ABSTRACT
Deregulated activation of RAS/extracellular signal-regulated kinase (ERK) signaling and defects in retinoic acid receptor (RAR) signaling are both implicated in many types of cancers. However, interrelationships between these alterations in regulating cancer cell fates have not been fully elucidated. Here, we show that RAS/ERK and RAR signaling pathways antagonistically interact with each other to regulate colorectal cancer (CRC) cell fates. We show that RAR signaling activation promotes spontaneous differentiation of CRC cells, while ERK activation suppresses it. Our microarray analyses identify genes whose expression levels are upregulated by RAR signaling. Notably, one of these genes, MKP4, encoding a member of dual-specificity phosphatases for mitogen-activated protein (MAP) kinases, mediates ERK inactivation upon RAR activation, thereby promoting the differentiation of CRC cells. Moreover, our results also show that RA induction of RAR target genes is suppressed by the ERK pathway activation. This suppression results from the inhibition of RAR transcriptional activity, which is shown to be mediated through an RIP140/histone deacetylase (HDAC)-mediated mechanism. These results identify antagonistic interactions between RAS/ERK and RAR signaling in the cell fate decision of CRC cells and define their underlying molecular mechanisms.
KEYWORDS: ERK MAP kinase, colorectal cancer, retinoic acid receptor
INTRODUCTION
Retinoic acid (RA), an active derivative of vitamin A, plays an important role in the development and homeostasis of many vertebrate tissues through its regulatory effects on cell proliferation, differentiation, and apoptosis. Most of these effects are mediated by nuclear retinoid receptors, RA receptors (RARs), and retinoid X receptors (RXRs) (1, 2). The basic mechanism of RAR/RXR-mediated transcriptional regulation relies on the ability of RAR/RXR to recruit transcriptional corepressors and coactivators and involves a series of steps, as follows (2–4). (i) In the absence of ligand, RAR associates with the corepressor complex that mediates transrepression by the recruitment of histone deacetylase (HDAC). (ii) Ligand binding to RAR causes the dissociation of the corepressor complex and the recruitment of the coactivator complexes, which contain histone acetyltransferase, methyltransferase, and kinase and/or ATP-dependent chromatin remodeling activities that decompact repressive chromatin. (iii) Then, the coactivators dissociate, and the mediator complex assembles to recruit RNA polymerase II and the general transcription factors to the promoter, resulting in transcriptional initiation.
Transactivation of RAR target genes often induces cell proliferation arrest, cell differentiation, and apoptosis in a wide variety of biological processes. Consequently, retinoids have tumor-suppressive activity, and loss of normal RAR function in the presence of physiological levels of RA is associated with the progression of a diverse range of cancers. In the case of acute promyelocytic leukemia (APL), chromosomal translocation generates the PML-RARα fusion gene that suppresses normal RAR functions (5–7). Administration of RA causes degradation of PML-RARα and restores normal RAR activity, which induces differentiation of leukemia cells and complete remission of APL (8). Retinoids also show suppressive effects on various solid tumors, including breast, lung, and colorectal carcinomas (3, 4, 8). For instance, previous studies have shown that RA-induced expression of RARβ promotes differentiation and apoptosis of several types of cancer cells (9). It has been also shown that RA promotes differentiation of intestinal tumor (adenoma) cells through inducing expression of HOXA5 (10). Despite a large number of reports, however, entire mechanisms by which RAR signaling exerts tumor-suppressive effects on many types of tumor have remained elusive. In addition, it has also remained unclear how normal RAR function is abolished during progression of these tumors.
Among genetic mutations that frequently occur in many solid tumors are those in the RAS and RAF genes (11–14). In colorectal cancers (CRCs), K-RAS or B-RAF mutations usually occur following the adenomatous polyposis coli gene mutations, the earliest genetic mutations found in colorectal tumorigenesis, and promote tumor progression (15–20). RAS/RAF mutations cause constitutive activation of downstream ERK/mitogen-activated protein (MAP) kinase signaling, which plays a critical role in cell proliferation (21). Thus, deregulated activation of RAS/RAF/ERK signaling and defects in RAR signaling are both characteristics of many solid cancers. However, it remains elusive whether there are any causal relationships between these alterations in many cancers.
In this study, we investigated a role of RAR signaling in regulating differentiation of CRC cells. We show that RAR signaling is enhanced during the spontaneous differentiation of Caco-2 CRC cells and that the enhanced RAR signaling promotes the differentiation. Our microarray analyses identify genes whose expression levels are regulated by RAR signaling. Remarkably, RAR signaling causes a reduction in ERK activity by inducing MAP kinase phosphatase 4 (MKP4), thereby promoting differentiation. Moreover, our results show that RA-induced upregulation of the downstream target genes of RAR signaling is suppressed by ERK activation through an RIP140/HDAC-mediated mechanism. Thus, RAR signaling and ERK signaling interact antagonistically to regulate the differentiation of CRC cells.
RESULTS
RAR signaling promotes the spontaneous differentiation of CRC cells.
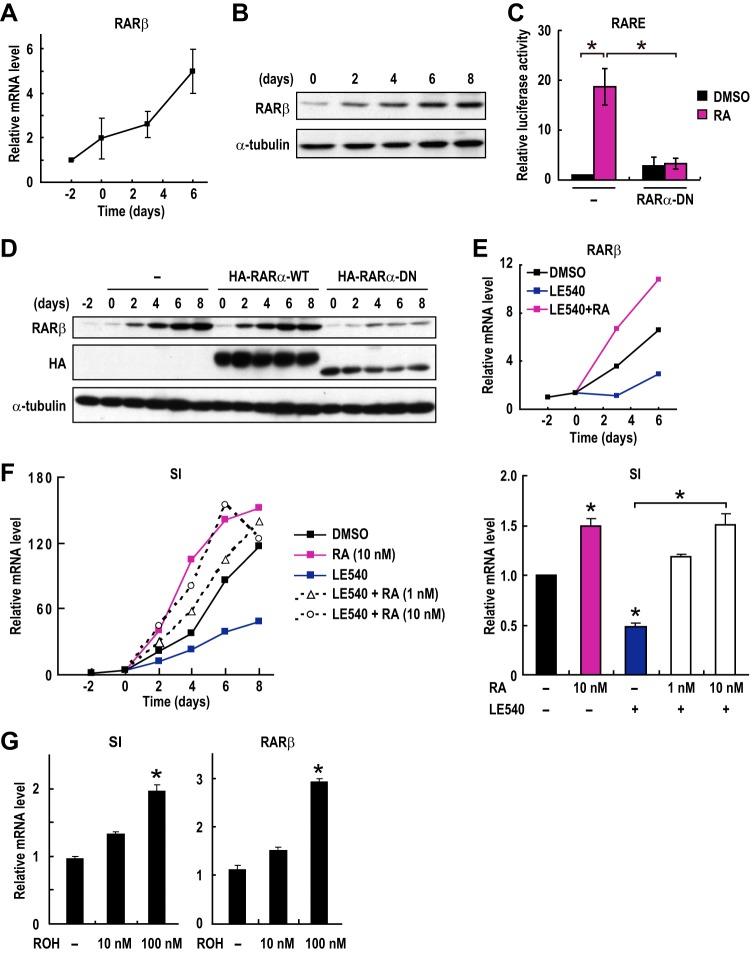
To investigate a mechanism by which RAR signaling exerts tumor-suppressive activity on CRC cells, we first examined the potential involvement of RAR signaling in the differentiation of CRC cells. As a model system for differentiation, we used Caco-2 CRC cells, which spontaneously differentiate into enterocytes upon reaching confluence (22). In fact, a molecular marker for enterocytic differentiation, sucrase-isomaltase (SI), was induced to be expressed after confluence (see below). We then investigated the expression levels of RARβ, a well-known transcriptional target gene of RAR itself. Both the mRNA level and the protein level of RARβ were found to be dramatically increased during differentiation (Fig. 1A and B). We then examined whether RARβ induction during differentiation was RAR dependent. To this end, we used the dominant negative form of RARα (RARα-DN), which lacks the C terminus ligand-binding domain. This RARα-DN does not have transactivation activity but is able to bind to the RA-responsive element (RARE), thereby competing with endogenous RARs for binding to the target promoters. In fact, RARE-dependent transcription of a reporter gene is suppressed by the expression of RARα-DN (Fig. 1C). The stable expression of RARα-DN suppressed RARβ induction during the differentiation of Caco-2 cells (Fig. 1D), suggesting that RARβ is induced by RAR during differentiation. Consistent with this, treatment with a synthetic RAR antagonist, LE540, was also found to suppress RARβ induction (Fig. 1E). The inhibitory effect of LE540 was canceled by RA (Fig. 1E), confirming the specific action of LE540 on RAR. These results suggest that RAR signaling is activated during the differentiation of CRC cells.
FIG 1.
RAR signaling promotes the differentiation of CRC cells. (A and B) The expression levels of RARβ mRNA (A) and protein (B) were evaluated in differentiating Caco-2 cells. Cells were incubated for the indicated days after reaching confluence at day zero. (C) Transcriptional activation of a reporter plasmid expressing luciferase under the control of the RAREs (RARE3-Luc) was evaluated in Caco-2 cells transfected with empty vectors or with expression plasmids for the dominant negative form of RARα (RARα-DN). Cells were treated with 100 nM RA for 12 h prior to the measurements (means ± standard errors of the means; n = 4). (D) Caco-2 cells were infected with a lentivirus expressing the HA-tagged wild-type (WT) or dominant negative form of RARα (DN) 2 days before confluence (day −2). Cell lysates were prepared at the indicated time points and analyzed by immunoblotting. (E to G) Caco-2 cells were treated with vehicle, RA (1 nM), LE540 (100 nM), or retinol (ROH) for the indicated days after reaching confluence (day 0), and the expression levels of the indicated mRNAs were analyzed by RT-PCR. Values in bar graphs are means ± standard errors of the means at day 6 (n = 3). *, P < 0.05, for differences in results from rom control samples (vehicle) or between indicated samples (Student's t test).
We treated Caco-2 cells with RA or the RAR antagonist LE540 to examine the importance of RA signaling for CRC cell differentiation. Consistent with findings of a previous report (23), RA treatment promoted markedly the differentiation of Caco-2 cells (Fig. 1F). LE540 treatment significantly suppressed differentiation (Fig. 1F). RA treatment cancelled the differentiation-suppressing effect of the RAR antagonist LE540 in a dose-dependent manner (Fig. 1F), confirming the specificity of the action of LE540. Moreover, treatment of cells with retinol, a precursor of RA, was found to enhance RARβ expression and promote differentiation in a dose-dependent manner (Fig. 1G). All of these results demonstrated that RAR signaling is enhanced during the differentiation of CRC cells and that the increased RAR signaling promotes differentiation.
Identification of gene expression programs regulated by RAR signaling in CRC cells.
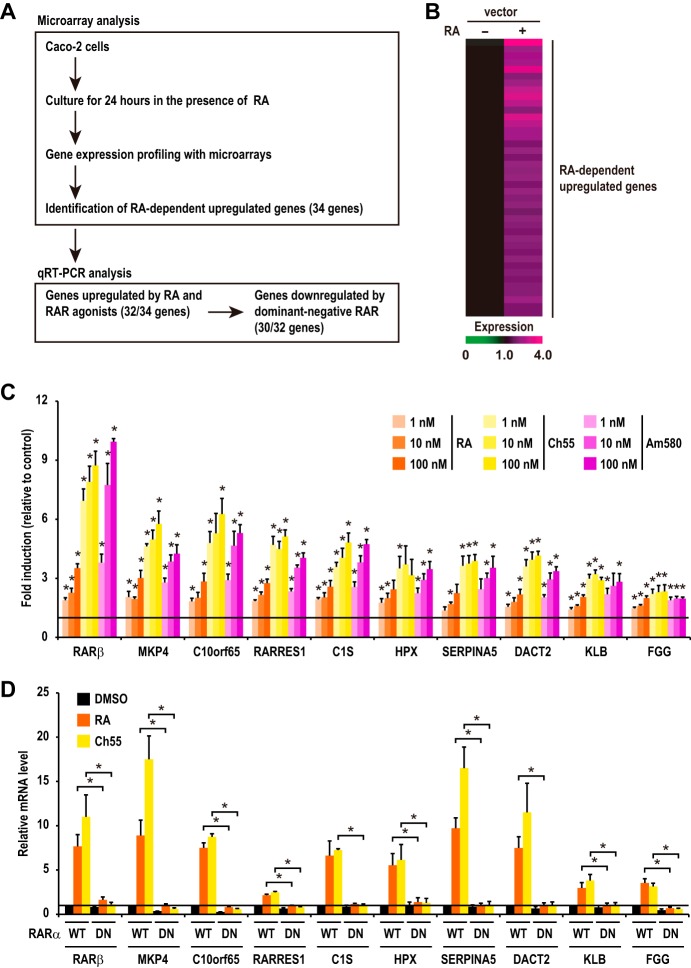
To further understand the role of RAR signaling in CRC cells, we have undertaken a large-scale analysis of gene expression profiles by using microarrays (Fig. 2A). We identified 41 probe sets (containing 34 genes) whose expression levels in RA-treated cells were upregulated by more than 1.5-fold compared to levels in vehicle-treated cells with statistical significance in replicate experiments (Fig. 2B). Quantitative reverse transcription-PCR (RT-PCR) analysis confirmed the induction of most of these genes by RA treatment (32 out of 34 genes) (Fig. 2C; see also Fig. S1 in the supplemental material). To examine whether the induction of these RA-responsive genes is mediated by RAR, we first used RAR-specific agonists, Ch55 and Am580. The results show that all of the 32 upregulated genes were induced by these agonists in a dose-dependent manner (Fig. 2C and S1), suggesting that RAR mediates the RA-induced upregulation of the expression of these genes. We then examined whether the expression of these RA-responsive genes was dependent on transactivation activity of RAR by using the dominant negative form of RARα (RARα-DN). The results showed that the expression of RARα-DN suppresses RA induction of almost all the RA-responsive genes (30 out of 32 genes) (Fig. 2D and S2). We thus identified these genes as downstream transcriptional targets of RAR signaling in CRC cells (Table S1). It has previously been shown that two genes (i.e., CA2 and RARRES1) are expressed strongly in terminally differentiated cells of normal mucosal tissues and adenoma tissues but are absent from poorly differentiated CRC cells (24, 25). These observations are in good agreement with our results that RAR signaling is activated during the differentiation of CRC cells (Fig. 1A to E).
FIG 2.
Identification of downstream target genes of RAR signaling in CRC cells. (A) Schematic representation of the strategy to identify downstream target genes of RAR signaling. We first performed microarray experiments to identify genes whose expression levels were upregulated by RA treatment in Caco-2 cells. Then, the effects of RAR agonists and dominant negative RAR on the expression levels of the identified upregulated genes were investigated to examine whether expression of these genes is indeed regulated by RAR. (B) Expression profiles of RA-dependently upregulated genes are shown. Cells were treated with DMSO or RA (100 nM) for 24 h. Each horizontal line displays the expression data for one gene, where the ratio of the mRNA level to its level in the control cells (DMSO) is represented by color according to the color scale. (C) Confluent Caco-2 cells were treated with RA, Ch55, or Am580 for 24 h. The mRNA levels of the RA-dependently upregulated genes were analyzed by RT-PCR. Data shown are relative mRNA levels compared to those in control (vehicle-treated) cells (set to 1). The data of individual genes are ranked from left to right according to the fold increase induced by treatment with 100 nM RA. Note that the expression levels of only the top 10 genes among the upregulated genes are shown in this graph. The expression levels of the remaining genes are shown in Fig. S1. *, P < 0.05, for differences in results from those of the control group (vehicle) (Student's t test). (D) Caco-2 cells were infected with the lentivirus expressing wild-type (WT) or a dominant negative form of RARα (DN). At 48 h after infection, cells were treated with DMSO, RA (100 nM), or Ch55 (1 nM) and incubated for 24 h. Total RNA was prepared and analyzed by RT-PCR. The data of individual genes are ranked as described for panel C. Note that the expression levels of only 10 genes among the upregulated genes are shown in this graph. The expression levels of the remaining genes are shown in Fig. S2. Values shown in panels C and D are means ± standard errors of the means (n = 3). *, P < 0.05 (Student's t test).
RAR promotes MKP4 expression through directly binding to its promoter region.
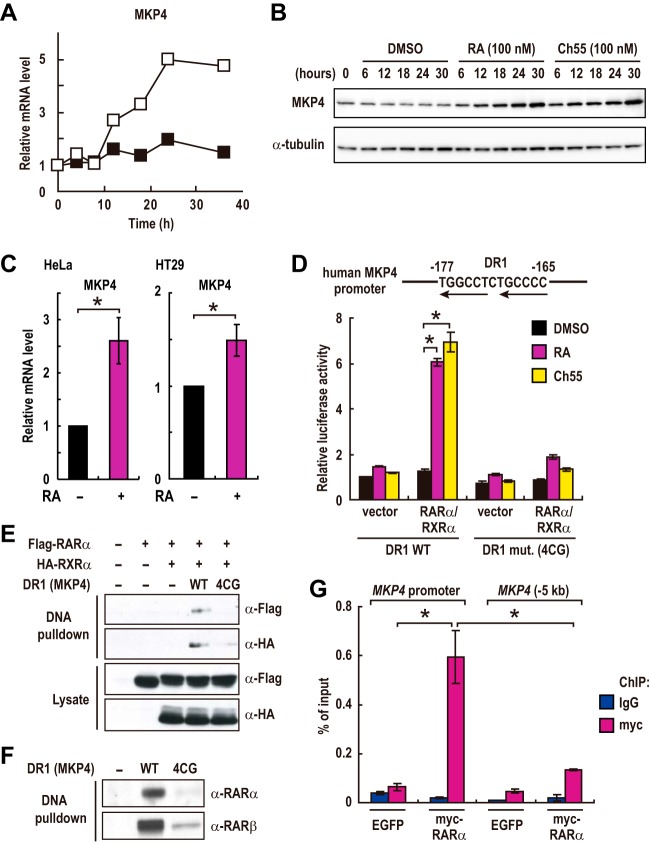
Among the identified target genes of RAR signaling, we noticed that the MAP kinase phosphatase 4 gene (MKP4) as it was one of the most strongly upregulated genes (second most upregulated upon treatment with 100 nM RA) (Fig. 2C), and its induction by RA was strongly suppressed by expression of RARα-DN (Fig. 2D). Since MKP4 is a phosphatase specific for ERK MAP kinases (26, 27) that often play a crucial role in controlling cell proliferation and differentiation, we considered that induction of MKP4 by RA might have some effects on the differentiation of colorectal cancer cells. Quantitative RT-PCR analysis showed that the mRNA level of MKP4 was gradually increased upon treatment with RA (Fig. 3A). Consistent with the increased mRNA level, the protein level of MKP4 was also increased by treatment with RA or an RAR agonist (Fig. 3B). The expression of other members of the MKPs was not affected by RA treatment (Fig. S3). RA treatment also increased the expression levels of MKP4 in HT29 and HeLa cells (Fig. 3C). These results suggest that RAR signaling promotes the expression of MKP4 in these cells.
FIG 3.
RAR promotes MKP4 expression through directly binding to its promoter region. (A and B) The mRNA and protein levels of MKP4 in Caco-2 cells treated with DMSO (filled squares), 100 nM RA (open squares), or 100 nM Ch55 were analyzed by RT-PCR (A) and immunoblotting (B), respectively. Data shown are representative of two independent experiments. (C) HeLa and HT29 cells were treated with 100 nM RA for 24 h or 1 μM RA for 3 days, respectively. The MKP4 mRNA levels were analyzed by RT-PCR (means ± standard errors of the means; HT29 cell experiments, n = 3; HeLa cell experiments, n = 4). *, P < 0.01. (D) Transcriptional activation of reporter plasmids expressing luciferase under the control of the MKP4 promoter containing a wild-type (WT) or mutated (4CG) DR1 element was evaluated in Caco-2 cells transfected with empty vectors or with expression plasmids for RARα and RXRα. Cells were treated with 1 μM RA or Ch55 for 36 h prior to the measurements (means ± standard errors of the means; n = 3). *, P < 0.01. (E and F) Lysates from HEK293 cells transfected with empty vectors or with expression plasmids for RARα and RXRα (E) and lysates from Caco-2 cells treated with 100 nM RA for 36 h (F) were subjected to DNA pulldown assays with biotinylated oligonucleotides corresponding to 60-bp fragments (−200 to −141) of the MKP4 promoter containing the WT or the mutated (4CG) DR1 and analyzed by immunoblotting. (G) Chromatin fragments were prepared from Caco-2 cells stably expressing myc-tagged RARα or enhanced green fluorescent protein (EGFP) and subjected to ChIP analyses with the indicated antibodies. Values are means ± standard errors of the means from two or three experiments.
To examine whether MKP4 is a direct transcriptional target of RAR and RXR, we searched the first 1,000 bp of the MKP4 promoter region for a putative RA-responsive element. We found an inverted direct repeat separated by 1 bp (DR1), which was shown to be a consensus sequence for the RAR/RXR heterodimer, within the MKP4 promoter region (−177 to −165) (Fig. 3D). Then, a reporter assay with constructs expressing luciferase (Luc) under the control of 60-bp fragments of the MKP4 promoter (−200 to −141) containing either the wild-type (WT) or the mutated (4CG) DR1 element, in which residues at positions −174C, −173C, −167C, and −166C were replaced by G, was performed to examine the importance of this DR1 element. Treatment with RA or the synthetic RAR agonist Ch55 significantly increased the activity of the MKP4 promoter containing the wild-type DR1 but not the mutated (4CG) DR1 in cells transfected with both RARα and RXRα (Fig. 3D). The effect of RA was specific to the DR1 element because RA did not show any activity on the parental pGL3 reporter in cells transfected with both RARα and RXRα (Fig. S4A). Moreover, treatment with the RAR antagonist LE540 inhibited the RA-induced activation of the DR1 element (Fig. S4B). These results suggest that transcriptional activity of this DR1 element is regulated by the RAR/RXR complex. We then examined whether RAR and RXR could bind directly to this DR1 element by DNA pulldown assay. We found that Flag-tagged RARα and hemagglutinin (HA)-tagged RXRα expressed in HEK293 cells were precipitated with biotinylated oligonucleotides corresponding to 60-bp fragments (−200 to −141) of the MKP4 promoter containing the wild-type (WT) but not the mutated (4CG) DR1 (Fig. 3E). Moreover, endogenous RARα and RARβ were also precipitated from Caco-2 cell lysates by oligonucleotides in an intact DR1 element-dependent manner (Fig. 3F), suggesting that the RAR/RXR heterodimer could directly bind to this DR1 element. Finally, we examined whether RAR binds to the MKP4 promoter in vivo by chromatin immunoprecipitation (ChIP) experiments. The results demonstrated the specific binding of RARα to the MKP4 promoter (Fig. 3G). These results, taken together, show that RA-induced transactivation of RAR directly upregulates the MKP4 expression level through this DR1 element.
RAR signaling inactivates ERK through induction of MKP4 during the differentiation of CRC cells.
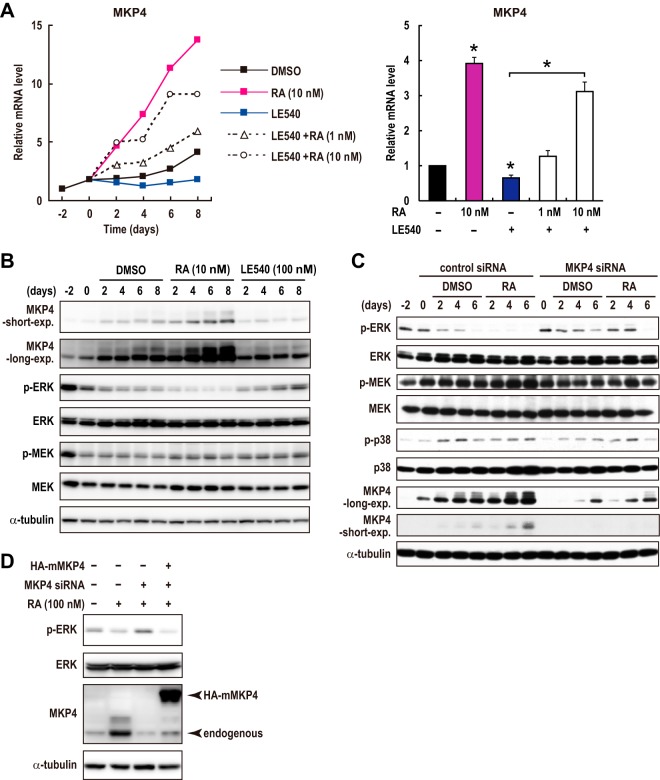
We then examined whether MKP4 is involved in RAR signaling-induced differentiation of CRC cells. We found that the expression level of MKP4 was gradually increased during the differentiation of Caco-2 cells at both the mRNA and the protein levels (Fig. 4A and B). This was likely due to the increase in RAR activity during differentiation (Fig. 1A) as treatment with an RAR-specific antagonist, LE540, suppressed the increase in MKP4 expression (Fig. 4A and B). The effect of LE540 was cancelled by the addition of RA, indicating again the specificity of the action of the RAR antagonist LE540 (Fig. 4A). Moreover, treatment with RA promoted an increase in MKP4 expression during differentiation (Fig. 4A and B). These results suggest that RAR signaling promotes MKP4 expression during the differentiation of CRC cells. Consistent with the increase in the MKP4 expression level, ERK activity was gradually decreased during differentiation (Fig. 4B). Remarkably, RA treatment promoted ERK inactivation during differentiation, whereas LE540 treatment suppressed it (Fig. 4B). Thus, the increase in the MKP4 protein level was inversely correlated with the phosphorylation level of ERK. Importantly, however, the state of MEK activation was not changed significantly by RA or LE540 treatment (Fig. 4B), suggesting that RAR signaling suppresses ERK without affecting MEK during differentiation.
FIG 4.
RAR signaling causes ERK inactivation through induction of MKP4 during the differentiation of CRC cells. (A and B) Caco-2 cells were treated as described in the legend to Fig. 1F. Total mRNA and cell lysates were analyzed by RT-PCR (A) and immunoblotting (B), respectively. Values in bar graphs are means ± standard errors of the means at day 6 (n = 3). *, P < 0.05, for differences in results from those of control samples (vehicle) or between indicated samples (Student's t test). (C) Caco-2 cells were transfected with an siRNA specific for MKP4 or a control siRNA 2 days before confluence (day −2). Then, cells were treated with DMSO vehicle or 10 nM RA after reaching confluence (day 0). Cell lysates were prepared and analyzed by immunoblotting. The data for MKP4 represent two distinct blots of the same samples taken by short or long exposure. (D) Caco-2 cells were transfected with the siRNAs as described for panel C. The next day, cells were infected with a control lentivirus or the virus expressing HA-tagged mouse MKP4. From 24 h after infection, cells were treated with DMSO or RA (100 nM) for 48 h. Cell lysates were prepared and analyzed by immunoblotting.
We then examined the role of MKP4 in ERK inactivation during differentiation. To this end, we inhibited MKP4 expression by a small interfering RNA (siRNA). Our MKP4 siRNA suppressed MKP4 expression by about 70% at the protein level (Fig. 4C). Since the effects of transfected siRNAs were gradually weakened by degradation and also since MKP4 expression was promoted by endogenous RAR signaling during differentiation, the MKP4 protein level was gradually upregulated even in cells transfected with the MKP4 siRNA (Fig. 4C). In these cells, however, the MKP4 protein level was still significantly lower than that in the control cells that were transfected with the control siRNA and treated similarly for the same days (Fig. 4C). The suppression of MKP4 significantly inhibited the reduction in ERK activity in both the absence and presence of RA (Fig. 4C). The state of MEK activation was not affected by either the MKP4 siRNA or retinoic acid (Fig. 4C). Although overexpressed MKP4 was previously reported to be able to inhibit both ERK and p38 MAP kinase (27), downregulation of MKP4 in Caco-2 cells affected only ERK activation and did not affect p38 activation (Fig. 4C). These results suggest that RAR signaling suppresses ERK activity without affecting MEK by inducing expression of MKP4. Importantly, however, the increase in the MKP4 protein level and the decrease in the phosphorylation level of ERK were not completely matched when samples cultured for different numbers of days were compared (Fig. 4C). Indeed, ERK activity was slightly decreased over the time course, independently of MKP4 expression (Fig. 4C, 2nd lane versus 11th lane). We thus think that the contribution of MKP4 to ERK inactivation during differentiation is partial and that there might be other, additional mechanisms responsible for ERK inactivation.
To confirm the specificity of the effect of the MKP4 knockdown, we then performed rescue experiments by using HA-tagged mouse MKP4, which is resistant to the MKP4 siRNA. MKP4 knockdown by siRNA suppressed the RA-promoted inhibition of ERK (Fig. 4D). In cells expressing mouse MKP4, however, the inhibition of ERK occurred (Fig. 4D). These results clearly indicate that the reduction in ERK activity occurs, at least in part, through MKP4 upregulation during the differentiation of CRC cells.
MKP4 mediates the differentiation-promoting effect of RAR signaling in CRC cells.
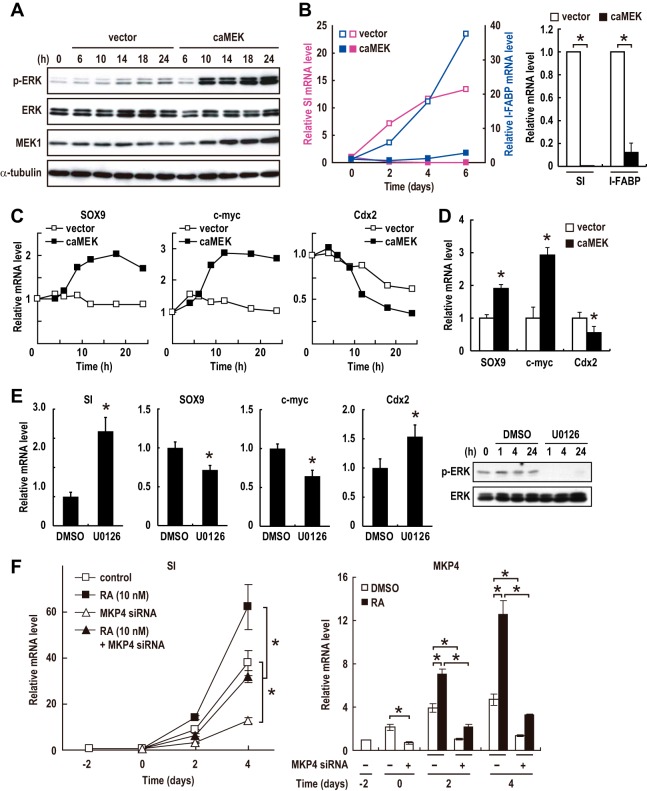
To assess the role of the reduction in ERK activity during differentiation of CRC cells, we examined the effect of constitutive activation of ERK on differentiation. To this end, we infected Caco-2 cells with an adenovirus which expresses constitutively active MEK. In the infected cells, ERK was constitutively activated 10 h after infection (Fig. 5A), and differentiation was almost completely suppressed (Fig. 5B). Therefore, inhibition of ERK is necessary for the differentiation of Caco-2 CRC cells.
FIG 5.
ERK inactivation caused by RAR-dependent upregulation of MKP4 promotes the differentiation of CRC cells. (A to C) Caco-2 cells were infected with an adenovirus expressing constitutively active MEK (caMEK) or a control virus (vector). Lysates and total RNA were prepared and analyzed by immunoblotting (A) and RT-PCR (B and C), respectively. Values in bar graphs are means ± standard errors of the means at day 6 (n = 3). *, P < 0.05. (D) Cells were infected with the virus expressing constitutively active MEK or a control virus as described for panel A. At 24 h postinfection, total RNA was prepared and analyzed by RT-PCR (means ± standard errors of the means; n = 4). *, P < 0.05. (E) Preconfluent Caco-2 cells were treated with an MEK-specific inhibitor, U0126 (10 μM), or DMSO vehicle for 24 h. Total RNA and cell lysates were analyzed by RT-PCR (means ± standard errors of the means; n = 9) and immunoblotting, respectively. *, P < 0.05. (F) Caco-2 cells were treated as described in the legend to Fig. 4C. Cells were transfected with an siRNA specific for MKP4 or a control siRNA 2 days before confluence (day −2) and then treated with DMSO vehicle or 10 nM RA after reaching confluence (day 0). The data confirming the reduction of the MKP4 protein was shown in Fig. 4C. Total RNA was prepared and analyzed by RT-PCR (means ± standard errors of the means; n = 3). *, P < 0.05.
Previous studies have revealed the involvement of several transcription factors in the differentiation of normal intestinal epithelial cells and CRC cells. Sox9 and c-Myc, which are subject to regulation by the Wnt signaling pathway, have been shown to be important for the proliferative/undifferentiated phenotypes in both normal intestinal epithelial cells and CRC cells (24, 28, 29). In contrast, Cdx2, which is expressed in differentiated villus cells, was shown to be essential for differentiation (30–32). We then examined the effect of activation or inhibition of ERK on the expression of these transcription factors. ERK activation by the expression of constitutively active MEK rapidly induced the upregulation of Sox9 and c-Myc expression and the downregulation of Cdx2 expression (Fig. 5C and D). The inhibition of ERK by treatment with the specific MEK inhibitor U0126 induced the reverse effect: the expression levels of Sox9 and c-Myc were decreased, and the Cdx2 level was increased in preconfluent/undifferentiated Caco-2 cells (Fig. 5E). As a result, the expression of the differentiation marker SI was increased (Fig. 5E). Thus, ERK activity regulates the expression of these transcription factors, and ERK inhibition induces changes in their expression profiles which are similar to those in the physiological differentiation of intestinal epithelial cells.
We then inquired whether MKP4, which mediates ERK inactivation induced by RAR signaling, is required for the differentiation of CRC cells. The siRNA-mediated knockdown of MKP4 significantly retarded the differentiation of Caco-2 cells in both the presence and absence of RA (Fig. 5F). Similar results were obtained with another siRNA that targets the distinct sequence of MKP4 (data not shown), confirming the specific effects of MKP4 siRNAs. These results taken together demonstrate that RAR signaling is enhanced during the differentiation of CRC cells and that the enhanced RAR signaling causes ERK inactivation, at least in part, through MKP4 upregulation to promote differentiation.
ERK activation suppresses RAR signaling through an RIP140/HDAC-mediated mechanism.
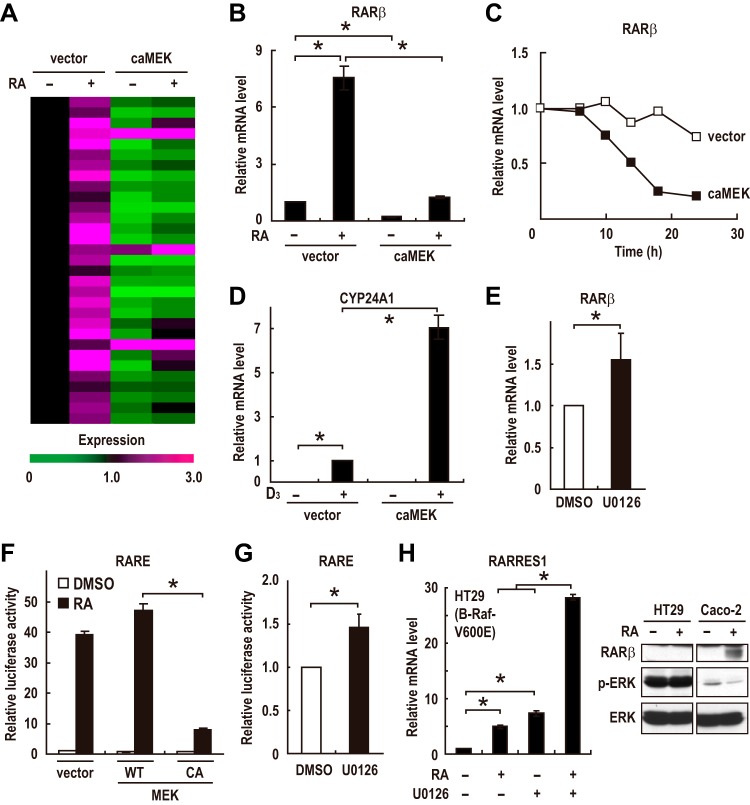
While investigating the effects of ERK activation on Caco-2 cells, we found that constitutively active MEK-induced ERK activation significantly suppresses RA induction of most of the RAR-dependent upregulated genes (Fig. 6A and B). ERK activation also induced a rapid decrease in the steady-state mRNA levels of RARβ (Fig. 6C). This is not a global effect on nuclear receptor-mediated gene expression as the ligand-dependent expression of a vitamin D receptor target gene, CYP24A1, was not suppressed; rather, it was enhanced by ERK activation (Fig. 6D). These results suggest that ERK activation specifically suppresses RAR target gene expression. Consistent with these results, the inhibition of ERK by U0126 treatment had the reverse effect on the expression levels of RARβ (Fig. 6E). To directly assess the effect of ERK activity on RAR transcriptional activity, we performed a reporter assay with a construct containing tandem repeats of an RARE (33). ERK activation by the expression of constitutively active MEK markedly suppressed RA-stimulated RAR transcriptional activity (Fig. 6F), and the inhibition of ERK by U0126 treatment enhanced it (Fig. 6G), indicating that ERK activation suppresses RAR transcriptional activity. As in Caco-2 cells, ERK activation suppressed RAR target gene expression and RAR transcriptional activity in HeLa, HEK293, and MCF7 cells (Fig. S5A to C). These results suggest that ERK activation antagonizes RAR target gene expression in many types of cells.
FIG 6.
ERK activation suppresses transcriptional activity of RAR. (A) Expression profiles of the RAR target genes in CRC cells are shown. Cells were infected with an adenovirus expressing constitutively active MEK (caMEK) or a control virus (vector) 12 h before treatment with DMSO or RA (100 nM). Cells were incubated for 24 h, and total RNA was prepared and subjected to RT-PCR analysis. Each horizontal line displays the expression data for one gene, where the ratio of the mRNA level to its level in the control cells, which were infected with a control virus (vector) and treated with DMSO, is represented by color according to the color scale. (B) Caco-2 cells were treated as described for panel A. The mRNA level of RARβ was analyzed by RT-PCR. (C) The expression levels of the RARβ mRNA in confluent Caco-2 cells infected with an adenovirus expressing constitutively active MEK (caMEK) or a control virus (vector) were evaluated by RT-PCR. (D) Confluent Caco-2 cells were infected with an adenovirus expressing constitutively active MEK (caMEK) or a control virus (vector). At 12 h postinfection, cells were treated with DMSO or 1,25-(OH)2-vitamin D3 (100 nM) for 24 h. Total RNA was prepared and analyzed by RT-PCR. (E) The expression levels of the RARβ mRNA in preconfluent Caco-2 cells treated with U0126 (10 μM) or DMSO vehicle for 24 h were analyzed by RT-PCR. (F and G) Transcriptional activation of a reporter plasmid expressing luciferase under the control of the RA-responsive elements (RARE3-Luc) was evaluated in Caco-2 cells transfected with empty vectors or with expression plasmids for wild-type (WT) or constitutively active (CA) MEK. Cells were treated with 100 nM RA for 12 h (F) or with 10 μM U0126 for 24 h (G) prior to the measurements. (H) The expression levels of the RARRES1 mRNA in HT29 cells treated with 1 μM RA and/or 10 μM U0126 for 72 h were analyzed by RT-PCR (left). HT29 and Caco-2 cells were treated with DMSO or 100 nM RA for 24 h, and cell lysates were prepared and analyzed by immunoblotting (right). Values in panels B and D to H are means ± standard errors of the means (n = 3). *, P < 0.05.
Activating mutations in K-RAS and B-RAF occur frequently in human cancers (13, 14). Therefore, we reasoned that constitutive activation of the ERK pathway by these mutations is responsible for the defects in RAR signaling in cancer cells. HT29 cells harbor an activating V600E mutation in B-RAF (13) and have high basal ERK activity (Fig. 6H, right). We then found that the inhibition of ERK by U0126 treatment in HT29 cells strongly upregulates both the basal and the RA-induced expression levels of an RAR target gene, RARRES1 (Fig. 6H, left). These results indicate that constitutive activation of the ERK pathway mediates the suppression of RAR transcriptional activity in cancer cells bearing activating mutations in B-RAF. It should be noted that treatment with RA did not decrease ERK activity in HT29 cells (Fig. 6H, right). Since induction of MKP4 by RA was moderate compared to that in Caco-2 cells and also since ERK is constitutively and strongly activated by a B-RAF mutation in these cells, such a moderate increase in the MKP4 level might not be sufficient to significantly suppress ERK activity in these cells.
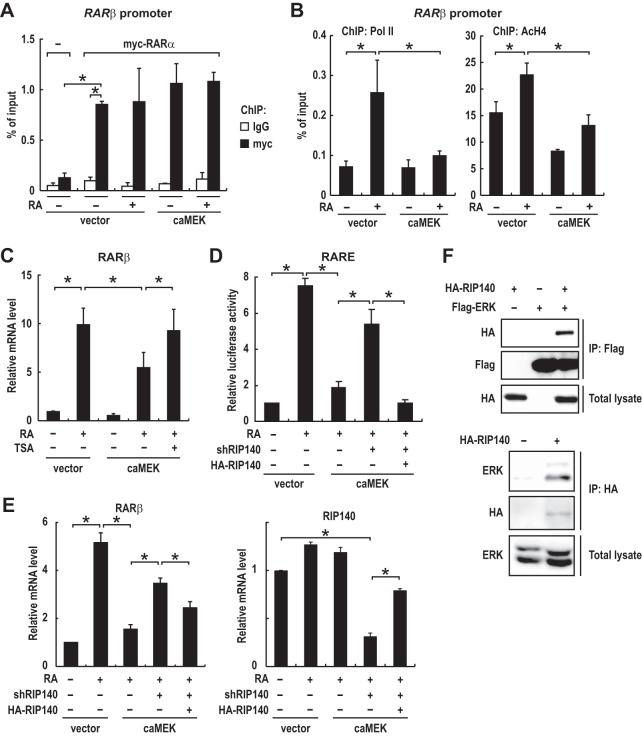
To address the mechanism by which ERK activation suppresses RAR signaling, we examined the effect of ERK activation on RAR binding to the RARβ promoter by using chromatin immunoprecipitation (ChIP) experiments. To this end, we generated a cell line stably expressing myc-tagged RARα and performed ChIP assays with an anti-myc antibody. The results showed that ERK activation did not inhibit the binding of RARα to the RARβ promoter (Fig. 7A). We then performed ChIP assays with antibodies to RNA polymerase II (Pol II) and acetylated histone H4 (AcH4) to examine which step in RAR target gene transcription is inhibited. Consistent with previous reports (2–4), RA treatment induced H4 acetylation at the RARβ promoter, and as a result, Pol II was recruited to the promoter (Fig. 7B). Remarkably, ERK activation by expression of constitutively active MEK markedly decreased RA-induced H4 acetylation as well as Pol II recruitment (Fig. 7B). We then examined the effect of the HDAC inhibitor trichostatin A (TSA) on the ERK-induced suppression of RARβ expression. The results showed that pretreatment with TSA partially but significantly suppressed the inhibitory effect of ERK activation on RARβ expression (Fig. 7C). In addition, combined treatment with TSA and RA increased RARβ expression more potently than treatment with either drug alone in HT29 cells harboring high ERK activity (Fig. S6) though expression of MKP4 was not increased in these cells (Fig. S6) (also see Discussion). These results suggest that ERK activation suppresses RAR target gene transcription partly by preventing RA-induced histone acetylation in the promoter region at a step prior to Pol II recruitment.
FIG 7.
ERK activation suppresses RAR transcriptional activity through an RIP140/HDAC-mediated mechanism. (A and B) Caco-2 cells stably expressing myc-tagged RARα, enhanced green fluorescent protein (−), and/or constitutively active MEK (caMEK) were treated with DMSO or RA (100 nM) for 90 min. Chromatin fragments were prepared and subjected to ChIP analyses with the indicated antibodies (means ± standard errors of the means; panel A, n = 3; panel B, n = 4). (C) Caco-2 cells were infected with an adenovirus expressing constitutively active MEK or a control virus (vector) 5 h after treatment with DMSO, RA (100 nM), and/or trichostatin A (TSA) (200 nM). Total RNA was prepared 19 h after infection and analyzed by RT-PCR. (D) Caco-2 cells stably expressing RIP140 shRNAs or a control shRNA (shRNA targeting green fluorescent protein) were generated by using lentivirus, and transcriptional activation of the reporter plasmid (RARE3-Luc) was evaluated in these cells. Cells were transfected with the expression plasmid for HA-RIP140 or a control plasmid. At 48 h after transfection, cells were infected with an adenovirus expressing constitutively active MEK or a control virus (vector) and incubated for 12 h. Then, cells were treated with 100 nM RA or DMSO for 12 h prior to the measurements. (E) Caco-2 cells were infected with lentiviruses expressing a control shRNA (shRNA targeting green fluorescent protein), RIP140 shRNAs, and/or HA-RIP140. At 72 h after lentiviral infection, cells were treated as described in the legend to Fig. 6A. Total RNA was analyzed by qRT-PCR. Note that HA-RIP140 is expressed at a level comparable to that of endogenous RIP140 (right). (F) Lysates of HEK293 cells transfected with expression plasmids for HA-RIP140 and/or Flag-ERK2 were subjected to immunoprecipitation (IP) with an anti-Flag antibody (top). Caco-2 cells stably expressing HA-RIP140 at a level comparable to that of endogenous RIP140 were generated by using lentivirus as described for panel E. Cell lysates were subjected to immunoprecipitation with an anti-HA antibody. Values in panels C to E are means ± standard errors of the means (n = 3). *, P < 0.05.
Since classical nuclear receptor corepressors such as the nuclear receptor corepressor (NCoR) and the silencing mediator of retinoid and thyroid receptor (SMRT), which recruit HDACs to RAR, do not associate with RAR in the presence of RA (2), we thought that other atypical corepressors might be involved in the HDAC-dependent suppression of RAR by ERK. We then hypothesized that members of a group of ligand-dependent corepressors that associate with and recruit HDACs to RA-bound RARs (34) could be candidate mediators of the suppression of RAR. Among these ligand-dependent corepressors, RIP140 and LCoR, but not PRAME, have been shown to be expressed in intestinal epithelial cells (35, 36). To test if RIP140 and LCoR are involved in the ERK activation-induced suppression of RAR activity, we inhibited their expression by short hairpin RNA (shRNA). Inhibition of LCoR expression by shRNA had essentially no effect on the ERK activation-induced suppression of RAR activity (Fig. S5D). In contrast, inhibition of RIP140 expression by shRNA significantly suppressed the inhibitory effects of ERK activation on RAR transcriptional activity and RARβ expression (Fig. 7D and E). The effects of RIP140 knockdown were reversed by restoring expression of RIP140 (Fig. 7D and E), showing the specificity of the effects. Thus, RIP140 should be required for the HDAC-dependent suppression of RAR transcriptional activity by ERK. Notably, we found that HA-tagged RIP140 was coimmunoprecipitated with Flag-tagged ERK2 from lysates of HEK293 cells (Fig. 7F, upper panel), and that endogenous ERK1/2 were coimmunoprecipitated with HA-tagged RIP140 that was stably expressed in Caco-2 cells to the level comparable to that of endogenous RIP140 (Fig. 7F, lower panel), demonstrating the interaction between ERK and RIP140 in CRC cells. Collectively, these results strongly suggest that ERK activation suppresses RAR transcriptional activity by inducing RIP140/HDAC-mediated histone deacetylation at the RAR target gene promoters (also see Discussion).
DISCUSSION
The RAR and RAS/ERK signaling pathways play a crucial role in regulating proliferation and differentiation of many types of cells. As transcriptional activation of RAR often leads to the inhibition of cell proliferation, loss of normal RAR function in the presence of physiological levels of RA is implicated in many types of cancers (3, 4, 8, 25). On the other hand, the activation of the ERK pathway has been shown to be required for cell proliferation, and the constitutive activation of this pathway by the mutational activation of K-RAS and B-RAF is associated with a diverse range of cancers (11–14). To date, interrelationships between these signaling pathways in regulating cancer cell fates have not been fully elucidated. Our results here indicate that RAR signaling promotes the differentiation of CRC cells, while ERK activation suppresses it. Our microarray analysis identified downstream target genes of RAR signaling in CRC cells. Notably, our results have demonstrated that RAR signaling activation causes ERK inactivation through the upregulation of MKP4, which is one of the RAR-dependently upregulated genes. Moreover, our analyses have revealed that ERK activation suppresses the expression of RAR target genes through an RIP140/HDAC-mediated mechanism. Taken together, our results have revealed novel antagonistic interactions between the RAS/ERK and RAR signaling pathways.
It should be noted that treatment with pharmacological doses of retinoids, which results in the activation of RAR signaling, has been considered a promising strategy for cancer therapy and prevention (3, 4, 8). However, how retinoids achieve these biological effects has not been fully elucidated. Rather surprisingly, our microarray analyses revealed that only a small number of genes are significantly upregulated by RAR signaling in CRC cells. Identification of the RAR-dependent upregulated genes here would facilitate our understanding of the molecular mechanisms of action of retinoids in cancer therapy and prevention. An important example is the present finding that RAR signaling induces MKP4 expression to inactivate ERK as the deregulated activation of ERK is among general mechanisms for cancer.
While many reports have shown the beneficial clinical effects of retinoids, accumulating evidence suggests that solid tumors acquire intrinsic resistance to retinoids during carcinogenesis, which has prevented retinoids from further application (4, 8). Strategies to overcome this resistance have long been discussed. Our finding that ERK activation, which is often caused by RAF or RAS mutations in cancer cells, suppresses RAR transcriptional activity through an RIP140/HDAC-mediated mechanism might provide a clue to the elucidation of underlying mechanisms of the retinoid resistance. In line with this idea, combinatorial treatment with RA and an HDAC inhibitor (TSA) restored expression of RARβ in HT29 colorectal cancer cells, in which ERK is constitutively activated by a B-RAF mutation (see Fig. S6 in the supplemental material). On the other hand, however, the expression level of MKP4 was not increased significantly by combinatorial treatment with RA and TSA (Fig. S6). Since the contribution of the RIP140/HDAC-mediated mechanism to ERK activation-induced suppression of RAR activity is partial (as shown by partial recovery of RAR activity after RIP140 knockdown) (Fig. 7D and E), other mechanisms might suppress expression of MKP4 in HT29 cells. Elucidation and pharmacological inhibition of such mechanisms would be the next important challenge in future studies. Notably, overexpression of PRAME, another ligand-dependent corepressor of RAR, has also been implicated in retinoid resistance of cancer cells (35). Based on our results and those of other investigators, it can be speculated that cancer cells often escape from the tumor-suppressive effect of RAR signaling by promoting the function of these ligand-dependent corepressors. An important difference between RIP140 and PRAME is that the function of RIP140 appears to be regulated posttranslationally by ERK activation whereas that of PRAME is regulated at the transcription level. Indeed, previous studies have shown that ERK phosphorylates RIP140 on Thr202/Thr207, which facilitates the transcription-suppressing activity of RIP140 by promoting its interaction with HDAC3 and CDK8-G9a (37, 38). Given that both HDACs and RIP140 are required for ERK activation-induced suppression of RAR and that ERK interacts with RIP140 in CRC cells, it is plausible that ERK activation confers resistance to retinoids on CRC cells by enhancing the function of RIP140.
In summary, our results identify novel antagonistic interactions between RAR and ERK signaling in CRC cells. These findings provide crucial insights into the mechanism of action of retinoids in cancer treatment and the retinoid resistance of cancer cells, two important issues related to the clinical use of retinoids. More generally, the elucidation of the mechanisms for novel interactions between the two important signaling pathways would shed light on the molecular bases for many physiological and pathological processes.
MATERIALS AND METHODS
Cell culture.
Caco-2, HT29, HeLa, and HEK293 cells were obtained from the American Type Culture Collection and maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 2 mM glutamine, and antibiotics (100 U/ml of penicillin and 0.2 mg/ml of kanamycin). For spontaneous differentiation experiments, the time when Caco-2 cells first reached confluence was designated day 0, and cells were treated with RA, LE540, and retinol from this time point. Medium was replaced daily during the experiments. For siRNA experiments, Caco-2 cells were transfected with a control siRNA or siRNAs that target human MKP4 (Invitrogen). Transfection of siRNAs was performed with siPORT Amine transfection reagent (Ambion).
Plasmids, antibodies, and reagents.
To obtain the RARE3-Luc reporter plasmid, a trimer of the RA-responsive element (RARE3) in the RARβ gene was inserted into the pGL3 luciferase (Luc) reporter vector (Promega) as described previously (33). For MKP4 promoter analysis, a region spanning −200 to −141 in reference to the transcriptional start site (ENST00000342782; Database of Transcriptional Start Sites [DBTSS]) of the MKP4 promoter, which contains an inverted direct repeat separated by 1 bp (DR1) (wild-type [WT], 5′TGGCCTCTGCCCC3′; mutant [4CG], 5′TGGGGTCTGCGGC3′), was inserted into the pGL3 luciferase reporter vector (Promega). Expression plasmids for wild-type and constitutively active MEK (LA-SDSE) have been described previously (39). To construct expression plasmids for RARα and RXRα, cDNAs of human RARα and RXRα were PCR amplified and cloned into a pcDNA3 vector (Invitrogen). Antibodies specific for RARβ (Santa Cruz), MKP4 (Santa Cruz), ERK1/2 (Santa Cruz), phospho-ERK1/2 (Cell Signaling), p38 (Santa Cruz), phospho-p38 (Cell Signaling), MEK1 (Santa Cruz), phospho-MEK1 (Cell Signaling), HA (Covance), Flag (Sigma), and α-tubulin (Sigma) were used for immunoblotting. U0126 was purchased from Promega. All-trans-retinoic acid and all-trans-retinol were purchased from Sigma. LE540 was purchased from Wako Pure Chemical. Retinol was stocked in ethanol, and the other drugs were stocked in dimethyl sulfoxide (DMSO).
Luciferase assay.
Caco-2 cells (1 × 105 cells/well) were seeded in 12-well plates 24 h before transfection. LipofectAMINE Plus transfection reagent (Invitrogen) was used according to the manufacturer's instructions. Cells were transfected with 0.3 μg of RARE3-Luc reporter plasmid and 1 μg of test plasmid and incubated for 24 h. Cells were treated with 100 nM RA for 12 h or 10 μM U0126 for 24 h prior to the measurement. For MKP4 promoter analysis, cells were transfected with 0.2 μg of pGL3 reporter vector containing a wild-type (WT) or mutated (4CG) DR1 element of the MKP4 promoter and 0.3 μg of expression plasmids for RARα and RXRα and incubated for 24 h. Cells were then treated with 1 μM RA or Ch55 for 36 h prior to the measurement. Luciferase activity in cell lysates was measured by using a luciferase assay system (Promega). We normalized the relative luciferase activity to the activity of coexpressed β-galactosidase.
RT-PCR analysis.
Total RNA was extracted by using an RNeasy minikit (Qiagen) according to the manufacturer's instructions and was then reverse transcribed into cDNA by Moloney murine leukemia virus (M-MLV) reverse transcriptase (Invitrogen) with oligonucleotide random hexamers. Prepared cDNA was purified and subjected to quantitative PCR analysis by using a Light Cycler (Roche Diagnostics) or ABI 7300 Real Time PCR System (Applied Biosystems) with a SYBR green PCR kit (Applied Biosystems). Each value obtained was normalized to that of glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
DNA pulldown and ChIP assays.
For DNA pulldown experiments, cell lysates were incubated with 2 μg of biotinylated double-stranded oligonucleotides (equivalent to the MKP4 promoter region [−200 to −141] containing the WT or mutated [4CG] DR1) and 5 μg of poly(dI-dC) at 4°C for 1 h in binding buffer (20 mM HEPES [pH 7.8], 100 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 10 mM Na4P2O4, 1% NP-40, and 10% glycerol). DNA-bound proteins were collected with streptavidin-agarose beads (Invitrogen) for 1 h, washed extensively with binding buffer, and resuspended in protein loading buffer. The samples were analyzed by immunoblotting. Chromatin immunoprecipitation (ChIP) assays were performed using a ChIP-IT Express kit (Active Motif) according to the manufacturer's instructions. Briefly, cells were fixed with 1% formaldehyde for 10 min at room temperature, and then chromatin was prepared and sonicated extensively. Chromatin fragments were immunoprecipitated with anti-RNA polymerase II (Covance), anti-myc (Santa Cruz), and anti-acetylated H4 antibodies (Upstate). Purified rabbit IgG (Santa Cruz) was used as a control. Immunoprecipitated DNA and input DNA were analyzed by quantitative PCR.
Microarray analysis.
For microarray analysis, we performed two independent series of experiments. Caco-2 cells were treated with 100 nM RA or DMSO vehicle for 24 h, and total RNA was extracted by using an RNeasy minikit (Qiagen). Synthesis of cDNA, in vitro transcription and biotin labeling of cRNA, and hybridization to the human genome U133 Plus 2.0 array (Affymetrix) were performed according to Affymetrix protocols. Hybridized arrays were scanned using an Affymetrix GeneChip scanner. Images from scanned chips were processed by using the default settings of GeneChip Operating Software (GCOS), version 1.4. The CELL files created by GCOS were imported into the GeneSpring GX, version 7.3 (Agilent Technologies), microarray analysis software for statistical analysis and presentation of the expression profiles and average-expression profiles. Probe intensities were normalized, and expression signals of all genes (probe sets) were calculated using GeneChip robust multiarray averaging (GCRMA). The genes whose signal intensities were extremely low were excluded from further analysis. RA-dependently upregulated genes were identified by statistical analysis and fold change values. Statistical analysis was performed by one-way analysis of variance (ANOVA) with a Benjamini-Hochberg false discovery rate (equal to 0.05) multiple testing correction followed by Tukey's post hoc tests.
Supplementary Material
ACKNOWLEDGMENTS
We thank Shin Yonehara and Yohei Kobayashi for lentiviral experiments.
This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to E.N.). M.I. was also supported by a Grant-in-Aid for Young Scientists (B) (25870363).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00012-17.
REFERENCES
- 1.Mark M, Ghyselinck NB, Chambon P. 2006. Function of retinoid nuclear receptors: lessons from genetic and pharmacological dissections of the retinoic acid signaling pathway during mouse embryogenesis. Annu Rev Pharmacol Toxicol 46:451–480. doi: 10.1146/annurev.pharmtox.46.120604.141156. [DOI] [PubMed] [Google Scholar]
- 2.Bastien J, Rochette-Egly C. 2004. Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene 328:1–16. doi: 10.1016/j.gene.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 3.Altucci L, Gronemeyer H. 2001. The promise of retinoids to fight against cancer. Nat Rev Cancer 1:181–193. doi: 10.1038/35106036. [DOI] [PubMed] [Google Scholar]
- 4.Freemantle SJ, Spinella MJ, Dmitrovsky E. 2003. Retinoids in cancer therapy and chemoprevention: promise meets resistance. Oncogene 22:7305–7315. doi: 10.1038/sj.onc.1206936. [DOI] [PubMed] [Google Scholar]
- 5.Kakizuka A, Miller WH, Umesono K, Warrell RP, Frankel SR, Murty VV, Dmitrovsky E, Evans RM. 1991. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RARα with a novel putative transcription factor, PML. Cell 66:663–674. doi: 10.1016/0092-8674(91)90112-C. [DOI] [PubMed] [Google Scholar]
- 6.de Thé H, Lavau C, Marchio A, Chomienne C, Degos L, Dejean A. 1991. The PML-RARα fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell 66:675–684. doi: 10.1016/0092-8674(91)90113-D. [DOI] [PubMed] [Google Scholar]
- 7.Rousselot P, Hardas B, Patel A, Guidez F, Gäken J, Castaigne S, Dejean A, de Thé H, Degos L, Farzaneh F. 1994. The PML-RARα gene product of the t(15;17) translocation inhibits retinoic acid-induced granulocytic differentiation and mediated transactivation in human myeloid cells. Oncogene 9:545–551. [PubMed] [Google Scholar]
- 8.Uray IP, Dmitrovsky E, Brown PH. 2016. Retinoids and rexinoids in cancer prevention: from laboratory to clinic. Semin Oncol 43:49–64. doi: 10.1053/j.seminoncol.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alvarez S, Germain P, Alvarez R, Rodríguez-Barrios F, Gronemeyer H, de Lera AR. 2007. Structure, function and modulation of retinoic acid receptor beta, a tumor suppressor. Int J Biochem Cell Biol 39:1406–1415. doi: 10.1016/j.biocel.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 10.Ordóñez-Morán P, Dafflon C, Imajo M, Nishida E, Huelsken J. 2015. HOXA5 counteracts stem cell traits by inhibiting Wnt signaling in colorectal cancer. Cancer Cell 28:815–829. doi: 10.1016/j.ccell.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 11.Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, Tuveson DA, Jacks T. 2001. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature 410:1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- 12.Janssen KP, el-Marjou F, Pinto D, Sastre X, Rouillard D, Fouquet C, Soussi T, Louvard D, Robine S. 2002. Targeted expression of oncogenic K-ras in intestinal epithelium causes spontaneous tumorigenesis in mice. Gastroenterology 123:492–504. doi: 10.1053/gast.2002.34786. [DOI] [PubMed] [Google Scholar]
- 13.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JW, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA. 2002. Mutations of the BRAF gene in human cancer. Nature 417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 14.Rajagopalan H, Bardelli A, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. 2002. RAF/RAS oncogenes and mismatch-repair status. Nature 418:934. doi: 10.1038/418934a. [DOI] [PubMed] [Google Scholar]
- 15.Sansom OJ, Meniel V, Wilkins JA, Cole AM, Oien KA, Marsh V, Jamieson TJ, Guerra C, Ashton GH, Barbacid M, Clarke AR. 2006. Loss of Apc allows phenotypic manifestation of the transforming properties of an endogenous K-ras oncogene in vivo. Proc Natl Acad Sci U S A 103:14122–14127. doi: 10.1073/pnas.0604130103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinzler KW, Vogelstein B. 1996. Lessons from hereditary colorectal cancer. Cell 87:159–170. doi: 10.1016/S0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 17.Vries RG, Huch M, Clevers H. 2010. Stem cells and cancer of the stomach and intestine. Mol Oncol 4:373–384. doi: 10.1016/j.molonc.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fearon ER, Vogelstein B. 1990. A genetic model for colorectal tumorigenesis. Cell 61:759–767. doi: 10.1016/0092-8674(90)90186-I. [DOI] [PubMed] [Google Scholar]
- 19.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. 2013. Cancer genome landscapes. Science 339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fearon ER. 2011. Molecular genetics of colorectal cancer. Annu Rev Pathol 6:479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- 21.Samatar AA, Poulikakos PI. 2014. Targeting RAS-ERK signalling in cancer: promises and challenges. Nat Rev Drug Discov 13:928–942. doi: 10.1038/nrd4281. [DOI] [PubMed] [Google Scholar]
- 22.Chantret I, Barbat A, Dussaulx E, Brattain MG, Zweibaum A. 1988. Epithelial polarity, villin expression, and enterocytic differentiation of cultured human colon carcinoma cells: a survey of twenty cell lines. Cancer Res 48:1936–1942. [PubMed] [Google Scholar]
- 23.Baltes S, Nau H, Lampen A. 2004. All-trans retinoic acid enhances differentiation and influences permeability of intestinal Caco-2 cells under serum-free conditions. Dev Growth Differ 46:503–514. doi: 10.1111/j.1440-169x.2004.00765.x. [DOI] [PubMed] [Google Scholar]
- 24.van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, van der Horn K, Batlle E, Coudreuse D, Haramis AP, Tjon-Pon-Fong M, Moerer P, van den Born M, Soete G, Pals S, Eilers M, Medema R, Clevers H. 2002. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell 111:241–250. doi: 10.1016/S0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 25.Wu CC, Shyu RY, Chou JM, Jao SW, Chao PC, Kang JC, Wu ST, Huang SL, Jiang SY. 2006. RARRES1 expression is significantly related to tumour differentiation and staging in colorectal adenocarcinoma. Eur J Cancer 42:557–565. doi: 10.1016/j.ejca.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 26.Farooq A, Zhou MM. 2004. Structure and regulation of MAPK phosphatases. Cell Signal 16:769–779. doi: 10.1016/j.cellsig.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 27.Dickinson RJ, Keyse SM. 2006. Diverse physiological functions for dual-specificity MAP kinase phosphatases. J Cell Sci 119:4607–4615. doi: 10.1242/jcs.03266. [DOI] [PubMed] [Google Scholar]
- 28.Muncan V, Sansom OJ, Tertoolen L, Phesse TJ, Begthel H, Sancho E, Cole AM, Gregorieff A, de Alboran IM, Clevers H, Clarke AR. 2006. Rapid loss of intestinal crypts upon conditional deletion of the Wnt/Tcf-4 target gene c-Myc. Mol Cell Biol 26:8418–8426. doi: 10.1128/MCB.00821-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blache P, van de Wetering M, Duluc I, Domon C, Berta P, Freund JN, Clevers H, Jay P. 2004. SOX9 is an intestine crypt transcription factor, is regulated by the Wnt pathway, and represses the CDX2 and MUC2 genes. J Cell Biol 166:37–47. doi: 10.1083/jcb.200311021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lorentz O, Duluc I, Arcangelis AD, Simon-Assmann P, Kedinger M, Freund JN. 1997. Key role of the Cdx2 homeobox gene in extracellular matrix-mediated intestinal cell differentiation. J Cell Biol 139:1553–1565. doi: 10.1083/jcb.139.6.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suh E, Traber PG. 1996. An intestine-specific homeobox gene regulates proliferation and differentiation. Mol Cell Biol 16:619–625. doi: 10.1128/MCB.16.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chawengsaksophak K, James R, Hammond VE, Köntgen F, Beck F. 1997. Homeosis and intestinal tumours in Cdx2 mutant mice. Nature 386:84–87. doi: 10.1038/386084a0. [DOI] [PubMed] [Google Scholar]
- 33.de Thé H, Vivanco-Ruiz MM, Tiollais P, Stunnenberg H, Dejean A. 1990. Identification of a retinoic acid responsive element in the retinoic acid receptor beta gene. Nature 343:177–180. doi: 10.1038/343177a0. [DOI] [PubMed] [Google Scholar]
- 34.White JH, Fernandes I, Mader S, Yang XJ. 2004. Corepressor recruitment by agonist-bound nuclear receptors. Vitam Horm 68:123–143. doi: 10.1016/S0083-6729(04)68004-6. [DOI] [PubMed] [Google Scholar]
- 35.Epping MT, Wang L, Edel MJ, Carlée L, Hernandez M, Bernards R. 2005. The human tumor antigen PRAME is a dominant repressor of retinoic acid receptor signaling. Cell 122:835–847. doi: 10.1016/j.cell.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 36.Fernandes I, Bastien Y, Wai T, Nygard K, Lin R, Cormier O, Lee HS, Eng F, Bertos NR, Pelletier N, Mader S, Han VK, Yang XJ, White JH. 2003. Ligand-dependent nuclear receptor corepressor LCoR functions by histone deacetylase-dependent and -independent mechanisms. Mol Cell 11:139–150. doi: 10.1016/S1097-2765(03)00014-5. [DOI] [PubMed] [Google Scholar]
- 37.Gupta P, Huq MD, Khan SA, Tsai NP, Wei LN. 2005. Regulation of co-repressive activity of and HDAC recruitment to RIP140 by site-specific phosphorylation. Mol Cell Proteomics 4:1776–1784. doi: 10.1074/mcp.M500236-MCP200. [DOI] [PubMed] [Google Scholar]
- 38.Ho PC, Gupta P, Tsui YC, Ha SG, Huq M, Wei LN. 2008. Modulation of lysine acetylation-stimulated repressive activity by Erk2-mediated phosphorylation of RIP140 in adipocyte differentiation. Cell Signal 20:1911–1919. doi: 10.1016/j.cellsig.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fukuda M, Gotoh I, Adachi M, Gotoh Y, Nishida E. 1997. A novel regulatory mechanism in the mitogen-activated protein (MAP) kinase cascade. Role of nuclear export signal of MAP kinase kinase. J Biol Chem 272:32642–32648. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.