Abstract
Maintaining cellular redox status to allow cell signaling to occur requires modulation of both the controlled production of oxidants and the thiol reducing networks to allow specific regulatory post-translational modification of protein thiols. The oxidative stress hypothesis captures the concept that over production of oxidants can be proteotoxic but failed to predict the recent findings that hyper-activation of the KEAP1-NRF2 system also leads to proteotoxicity. Further, sustained activation of thiol redox networks by KEAP1-NRF2 induces a reductive stress, by decreasing the lifetime of necessary oxidative post-translational modifications required for normal metabolism or cell signaling. In this context it is now becoming clear why antioxidants or hyper-activation of antioxidant pathways with electrophilic therapeutics can be deleterious. Further it suggests that the autophagy-lysosomal pathway is particularly important in protecting the cell against redox stress-induced proteotoxicity, since it can degrade redox damaged proteins without causing aberrant changes to the redox network needed for metabolism or signaling. In this context, it is important to understand (i) how NRF2-mediated redox signaling or (ii) the autophagy-mediated anti-oxidant/reductant pathways sense cellular damage in the context of cellular pathogenesis. Recent studies indicate that the modification of protein thiols plays an important role in the regulation of both the KEAP1-NRF2 and autophagy pathways. Here we discuss evidence demonstrating that the KEAP1-NRF2 pathway and autophagy act in concert to combat the deleterious effects of proteotoxicity. These findings will be discussed with a special emphasis on their impact on cardiovascular disease and neurodegeneration.
Keywords: autophagy, mitophagy, post-translational modification, thiol, reactive oxygen species, antioxidants, redox signaling, NRF2, KEAP1, p62, NDP52
Introduction
The redox dependent post-translational modification of proteins, most notably at thiol residues, are important cell regulatory and signaling mechanisms (1-4). In contrast, non-specific oxidative or nitrative damage to DNA, lipids or proteins is associated with diverse pathologies. However, contrary to the predictions of the oxidative stress hypothesis, that implies an oxidant-anti-oxidant imbalance causes non-specific accumulation of oxidative modification of protein, lipid and DNA this is not generally observed. In contrast, in living organisms, the level of modification of these biomolecules is often minor, with modifications occurring in the presence of cellular antioxidants rather than after their depletion. Importantly, non-specific accumulation of protein, lipid or DNA oxidative modifications rarely provides a specific molecular mechanism to explain toxicity or other biological effects. Furthermore, subtracting the total anti-oxidants from the total oxidants in a tissue is not predictive since biological redox regulation is inherently specific with respect both to the molecular species of antioxidant, their sub-cellular distribution and interactomes (5). These findings have led us and others to suggest that it is the loss of control of redox signaling and the gain of function associated with hyper-activation or suppression of redox cell signaling pathways or metabolism that underlies the cause of toxicity (5). Since the nuclear factor (erythroid-derived 2)-like 2 (NRF2) is important for transcriptional activation of electrophile response element-containing target genes, many encode antioxidant enzymes, a tight control of the NRF2 level itself is paramount. Indeed, the hyper-activation of the kelch-like ECH-associated protein 1 (KEAP1)-NRF2 system is not neutral or protective, as suggested by the oxidative stress hypothesis, but deleterious (6-9). Interestingly, proteotoxicity is a feature of both oxidative and reductive stress (10-12). This brings into focus the mechanisms in the cell which are designed to combat proteotoxic stress. The two most important of these are the proteasome and autophagy-lysosomal pathways (13-17). The mechanisms through which the proteasome processes oxidized proteins are well understood and have been discussed in a number of recent reviews (18-22). Here we will focus on the autophagy-lysosomal pathway and explore the recent concept that it contributes to the antioxidant protection of the cell through the removal of oxidatively damaged organelles and proteins (5;13;14;17;23) and extend this paradigm to the proteotoxicity associated with reductive stress (10-12).
How these antioxidant or anti-oxidative signals sense cellular damage is then critical to our understanding of cellular pathogenesis. It has been shown that in healthy cells, KEAP1 binds NRF2, and targets NRF2 to be ubiquitinated and degraded by the proteasome (24;25). Then in response to increased oxidative stress, cysteine thiols on KEAP1 are modified, allowing NRF2 to be released from KEAP1 and activate transcription of target genes (26). Although less well understood, it has been proposed that autophagy related proteins can also be redox regulated by cysteine thiol modifications. Recent evidence has also demonstrated significant cross regulation between KEAP1-NRF2 and autophagy activities (27-36). The redox regulation and cross regulation of KEAP1-NRF2 and autophagy activities will be reviewed below.
Redox regulation of the KEAP1-NRF2 pathway
Redox regulation of cysteine thiols has been implicated in controlling the function of a variety of regulatory processes mediated by thiol dependent signaling pathways, including electrophiles (2-5;37;38). Due to the reactive nature of the thiolate (R-S-) group on these residues, even subtle changes to the redox balance of the cell can result in cysteine modifications that can alter protein function (2-5;37;38). One of the major sensors of cellular redox status is the KEAP1 protein, which binds NRF2 and constantly promotes its degradation (5;24;26;39). Under basal homeostatic redox conditions, NRF2 remains in the cytoplasm, where it is degraded by the proteasome with an approximate half-life of 20 minutes (25;40). NRF2 is signaled for degradation by ubiquitination via a complex consisting of KEAP1 and Cullen 3 (CUL3) E3 ligase. KEAP1 serves as a crucial adapter in this process, locking NRF2 in the cytoplasm via one high affinity and one low affinity binding site, while CUL3 serves to ubiquitinate NRF2 lysine residues between these KEAP1 binding sites, and shuttle NRF2 to the ubiquitin proteasome system (25;41;42). In response to both oxidant and electrophilic stress, the KEAP1-NRF2-CUL3 interaction is disrupted, allowing NRF2 to translocate to the nucleus and activate target genes containing electrophile response elements (EpRE also termed antioxidant response elements, AREs), including key antioxidant genes such as glutathione S-transferase (GST), NAD(P)H oxidoreductase (NQO1), and glutamate cysteine ligase (GCL) (43-46) (Figure 1).
Figure 1. Redox regulation of the KEAP1-NRF2 pathway.
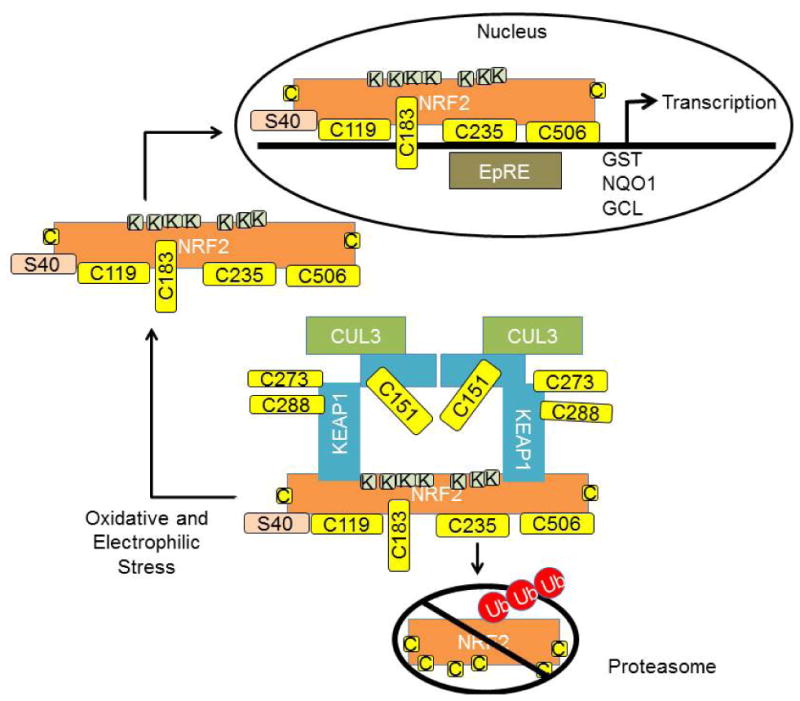
Under basal conditions NRF2 is bound in the cytosol by interaction with KEAP1. This interaction is modulated via modification of cysteine thiol groups (yellow boxes) on both proteins. KEAP1 serves as an adaptor, allowing NRF2 to be ubiquitinated by Cullen 3 (CUL3). Once ubiquitinated, NRF2 is sent to the proteasome for degradation. Cysteine modification at C151, C273 and C288 impacts CUL3 or NRF2 binding at the lower affinity sites, thereby impacting NRF2 ubiquitination and degradation. Unbound NRF2 translocates to the nucleus and activates transcription of genes with AREs or EpRE, including glutathione S-transferase (GST), NAD(P)H oxidoreductase (NQO1), and glutamate cysteine ligase (GCL). NRF2 C183 modification is important for nuclear localization, C119, 235 and 506 are required for NRF2 binding to EpREs. Ser40 phosphorylation is important for disassociation from KEAP1.
Regulation of the NRF2 pathway is complex. Human KEAP1 contains 27 cysteine residues, and depending on specific electrophiles or thiol reactive species, unique sets of cysteine residues are modified to alter the protein's function (43;47-52). For instance, Cys151 has been shown to be important for electrophile and oxidant-induced KEAP1-mediated ubiquitination and degradation of NRF2 via a mechanism that disrupts KEAP1-CUL3 interaction by modification at this residue (53;54). Conformational changes in other cysteine residues, Cys273 and Cys288, are required for constitutive KEAP1-mediated ubiquitination of NRF2 via a mechanism that disrupts the “latch” of KEAP1-NRF2 binding at the low affinity site, thus preventing ubiquitination and degradation of NRF2, and freeing de novo synthesized NRF2 free from KEAP1 binding, allowing it to translocate to the nucleus (44;53;55-57). Stabilization, translocation and activation of NRF2 are highly dynamic, as both KEAP1 and NRF2 contain nuclear localization and export signals to facilitate their translocations in and out of the nucleus, which are important for initiation and termination of NRF2-dependent transcriptional activities (58-64).
Interestingly, NRF2 can also be regulated by redox-dependent post-translational modifications. There are 6 cysteine residues in human NRF2, and 7 in mouse and rat. Mutations in cysteine residues of NRF2 led to decreased reaction with electrophiles, an enhanced KEAP1-NRF2 interaction and decreased NRF2 half-life. Cys183 has been shown to be localized in the transactivation domain with a functional nuclear exporting signal sequence, its modification with oxidants inactivates its nuclear exporting signaling activity (65). Cys119, 235 and 506 are also required for NRF2 binding to EpREs (66;67). In addition to thiol modification on cysteine residues, phosphorylation of NRF2 on serine or threonine residues also plays a crucial role in binding to KEAP1 or EpREs, as well as in mediating the transcriptional activity of NRF2. For example, a NRF2 Ser40 mutation led to enhanced association with KEAP1 and therefore impaired EpRE-mediated transcription (68). Phosphorylation of NRF2 at multiple sites by MAP kinases does not significantly contribute to NRF2 stability, while moderately affecting NRF2 transcriptional activity (69).
It is clear that different electrophiles modify unique sets of cysteines on these proteins, and how cysteine modifications code for their function in different cellular contexts is still an important area of investigation.
Redox regulation of autophagy
Complementing the NRF2-dependent regulation of antioxidant responses, the autophagy-lysosomal pathway facilitates additional defense mechanisms against cellular oxidative stress by selectively removing misfolded or damaged intracellular proteins and organelles, and thereby attenuating and reversing the injury caused by oxidative stress. Mechanisms by which the autophagy pathway responds to oxidative and nitrosative stress signals are still largely unclear. Studies have suggested that mTOR and PI3K activities mediate some of the autophagy response to oxidative stress (5;14;16;17;70-72), although it is also highly conceivable that modification of autophagy proteins at cysteine resides may also play a role in modulating autophagy.
There are a number of autophagy proteins that can be modified at key cysteines. Recombinant ATG4 activity is regulated by hydrogen peroxide modification of Cys81 residue near the active site of this protease (70). TOR has conserved cysteines that can form a disulfide bond which decreases structural flexibility and may control its stability and function (73). High-mobility group protein B1 (HMGB1), which is normally involved in chromatin remodeling, has also been shown to have a cysteine (Cys106) that when modified by reactive species or mutated, localizes to the cytosol and interacts with the Beclin/VPS34 complex to activate autophagy (74). The tumor suppressor protein p53, which has been shown to play a role in cell cycle arrest, autophagy activation, and apoptosis, also has two core cysteines (Cys182 and Cys277) that are prone to oxidative modification (75). Proteins implicated in removal of mitochondria via autophagy or mitophagy, which include PARKIN and DJ-1, have also been shown to contain conserved cysteines that can be modified to alter protein function (76-79) (Figure 2).
Figure 2. Redox regulation of autophagy.
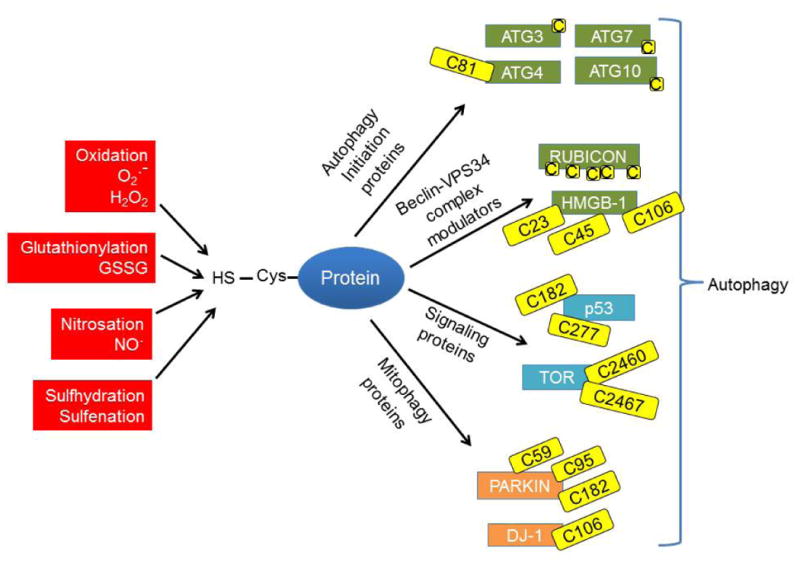
Proteins containing cysteine residues with a free thiol group can be modified through oxidation, glutathionylation, nitrosation, sulfhydration or sulfenation to modulate protein function. Many autophagy proteins have key modifiable cysteines including: 1) Autophagy initiation proteins (ATG 3, 4, 7 and 10); Recombinant ATG4 activity has been shown to be regulated by hydrogen peroxide modification of Cys81. 2) Modulators of the Beclin-VPS34 complex. RUBICON is a cysteine rich protein, and HMGB-1 is able to translocate to the cytosol, Cys106 modification is important for its translocation, Cys23-Cys45 disulfide bridge is important for its binding to BECN1. 3) The upstream autophagy signaling protein p53 has 2 cysteines that are prone to oxidative modification; and conserved cysteines in TOR can be oxidized. 4) Mitophagy-related proteins such as PARKIN, which is modifiable by nitrosation or sulfhydration, and DJ-1, which can be modified by oxidation. Modification of cysteines on these proteins may present a redox based regulation of the autophagy pathway during diseases associated with increased oxidative or nitrosative stress.
As such, increased formation of reactive species could cause modifications of key cysteine residues in autophagy proteins, and provide a redox-based regulation of the autophagy pathway. Three additional autophagy related proteins, ATG3, ATG7, and ATG10, which are involved in the processing of microtubule associated protein light chain (LC3) and autophagosome synthesis, have all been shown to have key regulatory cysteines (70;80-83). RUBICON, a Beclin/VPS34 complex-interacting protein, has a cysteine rich domain that could be modified during increased oxidative or nitrosative stress to affect autophagy initiation (84). Depending on the orientation of reactive cysteine residues in the active site or binding motifs of ATG proteins, the degree of susceptibility to conformational changes in response to oxidative or nitrosative modification of the cysteine residues, could greatly alter the ability of the cell to initiate autophagy, thus warranting further investigation of these modifications (Figure 2).
Cross regulation between the KEAP1-NRF2 pathway and autophagy
Recent evidence indicates a strong connection between the KEAP1-NRF2 antioxidant pathway and autophagy. p62/SQSTM1 (sequestosome 1), and NDP52 (nuclear dot protein 52) are adaptor proteins that possess both ubiquitin and LC3 binding sequences through a ubiquitin association domain (UBA) and an LC3-interaction region (LIR), and are therefore able to recruit ubiquitinated autophagy substrates to the autophagosome (13;85;86). Recent studies have also demonstrated that both p62 and NDP52 are NRF2 targets (27;33). Genetic ablation of KEAP1 led to the accumulation of ubiquitin aggregates and defective activation of autophagy, suggesting that KEAP1 binding to p62 may also be involved in p62 mediated autophagy of ubiquitinated proteins (31).
In addition to induced accumulation of p62 by KEAP1 ablation, inhibition of autophagy also results in accumulation of p62 and formation of cytoplasmic protein inclusions (32). Through a KEAP1 interacting region (KIR), p62 can bind and sequester KEAP1, blocking KEAP1-NRF2 binding, and allowing NRF2 to translocate to the nucleus, resulting in activation of EpRE containing genes, including antioxidants genes, p62 and NDP52 (Figure 3) (27-30;33). The further increase in p62 protein levels creates a positive feedback loop which leads to persistent activation of NRF2 (33;34).
Figure 3. Cross regulation of autophagy and KEAP1-NRF2.
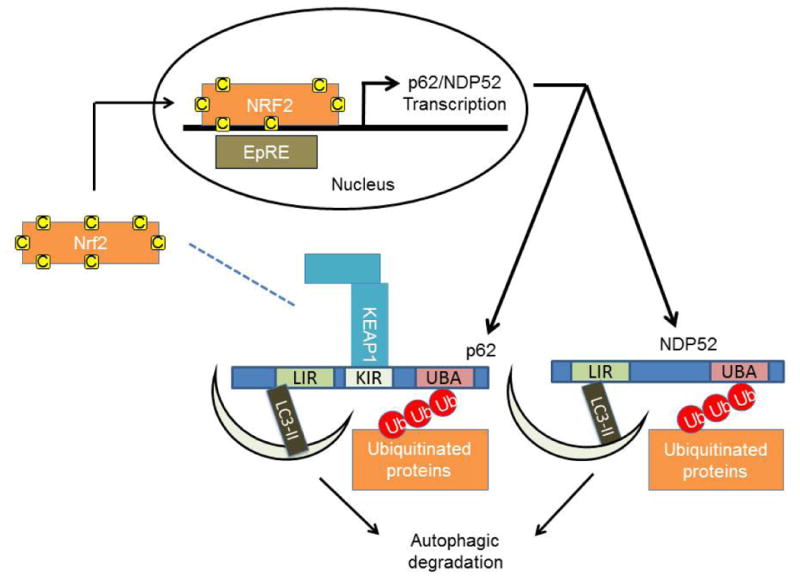
KEAP1 can also interact with p62. This interaction sequesters KEAP1 away from NRF2, preventing NRF2 ubiquitination and degradation. NRF2 stabilization and translocation to the nucleus allows its transcriptional activity. One of the NRF2 target genes is p62 which sequesters KEAP1 and brings it to be degraded by autophagy. This positive loop plays a role in coordinating autophagy and NRF2 activities.
p62 can also be phosphorylated in an mTOR-dependent manner, and phosphorylation of p62 increases its binding affinity to KEAP1 and subsequent induction of the expression of NRF2 target genes (35). The sulfinic acid reductases Sestrin1 and 2, have been shown to play an important role in promoting p62-dependent autophagic degradation of KEAP1 and upregulation of NRF2-dependent transcription (36). Knockdown or overexpression of p62 in a number of different mammalian cell lines, increased and decreased KEAP1 protein levels, respectively (30).
The integration of the KEAP1-NRF2 and autophagy pathways in response to oxidative and nitrosative stress may not be limited to p62 and NDP52 interactions. How exactly these two antioxidant pathways coordinate their responsibilities in maintaining cellular function and survival under different physiological and pathological conditions still needs further investigation.
Impact of redox regulation and cross-regulation of antioxidant and autophagy pathways on pathophysiology
The redox regulation and coordination of KEAP1-NRF2 and the autophagy pathway are essential for cell growth, differentiation, redox signaling and survival. Nrf2 knockout mice exhibit blunted induction of EpRE containing genes encoding cytoprotective enzymes, as well as increased susceptibility to toxic chemicals in various tissues (87-93). Transgenic overexpression of Nrf2 in cardiomyocytes suppressed cardiac proteotoxicity in a pressure overload study by transverse aortic arch constriction in mice, and the protective role may be mediated partially by upregulation of autophagy (94). Overexpression of Nrf2 in astrocytes protects against MPTP toxicity (95) (Figure 4).
Figure 4. NRF2, KEAP1 and p62 in disease pathogenesis.
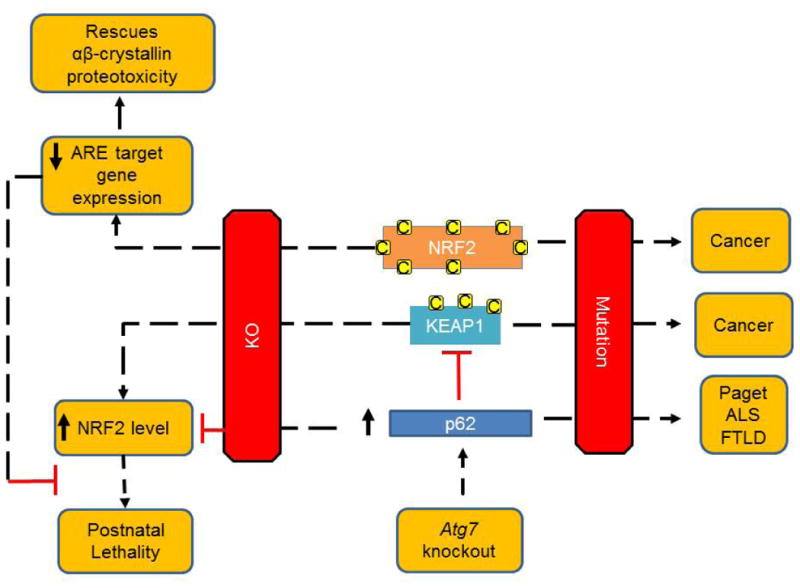
NRF2, KEAP1 and p62 play important roles in cellular redox, protein and organelle homeostasis. Mutations in NRF2 and KEAP1 are associated with human cancers, and mutations in p62 are associated with adult onset Paget disease of the bone, amyotrophic lateral sclerosis and frontotemporal lobar degeneration. Nrf2 knockout mice exhibit blunted induction of EpRE containing genes encoding cytoprotective enzymes, and increased susceptibility to toxic chemicals in various tissues, but also exhibit decreased proteotoxicity induced by overexpression of mutant αβ-crystallin through a redox-mediated mechanism. Keap1 knockout in mice is also detrimental and leads to constitutive NRF2 activity and postnatal lethality, this lethality can be rescued by a concurrent ablation of Nrf2, further demonstrating the mechanisms of KEAP1 in regulating NRF2 stability and function. Autophagy deficit-induced liver damage can be ameliorated by p62 or Nrf2 deletion, supporting the interaction of p62-NRF2 pathways. Overexpression of Nrf2 or p62 has been shown to lead to tissue specific and disease model specific consequences.
While the concept that loss of function of antioxidant enzymes can be deleterious is consistent with the oxidative stress hypothesis in disease pathogenesis, the finding that over-expression of antioxidants can be deleterious is not (7). In support for a detrimental role of constitutive NRF2 activity, Keap1 knockout in mice leads to constitutive NRF2 activity and postnatal lethality, this lethality can be rescued by a concurrent ablation of Nrf2 (96). With the generation of Keap1 knockout mice, the role of KEAP1 thiol modification in mediating its function has also been demonstrated by transgenic complementation rescue of Keap1 knockout mice with transgenic expression of wildtype versus cysteine mutated KEAP1 (97).
One of the first indications that the reductive stress is intimately linked to proteotoxicity was found in mice overexpressing mutant αβ-crystallin. These animals exhibit altered glutathione homeostasis due to changes in glutathione-6-phosphate dehydrogenase, glutathione reductase, and glutathione peroxidase activities, resulting in increased recycling of oxidized glutathione (GSSG) to the reduced form (GSH) (10). It has been shown that protein aggregates in these mice sequester KEAP1 and allow NRF2 activation of EpRE regulated genes (11). The role of NRF2-mediated reductive stress was demonstrated by the observation that crossing these mice to an Nrf2-deficient background significantly decreased proteotoxicity (12). These results indicate that protein aggregation can result in not only increased oxidative stress, but also increased reductive stress as a result of altered glutathione homeostasis and prolonged NRF2 activation and thus can be deleterious.
Mutations in NRF2 and KEAP1 are associated with human cancers, including cancers of the lung, head and neck, breast, liver, and skin (26;98), and mutations in p62 are found in adult onset Paget disease of the bone, amyotrophic lateral sclerosis and frontotemporal lobar degeneration (99). Constitutively high levels of NRF2 are associated with aggressive proliferation of cancer cells (100-102), and one important mechanism of NRF2 regulated cancer cell proliferation may be mediated by reprogramming metabolic activities (103). Highlighting the cross talk between the NRF2-KEAP1 and the p62 pathway is the recent demonstration that accumulation of p62 is also associated with poor cancer prognosis (102). When autophagy is blocked, for example due to Atg5, Atg7, or Vps34 deficiency, p62 accumulates and is proposed to lead to KEAP1 sequestration and persistent NRF2 activation (104-106). While p62 knockout mice develop adult-onset obesity and insulin resistance (107), impaired mitochondrial function with accelerated aging (108), high levels of p62 due to autophagy deficits have also been shown to be detrimental, as knockout of Atg7 in the liver and brain results in the accumulation of p62 and ubiquitin conjugates, and liver tumors, which can be reversed by knockout of p62 or Nrf2 in the Atg7 knockout background (29;32;109;110). p62 function also depends on the overall cellular context and circumstances. For example, p62 ablation is beneficial in a Huntington's mouse model, as it decreased nuclear inclusions and increased life span (111). On the other hand, p62 ablation in a spinal and bulbar muscular atrophy (SBMA) mouse model increased the levels of monomeric androgen receptor with polyQ expansion and exacerbated behavioral deficits such as performance on the rotarod, spontaneous motor activities and grip strength (112). p62 overexpression in transgenic mice increased inclusion body formation and ameliorated behavioral deficits (112), increased mitochondrial energy output, and improved behaviors associated with affective spectrum and anxiety disorders (113).
The equilibrium between molecular couples, namely the oxidation-reduction ratio of nicotinamide adenine dinucleotide phosphate (NADPH to NADP), glutathione (GSH/GSSG) and cysteine:cystine (disulfide), control numerous signaling mechanisms including the activity of transcription factors (114-118). Since oxidative stress is no longer adequate to explain redox related pathologies then attention is now focusing on the broader concept of redox stress which includes the conditions or a hyper-reductive cellular state. For example in cardiac hypertrophy due to protein aggregation (10;114) increased activity of G6PD enhanced the levels of NADPH, leading to hyper-reduction of glutathione (GSH). In addition, a recent study has also described increased G6PD activity stimulating a hyper-reductive state in the distinct cortical regions of Alzheimer's brain from affected individuals (119). The inability to maintain thiols in the regulated oxidized state necessary for cell signaling or metabolic function in a highly reductive environment appears to be as deleterious as hyper-oxidation under conditions of oxidative stress. Remarkably, these findings may reconcile inconsistent data in the literature and also explain the failure of antioxidant therapies in different pathologies including those with cardiovascular or neurodegenerative diseases.
Conclusions and Future Directions
Many proteins contain redox modulatable cysteine residues that regulate protein conformation and function. This review highlights the significance of the KEAP1-NRF2 as a sensor and regulator of redox homeostasis and its cross-talk with the autophagy pathway. Although extensive studies have been performed in the past decades, 3 major gaps still persist concerning our knowledge of redox signaling, metabolism and their dysfunction which lead to redox pathologies. 1) Our understanding of the regulatory mechanisms and impact of KEAP1-NRF2 pathway in various forms of diseases are still incomplete; 2) Even less is known regarding the redox regulatory mechanisms that affect the key autophagy proteins and/or autophagy substrates such as damaged proteins with modifiable cysteines; and 3) It's not obvious whether there could be novel, but undiscovered cellular antioxidant mechanisms besides KEAP1-NRF2 and autophagy pathways, and how these mechanisms are coordinated in different cellular and disease contexts which may also provide new insights into disease mechanisms and therapeutic strategies.
Acknowledgments
This work was supported by NIHR01-NS064090 (to JZ), NIHR01-HL118067), NIHR03-AG042860), AHA--BGIA# 0865015F (to NSR).
Abbreviations
- ARE
antioxidant responsive element
- NRF2
nuclear factor (erythroid-derived 2)-like 2
- KEAP1
kelch-like ECH-associated protein 1
- P62
P62/SQSTM1, sequestosome 1
Reference List
- 1.Winterbourn CC, Hampton MB. Thiol chemistry and specificity in redox signaling. Free Radic Biol Med. 2008;45:549–561. doi: 10.1016/j.freeradbiomed.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Higdon A, Diers AR, Oh JY, Landar A, Darley-Usmar VM. Cell signalling by reactive lipid species: new concepts and molecular mechanisms. Biochem J. 2012;442:453–464. doi: 10.1042/BJ20111752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mieyal JJ, Chock PB. Posttranslational modification of cysteine in redox signaling and oxidative stress: Focus on s-glutathionylation. Antioxid Redox Signal. 2012;16:471–475. doi: 10.1089/ars.2011.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murphy MP. Mitochondrial thiols in antioxidant protection and redox signaling: distinct roles for glutathionylation and other thiol modifications. Antioxid Redox Signal. 2012;16:476–495. doi: 10.1089/ars.2011.4289. [DOI] [PubMed] [Google Scholar]
- 5.Levonen AL, Hill BG, Kansanen E, Zhang J, Darley-Usmar VM. Redox regulation of antioxidants, autophagy, and the response to stress: Implications for electrophile therapeutics. Free Radic Biol Med. 2014;71C:196–207. doi: 10.1016/j.freeradbiomed.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Badia MC, Giraldo E, Dasi F, Alonso D, Lainez JM, Lloret A, Vina J. Reductive stress in young healthy individuals at risk of Alzheimer disease. Free Radic Biol Med. 2013;63:274–279. doi: 10.1016/j.freeradbiomed.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 7.Brewer AC, Mustafi SB, Murray TV, Rajasekaran NS, Benjamin IJ. Reductive stress linked to small HSPs, G6PD, and Nrf2 pathways in heart disease. Antioxid Redox Signal. 2013;18:1114–1127. doi: 10.1089/ars.2012.4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang H, Limphong P, Pieper J, Liu Q, Rodesch CK, Christians E, Benjamin IJ. Glutathione-dependent reductive stress triggers mitochondrial oxidation and cytotoxicity. FASEB J. 2012;26:1442–1451. doi: 10.1096/fj.11-199869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X, Min X, Li C, Benjamin IJ, Qian B, Zhang X, Ding Z, Gao X, Yao Y, Ma Y, et al. Involvement of reductive stress in the cardiomyopathy in transgenic mice with cardiac-specific overexpression of heat shock protein 27. Hypertension. 2010;55:1412–1417. doi: 10.1161/HYPERTENSIONAHA.109.147066. [DOI] [PubMed] [Google Scholar]
- 10.Rajasekaran NS, Connell P, Christians ES, Yan LJ, Taylor RP, Orosz A, Zhang XQ, Stevenson TJ, Peshock RM, Leopold JA, et al. Human alpha B-crystallin mutation causes oxido-reductive stress and protein aggregation cardiomyopathy in mice. Cell. 2007;130:427–439. doi: 10.1016/j.cell.2007.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajasekaran NS, Varadharaj S, Khanderao GD, Davidson CJ, Kannan S, Firpo MA, Zweier JL, Benjamin IJ. Sustained activation of nuclear erythroid 2-related factor 2/antioxidant response element signaling promotes reductive stress in the human mutant protein aggregation cardiomyopathy in mice. Antioxid Redox Signal. 2011;14:957–971. doi: 10.1089/ars.2010.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kannan S, Muthusamy VR, Whitehead KJ, Wang L, Gomes AV, Litwin SE, Kensler TW, Abel ED, Hoidal JR, Rajasekaran NS. Nrf2 deficiency prevents reductive stress-induced hypertrophic cardiomyopathy. Cardiovasc Res. 2013;100:63–73. doi: 10.1093/cvr/cvt150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J. Teaching the basics of autophagy and mitophagy to redox biologists-Mechanisms and experimental approaches. Redox Biol. 2015;4C:242–259. doi: 10.1016/j.redox.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giordano S, Darley-Usmar V, Zhang J. Autophagy as an essential cellular antioxidant pathway in neurodegenerative disease. Redox Biol. 2014;2:82–90. doi: 10.1016/j.redox.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J. Autophagy and Mitophagy in Cellular Damage Control. Redox Biol. 2013;1:19–23. doi: 10.1016/j.redox.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dodson M, Darley-Usmar V, Zhang J. Cellular metabolic and autophagic pathways: traffic control by redox signaling. Free Radic Biol Med. 2013;63:207–221. doi: 10.1016/j.freeradbiomed.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee J, Giordano S, Zhang J. Autophagy, mitochondria and oxidative stress: cross-talk and redox signalling. Biochem J. 2012;441:523–540. doi: 10.1042/BJ20111451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hohn A, Jung T, Grune T. Pathophysiological importance of aggregated damaged proteins. Free Radic Biol Med. 2014;71:70–89. doi: 10.1016/j.freeradbiomed.2014.02.028. [DOI] [PubMed] [Google Scholar]
- 19.Hohn TJ, Grune T. The proteasome and the degradation of oxidized proteins: Part III-Redox regulation of the proteasomal system. Redox Biol. 2014;2:388–394. doi: 10.1016/j.redox.2013.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jung T, Grune T. The proteasome and the degradation of oxidized proteins: Part I-structure of proteasomes. Redox Biol. 2013;1:178–182. doi: 10.1016/j.redox.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grune T, Reinheckel T, Davies KJ. Degradation of oxidized proteins in mammalian cells. FASEB J. 1997;11:526–534. [PubMed] [Google Scholar]
- 22.Grimm S, Hoehn A, Davies KJ, Grune T. Protein oxidative modifications in the ageing brain: consequence for the onset of neurodegenerative disease. Free Radic Res. 2011;45:73–88. doi: 10.3109/10715762.2010.512040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hill BG, Benavides GA, Lancaster JR, Jr, Ballinger S, Dell'italia L, Zhang J, Darley-Usmar VM. Integration of cellular bioenergetics with mitochondrial quality control and autophagy. Biol Chem. 2012;393:1485–1512. doi: 10.1515/hsz-2012-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McMahon M, Itoh K, Yamamoto M, Hayes JD. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J Biol Chem. 2003;278:21592–21600. doi: 10.1074/jbc.M300931200. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taguchi K, Motohashi H, Yamamoto M. Molecular mechanisms of the Keap1-Nrf2 pathway in stress response and cancer evolution. Genes Cells. 2011;16:123–140. doi: 10.1111/j.1365-2443.2010.01473.x. [DOI] [PubMed] [Google Scholar]
- 27.Jo C, Gundemir S, Pritchard S, Jin YN, Rahman I, Johnson GV. Nrf2 reduces levels of phosphorylated tau protein by inducing autophagy adaptor protein NDP52. Nat Commun. 2014;5:3496. doi: 10.1038/ncomms4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lau A, Wang XJ, Zhao F, Villeneuve NF, Wu T, Jiang T, Sun Z, White E, Zhang DD. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol. 2010;30:3275–3285. doi: 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 30.Copple IM, Lister A, Obeng AD, Kitteringham NR, Jenkins RE, Layfield R, Foster BJ, Goldring CE, Park BK. Physical and functional interaction of sequestosome 1 with Keap1 regulates the Keap1-Nrf2 cell defense pathway. J Biol Chem. 2010;285:16782–16788. doi: 10.1074/jbc.M109.096545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fan W, Tang Z, Chen D, Moughon D, Ding X, Chen S, Zhu M, Zhong Q. Keap1 facilitates p62-mediated ubiquitin aggregate clearance via autophagy. Autophagy. 2010;6 doi: 10.4161/auto.6.5.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata JI, Ezaki J, Murata S, et al. Homeostatic Levels of p62 Control Cytoplasmic Inclusion Body Formation in Autophagy-Deficient Mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 33.Jain A, Lamark T, Sjottem E, Larsen KB, Awuh JA, Overvatn A, McMahon M, Hayes JD, Johansen T. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem. 2010;285:22576–22591. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inami Y, Waguri S, Sakamoto A, Kouno T, Nakada K, Hino O, Watanabe S, Ando J, Iwadate M, Yamamoto M, et al. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J Cell Biol. 2011;193:275–284. doi: 10.1083/jcb.201102031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ichimura Y, Waguri S, Sou YS, Kageyama S, Hasegawa J, Ishimura R, Saito T, Yang Y, Kouno T, Fukutomi T, et al. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol Cell. 2013;51:618–631. doi: 10.1016/j.molcel.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 36.Bae SH, Sung SH, Oh SY, Lim JM, Lee SK, Park YN, Lee HE, Kang D, Rhee SG. Sestrins activate Nrf2 by promoting p62-dependent autophagic degradation of Keap1 and prevent oxidative liver damage. Cell Metab. 2013;17:73–84. doi: 10.1016/j.cmet.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 37.Cummins TD, Higdon AN, Kramer PA, Chacko BK, Riggs DW, Salabei JK, Dell'italia LJ, Zhang J, Darley-Usmar VM, Hill BG. Utilization of fluorescent probes for the quantification and identification of subcellular proteomes and biological processes regulated by lipid peroxidation products. Free Radic Biol Med. 2013;59:56–68. doi: 10.1016/j.freeradbiomed.2012.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Higdon AN, Dranka BP, Hill BG, Oh JY, Johnson MS, Landar A, Darley-Usmar VM. Methods for imaging and detecting modification of proteins by reactive lipid species. Free Radic Biol Med. 2009;47:201–212. doi: 10.1016/j.freeradbiomed.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Itoh K, Wakabayashi N, Katoh Y, Ishii T, O'Connor T, Yamamoto M. Keap1 regulates both cytoplasmic-nuclear shuttling and degradation of Nrf2 in response to electrophiles. Genes Cells. 2003;8:379–391. doi: 10.1046/j.1365-2443.2003.00640.x. [DOI] [PubMed] [Google Scholar]
- 41.Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol. 2004;24:8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci U S A. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang DD, Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol. 2003;23:8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Levonen AL, Landar A, Ramachandran A, Ceaser EK, Dickinson DA, Zanoni G, Morrow JD, Darley-Usmar VM. Cellular mechanisms of redox cell signalling: role of cysteine modification in controlling antioxidant defences in response to electrophilic lipid oxidation products. Biochem J. 2004;378:373–382. doi: 10.1042/BJ20031049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kobayashi A, Kang MI, Watai Y, Tong KI, Shibata T, Uchida K, Yamamoto M. Oxidative and electrophilic stresses activate Nrf2 through inhibition of ubiquitination activity of Keap1. Mol Cell Biol. 2006;26:221–229. doi: 10.1128/MCB.26.1.221-229.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eggler AL, Liu G, Pezzuto JM, van Breemen RB, Mesecar AD. Modifying specific cysteines of the electrophile-sensing human Keap1 protein is insufficient to disrupt binding to the Nrf2 domain Neh2. Proc Natl Acad Sci U S A. 2005;102:10070–10075. doi: 10.1073/pnas.0502402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holland R, Hawkins AE, Eggler AL, Mesecar AD, Fabris D, Fishbein JC. Prospective type 1 and type 2 disulfides of Keap1 protein. Chem Res Toxicol. 2008;21:2051–2060. doi: 10.1021/tx800226m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu C, Eggler AL, Mesecar AD, van Breemen RB. Modification of keap1 cysteine residues by sulforaphane. Chem Res Toxicol. 2011;24:515–521. doi: 10.1021/tx100389r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hong F, Sekhar KR, Freeman ML, Liebler DC. Specific patterns of electrophile adduction trigger Keap1 ubiquitination and Nrf2 activation. J Biol Chem. 2005;280:31768–31775. doi: 10.1074/jbc.M503346200. [DOI] [PubMed] [Google Scholar]
- 51.Hong F, Freeman ML, Liebler DC. Identification of sensor cysteines in human Keap1 modified by the cancer chemopreventive agent sulforaphane. Chem Res Toxicol. 2005;18:1917–1926. doi: 10.1021/tx0502138. [DOI] [PubMed] [Google Scholar]
- 52.Fujii S, Sawa T, Ihara H, Tong KI, Ida T, Okamoto T, Ahtesham AK, Ishima Y, Motohashi H, Yamamoto M, et al. The critical role of nitric oxide signaling, via protein S-guanylation and nitrated cyclic GMP, in the antioxidant adaptive response. J Biol Chem. 2010;285:23970–23984. doi: 10.1074/jbc.M110.145441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sekhar KR, Rachakonda G, Freeman ML. Cysteine-based regulation of the CUL3 adaptor protein Keap1. Toxicol Appl Pharmacol. 2010;244:21–26. doi: 10.1016/j.taap.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rachakonda G, Xiong Y, Sekhar KR, Stamer SL, Liebler DC, Freeman ML. Covalent modification at Cys151 dissociates the electrophile sensor Keap1 from the ubiquitin ligase CUL3. Chem Res Toxicol. 2008;21:705–710. doi: 10.1021/tx700302s. [DOI] [PubMed] [Google Scholar]
- 55.Tong KI, Kobayashi A, Katsuoka F, Yamamoto M. Two-site substrate recognition model for the Keap1-Nrf2 system: a hinge and latch mechanism. Biol Chem. 2006;387:1311–1320. doi: 10.1515/BC.2006.164. [DOI] [PubMed] [Google Scholar]
- 56.Kobayashi M, Li L, Iwamoto N, Nakajima-Takagi Y, Kaneko H, Nakayama Y, Eguchi M, Wada Y, Kumagai Y, Yamamoto M. The antioxidant defense system Keap1-Nrf2 comprises a multiple sensing mechanism for responding to a wide range of chemical compounds. Mol Cell Biol. 2009;29:493–502. doi: 10.1128/MCB.01080-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Holland R, Fishbein JC. Chemistry of the cysteine sensors in Kelch-like ECH-associated protein 1. Antioxid Redox Signal. 2010;13:1749–1761. doi: 10.1089/ars.2010.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nguyen T, Sherratt PJ, Nioi P, Yang CS, Pickett CB. Nrf2 controls constitutive and inducible expression of ARE-driven genes through a dynamic pathway involving nucleocytoplasmic shuttling by Keap1. J Biol Chem. 2005;280:32485–32492. doi: 10.1074/jbc.M503074200. [DOI] [PubMed] [Google Scholar]
- 59.Velichkova M, Hasson T. Keap1 regulates the oxidation-sensitive shuttling of Nrf2 into and out of the nucleus via a Crm1-dependent nuclear export mechanism. Mol Cell Biol. 2005;25:4501–4513. doi: 10.1128/MCB.25.11.4501-4513.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun Z, Zhang S, Chan JY, Zhang DD. Keap1 controls postinduction repression of the Nrf2-mediated antioxidant response by escorting nuclear export of Nrf2. Mol Cell Biol. 2007;27:6334–6349. doi: 10.1128/MCB.00630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun Z, Wu T, Zhao F, Lau A, Birch CM, Zhang DD. KPNA6 (Importin {alpha}7)-mediated nuclear import of Keap1 represses the Nrf2-dependent antioxidant response. Mol Cell Biol. 2011;31:1800–1811. doi: 10.1128/MCB.05036-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jain AK, Bloom DA, Jaiswal AK. Nuclear import and export signals in control of Nrf2. J Biol Chem. 2005;280:29158–29168. doi: 10.1074/jbc.M502083200. [DOI] [PubMed] [Google Scholar]
- 63.Theodore S, Cao S, McLean PJ, Standaert DG. Targeted overexpression of human alpha-synuclein triggers microglial activation and an adaptive immune response in a mouse model of Parkinson disease. J Neuropathol Exp Neurol. 2008;67:1149–1158. doi: 10.1097/NEN.0b013e31818e5e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li W, Jain MR, Chen C, Yue X, Hebbar V, Zhou R, Kong AN. Nrf2 Possesses a redox-insensitive nuclear export signal overlapping with the leucine zipper motif. J Biol Chem. 2005;280:28430–28438. doi: 10.1074/jbc.M410601200. [DOI] [PubMed] [Google Scholar]
- 65.Li W, Yu SW, Kong AN. Nrf2 possesses a redox-sensitive nuclear exporting signal in the Neh5 transactivation domain. J Biol Chem. 2006;281:27251–27263. doi: 10.1074/jbc.M602746200. [DOI] [PubMed] [Google Scholar]
- 66.He X, Ma Q. NRF2 cysteine residues are critical for oxidant/electrophile-sensing, Kelch-like ECH-associated protein-1-dependent ubiquitination-proteasomal degradation, and transcription activation. Mol Pharmacol. 2009;76:1265–1278. doi: 10.1124/mol.109.058453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bloom D, Dhakshinamoorthy S, Jaiswal AK. Site-directed mutagenesis of cysteine to serine in the DNA binding region of Nrf2 decreases its capacity to upregulate antioxidant response element-mediated expression and antioxidant induction of NAD(P)H:quinone oxidoreductase1 gene. Oncogene. 2002;21:2191–2200. doi: 10.1038/sj.onc.1205288. [DOI] [PubMed] [Google Scholar]
- 68.Huang HC, Nguyen T, Pickett CB. Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. J Biol Chem. 2002;277:42769–42774. doi: 10.1074/jbc.M206911200. [DOI] [PubMed] [Google Scholar]
- 69.Sun Z, Huang Z, Zhang DD. Phosphorylation of Nrf2 at multiple sites by MAP kinases has a limited contribution in modulating the Nrf2-dependent antioxidant response. PLoS ONE. 2009;4:e6588. doi: 10.1371/journal.pone.0006588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26:1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Alexander A, Cai SL, Kim J, Nanez A, Sahin M, MacLean KH, Inoki K, Guan KL, Shen J, Person MD, et al. ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proc Natl Acad Sci U S A. 2010;107:4153–4158. doi: 10.1073/pnas.0913860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wani WY, Boyer-Guittaut M, Dodson M, Chatham J, rley-Usmar V, Zhang J. Regulation of autophagy by protein post-translational modification. Lab Invest. 2015;95:14–25. doi: 10.1038/labinvest.2014.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dames SA, Mulet JM, Rathgeb-Szabo K, Hall MN, Grzesiek S. The solution structure of the FATC domain of the protein kinase target of rapamycin suggests a role for redox-dependent structural and cellular stability. J Biol Chem. 2005;280:20558–20564. doi: 10.1074/jbc.M501116200. [DOI] [PubMed] [Google Scholar]
- 74.Tang D, Kang R, Livesey KM, Cheh CW, Farkas A, Loughran P, Hoppe G, Bianchi ME, Tracey KJ, Zeh HJ, III, et al. Endogenous HMGB1 regulates autophagy. J Cell Biol. 2010;190:881–892. doi: 10.1083/jcb.200911078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Scotcher J, Clarke DJ, Weidt SK, Mackay CL, Hupp TR, Sadler PJ, Langridge-Smith PR. Identification of two reactive cysteine residues in the tumor suppressor protein p53 using top-down FTICR mass spectrometry. J Am Soc Mass Spectrom. 2011;22:888–897. doi: 10.1007/s13361-011-0088-x. [DOI] [PubMed] [Google Scholar]
- 76.Redmann M, Dodson M, Boyer-Guittaut M, Darley-Usmar V, Zhang J. Mitophagy mechanisms and role in human diseases. Int J Biochem Cell Biol. 2014;53C:127–133. doi: 10.1016/j.biocel.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Meng F, Yao D, Shi Y, Kabakoff J, Wu W, Reicher J, Ma Y, Moosmann B, Masliah E, Lipton SA, et al. Oxidation of the Cysteine-Rich Regions of Parkin Perturbs Its E3 Ligase Activity and Contributes to Protein Aggregation. Mol Neurodegener. 2011;6:34. doi: 10.1186/1750-1326-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vandiver MS, Paul BD, Xu R, Karuppagounder S, Rao F, Snowman AM, Ko HS, Lee YI, Dawson VL, Dawson TM, et al. Sulfhydration mediates neuroprotective actions of parkin. Nat Commun. 2013;4:1626. doi: 10.1038/ncomms2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Choi MS, Nakamura T, Cho SJ, Han X, Holland EA, Qu J, Petsko GA, Yates JR, III, Liddington RC, Lipton SA. Transnitrosylation from DJ-1 to PTEN attenuates neuronal cell death in parkinson's disease models. J Neurosci. 2014;34:15123–15131. doi: 10.1523/JNEUROSCI.4751-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Scherz-Shouval R, Elazar Z. Regulation of autophagy by ROS: physiology and pathology. Trends Biochem Sci. 2010 doi: 10.1016/j.tibs.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 81.Sakoh-Nakatogawa M, Matoba K, Asai E, Kirisako H, Ishii J, Noda NN, Inagaki F, Nakatogawa H, Ohsumi Y. Atg12-Atg5 conjugate enhances E2 activity of Atg3 by rearranging its catalytic site. Nat Struct Mol Biol. 2013;20:433–439. doi: 10.1038/nsmb.2527. [DOI] [PubMed] [Google Scholar]
- 82.Tanida I, Mizushima N, Kiyooka M, Ohsumi M, Ueno T, Ohsumi Y, Kominami E. Apg7p/Cvt2p: A novel protein-activating enzyme essential for autophagy. Mol Biol Cell. 1999;10:1367–1379. doi: 10.1091/mbc.10.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shintani T, Mizushima N, Ogawa Y, Matsuura A, Noda T, Ohsumi Y. Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. EMBO J. 1999;18:5234–5241. doi: 10.1093/emboj/18.19.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, Heintz N, Yue Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 87.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 88.Chan K, Kan YW. Nrf2 is essential for protection against acute pulmonary injury in mice. Proc Natl Acad Sci U S A. 1999;96:12731–12736. doi: 10.1073/pnas.96.22.12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Aoki Y, Sato H, Nishimura N, Takahashi S, Itoh K, Yamamoto M. Accelerated DNA adduct formation in the lung of the Nrf2 knockout mouse exposed to diesel exhaust. Toxicol Appl Pharmacol. 2001;173:154–160. doi: 10.1006/taap.2001.9176. [DOI] [PubMed] [Google Scholar]
- 90.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 91.Cho HY, Kwak MK, Pi J. Nrf2 in host defense: over the rainbow. Oxid Med Cell Longev. 2013;2013:975839. doi: 10.1155/2013/975839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cho HY, Kleeberger SR. Noblesse oblige: NRF2 functions in the airways. Am J Respir Cell Mol Biol. 2014;50:844–847. doi: 10.1165/rcmb.2014-0116PS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ma Q. Role of nrf2 in oxidative stress and toxicity. Annu Rev Pharmacol Toxicol. 2013;53:401–426. doi: 10.1146/annurev-pharmtox-011112-140320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang W, Li S, Wang H, Li B, Shao L, Lai Y, Horvath G, Wang Q, Yamamoto M, Janicki JS, et al. Nrf2 enhances myocardial clearance of toxic ubiquitinated proteins. J Mol Cell Cardiol. 2014;72:305–315. doi: 10.1016/j.yjmcc.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen PC, Vargas MR, Pani AK, Smeyne RJ, Johnson DA, Kan YW, Johnson JA. Nrf2-mediated neuroprotection in the MPTP mouse model of Parkinson's disease: Critical role for the astrocyte. Proc Natl Acad Sci U S A. 2009;106:2933–2938. doi: 10.1073/pnas.0813361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wakabayashi N, Itoh K, Wakabayashi J, Motohashi H, Noda S, Takahashi S, Imakado S, Kotsuji T, Otsuka F, Roop DR, et al. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat Genet. 2003;35:238–245. doi: 10.1038/ng1248. [DOI] [PubMed] [Google Scholar]
- 97.Yamamoto T, Suzuki T, Kobayashi A, Wakabayashi J, Maher J, Motohashi H, Yamamoto M. Physiological significance of reactive cysteine residues of Keap1 in determining Nrf2 activity. Mol Cell Biol. 2008;28:2758–2770. doi: 10.1128/MCB.01704-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kansanen E, Kuosmanen SM, Leinonen H, Levonen AL. The Keap1-Nrf2 pathway: Mechanisms of activation and dysregulation in cancer. Redox Biol. 2013;1:45–49. doi: 10.1016/j.redox.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rea SL, Majcher V, Searle MS, Layfield R. SQSTM1 mutations--bridging Paget disease of bone and ALS/FTLD. Exp Cell Res. 2014;325:27–37. doi: 10.1016/j.yexcr.2014.01.020. [DOI] [PubMed] [Google Scholar]
- 100.Shibata T, Ohta T, Tong KI, Kokubu A, Odogawa R, Tsuta K, Asamura H, Yamamoto M, Hirohashi S. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc Natl Acad Sci U S A. 2008;105:13568–13573. doi: 10.1073/pnas.0806268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Singh A, Boldin-Adamsky S, Thimmulappa RK, Rath SK, Ashush H, Coulter J, Blackford A, Goodman SN, Bunz F, Watson WH, et al. RNAi-mediated silencing of nuclear factor erythroid-2-related factor 2 gene expression in non-small cell lung cancer inhibits tumor growth and increases efficacy of chemotherapy. Cancer Res. 2008;68:7975–7984. doi: 10.1158/0008-5472.CAN-08-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang P, Singh A, Yegnasubramanian S, Esopi D, Kombairaju P, Bodas M, Wu H, Bova SG, Biswal S. Loss of Kelch-like ECH-associated protein 1 function in prostate cancer cells causes chemoresistance and radioresistance and promotes tumor growth. Mol Cancer Ther. 2010;9:336–346. doi: 10.1158/1535-7163.MCT-09-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mitsuishi Y, Taguchi K, Kawatani Y, Shibata T, Nukiwa T, Aburatani H, Yamamoto M, Motohashi H. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell. 2012;22:66–79. doi: 10.1016/j.ccr.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 104.Komatsu M, Waguri S, Chiba T, Murata S, Iwata JI, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 105.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 106.Jaber N, Dou Z, Chen JS, Catanzaro J, Jiang YP, Ballou LM, Selinger E, Ouyang X, Lin RZ, Zhang J, et al. Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proc Natl Acad Sci U S A. 2012;109:2003–2008. doi: 10.1073/pnas.1112848109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rodriguez A, Duran A, Selloum M, Champy MF, Diez-Guerra FJ, Flores JM, Serrano M, Auwerx J, Diaz-Meco MT, Moscat J. Mature-onset obesity and insulin resistance in mice deficient in the signaling adapter p62. Cell Metab. 2006;3:211–222. doi: 10.1016/j.cmet.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 108.Kwon J, Han E, Bui CB, Shin W, Lee J, Lee S, Choi YB, Lee AH, Lee KH, Park C, et al. Assurance of mitochondrial integrity and mammalian longevity by the p62-Keap1-Nrf2-Nqo1 cascade. EMBO Rep. 2012;13:150–156. doi: 10.1038/embor.2011.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Taguchi K, Fujikawa N, Komatsu M, Ishii T, Unno M, Akaike T, Motohashi H, Yamamoto M. Keap1 degradation by autophagy for the maintenance of redox homeostasis. Proc Natl Acad Sci U S A. 2012;109:13561–13566. doi: 10.1073/pnas.1121572109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, Eishi Y, Hino O, Tanaka K, Mizushima N. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kurosawa M, Matsumoto G, Kino Y, Okuno M, Kurosawa-Yamada M, Washizu C, Taniguchi H, Nakaso K, Yanagawa T, Warabi E, et al. Depletion of p62 reduces nuclear inclusions and paradoxically ameliorates disease phenotypes in Huntington's model mice. Hum Mol Genet. 2015;24:1092–1105. doi: 10.1093/hmg/ddu522. [DOI] [PubMed] [Google Scholar]
- 112.Doi H, Adachi H, Katsuno M, Minamiyama M, Matsumoto S, Kondo N, Miyazaki Y, Iida M, Tohnai G, Qiang Q, et al. p62/SQSTM1 differentially removes the toxic mutant androgen receptor via autophagy and inclusion formation in a spinal and bulbar muscular atrophy mouse model. J Neurosci. 2013;33:7710–7727. doi: 10.1523/JNEUROSCI.3021-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Seibenhener ML, Zhao T, Du Y, Calderilla-Barbosa L, Yan J, Jiang J, Wooten MW, Wooten MC. Behavioral effects of SQSTM1/p62 overexpression in mice: support for a mitochondrial role in depression and anxiety. Behav Brain Res. 2013;248:94–103. doi: 10.1016/j.bbr.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rajasekaran NS, Firpo MA, Milash BA, Weiss RB, Benjamin IJ. Global expression profiling identifies a novel biosignature for protein aggregation R120GCryAB cardiomyopathy in mice. Physiol Genomics. 2008;35:165–172. doi: 10.1152/physiolgenomics.00297.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shaikhali J, Noren L, de DBL, Srivastava V, Konig J, Sauer UH, Wingsle G, Dietz KJ, Strand A. Redox-mediated mechanisms regulate DNA binding activity of the G-group of basic region leucine zipper (bZIP) transcription factors in Arabidopsis. J Biol Chem. 2012;287:27510–27525. doi: 10.1074/jbc.M112.361394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ishikawa M, Numazawa S, Yoshida T. Redox regulation of the transcriptional repressor Bach1. Free Radic Biol Med. 2005;38:1344–1352. doi: 10.1016/j.freeradbiomed.2005.01.021. [DOI] [PubMed] [Google Scholar]
- 117.Wang K, Zhang T, Dong Q, Nice EC, Huang C, Wei Y. Redox homeostasis: the linchpin in stem cell self-renewal and differentiation. Cell Death Dis. 2013;4:e537. doi: 10.1038/cddis.2013.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liu H, Colavitti R, Rovira II, Finkel T. Redox-dependent transcriptional regulation. Circ Res. 2005;97:967–974. doi: 10.1161/01.RES.0000188210.72062.10. [DOI] [PubMed] [Google Scholar]
- 119.Russell RL, Siedlak SL, Raina AK, Bautista JM, Smith MA, Perry G. Increased neuronal glucose-6-phosphate dehydrogenase and sulfhydryl levels indicate reductive compensation to oxidative stress in Alzheimer disease. Arch Biochem Biophys. 1999;370:236–239. doi: 10.1006/abbi.1999.1404. [DOI] [PubMed] [Google Scholar]


