ABSTRACT
Canonical translation initiation involves ribosomal scanning, but short 5′ untranslated region (5′UTR) mRNAs are translated in a scanning-independent manner. The extent and mechanism of scanning-independent translation are not fully understood. Here we report that short 5′UTR mRNAs constitute a substantial fraction of the translatome. Short 5′UTR mRNAs are enriched with TISU (translation initiator of short 5′UTR), a 12-nucleotide element directing efficient scanning-independent translation. Comprehensive mutagenesis revealed that each AUG codon-flanking nucleotide of TISU contributes to translational strength, but only a few are important for accuracy. Using site-specific UV cross-linking of ribosomal complexes assembled on TISU mRNA, we demonstrate specific binding of TISU to ribosomal proteins at the E and A sites. We identified RPS3 as the major TISU binding protein in the 48S complex A site. Upon 80S complex formation, RPS3 interaction is weakened and switched to RPS10e (formerly called RPS10). We further demonstrate that TISU is particularly dependent on eukaryotic initiation factor 1A (eIF1A) which interacts with both RPS3 and RPS10e. Our findings suggest that the cap-recruited ribosome specifically binds the TISU nucleotides at the A and E sites in cooperation with eIF1A to promote scanning arrest.
KEYWORDS: translation initiation, TISU, RPS3, RPS10e, eIF1A, short 5′UTR, RPS10
INTRODUCTION
Canonical translation initiation in eukaryotes starts with binding of eukaryotic initiation factor 4F (eIF4F) to the m7G-capped 5′ end of the mRNA, which is then followed by the recruitment of the 43S preinitiation complex (PIC) consisting of the 40S small ribosomal subunit, initiator Met-tRNA, and a subset of initiation factors (eIF2, eIF1, eIF1A, eIF3, and eIF5). The 43S PIC then scans the mRNA 5′ untranslated region (5′UTR) and inspects it for an AUG start codon. During scanning, the 43S PIC is in an “open” state, and following AUG recognition, the 48S initiation complex is switched to a “closed” conformation that arrests scanning and promotes joining of the 60S large subunit to create the 80S elongation-competent complex (reviewed in references 1, 2, 3, 4, and 5). Translation usually initiates at the first 5′-proximal AUG codon, but in certain cases, translation initiates at a downstream AUG (DS-AUG) codon, a phenomenon known as leaky scanning. The extent of leaky scanning depends on the 5′UTR length and on the AUG nucleotide context (6, 7, 8, 9). For mRNA with a 5′UTR that is more than 30 nucleotides (nt) long, the optimal AUG context is the Kozak element, RCCAUGG, in which the most significant nucleotides (shown in boldface type) are the purine R in position −3 and G in position +4 relative to the A (+1) of the AUG codon. With this context, accurate initiation requires 5′UTR length of at least 20 nucleotides (10). Shorter 5′UTRs generally exhibit leaky translation initiation (6, 10–13). Therefore, translation of eukaryotic mRNAs with a short 5′UTR is usually inefficient. The translation of a class of short 5′UTR of mRNAs that is governed by TISU (translation initiator of short 5′UTR) deviates from this canonical scanning mechanism. TISU is located near the 5′ end of the mRNA and is capable of directing efficient and accurate translation initiation from very short 5′UTRs (6, 13, 14). TISU is present in ∼4% of all mammalian genes and controls not only translation but also transcription (13). This element is highly prevalent among genes with “housekeeping” functions such as protein synthesis, mitochondrial activities, and energy metabolism. A recent study revealed that TISU confers translational resistance to global inhibition of translation in response to energy stress but not to other stresses (15). In addition, it was recently found that circadian and feeding rhythms differentially affect translation of TISU mRNAs (16) and TISU is highly sensitive to mammalian target of rapamycin (mTOR) inhibition (17). The core of TISU consists of an AUG sequence, which also serves as the exclusive translation initiation codon in most TISU genes. Besides the A (−3) and G (+4) nucleotides, additional flanking sequences are unique to TISU and cooperate to direct accurate and efficient translation initiation from short 5′UTR. Importantly, TISU activity requires the cap but does not require scanning (6).
Interestingly, efficient AUG recognition of TISU does not require a specific factor and is intrinsic to the basal translation machinery (15). Omission of individual translation initiation factors in vitro and in a cell context revealed differential requirements for eIF1 and eIF4G1 compared with canonical AUG with a short 5′UTR (15). Additional studies revealed that a key step in start codon selection of TISU is the release of eIF4F, following AUG recognition by the 48S complex (15). The detachment of the cap complex facilitates translation from short 5′UTR mRNA by preventing a clash between the 48S initiation complex and the nearby eIF4F-cap complex. In spite of the sensitivity of TISU to the availability of various eIFs and stress conditions, TISU retains its resistance to leaky scanning. How TISU avoids leaky scanning under various translational constraints is not known.
In the present study, we investigated the extent of scanning-dependent and -independent translation in mammalian cells and the mechanistic basis for TISU recognition by the basal translation machinery. Our findings revealed that scanning-independent translation of short 5′UTR is highly prevalent in mammalian cells. In addition, we found that TISU forms sequence-specific contacts with A-site ribosomal proteins RPS3 and RPS10e. TISU activity is particularly dependent on eIF1A, which binds RPS3 and RPS10e, suggesting their cooperation in promoting scanning arrest.
RESULTS
Scanning-independent translation is highly prevalent in mammalian cells.
Many mammalian genes have more than one promoter and/or multiple transcription start sites, thus encoding transcripts that vary in their 5′UTR length and composition. As the length of the 5′UTR influences the extent of scanning, we determined the scope of scanning-dependent and -independent translation in mammalian cells in a global manner. To this end, we analyzed the CapSeq data of mRNA extracted from a polysomal profiling experiment of mouse embryonic fibroblasts (MEFs). In this experiment, cell lysates were subjected to sucrose gradient sedimentation. RNA extracted from the gradient fractions was used for the CapSeq analysis. In this method, capped 5′ ends of mRNA sequence tags are particularly enriched, allowing for accurate determination of the 5′UTR length of the mRNA molecules along with their translation (the exact details of the experiment are described by Tamarkin-Ben-Harush et al. [18]). The data were divided into two major pools: polysome-free, from the top of the gradient to a single ribosome; polysomal, two or more ribosomes per mRNA. Global analysis of 5′UTR lengths relative to the AUG of the annotated open reading frame (ORF) (rather than the first AUG, to avoid overestimation of mRNAs with short 5′UTR) and their relative abundance in the free and polysomal pools is shown in Fig. 1A. This analysis revealed that when considering overall transcript copy number, those transcripts with short 5′UTR mRNAs (up to 40 nt) are abundant in the transcriptome (free plus polysome-associated transcripts) and the translatome (polysome-associated transcripts). We also analyzed a recently published CapSeq data set of polysome profile with data from each ribosomal fraction (19) and similarly found that short 5′UTR mRNAs constitute a significant fraction of the translatome (see Fig. S1 in the supplemental material). Thus, translation without or with minimal scanning appears to be much more prevalent than anticipated.
FIG 1.
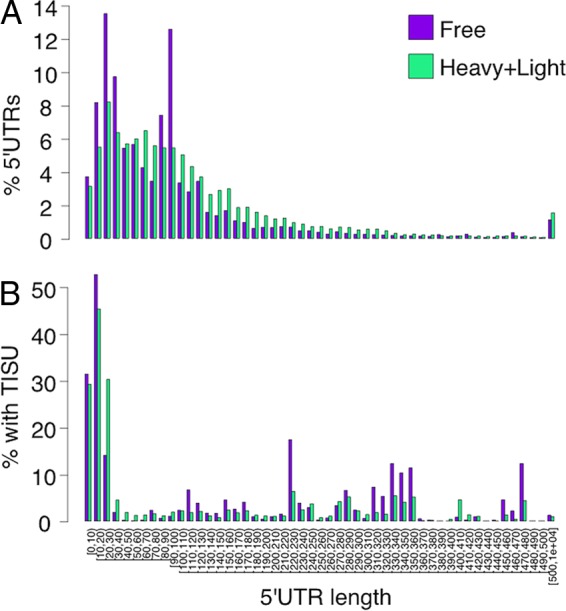
Distribution of 5′UTR lengths and TISU usage of transcripts in mouse embryonic fibroblasts (MEFs). (A) Each genomic position with CapSeq reads was associated with a protein-coding transcript from the Ensembl 82 annotations if it was within 100 nt of its 5′ end or overlapped one of its 5′UTR exons, and the corresponding 5′UTR length was computed. If multiple transcripts were relevant, the average 5′UTR length was used. Then, for each 5′UTR length, we summed the total transcript abundance, normalized the abundances, and binned lengths into bins of 10 nt. (B) Same as panel A but including only transcripts that match the TISU motif within the first 40 nt of the predicted transcript.
The global analysis confirms that translation of short 5′UTR is relatively inefficient. TISU is an element present in a subset of short 5′UTR mRNAs that can drive accurate translation. We analyzed the sequences of the 5′-end mRNA tags for the presence of the TISU motif (SAASATGGCGGC, with up to two mismatches). The results confirm that transcripts bearing the TISU element are highly enriched among short 5′UTR mRNAs, constituting between 30 and 50% of these transcripts, which is consistent with a previous report (13), and their translation appears to be more efficient, particularly for 5′UTR lengths of 10 to 30 nucleotides (Fig. 1B, compare to panel A).
TISU element sequence requirement.
The consensus sequence of the TISU element was initially determined by computational and block mutational analyses (13). To determine the sequence requirements of TISU in greater detail, each position of the element that flanks the central AUG codon was substituted to all the other three possible nucleotides along with mutations of the A (+1) position, yielding 29 mutants. We have previously verified that mutations in TISU do not change the decay rates of mRNA and protein (6, 13), so the effect of TISU mutants is expected to be primarily on translation. Wild-type (WT) TISU and mutants were placed upstream and in frame with the initiating AUG codon of a green fluorescent protein (GFP) reporter and downstream of the T7 promoter (Fig. 2A). In all the constructs, the core TISU AUG is preceded by a 5-nucleotide-long 5′UTR (Fig. 2A). Translation that begins from the TISU AUG generates an ∼30-kDa GFP protein (US-AUG). In a case of leaky scanning, translation begins from the downstream GFP AUG and produces an ∼27-kDa protein (DS-AUG). In vitro-transcribed, capped, and poly(A)-tailed GFP mRNAs (Fig. S2) were cotransfected into HeLa cells together with a luciferase mRNA that served as an internal control for transfection efficiency. The in vivo-translated GFP proteins were analyzed by Western blotting. The results show that the vast majority of the TISU mutants decreased translation from the upstream AUG (US-AUG) (Fig. 2B), indicating that AUG-flanking nucleotides contribute to the efficient translation initiation driven by TISU. From this analysis, it appears that the best TISU consensus sequence is CAAGAUGGCGGC. Of particular interest are mutations of the A at the −3 position in which substitutions to pyrimidine caused substantial leakage to the downstream AUG and substitution to the other purine G retained selective US-AUG translation but significantly decreased strength. Mutations of the G (+4) to U/C/A, C (+5) to U, and G (+6) to C also enhanced leaky scanning but to a lesser extent (Fig. 2B), indicating that these nucleotides cooperate to prevent leaky scanning. Consistent with that substitution of +4, +5, and +6 altogether resulted in substantial leaky translation (13). Mutations C (+5) to A and C (+8) to A caused a slight retardation in the mobility of the GFP protein, which is likely to be the consequence of alteration of the encoded amino acid from alanine to glutamic acid in these two mutants. Remarkably, only 7 out of 27 AUG-flanking mutants exhibit decreased translation initiation accuracy, highlighting TISU robustness and suggesting that many single nucleotide deviations from the consensus sequence are functional. On the other hand, the flanking sequences cannot support initiation from near cognate AUG codons such as CUG or GUG (Fig. 2B), as these are bypassed and translation initiates predominantly from the DS-AUG.
FIG 2.
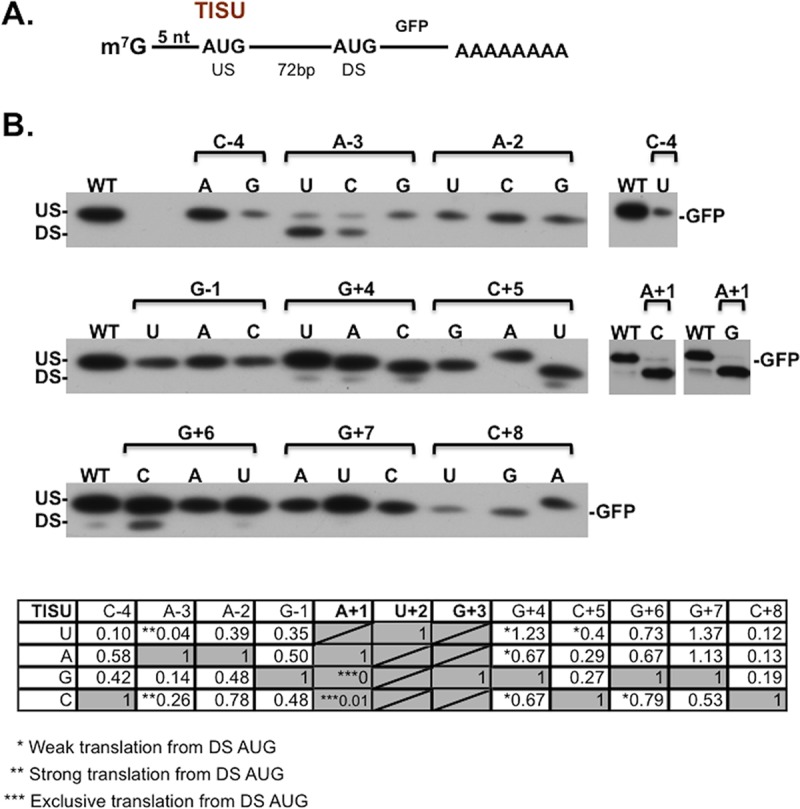
The exact sequence requirement for TISU activity. (A) A schematic representation of the reporter gene in which the TISU element is cloned downstream of the T7 promoter with a 5′UTR length of 5 nt and its AUG codon is in frame with the downstream (DS) AUG codon of GFP. (B) A total of 29 mutants of TISU, either in AUG-flanking nucleotides or in the AUG, were transcribed and capped in vitro, and the resultant mRNAs were then transfected into HeLa cells where they underwent in vivo translation. Cells were also transfected with a luciferase mRNA that served as an internal control for transfection efficiency. GFP expression was analyzed by immunoblotting using anti-GFP antibody. US and DS denote upstream and downstream initiation sites, respectively. The table represents quantification of translation from the US-AUG. The number of asterisks in the table denotes the extent of translation from the downstream AUG in a qualitative manner.
Sequence-specific binding of TISU.
As TISU associates with the 48S ribosome with high affinity (6, 15), we next wished to identify the proteins that directly interact with the TISU sequence. For this purpose, we employed site-specific UV cross-linking experiments. We synthesized a series of [32P]CTP-labeled TISU and canonical AUG mRNAs (Fig. 3, top) in which each position of the element is modified with a thio-UTP as shown in Fig. S3A. The thio-UTP can be specifically cross-linked to adjacent interacting proteins by low-energy UV irradiation (365 nm), yielding zero-length cross-links that represent direct contacts with proteins (20, 21, 22). The radiolabeled mRNAs were incubated with rabbit reticulocyte lysate in the presence of either GMP-PNP (a nonhydrolyzable analog of GTP) to stall the 48S complex or cycloheximide to form 80S complex. After complex formation, the reactions were subjected to UV cross-linking, RNase treatment, and SDS-PAGE. Following GMP-PNP treatment (48S formation), we observed specific cross-linked proteins in positions −4, +5, and +6 (Fig. S3B, red arrows) which have low molecular sizes ranging from 25 to 15 kDa. Likewise, with the 80S complex, we detected specific cross-linkings of small polypeptides at positions −3, −2, +4, +5, +6, +7, and +8 (Fig. S3C). As the vast majority of translation initiation factors are larger in their size, the cross-linked proteins are most likely ribosomal proteins that belong to the small subunit, known to have low molecular weights (MWs). It was previously shown that the canonical initiation sequence interacts with a 23-kDa RPS7 (formerly called RPS5) at the −3 position and with a 17-kDa RPS19 (formerly RPS15) at position +4 (22). We also observed cross-linked proteins with these exact sizes at those positions in both the canonical sequences and TISU elements (Fig. S3C), indicating that these are the same proteins.
FIG 3.
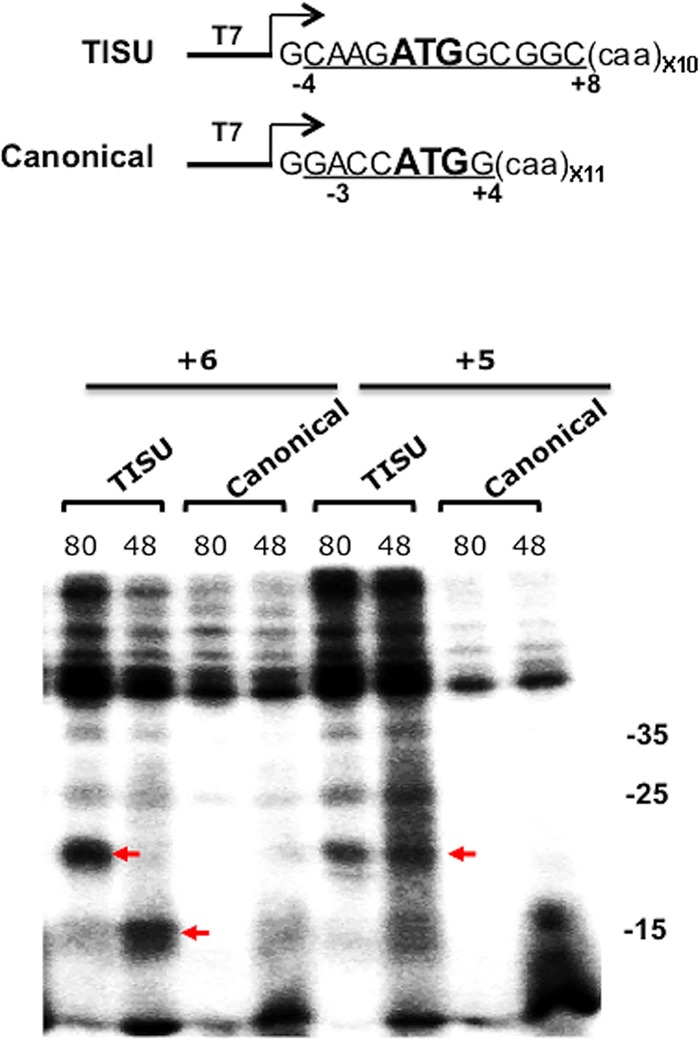
TISU forms specific contacts with ribosomal proteins. (Top) Schematic representation of the fragments used for in vitro transcription and UV cross-linking. (Bottom) Comparison of the cross-linking profile of +5 and +6 positions of the canonical and TISU AUG codons. These mRNAs were modified with a thio-U at +5 and +6 positions and were incubated with rabbit reticulocyte lysate in the presence of GMP-PNP (48S) or cycloheximide (80S) and then were subjected to 365-nm UV irradiation followed by RNase treatment. The cross-linked proteins were analyzed by 15% SDS-PAGE and autoradiography. Red arrows denote specific cross-linked polypeptides. The numbers on the right denote molecular masses in kilodaltons.
To examine the specificity of the cross-linked polypeptides of positions +5 and +6, we compared them to the canonical AUG mRNA bearing the thio-U at positions +5 and +6, both in the 48S and 80S complex. The cross-linked proteins of the 48S and 80S complexes at positions +5 and +6 are clearly seen with TISU mRNA but are barely detected with the canonical AUG mRNA (Fig. 3, bottom). These results are consistent with sequence-specific recognition of TISU.
Identification of RPS3 and RPS10e as TISU binding ribosomal proteins.
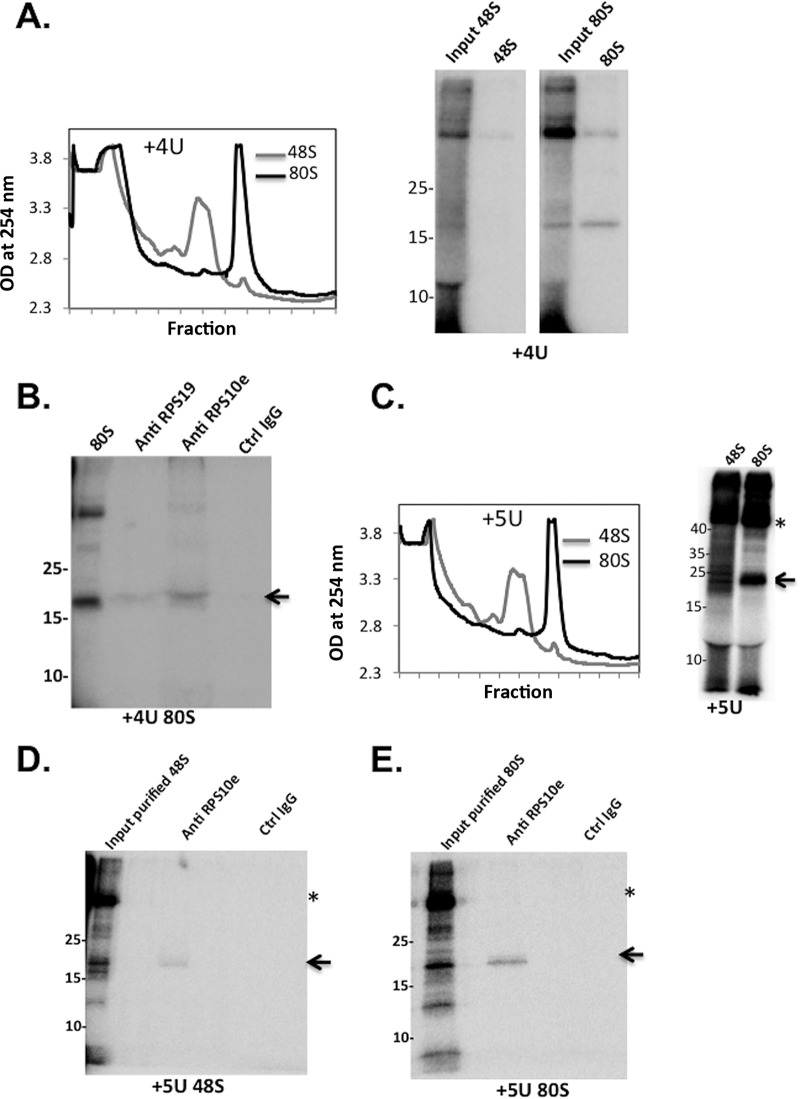
To further investigate the ribosomal proteins interacting with the TISU-specific AUG downstream nucleotides +4, +5, and +6, the cross-linked 48S and 80S complexes were resolved on sucrose gradients. To validate the formation of the 48S and 80S complexes under these conditions, fractions from similar sucrose gradients of TISU mRNA-ribosomal complex assemblies were analyzed by immunoblotting for the presence of ribosomal proteins that belong to the small (RPS5) and large (RPL11) subunits (Fig. S4). The peak fraction of the GMP-PNP-treated extract is enriched for RPS5 but not RPL11, while the peak of the cycloheximide (CHX)-treated extract is enriched for both RPS5 and RPL11, as expected for 48S and 80S complexes. The major cross-linked polypeptide detected with the 48S complex is seen with the +6 position and is of ∼35 kDa. This polypeptide is highly enriched in the 48S complex, and it binds TISU more strongly than the 80S complex (Fig. 4A). With the +4 and +5 positions, a major cross-linked polypeptide of 18 to 20 kDa is seen with both the 48S and 80S complexes, but it is much stronger with the latter (Fig. 5A and C). To identify the cross-linked polypeptides, we initially subjected the reaction mixtures to SDS-PAGE and mass spectroscopy and obtained multiple ribosomal proteins that are similar in size, most of which are far from the mRNA channel. We therefore turned to analyzing specific ribosomal protein candidates on the basis of their molecular size and position in the ribosome. We envisaged that the AUG downstream nucleotides must interact with ribosomal proteins that are proximal to the ribosomal A site. According to the structures of Saccharomyces cerevisiae and mammalian ribosomes (23, 24), the A-site ribosomal proteins RPS3, RPS19 (formerly called RPS15), and RPS10e (formerly RPS10) have the potential to interact with the RNA and the appropriate MW. Using antibodies against these ribosomal proteins, we first confirmed that they could be used for immunoprecipitation of the corresponding protein using cell lysates and Western blotting (Fig. S5). Next, we used these antibodies to immunoprecipitate the cross-linked polypeptides. With the 48S +6 position, the cross-linked protein was efficiently and specifically immunoprecipitated with RPS3 antibodies, indicating that this ribosomal protein specifically binds TISU +6 position in the 48S complex (Fig. 4B). With the +4 position we found weak immunoprecipitation of the cross-linked polypeptide with RPS19 antibodies and more-efficient precipitation by the RPS10e antibodies (Fig. 5B). The RPS10e antibodies also specifically immunoprecipitated the +5 cross-linked polypeptide seen with both the 48S and 80S complexes (Fig. 5D and E), but the signal with the 80S complex is stronger. As RPS3, RPS10e, and RPS19 are adjacent to each other in the A site (25, 26), we conclude that during 48S preinitiation complex formation, TISU specifically binds RPS3 and weakly binds RPS10e. Following 80S complex formation, the interaction with RPS3 is weakened and switched to RPS10e and perhaps RPS19. These findings are consistent with the idea that the cap-recruited ribosome specifically binds the TISU nucleotides at the A and E sites to arrest scanning.
FIG 4.
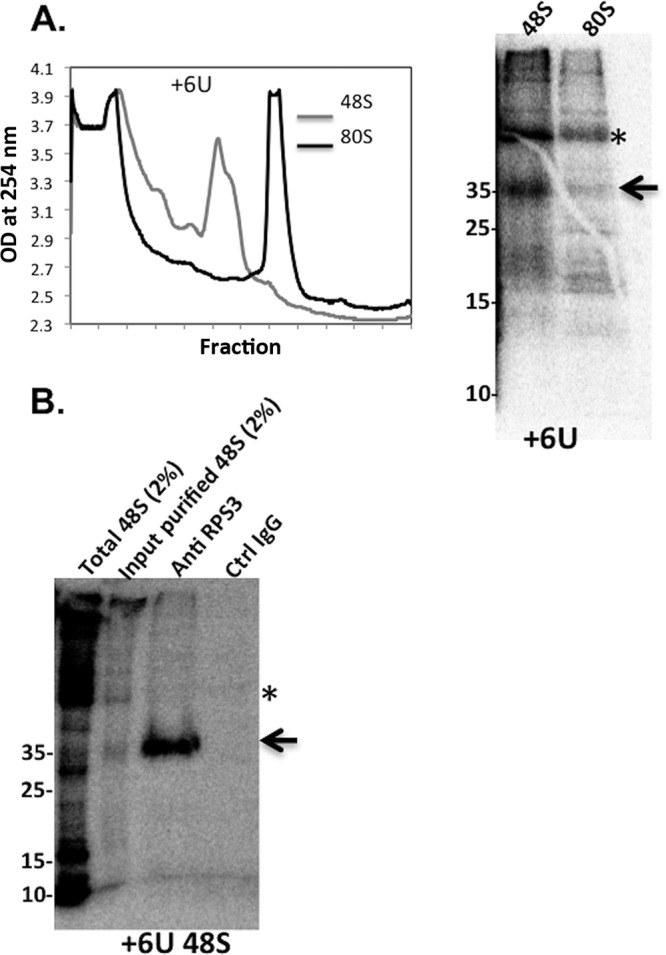
Identification of RPS3 as TISU-interacting ribosomal protein. (A) 48S and 80S complexes were assembled with TISU-labeled mRNA modified with thio-UTP at +6 position. The ribosomal complexes were UV cross-linked and then subjected to 8 to 32% sucrose gradient sedimentation and fraction collection (left panel). The peak fractions were TCA precipitated and analyzed by SDS-PAGE followed by autoradiography (right panel). The asterisk denotes a nonspecific polypeptide. The arrow points to a specific cross-linked polypeptide. OD, optical density. (B) The pooled peak fractions of the 48S complex were pooled, treated with RNase, and subjected to immunoprecipitation with anti-RPS3 and control antibodies as indicated, followed by 15% SDS-PAGE and autoradiography. The asterisk denotes a nonspecific polypeptide, and the arrow points to the position of RPS3. Ctrl, control.
FIG 5.
Identification of RP10Se as TISU-interacting ribosomal protein of the 80S complex. (A) 48S and 80S complexes were assembled with TISU-labeled mRNA modified with thio-UTP at the +4 position. The ribosomal complexes were UV cross-linked and then subjected to a sucrose gradient sedimentation (left panel) followed by SDS-PAGE and autoradiography (right panel). (B) The peak fractions of the 80S complex described in panel A were pooled, treated with RNase, and subjected to immunoprecipitation with anti-RPS19, anti-RPS10e, and control antibodies as indicated followed by 15% SDS-PAGE and autoradiography. Size markers (in kilodaltons) are shown to the left of the gel. (C) 48S and 80S complexes were assembled with +5 thio-UTP-modified and labeled TISU mRNA followed by UV cross-linked and sucrose gradient sedimentation (left panel) followed by SDS-PAGE and autoradiography (right panel). (D) The peak fractions of the 48S complex were subjected to immunoprecipitation with anti-RPS10e and control antibodies as indicated, followed by 15% SDS-PAGE and autoradiography. Size markers are shown to the left of the gel. (E) Same as panel D but with the 80S fractions. The asterisks in panels C, D, and E denote nonspecific polypeptides.
TISU binding by RPS3 and RPS10e may be linked to a specific requirement of eIF1A.
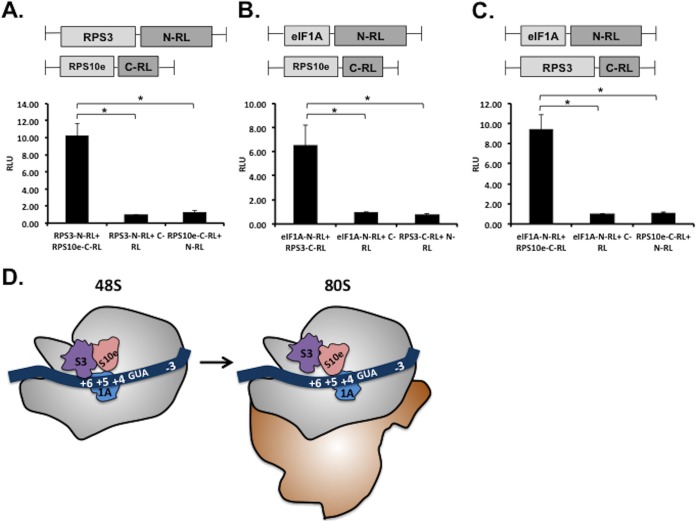
Addressing the significance of RPS3 and/or RPS10e binding to TISU is not practical, since both are highly conserved core ribosomal proteins and their downregulation is expected to affect 40S ribosome integrity and translation in general. The cryo-electron microscopy (cryo-EM) structure of the 48S complex in the presence of eIF1 and eIF1A suggests that eIF1A is located near the A site and the mRNA channel around +4 and +5 positions proximal to RPS3 and RPS10e (27). Interestingly, binding of eIF1A to the ribosome has an allosteric effect on RPS3 conformation (26). Given the structural evidence for a functional link between RPS3 and eIF1A and their proximity to the mRNA entry channel, we sought to examine the role of eIF1A in TISU-mediated translation. For this purpose, eIF1A was knocked down in HEK293T cells using small interfering RNA (siRNA). Forty-eight hours later, cells were transfected again with eIF1A siRNA together with GFP reporter genes. Two genes with short 5′UTR bearing either TISU or canonical AUG context (US-AUG) that is in frame with the authentic downstream AUG of GFP (DS-AUG), serving as readout of leaky scanning. The third gene contains a long unstructured 5′UTR, and its translation is scanning dependent. This knockdown approach resulted in undetectable eIF1A levels (Fig. 6, right panel). Remarkably, translation from TISU-AUG was dramatically decreased without concomitant increase in leaky scanning. In contrast, translation from the canonical AUG context at the same 5′UTR position was only slightly affected, and translation from the long 5′UTR reporter was unaffected by eIF1A depletion (Fig. 6). Thus, eIF1A plays a nonredundant essential role in TISU-mediated translation. To investigate further the differential requirement of eIF1A for TISU, we tested whether it binds RPS3 and/or RPS10e. We employed a protein-protein interaction assay that is based on the split Renilla luciferase (RL) complementation assay (28). In this assay, RL is divided into two inactive N- and C-terminal parts (N-RL and C-RL, respectively) and fused to target proteins. Interaction of the target proteins brings the N- and C-terminal fragments of the RL in close proximity, resulting in enhanced enzymatic activity. First, we fused full-length RPS3 and RPS10e to N- and C-RL, respectively (Fig. 7A). These plasmid pairs were transfected into cells and analyzed for RL activity. The RPS3-RPS10e pair conferred strong RL activity relative to the empty counterparts (Fig. 7A), consistent with the structural evidence of their interaction (24). We next analyzed the ability of eIF1A fused to N-RL to interact with RPS10e and RPS3, each fused to C-RL. Both eIF1A-RPS10e and eIF1A-RPS3 pairs drive stronger RL activity compared to the controls (Fig. 7B and C), suggesting that eIF1A forms a subcomplex with RPS10e and RPS3. Considering the selective involvement of these proteins in TISU activity, their interactions are likely to promote accurate initiation from this element (Fig. 7D).
FIG 6.
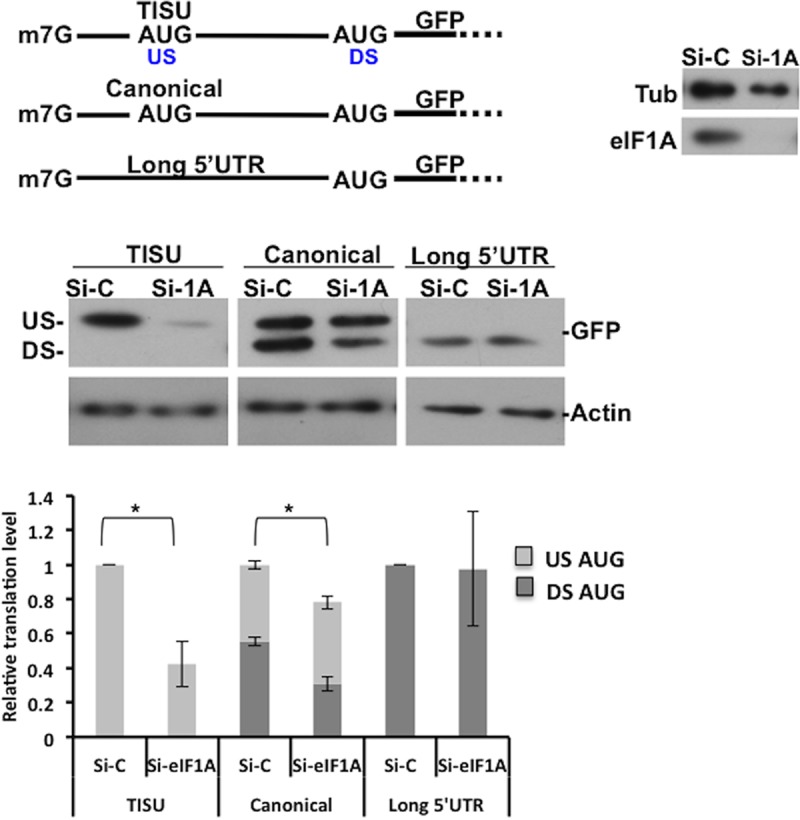
Effect of eIF1A knockdown on translation initiation of cap-proximal AUG and long 5′UTR. HEK293T cells were transfected with a smart pool of either nontargeting (control) or eIF1A siRNA (50 nM). Forty-eight hours later, cells were cotransfected again with the siRNA pool with short 5′UTR together with GFP reporter genes bearing TISU or canonical AUG context or with a long 5′UTR GFP reporter as shown schematically in the diagram at the top of the figure. Cells were harvested 24 h after the second transfection, and Western blot analysis was performed. Representative immunoblots are shown. Knockdown efficiency compared to si-control (Si-C) is presented in the right panel. The graph presents densitometric analysis of GFP levels of five or six independent experiments (average ± standard error [SE] [error bar]). The overall translation in each control was set to one. The relative translation from the US-AUG codon and the DS-AUG is presented. Values that are statistically significant different (P < 0.05) are indicated by a bar and asterisk.
FIG 7.
eIF1A interacts with RPS3 and RPS10e. (A) The N and C termini of the RL were fused to RPS3 and RPS10e, respectively, as shown in the schematic representation above the graph. Cells were transfected with the indicated plasmids together with miR-22-firefly luciferase (FL), which served as a measure for transfection efficiency. For controls, each of the fusion proteins was transfected with the empty counterpart. RL and FL activities (in relative light units [RLU]) were measured after 24 h. The bars represent the means plus standard errors of the means (SEM) (error bars) of four independent experiments. The asterisks denote statistically significant difference (P < 0.05). (B and C) A split RL assay as described in panel A for eIF1A-RPS10e (B) and eIF1A-RPS3 (C) pairs. (D) Illustration showing TISU-interacting ribosomal proteins RPS3 and RPS10e in the 48S and 80S complexes and their interaction with eIF1A. RPS3 and RPS10 undergo conformational change upon 48S to 80S transition; binding of RPS3 to TISU is diminished, while RPS10e is strengthened following this transition.
DISCUSSION
The most prevalent form of translation initiation in eukaryotes involves m7G cap binding and ribosomal scanning. Our findings revealed that translation initiation that operates without or with minimal scanning constitute a significant part of the translatome, highlighting the importance of this form of translation. Using TISU as a model for a robust scanning-free translation, we demonstrate that its strength and ability to prevent leaky scanning are linked to sequence-specific interactions with ribosomal proteins positioned at the E and A sites. We further identified the A-site ribosomal proteins RPS3 and RPS10e as TISU binding proteins in the 48S and 80S complexes, respectively (Fig. 7D). The RPS3 interaction with the +6 position of TISU fits well with the results of cryo-EM studies of the 48S complex, which revealed several basic residues of RPS3 facing the mRNA entry channel (25, 26). This finding is consistent with the requirement of AUG upstream and downstream nucleotides for TISU activity. Whether the AUG-flanking sequences of TISU also interact with the 18S rRNA is yet to be determined. Considering that TISU resists leaky scanning upon depletion or inactivation of most eIFs (15; this study), we propose that the interaction of ribosomal proteins with TISU triggers structural rearrangements that lead to a “closed”scanning-arrested conformation independently of eIFs. This can explain the resistance of TISU to leaky scanning under various conditions of translation initiation factor constraints.
The specific requirement of eIF1A (this study) and eIF1 (15) for TISU is unexpected, considering that both cooperate to promote the “open” scanning-competent form of the 48S complex (29–31). The “open” conformation involves the opening of the mRNA channel latch formed by helices 18 and 34 of the 18S rRNA. This conformational change is facilitated by a new interaction of RPS3 with helix 16 of the 18S rRNA (26). Following AUG recognition via base pairing with Met-tRNA interference (tRNAi) anticodon, eIF1A together with other initiation factors stimulates the reverse conformational change of the 40S rRNA to a “closed” state (26, 32, 33). Considering the importance of eIF1 and eIF1A in establishing the “open” conformation, we propose that the explicit requirement for each of these factors for the scanning-free translation directed by TISU may be for the initial opening of the mRNA entry channel latch in the empty 40S subunit to allow mRNA binding. Subsequently, eIF1A interaction with RPS3 and RPS10e may also promote their binding to TISU, perhaps by displacing h16 binding, resulting in closing of the latch and scanning arrest.
The detailed analysis of TISU sequence requirements highlights its robustness and further establishes it as an optimal initiator of short 5′UTR mRNAs which is distinguished from the Kozak element in its sequence requirements. Our findings revealed that the most critical nucleotides for the high precision of TISU are purine at position −3 and nucleotides +4, +5, and +6, which cooperate to support initiation accuracy and translational strength. Interestingly, in yeast, the purine at position −3 is also critical for start codon recognition (34, 35), and a comparative analysis of orthologous sets of mammalian and yeast mRNAs revealed high preference for purine at position −3 (36). Thus, it seems that the common preference for purine at position −3 is a conserved feature of eukaryotic mRNAs. Besides the sequence requirement, there are additional features that discriminate TISU from the Kozak element, including the strict location of TISU, its dual function in transcription and translation, and its mode of translation which is scanning free (37). It would be interesting to determine how these TISU mutants influence its activity in transcription.
MATERIALS AND METHODS
Plasmid construction and preparation of mRNA for in vivo translation.
The TISU construct used for mRNA preparation was described previously (6, 13). Using the transfer PCR (T-PCR) cloning method (38), we generated TISU mutants. For synthesis of capped mRNA, the constructs containing the T7 promoter were linearized and used with the T7 RiboMAX large-scale RNA production system (Promega) supplemented with Ribo m7G cap analog (New England BioLabs [NEB]). The reaction was stopped by the addition of RQ1 RNase-free DNase I (Promega), and the mRNA was extracted with phenol-chloroform and precipitated with ethanol. The capped mRNAs were denatured at 65°C for 10 min and then placed on ice for 2 min. The concentration of the synthesized mRNAs was determined, and their integrity was confirmed by agarose gel electrophoresis.
Cells, in vivo translation assays, and antibodies.
HEK293T and HeLa cell lines were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum, 100 U/ml penicillin, and 100 μg/ml streptomycin. For the in vivo translation assays, 0.5 to 1 μg of the in vitro-transcribed GFP mRNA together with luciferase mRNA that serves as an internal control for transfection efficiency were transfected into HeLa cells that had been previously seeded on 12-well plates, using ICAFectin441 transfection reagent (In-Cell-Art). The cells were harvested, and cell lysates were subjected to 12% SDS-polyacrylamide gel electrophoresis. The levels of GFP protein were determined using the mouse anti-GFP monoclonal antibody (ab290; Abcam). GFP protein bands were quantified using ImageQuant software (GE). Antibodies against RPS10e are from Antibody Verify (AAS52645C), anti-RPS3 and eIF1A are from Abcam, and antiactin is from Santa Cruz (sc-1616-R).
Site-specific UV-cross-linking assays.
Partially overlapping oligonucleotides were used to generate a 68-bp fragment using PCR. This fragment consists of the T7 promoter upstream of either TISU or canonical AUG followed by multiple CAA codons. Each position in the oligonucleotides, upstream and downstream relative to the AUG codon, was changed to a T nucleotide in the forward oligonucleotide and to an A nucleotide in the complementary position of the reverse oligonucleotide. The wild-type (WT) oligonucleotide sequences are CCCCCgtaatacgactcactataggGCAAGATGGCGGCcaa (forward) and ttgttgttgttgttgttgttgttgttgttgGCCGCCATCTTGCcct (reverse) or CCCCCgtaatacgactcactataggGGACCATGGcaacaa (forward) and ttgttgttgttgttgttgttgttgttgttgttgCCATGGTCCcct (reverse). The uppercase letters denote the AUG initiation codon and its flanking sequence.
RNA was synthesized with T7 enzyme in the presence of a thio-UTP (catalog no. NU-1156; Jena Bioscience) and [32P]CTP and capped using either a ScriptCap kit (C-SCCE0610) or vaccinia capping system (catalog no. M2080S; NEB). RNA was isolated with BioTri (catalog no. 959758027100; Bio-Lab) and RNA purification kit (catalog no. R2050; Zymo Research). Equal amounts of RNA (∼200,000 cpm) were incubated with buffer (10 mM HEPES [pH 7.6], 75 mM KCl, 1 mM dithiothreitol [DTT], and 1 mM magnesium acetate [MgAc]), 4 mM GMP-PNP or 2 mg/ml cycloheximide, and 2 μl rabbit reticulocyte lysate (RRL) for 5 min at 30°C. The reaction mixture was then subjected to 30 min of 365-nm UV light on ice followed by RNase A treatment for 30 min at 37°C. Half of the samples was loaded onto a 15% acryl amide gel, and the gel was dried and subjected to overnight exposure. For purification of the cross-linked complexes, 50 reaction mixtures after UV cross-linking were pooled and loaded onto 8 to 32% sucrose gradient made with buffer containing 20 mM Tris (pH 8), 100 mM potassium acetate (KAc), 2 mM DTT, 2.5 mM MgAc (for 48S) or 5 mM MgCl2 and 0.1 mg/ml cycloheximide (for 80S complex). Gradients were centrifuged at 40,000 rpm in a SW41 rotor for 3.5 h (for 48S complex) or 3 h (for 80S complex) at 4°C. Gradients were fractionated, and the optical density at 254 nm was continuously recorded using ISCO absorbance detector UA-6. The 48S or 80S peak fractions were pooled, treated with RNase A, and then were either precipitated by tricarboxylic acid (TCA) and subjected to immunoprecipitation (RPS10 and RPS15) or used directly for immunoprecipitation (RPS3).
Knockdown of eIF1A.
To knock down eIF1A, HEK293T cells were seeded on a six-well plate and transfected with eIF1A siRNA or a nontargeting siRNA 3 (Dharmacon). Forty-eight hours after the initial transfection, cells were cotransfected with the siRNAs and the GFP reporter plasmids. Cells were harvested 24 h after the second transfection.
Split Renilla complementation assay.
The split Renilla luciferase (RL) fusion plasmids were constructed by restriction-free two-step PCR cloning method using the RSV-ΔC/ΔN (RSV stands for Rous sarcoma virus) Renilla luciferase (pRL-ΔC/ΔN) plasmids as previously described (39). The N terminus of the Renilla luciferase (N-RL) contains positions 1 to 229, and the C terminus (C-RL) contains positions 230 to 311. In each construct, the split Renilla sequence and the target protein sequence are separated by a linker corresponding to amino acids GGGGS. Full-length eIF1A was amplified by PCR using pCRUZ HA-eIF1A (HA stands for hemagglutinin) as a template. RPS3 and RPS10e cDNAs were obtained by reverse transcription-PCR (RT-PCR) using HEK293T RNA. All constructs were verified by sequencing. HEK293T cells seeded on 24-well plate (70,000 cells/well) were cotransfected with 200 ng of each of the indicated split renilla constructs together with 50 ng of miR 22-firefly luciferase (FL) reporter, which served as a control for transfection efficiency. Twenty-four hours after transfection, the cells were harvested, and RL and FL activities were measured.
Statistical analysis.
We performed two-tailed Student t test for determining the significance of the differences in GFP levels as indicated in the figure legends.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Minerva Foundation (grant 712278) (R.D.), Israel Science Foundation (1168/13) (R.D.), and Agence Nationale pour la Recherche (ANR-11-SVSE802501) (F.M.), and Chateaubriand Fellowships from the Ambassade de France en Israël (H.S.). R.D. is the incumbent of the Ruth and Leonard Simon Chair of Cancer Research.
R.D, O.H., and H.S. conceived and designed the study. H.S. and O.H carried out most of the experiments. Some of the site-specific UV cross-linking experiments were done in the F.M. lab under his guidance. I.U. performed the bioinformatic analysis of the CapSeq data. R.E. generated the A(+1) mutants in TISU. A.T.-B.-H.. generated the Cap-seq data. A.V. performed the split-RL experiments. R.D and O.H wrote the paper.
We declare that we have no conflict of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00150-17.
REFERENCES
- 1.Aitken CE, Lorsch JR. 2012. A mechanistic overview of translation initiation in eukaryotes. Nat Struct Mol Biol 19:568–576. doi: 10.1038/nsmb.2303. [DOI] [PubMed] [Google Scholar]
- 2.Hinnebusch AG. 2014. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem 83:779–812. doi: 10.1146/annurev-biochem-060713-035802. [DOI] [PubMed] [Google Scholar]
- 3.Marintchev A, Wagner G. 2004. Translation initiation: structures, mechanisms and evolution. Q Rev Biophys 37:197–284. doi: 10.1017/S0033583505004026. [DOI] [PubMed] [Google Scholar]
- 4.Sonenberg N, Hinnebusch AG. 2009. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asano K. 2014. Why is start codon selection so precise in eukaryotes? Translation (Austin) 2:e28387. doi: 10.4161/trla.28387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elfakess R, Sinvani H, Haimov O, Svitkin Y, Sonenberg N, Dikstein R. 2011. Unique translation initiation of mRNAs-containing TISU element. Nucleic Acids Res 39:7598–7609. doi: 10.1093/nar/gkr484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kozak M. 2002. Pushing the limits of the scanning mechanism for initiation of translation. Gene 299:1–34. doi: 10.1016/S0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang XQ, Rothnagel JA. 2004. 5′-untranslated regions with multiple upstream AUG codons can support low-level translation via leaky scanning and reinitiation. Nucleic Acids Res 32:1382–1391. doi: 10.1093/nar/gkh305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haimov O, Sinvani H, Dikstein R. 2015. Cap-dependent, scanning-free translation initiation mechanisms. Biochim Biophys Acta 1849:1313–1318. doi: 10.1016/j.bbagrm.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 10.Kozak M. 1991. A short leader sequence impairs the fidelity of initiation by eukaryotic ribosomes. Gene Expr 1:111–115. [PMC free article] [PubMed] [Google Scholar]
- 11.Kozak M. 1991. Effects of long 5′ leader sequences on initiation by eukaryotic ribosomes in vitro. Gene Expr 1:117–125. [PMC free article] [PubMed] [Google Scholar]
- 12.Sedman SA, Gelembiuk GW, Mertz JE. 1990. Translation initiation at a downstream AUG occurs with increased efficiency when the upstream AUG is located very close to the 5′ cap. J Virol 64:453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elfakess R, Dikstein R. 2008. A translation initiation element specific to mRNAs with very short 5′UTR that also regulates transcription. PLoS One 3:e3094. doi: 10.1371/journal.pone.0003094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barth-Baus D, Bhasker CR, Zoll W, Merrick WC. 2013. Influence of translation factor activities on start site selection in six different mRNAs. Translation (Austin) 1:e24419. doi: 10.4161/trla.24419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sinvani H, Haimov O, Svitkin Y, Sonenberg N, Tamarkin-Ben-Harush A, Viollet B, Dikstein R. 2015. Translational tolerance of mitochondrial genes to metabolic energy stress involves TISU and eIF1-eIF4GI cooperation in start codon selection. Cell Metab 21:479–492. doi: 10.1016/j.cmet.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 16.Atger F, Gobet C, Marquis J, Martin E, Wang J, Weger B, Lefebvre G, Descombes P, Naef F, Gachon F. 2015. Circadian and feeding rhythms differentially affect rhythmic mRNA transcription and translation in mouse liver. Proc Natl Acad Sci U S A 112:E6579–E6588. doi: 10.1073/pnas.1515308112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gandin V, Masvidal L, Hulea L, Gravel SP, Cargnello M, McLaughlan S, Cai Y, Balanathan P, Morita M, Rajakumar A, Furic L, Pollak M, Porco JA Jr, St-Pierre J, Pelletier J, Larsson O, Topisirovic I. 2016. nanoCAGE reveals 5′ UTR features that define specific modes of translation of functionally related MTOR-sensitive mRNAs. Genome Res 26:636–648. doi: 10.1101/gr.197566.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tamarkin-Ben-Harush A, Vasseur JJ, Debart F, Ulitsky I, Dikstein R. 2017. Cap-proximal nucleotides via differential eIF4E binding and alternative promoter usage mediate translational response to energy stress. Elife 6:e21907. doi: 10.7554/eLife.21907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, Hou J, Quedenau C, Chen W. 2016. Pervasive isoform-specific translational regulation via alternative transcription start sites in mammals. Mol Syst Biol 12:875. doi: 10.15252/msb.20166941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanna MM, Dissinger S, Williams BD, Colston JE. 1989. Synthesis and characterization of 5-[(4-azidophenacyl)thio]uridine 5′-triphosphate, a cleavable photo-cross-linking nucleotide analogue. Biochemistry 28:5814–5820. doi: 10.1021/bi00440a017. [DOI] [PubMed] [Google Scholar]
- 21.Dissinger S, Hanna MM. 1990. Active site labeling of Escherichia coli transcription elongation complexes with 5-[4-azidophenacyl)thio)uridine 5′-triphosphate. J Biol Chem 265:7662–7668. [PubMed] [Google Scholar]
- 22.Pisarev AV, Kolupaeva VG, Pisareva VP, Merrick WC, Hellen CU, Pestova TV. 2006. Specific functional interactions of nucleotides at key -3 and +4 positions flanking the initiation codon with components of the mammalian 48S translation initiation complex. Genes Dev 20:624–636. doi: 10.1101/gad.1397906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. 2011. The structure of the eukaryotic ribosome at 3.0 A resolution. Science 334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 24.Anger AM, Armache JP, Berninghausen O, Habeck M, Subklewe M, Wilson DN, Beckmann R. 2013. Structures of the human and Drosophila 80S ribosome. Nature 497:80–85. doi: 10.1038/nature12104. [DOI] [PubMed] [Google Scholar]
- 25.Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. 2011. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science 331:730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- 26.Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V. 2007. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell 26:41–50. doi: 10.1016/j.molcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 27.Hussain T, Llacer JL, Fernandez IS, Munoz A, Martin-Marcos P, Savva CG, Lorsch JR, Hinnebusch AG, Ramakrishnan V. 2014. Structural changes enable start codon recognition by the eukaryotic translation initiation complex. Cell 159:597–607. doi: 10.1016/j.cell.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paulmurugan R, Gambhir SS. 2003. Monitoring protein-protein interactions using split synthetic renilla luciferase protein-fragment-assisted complementation. Anal Chem 75:1584–1589. doi: 10.1021/ac020731c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hinnebusch AG, Lorsch JR. 2012. The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harb Perspect Biol 4:a011544. doi: 10.1101/cshperspect.a011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jackson RJ, Hellen CU, Pestova TV. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asano K, Sachs MS. 2007. Translation factor control of ribosome conformation during start codon selection. Genes Dev 21:1280–1287. doi: 10.1101/gad.1562707. [DOI] [PubMed] [Google Scholar]
- 32.Obayashi E, Luna RE, Nagata T, Martin-Marcos P, Hiraishi H, Singh CR, Erzberger JP, Zhang F, Arthanari H, Morris J, Pellarin R, Moore C, Harmon I, Papadopoulos E, Yoshida H, Nasr ML, Unzai S, Thompson B, Aube E, Hustak S, Stengel F, Dagraca E, Ananbandam A, Gao P, Urano T, Hinnebusch AG, Wagner G, Asano K. 2017. Molecular landscape of the ribosome pre-initiation complex during mRNA scanning: structural role for eIF3c and its control by eIF5. Cell Rep 18:2651–2663. doi: 10.1016/j.celrep.2017.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitchell SF, Lorsch JR. 2008. Should I stay or should I go? Eukaryotic translation initiation factors 1 and 1A control start codon recognition. J Biol Chem 283:27345–27349. doi: 10.1074/jbc.R800031200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cavener DR, Ray SC. 1991. Eukaryotic start and stop translation sites. Nucleic Acids Res 19:3185–3192. doi: 10.1093/nar/19.12.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kozak M. 1986. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 36.Shabalina SA, Ogurtsov AY, Rogozin IB, Koonin EV, Lipman DJ. 2004. Comparative analysis of orthologous eukaryotic mRNAs: potential hidden functional signals. Nucleic Acids Res 32:1774–1782. doi: 10.1093/nar/gkh313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dikstein R. 2012. Transcription and translation in a package deal: the TISU paradigm. Gene 491:1–4. doi: 10.1016/j.gene.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 38.Erijman A, Dantes A, Bernheim R, Shifman JM, Peleg Y. 2011. Transfer-PCR (TPCR): a highway for DNA cloning and protein engineering. J Struct Biol 175:171–177. doi: 10.1016/j.jsb.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 39.Ashkenazi S, Plotnikov A, Bahat A, Ben-Zeev E, Warszawski S, Dikstein R. 2016. A novel allosteric mechanism of NF-kappaB dimerization and DNA binding targeted by an anti-inflammatory drug. Mol Cell Biol 36:1237–1247. doi: 10.1128/MCB.00895-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.