration assay using nano-ridges/grooves constructed of transparent poly (urethane acrylate) (PUA) as previously described (Garzon-Muvdi et al., 2012, Smith et al., 2016, Kondapalli et al., 2015, Tilghman et al., 2016). Nanopattern surfaces were coated with laminin (3 μg/cm2). Cell migration was quantified using timelapse microscopy. Long-term observation was done on a motorized inverted microscope (Olympus IX81) equipped with a Cascade 512B II CCD camera and temperature and gas controlling environmental chamber. Phase-contrast and epi-fluorescent cell images were automatically recorded under 10 × objective (NA = 0.30) using the Slidebook 4.1 (Intelligent Imaging Innovations, Denver, CO) for 15 h at 10–20 min intervals. We performed time-lapse videomicroscopy in order to measure cell migration speed, distance, and directional persistence. The obtained images were analyzed using Matlab with a previously written cell tracking script developed in our laboratory (Garzon-Muvdi et al., 2012). Videos consisted of 60 timeframes reflecting a total time of 20 min. Cells were selected manually for tracking and cells were discarded from analysis if they went into apoptosis, mitosis or migrated out of view.
2.6. Immunoprecipitation
For the measuring RhoGTPase activity we performed a pulldown assay of Rac1-GTP, RhoA-GTP using the RhoA/Rac1/Cdc42 Activation Assay Kit (Cell Biolabs, Inc.). The methodology used was followed according to the manufacturer's instructions. Briefly, the adherent cultured cells were lysed, centrifuged and the supernatant was collected for further steps. PAK PBD Agarose beads were added to an aliquot of the supernatant. After incubation, the beads were pelleted by centrifugation. The supernatant was discarded and the bead pellet was washed thrice with the supplied assay buffer, centrifuging and discarding the supernatant each time. After the third wash, the mixture was centrifuged again, the supernatant was removed and the bead pellet was re-suspended in 2 × reducing SDS-PAGE sample buffer. The samples were boiled and centrifuged. The samples were loaded to a polyacrylamide gel for electrophoresis. Immunoblotting and detection was performed as described in the immunoblotting methods. For GFP pulldown we utilized the GFP-Trap® system from Cromotek (Ref: GPF-Trap_A, gta-10) according to the manufacturer's instructions. HEK293 cells where used in these experiments as an efficient system to express the PCDNA-EGFP and PCDNA-NKCC1-EGFP plasmid using the lipofectamine-LTX transfection system. Briefly, HEK293 cells transfected with control-GFP and NKCC1-GFP plasmids. Cells where lysed with RIPA buffer, GFP-A trap beads where wash and equilibrated in dilution buffer (10 mM Tris/Cl pH 7.5; 150 mM NaCl; 0.5 mM EDTA) and protein lysate was incubated with GFP-A trap beads for 1 h at 4C with continuous mixing. After incubation, beads where spun down, supernatant was discarded, beads where washed and the bead pellet was suspended in 0.2 M glycine pH 2.5 to dissociate the immunocomplexes form the agarose beads. The eluate was further analyzed by silver staining; protein identification was performed by LC MS/MS (see below).
2.7. Protein Identification
Protein interactions were determined through liquid chromatography tandem mass spectrometry (LC-MS/MS) at the Johns Hopkins Mass Spectrometry & Proteomics core facility. Protein identification by mass spectrometry workflow: Proteins in solution (0.2 M glycine HCl, Tris base) were reduced with dithiothreitol (50 mM, 60 °C for 45 mins), and alkylated with iodoacetamide (100 mM, RT for 15 mins in the dark) prior to digestion with trypsin overnight. The samples were dried and subsequently desalted over a C18 stage-tip and eluted with 60%ACN/0.1%TFA × 2. Samples were dried and re-constituted in 8ul 2%ACN/0.1%FA. Protein identification by liquid chromatography FTFT tandem mass spectrometry (LCMS/MS): Digested peptides (25%, or 2/8uL) were analyzed by liquid chromatography/tandem mass spectrometry (LCMS/MS) using LTQ Orbitrap Velos MS (Thermo Fisher Scientific, www.thermofisher.com) interfaced with nano-Acquity LC system from Waters.
Peptides were loaded on a 75 um × 2.5 cm C18 (YMC*GEL ODS-A 12 nm S-10 um) trap at 600 nl/min 0.1% FA (solvent A) and fractionated at 300 nL/min on a 75 μm × 150 mm ProntoSIL-120-5-C18 H reverse-phase column with spherical particle 5 μm, pore size 120 Å, from BISCHOFF Chromatography (http://www.bischoff-chrom.com/hplc-prontosil-c18-h-c18-phasen.html), using a 3–10% solvent B (90% acetonitrile in 0.1% formic acid) gradient over first 10 min, then up to 30%B by 55 min, 45%B by 65 min and 100%B by 70 min. Eluting peptides were sprayed into an LTQ Orbitrap Velos mass spectrometer (ThermoScientific, www.thermo.com/orbitrap) through 1 μm emitter tip (New Objective, www.newobjective.com) at 2.0 kV. Survey scans (full ms) were acquired within 350–1800 m/z with up to 8 peptide masses (precursor ions) individually isolated at IW1.9 Da, and fragmented (MS/MS) using HCD 35 activation collision energy. Precursor and the fragment ions were analyzed at resolution 30,000 and 7500, respectively. Dynamic exclusion of 30 s, repeat count 1, MIPS (monoisotopic ion precursor selection) “on”, m/z option “off”, lock mass “on” (silocsane 371 Da) were used. Data Analysis: Tandem MS2 mass spectra were processed by Proteome Discoverer (v1.4 Thermo Fisher Scientific) in two ways, using Nodes: common, and MS2 Processor. MS/MS spectra were analyzed with Mascot v.2.2.2 Matrix Science, London, UK (www.matrixscience.com) using the RefSeqComplete2012 database, specifying Human species, trypsin as enzyme, missed cleavage 2, precursor mass tolerance 30 ppm, fragment mass tolerance 0.03 Da, and Oxidation (M), Carbamidomethyl (C) and Deamidation (NQ) as variable modifications.
For each sample, Mascot search result *.dat files for the two nodes were processed in Scaffold 3 (www.proteomesoftware.com) to validate protein and peptide identifications.
2.8. Immunofluorescence
GBM cells were plated on Lab-Tek® Chamber Slide coated with laminin (1 μg/cm2). Cells were fixed in 4% paraformaldehyde (pH 7.4) for 30 mins, washed 3 times and blocked with 10% normal donkey serum in PBS for 1 h. Subsequently, fixed cells were incubated with primary antibody at 4 °C overnight. Primary antibodies include: Cofilin 1 (ref: 5175, Cell signaling), Beta1 Sodium Potassium ATPase (ref: ab2873, AbCAM) and NKCC1 (ref: 8351, Cell signaling). After primary antibody, slides where incubated with Alexa Fluor-conjugated 594 and 488 secondary antibodies (1:500, Invitrogen), counter stained with DAPI and mounted using Aquamount (VWR). The cells were imaged using an Olympus IX81 epifluorescence microscope. Co-localization analysis was performed using Image J Colocalization Colormap plugin as previously described (Jaskolski et al., 2005).
2.9. Phalloidin Staining
GBM cells were plated on 15 mm glass coverslips (Warner Instruments) coated with laminin (1 μg/cm2). Cells were fixed in 4% paraformaldehyde (pH 7.4). Filamentous actin was stained using the Alexa Fluor® 594 Phalloidin probe (ref: A12381, Invitrogen). Briefly, cells were permeabilized with PBS-0.1% triton for 5 mins and incubated with 1:50 dilution of Alexa fluor® 594 Phalloidin in PBS-1% BSA for 1 h at RT. Slides are washed, counterstained with DAPI and mounted using Aquamount (VWR). The cells were imaged using an Olympus IX81 epifluorescence microscope. Filamentous actin content from the cells was determined with ImageJ, calculated as Corrected Total Cell Content Fluorescence (CTCF). CTCF = Integrated density −(Area of selected cell × Mean fluorescence of background signal).
2.10. Actin Fractionation
For actin fractionation, we utilized the G-Actin/F-actin In Vivo Assay Biochem Kit (ref: BK037, Cytoskeleton, Inc.). Prior to fractionation, cells were subjected to different conditions, namely transduction with NKCC1 shRNA or Control shRNA and EGF stimulation. Lysis of the cells and actin fractionation were performed according to manufacturer's instructions. F/G fractions were resolved using the NuPAGE 4–12% Bis-Tris gradient gel, immunoblotted using Actin antibody (ref: ab8227, AbCAM). Intensity of the bands from the different fractions where quantified by densitometry using ImageJ.
2.11. Interference Reflection Microscopy
Interference reflection microscopy (IRM) was used to detect surface-to-surface interference between light rays reflected from the substrate/medium interface and those from the medium/cell membrane interface as described in our previous work (Norman et al., 2011, Norman et al., 2010a, Norman et al., 2010b). The intensity of the light is a measure of the proximity of the cell membrane to the glass surface, so the membrane closest to the surface appears darker and those further away appear brighter. Therefore, IRM is an optimal method when evaluating cellular attachment, adhesion, and spreading behavior. Cells from lines 318 or 612 were plated onto polylysine-D or laminin coated coverslips, respectively. For image capture, we used an inverted microscope (Olympus IX71) with a 60 ×/1.42 NA oil objective lens and a 100 W mercury lamp (Olympus; used at wavelength 561), in combination with a CCD camera (Retiga SRV camera, QImaging). Experiments were performed in an enclosed microscope chamber which maintained culture conditions at 37 °C, 50% humidity, and 5% CO2. During cell spreading, 10–20 images were captured of single cells around the dish at time points 15 min, 30 min, 45 min, 60 min, 2 h, 4 h, 6 h, and 12 h. For statistical evaluations of contact areas, images were analyzed by using built-in ImageJ plugins (National Institutes of Health) software.
2.12. Statistical Analysis
Statistical analysis was performed using GraphPad Prism 5. It consisted of one-way ANOVA, followed by Kruskal-Wallis, Bonferroni, or Tukey, depending on the distribution of our data. Data distribution was determined using the D'agostino test, Graphs represent the mean ± SEM. Statistical significance is represented by *P < 0.05, **P < 0.01, and ***P < 0.001. All experiments were done in triplicate.
3. Results
3.1. NKCC1 Expression Regulates Cell Speed and Delays Cell Spreading
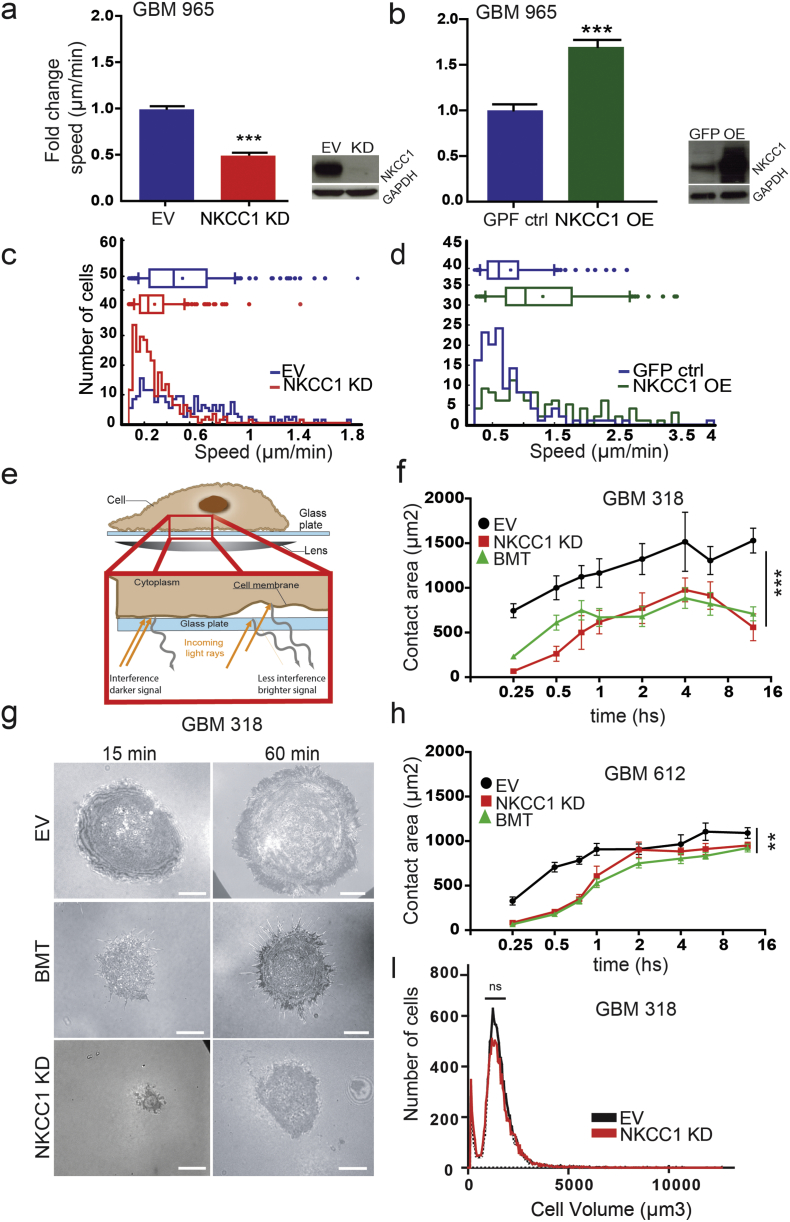
We have previously described that NKCC1 genetic or pharmacological inhibition decreases GBM cell migration in vitro and in vivo (Garzon-Muvdi et al., 2012). To increase our understanding of NKCC1's role in cell migration, we first evaluated how NKCC1 modulation can affect cell migration using single-cell subpopulation analysis previously described by our group (Smith et al., 2016) (Fig. 1a–d, Fig. S3A). In the whole population analysis of cell line GBM 965 we observed that NKCC1 knockdown (NKCC1 KD) decreases migration (Fig. 1a, Fig. S1D). Conversely, NKCC1 overexpression (NKCC1 OE) increases migration (Fig. 1b), confirming and further expanding our previously reported observations (Garzon-Muvdi et al., 2012). Moreover, the effects of NKCC1 changes in expression are more evident in the fastest cell subgroup (Fig. 1c–d). These results suggest that targeting NKCC1 can impact GBM subpopulations with the highest motility or aggressiveness, potentially resulting in less tumor spreading into healthy tissue. Another important aspect of cell dispersal is cell spreading. To evaluate spreading we utilized Internal Reflection Microscopy (IRM), an optimal method when evaluating cellular adhesion, and spreading behavior (Fig. 1e) (Norman et al., 2010a, Norman et al., 2011). We found that NKCC1 knockdown or inhibition (with bumetanide) in GBM cells yields significant delay in cell spreading (Fig. 1f–h, Fig. S1B). These results confirm our previous observation that NKCC1-KD cells display reduced traction forces (contractility) and displayed smaller projected cell area (Garzon-Muvdi et al., 2012). Remarkably, these observations were not due to cell volume changes, as volume sizes of EV (empty virus control) and KD cells were similar (Fig. 1i, Fig. S1C). Next, we wanted to understand the mechanisms by which NKCC1 affects spreading and migration.
Fig. 1.
NKCC1 inhibition reduces cell migration and delays cell spreading. a–b) NKCC1 expression regulates cell migration. a–b) NKCC1 knockdown (KD) and overexpression (OE) reduced or increased migration speed respectively in GBM 965 (EV: empty virus ctrl; GFP-ctrl: NKCC1-GFP plasmid), > 100 cells were analyzed in each experiment n = 3. c–d) Histogram representing the distribution of speed in the GBM subpopulations upon NKCC1 knockdown (KD) or overexpression (OE). e) Cartoon explaining interference reflection microscopy (IRM) technique used to determine cell spreading. f–h) Quantification of cell spreading area at different time points in two different cell lines: GBM 318 and 612 (EV: empty virus ctrl; BMT: Bumetanide), n = 3. g) Sample images of GBM 318 spreading at different time points from panel f. i) Cell volume measured by a Multisizer Coulter counter using GBM 318, n = 2. Bars represent mean ± SEM. **P-value < 0.01; ***P-value < 0.001, Scale bar: 10 μm.
3.2. NKCC1 Regulates Filamentous Actin Content and Actin Dynamics
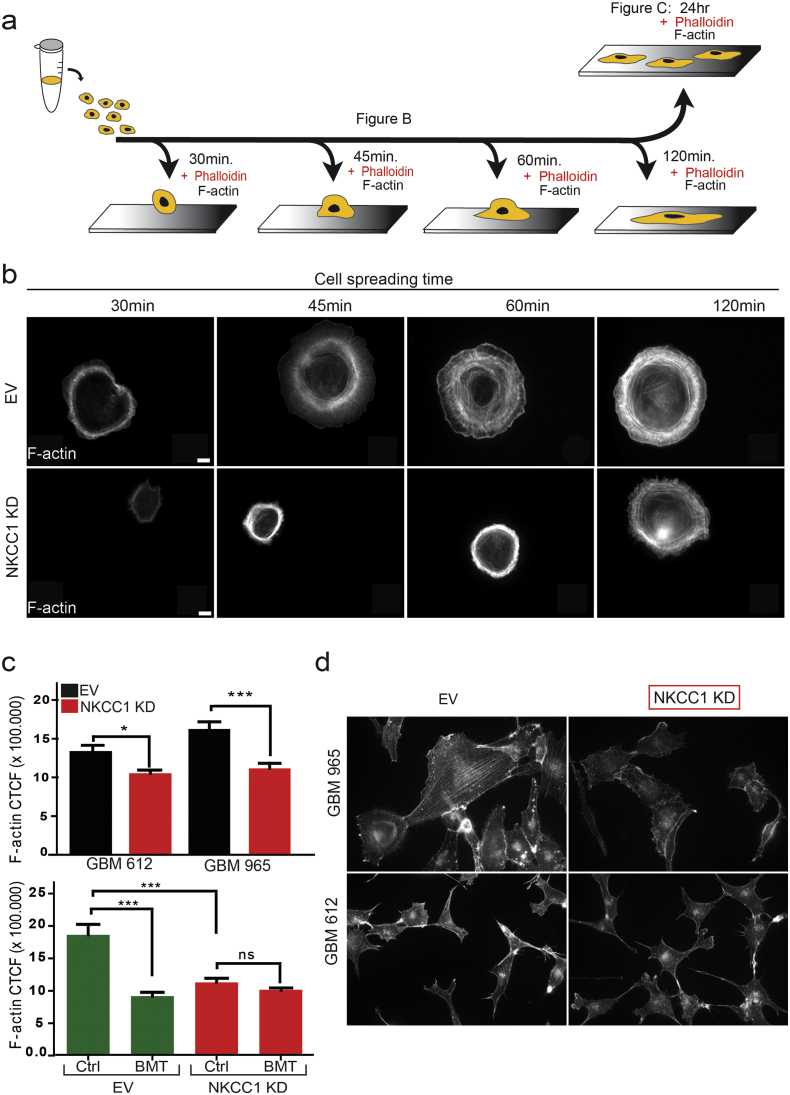
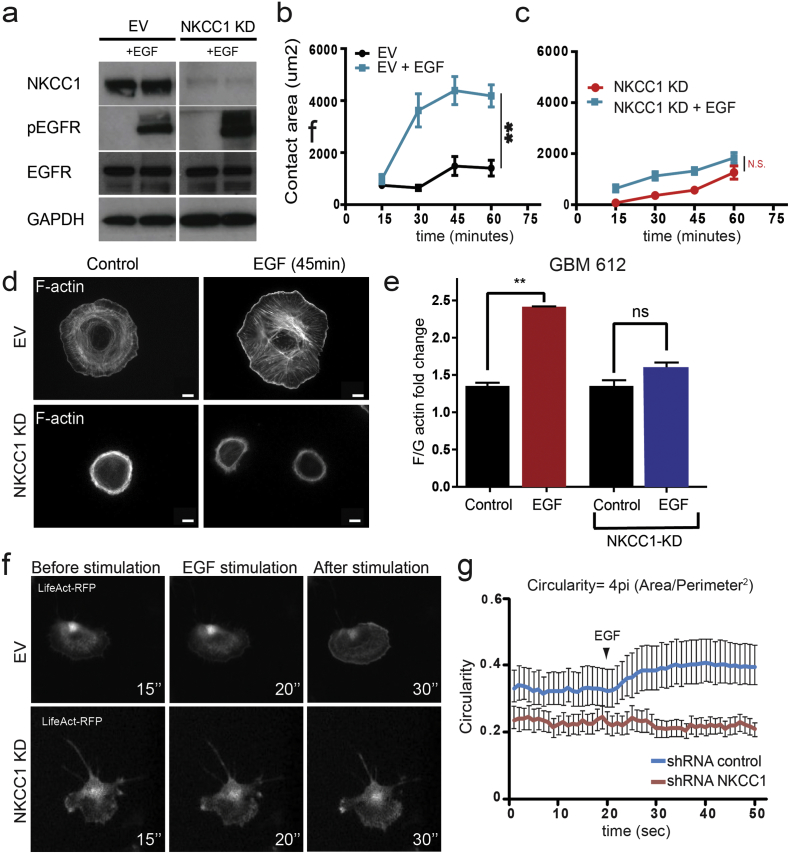
In order to investigate how NKCC1 affects cell spreading and migration, we stained filamentous actin (F-actin) in cells undergoing cell spreading at different time points (Fig. 2a–b). Phalloidin staining showed that GBM cells increase the number and organization of actin fibers as the cells spread during a 120-min course (Fig. 2b). Conversely, NKCC1-KD cells show a delay in the formation of the actin fibers, which seem to be less organized, acquiring a ring-shaped form. This difference in actin organization remained after 24 h of plating (Fig. 2c, d). In addition, NKCC1 pharmacological inhibition using bumetanide also significantly decreased filamentous actin content (Fig. 2c lower panel). After showing that NKCC1 down-regulation decreases cell migration and filamentous actin content, we decided to study how NKCC1 can affect actin dynamics. We stimulated GBM cells with EGF and evaluated cell spreading and actin fractionation on EV and NKCC1-KD cells. EGF receptor phosphorylation was increased upon EGF stimulation in both NKCC1 EV and KD cells (Fig. 3a). Cell spreading was evaluated by IRM for 1 h (Fig. 3b–c). We observed that EGF accelerates the spreading dynamics and increases filamentous actin of EV cells while KD cells remain unaffected (Fig. 3b–d). In accordance to this data we found that the ratio of filamentous to globular actin (f to g) is increased by 2-fold in EV cells after 1 min of EGF stimulation but not in NKCC1-KD cells (Fig. 3e). Finally, we transduced GBM cells with the Lifeact-RFP construct for live imaging of F-actin. We observed that upon EGF stimulation, EV cells responded by uniform spreading, as reflected by an increased cell circularity within seconds after stimulation, while NKCC1 KD cells did not respond (Fig. 3f–g, Supplementary video 1). These results suggest that the dynamic response of the actin cytoskeleton to external stimuli is decreased when NKCC1 is down-regulated. Next, we decided to study how NKCC1 regulates the actin cytoskeleton.
Fig. 2.
NKCC1 regulates filamentous actin content. a) Diagram representing the experimental design to study cell spreading and actin staining for figures b and c. b) Phalloidin staining of GBM 318 cells EV and NKCC1-KD cells undergoing spreading at different time points. c) Quantification of filamentous actin 24 h after cell plating. CTCF: corrected total cell fluorescence (CTCF = Integrated density −(Area of selected cell × Mean fluorescence of background signal)). Upper panel staining for 965 and 612 EV and NKCC1-KD cells, lower panel 965 EV and NKCC1-KD cells treated with or without bumetanide, n = 3, > 100 cells per group analyzed. d) Images of the cells used for F-actin quantification. Bars represent mean ± SEM. *P-value < 0.05; ***P-value < 0.001, Scale bar: 10 μm.
Fig. 3.
NKCC1 regulates actin dynamics. a) Western blot images from GBM 612 showing EGFR activation by phosphorylation after 1 min of EGF treatment in EV and NKCC1 KD cells n = 3. b—c) Cell spreading quantification by IRM using GBM 318 after EGF stimulation in EV and NKCC1 KD cells, n = 3. d) Phalloidin staining of GBM 318 EV and NKCC1-KD cells after EGF stimulation. e) Quantification of filamentous (F) and globular (G) actin after EGF stimulation, n = 3. f) Timelapse images of GBM 612 cells transduced with the f-actin sensor Lifeact before and after EGF stimulation (timestamp in seconds). g) Quantification of cell circularity of GBM 612 cells before and after EGF stimulation, n = 3. Bars represent mean ± SEM. **P-value < 0.01. Scale bar: 10 μm.
3.3. Actin-regulating Protein Cofilin 1 Associates with NKCC1, which Controls Its Membrane Localization
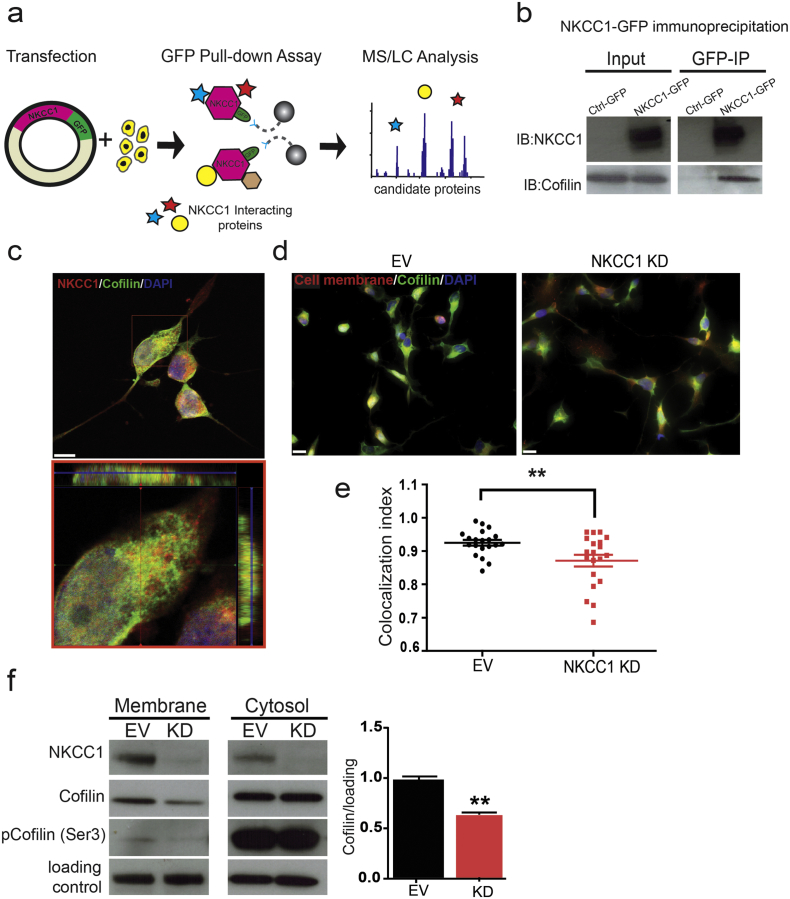
Given the effects of NKCC1 inhibition on cytoskeleton phenotypes we decided to study possible NKCC1 protein-protein interactions. We expressed a human NKCC1-GFP fusion protein on HEK293 cells, and purified the possible interacting partners by GFP immunoprecipitation (Fig. 4a, Fig. S2A—B). The samples were analyzed by silver staining (Fig. S2B) and liquid chromatography tandem mass spectrometry (LC-MS/MS). We identified several relevant NKCC1-interacting candidates (Table S2, Fig. 4b, and Fig. S2C) that play key roles in the cytoskeleton organization, the most significant one being the actin-interacting protein Cofilin 1. We first validated Cofilin 1-NKCC1 interaction by co-immunoprecipitation and IHC (Fig. 4b–c). Then, we studied the effects of NKCC1 expression on Cofilin 1 localization. First, we analyzed the amount of co-localization of Cofilin 1 at the plasma membrane in control and NKCC1-KD cells by immunohistochemistry of Cofilin 1 and membrane marker Sodium Potassium ATPase (Fig. 4d). We observed that there was a significant decrease of Cofilin 1 co-localization with the plasma membrane marker (Fig. 4e). To validate these results we performed sub-cellular fractionation and quantified the amount of Cofilin 1 in plasma membrane and cytosolic fractions (Fig. 4f), confirming the observations obtained in Fig. 4e–d. We corroborated that the active portion of Cofilin 1 is the one found in the membrane fraction (compared to the cytosol), as the majority of Cofilin 1 in the membrane was in the dephosphorylated from (Fig. 4f). These results suggest that NKCC1's interaction with Cofilin 1 can determine its localization, and given that the active fraction of the protein is on the plasma membrane this change in localization decreases Cofilin 1 activity in the NKCC1 KD cells. Given that Cofilin 1 is in charge of severing actin filaments to promote actin polymerization, the decreased actin content in NKCC1-KD cells could be explained by a decrease in Cofilin 1 activity. Given that actin polymerization is regulated by an intricate array of proteins, we decided to analyze if other actin-regulating proteins are affected by NKCC1 activity.
Fig. 4.
NKCC1 interacts with Cofilin 1 at the plasma membrane and determines its localization. a) Diagram showing the experimental design to study NKCC1 protein-protein interactions. Cells are transfected with NKCC1-GFP and proteins are purified by GFP pulldown. Proteins co-immunoprecipitating with NKCC1 are identified by mass spectometry. b) Co-immunoprecipitation (co-IP) of NKCC1 and Cofilin1. Cells are transfected either with a GFP-control plasmid (Crtl-GFP) or NKCC1-GFP (NKCC-GFP) GFP-IP is performed in the lysates. NKCC1 and Cofilin 1 are detected by western blot in the input (whole lysate) and IP samples. c) IHC images showing Cofilin and NKCC1 staining in GBM 612. Red square in the lower panel shows orthogonal view. d) IHC images showing Cofilin 1 and Sodium Potasium ATPase (NaK-ATPase, membrane marker) in GBM 612 EV and NKCC1-KD cells. d) Quantification of Cofilin 1 and NaK-ATPase co-localization by IHC. Colocalization index of red and green pixels were analyzed by ImageJ, n = 3, > 20 cells analyzed. e) Detection by western blot of Cofilin and phosphorylated Cofilin (Ser3) in protein fractions from membrane and cytosol portions in GBM 612 EV and NKCC1-KD cells, n = 3. In the membrane portion there is a decrease in the amount of Cofilin, in this fraction the majority of Cofilin is in the dephosphorylated (active). There is no change on the amount of Cofilin on the cytosolic fraction, here the majority of Cofilin is in the phosphorylated form (inactive). f) Quantification of Cofilin 1 on plasma membrane and cytosolic fractions obtained from GBM 612, n = 3. Bars represent mean ± SEM. **P-value < 0.01. Scale bar panel c: 10 μm, Scale bar panel d: 20 μm.
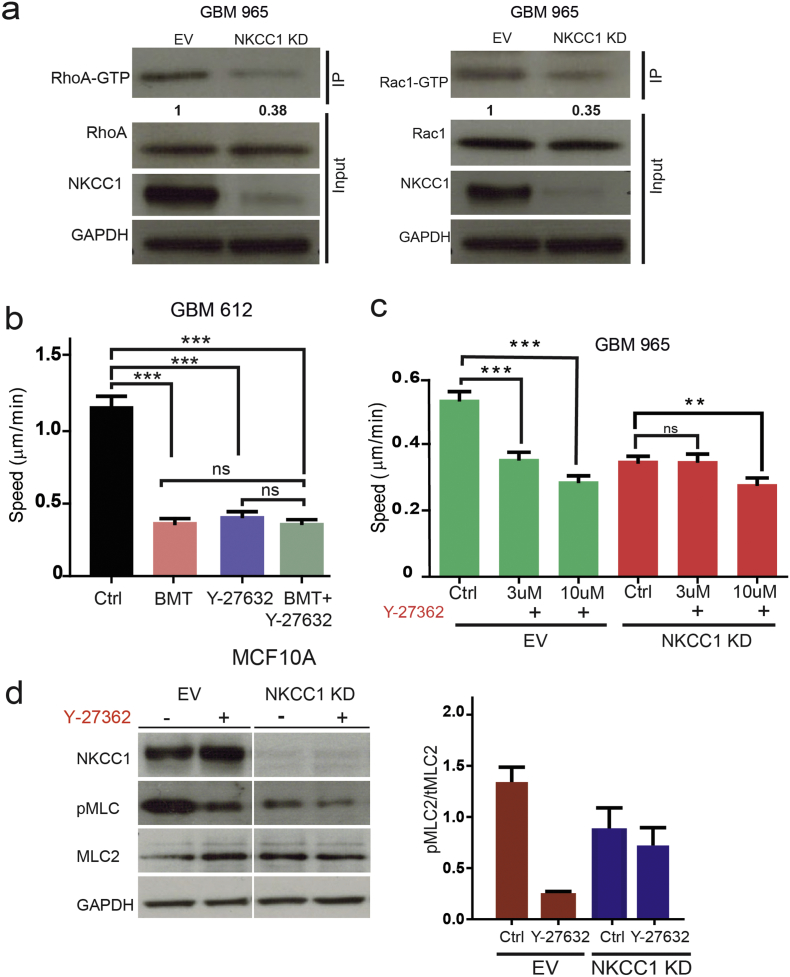
3.4. RhoGTPases Rac1 and RhoA Decreased Activity in NKCC1 KD Cells
To determine the actin-regulatory mechanisms affected by NKCC1 inhibition we studied the small Rho-GTPases, RhoA and Rac1. We observed that the levels of active (GTP-bound) forms of RhoA and Rac1 were decreased in NKCC1 KD cells (Fig. 5a, Fig. S3B). Given the decrease in RhoA-GTP levels, we examined the activity of ROCK1 (downstream substrate of RhoA). We observed that both bumetanide and Y-27632 (a ROCK1 inhibitor) decreased GBM cell migration, and combination of treatments does not seem to have a synergistic effect (Fig. 5b). These results suggest that RhoA activity seems to be downstream of NKCC1. Moreover, when GBMs were treated with the ROCK1 inhibitor they showed a dose-dependent decrease in cell migration. In addition, we studied phosphorylation status of MLC2 (ROCK substrate) in MCF10 control and NKCC1-KD cells. We found that NKCC1 inhibition decreases the amount of phospho MLC2 and no further inhibition is achieved in KD cells with Y-27632 (Fig. 5d). These effects were significantly weaker when the ROCK inhibitor was applied on NKCC1 KD or BMT-treated cells (Fig. 5c–d), suggesting that decreased RhoA and ROCK1 activity mediates the effects of NKCC1 inhibition.
Fig. 5.
NKCC1 regulates RhoGTPase activity. a) Analysis of RhoA and Rac1 activation levels after NKCC1 knockdown in GBM 965 by immunoprecipitation of their GTP active forms. Densitometry of the RhoA-GTP and Rac1-GTP band is shown below, n = 2. b) Cell speed of GBM 612 cells using bumetanide, ROCK inhibitor Y-27632 and combination. c) Cell speed of GBM 965 control and NKCC1-KD cells using different concentrations of the inhibitor Y-27632, n = 3. d) Western blot of showing the effects of ROCK inhibition on the MLC2 (ROCK substrate) in control and NKCC1-KD cells on MCF10A cells, n = 2. Bars represent mean ± SEM. **P-value < 0.01; *** P-value < 0.001.
4. Discussion
Cell migration plays a fundamental role in development, inflammatory responses and wound healing (Ridley et al., 2003, Weijer, 2009, Friedl and Weigelin, 2008). Cancer cells exploit multiple mechanisms to enhance their migratory and invasive behavior; this is especially the case of invasive brain cancers such as GBM. Unlike other types of tumors that disseminate through the blood, GBM cells move within the extracellular spaces of the brain following blood vessels and white matter tracts (Scherer, 1938, Watkins et al., 2014, Farin et al., 2006). One of the reasons of GBM therapy failure is the extensive infiltration of glioma cells away from the main tumor mass, which is a major obstacle for surgical removal (Almeida et al., 2015, Chaichana et al., 2014). Hence, targeting cell migration is a key factor to improve patient outcomes and understanding the molecular basis of GBM migration is a critical step towards achieving this goal.
In this context, NKCC1 is an interesting candidate to target GBM migration and invasion. This ion co-transporter regulates the cell intracellular chloride (Cl−) concentration, which is higher in glioma cells compared to non-cancer cells (Habela et al., 2008). In glioma patient derived tissues, we found that NKCC1 expression is upregulated in anaplastic astrocytomas and GBM and its expression is mostly localized to the extending processes of migrating GBM cells (Garzon-Muvdi et al., 2012, Haas and Sontheimer, 2010). In addition, pharmacological and genetic inhibition of NKCC1 decreases GBM cell migration in vitro and in vivo (Garzon-Muvdi et al., 2012). The mechanisms of how NKCC1 regulates GBM migration are not fully understood. Sontheinmer et al., proposed a hydrodynamic model of GBM invasion where cells use the concerted movement of ions and obligated water to dynamically change cell volume (Watkins and Sontheimer, 2011). In this model, NKCC1 is proposed as the main transporter that establishes a Cl− gradient that can prompt rapid changes in cell volume working together with several Cl− and K+ channels. Yet for cells to move and invade the brain, other mechanisms may be needed to induce a pathological invasive phenotype.
In the present study, we show that NKCC1 inhibition (in addition to decreasing cell migration) disrupts the regular formation and amount of bundled actin filaments in human primary derived-GBM cells. In addition, we observed that EGF treatment (which activates NKCC1 phosphorylation and actin bundling (Chan et al., 1998, Garzon-Muvdi et al., 2012)) stimulates cell spreading and actin polymerization in control cells but not in NKCC1-KD cells. These results imply that NKCC1 does not only regulate migration by hydrodynamic changes but also alters the actin cytoskeleton, the migratory engine of GBM cells. Actin polymerization is a highly dynamic process that drives membrane protrusion, cell migration and cytokinesis (Pollard and Borisy, 2003, Lauffenburger and Horwitz, 1996). Blockage of actin polymerization-depolymerization by the classic toxins Phalloidin and Cytochalasin D (Sampath and Pollard, 1991) greatly decreases motility and proliferation (Trendowski, 2014). Targeting the cytoskeleton is a promising strategy for cancer therapy, but finding a specific target can be elusive. In this context, our findings may help design such a strategy due to both the overexpression of NKCC1 in GBM and its apparent interactions with actin-regulating proteins.
In an effort to dissect NKCC1's role in actin regulation, we used LC-MS/MS and co-IP analysis to study NKCC1 protein interacting partners. We found that NKCC1 can interact with Cofilin 1, an actin-regulating protein that plays a key role in actin polymerization/depolymerization and cell migration in both normal and cancer cells (Sidani et al., 2007, Desmarais et al., 2005, Chen et al., 2001). We found that NKCC1-KD cells show decreased co-localization of Cofilin 1 at the cell membrane. Furthermore, western blot analysis of membrane and cytosol portions of GBM cells show that Cofilin 1 expression is decreased at the cell membrane upon NKCC1-KD. Cofilin 1 has been reported to be upregulated in multiple highly invasive cancers including breast, colon and glioma where it plays a crucial role in cell motility and invasion (Sidani et al., 2007, Park et al., 2014, van Rheenen et al., 2009). Several studies have investigated the role of Cofilin 1 activity and its regulation in GBM, revealing that Cofilin 1 is overexpressed in gliomas and can alter GBM migration (Starinsky-Elbaz et al., 2009, Hou et al., 2016, Yap et al., 2005, Park et al., 2014). The outcome of Cofilin inhibition on cell migration is variable because it can modulate migration in opposite directions. Yap et al. demonstrated that there is a fine balance between Cofilin 1 concentration and migratory behavior in GBM. Different levels of overexpression can reduce or increase cell migration and invasion (Yap et al., 2005). In addition, there are some reports in GBM that show opposite effects in cell migration outcome depending of the level of phosphorylated Cofilin (inactive form) (Park et al., 2014, Hou et al., 2016, Zhou et al., 2016, Jin et al., 2016). This finding might be due to the fact that altering different components of the Cofilin pathway in heterogeneous tumors can have diverse effects on invasive potential (van Rheenen et al., 2009).
Our results suggest that the expression of NKCC1 regulates the localization of Cofilin 1, perhaps by acting as a scaffold protein to the plasma membrane. Our hypothesis is that the decreased levels of filamentous actin detected upon NKCC1-KD are due to the change in Cofilin localization. In our previous report, we found that NKCC1 expression could alter focal adhesion maturation, as NKCC1-KD cells displayed bigger focal adhesions. In addition, we observed that NKCC1 localized to the extending processes of the cells during cell migration (Garzon-Muvdi et al., 2012). These results suggest that NKCC1 polarizes to the cell edge and can act as an actin anchor or scaffold during cell migration, affecting focal adhesion turnover and through its interaction with Cofilin1 activating actin remodeling at the leading edge. Further analyses are needed to determine how the association and activity of NKCC1 regulates Cofilin severing activity. Mutational and computational analyses are needed to determine which domains are involved in this interaction. This information will be critical to dissect the mechanism of NKCC1's regulation of the actin cytoskeleton dynamics and to develop novel targeted therapies to halt GBM migration.
In addition, our LC-MS/MS co-IP analysis also indicates that NKCC1 can bind to Tubulin beta-4B and Tubulin alpha-1C chain, suggesting that NKCC1 is potentially part of a targetable signaling hub that regulates cell migration, integrating signaling that affects actin and potentially microtubule organization. Moreover, more studies to describe the role of other candidate proteins binding to NKCC1 such as the heat shock protein 90-beta, heta shock protein 70 1A/1B and heat shock protein 60 are needed.
Cofilin 1 partners with the Arp2/3 and WAVE complex to stimulate the formation of dendritic actin, which is an essential step for cell protrusion and invasion (Ichetovkin et al., 2002, Desmarais et al., 2005, Beaty and Condeelis, 2014, Bravo-Cordero et al., 2013). Both pathways are activated by the small Rho GTPases (Heasman and Ridley, 2008, Ridley, 2011). For these reasons, we decided to study the activation levels of RhoA and Rac1 (bound to GTP) in GBM cells. We found that NKCC1-KD cells show decreased activity of the RhoA and Rac1 GTPases. These results might also explain the change in actin cytoskeleton dynamics upon NKCC1-KD, both by Cofilin 1 localization and decreased RhoGTPase activity leading to reduced GBM migration. Our previous data showed that NKCC1 inhibition increases the size of focal adhesions, while decreasing cell contractility and projected cell area adhesion (Garzon-Muvdi et al., 2012). Decreased RhoA activity could explain this contractility defect; perhaps NKCC1 inhibition plays a role in focal adhesion turnover by decreasing RhoA and Rac1 activity. Further analysis of NKCC1-interacting partners and changes in ion concentration and volume are needed to dissect this mechanism.
In summary, we have found that NKCC1 not only controls cell volume and Cl− concentration, but it can also regulate the actin cytoskeleton through Cofilin 1. NKCC1 can be targeted with the FDA approved drug Bumetanide (BMT), which has shown to decrease GBM migration in vitro and in vivo (Garzon-Muvdi et al., 2012, Haas and Sontheimer, 2010). BMT has poor blood brain barrier penetration (BBB); therefore, alternative delivery methods, such as convention enhanced delivery, nanoparticles, or BBB disruption will be necessary to achieve therapeutic concentrations in the brain. Given the unmet need of innovative therapies for GBM, targeting NKCC1 with BMT in combination with chemotherapy may reduce GBM infiltration and decrease tumor recurrence.
The following are the supplementary data related to this article.
EGF stimulation on control and NKCC1 KD cells expressing the fibrilary actin sensor lifeact.
Supplementary material
Funding Sources
This work was funded by NIH RO1 NS070024 (AQH).
Conflict of Interest
The authors declare no conflict of interest associated with this publication.
Author Contributions
Conceptualization: PS, HGC, HAE, AQH. Methodology: PS, HGC, RMM, CHH, HAE. Software: CHH. Formal Analysis: PS, HGC, SMH, RMM, CHH. Investigation: PS, RMM, SMH, EGLF, SG. Resources: AQH, HAE, PD, CHH. Writing: PS, RMM, HGC, AQH, EGLF. Visualization: PS, HGC, SG, EGLF. Supervision: PS, HGC, AQH. Funding Acquisition: AQH.
Acknowledgments
We will like to thank Juan Carlos Martinez-Gutierrez for help with actin staining and immunoprecipitation troubleshooting; Douglas Robinson for help with the actin polymerization protocols; Mingjie Wang for providing the Lifeact lentiviruses and help in time-lapse microscopy; Bob Cole and Lauren De Vine from the JHU mass spectrometry facility for their assistance with the LC-MS/MS experiments.
Contributor Information
Paula Schiapparelli, Email: Schiapparelli.paula@mayo.edu.
Alfredo Quinones-Hinojosa, Email: Quinones-Hinojosa.Alfredo@mayo.edu.
References
- Almeida J.P., Chaichana K.L., Rincon-Torroella J., Quinones-Hinojosa A. The value of extent of resection of glioblastomas: clinical evidence and current approach. Curr. Neurol. Neurosci. Rep. 2015;15:517. doi: 10.1007/s11910-014-0517-x. [DOI] [PubMed] [Google Scholar]
- Beaty B.T., Condeelis J. Digging a little deeper: the stages of invadopodium formation and maturation. Eur. J. Cell Biol. 2014;93:438–444. doi: 10.1016/j.ejcb.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo-Cordero J.J., Magalhaes M.A., Eddy R.J., Hodgson L., Condeelis J. Functions of cofilin in cell locomotion and invasion. Nat. Rev. Mol. Cell Biol. 2013;14:405–415. doi: 10.1038/nrm3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaichana K.L., Chaichana K.K., Olivi A., Weingart J.D., Bennett R., Brem H., Quinones-Hinojosa A. Surgical outcomes for older patients with glioblastoma multiforme: preoperative factors associated with decreased survival. Clinical article. J. Neurosurg. 2011;114:587–594. doi: 10.3171/2010.8.JNS1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaichana K.L., Jusue-Torres I., Navarro-Ramirez R., Raza S.M., Pascual-Gallego M., Ibrahim A., Hernandez-Hermann M., Gomez L., Ye X., Weingart J.D., Olivi A., Blakeley J., Gallia G.L., Lim M., Brem H., Quinones-Hinojosa A. Establishing percent resection and residual volume thresholds affecting survival and recurrence for patients with newly diagnosed intracranial glioblastoma. Neuro-Oncology. 2014;16:113–122. doi: 10.1093/neuonc/not137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A.Y., Bailly M., Zebda N., Segall J.E., Condeelis J.S. Role of cofilin in epidermal growth factor-stimulated actin polymerization and lamellipod protrusion. J. Cell Biol. 2000;148:531–542. doi: 10.1083/jcb.148.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A.Y., Raft S., Bailly M., Wyckoff J.B., Segall J.E., Condeelis J.S. EGF stimulates an increase in actin nucleation and filament number at the leading edge of the lamellipod in mammary adenocarcinoma cells. J. Cell Sci. 1998;111(Pt 2):199–211. doi: 10.1242/jcs.111.2.199. [DOI] [PubMed] [Google Scholar]
- Chen J., Godt D., Gunsalus K., Kiss I., Goldberg M., Laski F.A. Cofilin/ADF is required for cell motility during drosophila ovary development and oogenesis. Nat. Cell Biol. 2001;3:204–209. doi: 10.1038/35055120. [DOI] [PubMed] [Google Scholar]
- Desmarais V., Ghosh M., Eddy R., Condeelis J. Cofilin takes the lead. J. Cell Sci. 2005;118:19–26. doi: 10.1242/jcs.01631. [DOI] [PubMed] [Google Scholar]
- Farin A., Suzuki S.O., Weiker M., Goldman J.E., Bruce J.N., Canoll P. Transplanted glioma cells migrate and proliferate on host brain vasculature: a dynamic analysis. Glia. 2006;53:799–808. doi: 10.1002/glia.20334. [DOI] [PubMed] [Google Scholar]
- Fortin Ensign S.P., Mathews I.T., Symons M.H., Berens M.E., Tran N.L. Implications of rho GTPase signaling in glioma cell invasion and tumor progression. Front. Oncol. 2013;3:241. doi: 10.3389/fonc.2013.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P., Weigelin B. Interstitial leukocyte migration and immune function. Nat. Immunol. 2008;9(9):960. doi: 10.1038/ni.f.212. [DOI] [PubMed] [Google Scholar]
- Garzon-Muvdi T., Schiapparelli P., Ap Rhys C., Guerrero-Cazares H., Smith C., Kim D.H., Kone L., Farber H., Lee D.Y., An S.S., Levchenko A., Quinones-Hinojosa A. Regulation of brain tumor dispersal by NKCC1 through a novel role in focal adhesion regulation. PLoS Biol. 2012;10 doi: 10.1371/journal.pbio.1001320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giese A., Bjerkvig R., Berens M.E., Westphal M. Cost of migration: invasion of malignant gliomas and implications for treatment. J. Clin. Oncol. 2003;21:1624–1636. doi: 10.1200/JCO.2003.05.063. [DOI] [PubMed] [Google Scholar]
- Guerrero-Cazares H., Attenello F.J., Noiman L., Quinones-Hinojosa A. Stem cells in gliomas. Handb. Clin. Neurol. 2012;104:63–73. doi: 10.1016/B978-0-444-52138-5.00006-2. [DOI] [PubMed] [Google Scholar]
- Haas B.R., Sontheimer H. Inhibition of the sodium-potassium-chloride cotransporter isoform-1 reduces glioma invasion. Cancer Res. 2010;70:5597–5606. doi: 10.1158/0008-5472.CAN-09-4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habela C.W., Olsen M.L., Sontheimer H. ClC3 is a critical regulator of the cell cycle in normal and malignant glial cells. J. Neurosci. 2008;28:9205–9217. doi: 10.1523/JNEUROSCI.1897-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heasman S.J., Ridley A.J. Mammalian rho GTPases: new insights into their functions from in vivo studies. Nat. Rev. Mol. Cell Biol. 2008;9:690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- Hou M., Liu X., Cao J., Chen B. SEPT7 overexpression inhibits glioma cell migration by targeting the actin cytoskeleton pathway. Oncol. Rep. 2016;35:2003–2010. doi: 10.3892/or.2016.4609. [DOI] [PubMed] [Google Scholar]
- Ichetovkin I., Grant W., Condeelis J. Cofilin produces newly polymerized actin filaments that are preferred for dendritic nucleation by the Arp2/3 complex. Curr. Biol. 2002;12:79–84. doi: 10.1016/s0960-9822(01)00629-7. [DOI] [PubMed] [Google Scholar]
- Insall R.H., Machesky L.M. Actin dynamics at the leading edge: from simple machinery to complex networks. Dev. Cell. 2009;17:310–322. doi: 10.1016/j.devcel.2009.08.012. [DOI] [PubMed] [Google Scholar]
- Jaskolski F., Mulle C., Manzoni O.J. An automated method to quantify and visualize colocalized fluorescent signals. J. Neurosci. Methods. 2005;146:42–49. doi: 10.1016/j.jneumeth.2005.01.012. [DOI] [PubMed] [Google Scholar]
- Jin S.G., Ryu H.H., Li S.Y., Li C.H., Lim S.H., Jang W.Y., Jung S. Nogo-A inhibits the migration and invasion of human malignant glioma U87MG cells. Oncol. Rep. 2016;35:3395–3402. doi: 10.3892/or.2016.4737. [DOI] [PubMed] [Google Scholar]
- Kondapalli K.C., Llongueras J.P., Capilla-Gonzalez V., Prasad H., Hack A., Smith C., Guerrero-Cazares H., Quinones-Hinojosa A., Rao R. A leak pathway for luminal protons in endosomes drives oncogenic signalling in glioblastoma. Nat. Commun. 2015;6:6289. doi: 10.1038/ncomms7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowska A., Symons M. Signaling determinants of glioma cell invasion. Adv. Exp. Med. Biol. 2013;986:121–141. doi: 10.1007/978-94-007-4719-7_7. [DOI] [PubMed] [Google Scholar]
- Lang F., Busch G.L., Ritter M., Volkl H., Waldegger S., Gulbins E., Haussinger D. Functional significance of cell volume regulatory mechanisms. Physiol. Rev. 1998;78:247–306. doi: 10.1152/physrev.1998.78.1.247. [DOI] [PubMed] [Google Scholar]
- Lathia J.D., Mack S.C., Mulkearns-Hubert E.E., Valentim C.L., Rich J.N. Cancer stem cells in glioblastoma. Genes Dev. 2015;29:1203–1217. doi: 10.1101/gad.261982.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauffenburger D.A., Horwitz A.F. Cell migration: a physically integrated molecular process. Cell. 1996;84:359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- Nakada M., Nakada S., Demuth T., Tran N.L., Hoelzinger D.B., Berens M.E. Molecular targets of glioma invasion. Cell. Mol. Life Sci. 2007;64:458–478. doi: 10.1007/s00018-007-6342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman L., Sengupta K., Aranda-Espinoza H. Blebbing dynamics during endothelial cell spreading. Eur. J. Cell Biol. 2011;90:37–48. doi: 10.1016/j.ejcb.2010.09.013. [DOI] [PubMed] [Google Scholar]
- Norman L.L., Brugues J., Sengupta K., Sens P., Aranda-Espinoza H. Cell blebbing and membrane area homeostasis in spreading and retracting cells. Biophys. J. 2010;99:1726–1733. doi: 10.1016/j.bpj.2010.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman L.L., Oetama R.J., Dembo M., Byfield F., Hammer D.A., Levitan I., Aranda-Espinoza H. Modification of cellular cholesterol content affects traction force, adhesion and cell spreading. Cell. Mol. Bioeng. 2010;3:151–162. doi: 10.1007/s12195-010-0119-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.B., Agnihotri S., Golbourn B., Bertrand K.C., Luck A., Sabha N., Smith C.A., Byron S., Zadeh G., Croul S., Berens M., Rutka J.T. Transcriptional profiling of GBM invasion genes identifies effective inhibitors of the LIM kinase-cofilin pathway. Oncotarget. 2014;5:9382–9395. doi: 10.18632/oncotarget.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T.D., Borisy G.G. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- Quinones-Hinojosa A., Chaichana K. The human subventricular zone: a source of new cells and a potential source of brain tumors. Exp. Neurol. 2007;205:313–324. doi: 10.1016/j.expneurol.2007.03.016. [DOI] [PubMed] [Google Scholar]
- Ridley A.J. Life at the leading edge. Cell. 2011;145:1012–1022. doi: 10.1016/j.cell.2011.06.010. [DOI] [PubMed] [Google Scholar]
- Ridley A.J., Schwartz M.A., Burridge K., Firtel R.A., Ginsberg M.H., Borisy G., Parsons J.T., Horwitz A.R. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- Riedl J., Crevenna A.H., Kessenbrock K., Yu J.H., Neukirchen D., Bista M., Bradke F., Jenne D., Holak T.A., Werb Z., Sixt M., Wedlich-Soldner R. Lifeact: a versatile marker to visualize F-actin. Nat. Methods. 2008;5(7):605. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath P., Pollard T.D. Effects of cytochalasin, phalloidin, and pH on the elongation of actin filaments. Biochemistry. 1991;30:1973–1980. doi: 10.1021/bi00221a034. [DOI] [PubMed] [Google Scholar]
- Scherer H.J. Structural development in gliomas. J. Cancer. 1938;34:333–351. [Google Scholar]
- Sidani M., Wessels D., Mouneimne G., Ghosh M., Goswami S., Sarmiento C., Wang W., Kuhl S., El-Sibai M., Backer J.M., Eddy R., Soll D., Condeelis J. Cofilin determines the migration behavior and turning frequency of metastatic cancer cells. J. Cell Biol. 2007;179:777–791. doi: 10.1083/jcb.200707009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R., Naishadham D., Jemal A. Cancer statistics, 2013. CA Cancer J. Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- Smith C.L., Kilic O., Schiapparelli P., Guerrero-Cazares H., Kim D.H., Sedora-Roman N.I., Gupta S., O'Donnell T., Chaichana K.L., Rodriguez F.J., Abbadi S., Park J., Quinones-Hinojosa A., Levchenko A. Migration phenotype of brain-cancer cells predicts patient outcomes. Cell Rep. 2016;15:2616–2624. doi: 10.1016/j.celrep.2016.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starinsky-Elbaz S., Faigenbloom L., Friedman E., Stein R., Kloog Y. The pre-GAP-related domain of neurofibromin regulates cell migration through the LIM kinase/cofilin pathway. Mol. Cell. Neurosci. 2009;42:278–287. doi: 10.1016/j.mcn.2009.07.014. [DOI] [PubMed] [Google Scholar]
- Stupp R., Mason W.P., van Den Bent M.J., Weller M., Fisher B., Taphoorn M.J., Belanger K., Brandes A.A., Marosi C., Bogdahn U., Curschmann J., Janzer R.C., Ludwin S.K., Gorlia T., Allgeier A., Lacombe D., Cairncross J.G., Eisenhauer E., Mirimanoff R.O. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- Tilghman J., Schiapparelli P., Lal B., Ying M., Quinones-Hinojosa A., Xia S., Laterra J. Regulation of glioblastoma tumor-propagating cells by the integrin partner tetraspanin CD151. Neoplasia. 2016;18:185–198. doi: 10.1016/j.neo.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trendowski M. Exploiting the cytoskeletal filaments of neoplastic cells to potentiate a novel therapeutic approach. Biochim. Biophys. Acta. 2014;1846:599–616. doi: 10.1016/j.bbcan.2014.09.007. [DOI] [PubMed] [Google Scholar]
- van Rheenen J., Condeelis J., Glogauer M. A common cofilin activity cycle in invasive tumor cells and inflammatory cells. J. Cell Sci. 2009;122:305–311. doi: 10.1242/jcs.031146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins S., Robel S., Kimbrough I.F., Robert S.M., Ellis-Davies G., Sontheimer H. Disruption of astrocyte-vascular coupling and the blood-brain barrier by invading glioma cells. Nat. Commun. 2014;5:4196. doi: 10.1038/ncomms5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins S., Sontheimer H. Hydrodynamic cellular volume changes enable glioma cell invasion. J. Neurosci. 2011;31:17250–17259. doi: 10.1523/JNEUROSCI.3938-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijer C.J. Collective cell migration in development. J. Cell Sci. 2009;122:3215–3223. doi: 10.1242/jcs.036517. [DOI] [PubMed] [Google Scholar]
- Yang J.M., Schiapparelli P., Nguyen H.N., Igarashi A., Zhang Q., Abbadi S., Amzel L.M., Sesaki H., Quinones-Hinojosa A., Iijima M. Characterization of PTEN mutations in brain cancer reveals that pten mono-ubiquitination promotes protein stability and nuclear localization. Oncogene. 2017 doi: 10.1038/onc.2016.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap C.T., Simpson T.I., Pratt T., Price D.J., Maciver S.K. The motility of glioblastoma tumour cells is modulated by intracellular cofilin expression in a concentration-dependent manner. Cell Motil. Cytoskeleton. 2005;60:153–165. doi: 10.1002/cm.20053. [DOI] [PubMed] [Google Scholar]
- Zhou T., Wang C.H., Yan H., Zhang R., Zhao J.B., Qian C.F., Xiao H., Liu H.Y. Inhibition of the Rac1-WAVE2-Arp2/3 signaling pathway promotes radiosensitivity via downregulation of cofilin-1 in U251 human glioma cells. Mol. Med. Rep. 2016;13:4414–4420. doi: 10.3892/mmr.2016.5088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
EGF stimulation on control and NKCC1 KD cells expressing the fibrilary actin sensor lifeact.
Supplementary material