Abstract
We used next generation DNA sequencing to profile the microbiome of infected Diabetic Foot Ulcers (DFUs). The microbiota was correlated to clinical parameters and treatment outcomes to determine if directed antimicrobial therapy based on conventional microbiological cultures are relevant based on genomic analysis. Patients ≥ 18 years presenting with a new Diabetic Foot Infection (DFI) who had not received topical or oral antimicrobials in the two weeks prior to presentation, were eligible for enrolment. Tissue punch biopsies were obtained from infected DFUs for analysis. Demographics, clinical and laboratory data were collected and correlated against microbiota data. Thirty-nine patients with infected DFUs were recruited over twelve-months. Shorter duration DFUs (< six weeks) all had one dominant bacterial species (n = 5 of 5, 100%, p < 0.001), Staphylococcus aureus in three cases and Streptococcus agalactiae in two. Longer duration DFUs (≥ six weeks) were diversely polymicrobial (p < 0.01) with an average of 63 (range 19–125) bacterial species. Severe DFIs had complex microbiomes and were distinctly dissimilar to less severe infections (p = 0.02), characterised by the presence of low frequency microorganisms. Nineteen patients (49%) during the study period experienced antimicrobial treatment failure, but no overall differences existed in the microbiome of patients who failed therapy and those who experienced treatment success (p = 0.2). Our results confirm that short DFUs have a simpler microbiome consisting of pyogenic cocci but chronic DFUs have a highly polymicrobial microbiome. The duration of a DFU may be useful as a guide to directing antimicrobial therapy.
Keywords: Microbiome, Diabetic foot ulcers, Diabetic foot infections, 16S rRNA, Next generation DNA sequencing
Highlights
-
•
Next generation DNA sequencing of infected diabetic foot ulcers identifies similar pathogens of infection from culture-dependent data
-
•
Aerobic Gram-positive cocci are the predominant pathogens of infection in diabetic foot ulcers from a molecular perspective.
-
•
Anaerobes are frequently observed in high proportions in infected diabetic foot ulcers and may act synergistically with aerobic counterparts.
-
•
The duration of diabetic foot ulcer prior to a new infective episode is important to consider when choosing antimicrobial therapy.
Several publications utilising molecular centred approaches have reported on the microbiome of non-infected wounds. These studies have revealed a complex array of microbial communities, identifying many “hidden” microorganisms not identified by culture methods. This has contributed to understanding the microbiome of non-infected wounds, but the significance of this extended ‘microbial” view in Diabetic Foot Ulcers with clinical infection, and if changes to clinical care as a result of molecular data are required remains unclear. The outcomes of this study using DNA techniques provide clarity and support for the continued use of current antibiotic/therapeutic guidelines based on culture-dependent evidence.
1. Introduction
In a person with diabetes a “triad” of factors that include peripheral neuropathy, peripheral vascular disease and trauma (Lipsky et al., 2012), place the foot at risk of developing a wound. Infections of the feet in people with diabetes are the primary pathway to lower extremity amputation (Lavery et al., 2003). The management of diabetes foot infection (DFI) is underpinned by the requirement to identify the pathogen/s of infection and thus direct antimicrobial therapy. Laboratory based methods that are culture-dependent have been utilised to identify planktonic microorganisms that are potential pathogens of infection, in addition to examining their density through qualitative and quantitative measures. This has shown acute ulcers are usually colonised by Staphylococcus aureus and/or Streptococcus agalactiae (Group B Streptococcus), and chronic ulcers have a more diverse microbiome, with anaerobic organisms and Pseudomonas aeruginosa becoming more important (Lipsky et al., 2016a, Lipsky et al., 2016b). Culture-dependent techniques select for species that flourish under the typical conditions of the diagnostic microbiology laboratory, and this may not necessarily reflect the most abundant or clinically important microorganisms in DFIs especially anaerobes and species not detected under standard clinical microbiology laboratory protocols (Grice et al., 2008).
Molecular DNA based techniques that are culture-independent have identified the limitations of traditional cultivation based methods when examining the microbiome of wounds. Using amplification and sequence analysis of 16s rDNA, a highly-conserved gene present in all prokaryotes (bacteria) but not eukaryotes (humans), has revealed a vastly more complex array of bacterial communities in non-infected chronic wounds (Dowd et al., 2008a, Dowd et al., 2008b, Gardner et al., 2013, Rhoads et al., 2012a, Rhoads et al., 2012b, Smith et al., 2016). No data exists for acutely infected DFUs using this methodology.
We explored the microbiome of infected DFUs using next generation DNA sequencing. Data is presented on the microbial diversity, community structure, bacterial load and presence of likely pathogens from diabetic foot infections. Molecular findings are correlated against clinical factors and treatment outcomes.
2. Materials and Methods
2.1. Patients, Samples and Ethics
Individuals presenting to a tertiary referral hospital (Liverpool Hospital High Risk Foot Service and Liverpool Hospital Emergency Department) with a newly infected diabetic foot ulcer occurring below the malleolus (Lipsky et al., 2012) were recruited consecutively over a twelve-month study period between January 2015 and December 2015. A 3 mm (width) × 10 mm (depth) tissue punch biopsy was obtained from the edge of each DFU after debriding and cleansing the wound with NaCl 0.9%. Patients who had received any systemic or topical antimicrobial therapy two weeks prior to enrolment were excluded. Ethics approval for this study was granted by the South West Sydney Local Health District Research and Ethics Committee (HREC/14/LPOOL/487, SSA/14/LPOOL/489). The study methodology was designed in guidance by STROME-ID and our molecular surveillance data are reported in keeping with this (Field et al., 2014).
2.2. Patient Demographic, Laboratory and Clinical Data
Patient demographics, laboratory and clinical data were collected through patient charts and the electronic medical records for correlation against microbiome data. Clinical data and wound metrics of interest that were collected included; present or absent foot pulses, foot doppler waveforms, toe brachial indices (TBI) and completion of the modified neuropathic disability score (Abbott et al., 2002). DFU location, duration of DFU prior to presentation, size (length x width in mm), depth (mm) and tissue type (granulation, slough, necrosis). Laboratory data included; full blood count, inflammatory markers (White cell count [WCC], Erythrocyte sedimentation rate [ESR], C-reactive protein [CRP]), glycosylated haemoglobin (HbA1c) and estimated glomerular filtration rate (eGFR). All newly infected DFUs were diagnosed clinically, and their severity graded using the Infectious Disease Society of America Guidelines for DFI (Lipsky et al., 2012). Acute infections were defined based on new presenting symptoms (classic signs of infection) being present and untreated of less than fourteen days duration.
DFUs were classified based on their duration, with shorter duration DFUs (Acute) being less than six weeks and longer duration DFUs (Chronic) defined as those greater than six weeks. Treatment failure during the study period were defined as no resolution of infective symptoms over an appropriate treatment period (> 28 days) despite directed anti-infective treatment (Lipsky et al., 2012), a requirement to replace oral antimicrobial therapy with parenteral delivery due to deterioration of infective symptoms, or the need for surgical intervention.
2.3. Specimen Collection, Sampling Processing and the Work Flow for Undertaking Molecular and Culture Dependent Approaches
Specimen collection, storage and the work flows for performing DNA extraction, next generation DNA sequencing, sequence analysis and quality control of DNA reads and qPCR to determine the microbial load, were performed as previously described by our group (Malone et al., 2017) and can be found in supplementary appendix (S1). Culture-dependent bacteriological enumeration and identification from tissue cultures was performed by a hospital pathology service (Sydney South West Pathology Service) using methods previously described (Oates et al., 2014).
2.4. Statistics
CLC genomics workbench version 8.5.1 in combination with the microbial genome-finishing module (CLC bio, Qiagen Aarhus, Denmark) were used to analyse DNA sequence data. Operational taxonomic units (OTU) clustering were based on previously reported wound microbiome analysis (Gardner et al., 2013). OTUs were defined as molecular proxies for describing organisms based on their phylogenetic relationships to other organisms. Associations between microbiome community structure and membership were compared using permutational multivariate analysis of variance (PERMANOVA) in combination with principal coordinate analysis PCoA Bray-curtis dissimilarity matrix. Patient demographics, laboratory and clinical data were examined using Chi-square and Spearman correlation coefficients. Kappa coefficients were used to determine the level of agreement between culture-dependent approaches and DNA sequencing. Independent predictors of treatment failure were explored using general linear model (GLM). Mann Whitney U test for non-parametric data were undertaken when analysing the subgroups of neuropathic or neuroischemic lesions. Analysis was performed using Statistical Package for Social Sciences (SPSS) Version 23, SPSS Inc., Chicago, Illinois, USA. For all comparisons and modelling, the level of significance was set at p < 0.05. Molecular data analysed through Bray-curtis and PERMNOVA incorporated a Bonferroni correction. Data are given as mean, median and standard deviation (±).
3. Results
39 patients (39 tissue specimens) with newly infected DFUs were recruited over the 12-month study period. Broad demographic, clinical and laboratory data are shown in Table 1. Next generation DNA sequencing generated 1,028,895 sequences, which were clustered and aligned at 97% similarity to reveal 1139 unique OTUs. A total of seven major phyla were identified including Firmicutes (48%), Proteobacteria (26%), Actinobacteria (12%), Bacteroidetes (8%), Fusobacteria (2%) and Cyanobacteria (1%). The clustering of OTUs contributing to greater than 10% within each DFU sample at the genera/species-level is noted in Table 2 and those contributing to < 10% are noted in (S2). Staphylococcus spp. was the most commonly sequenced microorganism in infected DFUs. This was followed by Corynebacterium spp., Finegoldia spp., Peptoniphilus spp., Acinetobacter spp., Anaerococcus spp., and Streptococcus spp., We further categorized microorganisms based on their residing niche (environmental, skin, oral and gut) to better define the site of origin of microorganisms that colonize DFUs (S3). Microorganisms commensal to the skin were predominant in half of patients (50.6%) followed by environmental (29.1%), gut (14%) and oral (6.3%) microorganisms.
Table 1.
Patient demographics, clinical and laboratory data for 39 patients presenting with diabetic foot infection.
| Characteristics | n = patients (%) (± SD) |
|---|---|
| Demographics | |
| Mean age | 57.4 years (± 11.5) |
| Male/female | 28 (71%)/11 (29%) |
| Type of diabetes: type 1/type2 | 4 (10%)/35 (90%) |
| Duration of diabetes | 12.8 years (± 6.5) |
| Chronic kidney disease stage 5 | 16 (27%) |
| Duration of ulcer prior to presentation | 15.7 weeks (± 13.7) |
| Co-morbidities | |
| Loss of protective sensation | 39 (100%) |
| Peripheral arterial disease | 15 (38.5%) |
| Toe brachial index | 0.5 (± 0.1) |
| Laboratory data | |
| Glycosylated haemoglobin (HbA1c) (%) | 8.5 (± 2.5) |
| Erythrocyte sedimentation rate (mmol/L) | 54.3 (± 33) |
| C-reactive protein (mg/l) | 28.1 (± 25) |
| White cell count | 9.2 (± 2.4) |
| Infection grading and classification (IDSA) | |
| Mild | 5 (13%) |
| Moderate | 25 (64%) |
| Severe | 9 (23%) |
| Systemic antimicrobial/route of delivery | |
| Cephalexin/oral | 6 (15%) |
| Amoxycillin + clavulanic acid/oral | 13 (33.5%) |
| Flucoxacillin/oral | 3 (8%) |
| Clindamycin/oral | 1 (2.5%) |
| Ciprofloxacin/oral | 1 (2.5%) |
| Rifampin + fusidic acid/oral | 2 (5%) |
| Sulfamethoxazole + trimethoprim/oral | 1 (2.5%) |
| Combination therapy/oral | 3 (8%) |
| Piperacillin + tazobactam/intravenous | 6 (15%) |
| Cephazolin/intravenous | 3 (8%) |
Table 2.
Microorganisms contributing ≥ 10% in each sample (representing the dominant taxa) (Rhoads et al., 2012a, Rhoads et al., 2012b).
| Genera/species | Samples | Avg abundance % | SD | Min-max avg. abu % | Aerotolernace |
|---|---|---|---|---|---|
| Staphylococcus spp.: | 15 | 40.7 | 30.3 | 12 to 92 | Facultative |
| Staphylococcus aureusa | 8 | 43.1 | 32.9 | 12 to 92 | Facultative |
| Unclassified Staphylococcus sppa | 7 | 31.8 | 28.2 | 12 to 81 | Facultative |
| Staphylococcus pettenkoferia | 2 | 26 | 3 | 23 to 29 | Facultative |
| Corneybacterium striatum | 8 | 32 | 16.6 | 12 to 59 | Facultative |
| Finegoldia spp. | 7 | 12 | 2.8 | 10 to 18 | Anaerobe |
| Peptoniphilus spp. | 7 | 14.5 | 5.1 | 10 to 22 | Anaerobe |
| Acinetobacter baumannii | 7 | 30.5 | 18.7 | 16 to 69 | Facultative |
| Anaerococcus spp. | 6 | 14.3 | 5.1 | 12 to 24 | Anaerobe |
| Streptococcus agalactiae | 5 | 45.2 | 39 | 16 to 89 | Facultative |
| Enterobacter spp. | 5 | 19.6 | 8.1 | 10 to 28 | Facultative |
| Proteus spp. | 4 | 22.7 | 4.5 | 19 to 23 | Facultative |
| Prevotella spp. | 4 | 14.3 | 4 | 10 to 18 | Anaerobe |
| Haemophilus spp. | 4 | 21 | 14 | 12 to 42 | Facultative |
| Blastocatella fastidiosa | 3 | 24 | 11 | 12 to 32 | Facultative |
| Pseudomonas aeruginosa | 2 | 12.5 | 3.5 | 10 to 15 | Aerobe |
| Porphymonas spp. | 2 | 11.5 | 2 | 10 to 13 | Anaerobe |
Refers to the species level identification of Staphylococcus genus level data.
3.1. Community Structure of DFIs are Heterogeneous
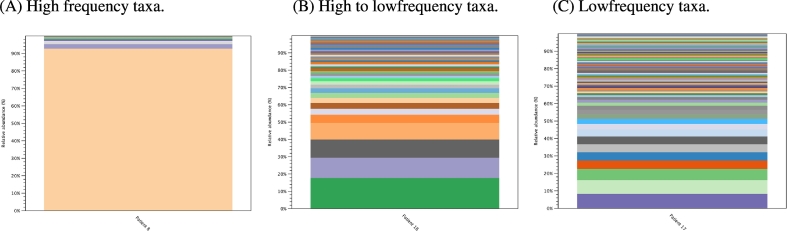
The community structures of DFIs were depicted using rarefaction and Shannon Weaver index plots, which explore the richness, and diversity of individual infected DFUs (Fig. 1). Most DFUs had complex polymicrobial communities with great heterogeneity between patients. Rarefaction identified a mean of 56 OTUs (± 31.2, range 4 to 125) per DFU, and Shannon Weaver index identified a mean indices of 2.3 (± 0.9, range 0.4–4.1). Descriptive statistics allowed for a more clinically relevant picture to be composed of the overall community structure. We identified three general profiles that sub-divided DFUs based on their community structure (Fig. 2). High frequency taxa mostly comprised of a single microorganism (± 3) (i.e. monomicrobial infection), high to low frequency taxa were comprised of between one to five (± 2) dominant microorganisms followed by many low frequency taxa (i.e. polymicrobial infection) and low frequency taxa comprised on average of ≥ 20 (±) minor microorganisms (complex polymicrobial infection).
Fig. 1.
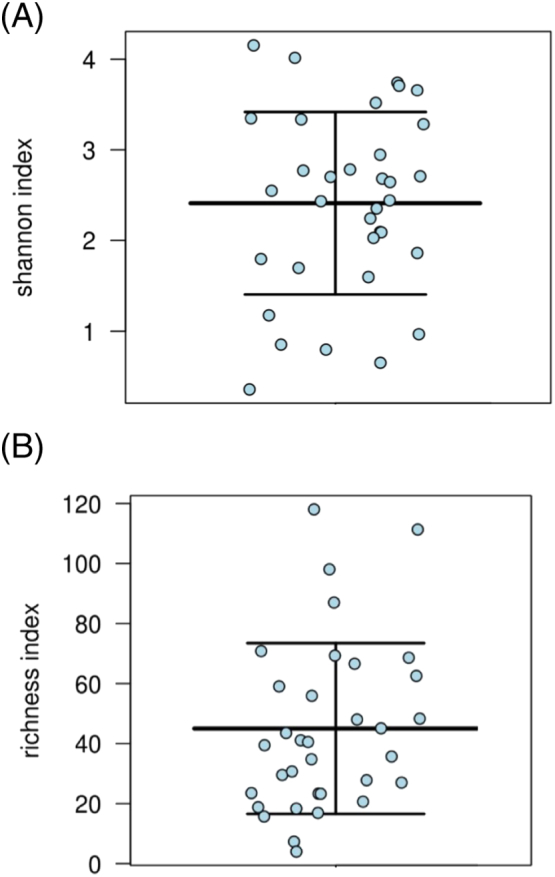
Community diversity and richness reported for 39 patients with DFI. (A) Community diversity of DFUs presented using the Shannon-Weaver index at maximum read length of 300 (Price et al., 2009). Shannon Weaver Index is a measure of diversity that includes the number of unique microbial taxa and their relative evenness within each sample. Thus, a higher Shannon Weaver Index correlates to a greater diversity. (B) Community richness of DFUs presented using richness index reporting the number of unique OTUs in each wound sample. Data sets were normalised to remove low abundance OTUs contributing too less then 1% within each wound sample.
Fig. 2.
Bar chart represents relative abundances (%) of taxa profiles for 39 DFUs. Each bar represents individual genera/species. (A) High frequency taxa were observed in ten patients (26%), mostly comprised of a single microorganism (± 3) (i.e. monomicrobial infection) contributing to ≥ 88% (± 5.4%) of total abundance. (B) High to low frequency taxa were the most common profile and were observed in 25 patients (64%). Low frequency taxa comprised on average of ≥ 20 (±) minor microorganisms each contributing < 1%–5% abundance and no single microorganism contributing > 10% (complex polymicrobial infection). (C) Low frequency taxa were infrequently observed in only four patients (10%) and contained higher relative abundances of environmental microorganisms (p < 0.01).
3.2. The Duration of DFU Prior to Infection Presentation may Present a Major Driver Behind the Microbiome
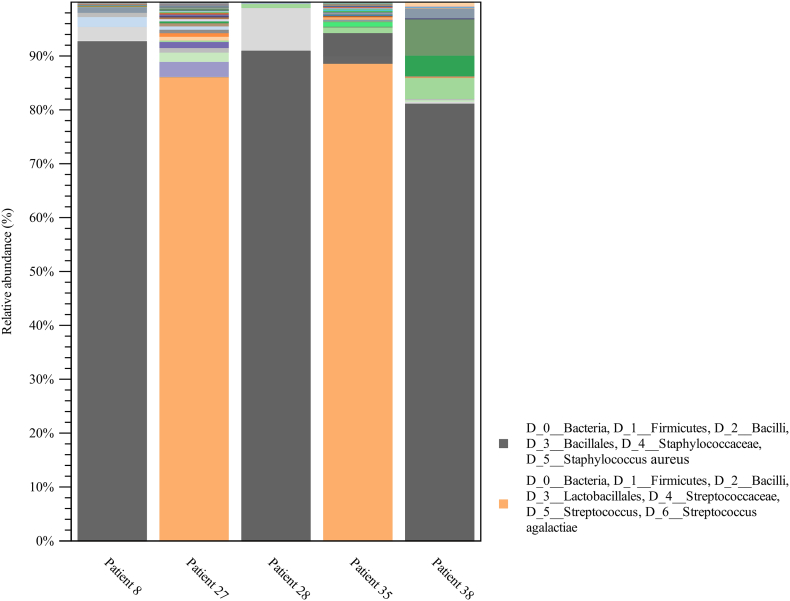
Five (13%) DFUs at the time of presentation were less than six-weeks in duration, and were composed of high frequency taxa with one predominant microorganism (Fig. 3). These were S. aureus in three cases and S. algalactiae in two cases. The relative abundance of Staphylococcus spp., was far greater in DFUs < six-weeks then DFUs of longer duration where it was present but at significantly lower relative abundances (Fig. 4).
Fig. 3.
Bar chart representing relative abundance of taxa in acute diabetic foot ulcers (< 6 weeks duration).
Fig. 4.
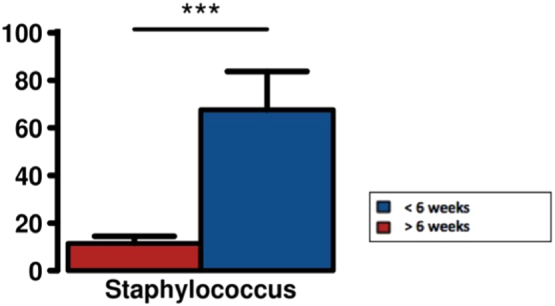
Analysis of variance between Staphylocci spp., relative abundance (%) in DFUs based on duration. In DFUs < six weeks Staphyloccci spp., were present as the dominant taxa (high frequency). The average relative abundance of Staphyloccci in DFUs > six-weeks is far less and this is because DFUs of longer duration are typically polymicrobial.
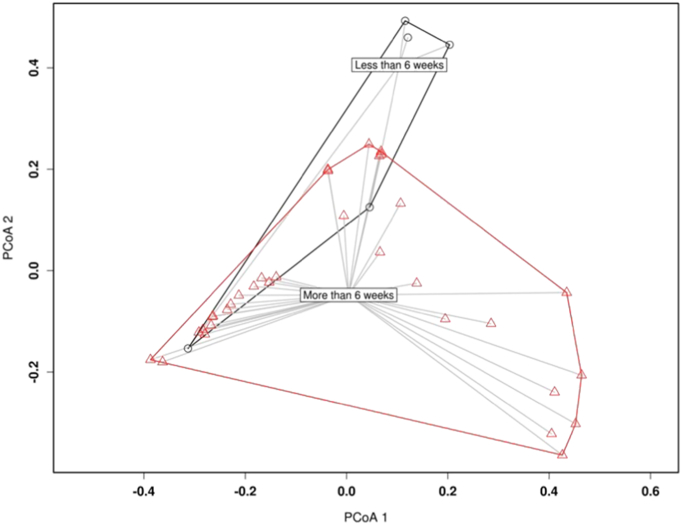
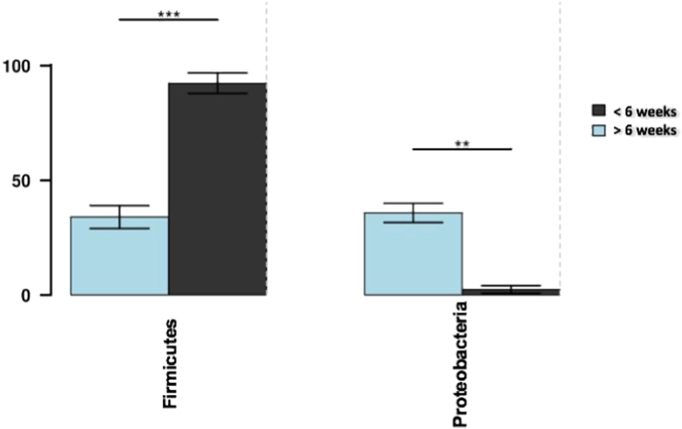
Longer duration DFUs (≥ 6 weeks) with a new acute infective episode (n = 34, 87%) were the most common presentation. PCoA bray-curtis plots with PERMANOVA identified the community structures between longer and shorter duration DFUs were dissimilar (p < 0.003) (Fig. 5 and S4). Furthermore, analysis of variance identifies that longer duration DFUs (> 6 weeks) were associated with a greater relative abundance of Proteobacteria (p < 0.05), whilst shorter duration DFUs (< 6 weeks) were associated with greater relative abundance of Firmicutes (p < 0.001). (Fig. 6) Closer examination of OTUs revealed that Staphylocci spp., were contributing to the positive correlation detected between Firmicutes and relative abundance and ulcer duration (p < 0.05).
Fig. 5.
PCoA bray-curtis plots identify that differences are present in the community structures between longer and shorter duration DFUs.
Fig. 6.
Analysis of variance identifies that ulcer duration > 6 weeks was associated with a greater relative abundance of Proteobacteria (p < 0.05), whilst ulcer duration <6 weeks was associated with greater relative abundance of Firmicutes (p < 0.001). The genera responsible for the high relative abundance of firmicutes in DFUs < 6 weeks were Staphylococcus spp., and Streptococcus spp predominantly.
Spearman's correlation coefficients further clarified that DFUs of longer duration were polymicrobial, typically having greater number of OTUs and were broader in diversity (p < 0.01). This statistical approach further correlated higher frequencies of DFUs containing obligate anaerobes that constituted greater than 30% of the total abundance in DFUs of greater duration (p < 0.03) (S5).
3.3. Wound Observations and Clinical Factors Lack Association With the Microbiome
Associations between clinical factors and DFI microbiome were compared using PERMANOVA and spearman rank correlation coefficients. The location, depth and the level of glycosylated haemoglobin (HbA1C ≥ 7%) were not associated to any significant microbiota. The presence of slough or malodour within an infected DFU were independently associated with community structure, but were not inversely correlated to each other (p = 0.7). Slough in an infected DFU was associated with higher abundances of obligate anaerobes (slough present and ≥ 30% anaerobe present = 13 of 39, 33%, p < 0.01), as was malodour (malodour DFUs = 15, mean anaerobe abundance 34%, SD25.3 versus no malodour of DFUs, mean anaerobe abundance 15%, SD 18.4).
3.4. Infection Severity of Diabetic Foot Infections are Associated with Altered Community Structures
PERMANOVA identified some disparity between the community structure and infection severity. Mild DFIs were different from both moderate infection (p < 0.01) and severe infection (p < 0.001) (Fig. 7), and were positively correlated to fewer OTUs and were less diverse. In contrast, severe infections often presented exclusively with low frequency taxa profiles (n = 3 of 4, p = 0.02). Obligate anaerobes and their abundance within each DFU were explored for relationships between infection severities. The abundance of anaerobes was similar across mild DFIs (abundance = 29.5%, ± 31) moderate DFIs (abundance = 20.5%, ± 22.3) and severe DFIs (abundance = 27.3%, ± 21), indicating there exist no differences between patients presenting with more severe infections and a greater abundance of anaerobes (p = 0.6).
Fig. 7.
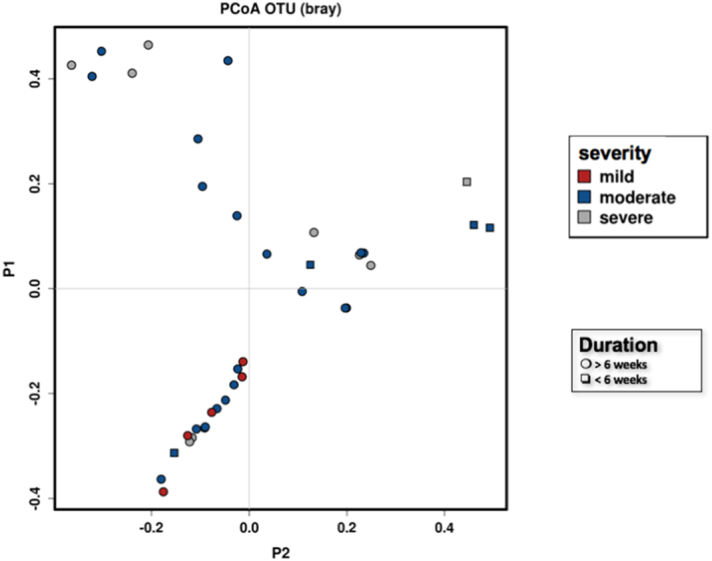
PCoA Bray-curtis plot demonstrates the community structure difference between infection severities in addition to defining the duration of DFU.
3.5. Neuropathic and Neuroischaemic Diabetic Foot Ulcers Harbor Similar Microbiomes
Twenty-three patients were classified as having neuropathic DFUs (normal TBI ranges ≥ 0.9–<1.2 and MNDS ≥ 6) and sixteen patients were classified as having neuroischaemic DFUs (TBI < 0.7 to 0.3, and MNDS ≥ 6). Non-parametric approaches identified no difference in the mean average abundances between neuropathic and neuroischaemic DFUs (S6).
3.6. Culture Dependent Methods Underestimate Anaerobic Microorganisms
Kappa coefficients were used to determine the level of agreement between culture-dependent methods and DNA sequencing. Agreement in the identification of obligate anaerobes was poor between culture and DNA sequencing (p = 0.4). Culture underestimated obligate anaerobe presence in 90% of samples (detection of obligate anaerobes by culture = 4 of 39, 10% vs detection of obligate anaerobes by DNA sequencing = 34 of 39, 79%).
3.7. Treatment Parameters and Outcomes
In total, nineteen patients (49%) during the study period experienced treatment failure. Of the thirty-three patients who had DFUs > 6 weeks, fifteen (45%) with moderate to severe IDSA infections experienced treatment failure. In the group of five patients with DFUs < 6 weeks, four patients (80%) with moderate IDSA infections experienced treatment failure. These infections were mono-microbial and were predominated by high frequency taxa of either Staphylococcus spp. and Streptococcus spp. Correlation coefficients were used to explore if DFUs containing high relative abundances of commonly cited pathogens of infection in DFI (S. aureus, S. agalactiae and A. baumannii) were at greater risk of treatment failure. This revealed the presence of S. agalactiae in DFUs (regardless of duration of DFU) were associated with greater treatment failures (P < 0.007). PERMANOVA revealed no further differences in the community structures between patients who failed therapy and those who experienced treatment success (p = 0.2).
In patient samples where obligate anaerobes were identified as contributing to the overall wound microbiome at levels greater than both 30% and 50%, there were no increased tendency towards failing therapy. The type of ant-infective therapy provided to patients in this study provided adequate anaerobe cover (25 of 39 wounds received antimicrobials with anaerobe cover, 64%) and this may explain the lack of significance between high relative abundance of anaerobes in DFUs and no increased tendency to fail therapy.
Thirteen patients (33.3%) received narrow spectrum antimicrobials with nine (23.1%) of these patients having DFUs > 6 weeks duration. Four of these nine patients (44.4%) experienced treatment failure whilst receiving narrow spectrum antimicrobials, but the five remaining patients with DFUs > 6 weeks on narrow spectrum antimicrobials experienced treatment success. The number of cases were too low for statistical analysis. Twenty-five patients received broad-spectrum antimicrobials with eleven patients (44%) experiencing treatment failure. There were no correlations between a tendency to fail therapy whether on narrow spectrum or broad-spectrum antimicrobials.
A GLM was performed to identify any predictors of treatment failure independent of the microbiome. These identified patients having a low TBI (< 0.7) as being the only predictor of failure regardless of the microbiome (p = 0.01). No other clinical factors such as a level of glycosylated haemoglobin greater than ≥ 7% (p = 0.72) or the severity of infection were correlated to treatment failure (Mild, p = 0.13, Moderate, p = 0.65, Severe, p = 0.26).
4. Discussion
In the context of managing diabetes foot infections from an infectious disease viewpoint, current guidelines based on culture-dependent data, are now subject to the scrutiny of molecular DNA based approaches. Furthermore, studies employing amplification and sequence analysis of the 16S rRNA gene to characterize the microorganisms involved in DFI, none to date have sampled participants with overt clinical signs of infection. Instead the available data report on chronic, new or recurrent DFUs that are clinically non-infected (Dowd et al., 2008a, Dowd et al., 2008b, Gardner et al., 2013, Price et al., 2009, Smith et al., 2016, Wolcott et al., 2015). Given the increasing utilisation of genomic analysis from both a clinical and research domain, it is essential to understand the microbiota of clinically infective DFUs and if current anti-infective practices can be improved through the translation of complex bioinformatics arising from the DNA analysis of microbial communities. We analysed robust microbiota datasets from Infections of the feet in people with diabetes, and detailed their clinical outcomes, relating this back to current anti-infective practices. We found that the duration of a DFU prior to presenting with a new clinical infection may help clinicians decide on the antimicrobial regimen of choice.
We identify Staphylococci spp. as the most commonly sequenced dominant bacteria in approximately one third of DFUs in this study, followed closely by Corynebacterium spp. In a recent review by our group on the bacteriology of DFUs from both a molecular and culture based approach (Malone et al., 2016), the predominant pathogen/s of infection for DFI was S. aureus. Additionally, Corynebacterium spp., Streptococcus spp., and obligate anaerobes belonging to Clostridiales family XI all identified as major players in this study were similarly identified in studies of chronic non-infected wounds. Based on our molecular data and that of previous molecular and culture based publications, current guidelines that promote the use of antimicrobials targeting Gram-positive aerobic cocci as a first line treatment are appropriate.
Corynebacterium spp. has provided a continuing source of debate regarding its role as a non-pathogenic skin commensal (Citron et al., 2007), or as a pathogen of infection in the presence of an in immunocompromised patient (Dowd et al., 2008a, Dowd et al., 2008b, Uçkay et al., 2015). In this study, we seldom identified the presence Corynebacterium spp. as a sole pathogen (High frequency taxa), with it almost exclusively occurring in combination with other known pathogens of DFI. This suggests that there may be a role for Corynebacterium spp. as part of a polymicrobial infection. Given that many first line antimicrobials of choice for DFI are active against this microorganism, there may not be a requirement to target this sole microorganism unless a mono-microbial culture is identified.
Community structure is essentially the composition of a community, including the number of species in that community, their relative numbers (Richness) and their complexity (Diversity). We identify that the duration of DFU is a major driver behind the microbiome, with longer duration DFUs typically having greater species richness and diversity. This correlated to increased relative abundances of Gram-negative proteobacteria and reduced relative abundances of firmicutes in a pattern previously described by Gardener et al. on neuropathic non-infected DFUs (Gardner et al., 2013). Proteobacteria are commonly identified in wounds (Dowd et al., 2008a, Wolcott et al., 2015) and largely belong to the Pseudomonadaceae and Enterobacteriaceae families. It is unclear from our data if these microorganisms require special attention, for example P. aeruginosa was present as minor taxa in only one quarter of samples (eight DFUs), thus supporting the general consensus (Lipsky et al., 2012) that P. aeruginosa is not a typical pathogen of infection in DFI (excluding southern hemisphere locations) (Sivanmaliappan and Sevanan, 2011), and may not require additional tailored therapy should it be identified through cultivation based methods.
Obligate anaerobes were also identified in 90% of DFUs, but great heterogeneity existed between patients with regards to their relative abundances. In most DFUs, obligate anaerobes co-existed in high abundances with aerobic microorganisms, suggesting that obligate anaerobes likely play a role as co-pathogens in DFI. Current microbiology laboratories do not employ enhanced culturing methods to isolate many of the obligate anaerobes identified in this study through DNA sequencing. However, even in the absence of culture-dependent guidance, many commonly utilised antimicrobials for DFI are active against obligate anaerobes.
Furthermore, there are no studies exploring if additional anti-anaerobic therapy improves DFI outcome, and in this study, we find no correlation between the high relative abundance of obligate anaerobes and a greater tendency to fail antimicrobial therapy. The decision to use targeted antimicrobials against obligate anaerobes by clinicians should be administered under the guidance of antimicrobial stewardship (Lipsky et al., 2016a, Lipsky et al., 2016b). The pattern of antimicrobial therapy prescribing in this study were based on specialist Infectious disease physicians with experience of managing these complex patients, but these results may reflect differently when managed by non-specialist clinicians with limited exposure to these wounds. Further work in this area is required.
The current guidance materials available to clinicians managing DFIs are predominantly based on culture-dependent data, yet this study employing DNA sequencing techniques re-enforces most of this data as being clinically relevant (Lipsky et al., 2012, Lipsky et al., 2016a, Lipsky et al., 2016b). Pyogenic cocci were predominant in acute DFUs in this study, and thus, supports directed narrow spectrum antimicrobial regimens (with consideration for culture sensitivities looking for MRSA). DNA sequencing methods however, highlighted the limitation of conventional bacterial cultures with regards to the microbial diversity and ability to isolate microorganisms not detected under standard clinical microbiology laboratory protocols. Many of these microorganisms were found in chronic wounds that harboured flora similar to the environment, suggesting patients expose their wounds to an array of environments (i.e. barefoot walking, showering with no wound dressing, gardening etc.). Therefore, patient education is vital in order of minimizing exposure of DFUs to environmental contaminants and opportunistic pathogens of infection.
Current guidelines for classifying and managing infected DFUs provide guidance (in conjunction with local policies and patterns of microbial sensitivities for resistance) on the duration and route of delivery of antimicrobials based on infection severity (Lipsky et al., 2012, Lipsky et al., 2016a, Lipsky et al., 2016b). The use of broad-spectrum antimicrobials delivered parenterally is promoted for severe DFIs, owing to the polymicrobial nature of infection. We confirm severe DFIs are extremely diverse, polymicrobial and complex, and our data supports current clinical practice by parenteral, broad-spectrum antimicrobials is warranted. Exploration from a larger sub-set of patients with severe DFI composing of low frequency taxa profiles, may provide greater insight into managing these challenging infections.
Previous reports have suggested that DFUs complicated by peripheral arterial disease (i.e. ischemic or neuroischemic) likely lead to an altered wound microenvironment and thus microbiota (Gardner et al., 2013). Additionally, the presence of peripheral vascular disease as a comorbid variable in the presence of an infected foot in a person with diabetes has been reported as well known independent predictor of poor outcome (Hinchliffe et al., 2016, Prompers et al., 2008). Sixteen patients in this study had neuroischemic ulcers (TBI < 0.7 to 0.3) with most DFUs presenting with mild to moderate peripheral arterial disease. We ascertained that both neuropathic and neuroischemic (patients with mild to moderate PAD) DFUs harbor similar microbiomes and the requirement to segregate these differing wound aetiologies may not be required for microbiota analysis when using a TBI cut off value of 0.5 as an arbitrary marker.
Nineteen patients during the study period experienced antimicrobial treatment failure, but no differences existed in the microbiome of patients who failed therapy and those who experienced treatment success. Furthermore patients who were treated with either narrow spectrum or broad-spectrum antimicrobials experienced similar failure rates (44.4% versus 44%) and this suggests that other factors are likely at play including the host immune response to infection, patient compliance in adhering to therapy and or peripheral perfusion. A general linear model approach identified that the presence of a TBI < 0.7 was an independent predictor of treatment failure regardless of the microbiota, or antimicrobial, emphasising the difficulties in managing these complex infected wounds.
Whilst our microbiome data identifies DFUs of greater than six-weeks duration presenting with a new clinical infection (includes mild-moderate-severe, with no discrimination) are often polymicrobial, with exception to nearly always targeting aerobic Gram-positive cocci, the requirement to also target provide additional anti-anaerobe therapy requires further research.
Furthermore, whilst DNA sequencing provides an extended view of the microbiome, it is limited in providing information on “which microorganisms” maybe directly contributing to infection. This is increasingly important when analysing our data set where regardless of the spectrum of activity of antimicrobials (i.e. Narrow or broad-spectrum), patients experienced similar outcomes. It may be possible in a highly diverse microbiota, that narrow spectrum antimicrobials targeting pyogenic cocci alone, is enough to reduce the virulence/pathogenicity of infective symptoms without the requirement to use a scatter gun approach to target everything broadly. The use of whole genome sequencing may allow us to better understand this question (Malone et al., 2016).
This paper provides a useful insight into the bacterial communities in infected DFUs and reflects on treatment outcomes of anti-infective therapy and if molecular based data would have altered therapeutic regimens. The paper is limited by the sample size and thus recommendations based on molecular data are not possible. A larger cohort of patients would provide greater detail and where possible analysis of small subsets of interest. This paper also identifies the difficulties with obtaining species level data when using the 16s rRNA subunit. Furthermore, what is strikingly apparent from our data is that whilst we provide an extended view of “which microorganism/s” are present, we cannot be definitive on “which microorganism/s” are responsible as contributing as pathogens of infection. The era of “metagenomics” and whole genome sequencing that can analyse genes responsible for virulence or pathogenicity may unveil these answers.
Funding Source
M. Malone received a $45,000 Early Career Research Grant from South West Sydney Local Health District to undertake this project.
Conflict of Interests
MM is a paid consultant for Smith and Nephew LTD and has received grant funding from Mundi Pharmaceuticals.
Author Contributions
MM, HD, SJ, IG, KV conceived the study, and MM, HD and IG acted as principal site investigators and were responsible for patient care and acquisition and quality of the data. SJ, KV, KJ, HH were responsible for performing laboratory based activities for DNA methods. MM, SJ and KJ undertook molecular data analysis. All authors were involved in interpretation of the data and participated in writing of the manuscript.
Acknowledgements
The work was supported by a $45,000 Early Career Research Grant from South West Sydney Local Health District (MM). The authors thank the Department of Microbiology and Infectious Diseases, Sydney South West Pathology Service, New South Wales Health Pathology, Liverpool, Sydney, Australia, for performing semi-quantitative bacterial cultures of the tissue biopsy specimens.
Footnotes
Supplemental information includes a detailed methodology of DNA sequence workflow and statistical analysis. Supplementary data associated with this article can be found in the online version, at doi: http://dx.doi.org/10.1016/j.ebiom.2017.06.026.
Appendix A. Supplementary Data
Supplementary material
References
- Abbott C.A., Carrington A.L., Ashe H., Bath S., Every L.C., Griffiths J., Boulton A.J.M. The north-west diabetes foot care study: incidence of, and risk factors for, new diabetic foot ulceration in a community-based patient cohort. Diabet. Med. 2002;19(5):377–384. doi: 10.1046/j.1464-5491.2002.00698.x. [DOI] [PubMed] [Google Scholar]
- Citron D.M., Goldstein E.J., Merriam C.V., Lipsky B.A., Abramson M.A. Bacteriology of moderate-to-severe diabetic foot infections and in vitro activity of antimicrobial agents. J. Clin. Microbiol. 2007;45 doi: 10.1128/JCM.00551-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd S., Sun Y., Secor P., Rhoads D., Wolcott B., James G., Wolcott R. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 2008;8(1):43. doi: 10.1186/1471-2180-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd S.E., Wolcott R., Sun Y., McKeehan T., Smith E., Rhoads D. Polymicrobial nature of chronic diabetic foot ulcer biofilm infections determined using bacterial tag encoded FLX amplicon pyrosequencing (bTEFAP) PLoS One. 2008;3(10):3326. doi: 10.1371/journal.pone.0003326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field N., Cohen T., Struelens M.J., Palm D., Cookson B., Glynn J.R., Abubakar I. Strengthening the reporting of molecular epidemiology for infectious diseases (STROME-ID): an extension of the STROBE statement. Lancet Infect. Dis. 2014;(4):341–352. doi: 10.1016/S1473-3099(13)70324-4. [DOI] [PubMed] [Google Scholar]
- Gardner S.E., Hillis S.L., Heilmann K., Segre J.A., Grice E.A. The neuropathic diabetic foot ulcer microbiome is associated with clinical factors. Diabetes. 2013;62(3):923–930. doi: 10.2337/db12-0771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice E.A., Kong H.H., Renaud G., Young A.C., Bouffard G.G., Blakesley R.W., Segre J.A. A diversity profile of the human skin microbiota. Genome Res. 2008;18 doi: 10.1101/gr.075549.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchliffe R.J., Brownrigg J.R.W., Apelqvist J., Boyko E.J., Fitridge R., Mills J.L., International Working Group on the Diabetic Foot IWGDF guidance on the diagnosis, prognosis and management of peripheral artery disease in patients with foot ulcers in diabetes. Diabetes Metab. Res. Rev. 2016;32:37–44. doi: 10.1002/dmrr.2698. [DOI] [PubMed] [Google Scholar]
- Lavery L.A., Armstrong D.G., Wunderlich R.P., AJM Boulton, Tredwell J.L. Diabetic foot syndrome: evaluating the prevalence and incidence of foot pathology in Mexican Americans and non-Hispanic whites from a diabetes disease management cohort. Diabetes Care. 2003;26(5):1435–1438. doi: 10.2337/diacare.26.5.1435. [DOI] [PubMed] [Google Scholar]
- Lipsky B.A., Aragón-Sánchez J., Diggle M., Embil J., Kono S., Lavery L., International Working Group on the Diabetic Foot IWGDF guidance on the diagnosis and management of foot infections in persons with diabetes. Diabetes Metab. Res. Rev. 2016;32:45–74. doi: 10.1002/dmrr.2699. [DOI] [PubMed] [Google Scholar]
- Lipsky B.A., Berendt A.R., Cornia P.B., Pile J.C., Peters E.J.G., Armstrong D.G., Senneville E. 2012 infectious diseases society of America clinical practice guideline for the diagnosis and treatment of diabetic foot infections. Clin. Infect. Dis. 2012;54(12):e132–e173. doi: 10.1093/cid/cis346. [DOI] [PubMed] [Google Scholar]
- Lipsky B.A., Dryden M., Gottrup F., Nathwani D., Seaton R.A., Stryja J. Antimicrobial stewardship in wound care: a position paper from the British Society for antimicrobial chemotherapy and European wound management association. J. Antimicrob. Chemother. 2016 doi: 10.1093/jac/dkw287. [DOI] [PubMed] [Google Scholar]
- Malone M., Gosbell I.B., Dickson H.G., Vickery K., Espedido B.A., Jensen S.O. Can molecular DNA-based techniques unravel the truth about diabetic foot infections? Diabetes Metab. Res. Rev. 2016 doi: 10.1002/dmrr.2834. [DOI] [PubMed] [Google Scholar]
- Malone M., Johani K., Jensen S.O., Gosbell I.B., Dickson H.G., McLennan S., Hu H., Vickery K. The effect of cadexomer iodine on the microbial load and diversity of chronic non-healing DFUs complicated by biofilm in vivo. J. Antimicrob. Chemother. 2017 doi: 10.1093/jac/dkx099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oates A., Bowling F.L., Boulton A.J.M., Bowler P.G., Metcalf D.G., McBain A.J. The visualization of biofilms in chronic diabetic foot wounds using routine diagnostic microscopy methods. J. Diabetes Res. 2014;2014:8. doi: 10.1155/2014/153586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price L.B., Liu C.M., Melendez J.H., Frankel Y.M., Engelthaler D., Aziz M., Zenilman J.M. Community analysis of chronic wound bacteria using 16S rRNA gene-based pyrosequencing: impact of diabetes and antibiotics on chronic wound microbiota. PLoS One. 2009;4(7):e6462. doi: 10.1371/journal.pone.0006462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prompers L., Schaper N., Apelqvist J., Edmonds M., Jude E., Mauricio D., Huijberts M. Prediction of outcome in individuals with diabetic foot ulcers: focus on the differences between individuals with and without peripheral arterial disease. The EURODIALE study. Diabetologia. 2008;51(5):747–755. doi: 10.1007/s00125-008-0940-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads D.D., Cox S.B., Rees E.J., Sun Y., Wolcott R.D. Clinical identification of bacteria in human chronic wound infections: culturing vs. 16S ribosomal DNA sequencing. BMC Infect. Dis. 2012;12:321. doi: 10.1186/1471-2334-12-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads D.D., Wolcott R.D., Sun Y., Dowd S.E. Comparison of culture and molecular identification of bacteria in chronic wounds. Int. J. Mol. Sci. 2012;13(3):2535–2550. doi: 10.3390/ijms13032535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivanmaliappan T.S., Sevanan M. Antimicrobial susceptibility patterns of Pseudomonas aeruginosa from diabetes patients with foot ulcers. Int. J. Microbiol. 2011;2011:605195. doi: 10.1155/2011/605195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K., Collier A., Townsend E.M., O'Donnell L.E., Bal A.M., Butcher J., Williams C. One step closer to understanding the role of bacteria in diabetic foot ulcers: characterising the microbiome of ulcers. BMC Microbiol. 2016;16:54. doi: 10.1186/s12866-016-0665-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uçkay I., Aragón-Sánchez J., Lew D., Lipsky B.A. Diabetic foot infections: what have we learned in the last 30 years? Int. J. Infect. Dis. 2015;40:81–91. doi: 10.1016/j.ijid.2015.09.023. [DOI] [PubMed] [Google Scholar]
- Wolcott R.D., Hanson J.D., Rees E.J., Koenig L.D., Phillips C.D., Wolcott R.A., White J.S. Analysis of the chronic wound microbiota of 2,963 patients by 16S rDNA pyrosequencing. Wound Repair Regen. 2015 doi: 10.1111/wrr.12370. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material