SUMMARY
G protein-coupled receptors (GPCRs) are the largest family of membrane receptors in humans, and they regulate processes ranging from neurotransmission to cardiovascular biology. Although GPCRs have been studied for decades, current methods for tracking GPCR signaling often suffer from low throughput, modification or overexpression of effector proteins, and low temporal resolution. Here, we introduce a new approach using peroxidase-catalyzed proximity labeling to track GPCR signaling and internalization in living cells. Combination of this technique with isobaric labeling and triple-stage mass spectrometry enables precise, quantitative, and time-resolved measurement of thousands of receptor-proximal proteins at their native levels to comprehensively track GPCR agonist response. Using this technique, we examine the response of the angiotensin II type 1 receptor to both balanced and biased agonists. In addition, we extend the approach to the β2 adrenergic receptor, underscoring the generalizability of this technology.
Graphical Abstract

INTRODUCTION
GPCRs comprise the largest family of transmembrane receptors in humans, and they serve as critical regulators of most aspects of physiology. As a consequence of their profound biological importance, GPCRs have become the most successful target class for therapeutic drug development, and GPCR-targeted drugs include most treatments for cardiovascular, neuropsychiatric, and metabolic diseases. GPCR research has seen major advances in recent years, particularly in terms of studies of receptor structure, dynamics, and pharmacology. Nonetheless, many major aspects of GPCR signaling remain incompletely characterized, and it has become increasingly clear that GPCRs possess an unexpectedly rich and complex signaling biology that is only beginning to be fully understood.
The classical paradigm for GPCR signaling through G proteins was elucidated decades ago (Gilman, 1995) and is now relatively well understood. Upon binding an activating ligand (agonist) a GPCR undergoes conformational changes and catalyzes the exchange of GDP for GTP in Gα subunits of G protein heterotrimers. Activated G proteins subsequently activate or inhibit downstream effectors like adenylyl cyclase, leading to a plethora of cellular responses. Shortly following G protein activation, GPCRs are phosphorylated by GPCR kinases (GRKs), leading to the recruitment of β-arrestins and consequent endocytosis. Initially, β-arrestins were considered to be nothing more than silencers of GPCR signaling, but it is now clear that GPCR activation results in G protein-independent signaling mediated primarily by β-arrestins in addition to G protein-mediated responses (Lefkowitz, 2013).
Existing GPCR signaling assays are informative, but suffer from limitations including single pathway readout, limited time resolution, and overexpression or modification of the very effectors they aim to study. Moreover, most signaling assays report on events far downstream of ligand-mediated changes in receptor activity. In addition, studies of GPCR signaling are complicated by the fact that many ligands exhibit varying degrees of “biased” signaling, preferentially activating some pathways more than others (e.g., activation of G proteins but not β-arrestins). The phenomenon of biased signaling holds the potential for transformative innovations in medicine as it allows pharmacological separation of G protein signaling from β-arrestin signaling, there by achieving clinical benefit with greatly reduced side effects (Violin et al., 2014). Despite this, the molecular details underlying biased signaling remain poorly characterized, and even quantification of signaling bias has been intractable due to limitations with existing methods.
To address these challenges, we sought to develop a method that allows parallel quantification of GPCR interactions with all effectors within a native cellular environment at endogenous expression levels. One candidate approach to achieve this was fusion to a promiscuous biotin ligase (Roux et al., 2012), but the slow labeling kinetics made this approach unsuitable for studying GPCR activation with high temporal resolution – an essential requirement in light of the fact that most GPCRs regulate physiological responses on timescales ranging from seconds to minutes. In contrast, the engineered ascorbate peroxidase APEX2 rapidly produces short-lived (<1 ms) biotin-phenoxy radicals that have a limited (~20 nm) labeling radius, thereby overcoming these problems (Hung et al., 2016). We reasoned that combining GPCR-APEX2 fusion with quantitative mass spectrometry could allow observation and quantification of GPCR-proximal proteins with high temporal resolution. To enable accurate relative quantification of protein abundance, we chose to use isobaric tandem mass tags to label samples (Thompson et al., 2003). This approach allows all labeled samples to be pooled and analyzed in a single mass spectrometry experiment so that relative abundances of thousands of proteins from up to ten different samples are measured in a highly reproducible and quantitative manner.
To test the suitability of APEX2 as a potential tool to study GPCR signaling, we chose the angiotensin II type 1 receptor (AT1R) as our model. The AT1R is one of the principal regulators of blood pressure in humans, and is the target of widely used antihypertensive drugs including losartan (Cozaar), irbesartan (Avapro), telmisartan (Micardis) and several others. In addition to its profound importance in human health, the AT1R is one of the most extensively studied model GPCRs in terms of pharmacology and cell biology, and strongly β-arrestin biased ligands such as TRV027 are readily available and well characterized (Strachan et al., 2014; Violin et al., 2010). The AT1R robustly internalizes for an extended period following agonist treatment, making it a prototypical “class B” receptor with respect to its internalization properties (Oakley et al., 2000). Taken together, these features make the AT1R an ideal system for exploring both receptor internalization and signaling bias.
RESULTS
Design and assessment of GPCR-APEX platform
We envisioned that the AT1R fused to APEX2 at its carboxy-terminus would offer a straightforward approach to performing proximity-labeling (Figure 1A). The carboxy-terminus of AT1R is predicted to be intrinsically unstructured, and is not resolved in a recent crystal structure of the receptor bound to an antagonist (Zhang et al., 2015), indicating that fusion to APEX2 enzyme at its carboxy-terminus is unlikely to alter its functional properties. Indeed, the APEX2 fusion receptor showed signaling and internalization properties similar to those of wild-type receptor (Figure S1A–B). AT1R-APEX differed significantly from the wild-type receptor only in its expression level, which was reduced roughly 4-fold relative to the wild-type receptor.
Figure 1. Design and experiment procedure of proximity-labeling by AT1R-APEX.
(A) Schematic of APEX2-mediated protein labeling. The angiotensin receptor (AT1R) fused to APEX2 at its carboxy-terminus via a glycine-serine linker biotinylates proteins in proximity (< 20 nm) to the receptor in an unbiased manner, allowing identification and quantification of interactions between the receptor and its effectors at distinct time points after ligand treatment.
(B) Proximity-labeling and mass spectrometric analysis workflow for AT1R-APEX. Four different pairs of biological replicates were prepared to assess reproducibility of labeling/proteomic analysis in both short and long time points for agonist/antagonist treatment. Biotin-labeled proteins were purified by TCA precipitation followed by denaturing streptavidin pull-down and tandem mass tag labeling to allow quantitative mass spectrometric analysis of 10 different samples in parallel.
See also Figure S1.
We first sought to assess proximity-labeling efficiency and the reproducibility of protein quantification. To perform proximity-labeling experiments, HEK293T cells stably expressing AT1R-APEX were pre-incubated in medium containing biotin-phenol (the total length of incubation in biotin-phenol was 1 hour before H2O2 treatment), then treated with agonist (angiotensin II) or antagonist (losartan) at either 1 or 20 minutes before 1 min hydrogen peroxide incubation to start labeling. Similarly, samples without ligand-treatment or hydrogen peroxide treatment were prepared to assess changes in biotinylated proteins upon ligand treatment and endogenous biotinylation level, respectively. Biotin-labeling was quenched immediately after 1 min using described quench buffer (Hung et al., 2016). Afterwards, biotinylated proteins were enriched by denaturing streptavidin affinity purification, digested into tryptic peptides, labeled with isobaric tandem mass tags (TMT), fractionated by alkaline reversed phase chromatography, and analyzed by triple stage mass spectrometry (TMT SPS MS3; Figure 1B). Quantification by SPS MS3 rather than applying conventional tandem mass spectrometry (MS2) has the advantage of reducing signal ratio distortion and enhancing the accuracy of peptide (and by inference, protein) quantification (McAlister et al., 2014; Paulo et al., 2016b; Ting et al., 2011).
Western blotting confirmed robust biotinylation and revealed a different spectrum of labeled proteins in agonist- and antagonist-treated samples (Figure 2A). By mass spectrometry analysis, 8281 peptides were quantified, representing a total of 1242 proteins. For the four independent biological replicate pairs, isobaric tag-based measurements of protein abundance showed excellent correlation (R2 > 0.97), indicating that the procedure allows highly reproducible protein quantification regardless of experimental conditions (Figure 2B, Figure S2A–C). It is important to note that in these experiments a lack of observation of a given protein does not imply lack of association. Peptides may not be detected due to many factors including inherent unsuitability for mass spectrometry, steric obstruction of labeling, or lack of suitable reactive moieties in the peptide/protein sequence. Nonetheless, despite these caveats a broad collection of GPCR signaling effectors were observed, including β-arrestin 2 and a variety of G proteins, among many others.
Figure 2. Analysis of AT1R-APEX experiment reproducibility and changes in biotinylated proteome upon agonist-mediated receptor activation.
(A) Biotinylated proteins from a proximity-labeling experiment analyzed by streptavidin-HRP blot. Clear changes in band patterns were observed for agonist-treated samples in comparison to antagonist-treated/ligand-free samples.
(B) Representative linear regression analysis of biologically independent replicates treated with angiotensin II for 2 minutes. This analysis confirms high reproducibility in protein abundance measurements.
(C) Fold-enrichment of detected G-protein subunits and β-arrestin 2 relative to the background levels measured in H2O2-untreated cells. β-arrestin 2 is included as a reference to assess significant enrichment levels over background.
(D) Linear regression analysis of protein abundances in losartan-treated samples vs. ligand-free sample. No distinct change in biotinylated proteome was observed. Errors shown as means ± SEM for two biological replicates.
(E) Enrichment of selected factors in angiotensin II-treated samples relative to the ligand-free sample. Data shown as means ± SEM for two biological replicates.
(F) Volcano plots for 2 minute agonist vs. antagonist-treatment. Select proteins with effect size greater than 4 are marked based on gene ontology. Higher effect size represents higher enrichment upon angiotensin-treatment. P-values were Benjamini-Hochberg corrected (Hochberg and Benjamini, 1990).
One of the fundamental questions in GPCR signaling is whether GPCRs co-localize with G protein subunits in specific micro-domains or lipid rafts within the cell membrane to facilitate rapid signal transduction (Oates and Watts, 2011). However, characterization of such micro-domains has been challenging due to difficulties in their biochemical manipulation. To characterize the native local milieu of the AT1R in cells, we used AT1R-APEX to identify and quantify proteins within the APEX labeling radius. By comparing the abundance of peptides derived from AT1R-APEX cells with hydrogen peroxide treatment to those without (background), we could confirm that heterotrimeric G proteins were strongly enriched in the absence of ligand treatment, with the AT1R signal transducer Gαq showing the strongest enrichment among G proteins (Figure 2C). However, other Gα subunits, including those that do not couple with AT1R, were also strongly enriched, supporting the idea that GPCRs co-localize with multiple components of their signaling machinery in lipid rafts or other microdomains. β-arrestin 2 was also observed and quantified in both control and labeled samples, but unlike G proteins it showed relatively low enrichment in the absence of ligand treatment, consistent with its cytosolic localization prior to receptor activation.
These data additionally allowed straightforward assessment of the effects of the antagonist losartan relative to untreated samples. Interestingly, no strongly different enrichment is seen for any of the 1242 proteins measured, including G proteins and β-arrestins (Figure 2D). This suggests that losartan acts effectively as a neutral antagonist in these conditions, with no significant effect on receptor-effector interactions or subcellular localization.
Comparison of cells treated with the endogenous agonist angiotensin II to the untreated control cells was more revealing. At the two time points measured (2 minutes and 21 minutes), a diverse population of proteins showed strong enrichment in the presence of agonist, including known effectors and regulators of receptor internalization: β-arrestin 2, AP-2, clathrin, FCHo protein, and intersectins (Figure 2E). Importantly, the isobaric tagging approach allows simultaneous quantification of all proteins measured rather than just a select subset. Hence statistical analysis of relative enrichment between agonist-treated and antagonist-treated samples was possible across all proteins (Figure 2F). The analysis highlighted known effectors and components of the endocytosis machinery that had the largest and most significant divergence between samples. These data suggest that even in the absence of prior knowledge in signaling effectors, it may be possible to identify signal transduction components using receptor-APEX fusion together with isobaric labeling. Full experimental data are reported in Table S1.
Time-resolved AT1R-APEX
We next sought to extend these techniques to address a key limitation of most existing GPCR signaling assays: time resolution. Given the highly dynamic nature of GPCR signaling, the kinetics of signaling and receptor trafficking are of critical importance, but are difficult to measure using most conventional GPCR signaling assays. Using a modified APEX labeling protocol (Figure 3A), we tracked AT1R interactions at ten different time points, including 10 second intervals for the first 50 seconds, as well as sparser sampling at time points up to 30 minutes. One sample with no ligand treatment was included to serve as a reference for assessing relative peptide enrichment at other time points.
Figure 3. AT1R-APEX timecourse experiment with the balanced full agonist angiotensin II.
(A) Schematic of proximity-labeling experiment protocol for timecourse experiment
(B) Timecourse of AT1R internalization revealed by relative abundances of proximal proteins following agonist treatment. β-arrestin 2 (ARRB2), clathrin (CLTA), and early endosomal marker Rab5 (RAB5C) maximally enrich at 180 s, followed by maximum enrichments of late endosomal Rab7 (RAB7A) and lysosomal (LAMP1) markers.
(C) Enrichment pattern heatmap of G protein subunits, β-arrestin 2, and Rab5. Following internalization and the β-arrestin 2 enrichment peak at 180 s, receptor-proximal G protein levels drop substantially. Each protein’s enrichment level across different time points is normalized to its maximum enrichment signal.
(D) Linear plot of G alpha and beta subunits detected during short time points (0 – 80 s), showing initial drop in enrichment followed by a slow rise and then fall.
(E) Enrichment pattern heatmap of endosomal marker transferrin receptor (TFRC) and components of retromer complex VPS29 and SNX1.
The time-resolved analysis allowed tracking of AT1R proximity kinetics for 1034 proteins. Known interaction partners including G proteins and β-arrestin 2 were readily observed and tracked. At its peak, β-arrestin 2 reached levels that were 27-fold higher than in the agonist-free state. β-arrestin 2 association kinetics were almost identical to those for known clathrin-mediated endocytosis components such as AP-2, PICALM, and clathrin itself (Figure S3A). Surprisingly, β-arrestin enrichment level dropped after its peak at 180 seconds while endosomal markers including Rab5 and Rab7 persisted, indicating that arrestin at least partially dissociates from the receptor following clathrin-mediated endocytosis (Figure 3B). This observation contrasts with the current view of β-arrestin-class B GPCR interaction that the arrestin-receptor complex remains stable in endosomes (Pierce and Lefkowitz, 2001).
A similar kinetic analysis for heterotrimeric G protein α subunits showed an immediate drop in G protein labeling in the first 10 seconds. This was followed by a gradual rise and subsequent decline over the first minute, with peak labeling at 50 seconds. In light of recent reports that endosomal signaling may be important for some GPCRs (Irannejad et al., 2013; Thomsen et al., 2016), we sought to examine whether G proteins are internalized together with the AT1R. In fact, receptor-proximal G protein levels showed a significant decrease coincident with the arrival of arrestin and the clathrin-mediated endocytosis machinery (Figure 3C). This indicates that endocytosis selectively internalizes the receptor and largely sequesters it away from the G proteins of all classes observed. These results do not exclude the possibility of endosomal signaling entirely, however, as low levels of G protein from each class were still present following internalization.
In addition, GPCR-APEX provides a platform for tracking endocytosis and subcellular localization over longer timescales. As discussed above, recruitment of the clathrin machinery closely paralleled arrestin recruitment. Shortly thereafter, endosomal markers including Rab5, Rab7, and transferrin receptor all increased in enrichment. At later timepoints, receptor interaction with retromer complex proteins including VPS29 and SNX1 was observed beginning around 10 minutes. A portion of the receptor population also appears to enter lysosomes, indicated by the enrichment of LAMP1 at 10 minutes and 30 minute timepoints (Figure 3B, Figure 3E). Full quantification data for all 1034 proteins measured at each timepoint are included in Table S2.
Kinetics of AT1R response to a β-arrestin biased agonist
We next sought to extend this technique to examine the consequences of AT1R activation by the biased agonist TRV027. While the endogenous agonist angiotensin II is known to activate both Gq and β arrestin-mediated signaling, TRV027 was developed and designed to preferentially activate β-arrestin (Violin et al., 2010). This β-arrestin-biased signaling is thought to account for the fact that TRV027 administration both decreases blood pressure and increases cardiac output. To assess the signaling and internalization processes induced by TRV027, we employed AT1R-APEX in the same time-resolved approach described above.
In general, TRV027 led to receptor internalization on a time scale similar to that induced by angiotensin II. For both agonists, β-arrestin was robustly recruited beginning around 50 seconds and reached the maximum enrichment level at 180 seconds before declining slowly thereafter. Clathrin light and heavy chains as well as AP-2 subunits were similarly enriched at 180 seconds, showing that receptor internalization kinetics and the recruitment of proteins involved are mostly independent of ligand bias (Figure 4A, Figure S3B). As with angiotensin II treatment, β-arrestin 2 level declined over 5-fold following the arrival of the endosomal markers Rab5 and Rab7, indicating that arrestin is released from the receptor at later stages of endocytosis.
Figure 4. AT1R-APEX timecourse experiment with β-arrestin biased agonist TRV 027.
(A) Linear plot tracing receptor internalization and trafficking following TRV027 treatment. As in angiotensin II timecourse experiments, β-arrestin 2, clathrin, and Rab5 enrichment levels peak at 180 s, followed thereafter by maximal recruitment of late endosomal markers: Rab7 and V-type proton ATPase catalytic subunit A (ATP6V1A).
(B) Enrichment pattern heatmap of G protein subunits, β-arrestin 2, and Rab5. As in angiotensin II treatment, the recruitment of β-arrestin 2 and receptor internalization occurs commensurate with a loss of receptor-proximal G proteins. Each protein’s enrichment level across different time points is normalized to its maximum enrichment signal.
(C) Linear plot of G protein subunits detected during short time points (0 – 80 s).
(D) Enrichment pattern heatmap of endosomal marker transferrin receptor (TFRC) and components of retromer complex VPS29 and SNX1.
Similar to AT1R experiments with other ligands, in TRV027 treated samples many Gα subunits were quantified over all timepoints. This includes both G proteins that couple strongly (Gα11 and Gαq) and those couple less robustly or not at all (Gαi1, Gαi2, Gαi3, Gαs, Gα13, Gαo1) with AT1R. A detailed analysis of G protein kinetics revealed more similarities and differences in pharmacology between angiotensin II and TRV027. When cells were treated with angiotensin II, G protein labeling efficiency showed an immediate drop followed by a slow rise and then a decrease starting at the 50 seconds timepoint (Figure 3D). TRV027-treated cells showed a more moderate decline in all G protein enrichments starting from 10 seconds until the 30 second timepoint after which labeling efficiency recovered at 50 seconds (Figure 4C). However, decrease in enrichment at early timepoints (0 – 50 s) is challenging to interpret since the labeling events that occur within one timepoint also occur in the subsequent timepoints. G protein labeling efficiency stayed static afterward and dropped thereafter as the activated receptor internalized via clathrin-mediated endocytosis (Figure 4B–C). Later stages of the receptor internalization process appear largely similar between angiotensin II and TRV027-treated samples, with retromer components identified and quantified for both ligands (Figure 4A, Figure 4D).
β2AR-APEX for elucidating β2 adrenergic receptor signaling
A key question surrounding the GPCR-APEX technique presented here is its potential generalizability. In principle, fusion of APEX2 to the receptor carboxy-terminus should allow time-resolved multidimensional tracking of interactions for virtually any GPCR. To test this, we chose to extend our study to the human β2 adrenergic receptor (β2AR). This receptor is one of the most extensively investigated GPCRs in terms of its structure, conformational dynamics, and cell biology. It is the target of β agonists used in the treatment of asthma and other respiratory conditions, including the widely used drugs albuterol and salmeterol. The β2AR is also the prototypical “class A” GPCR (i.e., it internalizes and is recycled rapidly) with respect to its internalization behavior (Oakley et al., 2000). In contrast to the AT1R, the β2AR is rapidly recycled to the plasma membrane following its stimulation with agonists. In addition, internalized β2AR has been suggested to contribute to cell signaling through its persistent activation of G proteins and adenylyl cyclase in endosomes (Irannejad et al., 2013).
First, we designed a β2AR-APEX fusion construct in which the proximity-labeling enzyme is fused to the receptor at its carboxy terminus, as for the AT1R. Then we confirmed β2AR-APEX has signaling properties similar to those of the wild-type receptor (Figure S4A), and assessed its expression level along with that of AT1R-APEX (Figure S4B). As with AT1R-APEX, we generated a HEK293T cell line stably expressing the β2AR-APEX and treated the cells with the high-affinity full agonist BI167107 for varying time intervals (Figure S4C). We selected BI167107 rather than a conventional agonist such as isoproterenol due to the generation of biotin-phenoxy radicals by APEX for proximity-labeling. Catecholamines like isoproterenol are prone to oxidation by radicals, potentially complicating or precluding measurements at short timepoints in which ligands are treated after APEX starts generating biotin-phenoxy radicals. As with the AT1R, we chose to perform 10 second sampling for early events, and then sparser sampling extending to 30 minutes. In total 48,392 peptides were observed, accounting for 3,598 unique proteins that could be tracked throughout the timecourse (Table S3).
As in the case of the AT1R, receptor internalization could be tracked by following endosomal and lysosomal markers. Arrestin with AP-2 and then clathrin appeared in sequence, as did Rab5 and Rab7 followed by LAMP1 (Figure 5A, Figure S5A). Taken together, these data suggest that the receptor ultimately reaches the lysosome, although retromer complex components such as VPS29 are also robustly recruited at late timepoints and may salvage some of the receptor for recycling. However, an important caveat is that the carboxy-terminal fusion of APEX to the β2AR may disrupt endocytic retrieval and recycling by the PDZ domain protein EBP50/NHERF1 (Cao et al., 1999). EBP50 is in fact observed as a receptor-proximal protein, but is depleted commensurate with receptor internalization (Figure 5B).
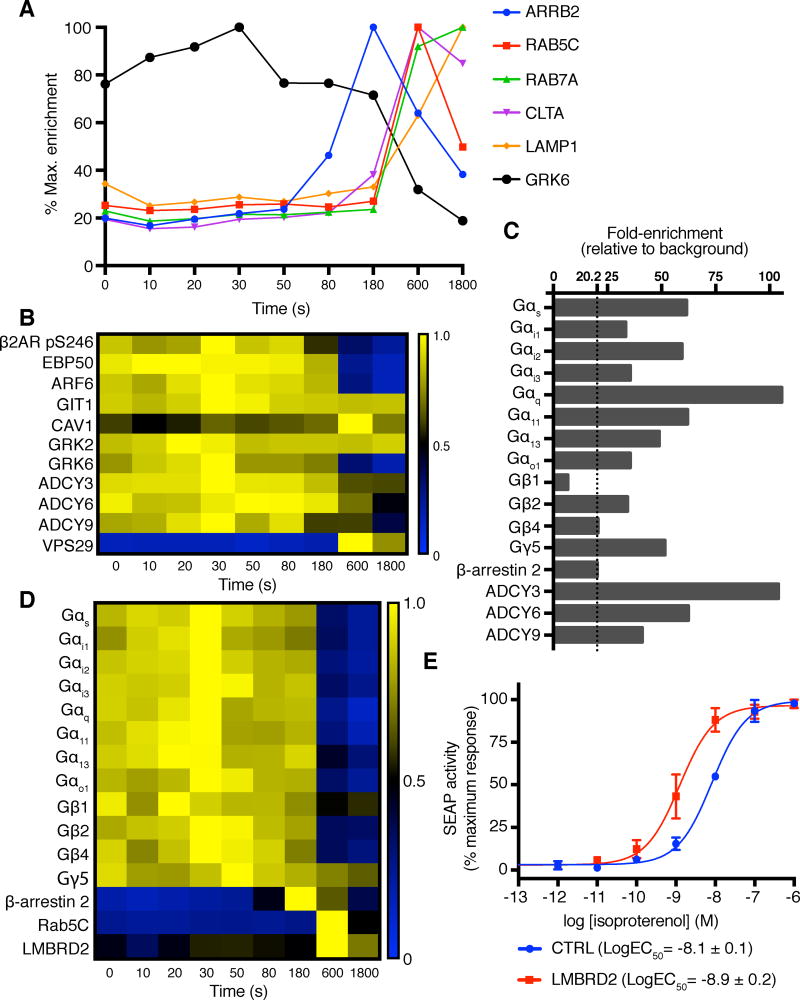
Figure 5. β2AR-APEX timecourse experiment with extremely potent β2AR agonist BI 167107.
(A) Internalization kinetics of β2AR (class A GPCR) are different from those of AT1R (class B GPCR). β-arrestin is recruited with the peak at 180 s, but clathrin recruitment is delayed to 600 s. Also, Rab5 enrichment peak is at 600 s, indicating arrestin-receptor complex formation and endosomal entry do not occur simultaneously. Late endosomal marker Rab7 and lysosomal marker LAMP1 enrichment peaks appear afterward.
(B) Enrichment pattern heatmap of selected proteins. Each protein’s enrichment level across different time points is normalized to its maximum enrichment signal. Receptor internalization results in a loss of receptor-proximal EBP50, ARF6, and GRK6. However, ARF6 activator GIT1, GRK2, and adenylyl cyclase 3 (ADCY3) enrichment levels remained constant. Adenylyl cyclases 6 and 9 (ADCY6 and ADCY9) show moderate decrease in enrichment levels upon receptor internalization. However, significant portion of these proteins remained in proximity of β2AR (>40% of maximum enrichment levels). Gs/adenylyl cyclase signaling attenuator caveolin-1 (CAV1) and retromer component VPS29 highly enrich during internalization (600 s). Heatmap of β2AR S246 phosphorylation level shows a drop in phosphorylation commensurate with receptor internalization.
(C) Fold-enrichment of detected G protein subunits, β-arrestin 2, and adenylyl cyclases prior to ligand treatment. Fold enrichment in each case is relative to levels in control samples that are not treated with hydrogen peroxide (background). β-arrestin 2 is included as a reference to assess significant enrichment levels over background.
(D) Heatmap of all G protein subunits detected shows almost all G proteins are excluded from endosomes regardless of their subtypes.
(E) β2AR signaling assay to examine LMBRD2 knockdown. HEK293T cells transfected with LMBRD2 siRNA showed a statistically significant difference (decrease) in EC50 in comparison to control siRNA-transfected cells (P < 0.0001). Data points for ligand-free cells are not shown. All measurements were performed in triplicate and data are representative of three independent experiments. Data shown as means ± SEM, with EC50 values reported as LogEC50 ± 95% confidence interval for the experiment shown here.
Interestingly, β2AR-APEX was able to capture differences in internalization kinetics of β2AR from AT1R. Robust β-arrestin 2 recruitment with its maximal enrichment at 180 seconds was observed for both receptors. However, maximum or near-maximum enrichment peaks for clathrin light/heavy chains and Rab5 that were at 180 seconds for AT1R all shifted to the 600 second timepoint for β2AR. For both GPCRs, the late endosomal marker Rab7 showed similar maximum/near-maximum enrichment at 600 seconds (Figure 3B, 5A). β-arrestin 2 enrichment level dropped after internalization, in agreement with the reported features of the β-arrestin-GPCR complex for class A receptors (Pierce and Lefkowitz, 2001). In addition, ADP-ribosylation factor 6 (ARF6) and its ARF GTPase-activating protein GIT1 that orchestrate β2AR internalization were quantified: ARF6 levels stayed constant until they dropped more than 4-fold upon internalization, while GIT1 levels remained static through all timepoints as shown in Figure 5B (Claing et al., 2001). Lastly, caveolin-1 (CAV1), which has been suggested to attenuate Gs/adenylyl cyclase signaling by assisting G protein separation from GPCRs (Allen et al., 2009), increased in enrichment starting at 180 seconds until its maximum at 600 seconds.
As for AT1R-APEX, β2AR-APEX analysis revealed a collection of G protein α subunits that were highly enriched relative to background at the 0 s (untreated) timepoint: Gαs, Gαi2, Gαq, and Gα11 were over 50-fold enriched with respect to the background (Figure 5C). Again, β-arrestin 2 showed relatively low enrichment at ligand-free state. Quantified adenylyl cyclases (ADCY3, ADCY6, and ADCY9) were all highly enriched over the background, suggesting that the β2AR is likely in proximity of these proteins even prior to ligand stimulation. Previous work using receptor G protein fusions has suggested a possible direct association between the β2AR and adenylyl cyclases, consistent with this observation (Seifert et al., 2002), although the APEX technique cannot distinguish between direct and indirect interactions. As in the case of the AT1R, these observations support the idea that GPCRs co-localize with the signaling machinery in lipid rafts or other micro-domains prior to activation.
In comparison to AT1R experiments, many more known signaling effectors were detected and quantified in the β2AR experiment. These include the GPCR kinases GRK2 and GRK6 (Figure 5A, B), which selectively phosphorylate activated GPCRs, a prerequisite for efficient β-arrestin recruitment (Pitcher et al., 1998). In our experiments, GRK6 was recruited to the receptor before β-arrestin, with enrichment level peaking at 30 s and then declining thereafter. In contrast, GRK2 levels peaked earlier at 20 s and showed relatively little decline at subsequent timepoints (Figure 5B). As in the case of the AT1R, kinetic analysis revealed that G protein labeling falls coincident with the arrival of endosomal markers such as Rab5, suggesting that relatively little G protein is internalized with the receptor to sustain signaling (Figure 5D). Unlike AT1R, however, β2AR-APEX does not show any significant decrease in G protein subunit labeling immediately (< 20 s) upon agonist binding (Figure S5B). Like G proteins, GRK6 levels decreased upon receptor internalization, falling more than 5-fold from peak levels. In contrast, adenylyl cyclase 3 showed a far more modest decline (< 40% from the peak to the lowest point), indicating that a significant fraction of this protein co-internalized with the β2AR even as the receptor separates from G protein subunits (Figure 5B). In addition to revealing kinetics of protein association, our experiments also allowed tracking of one post-translational modification: phosphorylation of Ser246. The ratio of phosphorylated to non-phosphorylated peptides remained static in early timepoints but dropped coincident with receptor internalization as shown in Figure 5B.
Many of the proteins quantified in timecourse experiments that clustered in the kinetic profiling with known signaling components, like β-arrestin 2, were largely uncharacterized not only in relation to GPCR biology, but also in any context. This was surprising since the biology of the AT1R and β2AR have been studied for decades. To see the potential of GPCR-APEX technique in discovering novel modulators of GPCR signaling, we examined the functional role of one of these proteins, LMBRD2, with respect to receptor signaling.
LMBRD2 is predicted to be a nine-pass transmembrane protein and was identified in both AT1R and β2AR timecourse experiments. Its biology is, to our knowledge, entirely uncharacterized in any context. The protein has relatively low enrichment level prior to agonist treatment, but it is robustly recruited and reached maximum enrichment at 180 seconds and clustered with β-arrestin 2 in AT1R experiments (Figure S3B). In the β2AR timecourse experiment, it showed delayed maximum enrichment at 600 seconds and recruitment kinetics similar to early endosomal marker Rab5 (Figure 5D). We hypothesized that LMBRD2 may have a role in receptor internalization. To examine functional association between LMBRD2 and β2AR, we performed a β2AR G protein signaling assay with HEK293T cells transfected either with a LMBRD2-targeting siRNA or a control siRNA. LMBRD2 knockdown resulted in roughly a seven-fold potentiation of signaling in response to the full-agonist isoproterenol (Figure 5E, Figure S5C). Hence, GPCR-APEX not only allows quantitative tracking of known effectors and regulators, but also holds the potential to enable the discovery of novel mediators of GPCR signaling.
Finally, we investigated the possibility of interactions between the β2AR and other GPCRs. There is evidence suggesting that receptor oligomerization may be important for some GPCRs (Gomes et al., 2016). Hence we sought to assess whether there are other seven-pass transmembrane receptors proximal to the β2AR in a native cellular environment. In total, 16 different seven transmembrane receptors were biotin-labeled and identified by β2AR-APEX: proteinase-activated receptor 2 (F2RL1), frizzled receptors (FZD3, FZD6, FZD7, and FZD8), retinoic acid-inducible receptors (GPRC5A, GPRC5C), an adhesion receptor (GPR126), and several others (GPR98, GPR107, GPR125, GPR180, CELSR1, CELSR2, LGR4, and LGR5; Figure S5D). Receptor enrichment levels remained constant throughout all time points for most of these proteins, indicating that the receptors largely remain associated with the activated receptor even upon internalization. In AT1R timecourse experiments, GPRC5A and GPRC5C were also detected through all time points. As in the case of the β2AR-associated receptors, GPRC5A and GPRC5C co-internalized with AT1R (Figure S5F–G). While the functional implications of this finding remain unknown, these results suggest that GPCR-APEX may also be a useful tool for the interrogation of GPCR interactions with other membrane proteins.
Discussion
The GPCR structural biology revolution has reinvigorated GPCR drug discovery and opened the door to fundamentally new approaches to GPCR modulation, including discovery and design of biased ligands with improved medical potential (Manglik et al., 2016). Despite these transformative innovations in GPCR structure and pharmacology, methods for tracking and quantifying GPCR signaling and internalization have not seen commensurate advances to elucidate remaining questions such as the extent and role of endosomal signaling, the collection of receptor-proximal proteins at baseline, and the similarities and differences of class A and class B GPCR internalization processes. Here, we showed that a new approach using GPCR-APEX fusion combined with isobaric peptide labeling enables massively parallel, time-resolved, and quantitative analysis of GPCR interactions to answer multiple important questions through a single experimental procedure.
The classical view of GPCR-mediated G protein signaling is that stimulated receptors desensitize upon internalization to downregulate signaling and return to the cell membrane after disappearance of stimulus (agonist). Recent reports of endosomal G protein signaling have challenged this view, suggesting that receptors may continue to signal even after endocytosis. However, the relative extent to which G proteins internalize with receptors has been difficult to assess quantitatively. AT1R-APEX and β2AR-APEX timecourse experiments provide a solution to this problem, revealing that almost all G proteins are separated from activated receptors during internalization. This suggests that endocytosis selectively sequesters receptors away from G proteins and allows only a small amount of G proteins to internalize together with the receptor. Surprisingly, however, adenylyl cyclase 3 labeling remains high following β2AR endocytosis, indicating that a significant fraction of this effector is endocytosed together with the receptor.
In addition, analysis of AT1R-APEX and β2AR-APEX in their ligand-free state provides insights into the local milieu of GPCRs in cell membrane. Pre-association between GPCRs and G proteins in lipid rafts has long been proposed, but it has been challenging to prove because of difficulties in isolating and identifying components of lipid rafts in a reproducible manner. GPCR-APEX provides a means to address the issue by labeling receptor proximal proteins in living cells. Both AT1R-APEX and β2AR-APEX confirmed the presence of a variety of G alpha subunits in vicinity of GPCRs even before ligand-treatment, confirming their co-localizations. In addition, lipid raft marker flotillins (FLOT1 and FLOT2) were labeled and detected by both receptors, as was another raft marker, caveolin-1 (Figure 5B, Figure S5E, Figure S5G). Caveolin-1 enrichment level increased upon stimulation and reached peak at 600 s, closely paralleling the drop in G protein enrichment levels. An additional observation is that several seven transmembrane receptors co-localized with AT1R and β2AR, and co-internalized with the activated receptors. While the biological significance of this observation remains unknown, the fact that receptor-receptor associations can be tracked opens the door to quantification of GPCR heteromerization in cells.
GPCR-APEX also allowed discrimination of internalization kinetics between prototypical class A and B GPCRs (β2AR and AT1R, respectively). In the case of AT1R, maximal recruitment of β-arrestin 2 was accompanied by peak enrichment of clathrin light/heavy chains and Rab5, indicating that the receptor enters the endosome as soon as the receptor-arrestin complex forms (Figure 3B). Similar robust recruitment of β-arrestin 2 at 180 seconds was observed for β2AR (Figure 5A). However, clathrin light/heavy chains and Rab5 showed delayed maximum enrichment at 600 seconds, suggesting that the receptor enters endosomes after the receptor-arrestin complex at least partially dissociates. Intriguingly, AT1R-APEX revealed that the receptor-arrestin complex for class B GPCR upon endosomal entry is not as stable as previously thought.
In addition to quantifying GPCR interaction kinetics, GPCR-APEX allowed analysis of one of the receptor’s post-translational modifications. For β2AR, Ser246 is located near the beginning of the unstructured third intracellular loop, a region of the receptor which is important for G protein coupling and other aspects of receptor signaling, but is disordered in the crystal structure of the β2AR-Gs heterotrimer complex (Rasmussen et al., 2011). Of all previously identified β2AR phosphorylation sites, Ser246 is reported as the only site that decreases in phosphorylation level upon agonist-mediated stimulation, while other sites including Ser355/Ser356 are phosphorylated by GRKs upon receptor activation (Nobles et al., 2011). Although a biological role for Ser246 phosphorylation has not yet been described, its relative phosphorylation levels show clear decrease coincident with receptor internalization. It is important to note, however, that the observation of this peptide was largely coincidental, because our protocol was not designed to enrich for phosphopeptides. Nonetheless, this observation shows that it is possible to simultaneously perform time-resolved tracking of both protein-protein interactions as well as receptor post-translational modifications.
Finally, a key value of the proteomics data generated by GPCR-APEX lies in the kinetic profiles of the thousands of receptor-proximal proteins that are detected and quantified. Due to the nature of proximity-labeling, the data necessarily include many bystander proteins that do not functionally associate with the GPCR, but are in the same subcellular locale. Nonetheless, statistical pairwise comparison of agonist-treated and untreated samples (Figure 2F) showed that known effectors are highly enriched in agonist-treated samples, indicating receptor-APEX may be useful in general to identify new signaling components in poorly understood systems even without prior information. Even in the case of the AT1R and β2AR, which have been studied for decades, GPCR-APEX allowed identification and characterization of LMBRD2 as a novel modulator of signaling. Moreover, GPCR-APEX allowed direct quantification of the extent to which G proteins, GRKs, and arrestins interact with the receptor as a function of time. Of particular note, our results show that in the conditions tested, receptors are largely sequestered away from heterotrimeric G proteins upon internalization, while at least one downstream effector, adenylyl cyclase 3, internalizes with the β2AR.
In principle, APEX-fusion with tandem mass tag labeling should be broadly applicable to the kinetic monitoring of virtually any signal transduction protein, such as non-GPCR receptors, β-arrestins, or heterotrimeric G proteins. Overall, GPCR-APEX provides a powerful new platform for quantitative analysis of GPCR signaling, allowing parallel time-resolved measurement of hundreds to thousands of protein-protein interactions and revealing new aspects of GPCR signaling biology.
CONTACT FOR REAGENT AND RESOURCE SHARING
Requests for further information or reagents may be directed to the Lead Contact, Andrew C. Kruse (Andrew_Kruse@hms.harvard.edu)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Flp-In™ T-REx™ 293 Cell Line purchased from Thermo Fisher Scientific was used for all proximity-labeling experiments. Unless noted otherwise, cells were grown at 37°C and 5% CO 2 in DMEM with 4.5 g/L glucose and L-glutamine, no pyruvate (VWR) supplemented with 10% FBS (USA scientific), 10 µg/mL Blasticidin (Invivogen), and 10 µg/mL Gentamicin (VWR). Prior to stable transfection, cell medium was supplemented with 100 µg/mL Zeocin (Thermo Fisher Scientific). All stable cell lines were maintained in medium supplemented with 100 µg/mL Hygromycin B Gold (Invivogen) to maintain selection pressure.
METHOD DETAILS
Design and cloning of GPCR-APEX constructs
pcDNA3.1/zeo/TO plasmid encoding angiotensin II type 1 receptor (AT1R) was a gift from the Lefkowitz lab (Duke University) and was used to PCR-amplify AT1R gene used for the study. β2 adrenergic receptor gene (β2AR) was PCR-amplified from the Roth Lab PRESTO-Tango GPCR Kit (Addgene). APEX2 DNA was PCR-amplified from pcDNA3 APEX2-NES, which was a gift from Alice Ting (Addgene). Each GPCR-APEX fusion protein has a C-terminal 8-residue glycine-serine linker (GGSSGGSS), followed by APEX2. β2AR-APEX also includes a Protein C affinity tag (EDQVDPRLIDGK) at the C-terminus of APEX2. All DNA constructs were cloned into pcDNA5/FRT/TO plasmid for mammalian expression (Invitrogen).
Preparation of stable cell lines for proximity-labeling experiments
Flp-In™ T-REx™293 Cell were seeded in 6-well plates in complete medium without zeocin and were grown until 80 – 90% confluency, at which point transfection was performed using Lipofectamine 2000 (Thermo Fisher Scientific). Plasmid encoding GPCR-APEX was co-transfected with pOG44 Flp-Recombinase Expression Vector (Thermo Fisher Scientific) for stable integration of GPCR-APEX into the genome.
24 hours after transfection, transfected cells were lifted using trypsin-EDTA (Corning) and plated into 10 cm culture dishes. 48 hours after transfection, stably transfected cells were selected by supplementing the medium with 100 µg/mL Hygromycin B Gold. After selection was complete, stable cell lines were grown to make frozen stocks in Bambanker (VWR).
Proximity-labeling experiments
Frozen stable cell line stocks were recovered in 15 cm culture dishes with complete cell medium without Blasticidin until reaching sufficient density for passaging into 10 cm culture dishes. Upon 15–20% confluency, cell medium was supplemented with 1 µg/mL doxycycline hyclate (Sigma Aldrich) to induce GPCR-APEX expression.
48 hours after doxycycline induction, cell medium was replaced with 10 mL of labeling medium (DMEM supplemented with 10% FBS, 10 µg/mL gentamicin, and 500 µM biotinyl tyramide purchased from Toronto Research Chemicals). All cells were incubated in labeling medium for 1 hr before adding hydrogen peroxide (H2O2; Sigma Aldrich) to a final concentration of 1 mM. 30% (v/v) H2O2 stock solution was freshly diluted to 1 M with Dulbecco’s Phosphate Buffered Saline (DPBS) immediately before each experiment. Ligands were added to the cell medium and incubated for the indicated length of time. In all cases, exposure to H2O2 was fixed to a 1 minute labeling interval. For example, 40 second ligand incubation means the ligand was added to the cell medium 20 seconds after adding H2O2 while 80 second ligand incubation means the ligand was added 20 seconds before adding H2O2. For all labeling experiments, H2O2 was added to 1 mM final concentration. Immediately after adding H2O2/ligand, the culture dish was rocked several times to mix. Angiotensin II, Losartan, TRV027, and BI167107 were added to final concentrations of 100 nM, 100 nM, 500 nM, and 100 nM, respectively. In each case, these values are well in excess of ligand binding KD in order to ensure receptor saturation.
Exactly 1 minute after H2O2 treatment, the labeling medium was decanted and cells were washed three times with quenching solution (DPBS supplemented with 10 mM sodium ascorbate, 5 mM trolox, and 10 mM sodium azide). Following washing, cells were re-suspended repeatedly with quenching solution containing 5 mM EDTA. After cells were harvested by centrifugation, quenching solution was aspirated and the pellet was flash-frozen and stored at −80 °C until strepta vidin pull-down.
AT1R-APEX and β2AR-APEX radioligand binding assay
To measure receptor expression levels for AT1R-APEX and β2AR-APEX during proximity-labeling experiments, stable cell lines used for the proximity-labeling experiment were seeded on 10 cm plates, cultured and doxycycline-induced in the conditions used for proximity-labeling experiments. Cells were washed in PBS and subsequently harvested by incubating with PBS with 0.05% EDTA, pH 7.4, for 10 minutes at 4 oC. Cells were collected and centrifuged at 1000 × RPM for 5 minutes and re-suspended in assay buffer (20 mM HEPES, 100 mM NaCl, pH 7.4).
AT1R-APEX expression was assayed using radioligand binding using 50 nM [3H]-olmesartan. Binding reactions were conducted in assay buffer with 1 mg/mL bovine serum albumin (Rockland), pH 7.4. Specific binding was determined by subtracting nonspecific binding (10 µM candesartan) from total binding (assay buffer). Reactions were incubated for 90 minutes at room temperature and subsequently washed (50 mM Tris, pH 7.4) and captured on GF-B glass fiber filters using a Brandel harvester (Brandel). β2AR-APEX expression level was assayed using [3H]-dihydroalprenolol at 10 nM and 10 µM propranolol was used to measure non-specific binding by same procedure. Protein concentrations were measured using Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific).
AT1R-APEX ERK signaling assay
Expi293F HEK293 cells (Thermo Fisher Scientific) (10 mL) stably expressing pcDNA™6/TR (Thermo Fisher Scientific) were transiently transfected and enhanced following the manufacturer’s protocol with pcDNA-AT1R–WT or AT1R-APEX under the control of a TET-inducible promoter. Receptor expression was induced 48 hours after transfection by addition of 5 mM sodium butyrate and 4 µg/mL doxycycline. To induce AT1R activation and downstream ERK phosphorylation, 24 hours post-transfection cells were left untreated (control) or treated with 10 µM angiotensin II. At the indicated time following angiotensin II stimulation, 200 µL of cells were removed and cell lysed by the addition of 200 µL 2x Laemmli buffer. Cell lysates were sonicated for 20 seconds and remaining cell debris was pelleted at 14,000 × RPM for 15 minutes. Cell lysates were subject to SDS PAGE and levels of phosphorylated ERK (p44/42, Thr202/Tyr204) and total cellular ERK (tERK) were determined by traditional western blotting techniques using anti-phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) antibody (Cell Signaling Technology) and anti-MAP Kinase 1/2 (Erk1/2) Antibody (EMD Millipore).
AT1R-APEX internalization assay
β-arrestin-dependent internalization of wild-type AT1R and AT1R-APEX was measured as described previously (Strachan et al., 2014) using the activated GPCR endocytosis assay (DiscoveRx), based on complementation of β-galactosidase “Enzyme Acceptor” and “ProLink” fragments. Briefly, U2OS cells with stably integrated Enzyme Acceptor-tagged β-arrestin 2 and an endosome-localized ProLink tag were transfected transiently with N-terminal Flag-tagged wild-type AT1R or AT1R-APEX constructs. Transfections were performed using FuGENE® 6 (Promega) using a 1:5 ratio of DNA/transfection reagent. One day post-transfection, cells were seeded into 96-well plates at a density of 35,000 cells/well using AssayComplete™ Cell Plating 5 Reagent. Two days post-transfection, cells were stimulated with a serial dilution of angiotensin II for 3 hours at 37°C and then treated with PathHunter® detection reagents. After an additional 1-hour incubation at room temperature, luminescence was detected using a NOVOstar microplate reader (BMG Labtech).
β2AR-APEX signaling assay
Signaling assays for β2AR-APEX and β2AR wild-type were performed as described previously with some modifications (Liberles and Buck, 2006). HEK293T (ATCC) cells were seeded in 96-well plates to achieve 70–80% confluency on the day of transfection. Each well was transfected with 20 ng each of a pcDNA5 encoding β2AR-APEX/β2AR wild-type and CRE-SEAP reporter plasmid (BD Biosciences). Transfection was performed using Lipofectamine 2000 (Thermo Fisher Scientific) following manufacturer’s instructions. After incubation of the cells with the transfection mixture and serum-free media for 5 hours at 37 °C , transfec tion mixtures were aspirated and replaced with fresh DMEM with 4.5 g/L glucose and L-glutamine, no pyruvate (VWR), supplemented with 10 µg/mL Gentamicin (VWR). Then transfected cells were treated with indicated final concentrations of isoproterenol (Sigma Aldrich) and incubated at 37 °C for 48 hours followed by 2 hours of incubation a t 70°C. Supernatant from each well was mixed with an equal volume of 0.12 mM 4-methylumbelliferyl phosphate (Sigma Aldrich) in 2 M diethanolamine bicarbonate (diethanolamine from Alfa Aesar diluted in Milli-Q and supplemented with dry ice), pH 10, and incubated for 9 minutes at room temperature. Lastly, the fluorescence was measured using EnVision 2103 Multilabel Reader (PerkinElmer).
LMBRD2 Knockdown assay
HEK293T cells (ATCC) were seeded in 15 cm plates to achieve 50 – 60% confluency on the day of first transfection. Each plate was transfected with either Silencer Select LMBRD2 siRNA (Thermo Fisher Scientific) or AllStars Negative Control siRNA (QIAGEN) using Lipofectamine RNAiMAX (Thermo Fisher Scientific) according to manufacturer’s protocol. The final siRNA concentration was 25 nM in both cases. 24 hours after siRNA transfection, cells were trypsinized and seeded in 96-well plates to achieve 70 – 80% confluency on the following day. Then each well was transfected with plasmid encoding wild-type β2AR and agonist-treated as described above for β2AR-APEX signaling assay. Transfected cells were incubated for 24 hours after agonist-treatment and were assayed for fluorescence measurement (72 hours after siRNA transfection). The difference in Log(EC50) values was assessed by an unpaired two-sided Student’s t-test.
For qPCR, cells transfected with siRNA was harvested using RNeasy Mini Kit (QIAGEN) according to manufacturer’s protocol. Extracted RNA was reverse-transcribed using SuperScript II Reverse Transcriptase (Thermo Fisher Scientific) with Random Primers (Thermo Fisher Scientific). Resulting cDNA was mixed with PowerUp SYBR Green Master Mix (Thermo Fisher Scientific) and either ACTB or LMBRD2 qPCR primers (Eurofins Genomics). qPCR was performed and analyzed using QuantStudio 6 Flex Real-Time PCR System (Thermo Fisher Scientific).
Streptavidin pull-down of biotinylated proteins
All stock solutions and buffers used for streptavidin pull-down experiments were prepared freshly and filtered through 0.22 µm filters. Frozen cells were lysed with lysis solution (2 M sodium hydroxide with 7.5% 2-mercaptoethanol in Milli-Q water) and left on ice for 15 minutes. Lysates were pipetted repeatedly to ensure complete cell lysis. For subsequent TCA precipitation, an equal volume of ice-cold 55% trichloroacetic acid (TCA) was added followed by incubation on ice for 15 minutes.
Proteins were precipitated by centrifugation at 21,130 × g at 4 °C for 10 min. The pellet was washed with −20 °C cold acetone, vortexe d, and centrifuged at 21,130 × g at 4 °C for 10 minutes using a tabletop centrifuge. Fo llowing centrifugation, acetone was aspirated and the pellet was acetone-washed again three more times. After the last washing step, re-suspension buffer (8M urea, 100 mM sodium phosphate pH 8, 100 mM NH4HCO3, 1% SDS (w/v), 10 mM TCEP in Milli-Q water titrated to pH 7 with NaOH) was added to the pellets. Tubes containing pellets and re-suspension buffer were sonicated in a Bioruptor sonifier bath (Diagenode) with 3 cycles for 30 seconds each, 100% duty cycle, highest power setting. Samples were then vortexed for 1 hour at room temperature to completely re-suspend proteins and allow for reduction of proteins by TCEP. Pellet dissolvation and alkaline pH of the solution was checked afterwards.
Re-suspended proteins were centrifuged at 21,130 × g at room temperature for 10 minutes and the clear supernatant was transferred into new microcentrifuge tubes. Freshly prepared 400 mM iodoacetamide stock solution in 50 mM ammonium bicarbonate was added to the supernatant to a final concentration of 20 mM, immediately vortexed, and incubated in the dark for 25 minutes at room temperature. After alkylation, freshly prepared dithiothreitol stock solution was added to 50 mM final concentration to quench alkylation. Water was added to each sample to reach final concentrations of 4 M urea and 0.5% (w/v) of SDS.
A 75 µL suspension equivalent per sample of streptavidin magnetic beads (Thermo Fisher Scientific) was washed twice with 4 M urea, 0.5% SDS (w/v), 100 mM sodium phosphate pH 8 and was added to each sample and tubes were rotated overnight at 4 °C . Aliquots containing 5% of protei ns in re-suspension buffer of each sample were saved prior to pull-down and 1.5% of total (inputs) were subjected to SDS PAGE and Western blotting analysis with StreptActin-HRP conjugate (Bio-Rad) to assess biotinylation of inputs. Following streptavidin pull-down, magnetic beads were washed three times with 4 M urea, 0.5% SDS (w/v), 100 mM sodium phosphate pH 8, and three times with the same buffer without SDS to avoid interference of SDS with downstream mass spectrometry analysis. Beads were transferred to fresh tubes after the first washing step.
Western blotting analysis of biotinylated proteins
Samples of cell lysates saved after TCA-precipitation and re-suspension (inputs) were analyzed by Western blotting. After standard SDS-PAGE, proteins were transferred to nitrocellulose membranes and stained with Ponceau S (Sigma Aldrich) prior to blocking to monitor equal protein loading. Membranes were blocked with 4% (w/v) non-fat milk powder in TBST (0.1% Tween-20 in Tris-buffered saline) at room temperature for 2 hours. Blocked membranes were incubated with StreptActin-HRP conjugate (Bio-Rad) diluted 1:50,000 in TBST with 4% non-fat milk powder overnight at 4 °C with constant shaking. Prior to chemiluminesce nt detection, blots were washed with TBST four times, 5 minutes each time. Western blots were developed using Amersham ECL Western Blotting Detection Kit (GE Healthcare).
On-bead digestion and TMT labeling
All liquid reagents used were HPLC quality grade. Washed beads were re-suspended in 50 ml of freshly prepared and filtered 2 M Urea, 50 mM ammonium bicarbonate, 8% acetonitrile (v/v) with 1ml of LysC stock solution (2 mg/ml, Wako), vortexed briefly and incubated at 37°C for 3 hours. Samples were then diluted 1:4 with 50 mM ammonium bicarbonate to lower the concentration of urea to 0.5 M and trypsin stock (Promega) was added 1:200 (v/v). After mixing, digests were continued for at least 6 hours at 37°C or overnight and beads were m agnetically removed. Digests were acidified with trifluoroacetic acid (TFA) and peptides were desalted and purified by C18 solid phase extraction. Peptides dried in a vacuum centrifuge (SpeedVac) to completion were re-suspended in 200 mM HEPES pH 8.5, 30% acetonitrile (v/v) and labeled with TMT 10-plex reagents (Thermo Fisher Scientific) for 1 hour. Reactions were quenched with hydroxylamine at a final concentration of 0.3% (v/v) for 15 minutes and 1% of labeled peptides were analyzed for efficiency of label incorporation and relative ratios by mass spectrometry.
Labeled and quenched peptides were pooled and 10% of the pool was separated to obtain unfractionated samples and samples were dried to near completion in a speed vac. Samples were re-suspended in water and acidified with TFA. Fractionation of labeled peptides was done by high pH reversed phase (Thermo Fisher Scientific). After loading of labeled peptides onto pre-conditioned columns and a single wash with water, excess unincorporated TMT label was removed by washing reverse phase columns once with 0.1% trimethylamine (TEA) buffer containing 5% acetonitrile. Samples were fractionated under alkaline conditions into 12 fractions with increasing concentrations of acetonitrile: 10%, 12.5%, 15%, 17.5%, 20%, 25%, 30%, 35%, 40%, 50%, 65% and 80%. Fractions 1 and 7, 2 and 8, 3 and 9, 4 and 10, 5 and 11, 6 and 12 were pooled to obtain 6 final pooled fractions for subsequent analysis. Pooled fractions and the unfractionated sample were dried to completion and further purified and desalted by acidic C18 solid phase extraction (StageTip). For the unfractionated sample, unincorporated TMT reagents were removed by an additional washing step with 5% acetonitrile (v/v) and 1% formic acid (v/v) after peptide binding. Labeled peptides were finally re-suspended in 5% formic acid (v/v) and 5% acetonitrile (v/v).
Mass spectrometry analysis
Data were collected by an SPS MS3 TMT method (McAlister et al., 2014) using Orbitrap Fusion Lumos mass spectrometers (Thermo Fisher Scientific) coupled to a Proxeon EASY-nLC 1000 liquid chromatography (LC) system (Thermo Fisher Scientific). The SPS MS3 method uses a notched waveform to select, isolate, and co-fragment the 10 most intense product ions from an MS2 spectrum to produce the reporter ions used for relative quantification. The capillary column was a 100 µm inner diameter microcapillary column packed with ~35 cm of Accucore C18 resin (2.6 µm, 150 Å, Thermo Fisher Scientific).
For each analysis, we loaded ~2 µg onto the column. Peptides of each fraction were separated in 2.5 hour acidic acetonitrile gradients by LC prior to MS injection. The scan sequence began with a MS1 spectrum (Orbitrap analysis; resolution 120,000; mass range 400–1400 Th). MS2 analysis followed collision-induced dissociation (CID, CE=35) with a maximum ion injection time of 120 ms and an isolation window of 0.7 Da. Precursors for MS2/MS3 analysis were selected using a Top10 method. To obtain quantitative information, MS3 precursors were fragmented by high-energy collision-induced dissociation (HCD, CE=65) and analyzed in the Orbitrap (resolution was 60,000 at 200 Th) with a maximum ion injection time of 150 ms and a charge state-dependent variable isolation window of 0.7 to 1.2 Da, as performed previously (Paulo et al., 2016a) Peptides were searched using a SEQUEST-based in-house software against a human proteome database with a target decoy database strategy. Spectra were converted to mzXML using a modified version of ReAdW.exe. Searches were performed using a 50 ppm precursor ion tolerance for total protein level profiling. The product ion tolerance was set to 0.9 Da. These wide mass tolerance windows were chosen to maximize sensitivity in conjunction with Sequest searches and linear discriminant analysis (Beausoleil et al., 2006; Huttlin et al., 2010). TMT tags on lysine residues and peptide N termini (+229.163 Da) and carbamidomethylation of cysteine residues (+57.021 Da) were set as static modifications, while oxidation of methionine residues (+15.995 Da) was set as a variable modification. Peptide-spectrum matches (PSMs) were identified, quantified, and collapsed to a 1% peptide false discovery rate (FDR) and then collapsed further to a final protein-level FDR of 1%. Moreover, protein assembly was guided by principles of parsimony to produce the smallest set of proteins necessary to account for all observed peptides.
Quantitative information on peptides was derived from MS3 scans. PSMs with poor quality, MS3 spectra with more than eight TMT reporter ion channels missing, MS3 spectra with TMT reporter summed signal-to-noise ratio less than 100, or no MS3 spectra were excluded from quantification. Proteins were quantified by summing reporter ion counts across all matching PSM. Data tables (Table S1–3) were generated requiring an MS2 isolation specificity of >70% for each peptide and a sum of TMT s/n of >100 across all channels for any given peptide and exported to Excel and further processed therein (Paulo et al., 2016b). A modified version of the Ascore algorithm was used to quantify the confidence assignment of phosphorylation sites. Phosphorylation localized to particular residues required Ascore values >13 (P≤0.05) for confident localization (Huttlin et al., 2010).
Data were normalized to Acetyl-CoA Carboxylase signal (ACACA), an endogenously biotinylated protein present in all experiments after streptavidin pulldown. Enrichment of proteins over background (no H2O2 control) was calculated from the sum of TMT s/n for all peptides quantified of a given protein. Quantification data were imported into interactive computable documents (Wolfram CDF) generated from in-house developed templates for analysis of complex TMT datasets. Changes in enrichment over time upon agonist exposure and clustering of proteins with similar patterns were visualized within CDFs. Statistical analysis was performed using Graphpad Prism 7 (Graphpad Software). Raw files are available upon request.
Supplementary Material
(A) ERK signaling assay confirms AT1R-APEX activates downstream ERK signaling similarly to wild-type AT1R.
(B) AT1R-APEX internalization assay to confirm AT1R-APEX undergoes β-arrestin-dependent internalization upon angiotensin II treatment similarly to wild-type AT1R. All measurements were performed in duplicate and repeated in three independent experiments. Errors shown as means ± SEM.
Linear regression analysis of biological replicates treated with (A) losartan for 2 minutes, (B) losartan for 21 minutes, and (C) angiotensin II for 21 minutes. (D–F) Representative MS1, MS2, and MS3 spectra of an ARRB2 tryptic peptide, respectively.
Proteins identified to show the most similar kinetic association pattern as a selection of representative proteins (in blue) following (A) angiotensin II or (B) TRV 027 treatment. Grouping was performed by hierarchical clustering of relative protein signals with Euclidian distance as clustering metric.
(A) β2AR-APEX signaling assay to confirm β2AR-APEX has G protein signaling property similar to the wild-type. Data points for ligand-free cells are not shown. All measurements were performed in triplicate and repeated in three independent experiments. Errors shown as means ± SEM.
(B) Radioligand saturation binding assay with AT1R-antagonist [3H]-olmesartan and β2AR antagonist [3H]-dihydroalprenolol to assess β2AR-APEX expression level relative to AT1R-APEX during proximity-labeling experiments. Errors shown as means ± SD, from two independent experiments. In each independent experiment, n=2 for total binding and n=1 for non-specific binding.
(C) Biotinylated proteins from the proximity-labeling experiment analyzed by streptavidin-HRP blot.
(A) Proteins identified to show the most similar kinetic enrichment pattern as the selected proteins (in blue) upon treatment with the agonist BI167107.
(B) Linear plot of G protein subunits detected during short time points (0 – 80 s), showing relatively stable levels of G protein enrichment in this timeframe. No decline in G protein labeling immediately upon agonist-treatment is observed for the β2AR, unlike the AT1R.
(C) Decrease in mRNA expression level of LMBRD2 upon siRNA-knockdown was confirmed by qPCR. All measurements were performed in triplicate and repeated in three independent experiments. Errors shown as means ± SEM.
(D) Enrichment pattern heatmap of β2AR-proximal seven transmembrane receptors and endosomal marker Rab5.
(E) Enrichment pattern heatmap of β2AR and lipid raft markers (FLOT1/2)
(F) Fold-enrichment of AT1R and AT1R-proximal GPCRs upon angiotensin II/losartan treatment. Fold enrichment in each case is relative to ligand-free sample. Errors shown as means ± SEM for two biological replicates.
(G) Enrichment pattern heatmap of AT1R, GPRC5A, and raft marker FLOT1/2 from angiotensin timecourse experiment.
Tab 1: proteins quantified from AT1R-APEX timecourse study with angiotensin II, with their enrichment levels at different time points. Tab 2: proteins from AT1R-APEX timecourse study with TRV 027, with their enrichment levels at different time points.
HIGHLIGHTS.
GPCR agonist response tracked by time-resolved labeling of receptor-proximal proteins
GPCRs co-localize with a variety of G proteins even before activation
G proteins are largely separated from receptors upon receptor endocytosis
In-cell time-resolved labeling technique is generalizable to multiple receptor types
In brief.
Using APEX proximity labeling to monitor GPCR signaling provides both spatial and temporal perspectives on the factors involved in biased signaling and receptor internalization.
Acknowledgments
We thank Robert J. Lefkowitz for insightful comments and critical reading of the manuscript. Funding was provided by the Smith Family Foundation (A.C.K), the Vallee Foundation (A.C.K.), the Esther A. and Joseph Klingenstein Fund (A.C.K.), Biogen Idec (S.P.G.) and NIH grants R01HL016037 and 1DP5OD021345. J.A.P. was funded by NIH grant K01DK098285.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
J.P. designed GPCR-APEX constructs, created stable cell lines, performed proximity-labeling experiments, performed adrenergic receptor functional assays, and assayed LMBRD2 knockdown. M.K. developed the biochemical purification scheme, processed samples from proximity-labeling experiments, assayed sample labeling, prepared fractionated TMT mass spectrometry samples, analyzed mass spectrometry data, and performed cluster analysis. M.K. and J.A.P. performed mass spectrometry method design. D.P.S. performed radioligand binding assays and ERK signaling assays. L. W. performed angiotensin receptor internalization assays. R.P. assisted in writing of the manuscript and in experimental design. S.P.G. wrote cluster analysis software and oversaw mass spectrometry procedures. A.C.K. designed the overall project. J.P. and A.C.K. wrote the manuscript with input from the other authors.
References
- Allen JA, Jiang ZY, Dave RH, Bhatnagar A, Roth BL, Rasenick MM. Caveolin-1 and lipid microdomains regulate Gs trafficking and attenuate Gs/adenylyl cyclase signaling. Mol Pharmacol. 2009;76:1082–1093. doi: 10.1124/mol.109.060160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beausoleil SA, Villén J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nature biotechnology. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- Cao TT, Deacon HW, Reczek D, Bretscher A, von Zastrow M. A kinase-regulated PDZ-domain interaction controls endocytic sorting of the β2-adrenergic receptor. Nature. 1999;401:286–290. doi: 10.1038/45816. [DOI] [PubMed] [Google Scholar]
- Claing A, Chen W, Miller WE, Vitale N, Moss J, Premont RT, Lefkowitz RJ. β-Arrestin-mediated ADP-ribosylation Factor 6 Activation and β2-Adrenergic Receptor Endocytosis. Journal of Biological Chemistry. 2001;276:42509–42513. doi: 10.1074/jbc.M108399200. [DOI] [PubMed] [Google Scholar]
- Gilman AG. G Proteins and Regulation of Adenylate Cyclase (Nobel Lecture) Angewandte Chemie International Edition in English. 1995;34:1406–1419. [Google Scholar]
- Gomes I, Ayoub MA, Fujita W, Jaeger WC, Pfleger KD, Devi LA. G protein-coupled receptor heteromers. Annu Rev Pharmacol Toxicol. 2016;56:403–425. doi: 10.1146/annurev-pharmtox-011613-135952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Statistics in medicine. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- Hung V, Udeshi ND, Lam SS, Loh KH, Cox KJ, Pedram K, Carr SA, Ting AY. Spatially resolved proteomic mapping in living cells with the engineered peroxidase APEX2. Nature protocols. 2016;11:456–475. doi: 10.1038/nprot.2016.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villén J, Haas W, Sowa ME, Gygi SP. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irannejad R, Tomshine JC, Tomshine JR, Chevalier M, Mahoney JP, Steyaert J, Rasmussen SG, Sunahara RK, El-Samad H, Huang B, et al. Conformational biosensors reveal GPCR signalling from endosomes. Nature. 2013;495:534–538. doi: 10.1038/nature12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefkowitz RJ. A Brief History of G-Protein Coupled Receptors (Nobel Lecture) Angewandte Chemie International Edition. 2013;52:6366–6378. doi: 10.1002/anie.201301924. [DOI] [PubMed] [Google Scholar]
- Manglik A, Lin H, Aryal DK, McCorvy JD, Dengler D, Corder G, Levit A, Kling RC, Bernat V, Hubner H, et al. Structure-based discovery of opioid analgesics with reduced side effects. Nature. 2016;537:185–190. doi: 10.1038/nature19112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberles SD, Buck LB. A second class of chemosensory receptors in the olfactory epithelium. Nature. 2006;442:645–650. doi: 10.1038/nature05066. [DOI] [PubMed] [Google Scholar]
- McAlister GC, Nusinow DP, Jedrychowski MP, Wuhr M, Huttlin EL, Erickson BK, Rad R, Haas W, Gygi SP. MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Analytical chemistry. 2014;86:7150–7158. doi: 10.1021/ac502040v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobles KN, Xiao K, Ahn S, Shukla AK, Lam CM, Rajagopal S, Strachan RT, Huang T-Y, Bressler EA, Hara MR, et al. Distinct Phosphorylation Sites on the β2-Adrenergic Receptor Establish a Barcode That Encodes Differential Functions of β-Arrestin. Science Signaling. 2011;4:ra51–ra51. doi: 10.1126/scisignal.2001707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley RH, Laporte SA, Holt JA, Caron MG, Barak LS. Differential affinities of visual arrestin, beta arrestin1, and beta arrestin2 for G protein-coupled receptors delineate two major classes of receptors. J Biol Chem. 2000;275:17201–17210. doi: 10.1074/jbc.M910348199. [DOI] [PubMed] [Google Scholar]
- Oates J, Watts A. Uncovering the intimate relationship between lipids, cholesterol and GPCR activation. Current opinion in structural biology. 2011;21:802–807. doi: 10.1016/j.sbi.2011.09.007. [DOI] [PubMed] [Google Scholar]
- Paulo JA, O’Connell JD, Everley RA, O’Brien J, Gygi MA, Gygi SP. Quantitative mass spectrometry-based multiplexing compares the abundance of 5000 S. cerevisiae proteins across 10 carbon sources. Journal of Proteomics. 2016a;148:85–93. doi: 10.1016/j.jprot.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulo JA, O’Connell JD, Gygi SP. A Triple Knockout (TKO) Proteomics Standard for Diagnosing Ion Interference in Isobaric Labeling Experiments. Journal of The American Society for Mass Spectrometry. 2016b;27:1620–1625. doi: 10.1007/s13361-016-1434-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce KL, Lefkowitz RJ. Classical and new roles of β-arrestins in the regulation of G-protein-coupled receptors. Nature reviews neuroscience. 2001;2:727–733. doi: 10.1038/35094577. [DOI] [PubMed] [Google Scholar]
- Pitcher JA, Freedman NJ, Lefkowitz RJ. G PROTEIN-COUPLED RECEPTOR KINASES. Annual Review of Biochemistry. 1998;67:653–692. doi: 10.1146/annurev.biochem.67.1.653. [DOI] [PubMed] [Google Scholar]
- Rasmussen SG, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux KJ, Kim DI, Raida M, Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. The Journal of Cell Biology. 2012;196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert R, Wenzel-Seifert K, Arthur JM, Jose PO, Kobilka BK. Efficient adenylyl cyclase activation by a beta2-adrenoceptor-G(i)alpha2 fusion protein. Biochemical and biophysical research communications. 2002;298:824–828. doi: 10.1016/s0006-291x(02)02569-x. [DOI] [PubMed] [Google Scholar]
- Strachan RT, Sun JP, Rominger DH, Violin JD, Ahn S, Rojas Bie Thomsen A, Zhu X, Kleist A, Costa T, Lefkowitz RJ. Divergent transducer-specific molecular efficacies generate biased agonism at a G protein-coupled receptor (GPCR) J Biol Chem. 2014;289:14211–14224. doi: 10.1074/jbc.M114.548131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A, Schafer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Johnstone R, Mohammed AK, Hamon C. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Analytical chemistry. 2003;75:1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- Thomsen AR, Plouffe B, Cahill TJ, 3rd, Shukla AK, Tarrasch JT, Dosey AM, Kahsai AW, Strachan RT, Pani B, Mahoney JP, et al. GPCR-G Protein-beta-Arrestin Super-Complex Mediates Sustained G Protein Signaling. Cell. 2016;166:907–919. doi: 10.1016/j.cell.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting L, Rad R, Gygi SP, Haas W. MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat Methods. 2011;8:937–940. doi: 10.1038/nmeth.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Violin JD, Crombie AL, Soergel DG, Lark MW. Biased ligands at G-protein-coupled receptors: promise and progress. Trends in Pharmacological Sciences. 2014;35:308–316. doi: 10.1016/j.tips.2014.04.007. [DOI] [PubMed] [Google Scholar]
- Violin JD, DeWire SM, Yamashita D, Rominger DH, Nguyen L, Schiller K, Whalen EJ, Gowen M, Lark MW. Selectively engaging beta-arrestins at the angiotensin II type 1 receptor reduces blood pressure and increases cardiac performance. J Pharmacol Exp Ther. 2010;335:572–579. doi: 10.1124/jpet.110.173005. [DOI] [PubMed] [Google Scholar]
- Zhang H, Unal H, Gati C, Han GW, Liu W, Zatsepin NA, James D, Wang D, Nelson G, Weierstall U, et al. Structure of the Angiotensin receptor revealed by serial femtosecond crystallography. Cell. 2015;161:833–844. doi: 10.1016/j.cell.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) ERK signaling assay confirms AT1R-APEX activates downstream ERK signaling similarly to wild-type AT1R.
(B) AT1R-APEX internalization assay to confirm AT1R-APEX undergoes β-arrestin-dependent internalization upon angiotensin II treatment similarly to wild-type AT1R. All measurements were performed in duplicate and repeated in three independent experiments. Errors shown as means ± SEM.
Linear regression analysis of biological replicates treated with (A) losartan for 2 minutes, (B) losartan for 21 minutes, and (C) angiotensin II for 21 minutes. (D–F) Representative MS1, MS2, and MS3 spectra of an ARRB2 tryptic peptide, respectively.
Proteins identified to show the most similar kinetic association pattern as a selection of representative proteins (in blue) following (A) angiotensin II or (B) TRV 027 treatment. Grouping was performed by hierarchical clustering of relative protein signals with Euclidian distance as clustering metric.
(A) β2AR-APEX signaling assay to confirm β2AR-APEX has G protein signaling property similar to the wild-type. Data points for ligand-free cells are not shown. All measurements were performed in triplicate and repeated in three independent experiments. Errors shown as means ± SEM.
(B) Radioligand saturation binding assay with AT1R-antagonist [3H]-olmesartan and β2AR antagonist [3H]-dihydroalprenolol to assess β2AR-APEX expression level relative to AT1R-APEX during proximity-labeling experiments. Errors shown as means ± SD, from two independent experiments. In each independent experiment, n=2 for total binding and n=1 for non-specific binding.
(C) Biotinylated proteins from the proximity-labeling experiment analyzed by streptavidin-HRP blot.
(A) Proteins identified to show the most similar kinetic enrichment pattern as the selected proteins (in blue) upon treatment with the agonist BI167107.
(B) Linear plot of G protein subunits detected during short time points (0 – 80 s), showing relatively stable levels of G protein enrichment in this timeframe. No decline in G protein labeling immediately upon agonist-treatment is observed for the β2AR, unlike the AT1R.
(C) Decrease in mRNA expression level of LMBRD2 upon siRNA-knockdown was confirmed by qPCR. All measurements were performed in triplicate and repeated in three independent experiments. Errors shown as means ± SEM.
(D) Enrichment pattern heatmap of β2AR-proximal seven transmembrane receptors and endosomal marker Rab5.
(E) Enrichment pattern heatmap of β2AR and lipid raft markers (FLOT1/2)
(F) Fold-enrichment of AT1R and AT1R-proximal GPCRs upon angiotensin II/losartan treatment. Fold enrichment in each case is relative to ligand-free sample. Errors shown as means ± SEM for two biological replicates.
(G) Enrichment pattern heatmap of AT1R, GPRC5A, and raft marker FLOT1/2 from angiotensin timecourse experiment.
Tab 1: proteins quantified from AT1R-APEX timecourse study with angiotensin II, with their enrichment levels at different time points. Tab 2: proteins from AT1R-APEX timecourse study with TRV 027, with their enrichment levels at different time points.