Abstract
Breast cancer represents a major health challenge. The majority of breast cancer deaths are due to cancer progression/recurrence for which no efficient therapies exist. Aggressive breast cancers are characterized by loss of cellular differentiation. Defining molecular mechanisms/targets contributing to cancer aggressiveness is needed to guide the design of new screening and targeted treatments. Here, we describe a novel tumor promoting function for the Cleavage and Polyadenylation Factor-6 (CPSF6). Importantly, aggressive breast cancer cells of luminal B, HER2-overexpressing and triple negative subtypes show dependency on CPSF6 for viability and tumorigenic capacity. Mechanistically, we found CPSF6 to interact with components of the A-to-I RNA editing machinery, paraspeckles and ADAR1 enzyme, and to be required for their physical integrity. Clinically, we found CPSF6 and all core paraspeckles proteins to be overexpressed in human breast cancer cases and their expression to correlate with poor patient outcomes. Finally, we found prolactin, a key mammary differentiation factor, to suppress CPSF6/RNA editing activity. Together, this study revealed CPSF6 as a molecular target with clinical relevance for prognosis and therapy in breast cancer.
Keywords: Paraspeckles, A-to-I RNA editing, Long non-coding RNA, Prolactin, Differentiation, Mammary
Highlights
-
•
Aggressive breast cancer cells require CPSF6 for viability and tumorigenesis.
-
•
CPSF6 and paraspeckles core proteins are associated with poor outcome in breast cancer.
-
•
Prolactin hormone reprograms cells to suppress CPSF6 interaction with paraspeckles and ADAR1, A-I RNA editing machinery.
Breast tumors of luminal B, HER2-enriched, and basal (triple negative) subtypes are associated with poor differentiation and aggressive behavior. Defining targets contributing to cancer aggressiveness may offer new modalities for prognosis and therapy. This study demonstrated the critical role of CPSF6 for survival and tumorigenic capacity of aggressive breast cancer cells. CPSF6 is required for the physical integrity of the A-to-I RNA editing complex, paraspeckles and ADAR1 enzyme. CPSF6 and core paraspeckles proteins are biomarkers of poor patient outcome. In addition, prolactin hormone was found to suppress CPSF6 function. This study highlights CPSF6 as a therapeutic target in breast cancer.
1. Introduction
Microarray-based gene profiling studies of breast tumors validated that breast cancer is a heterogonous disease and can be sub-classified into different subtypes exhibiting distinct molecular, phenotypic and clinical outcomes (Perou et al., 2000, Sorlie et al., 2001). Importantly, molecular subtyping has also emphasized the well-known role of tumor differentiation status in predicting tumor behavior and patient outcome (Prat and Perou, 2011, Sonnenblick et al., 2014, Voduc et al., 2010). Indeed, tumors of luminal A subtype, typically of low grade and well-moderately differentiated phenotype, are associated with a low risk of tumor progression and recurrence. In contrast, tumors of luminal B, HER2-enriched, and basal (triple negative) subtypes, predominantly of high grade and poor differentiation, are associated with high risk of local and regional tumor relapse. Therefore, it is essential to gain a better understanding of the biological role and clinical impact of differentiation pathways in tumorigenesis. Identifying these differentiation mechanisms and networks may offer new modalities for prognosis and therapy for breast cancer patients.
In the breast tissue, prolactin (PRL) hormone plays a key role in mammary gland development and differentiation (Goffin and Touraine, 2015, Liu et al., 2015). The role of PRL in breast tumorigenesis is not fully elucidated. Recent studies have revealed however, that expression of this hormonal differentiation pathway sub-classifies breast cancer patients with better survival outcomes and functionally this pathway may suppress breast tumorigenesis (Hachim et al., 2016a, Hachim et al., 2016b, Lopez-Ozuna et al., 2016) (Bonuccelli et al., 2012, Haines et al., 2009, Lopez-Ozuna et al., 2016, Nouhi et al., 2006, Nukumi et al., 2007, Peck et al., 2012, Yamashita et al., 2006). Elucidating the mechanisms underlying these protective effects of PRL may identify novel molecular targets for prevention, screening and treatment in breast cancer.
Cleavage factor I (CFIm) is an RNA binding protein complex identified originally as a central player in alternative cleavage and polyadenylation process (Brown and Gilmartin, 2003, Millevoi and Vagner, 2010, Ruegsegger et al., 1996). This protein complex is composed of a large subunit CFIm68 designated as CPSF6 and a small subunit CFIm25 designated as Nudt21 (CPSF5). CPSF6 and Nudt21 complex binds RNA and facilitates RNA looping allowing alternative mRNA 3′ end processing (Yang et al., 2011). CFIm protein complex, however, appears to be part of another ribonucleo-protein complex involved in RNA processing designated as paraspeckles (Dettwiler et al., 2004, Naganuma et al., 2012). The core proteins of paraspeckles are paraspeckles component 1 (PSPC1), splicing factor proline and glutamine-rich (SFPQ) and non-POU domain containing octamer binding (P54nrb). They are members of DBHS (Drosophila melanogaster behavior human splicing) family and they are built on long noncoding RNA designated as NEAT1 forming the paraspeckles RNA-protein complex (Bond and Fox, 2009). The specific function of paraspeckles remains unclear. However, paraspeckles are shown to be involved in regulating gene expression through nuclear retention of adenosine-to-inosine (A-to-I) RNA edited molecules. While these edited RNA molecules do not immediately produce proteins they are post-transcriptionally cleaved to rapidly release a translation-competent mRNA upon cellular stress (Prasanth et al., 2005). Importantly, recent elegant studies have revealed extensive A-to-I RNA editing and high expression of the adenosine deaminase enzymes (ADARs) in various cancers including breast cancer contributing to tumor transcriptomic diversity and tumorigenesis (Fumagalli et al., 2015, Han et al., 2015, Paz-Yaacov et al., 2015). Regulatory mechanisms and extracellular ligands controlling this pro-oncogenic A-to-I RNA editing process in breast cancer is still to be discovered.
Here we describe a previously unknown function for CPSF6 in breast cancer. Our results show that in contrast to luminal A, CPSF6 is critical for luminal B, HER-2 overexpressing and triple negative aggressive breast cancer cell viability and tumorigenic capacity. At the molecular level, we demonstrate that CPSF6 is a key component of the recently described pro-oncogenic A-to-I RNA editing machinery through physical interactions with paraspeckles and ADAR1. Moreover, we show that formation of this CPSF6/paraspeckles/ADAR1 ribo-nucleo-protein complex to be enriched in the aggressive breast cancer cells in comparison to the less aggressive cells. Significantly, the A-to-I RNA editing machinery displayed physical dependency on CPSF6 in aggressive breast cancer cells. Additionally, we found CPSF6 as well as core paraspeckles proteins to be highly expressed in breast cancer clinical cases and to be associated with poor patient outcomes including relapse and distant metastasis free survival. Finally, we show PRL hormone to suppress this pro-oncogenic pathway in aggressive breast cancer cells highlighting the important role of differentiation pathways in tumor suppression. Together, our study defined CPSF6 to play a vital role in breast cancer aggressiveness providing novel strategies for prognosis and therapy in aggressive breast cancer.
2. Material and Methods
2.1. Cell Culture
Human breast cancer cells: MDA-MB-231 obtained from Dr. Shafaat Rabbani, McGill University, MDA-MB-453, SKBR3 and BT474 obtained from Dr. Morag Park, McGill University. MDA-MB-231, MDA-MB-453, SKBR3 and MCF7 cells were maintained in DMEM media (Multicell Invitrogen) containing 10% fetal bovine serum (FBS) (Multicell Invitrogen). BT474 was maintained in RPMI–1640 (Multicell Invitrogen) containing 10% FBS. Normal mammary epithelial cells: mouse HC11 cells were obtained from N. Hynes (Friedrich Miescher Institute, Basel, Switzerland) and were maintained in RPMI–1640 containing 10% FBS.
2.2. CPSF6 Knock-Down in Human Breast Cancer Cells
Cells (MDA-MB-231, SKBR3, BT474 and MCF7) were infected with lentiviral particles expressing human shRNA against CPSF6 or scramble shRNA. The scramble shRNA in pLKO.1 vector was obtained from Addgene (Addgene plasmid #1864) and human CPSF6 MISSION shRNA Bacterial GlycerolStock (#TRCN0000237833) (CCGGGTTGTAACTCCATGCAATAAACTCGAGTTTATTGCATGGAGTTACAACTTTTTG) and (#TRCN0000244314) (CCGGGGTGATTATGGGAGTGCTATTCTCGAGAATAGCACT CCCATAATCACCTTTTTG) were obtained from Sigma. Stable cell lines were then generated using puromycin selection (InvivoGen) 1 μg/ml puromycin for MDA-MB-231 and SKBR3 cells and 2 μg/ml for BT474 and MCF7 cells.
2.3. Antibodies, Plasmids and Reagents
Antibodies used were: anti-CPSF6 rabbit monoclonal antibody (abcam #ab175237), anti-Nudt21 mouse monoclonal antibody (Santa-Cruz #sc-81109), anti-SFPQ rabbit polyclonal antibody (abcam#ab38148) anti-P54nrb rabbit polyclonal antibody (Santa-Cruz # sc-67016), anti-PSPC1 rabbit polyclonal antibody (Santa-Cruz # sc-84576), anti-ADAR1 rabbit polyclonal antibody (abcam #ab126755), and anti-GAPDH mouse polyclonal antibody (Santa-Cruz # sc-365062).
Secondary antibodies used were goat anti-rabbit IgG HRP (Santa-Cruz #sc-2004). As well, goat anti-mouse IgG-HRP (Santa-Cruz #sc-2005). Secondary antibodies for confocal immunofluorescence studies were: donkey anti-rabbit IgG (H + L) Fluor 546 (Invitrogen) and donkey anti-mouse Fluor 488 (Invitrogen).
The dilutions of antibodies for western blotting analysis are as indicated: 1: 1000 for all primary antibodies except for CPSF6 (1: 10,000). The dilutions for secondary antibodies for western blotting analysis are 1:5000. For immunofluorescence staining: 1:100 for primary antibodies and 1: 200 for secondary antibodies.
Other reagents used include: Recombinant human prolactin (rhPRL) (250 ng/ml) used for cell stimulation was purchased from Feldan Therapeutics (1F-02-008), SosoFast EvaGreen Supermix (Bio-Rad # 172-5201), protein A-Sepharose beads (Amersham Biosciences and GE Healthcare), protein A and G magnetic beads (Thermo Scientific # 88802), 12-well plates HTS multi-well insert system format (BD Falcon) and 96-well plates (Corning #3753 and Fisher #7201216).
2.4. Tissue Microarray
Tissue microarrays (TMA) (BIOMAX, BC081120) and (Pantomics, BRC1021) including 197 invasive ductal carcinoma cases and 15 normal and benign tissues were used. Both TMAs include information including; age, grade, stage and TNM. Additional information about ER, PR, HER-2 and Ki-67 status was also available for (Pantomics, BRC1021) TMA. The virtual H & E slides for those cases were available and were reviewed by a pathologist to confirm the diagnosis and that they are representative of the tumor.
2.5. Immunohistochemistry
Paraffin embedded slides were applied for immunohistochemical staining. Slides were baked for 30 min at 55C, followed by deparaffinization then rehydration. Antigen retrieval was performed in sodium citrate 10 mM, pH 6.0 buffer. The slides were incubated in hydrogen peroxide block for 10 min, followed by Ultra V Block for 5 min. Slides were incubated with a rabbit anti-CPSF6 monoclonal Antibody (abcam#ab175237) or rabbit anti-SFPQ polyclonal Antibody (abcam#ab38148). UltraVision LP Detection System HRP Polymer & DAP Plus Chromogen (Thermo Fisher Scientific, Fremont CA) was used for detection. The TMA slides were scanned using Aperio XT slide scanner (Leica Biosystems).
2.6. Immunohistochemistry Scoring
Quantitative IHC scoring systems were used to evaluate CPSF6 and SFPQ immunostaining. In brief, a representative pathologist-annotated malignant regions were selected for each core using images of 40 × magnification from digital IHC-stained TMA slides. The mean positive pixel count (PPC) for each representative region was obtained using positive pixel count (PPC) algorithm (Aperio). The lower PPC count indicates higher IHC staining intensity. The staining intensity was divided equally into 4 categories. For CPSF6 cases with PPC ≥ 95 considered + 3, 96–110 PPC considered + 2, 111–127 considered + 1 and cases with PPC ≤ 128 considered as score 0. For evaluation of SFPQ cases PPC score ≥ 132 considered + 3, 133–151 considered + 2, 152–170 considered + 1 and ≥ 170 considered as 0 score. ER, PR, HER-2 and Ki67 classification into molecular subtypes was done as previously described (Hachim et al., 2016a).
2.7. Gene Expression Analyses
Publically available (ONCOMINE) and (GOBO) databases were used to determine associations between CPSF6, P54NRB, PSPC1 and SFPQ m-RNA expression levels and different clinicopathological parameters in large human breast cancer cohorts. KM plotter and GOBO databases were used to determine associations of gene expression in relation to patient outcome. Affymetrix ID numbers: CPSF6: 202469_s_at, P54NRB: 200057_s_at, PSPC1: 222611_s_at and SFPQ: 201585_s_at.
2.8. Cell Lysis and Western Blotting Analyses
Total protein lysates were obtained by SDS lysis buffer (150 mM sodium chloride, 1 mM Na2 EDTA, 0.5% sodium deoxycholate, 1% SDS, 1 mM Na3VO4 and Protease inhibitors cocktail). 30 μg proteins were loaded onto SDS-PAGE gel and transferred onto nitrocellulose membranes. Membranes were then incubated with the relevant primary antibodies and secondary antibodies. 20 μg proteins were loaded onto SDS-PAGE gel.
2.9. Immunofluorescence
Cells were grown on coverslips coated with poly-d-lysine hydrobromide (Santa-Cruz) for 24 h. Cells were washed then fixed 15 min with 4% PFA at room temperature, followed by 5 min incubation with 0.1% Triton X-100 (Fisher) in PBS. Cells were subsequently immunostained with primary antibody for an overnight period at 4 °C. Cells were then, incubated with secondary antibody and Dapi for 1 h at room temperature in the dark. Coverslips were then mounted on slides with mounting media (Lerner # 13800) and stored at 4 °C. Experiments involving hPRL treatment, cells were grown on coverslips coated with poly-d-lysine for 24 h. Cells were then starved in media containing 2% FBS for an overnight period and then treated with Prolactin for 72 h. Confocal microscopy was performed using Zeiss LSM 780 confocal microscope equipped with a Plan-Apochromat × 63-1.4 oil immersion objective.
2.10. RNA–Fluorescence In Situ Hybridization
Cells were grown on coverslips coated with poly-d-lysine hydrobromide (Santa-Cruz) for 24 h. Cells were washed then fixed 15 min with 3% PFA at room temperature, followed by 5 min incubation with 0.1% Triton X-100 (Fisher) in PBS. Cells were washed twice in 2 × SSC for 5 min. Then cells were subsequently hybridized with human NEAT1 probe (BioSEARCH TECHNOLOGIES CAT # SMF-2036-1) overnight period at 4 °C. Cells were washed three in 2 × SSC for 5 min at 42 °C. Then cells were incubated with 2 × SSC containing 0.2 mg/ml DAPI. Then, Cells were washed twice in 2 × SSC for 5 min. Finally, mount the coverslips on a slide with mounting medium. Confocal microscopy was performed using Zeiss LSM 780 confocal microscope equipped with a Plan-Apochromat × 63-1.4 oil immersion objective.
2.11. Co-Immunoprecipitation
Total protein lysates were obtained by RIPA lysis buffer (50 mM Tris pH 8, 150 mM sodium chloride, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM Na3VO4 and Protease inhibitors cocktail). For endogenous proteins, 0.1 μg of anti-CPSF6 antibody were bounded to 20 μl mixed protein A/G beads and incubated with 0.6 ml of cell lysates for 3 h at 4 °C. The beads were washed three times with IP buffer. Eluted proteins were subjected to SDS-PAGE and detected using specific antibodies.
2.12. RNA-Immunoprecipitations
RNA IP was performed according to (Imprint – RNA immunoprecipitation protocol, SIGMA-ALDRICH). Cells (3.106) were washed with cold PBS. Then the cell pellets were resuspended in lysis buffer containing ribonuclease inhibitor and protease inhibitor Cocktail. Cell lysates were incubated with primary antibody (CPSF6) or antibody IgG for 3 h followed by 1 h with protein A and G magnetic beads. Then, Cells were lysed in 500 ml of trizol and RT-PCR amplification of cDNA was carried out using the following primer pairs: NEAT1 forward GCCTTGTAGATGGAGCTTGC, NEAT1 reverse TGTACCTCCCAGCGTTTAG. NEAT1_2 forward CTCTCCATTTCCCCATCTGA, NEAT1_2 reverse GCTGCTGCCAAACATCTACA. AZIN1 forward AGGGAGCCTTGGTTTGTTTT, AZIN1 reverse CCAGTGGGAATCTGTGTGTG.
2.13. RNA Isolation and RT-qPCR
HC11 cells were grown to confluence then allowed to undergo differentiation for 1 day in media containing 10% FBS, insulin and hydrocortisone. Cells were then starved or treated with ovine PRL (sigma) for 24 h. Cells were lysed in 1 ml of trizol. Total RNA was isolated as described by the manufacturer (Abcam, United States). RNA concentrations were quantified by Nanodrop at 260 nm. Total RNA 1 μg was used for reverse transcription by using (iScript Reverse Transcription supermix kit # 170–8841). Real-time monitoring of PCR amplification of cDNA was carried out using the following primer pairs:
GAPDH forward CCTCAACTACATGGTTTAC, GAPDH reverse GGGATTTCCATTGATGAC. CPSF6 forward CACCACAACCTTCACCTCC, CPSF6 reverse AGAGATGGGTGGATCCACCT. Csn2 (β-casein) forward GCTCTTGCAAGGGAGGTAT, Csn2 (β-casein) reverse GCATTTGGGGCACTATAGG.
2.14. MTT Assay
5.103 cells were seeded into 96-well plate and grown for a period 2 to 8 days. Then, cells were incubated with 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) at 37 °C for 2 h.
2.15. Caspase 3–7 Assay
5.103 cells were seeded into white 96-well plate and grown for a 72 h. Caspase-Glo® 3–7 Assay from Promega was used according to manufacturer's instructions protocol.
2.16. Soft Agar Transformation Assay
30.103 cells were seeded into 24-well plate coated with 1% agar gel and grown in growth media with 0.6% agar for 3 weeks. Colonies were stained by 0.05% crystal blue. Number of colonies was counted using low power lens microscopy.
2.17. NOD-SCID Mouse Xenografts
12 Female NOD-SCID mice were purchased from Charles River Laboratories (Sain-Constant, QC, Canada), housed and maintained under specific pathogen-free conditions (RI-MUHC animal facility). The mice were randomly divided into two groups (n = 6 mice per group). At 9 to 10 weeks of age, the first group was injected in the forth-left mammary fat pad with 1 × 106 MDA-MB-231-Scr and the second group was injected with MDA-MB-231-Sh-CPSF6. Tumor growth was monitored up to 16 weeks after injection. When tumors were detectable, tumor size was measured with a vernier caliper (Mitutoyo, Kawasaki, Japan) and calculated using the formula [length + width2] / 2. Mice were sacrificed by CO2 asphyxiation.
2.18. Statistical Analyses
Statistical analyses were performed using GraphPad prism 6 software using Student's t-test or one-way ANOVA analysis accordingly. In addition, the results of CPSF6 and SFPQ immunoreactivity were tabulated and the relation of their expression with different clinical and pathologic parameters was performed using Chi square test. Results were shown as mean ± SEM and P < 0.05 was considered as cut-off for significant association.
3. Results
3.1. Aggressive Breast Cancer Cells Show Dependency on CPSF6 for Survival and Tumorigenesis and Distinct Sub-nuclear Localization
Previous large gene profiling analysis of PRL-induced mammary epithelial cell differentiation program, identified components of CFIm both Nudt21 and CPSF6, to be novel PRL down-regulated target genes (Hachim et al., 2016b). Indeed, PRL treatment of mammary epithelial cells resulted in suppression of m-RNA expression of not only Nudt21 (Hachim et al., 2016b) but also CPSF6 (Supplementary Fig. S1A) implicating a possible role for CFIm protein complex in breast tumorigenesis. To address the role of CFIm in breast cancer we first examined the expression of CFIm proteins CPSF6 and Nudt21 in human breast cancer cells representative of the clinically relevant breast cancer molecular subtypes including, luminal A (MCF7), luminal B (BT474 cells), HER-2 overexpressing (SKBR3) and triple negative (MDA-MB-231). While we observed relatively equal levels of CPSF6 and Nudt21 protein expression in all breast cancer cell lines (Supplementary Fig. S1B), significantly, however, distinctive patterns of nuclear sub-localization of CPSF6 were observed (Fig. 1A). Indeed, CPSF6 was found to localize mainly in the nuclear periphery in the less aggressive luminal A cells, similar to normal mouse luminal mammary epithelial cells (Supplementary Fig. S1C). In contrast, CPSF6 showed diffuse nuclear localization in the more aggressive breast cancer subtypes including luminal B, HER-2 overexpressing and triple negative breast cancer cells. Nudt21, on the other hand, showed diffuse nuclear localization in all normal and cancerous cell lines tested (Fig. 1A and Supplementary Fig. S1C). Moreover, scatterplot analyses of CPSF6 and Nudt21 immunofluorescence signals in normal mammary and less aggressive cancer cells exhibited sporadic scatterplot (indicative of low co-localization) (coefficient of co-localization, R-value ~ 0.2), whereas, all aggressive breast cancer subtypes showed a linear scatterplot (indicative of high co-localization) (coefficient of co-localization, R-value ~ 0.8) (p < 0.0001) (Supplementary Fig. S1D and Fig. 1B). Moreover, using co-immunoprecipitation studies we found that aggressive breast cancer cells, MDA-MB-231 and SKBR3, displayed increased CPSF6/Nudt21 interaction in comparison to the less aggressive luminal A MCF7 cells (Fig. 1C). Together, these results indicate that CFIm complex formation, primarily determined by the sub-nuclear localization of CPSF6, is significantly enriched in aggressive breast cancer cells and may play a role in aggressive breast cancer behavior.
Fig. 1.
CPSF6 intracellular localization and role in breast cancer cells viability and tumorigenic capacity.
A. Confocal immunofluorescence images of CPSF6 (red), Nudt21 (green) and nucleus (Dapi) (blue) of breast cancer cells MCF7, BT474, SKBR3 and MDA-MB-231. Scale bars, 10 μm.
B. Pearson correlation coefficient (Correlation R) measurements of CPSF6 (red) and Nudt21 (green) by confocal microscopy in breast cancer cells as indicated in legend. Results are expressed as mean ± SEM of three independent experiments. ***p < 0.001.
C. MDA-MB-231, SKBR3 and MCF7 cells were lysed and immunoprecipitations using a mouse monoclonal antibody against Nudt21 or control normal mouse IGg were performed. Western blotting was carried out using a monoclonal antibody against CPSF6 (upper panel). Membrane was reprobed using a monoclonal antibody against Nudt21 (lower panel).
D. Immunoblot analysis of total cell lysates of MDA-MB-231-Scr (control) & MDA-MB-231-Sh1-CPSF6, SKBR3-Scr (control) & SKBR3-Sh1-CPSF6, BT474-Scr (control) & BT474-Sh1-CPSF6 and MCF7-Scr (control) & MC7-Sh1-CPSF6 cells using antibodies against CPSF6 and GAPDH.
E. MTT assays were performed in MDA-MB-231-Scr & MDA-MB-231-Sh1-CPSF6, SKBR3-Scr & SKBR3-Sh1-CPSF6, BT474-Scr & BT474-Sh1-CPSF6 and MCF7-Scr & MCF7-Sh1-CPSF6 for 2, 3, 4 and 8 days. Results are expressed as mean ± SEM of triplicates of three independent experiments. ***p < 0.001, ns: not significant.
F. Colony formation assays were performed using MDA-MB-231-Scr & MDA-MB-231-Sh1-CPSF6, SKBR3-Scr & SKBR3-Sh1-CPSF6, BT474-Scr & BT474-Sh1-CPSF6 and MCF7-Scr & MCF7-Sh1-CPSF6 for a period of three weeks. Results are expressed as mean ± SEM of triplicates of three independent experiments. ***p < 0.001.
G. Caspases 3–7 activity assays were performed in MDA-MB-231-Scr & MDA-MB-231-Sh1-CPSF6, SKBR3-Scr & SKBR3-Sh1-CPSF6, BT474-Scr & BT474-Sh1-CPSF6 and MCF7-Scr & MCF7-Sh1-CPSF6 following 72 h incubation in growth media. Results are expressed as mean ± SEM of triplicates of three independent experiments. **p < 0.01, *p < 0.05, ns: not significant.
H. Left panel, tumor volume measurements of NOD-SCID xenografts of MDA-MB-231-Scr and MDA-MB-231-Sh1-CPSF6 followed for a period of 70 days. Right panel, images of mice of MDA-MB-231-Scr xenografts (black arrow heads indicate tumor) (upper panel) and mice of MDA-MB-231-Sh1-CPSF6 xenografts showing no tumor development (lower panel). H & E staining of primary tumor of MDA-MB-231-Scr tumor (10 × and 20 ×).
Since CPSF6 but not Nudt21 showed differential sub-nuclear localization in relation to breast cancer aggressiveness we next focused on assessing the role of CPSF6 in regulating breast tumorigenesis. We employed lentiviral ShRNA technology, to suppress CPSF6 expression in the various human breast cancer cell lines using two independent shRNAs targeting CPSF6 (Fig. 1D and Supplementary Fig. S2A). Crucially, we found that loss of CPSF6 significantly decreased cell viability, suppressed colony formation capacity and induced apoptosis in all the aggressive breast cancer subtypes, including MDA-MB-231, SKBR3 and BT474 breast cancer cells. This was in contrast to the results observed in the less aggressive luminal A MCF 7 cell line, where loss of CPSF6 gene expression had no effect on cell viability, colony formation capacity neither induction of caspase activity (Fig. 1E–G and Supplementary Fig. S2B–D). These results highlight a central role for CPFS6 in regulating viability of aggressive breast cancer cells.
To address the role of CPSF6 in driving breast tumorigenesis, we next examined the effects of loss of CPSF6 gene expression on the tumorigenic potential of the highly aggressive MDA-MB-231 cells using a mammary fat-pad orthotopic mouse model. While 4/6 mice injected with the control MDA-MB-231 cells expressing the scrambled ShRNA developed large tumors within the mammary fat-pad, none of the (0/6) mice injected with MDA-MB-231 cells expressing the CPSF6 specific ShRNA developed tumors (Fig. 1H). These results clearly demonstrate the important role and contribution of CPFS6 to survival and tumor formation of aggressive breast cancer cells.
3.2. CPSF6 is a Clinically Relevant Marker of Breast Cancer Aggressiveness and Poor Patient Outcome
To evaluate the clinical relevance of this previously undescribed tumor promoting role of CPSF6 in human breast cancer, we next evaluated CPSF6 protein expression in human breast cancer TMAs. These TMAs are composed of 197 invasive ductal carcinomas (IDC) and 15 normal and benign cores (Supplementary Table S1 and Supplementary Fig. S3A–C). Interestingly, using a quantitative IHC scoring system based on positive pixel count (PPC) algorithm (Aperio), we found high CPSF6 expression (Scores + 2 & + 3) in 125/179 (69.8%) breast cancer cases compared to 4/12 (33.34%) normal and benign cases (p = 0.02) (Fig. 2A and B) suggesting increased expression of CPSF6 in breast cancer cases. To further expand this finding, next we analyzed CPSF6 mRNA expression levels in three different ONCOMINE breast cancer datasets including Curtis (1700 cases), TCGA (450 cases) and Sorlie (97 cases) (Rhodes et al., 2004). Importantly, we found CPSF6 mRNA levels to be significantly higher in invasive breast cancer cases compared to normal breast tissue (p = 3.07E − 35, p = 2.98E − 27 and p = 0.017 respectively) (Fig. 2C, D and E). Together, this data emphasizes the up-regulation of CPSF6 protein and m-RNA expression in human breast cancer clinical cases.
Fig. 2.
CPSF6 expression and prognostic value in breast cancer.
A. Positive immunohistochemical staining of CPSF6 in normal adjacent tissue, in situ and invasive breast cancer lesions (10 × and 40 ×).
B. Associations between CPSF6 protein expression and different clinicopathological parameters.
C. CPSF6 mRNA expression levels in 144 normal and 1556 invasive breast cancer cases using Curtis dataset of ONCOMINE database.
D. CPSF6 mRNA expression levels in 61 normal and 389 invasive breast cancer cases using TCGA dataset of ONCOMINE database.
E. CPSF6 mRNA expression levels in 4 normal and 93 invasive breast cancer cases using Sorlie dataset of ONCOMINE database.
F. Representative immunohistochemical staining of CPSF6 in breast cancer cases of grades I, II and III (10 × and 40 ×).
G. CPSF6 mRNA expression levels stratified according to tumor grade in a cohort of 1402 cases using GOBO database.
H. CPSF6 mRNA expression levels in association with breast cancer molecular subtypes stratified according to PAM50 sub-classification method in 1881 human breast cancer samples using GOBO database.
I. CPSF6 mRNA expression levels in association with breast cancer molecular subtypes stratified according to Hu et al. sub-classification method in 1881 human breast cancer samples using GOBO database.
J. Kaplan-Meier survival curves of CPSF6 gene expression in association with patient outcome (3554 patients, KM-plotter database) using RFS as an end point.
K. Kaplan-Meier survival curves of CPSF6 gene expression in association with patient outcome (1609 patients, KM-plotter database) using DMFS as an end point.
We next analyzed CPSF6 protein expression in relation to critical parameters related to tumor aggressiveness and progression, including tumor grade, tumor size and stage. In relation to tumor grade, interestingly, CPSF6 protein expression was found to be significantly higher in the poorly differentiated grade III tumors 19/21 (90.4%) compared to the moderately-differentiated grade II 67/106 (63.2%) and well-differentiated grade I 24/34 (70.5%) tumors (p = 0.04) (Fig. 2B and F). To further validate this finding using a large patient number, we examined CPSF6 mRNA levels in association with tumor grade in a cohort of 1411 breast cancer patients using GOBO database, a publicly available database allowing the study of gene expression levels in association with a range of clinicopathological parameters in human breast cancer samples(Ringner et al., 2011) (Fig. 2G). We found CPSF6 mRNA levels to be significantly higher in grade III tumors compared to grades I and II tumors (p = 1e − 05).
Other important independent prognostic parameters in breast cancer are tumor size and stage. Large tumor size and advanced stage are associated with poor patient outcome. Interestingly, using the TMAs described above, higher CPSF6 protein expression was also found to associate with larger tumor size, T3 31/35 (88.5%) compared to smaller T2 70/103 (67.96%) & T1, 8/13 (61.53%) tumors (p = 0.09) (Fig. 2B). This association with larger tumor size was further confirmed using 166 breast cancer cases of Sorlie dataset in ONCOMINE database showing higher CPSF6 mRNA expression in the larger size T4 tumors (Supplementary Fig. S3D). In addition, Sorlie dataset revealed a strong association between higher CPSF6 mRNA levels and tumor metastasis (Supplementary Fig. S3E). Further analysis of the TMA data and Curtis dataset (2136 cases) revealed higher CPSF6 gene expression in tumors of advanced stages (Fig. 2B and Supplementary Fig. S3F). Together, this data suggest that CPSF6 expression is associated with more advanced and aggressive tumors.
We then investigated whether CPSF6 expression correlated with the different molecular and histological subtypes of breast cancer. We found high CPSF6 protein expression in all the different histological and molecular subtypes of breast cancer, indicative of a broad and important role for CPFS6 in breast tumorigenesis (Supplementary Fig. S4A and B). Further analysis using large human breast cancer samples available in GOBO database (1881 patients) revealed that higher CPSF6 mRNA levels in the highly aggressive basal-like and HER-2 enriched subtypes compared with the luminal subtypes (Fig. 2H and I). Taken together, our data demonstrate an important association between higher CPSF6 and poor classical clinicopathological parameters in human breast cancer implicating CPSF6 as a clinically relevant marker of the aggressive breast cancer phenotype.
Finally, to address the clinical relevance of CPSF6 as a prognostic marker for breast cancer patient survival, we analyzed the association between CPSF6 mRNA levels and patient outcomes in relation to relapse free survival (RFS) and distant metastasis free survival (DMFS). For this we used Kaplan Meier plotter as well as GOBO databases which allow monitoring of survival of a large number of breast cancer patients for > 10 years(Gyorffy et al., 2010, Ringner et al., 2011). Interestingly, we found high CPSF6 mRNA levels to be significantly associated with reduced RFS and reduced DMFS (Fig. 2J, K and Supplementary Fig. S5A, B). To exclude any potential interference on the correlation between CPSF6 and patient outcomes that could result from treatment with chemotherapeutic agents, we analyzed the prognostic role of CPSF6 in systemically untreated patients. Interestingly, CPSF6 expression was also significantly associated with poor RFS and DMFS outcomes in untreated patients (Supplementary Fig. S5C and D). Altogether, these results highlight the clinical relevance of CPSF6 as a novel biomarker of poor prognosis and strongly underscore its role in promoting aggressive breast cancer behavior.
3.3. Paraspeckles Expression Associates With Aggressive Breast Cancer Phenotype and Poor Patient Outcome
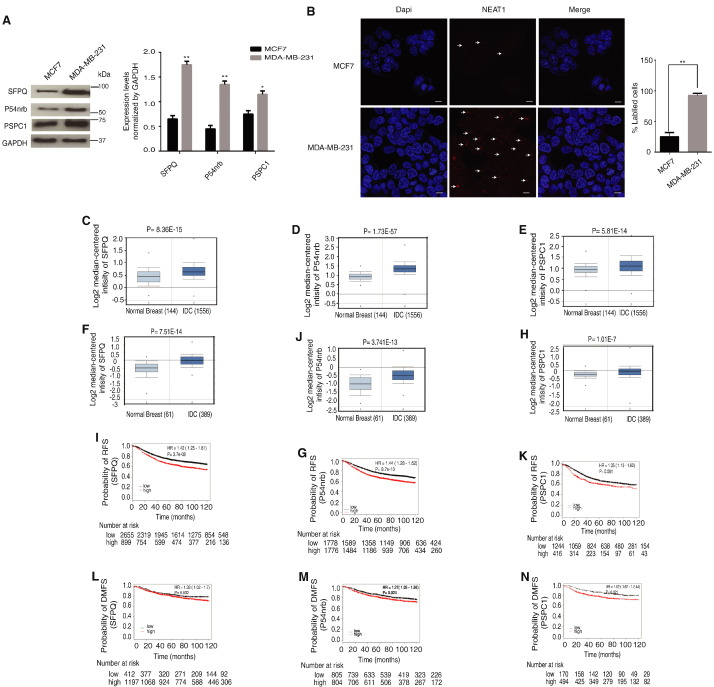
Paraspeckles is a nuclear subdomain consists of a protein complex built on lncRNA (NEAT1). The role of paraspeckles in breast tumorigenesis is still to be fully elucidated. However, NEAT 1 expression has been shown to contribute breast tumorigenesis and to correlate with poor patient outcome (Choudhry et al., 2015). Interestingly, a possible link between CFIm and this ribo-nuclear protein complex was previously proposed (Dettwiler et al., 2004, Naganuma et al., 2012). This data together promoted us to investigate whether paraspeckles are involved with CPSF6 in breast cancer. Therefore, we next examined the protein expression levels of the core paraspeckles proteins, SFPQ, P54nrb and PSPC1 as well as NEAT1 RNA in the less aggressive luminal A MCF7 cells in comparison to the more aggressive MDA-MB-231 cells. Interestingly, MDA-MB-231 cells showed higher expression levels of all paraspeckles proteins as well as NEAT1 RNA in comparison to MCF7 cells (Fig. 3A and B) implicating a possible role for paraspeckles in breast cancer. Next we aimed to establish the clinical relevance of paraspeckles in breast cancer clinical cases. To do this, we investigated the mRNA expression levels of the core paraspeckles proteins (SFPQ, P54nrb and PSPC1) in two different large datasets including Curtis (1700 cases) and TCGA (450 case) (Fig. 3C–H). Interestingly, m-RNA levels of all paraspeckles core proteins were significantly upregulated in invasive breast cancer compared to normal tissue and to be significantly associated with poor patient outcome in relation to RFS and DMFS (Fig. 3I–N).
Fig. 3.
Paraspeckles protein components are associated with poor outcome in breast cancer.
A. Immunoblot blot analysis of total cell lysates of breast cancer cells using antibodies against SFPQ, P54nrb, PSPC1 and GAPDH.
B. Confocal images of NEAT1 RNA–FISH (red) with nuclear DAPI counterstain (blue) in MCF7 and MDA-MB-231 cells. Results are expressed as mean ± SEM of triplicate of two independent experiments. **p < 0.01. Scale bars, 10.
C–E. SFPQ, P54nrb and PSPC1 and mRNA levels in 144 normal and 1556 invasive breast cancer cases using Curtis dataset of ONCOMINE database.
F–H. SFPQ, P54nrb and PSPC1 mRNA expression levels in 61 normal and 389 invasive breast cancer cases using TCGA dataset of ONCOMINE database.
I–K. Kaplan-Meier survival curves of SFPQ, P54nrb and PSPC1 mRNA levels in association with patient outcome using RFS as an end point in breast cancer patients (3554, 1660 and 3554 respectively) (KM-plotter database).
L–N. Kaplan-Meier survival curves SFPQ, P54nrb and PSPC1 mRNA levels in association with patient outcome using DMFS as an end point in breast cancer patients (1609, 664 and 1609 respectively) (KM-plotter database).
To further validate the clinical relevance of paraspeckles in breast cancer, next we examined the expression of a member of the paraspeckles proteins, SFPQ, using the TMA described above (Supplementary Fig. S3A (left panel)). Importantly, we found SFPQ to be up regulated in malignant tissues 54/95 (56.8%) compared to normal adjacent tissue 4/9 (44.4%) (Fig. 4A and B). Moreover, high SFPQ protein expression was found in poorly differentiated tumors 5/7 (71.42%) compared to well-differentiated tumors 7/18 (38.89%) (p = 0.09) (Fig. 4B and C). This was further confirmed using GOBO (1441 case) and Curtis (2154 case) datasets indicating higher SFPQ mRNA levels in the poorly differentiated grade III tumors compared to well-moderately differentiated tumors (p < 0.00001) (Fig. 4D and E). We also observed an important association between high SFPQ mRNA levels and advanced tumor stage in Curtis dataset (2154 cases) (Fig. 4F). Further analysis using GOBO database (1881 patients) revealed significantly higher SFPQ mRNA levels in the aggressive basal-like and HER-2 enriched subtypes compared with the luminal subtypes (Fig. 4G and H). These results indicate that SFPQ is highly expressed in breast cancer and is associated with poor clinicopathological parameters and aggressive breast cancer phenotype. Finally, we examined whether CPSF6 and SFPQ show co-expression in the breast cancer cases present in the TMAs used (Supplementary Fig. S3A (left panel)). Importantly, we found the majority 44/53 (77%) of CPSF6 positive breast cancer cases were also positive for SFPQ, whereas, only 11/38 (28.9%) of SFPQ positive cases in CPSF6 negative group (p = 4.1E-06) (Fig. 4I). Altogether, these results highlight paraspeckles proteins as biomarkers of poor prognosis and implicate a pro-oncogenic role for CPSF6-paraspeckleas in human breast cancer.
Fig. 4.
SFPQ protein expression in association with different clinicopathological parameters using large cohorts of breast cancer clinical cases.
A. Positive immunohistochemical staining of SFPQ in normal and invasive breast cancer lesions (10 × and 40 ×).
B. Associations between SFPQ protein expression and different clinicopathological parameters.
C. Representative immunohistochemical staining of SFPQ expression in breast cancer cases of different grades (10 × and 40 ×, 100 ×).
D. SFPQ mRNA levels stratified according to tumor grade in 1402 breast cancer patients using GOBO database.
E. SFPQ mRNA levels stratified according to tumor grade in 2136 breast cancer patients using Curtis dataset of ONCOMINE database.
F. SFPQ mRNA expression levels stratified according to tumor stage in 2136 human breast cancer cases using Curtis dataset of ONCOMINE database.
G. SFPQ mRNA expression levels in association with breast cancer molecular subtypes stratified according to Hu et al. sub-classification method in 1881 human breast cancer sample using GOBO database.
H. SFPQ mRNA expression levels in association with breast cancer molecular subtypes stratified according to PAM50 sub-classification method in 1881 human breast cancer sample using GOBO database.
I. Association between CPSF6 and SFPQ expression in TMA (BC081120) containing 100 IDC and 10 normal adjacent tissues.
3.4. CPSF6 Forms a Complex With A-to-I RNA Editing Machinery in Breast Cancer
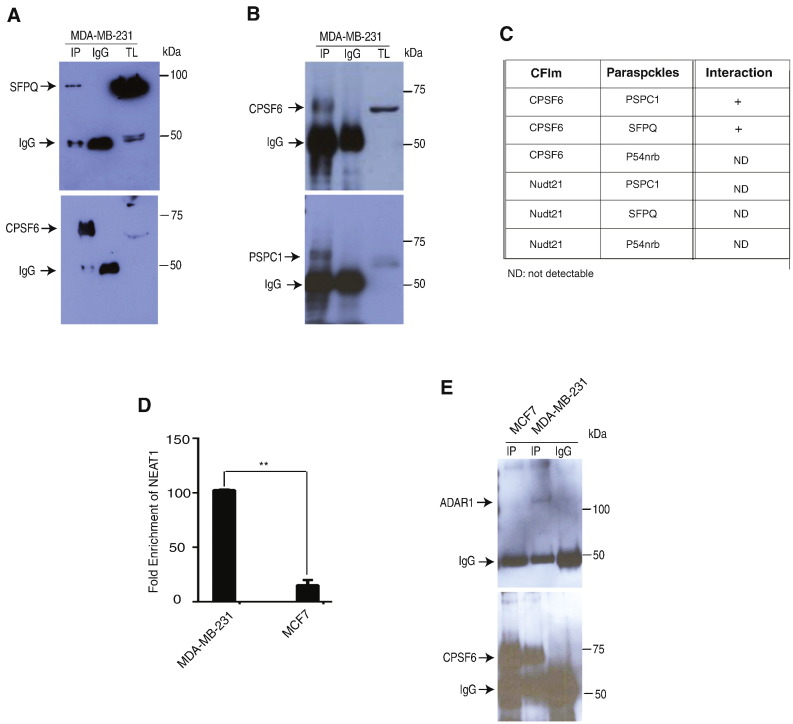
A-to-I RNA editing has recently been shown to be up-regulated in tumors of various types. Paraspeckles and ADAR1 enzyme are two known components of this pro-oncogenic process. Here we hypothesized that CPSF6 may interact and regulate this mechanism in aggressive breast cancer. Therefore, we screened for possible interactions between CPSF6 and Nudt21 with paraspeckles proteins (SFPQ, PSPC1 and P54nrb) by co-immunoprecipitation experiments. Using the aggressive MDA-MB-231 cells we found CPSF6 to physically interact with paraspeckles core proteins SFPQ and PSPC1 but not with p54nrb (Fig. 5A–C). On the other hand, we did not detect physical interactions between Nudt21 and the core paraspeckles proteins (Fig. 5C). To elaborate on this data, we next examined CPSF6 interaction with lncRNA NEAT1 using RNA-immunoprecipitation assays. Importantly, our results showed a significant enrichment of NEAT1 in immunoprecipitates of CPSF6 in the aggressive MDA-MB-231 cells in comparison to the less aggressive MCF7 cells (Fig. 5D). These results indicate that CFIm forms a complex with paraspeckles through CPSF6 in aggressive breast cancer cells. Next, we investigated whether CPSF6 can also physically interacts with the enzyme ADAR1. Indeed, we found a physical interaction between CPSF6 and ADAR1 protein in MDA-MB-231 cells but not in MCF7 luminal A cells (Fig. 5E). Together these results support the conclusion that CPSF6 is a component of the A-to-I RNA editing machinery. As well, our data emphasize that aggressive breast cancer cells display increased CPSF6, paraspeckles and ADAR1 assembly and is in agreement with the previously described tumor promoting role of A-to-I RNA editing in breast cancer.
Fig. 5.
CPSF6/Paraspeckles/ADAR1 protein complex in aggressive breast cancer.
A. MDA-MB-231 cells were lysed and immunoprecipitations using a rabbit monoclonal antibody against CPSF6 or control normal rabbit IGg were performed. Western blotting was carried out using a polyclonal antibody against SFPQ (upper panel). Membrane was reprobed using a monoclonal antibody against CPSF6 (lower panel). TL: total cell lysates.
B. MDA-MB-231 cells were lysed and immunoprecipitations using a rabbit polyclonal antibody against PSPC1 or control normal rabbit IGg were performed. Western blotting was carried out using a monoclonal antibody against CPSF6 (upper panel). Membrane was reprobed using a polyclonal antibody against PSPC1 (lower panel). TL: total cell lysates.
C. Summary of CFIm and paraspeckles proteins physical interactions in MDA-MB-231 cells.
D. CPSF6 immunoprecipitates of MDA-MB-231 and MCF7 cells were subjected to RT-qPCR using NEAT1 primers. Results are expressed as mean ± SEM of triplicates of two independent experiments. **p < 0.01.
E. MDA-MB-231 and MCF7 cells were lysed and immunoprecipitations using a monoclonal antibody against CPSF6 or control normal rabbit IGg were performed. Western blotting was carried out using polyclonal antibody against ADAR1 (upper panel). Membrane was reprobed using a monoclonal antibody against CPSF6 (lower panel).
3.5. CPSF6 is Required for Maintaining the Physical Integrity of Paraspeckles and ADAR1
To further characterize the role of CPSF6 in breast cancer, next we investigated whether CPSF6 exerts regulatory role on CFIm, paraspeckles protein complex and ADAR1 in breast cancer cells. Therefore, we analyzed the effects of loss of CPSF6 expression on the protein levels of Nudt21 and the components of the A-to-I RNA editing machinery paraspeckles and ADAR1. Remarkably, loss of CPSF6 in MDA-MB-231 cells led to a significant decrease in Nudt21 protein levels based on immunofluorescence and western blotting analyses (Fig. 6A). Importantly, loss of CPSF6 also resulted in a significant loss of gene expression of NEAT1 RNA and protein levels of all core paraspeckles proteins as well as ADAR1 enzyme (Fig. 6B–D). Notably, suppressing CPSF6 expression in the other aggressive breast cancer cell lines SKBR3 and BT474 also resulted in loss of protein levels of Nudt21, members of the paraspeckles core proteins, NEAT1 RNA levels as well as ADAR1 protein (Supplementary Fig. S6A–G). In contrast, there was no significant change in the levels of Nudt21 protein, NEAT1 RNA, paraspeckles proteins as well as ADAR1 following knock-down of CPSF6 in the luminal A MCF7 cells (Fig. 6E and Supplementary Fig. S6H). Together, these results indicate that CPSF6 plays a vital role in maintaining the physical integrity of CFIm, paraspeckles and ADAR enzyme in aggressive breast cancer cells.
Fig. 6.
CPSF6 is essential for maintaining physical integrity of CFIm, paraspeckles and ADAR1.
A. Confocal immunofluorescence images (left panel) and immunoblot analysis (right panel) of MDA-MB-231-Scr and MDA-MB-231-Sh1-CPSF6 immunodetected using antibodies to CPSF6 and Nudt21. Quantification of western blots was performed for three independent experiments and expressed as mean ± SEM. ***p < 0.001. Scale bars, 10 μm.
B. Confocal images of NEAT1 RNA–FISH (red) with nuclear DAPI counterstain (blue) in MDA-MB-231-Scr and MDA-MB-231-Sh1-CPSF6 cells. Scale bars, 10 μm.
C. Immunoblot analysis of MDA-MB-231-Scr and MDA-MB-231-Sh1-CPSF6 using antibodies against SFPQ, p54nrb, PSPC1 and GAPDH (control). Right panels represent quantification of protein expression levels normalized by the control. Results are expressed as mean ± SEM of three independent experiments. **p < 0.01.
D. Confocal immunofluorescence images (right panel) and immunoblot analysis (left panel) of MDA-MB-231-Scr and MDA-MB-231-Sh1-CPSF6 using antibody against ADAR1. Quantifications of western blots were performed for three independent experiments normalized by the control (GAPDH) and results are expressed as mean ± SEM. **p < 0.01, ***p < 0.01. Scale bars, 10 μm.
E. Confocal immunofluorescence images (upper panel) and immunoblot analyses (lower panel) of MCF7-Scr and MCF7-Sh1-CPSF6 using antibodies against CPSF6, Nudt21, PSPC1, ADAR1 and GAPDH. Quantifications of western blots were performed for three independent experiments normalized by the control and results are expressed as mean ± SEM. **p < 0.01 and ns: not significant. Scale bars, 10 μm.
3.6. Prolactin Regulation of CPSF6/A-to-I RNA Editing Process
The above data indicate that in aggressive breast cancer cells CPSF6 within CFIm is important in maintaining two A-to-I RNA processing mechanisms; A-to-I RNA editing and A-to-I RNA nuclear retention. Our results also indicate that under these conditions CPSF6 is indispensable for the survival and tumorigenic potential of aggressive breast cancer cells. This information renders CPSF6 a candidate for the development of cancer therapeutic modalities that can specifically inhibit survival of malignant cells. Due to the effects of PRL in suppressing CFIm gene expression in mammary epithelial cells and the recent evidence suggesting a tumor suppressor function of PRL in breast carcinogenesis, we hypothesized that PRL hormone may regulate CPSF6 function and thereby A-to-I RNA editing process. Importantly, our results showed that PRL treatment of aggressive breast cancer cells including MDA-MB-453, a representative of TNBC-PRLR subgroup(Lopez-Ozuna et al., 2016), SKBR3 and BT474 resulted in the re-localization of CPSF6 from the nucleoplasm to the nuclear periphery similar to that seen in luminal mammary epithelial cells and luminal A breast cancer cells (Fig. 7A). On the other hand, no change in the nuclear localization of Nudt21 was observed. This was also accompanied by a significant decrease in the co-localization of CPSF6 and Nudt21 (Fig. 7B). This data indicates that PRL signaling regulates CPSF6 sub-nuclear localization and suggests that indeed PRL may regulate A-to-I RNA editing process. Therefore, we next examined the ability of PRL to regulate the interaction between CPSF6 and NEAT1 RNA. Indeed, treatment of MDA-MB-453 and SKBR3 cells with PRL led to a significant reduction in CPSF6 and NEAT1 interaction (Fig. 7C). Similar results were obtained examining CPSF6 interaction with the long isoform of NEAT1 (NEAT1_2) (Fig. 7D). Moreover, PRL also suppressed CPSF6/ADAR1 complex formation (Fig. 7E). This data demonstrates that PRL suppresses CPSF6 interaction with paraspeckles complex and ADAR1 enzyme in breast cancer cells. To further elaborate on these findings, we examined the ability of PRL to regulate CPSF6 interaction with AZIN1 m-RNA, an important driver of tumorigenesis harboring a nonsynonymous A-to-I RNA editing event in breast cancer(Fumagalli et al., 2015, Han et al., 2015). Therefore, we performed RNA immunoprecipitation of CPSF6/AZIN1 in MDA-MB-453 and SKBR3 cell lines following PRL treatment. Crucially, we found significant down regulation of AZIN1 m-RNA levels in CPSF6 immunoprecipitations following PRL treatment of both MDA-MB-453 and SKBR3 (Fig. 7F). These results together indicate that PRL suppresses the function of CPSF6 in aggressive breast cancer cells resulting in loss of cellular A-to-I RNA editing activity further emphasizing the role of CPSF6 as a critical regulator of the pro-oncogenic A-to-I RNA editing in breast cancer.
Fig. 7.
Prolactin regulation of CPSF6/paraspeckles/ADAR1 complex assembly and A-to-I RNA editing activity.
A. Confocal immunofluorescence images of CPSF6 (red), Nudt21 (green) and nucleus (Dapi) (blue) of breast cancer cells MDA-MB-453, SKBR3 and BT474 following treatment or not with rhPRL for 72 h. Scale bars, 10 μm.
B. The correlation R measurements of co-localization of CPSF6 and Nudt21 in cells treated or not with rhPRL for 72 h. Results are expressed as mean ± SEM of three independent experiments. **p < 0.01.
C. CPSF6 immunoprecipitates of MDA-MB-453 and SKBR3 cells treated or not with hPRL for 72 h were subjected to RT-qPCR using NEAT1 primers (error bars indicate SEM of triplicate for two independent experiments), (**p < 0.01, *p < 0.05).
D. CPSF6 immunoprecipitates of MDA-MB-453 and SKBR3 cells treated or not with rhPRL for 72 h were subjected to RT-qPCR using NEAT1_2 primers (error bars indicate SEM of triplicates of two independent experiments) (*p < 0.05).
E. MDA-MB-453 and SKBR3 cells treated or not with hPRL for 72 h were lysed and subjected to immunoprecipitation using antibody against CPSF6 or normal rabbit IgG followed by immunoblot analysis using antibody against ADAR1 (upper panels). Membranes were reprobed with monoclonal antibody against CPSF6 (lower panels).
F. CPSF6 immunoprecipitates of MDA-MB-453 and SKBR3 cells treated or not with hPRL for 72 h were subjected to RT-qPCR using AZIN1 primers (error bars indicate S.E.M. normalized to control of triplicates of two independent experiments), (***p < 0.01, *p < 0.05).
4. Discussion
Here we describe a central tumor-promoting role for CPSF6 in luminal B, HER2-overexrepressing and TNBC subtypes associated with poor patient outcome. CPSF6 was found to be part of the pan-cancer pro-oncogenic A-to-I RNA editing machinery through interaction with paraspeckles nuclear subdomain involved in retention of A-to-I RNA edited molecules as well as ADAR1 enzyme thus influencing A-to-I RNA editing process and tumorigenesis (Fig. 8).
Fig. 8.
CPSF6 is a clinically relevant vulnerability target in aggressive breast cancer.
In luminal B, HER2-overexpressing and triple negative aggressive breast cancer cells CPSF6 is present within the nucleoplasm in association with Nudt21, paraspeckles and ADAR1 promoting A-to-I RNA editing, cell viability and tumorigenesis. Prolactin treatment disrupts CFIm/paraspeckles/ADAR1 ribonucleo-protein complex resulting in relocalization of CPSF6 to the nuclear periphery and suppression of A-to-I RNA editing, a phenotype similar to that seen in the less aggressive luminal A breast cancer cells and normal mammary epithelial cells. Thus, CPSF6 appears as a central component of the pro-oncogenic A-to-I RNA editing process and represents a molecular target for prognosis and therapy in aggressive breast cancer.
4.1. CPSF6 as a Regulator of A-to-I RNA Editing Process and its Implication in Tumorigenesis
A-to-I RNA editing is an epigenetic mechanism that allows for transcriptomic diversity and is well known to play a role in normal physiology (Miele et al., 1995). Recent advances have indicated that A-to-I RNA editing is also globally amplified in various cancers including breast cancer (Fumagalli et al., 2015, Han et al., 2015, Paz-Yaacov et al., 2015). This RNA processing mechanism was found to be clinically relevant major driver of tumorigenesis and contributing to tumor heterogeneity. Nuclear double stranded RNA (dsRNAs) has been shown to serve as substrates for A-to-I editing by members of the ADAR enzyme family (Murayama et al., 2015). The usage of secondary alternative polyadenylation signals has been suggested to play a role in generating RNA molecules containing inverted repeat elements allowing for intramolecular annealing resulting in dsRNA molecules (Murakami et al., 2016). These inosines containing dsRNA molecules are now recognized by the paraspeckles protein complex through p54nrb resulting in the nuclear retention of edited RNAs that are then subjected to cleavage and nuclear export. Mechanisms regulating cleavage and polyadenylation of nuclear-retained A-to-I edited RNA molecules are not known. Indeed, our results showed that in aggressive breast cancer cells CPSF6 interacts with ADAR1, paraspeckles as well as AZIN1 m-RNA, a known A-to-I RNA edited molecule. Therefore, we propose that CPSF6 as part of the CFIm protein complex to mediate alternative polyadenylation and cleavage of A-to-I RNA edited molecules allowing expression of the edited molecules important for tumorigenesis.
The interaction of CPSF6 with the editing machinery was found to be dependent on CPSF6 localization within the nucleoplasm in aggressive breast cancer cells. On the other hand, we found CPSF6 to be localized within the nuclear periphery in luminal mammary epithelial cells and less aggressive luminal A breast cancer cells precluding CPSF6 interaction with the RNA editing machinery. Moreover, CPSF6 was found to be required for maintaining paraspeckles proteins and ADAR1 enzyme levels. The details of CPSF6 regulation of the integrity of the RNA editing machinery is still to be discovered.
4.2. CPSF6 as a Bio-marker and a Vulnerability Target in Aggressive Breast Cancer
Identification of biomarkers with clinical value is needed to help in patient's prognosis and stratification. Here we describe CPSF6 and core paraspeckles proteins to be co-expressed and upregulated in human breast cancer cases and their expression correlate with aggressive phenotype such as high grade and poor patient survival outcomes. These results indicate that CPSF6 and paraspeckles can stratify breast cancer patients with poor prognosis.
The principles of next generation precision therapeutics against cancer are to exploit and target cancer vulnerabilities, sparing normal cells. Thus, it is fundamental to identify cancer vulnerabilities. To maintain the malignant phenotype cancer cells relay on adapting cellular pathways (the stress phenotype) for survival. This cellular adaptation represents a potential therapeutic intervention to kill tumor cells (Luo et al., 2009, Solimini et al., 2007). Cancer cells are known to exhibit extensive A-to-I RNA editing events. This information coupled with our findings that aggressive breast cancer cells are dependent on CPSF6 for survival and tumorigenesis suggest that cancer cells are experiencing A-to-I RNA editing stress. Thus, we propose that aggressive breast cancer cells hijack CPSF6 to interact with the editing machinery to promote A-to-I RNA editing providing a survival advantage for these aggressive cells. Therefore, CPSF6 represents a vulnerability target in aggressive breast cancer.
4.3. Prolactin Regulation of CPSF6/A-to-I RNA Editing in Tumor Suppression
Mechanisms involved in cellular differentiation are normally associated with tumor suppression. While PRL is a key regulator of mammary epithelial cell differentiation its role in breast cancer is debatable (Goffin and Touraine, 2015). Nevertheless, recent developments have indicated that PRL exerts a tumor suppressor function in the breast. Indeed, PRL was found to suppress cell viability, invasive capacity and tumorigenesis of human breast cancer cells (Haines et al., 2009, Lopez-Ozuna et al., 2016, Nouhi et al., 2006). Also, expression of PRL and components of tis signaling pathway correlate with favorable patient's survival outcomes (Hachim et al., 2016a, Hachim et al., 2016b). The mechanisms and pathways that mediate PRL suppressor role are still to be identified. Here we show that PRL pro-differentiation pathway identified in mammary epithelial cells can operate in breast cancer tumor suppression. Interestingly, our results showed that PRL regulates CPSF6 nuclear sub-localization in aggressive breast cancer cells. We found that PRL treatment reverses the localization of CPSF6 to the nuclear periphery similar to that seen in less aggressive cancer cells and luminal mammary cells. Importantly PRL was also found to suppress CPSF6 interactions with paraspeckles and ADAR1 resulting in loss of A-to-I RNA editing activity. Thus, we propose that PRL hormone provides a possible therapeutic strategy to suppress A-to-I RNA editing activity and tumorigenesis in breast cancer. Collectively, our study unveiled a key vulnerability mechanism promoting the survival and tumorigenesis of aggressive breast cancer that can be exploited for the development of anti-cancer therapeutics.
Conflicts of Interest
The authors declare no conflicts of interest.
Authors' Contributions
NB: Designed, performed experiments and drafting the article. IH: Performed and analyzed IHC experiments, bioinformatics data analyses and contributed to drafting the article, JJL: Contributed design and revising the article, SA: Principal research design and supervision of the project and drafting of the article.
Acknowledgements
We would like to thank the confocal Imaging Platform of the Research Institute, McGill University Health Centre. We would like to thank Dr. Vanessa M. López Ozuna for help with animal dissection. The authors declare no competing financial interests.
J.J. Lebrun is the recipient of the McGill Sir William Dawson Research Chair. This work was supported by the Canadian Institutes of Health Research (operating grants #233437 and 233438) granted to Suhad Ali.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2017.06.023.
Contributor Information
Najat Binothman, Email: Najat.Binothman@mail.mcgill.ca.
Ibrahim Y. Hachim, Email: ibrahim.hachim@mail.mcgill.ca.
Jean-Jacques Lebrun, Email: jj.lebrun@mcgill.ca.
Suhad Ali, Email: suhad.ali@mcgill.ca.
Appendix A. Supplementary data
Supplementary material
References
- Bond C.S., Fox A.H. Paraspeckles: nuclear bodies built on long noncoding RNA. J. Cell Biol. 2009;186:637–644. doi: 10.1083/jcb.200906113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonuccelli G., Castello-Cros R., Capozza F., Martinez-Outschoorn U.E., Lin Z., Tsirigos A., Xuanmao J., Whitaker-Menezes D., Howell A., Lisanti M.P. The milk protein α-casein functions as a tumor suppressor via activation of STAT1 signaling, effectively preventing breast cancer tumor growth and metastasis. Cell Cycle. 2012;11:3972–3982. doi: 10.4161/cc.22227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K.M., Gilmartin G.M. A mechanism for the regulation of pre-mRNA 3′ processing by human cleavage factor Im. Mol. Cell. 2003;12:1467–1476. doi: 10.1016/s1097-2765(03)00453-2. [DOI] [PubMed] [Google Scholar]
- Choudhry H., Albukhari A., Morotti M., Haider S., Moralli D., Smythies J., Schödel J., Green C.M., Camps C., Buffa F. Tumor hypoxia induces nuclear paraspeckle formation through HIF-2α dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene. 2015;34:4482–4490. doi: 10.1038/onc.2014.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettwiler S., Aringhieri C., Cardinale S., Keller W., Barabino S.M. Distinct sequence motifs within the 68-kDa subunit of cleavage factor Im mediate RNA binding, protein-protein interactions, and subcellular localization. J. Biol. Chem. 2004;279:35788–35797. doi: 10.1074/jbc.M403927200. [DOI] [PubMed] [Google Scholar]
- Fumagalli D., Gacquer D., Rothe F., Lefort A., Libert F., Brown D., Kheddoumi N., Shlien A., Konopka T., Salgado R. Principles governing A-to-I RNA editing in the breast cancer transcriptome. Cell Rep. 2015;13:277–289. doi: 10.1016/j.celrep.2015.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffin V., Touraine P. The prolactin receptor as a therapeutic target in human diseases: browsing new potential indications. Expert Opin. Ther. Targets. 2015;19:1229–1244. doi: 10.1517/14728222.2015.1053209. [DOI] [PubMed] [Google Scholar]
- Gyorffy B., Lanczky A., Eklund A.C., Denkert C., Budczies J., Li Q., Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1809 patients. Breast Cancer Res. Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- Hachim I.Y., Hachim M.Y., Lopez V.M., Lebrun J.J., Ali S. Prolactin receptor expression is an independent favorable prognostic marker in human breast cancer. Appl. Immunohistochem. Mol. Morphol. 2016;24:238–245. doi: 10.1097/PAI.0000000000000178. [DOI] [PubMed] [Google Scholar]
- Hachim I.Y., Shams A., Lebrun J.J., Ali S. A favorable role of prolactin in human breast cancer reveals novel pathway-based gene signatures indicative of tumor differentiation and favorable patient outcome. Hum. Pathol. 2016;53:142–152. doi: 10.1016/j.humpath.2016.02.010. [DOI] [PubMed] [Google Scholar]
- Haines E., Minoo P., Feng Z., Resalatpanah N., Nie X.M., Campiglio M., Alvarez L., Cocolakis E., Ridha M., Di Fulvio M. Tyrosine phosphorylation of Grb2: role in prolactin/epidermal growth factor cross talk in mammary epithelial cell growth and differentiation. Mol. Cell. Biol. 2009;29:2505–2520. doi: 10.1128/MCB.00034-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L., Diao L., Yu S., Xu X., Li J., Zhang R., Yang Y., Werner H.M., Eterovic A.K., Yuan Y. The genomic landscape and clinical relevance of A-to-I RNA editing in human cancers. Cancer Cell. 2015;28:515–528. doi: 10.1016/j.ccell.2015.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Pawliwec A., Feng Z., Yasruel Z., Lebrun J.J., Ali S. Prolactin/Jak2 directs apical/basal polarization and luminal linage maturation of mammary epithelial cells through regulation of the Erk1/2 pathway. Stem Cell Res. 2015;15:376–383. doi: 10.1016/j.scr.2015.08.001. [DOI] [PubMed] [Google Scholar]
- Lopez-Ozuna V.M., Hachim I.Y., Hachim M.Y., Lebrun J.J., Ali S. Prolactin pro-differentiation pathway in triple negative breast cancer: impact on prognosis and potential therapy. Sci Rep. 2016;6:30934. doi: 10.1038/srep30934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Solimini N.L., Elledge S.J. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136:823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele A., Bandera M., Goldstein B.P. Use of primers selective for vancomycin resistance genes to determine van genotype in enterococci and to study gene organization in VanA isolates. Antimicrob. Agents Chemother. 1995;39:1772–1778. doi: 10.1128/aac.39.8.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millevoi S., Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res. 2010;38:2757–2774. doi: 10.1093/nar/gkp1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami K., Yonezawa T., Matsuki N. Synovial fluid total protein concentration as a possible marker for canine idiopathic polyarthritis. J. Vet. Med. Sci. 2016;77:1715–1717. doi: 10.1292/jvms.15-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama M.A., Kakuta S., Inoue A., Umeda N., Yonezawa T., Maruhashi T., Tateishi K., Ishigame H., Yabe R., Ikeda S. CTRP6 is an endogenous complement regulator that can effectively treat induced arthritis. Nat. Commun. 2015;6:8483. doi: 10.1038/ncomms9483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naganuma T., Nakagawa S., Tanigawa A., Sasaki Y.F., Goshima N., Hirose T. Alternative 3′-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J. 2012;31:4020–4034. doi: 10.1038/emboj.2012.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouhi Z., Chughtai N., Hartley S., Cocolakis E., Lebrun J.J., Ali S. Defining the role of prolactin as an invasion suppressor hormone in breast cancer cells. Cancer Res. 2006;66:1824–1832. doi: 10.1158/0008-5472.CAN-05-2292. [DOI] [PubMed] [Google Scholar]
- Nukumi N., Iwamori T., Kano K., Naito K., Tojo H. Reduction of tumorigenesis and invasion of human breast cancer cells by whey acidic protein (WAP) Cancer Lett. 2007;252:65–74. doi: 10.1016/j.canlet.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Paz-Yaacov N., Bazak L., Buchumenski I., Porath H.T., Danan-Gotthold M., Knisbacher B.A., Eisenberg E., Levanon E.Y. Elevated RNA editing activity is a major contributor to transcriptomic diversity in tumors. Cell Rep. 2015;13:267–276. doi: 10.1016/j.celrep.2015.08.080. [DOI] [PubMed] [Google Scholar]
- Peck A.R., Witkiewicz A.K., Liu C., Klimowicz A.C., Stringer G.A., Pequignot E., Freydin B., Yang N., Ertel A., Tran T.H. Low levels of Stat5a protein in breast cancer are associated with tumor progression and unfavorable clinical outcomes. Breast Cancer Res. 2012;14:R130. doi: 10.1186/bcr3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou C.M., Sorlie T., Eisen M.B., van de Rijn M., Jeffrey S.S., Rees C.A., Pollack J.R., Ross D.T., Johnsen H., Akslen L.A. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Prasanth K.V., Prasanth S.G., Xuan Z., Hearn S., Freier S.M., Bennett C.F., Zhang M.Q., Spector D.L. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- Prat A., Perou C.M. Deconstructing the molecular portraits of breast cancer. Mol. Oncol. 2011;5:5–23. doi: 10.1016/j.molonc.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D.R., Yu J., Shanker K., Deshpande N., Varambally R., Ghosh D., Barrette T., Pandey A., Chinnaiyan A.M. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringner M., Fredlund E., Hakkinen J., Borg A., Staaf J. GOBO: gene expression-based outcome for breast cancer online. PLoS One. 2011;6:e17911. doi: 10.1371/journal.pone.0017911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegsegger U., Beyer K., Keller W. Purification and characterization of human cleavage factor Im involved in the 3′ end processing of messenger RNA precursors. J. Biol. Chem. 1996;271:6107–6113. doi: 10.1074/jbc.271.11.6107. [DOI] [PubMed] [Google Scholar]
- Solimini N.L., Luo J., Elledge S.J. Non-oncogene addiction and the stress phenotype of cancer cells. Cell. 2007;130:986–988. doi: 10.1016/j.cell.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Sonnenblick A., Fumagalli D., Sotiriou C., Piccart M. Is the differentiation into molecular subtypes of breast cancer important for staging, local and systemic therapy, and follow up? Cancer Treat. Rev. 2014;40:1089–1095. doi: 10.1016/j.ctrv.2014.07.005. [DOI] [PubMed] [Google Scholar]
- Sorlie T., Perou C.M., Tibshirani R., Aas T., Geisler S., Johnsen H., Hastie T., Eisen M.B., van de Rijn M., Jeffrey S.S. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. U. S. A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voduc K.D., Cheang M.C., Tyldesley S., Gelmon K., Nielsen T.O., Kennecke H. Breast cancer subtypes and the risk of local and regional relapse. J. Clin. Oncol. 2010;28:1684–1691. doi: 10.1200/JCO.2009.24.9284. [DOI] [PubMed] [Google Scholar]
- Yamashita H., Nishio M., Ando Y., Zhang Z., Hamaguchi M., Mita K., Kobayashi S., Fujii Y., Iwase H. Stat5 expression predicts response to endocrine therapy and improves survival in estrogen receptor-positive breast cancer. Endocr. Relat. Cancer. 2006;13:885–893. doi: 10.1677/erc.1.01095. [DOI] [PubMed] [Google Scholar]
- Yang Q., Coseno M., Gilmartin G.M., Doublie S. Crystal structure of a human cleavage factor CFI(m)25/CFI(m)68/RNA complex provides an insight into poly(A) site recognition and RNA looping. Structure. 2011;19:368–377. doi: 10.1016/j.str.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material