Abstract
Our groups have recently developed related approaches for sample preparation for super-resolution imaging within endogenous cellular environments using correlative light and electron microscopy (CLEM). Four distinct techniques for preparing and acquiring super-resolution CLEM datasets on aldehyde-fixed specimens are provided, including Tokuyasu cryosectioning, whole-cell mount, cell unroofing and platinum replication, and resin embedding and sectioning. Choice of the best protocol for a given application depends on a number of criteria that are discussed in detail. Tokuyasu cryosectioning is relatively rapid but is limited to small, delicate specimens. Whole-cell mount has the simplest sample preparation but is restricted to surface structures. Cell unroofing and platinum replica creates high-contrast, 3-dimensional images of the cytoplasmic surface of the plasma membrane, but is more challenging than whole-cell mount. Resin embedding permits serial sectioning of large samples, but is limited to osmium-resistant probes, and is technically difficult. Expected results from these protocols include super-resolution localization (~10–50 nm) of fluorescent targets within the context of electron microscopy ultrastructure, which can help address cell biological questions. These protocols can be completed in 2–7 days, are compatible with a number of super-resolution imaging protocols, and are broadly applicable across biology.
Introduction
Since the advent of microscopy, scientists have sought ways to provide contrast to tissues, cells and subcellular structures in order to enhance visualization. At first, stains that delineated specific cellular populations within tissues were developed (e.g., Golgi stain for neurons), followed by methods to visualize sub-cellular objects (e.g., hematoxylin for nucleic acids). Concomitant improvements in sample processing and preservation were additionally developed to maximize utility for biological specimens. The invention of electron microscopy (EM) provided unprecedented nanometer-level resolution of cellular structures; on the other hand, it has suffered from low throughput and the limitations of current methods for providing protein-specific information. Therefore, light and electron microscopy largely diverged as separate modalities, each with distinct staining, sample processing and imaging methods.
The discovery and optimization of fluorescent proteins (FPs) like green fluorescent protein (GFP)1,2 reinvigorated light microscopy owing to the superb specificity of the label (genetic encoding enabling easy targeting of specific proteins, organelles and cell populations in diverse organisms)3. A broad palette of FPs is available across the visible spectrum, and FPs routinely provide high contrast over specimen background autofluorescence. The main drawback to fluorescence microscopy compared to EM has historically been the resolution limit, i.e. the size of the smallest discernible structures in the sample. Using conventional microscopy under ideal conditions, fluorescence signals can only be localized to within ~200 nm, compared to the sub-nanometer resolution of electron microscopy.
Recent advances both in labels (i.e. the discovery and engineering of photoactivatable or photoswitchable FPs (paFPs)4) and in microscope design and hardware have given birth to a new generation of fluorescence microscopy approaches with dramatically improved spatial resolution (<50 nm, and as low as ~10 nm under ideal conditions), collectively dubbed “super-resolution imaging”5–10. A major super-resolution imaging modality is single-molecule localization microscopy (SMLM), which relies on the serial determination of the location of individual emitting molecules. Localization microscopy techniques include Photoactivatable Localization Microscopy (PALM)5 and Fluorescence PALM (FPALM)6, which rely on paFP labels, and Stochastic Optical Reconstruction Microscopy (STORM)7 and direct STORM (dSTORM)11, which use photoswitchable small molecule dyes brought into vicinity of the target structure via affinity reagents such as antibodies. Localization microscopy is slow relative to conventional microscopy. This typically necessitates the use of fixed tissue, precluding studying live-cell dynamic processes, but is ideal for combination with EM, another fixed-sample technique.
Capturing and correlating complementary datasets from fluorescence and electron microscopy channels has been possible for decades. However, until the advent of super-resolution imaging, the resolutions of light and electron microscopy data were not well matched (~1 nm for EM compared to ~200 nm for light), making such endeavors mostly useful only for general feature identification. Additionally, traditional CLEM approaches can suffer from degraded sample and/or image quality in either one or both imaging modalities. Examples include: weak tissue preservation protocols resulting in poor ultrastructure, e.g. mitochondrial cristae membranes not resolved; strong tissue preservation protocols that destroy target fluorescence or create unacceptable autofluorescence; or protocols that are only applicable to very small samples, precluding critical experiments such as those on neurons, cellular networks or whole organs.
In the last few years, with advances in FPs, microscopes and preservation protocols, these failure modes are beginning to be systematically addressed12–17. In this article, we provide protocols of several successful methods developed by our groups over the last few years for super-resolution fluorescence CLEM18–22. The better match between the resolution of super-resolution fluorescence microscopy (~10 – 50 nm) and that of EM permits quantitative studies that go beyond general feature identification to the elucidation of sub-cellular and macromolecular complexes.
Overview of the protocols
The protocols detailed here (summarized in Figure 1) were developed independently to accommodate various biological questions but share many steps. For a given biological question and specimen, the most suitable technique may be one or a combination of these techniques. Each protocol consists of four main stages: sample preparation for light microscopy, SMLM imaging, sample preparation for EM and EM, and data analysis/co-registration.
Figure 1.
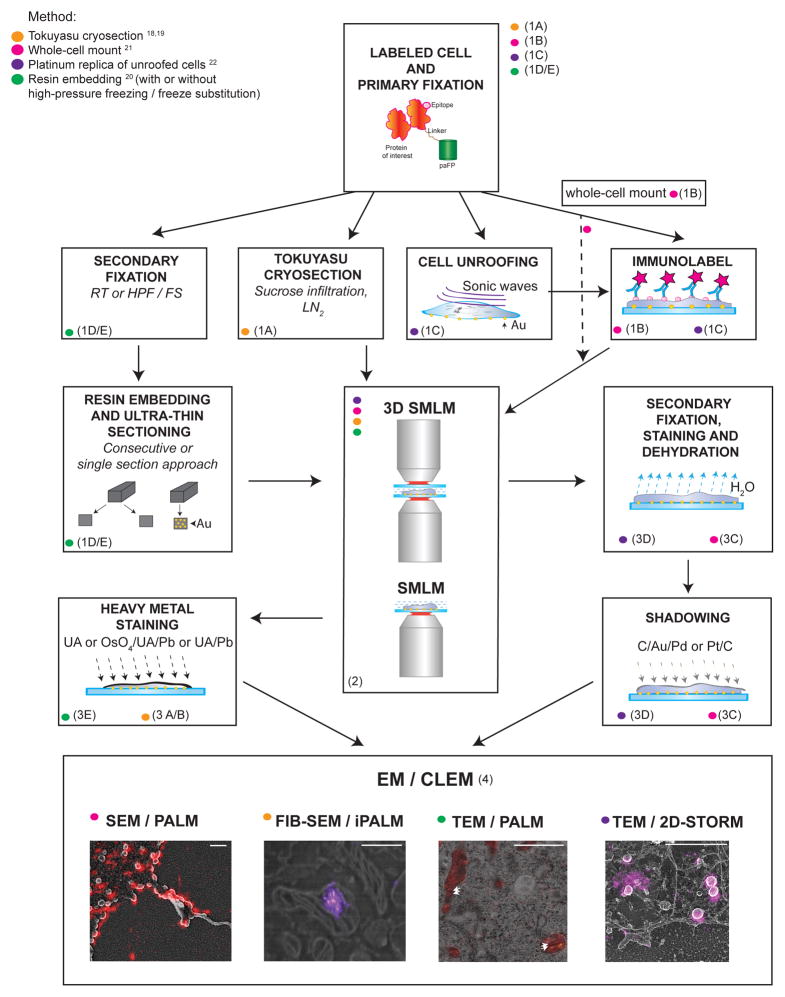
Flowchart of the protocols featured in this paper. See text and Table 1 for the details, advantages and limitations, and anticipated results for each procedure. Scale bars = 500 nm. FIB-SEM/iPALM image in bottom panel is reproduced from19. Arrows in TEM/PALM image indicate the locations of mitochondrial cristae. Images further discussed in Figure 6. Au, gold; C, carbon; HPF/FS, high-pressure freezing/freeze substitution; LN2, liquid nitrogen; OsO4, osmium tetroxide; Pb, lead; Pd, palladium; Pt, platinum; UA, uranyl acetate.
In the four protocols described here, the samples were initially fixed with aldehydes (paraformaldehyde and/or glutaraldehyde), to preserve sample integrity and prevent molecular movement. (There are alternatives to aldehyde fixation, discussed below.) After this, the protocols diverge depending on the specific targets and questions. The SMLM and image registration steps are quite similar across the four protocols; they differ largely in the sample preparation and EM steps. The protocols are typically performed with genetic targeting of FPs to specific proteins, with PALM or interferometric (iPALM) constituting the SMLM modality. Some of the protocols are equally compatible with fluorophores being brought in on affinity reagents, as in STORM and dSTORM. Each compatible protocol is marked with the optional step of (d)STORM imaging; the primary modification is the necessity of adding a reducing buffer to induce small molecule dye blinking11. Experimental workflows are essentially identical for PALM and (d)STORM; the sole differences are the addition of the labeled affinity reagent and reducing buffer in (d)STORM. Similarly, for 3-dimensional imaging, the setups for iPALM and interferometric (d)STORM are also identical. We use standard protocols for PALM23,24 and (d)STORM25. Other SMLM modalities are also possible (see below).
Choice of protocol
The choice of protocol depends both on the biological question and available equipment. There are hundreds of pipelines that can be combined from preparation and imaging options for super-resolution optical and electron microscopy. Here, we present a small but diverse sampling of optimized protocols that illustrate the relative advantages and limitations of these techniques. The protocols described here vary in sample preparation and the data they allow one to collect (summarized in Table 1).
Table 1.
Comparison of the correlative super-resolution fluorescence/electron microscopy protocols.
| Sample Process | SMLM Method | EM Method | Advantages | Limitations |
|---|---|---|---|---|
| Tokuyasu Cryosectioning (100 nm section thickness) | PALM | SEM or TEM | - Rapid | - Delicate hydrated sections - 2D |
| Tokuyasu Cryosectioning (500–750 nm section thickness) | iPALM | FIB-SEM | - 3D | - Technically demanding |
| Whole-cell mount | iPALM and/or dSTORM | FIB-SEM | - Aids iPALM interpretation | - Shrinkage limits registration quality - Limited to surface structures |
| Platinum Replica of Unroofed Cells | iPALM and/or dSTORM | ET or SEM | - 3D data - High-contrast visualization of membrane processes |
- Limited to surface structures |
| Resin embedding (with Os staining) | PALM or iPALM | SEM or TEM | - Excellent ultrastructure - Serial sectioning capability |
-Requires OsO4- tolerant FP - FP properties still somewhat compromised |
At one limit, the biology may present itself on the cellular surface and two protocols cover this situation well. One protocol is suited to image the outer morphology of the surface of a whole cultured cell. This approach of whole-cell mount PALM-scanning electron microscopy (PALM-SEM) is relatively simple to implement and yields qualitative assessment of membrane curvature as a function of probe localization. The other protocol images the rich morphology of a mechanically exposed surface of the inner cell membrane. This correlative 2D PALM-TEM of a platinum replica of the plasma membrane can be extended to 3D with iPALM-Electron Tomography (ET). Thus, it is a powerful tool for studies of endocytosis or exocytosis (and potentially other processes) on membranes. The membrane environment of isolated organelles such as nuclei and mitochondria could also be investigated.
If the biological target is not already or cannot be exposed on an open surface then some form of mechanical sectioning is needed to expose and image the interior of a cell or tissue by EM and localization microscopy techniques operating within the total internal reflection fluorescence (TIRF) regime (<200 nm from the coverslip surface). The two main alternatives are Tokuyasu cryosections and plasticized sections. The former is faster and simpler to implement (Tokuyasu protocol can be performed in 1–2 d), but the ultrastructure preservation might be not as good as with plasticized sections. The latter is also compatible with serial sectioning. Another very important consideration when deciding between these protocols is the labeling strategy. The Tokuyasu technique generally permits milder fixation and thus allows a wider choice of fluorescent labels. In order to have good ultrastructure preservation in plasticized sections, staining with heavy metals such as osmium tetroxide (OsO4) is recommended, which substantially limits the choice of labels that can preserve fluorescence under such stringent fixation. The choice of resin used for embedding may also be limited by how well fluorescence is preserved (In general, hydrophilic resins tend to better preserve fluorescence than hydrophobic ones). Acrylic resins, such as LR White26, GMA13,20 and Lowicryl27 have been successfully used for CLEM purposes, while epoxy resins, such as Epon, remain challenging.
Two protocols are based on Tokuyasu cryosectioning. One focuses on simplicity and speed of implementation, while retaining 2D correlative images with PALM-SEM. The other, with a significant extension of equipment sophistication, time and effort, achieves 3D correlative imaging on thick (~1 μm) cryosections. The final protocol illustrates how high-quality PALM imaging can be extended to sections of plastic embedded specimens, showing excellent EM preservation. This was enabled by developing a customized photoactivatable fluorescent protein that survives these harsher preparation conditions20.
In deciding which protocol to use or develop there are several further considerations:
x-y resolution
This protocol describes methods for super-resolution localization (<50 nm), although users may substitute typical confocal, epifluorescence or TIRF microscopy if lower resolution is sufficient. Additionally, if very precise registration between light and EM modes is required, then the same sample section should be used in both light and electron microscopy, ideally with fluorescent, EM-dense gold particles added, and with minimal manipulation (e.g. secondary fixation, staining) between light and EM imaging. If lower registration precision is acceptable then non-identical (i.e. consecutively cut, adjacent) sections can be used, or light microscopy (LM) can be performed before samples are further processed for EM, in which case samples should be processed in situ.
z-resolution
Many biological problems can be investigated in thin sections, effectively reducing CLEM to 2D. This is attractive because one could use simpler and more widely available equipment. Also, sample distortion is minimized. If high z-resolution is required in 3D samples such as thick sections, then iPALM or other 3D-super-resolution fluorescence methods can be combined with electron tomography, FIB-SEM, or serial sectioning of resin-embedded specimens (plasticized samples can also be imaged under FIB-SEM). iPALM provides the highest z-resolution (~10 nm) of 3D super-resolution fluorescence methods. Electron tomography and FIB-SEM provide high z-resolution (5–10 nm) in electron microscopy. The z-resolution of serial sectioning is dependent on the thickness of the sections (generally limited to >30 nm).
Sample size
Sample sectioning allows for greater fields of view than FIB milling, which is generally limited to 300 μm × 100 μm. Imaging tissues with a large z-depth (mm) is best performed through a serial sectioning procedure using resin-embedded specimens where one can cut reliable sections over a large volume. An array tomography-type approach28 can be taken whereby sections are placed on a coverslip, imaged in fluorescence mode, and then imaged by SEM (or possibly TEM). Commercial solutions to automate LM/EM image acquisition are becoming available, such as the Zeiss Shuttle & Find SEM system. Serial-sectioning could theoretically be used to image very large samples such as brain regions. Loss of material (or other physical damage) can occur upon mechanical sectioning, however, which can yield imperfect sampling along volumes of a specimen.
Sample shape
These protocols have been optimized on flat, adherent cultured cell lines like 3T3, HeLa and HEK293. These cells grow in a single layer coverslips coated with poly-L-lysine. Unroofing and whole-cell mount protocols require 2-dimensional samples. Tokuyasu cryosectioning and resin embedding could be used on 3D samples such as tissues.
Location of a target within cells or tissues
If targets are located at the outer surface of the plasma membrane, then using whole cells grown and imaged on coverslips may be the best option21. If the target is on the inner leaflet of the plasma membrane, then a specialized technique such as “unroofing”22, which removes the body of the cell, may be the best option. Purified nuclei or mitochondria could also be immobilized and observed at the tight coverslip interface. Visualizing internal sub-cellular components more broadly requires some form of sectioning19,20. All commonly investigated organelles are amenable to visualization with one or more of the protocols described here. Those at a membrane surface can be imaged with unroofing or whole-cell mount; any organelle is addressable by Tokuyasu cryosectioning or resin embedding.
Degree of ultrastructural preservation
Although all of the protocols here provide adequate ultrastructural preservation for most applications, the researcher must weigh whether it is sufficient for their specific application. Some targets with high-labeling density might withstand harsher fixatives and stains that better preserve ultrastructure, but low-copy number targets may require gentler methods to preserve fluorescence while retaining an appropriate level of ultrastructure.
Processing time
The Tokuyasu cryosectioning and whole-cell mount protocol achieve the fastest turnaround time (~2 days) of the methods discussed here. The other protocols take less than 1 week.
Alternative techniques
These protocols were developed using SMLM techniques utilizing irreversible probe photoconversion in the context of chemically fixed cells and tissue. This might not be ideally suited to all samples; below we describe other related methods and discuss the settings in which each could be appropriate.
A simple modification would be the use of reversible probe photoswitching, such as “PALM with independently running acquisition” (PALMIRA)29, Stochastic Optical Fluctuation Imaging (SOFI)30 or non-linear structured illumination microscopy31.
Alternatives to the SMLM imaging techniques employed in these protocols include spectral position determination microscopy (SPDM)32,33 and its extensions, such as SPDMphymod (“physically modifiable fluorophores”)34. The SPDM approaches can be compatible with traditional fluorophores (as opposed to overtly photoswitchable or photoactivatable probes), and as such are amenable to straightforward multi-color imaging; sometimes this is possible with a single illumination source.
Super-resolution imaging is also possible with non-SMLM modalities, such as those that sharpen the excitation spot, including stimulated emission depletion (STED)8, REversible Saturable Optical Linear Fluorescence Transitions (RESOLFT)35, ground state depletion (GSD)36, and 4pi microscopy37–39. Correlative 4pi/EM40 and correlative STED/EM13,41 have both been shown to work well.
Methods to develop correlative super-resolution fluorescence with cryo-electron tomography (CET) have recently been described42,43,44; in general, cryo-microscopy is restricted to very small samples. Cryo-temperature experiments are also hampered by technical considerations such as the lack of suitable objectives, which limits effective numerical aperture. Use of a high-pressure freeze/freeze substitution (HPF-FS) protocol incorporating the contrast agents uranyl acetate (UA) and tannic acid 45 improved the label contrast of fluorophores in SOFI, and allowed SMLM/EM correlation. However, tannic acid worsens EM ultrastructural preservation 45,46.
In addition to fluorescent protein-based methods, target proteins can be labeled by fusion with a self-labeling enzyme such as Halo tag47 and SNAP tag48, followed by incubation with an appropriate small molecule dye, fluorescence imaging and then EM. SNAP tag labeling has been shown to be compatible with STORM imaging with very little post-LM processing to prepare for EM16. Fluorescence emitters could also conceivably be generated by interaction-dependent fluorogenesis, e.g. Point Accumulation for Imaging in Nanoscale Topography (PAINT)49. The compatibility of these methods with resin embedding is untested.
For review of correlative SMLM methods, see 50 or 51.
We have left out of our discussion here a variety of other promising, but non-fluorescent, labeling techniques, such as electron density deposition by oxidases52–56, immunolabeling EM (e.g. with hyper-antigenic tags57), and fusion to electron dense proteins58,59.
Experimental Design
Protocol 1. Tokuyasu cryosectioning
The original PALM paper5 used Tokuyasu cryosectioning but fine ultrastructural details were not observable. We improved the original protocol, in both sample handling and ultrastructural preservation, through several modifications. First, we began by increasing the amount of glutaraldehyde in the initial fixation step from 0.1% to 2%. The high level of glutaraldehyde had no discernable effect on the fluorescent proteins or dyes used. However, the use of glutaraldehyde creates a prohibitive level of autofluorescence60 that can be mitigated by treating the samples with 0.5% sodium borohydride (an aldehyde “quencher”) prior to fluorescence imaging61. After primary fixation with aldehydes, the sample is prepared using standard procedures for Tokuyasu cryosectioning, including embedding in gelatin, infiltration with sucrose, sectioning the frozen sample, and retrieving the sample with a sucrose/methylcellulose solution. Sample sections are placed on 25 mm glass coverslips that: 1) have gold nanoparticles deposited, and 2) are coated with indium tin oxide (ITO) (Box 1). The gold nanoparticles (which are both fluorescent and electron-dense) are used to register fluorescence and EM data sets (Figure 2), and the ITO coating is required for charge conductance in scanning EM (SEM). Placing the sections on these glass coverslips allows optical and electron microscopy to be performed without transferring the sections to another substrate (i.e., EM grid), thus reducing sample handling damage and distortions. Using this method, we were able to correlate super-resolution fluorescence and electron microscopy images of diverse targets. Ultrastructure preservation by Tokuyasu cryosectioning is generally good.
BOX 1. Cleaning, coating, and addition of gold fiducial markers to coverslips.
Materials
#1.5 glass coverslips, 18 or 25 mm (Warner Instruments cat no. 64-0734 or 64-0735)
Poly-L-lysine hydrochloride (Sigma-Aldrich, cat. no. P2658-100MG)
Hydrogen peroxide, 50% (vol/vol) (Fisher Scientific, cat. no. H341-500)
-
Ammonium hydroxide, 29% (wt/wt) (Fisher Scientific, cat. no. A669-500)
!CAUTION ammonium hydroxide is toxic; use the reagent in a hood and wear gloves and goggles while handling.
Gold 80 nm nano-spheres (Corpuscular, cat. no. 790120-010)
Gold bare nanorods 25 × 57 nm (Nanopartz, cat. no. A12-25-600) or 25 × 75 nm (Nanopartz, cat. no. A12-25-700)
Reagent Setup
Basic Piranha (aka RCA clean) for coverslip preparation. Make solution using 5 parts deionized H2O, 1 part 50% (wt/vol) H2O2 and 1 part 29% (wt/wt) NH4OH. !CAUTION Piranha solution is extremely hazardous requiring special precautions for handling and disposal. Working in a chemical safety hood, wear a full-face shield, heavy-duty rubber gloves and a protective apron. Only use glass containers. Mix the solution in the chemical flow hood with the sash between you and the solution. Let the solution cool before handling and putting in a glass disposal container.
Clean the coverslips (#1.5, 18 or 25 mm, from Werner Instruments) in basic Piranha (RCA clean) at 80 °C, stirring, at least 2–3 hours (can be done overnight). Then rinse with deionized water and blow-dry.
Put coverslips into cover-slip support blocks in individual small Petri dishes. Write numbers near the edge of the top surface for later identification.
Cover the surface with 75 μl fresh-made 0.1% (wt/vol) poly-L-lysine for 15 minutes. It should cover most of the top surface. After 15 minutes rinse with deionized water and blow-dry.
Cover the surface with 75 μl 0.5% (1:200 diluted in deionized water) of stock Au nanoparticles or nanorods for 15 minutes. After 15 minutes rinse with water and blow-dry. Most versatile are bare Au nanorods 25 × 57 nm from Nanopartz. Sometimes larger 80 nm Au nanospheres are useful (they are brighter at 500–600 nm emission wavelength and are sometimes easier to see in SEM). If fluorescent imaging is done at 800 nm (such as with Alexa Fluor 750), then bare Au 25 × 75 nm nanorods from Nanopartz should be used. Depending on application and image area, the target surface density of nanoparticles should be 10,000–100,000 mm−2. The concentration of Au nanoparticles may need to be adjusted to achieve this.
Deposit silicon dioxide (SiO2) or indium tin oxide (ITO) using a sputtering deposition system. For SEM, deposit ITO (ITO is conductive and will help with avoiding charging during SEM imaging). ITO thickness should be selected so that the resistance of the coverslip surface (measured by connecting two multimeter probes near the opposing edges of the coverslip) is below 5 kOhm. For platinum replicas, deposit 20 nm of SiO2.
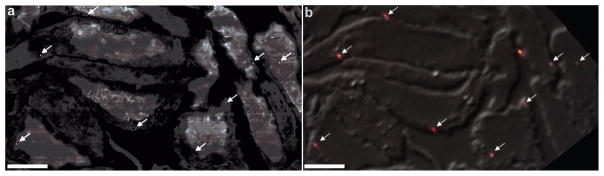
Figure 2.
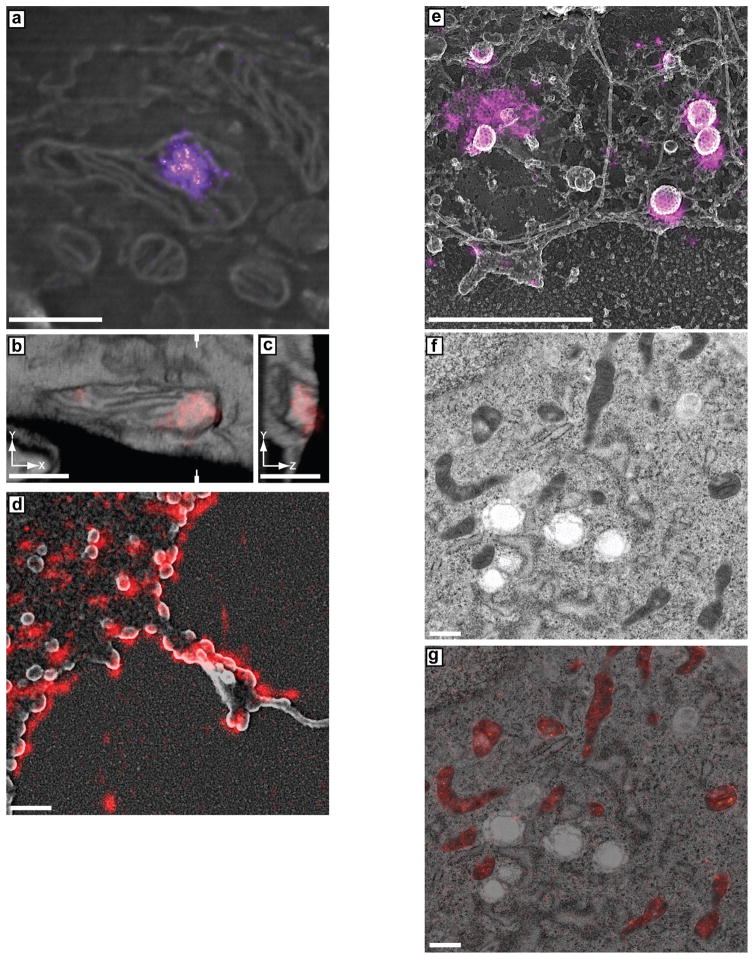
Example of using gold nanoparticles to align fluorescence and electron microscopy datasets. (a) Scanning electron micrograph where gold nanoparticles can be observed as white dots (arrows) due to their high electron densities. (b) Fluorescence signal from gold nanoparticles (arrows) with a DIC image showing cell outlines. Note this image shows diffraction-limited fluorescence for illustration purposes, as a super-resolution fluorescence rendering would make the gold nanoparticles difficult to see. Scale bars = 5 μm. Figure reproduced with permission from18.
For our experiments, we used the fluorescent proteins mEos262 and photoswitchable cyan fluorescent protein 2 (PS-CFP2)63. Both of these proteins worked well for our studies, but additional photoactivatable protein derivatives have been developed since our initial work that may work better20,64,65. We also found that caged small molecule dyes66,19 (e.g. attached to the actin-labeling molecule phalloidin) worked well. A quick and easy test, and necessary control, to determine if your fluorescent label withstands primary fixation is to grow and label the cells in chambered coverslips. Examining the labeled cells by fluorescence microscopy, both pre- and post-fixation, provides insight as to how the fluorescent label withstands primary fixation.
Tokuyasu cryosectioning requires a specialized ultramicrotome with a cryogen attachment. Cryosections are picked up with a droplet of methylcellulose/sucrose solution and placed on a glass coverslip. The gelatinous methylcellulose/sucrose solution prevents the samples from drying out (for time periods less than 24 hours) but must be washed away with buffer prior to imaging. From this point forward until the final drying steps it is imperative that the sections remain in an aqueous environment. Incubating inverted coverslips on drops of 0.5% sodium borohydride for 15 minutes minimizes autofluorescence from glutaraldehyde. Many standard protocols for Tokuyasu cryosectioning exist67,68.
3D iPALM69 uses dual objectives requiring that another coverslip be placed over the cryosection-containing coverslip and sealed with epoxy and Vaseline. A thin layer of buffer is trapped between the two coverslips to ensure that the sample remains hydrated. After SMLM image acquisition, the coverslip sandwich is separated and the sample is placed in a small dish with buffer. At this point, staining depends on whether 2D or 3D electron microscopy will be performed.
If thicker sections (>250 nm) were used for 3D SMLM imaging then serial focused ion beam milling/SEM (FIB-SEM) can be used to obtain 3D EM data from the sample18,70. The preparation of thick Tokuyasu cryosections for FIB-SEM is relatively straightforward. Drops of methylcellulose containing 0.5% uranyl acetate (UA) are placed on a piece of Parafilm attached to an aluminum plate sitting on ice. The coverslip is inverted such that the section faces down and placed on the methylcellulose drops. After an incubation of 15 minutes, the coverslip is dried by dragging the edge of the coverslip across filter paper, wicking away excess methylcellulose while leaving a thin layer on top. The coverslip may be held on its edge in contact with the filter paper until completely dry. Differential interference contrast (DIC) imaging helps to identify the SMLM-imaged region of the sample for FIB-SEM (Box 2, Figure 3). Prior to FIB-SEM, the addition of cyanoacrylate directly on top of the sections aids ion beam milling. This is followed by carbon coating to avoid charging. FIB-SEM operates in a two-step cycle where a focused beam of Ga+ ions mills a few-nanometer-thick layer to expose a new layer inside a sample, which is then imaged by SEM70. This procedure is repeated thousands of times to form a 3D EM image stack. This stack is then registered to a 3D fluorescent image stack using Au nanoparticles, which are localized with high precision in both EM and fluorescent images (Figure 2).
BOX 2. Identifying the same area in samples where EM fixation occurs after LM.
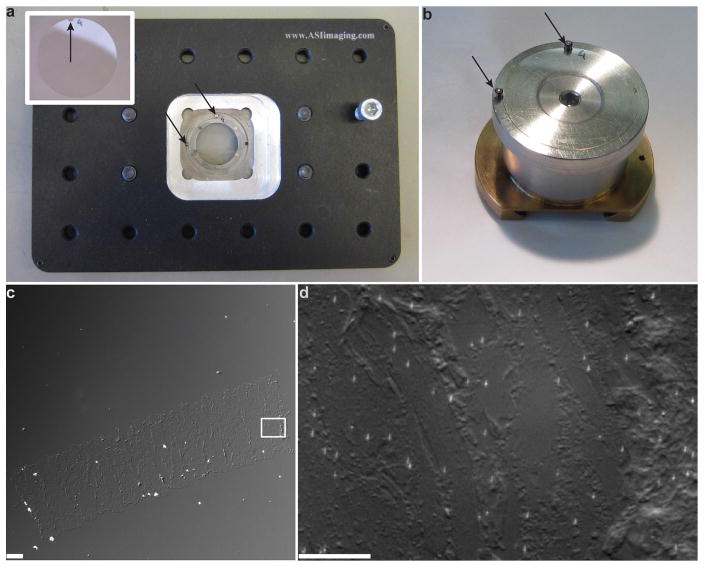
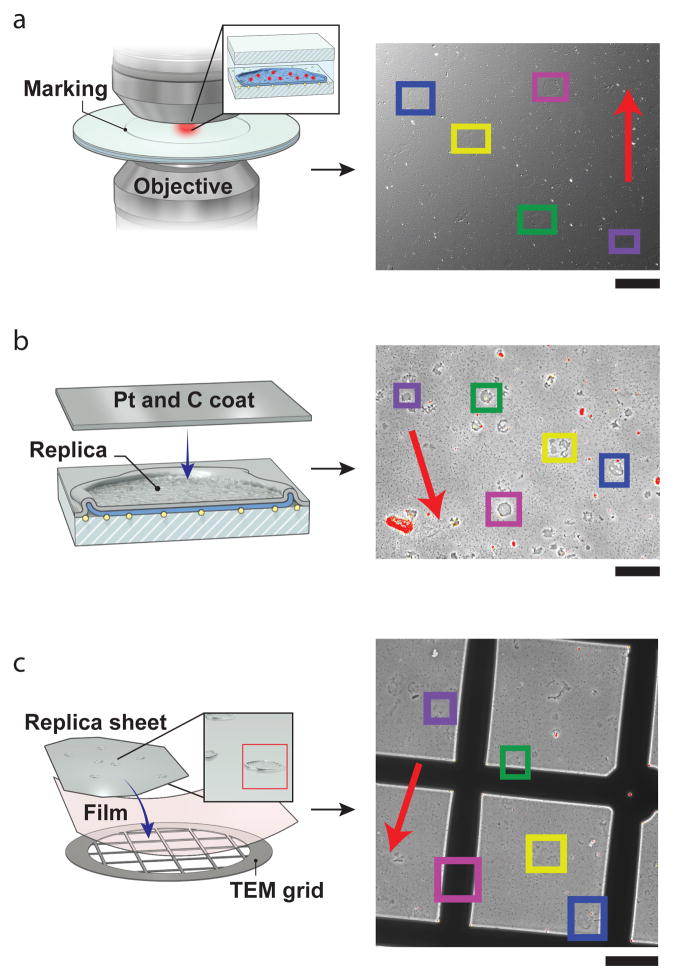
Correlative LM and EM imaging is usually performed sequentially: LM imaging is done first, followed by additional sample staining and treatment, and then EM imaging. One important and potentially challenging step is finding the areas where LM imaging was performed for subsequent EM imaging. “Blind” searching in high-resolution EM for a structure that resembles what was imaged during LM can be very time-consuming and ultimately unsuccessful. A better procedure for finding the previously imaged area must be established. This can be done in several possible ways. If both microscopes are equipped with sample coordinate registration software and sample holders (such as in Figure 4 a, b), and the sample did not need to change substrates (e.g., from coverslip to grid), then follow procedure A. If the microscopes are not equipped with sample coordinate registration software, then creating a set of intermediate-zoom optical image maps can be very helpful. Since most cells and thin sections are optically transparent, differential interference contrast (DIC) becomes very useful. It is highly desirable to have the DIC imaging modality available in the LM setup (it is very useful not only for this step, but in general to check sample quality before imaging). If DIC is not available, a fluorescence image map could also be used if all cells are fluorescently labeled with a membrane or similarly uniform stain. In this case, follow procedure B. An example of procedure B is shown in Figure 4 c, d, Figure 4c shows the 10× DIC image taken after the sample has been removed from the LM microscope, prior to post-staining and EM imaging. Figure 4d presents the 120× DIC image taken in the LM microscope after LM imaging. Having this map greatly simplifies searching for the areas that have been previously imaged. For the purposes of making a platinum replica for TEM, this process is also aided by a circle that is etched on the bottom of the coverslip with a diamond objective marker after fluorescence imaging (Figure 5). To do this follow procedure C.
A. Procedure if microscopes have sample coordinate registration software and sample holders
Record coordinates of every site imaged during LM.
Calculate the coordinates in EM sample reference from the coordinates of the imaged site in LM sample reference and the transformation coefficients (which can be established using a simple test sample with a few distinct features).
B. Procedure if microscopes do not have sample coordinate registration software and sample holders
Once each LM data acquisition is complete, take a DIC image of the LM imaged area (preferably DIC image of slightly expanded area if possible).
Once all LM acquisitions have been performed on a sample, take low-zoom DIC image of the sample to create a look-up map of the imaged sites.
Using this look-up map, search for the imaged sites in EM.
C. Procedure for platinum replicas
Use a diamond objective marker to etch a 4 mm circle around the region that was imaged in fluorescence and image a zoomed out view of the region in DIC to create a map of the cells imaged (Figure 5a).
Coat the sample with platinum and carbon and again map the region within the etched circle, this time with 10× phase contrast (Figure 5b).
Cut out the etched circular region and lift the replica onto a grid. Image the grid again with 10× phase contrast to identify the location of your cells on the grid. Colored rectangles indicate cells imaged in fluorescence (Figure 5c).
Figure 3.
Tools facilitating location of the regions of interest to be imaged using different microscopy techniques. (a,b) Sample holders for fluorescent microscopy (a) and for SEM (b) that allow for precise referencing of the imaged areas. The coverslip has a notch ground using a 1/16-inch diamond bit near its top (indicated by an arrow in the inset). Both sample holders have two fixed 1/16-inch posts (indicated by arrows), which ensure unambiguous sample placement, allowing for precise coordinate referencing. (c) 10× DIC image of a Tokuyasu cryosection of NIH 3T3 mouse fibroblast cells used for correlative PALM and EM imaging. This can be used as a look-up map to find the area imaged in PALM when performing EM. (d) Magnified (120×) DIC image of the area in the rectangle in c, imaged during the PALM step. Scale bars, 40 μm (c); 10 μm (d).
Methylcellulose/UA is the traditional staining method for Tokuyasu cryosectioning. However, for thin sections, methylcellulose can interfere with electron microscopy, causing the image to appear hazy. This is not an issue with FIB-SEM since the methylcellulose layer is cut through and the section is imaged from the side due to the perpendicular geometry of the ion gun (top) and electron beam (side). For non-FIB-SEM samples, though, the methylcellulose can degrade EM image quality; thus, we developed a complementary method. In this case, after PALM imaging, the coverslip is placed in a small Petri dish for additional electron-dense staining. The sample is treated with 2% osmium tetroxide (OsO4) reduced with 1.6% potassium ferrocyanide (K4[Fe(CN)6]) for 15 minutes. Next, the sample is incubated with 0.6% UA in polyvinyl alcohol (PVA) for 15 minutes. (PVA acts in a similar manner to methylcellulose, by reducing surface tension effects during sample drying that may result in artifacts71. Samples dried in the presence of polyvinyl alcohol do not have the haziness observed when methylcellulose is used.) Finally, the sample is incubated with 0.0075% Sato’s triple lead (citrate, acetate and nitrate salts of Pb2+) in polyvinyl alcohol. The coverslip is placed on a spinning slide drier and dried for 10 seconds at 5000 rpm. In general, the methylcellulose/UA protocol is quicker and easier, but the PVA/OsO4/ferrocyanide/UA technique is compatible with more diverse EM steps. PVA-prepared samples should be amenable to FIB-SEM imaging, although we did not test this yet.
Protocol 2. Whole-cell mount
The whole-cell mount correlative imaging protocol provides a relatively simple and convenient method for obtaining images of membrane surface topologies, while providing molecular specificity for surface molecular assemblies. This protocol was inspired by previous work using standard diffraction-limited fluorescence microscopy and correlative SEM imaging72. This protocol does not require mechanical sectioning of the specimen and as such does not require any special preparation of the sample prior to standard fluorescence microscopy imaging. As this protocol also does not necessitate strong fixation of the specimen prior to fluorescence imaging, many standard fluorescent proteins used for PALM can be utilized without significant loss of fluorescent signal. This protocol also requires very little sample preparation for SEM following fluorescence image acquisition. Briefly, the cellular specimen is placed on a coverslip, allowed to express the probes of interest, and fixed using paraformaldehyde and a low concentration of glutaraldehyde. The specimen is then imaged using fluorescence super-resolution protocols. Upon acquisition of super-resolution datasets, the specimen is post-fixed with glutaraldehyde and osmium. Optionally, the sample can be further fixed with successive rounds of OsO4 and thiocarbohydrazide (“OTOTO”) to improve membrane contrast and stability73. The specimen is dehydrated using an ethanol dehydration series and critical point drying (CPD)74. The surface topology of the specimen is then visualized using SEM. It is important to note, however, that SEM only provides surface information, precluding visualization of intracellular membranes and structures using the whole-cell mount procedure. This technique typically provides only 2D information about surface specimen features. An important consideration for implementation of this protocol comes from the observation that large cellular specimens (high mass objects) can be displaced due to the drying procedure for SEM observation. Care must be taken to meticulously follow each dehydration step in the protocol to prevent movement of the specimen. The optional OTOTO protocol can further mitigate specimen movement. This protocol describes the use of cellular (tissue culture) specimens for interrogating surface topologies and associated molecular assemblies involved in membrane remodeling.
Protocol 3. Platinum replica TEM/unroofing
Platinum replica TEM of unroofed cells is a high-contrast method well suited for observing the topography of the inner surface of the plasma membrane75,76. In these images the spatial organization of membrane events (e.g. endocytosis and exocytosis) can be viewed en face with high resolution. Immunolabeling, such as with gold nanoparticle-coupled antibodies, typically provides a straightforward way to visualize target protein localization within an EM image. Furthermore, in unroofed samples, the proteins of interest are very accessible to antibodies and self-labeling enzymes, typically resulting in a high labeling density. Unfortunately, using immunogold methods to locate proteins in these replicas is difficult due to the inherently high contrast of the platinum (Pt) coating, which thus necessitates the use of sterically bulky 15 nm gold particles to achieve visibility over the Pt coating. To overcome these issues, we developed a correlative super-resolution localization microscopy technique to provide an alternative to immunogold labeling in platinum replicas. This was especially important in our studies of clathrin-mediated endocytosis, where the clathrin meshwork and associated proteins likely produce a steric impediment to the use of large immunogold probes.
In this protocol, we grow cells on gold nanorod embedded coverslips, fix with paraformaldehyde (PFA), and gently sonicate the specimen to remove the top of the cells (unroofing). These thin membrane sheets have exposed and buffer-accessible cytoplasmic faces; they have been extensively used in the past for imaging cell cortices77. Super-resolution microscopy can then easily be performed on these samples with nearly any fluorescent label that is compatible with the user’s biological system. Alexa Fluor 647, Alexa Fluor 750, mEos364 or mEos420 and PS-CFP263 are our labels of choice. After fluorescence imaging, the sample is further stabilized with a previous platinum replica CLEM protocol74, which uses glutaraldehyde, tannic acid and UA prior to ethanol dehydration and CPD. The dry sample is coated with Pt and carbon to make a rigid replica of the sample that can be transferred to a TEM grid for imaging. This method provides highly reproducible correlation with 20 nm accuracy across the 20 μm-wide landscape, which can be directly observed in many cases if using immunofluorescence because the antibody assemblies are often large enough to be visible in the EM micrograph. This robust correlation owes to the tight adherence of the thin sample to the coverslip during the dehydration and CPD of the sample in preparation for EM and makes it an especially trusted method for finding unknown positions of single proteins. However, this method requires physical disruption of the cell by sonication during fixation, which disrupts the cytoskeleton and washes away cytoplasmic components. Therefore, this method is specific to membrane-bound systems or other thin systems tightly adhered to a coverslip.
Protocol 4. Resin embedding
Because thick volumetric samples are incompatible with cryosectioning, embedding into an easily sectioned plastic resin remains the best option for these samples. Many standard resin options exist from water-incompatible Epon epoxy resins to hydrophilic resins such as glycol methacrylate (GMA), LR Gold or LR White, and Lowicryl resins (e.g. K4M and HM20). Epon is typically preferred for EM because of its superior ultrastructure preservation and sectioning properties. Epon, however, requires complete sample dehydration and epoxy polymerization, which can extinguish fluorescence and render antibodies unable to bind antigens. In contrast, hydrophilic resins generally preserve the function of proteins including FPs and antigens better than hydrophobic resins but lack the strong cross-linking of Epon. This results in weaker samples that are more sensitive to electron beam damage.
Correlative super-resolution/EM imaging in plastic sections was first shown with PALM and stimulated emission depletion (STED) microscopy in both GMA and LR White resins13. In this study, GMA was selected as the optimal resin due to its retention of FP fluorescence and homogeneous polymerization. However, due to the weak fixation conditions used (0.1% potassium permanganate and 0.001% OsO4, no aldehydes) necessary to retain fluorescence, ultrastructure preservation was poor compared to that achievable with much higher OsO4 concentrations.
Other protocols have advanced the use of the acrylic resins. For instance, Lowicryl HM-20 infiltrated samples could be sectioned and antibody labeled for dSTORM-type super-resolution imaging followed by UA staining for SEM 78. A related protocol45 retains or even slightly improves switching of fluorescent proteins by optimizing the addition of stains such as tannic acid. Another protocol12 discusses STORM dyes in various resins.
We set out to optimize the protocol of Watanabe et al.13, both through the systematic exploration of fixative cocktails and through protein engineering of mEos2 to better resist fixatives, in particular OsO4, while retaining fluorescence and photoconvertibility20. We first began by decreasing the amount of water used in the GMA resin mix, which slightly improved ultrastructure preservation without affecting FP fluorescence. Second, we systematically explored combinations of primary fixatives, optimizing for EM ultrastructure, preservation of mEos2 fluorescence properties and low autofluorescence. This led to the selection of 4% PFA + 0.2% glutaraldehyde as primary fixatives before secondary fixation and embedding. Use of aldehyde quenchers such as borohydride before imaging decreased the background fluorescence attributable to glutaraldehyde polymerization61. Finally, we mutated surface residues on mEos2 to remove nucleophilic groups, which are involved in cross-linking with aldehydes and OsO479,80. This resulted in the selection of two mutants, mEos4a and mEos4b, each with significantly improved resistance to OsO4 fixation and fluorescence properties unchanged from the starting scaffold mEos2.
These proteins facilitated the development of two protocols: 1) a “consecutive-section” approach where adjacent ultra-thin sections cut from resin are separately split between PALM imaging and EM fixation and imaging; and 2) a “same-section” approach, where a single resin-cut section is subjected to both PALM and TEM and/or SEM. In both cases, plastic resin embedding dramatically decreases tissue distortion from dehydration and secondary fixation, and additionally improves performance of the specimen under the electron beam. We also found that this protocol was appropriate for use with HPF-FS (e.g. Figure 1D) or without it (e.g. Figure 1E). We found ultrastructure preservation to be comparable between the two20.
The resin embedding protocol is appropriate for all samples. It is a specialized technique and requires the use of fixation-resistant fluorophores such as mEos4 to achieve optimal ultrastructure preservation. The “same-section” approach requires no sample treatment after LM and prior to EM, and thus results in extremely low sample deformation, allowing for precise and quantifiable registration of LM and EM images. Resin embeeding is the only CLEM technique that is readily applicable to large samples.
Questions should be addressed to: B.G.K. for the Tokuyasu cryosectioning protocol, S.B.vE. for the whole-mount protocol, J.W.T. for the platinum replica/TEM unroofing protocol, L.L.L. for the fluorescent protein engineering/resin embedding protocol, and H.F.H. for iPALM and FIB-SEM.
Registering and transforming SMLM and EM data sets
The registration of data sets acquired in different modalities is essential for correlative SMLM and EM. EM images may have different nm/pixel scales as compared to SMLM data sets; in addition they may be shifted and tilted. The uncertainties due to imperfect image registration must be accounted for when estimating compound localization accuracy; usually these are treated as independent and are added in quadratures81: , where σloc1 and σloc2 are the localization accuracies in each data set, and σreg the accuracy of registration.
Since SMLM data sets are vector based, and EM images are typically pixel maps, it is easier to register and transform SMLM data to overlay it with EM data. The registration can be performed using the coordinates of objects (fiducials) that are detectable in both modalities. The procedure is fairly straightforward for 2D registration and we discuss this first.
2D SMLM and EM image registration
We found that Au nanoparticles are very good fiducial markers since they behave as bright, single-dipole emitters during fluorescent imaging and are also electron-dense, so they can be registered in both fluorescent microscopy and EM images. Once the SMLM and EM imaging have been performed, the coordinates of the same Au nanoparticles can be determined with accuracy on the order of 2–5 nm in both data sets. As a result, we have corresponding coordinate pairs { }, and { }. These two sets are used to establish the transformation procedure for the rest of the data. We found that bilinear mapping works very well for SMLM -EM registration:
where { }, and { } are as above, while Kxyij terms are the transformation coefficients19. The bilinear terms Kx11 and Ky11 are usually very small. In choosing imaging sites, we recommend selecting areas that contain as many fiducials as possible. The transformation coefficients Kx, yij can be determined using POLYWARP function in IDL or using CP2TFORM function in MATLAB. This step requires a minimum of 4 coordinate pairs to determine the transformation coefficients; more are desirable in order to reduce the registration error. Au nanoparticles occasionally form clusters, which tend to exhibit wavelength-dependent, higher order multi-pole radiation patterns, resulting in erroneous localization results. It is usually relatively easy to identify and exclude these fiducials during processing. But in order to perform this iterative procedure, the initial reference sets must exceed the minimum size. It is recommended to start with at least 20 fiducial points identifiable in PALM and EM data sets. Then, for a ~30 μm field of view, the achievable average registration error is typically below 10 nm.
3D SMLM and EM image registration
Registration of 3D SMLM and EM images is more complicated. The main problem is sample deformation during transfer from the aqueous environment of SMLM to the vacuum of an EM. This sample deformation can occur in any direction and is a serious limitation for CLEM in general. Making the sample as thin as possible helps minimize the lateral shrinkage. This can be achieved by working with relatively thin (<1 μm) sections or unroofed cells. We believe that the most promising approach to minimizing shrinkage of sections is plastic embedding. Other methods that we tried still suffer from some degree of vertical shrinkage. Fortunately, due to very high sample aspect ratio (tens of microns lateral to ~0.5 μm vertical), this shrinkage may be considered uniform, and the registration can be done with a constant scaling factor applied to the SMLM data set in the vertical direction (z-dimension). Ideally one would like to create fiducial points either throughout the volume of the sample or on both top and bottom surfaces of the sample. This would allow for precise quantitative 3D registration of multiple data sets. Unfortunately, we have not been able to create stable fiducial markers on the top surface of cryosections or unroofed cells. Au nanoparticles tend to detach and move during the SMLM experiments unless they have been covered by a layer of SiO2, ITO, or by a section itself (in all of these cases they create good fiducial points in the plane of the bottom surface of the sample).
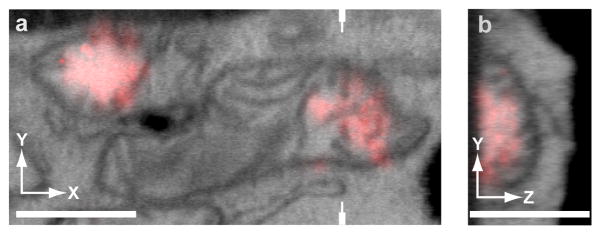
We have successfully used a feature-selection approach to establish the vertical scaling for registering 3D SMLM and EM data sets (Figure 4). Such features need to clearly define an object in the volume of the sample (ideally as close to the top surface of the sample as possible – or even better – the top surface itself) and must be identified in both imaging modalities with high vertical precision.
Figure 4.
Example 3D alignment of iPALM-FIB-SEM of thick cryosections. The scaling is adjusted so that fluorescence falls off at the same vertical location as the boundary of the section observed in the 3D EM data set. (a) x–y correlation, which is done using fiducial markers. (b) The y–z plane is cut through the volume, as indicated by the marks in a. A flat side in the fluorescence signal indicates the bottom plane of the section and can be used for alignment to the bottom of the section observed in the EM. The images can then be scaled so that the fluorescence falls within boundaries on the EM (i.e., the bottom edge of the section and the mitochondrial membrane). This feature-selection approach can be used for scaling and registration. Scale bars, 500 nm. Figure adapted from18.
MATERIALS
REAGENTS
Tokuyasu Cryosectioning Module
16% (wt/vol) Paraformaldehyde (PFA), aqueous (Electron Microscopy Sciences, cat. no. 15700) !CAUTION Paraformaldehyde is a tissue fixative. Handle it with appropriate protective clothing, gloves, and goggles. Work in an appropriate fume hood.
8% (wt/vol) Glutaraldehyde, aqueous (Electron Microscopy Sciences, cat. no. 16000) !CAUTION Glutaraldehyde is a strong tissue fixative. Handle it with appropriate protective clothing, gloves, and goggles. Work in an appropriate fume hood.
Sucrose (Sigma-Aldrich, cat. no. S7903)
Glycine (Sigma-Aldrich, cat. no. 50046)
Gelatin from porcine skin, Bloom 90-110 (Sigma-Aldrich, cat. no. G6144)
Potassium hexacyanoferrate(II) trihydrate, a.k.a. potassium ferrocyanide (Sigma-Aldrich, cat. no. P3289)
Methylcellulose, 25 centipoises (Sigma-Aldrich, cat. no. M6385)
Polyvinyl alcohol (Sigma-Aldrich, cat. no. S8045)
-
Lead citrate (Electron Microscopy Sciences, cat. no. 17810)
!CAUTION Lead citrate is toxic; use the reagent in a hood and wear gloves while handling.
-
Lead acetate (Electron Microscopy Sciences, cat. no. 17600)
!CAUTION Lead acetate is toxic; use the reagent in a hood and wear gloves while handling.
-
Lead nitrate (Electron Microscopy Sciences, cat. no. 17900)
!CAUTION Lead nitrate is toxic; use the reagent in a hood and wear gloves while handling.
Sodium citrate (Electron Microscopy Sciences, cat. no. 21140)
Sodium hydroxide (Sigma-Aldrich, cat. no. S8045)
4% (wt/vol) Osmium tetroxide, aqueous (Electron Microscopy Sciences, cat. no. 19150) !CAUTION Osmium tetroxide is extremely toxic; its high vapor pressure necessitates that it is handled only in an appropriate fume hood. Wear protective clothing, gloves and goggles. Post warning information in the working area.
Bovine serum albumin (BSA), lyophilized powder (Sigma-Aldrich, cat. no. A2153)
-
Uranyl acetate (Electron Microscopy Sciences, cat. no. 22400)
!CAUTION Uranyl acetate is an alpha emitter. Uranyl acetate is a cumulative toxin.
Wear protective clothing, gloves, and goggles.
Sodium borohydride (Sigma-Aldrich, cat. no. 480886)
Whatman 50 hardened filter paper (Sigma Aldrich, cat. no. WHA1450055)
Whole-cell Mount Module
16% (wt/vol) Paraformaldehyde (PFA), aqueous (Electron Microscopy Sciences, cat. no. 15700) !CAUTION Paraformaldehyde is a tissue fixative. Handle it with appropriate protective clothing, gloves, and goggles. Work in an appropriate fume hood.
-
8% (wt/vol) Glutaraldehyde (Electron Microscopy Sciences, cat. no. 16000)
!CAUTION Glutaraldehyde is a strong tissue fixative. Handle it with appropriate protective clothing, gloves, and goggles. Work in an appropriate fume hood.
4% (wt/vol) Osmium tetroxide, aqueous (Electron Microscopy Sciences, cat. no. 19150) !CAUTION Osmium tetroxide is extremely toxic; its low vapor pressure necessitates that it is handled only in an appropriate fume hood. Wear protective clothing, gloves and goggles. Post warning information in the working area.
Potassium hexacyanoferrate(II) trihydrate, a.k.a. potassium ferrocyanide (Sigma-Aldrich, cat. no. P3289) (optional)
Glycine (Sigma-Aldrich, cat. no. 50046)
Ethanol, 200 proof (Electron Microscopy Sciences, cat. no. 15055)
Molecular sieve (Sigma-Aldrich, cat. no. 208574)
Hexamethyldisilazane (EMS cat. no 16710; optional)
Carbon dioxide with syphon tube (United Oxygen Company cat. no. CO250ST)
Platinum Replica Module
16% (wt/vol) Paraformaldehyde (PFA), aqueous (Electron Microscopy Sciences, cat. no. 15700) !CAUTION Paraformaldehyde is a tissue fixative. Handle it with appropriate protective clothing, gloves, and goggles. Work in an appropriate fume hood.
-
8% (wt/vol) Glutaraldehyde (Electron Microscopy Sciences, cat. no. 16000)
!CAUTION Glutaraldehyde is a strong tissue fixative. Handle it with appropriate protective clothing, gloves, and goggles. Work in an appropriate fume hood.
Ethanol, 200 proof (Electron Microscopy Sciences, cat. no. 15055)
Tannic acid (Mallinckrodt, cat. no. 1764)
Potassium chloride (Sigma Aldrich, cat. no. P9541)
HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) (Sigma Aldrich, cat. no. H3375)
Magnesium chloride (Sigma Aldrich, cat. no. M8266)
Potassium hydroxide (Sigma Aldrich, cat. no. H3375)
EGTA (ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid) (Sigma Aldrich, cat. no. E3889)
-
49% (vol/vol) Hydrofluoric acid (Fisher Scientific, cat. no. A147-1LB)
!CAUTION. Hydrofluoric acid is toxic. Use in chemical hood with neoprene or nitrile gloves, goggle, acid-resistant apron. Contact with skin requires immediate medical attention.
Carbon dioxide with syphon tube (United Oxygen Company, cat. no. CO250ST)
Poly-L-lysine solution, 0.01% (Sigma, cat. no. P4832)
-
Uranyl acetate (Electron Microscopy Sciences, cat. no. 22400)
!CAUTION Uranyl acetate is an alpha emitter. Uranyl acetate is a cumulative toxin.
Wear protective clothing, gloves, and goggles.
10 nm gold particles if making a 3D EM tomogram. We use a 1 in 5 dilution of 10 nm gold anti-rabbit antibody conjugate (Cytodiagnostics, cat. no. AC-10-01).
Resin Embedding Module
16% (wt/vol) Paraformaldehyde (PFA), aqueous (Electron Microscopy Sciences, cat. no. 15700) !CAUTION Paraformaldehyde is a tissue fixative. Handle it with appropriate protective clothing, gloves, and goggles. Work in an appropriate fume hood.
-
50% (wt/vol) Glutaraldehyde (GA), aqueous (Electron Microscopy Sciences, cat. no. 16320)
!CAUTION Glutaraldehyde is a tissue fixative. Handle it with appropriate protective clothing, gloves, and goggles. Work in an appropriate fume hood.
Sodium borohydride (Sigma-Aldrich, cat. no. 480886)
Bovine serum albumin (BSA), lyophilized powder (Sigma-Aldrich, cat. no. A2153)
4% (wt/vol) Osmium tetroxide, aqueous (Electron Microscopy Sciences, cat. no. 19150) !CAUTION Osmium tetroxide is extremely toxic; its low vapor pressure necessitates that it is handled only in an appropriate fume hood. Wear protective clothing, gloves and goggles. Post warning information in the working area.
Ethanol, 200 proof (Electron Microscopy Sciences, cat. no. 15055)
-
Osmium tetroxide, crystalline (Electron Microscopy Sciences, cat. no. 19110)
!CAUTION Osmium tetroxide is extremely toxic; its low vapor pressure necessitates that it is handled only in an appropriate fume hood. Wear protective clothing, gloves and goggles. Post warning information in the working area.
Acetone (Electron Microscopy Sciences, cat. no. 10012)
Methanol (Electron Microscopy Sciences, cat. no. 18510)
Glycol methacrylate resin (SPI, cat. no. 02626-AB)
Butyl methacrylate resin (SPI, cat. no. 02822-BA)
-
Benzoyl peroxide catalyst (SPI, cat. no. 02825-CA)
!CAUTION Benzoyl peroxide is a hazardous material and potentially explosive when dry. Wear protective gloves and goggles.
-
N,N-Dimethyl-p-toluidine (Sigma-Aldrich, cat. no. D189006-100ml)
!CAUTION N,N-Dimethyl-p-toluidine is a hazardous material and should be handled under a chemical fume hood. Wear protective gloves.
-
Uranyl acetate (Electron Microscopy Sciences, cat. no. 22400)
!CAUTION Uranyl acetate is an alpha emitter. Uranyl acetate is a cumulative toxin.
Wear protective clothing, gloves, and goggles.
Hexadecene (Sigma-Aldrich, cat. no. H2131)
Pioloform (Ted Pella cat no., 19244)
-
1,2-Dichloroethane (Sigma-Aldrich, cat. no. 284505)
!CAUTION 1,2-Dichloroethane is a hazardous material and potential carcinogen. Wear protective clothing, gloves and goggle and handle under a chemical fume hood.
-
Lead citrate (Electron Microscopy Sciences, cat. no. 17810)
!CAUTION Lead citrate is toxic; use the reagent in a hood and wear gloves while handling.
-
Lead acetate (Electron Microscopy Sciences, cat. no. 17600)
!CAUTION Lead acetate is toxic; use the reagent in a hood and wear gloves while handling.
-
Lead nitrate (Electron Microscopy Sciences, cat. no. 17900)
!CAUTION Lead nitrate is toxic; use the reagent in a hood and wear gloves while handling.
Sodium citrate (Electron Microscopy Sciences, cat. no. 21140)
Poly-L-lysine hydrochloride (Sigma-Aldrich, cat. no. P2658-100MG)
-
49% (vol/vol) Hydrofluoric acid (Fisher Scientific, cat. no. A147-1LB)
!CAUTION. Hydrofluoric acid is toxic. Use in chemical hood with gloves. Contact with skin requires immediate medical attention.
dSTORM Buffer
Glucose oxidase (Fisher Scientific, cat. no. ICN19519650)
Catalase (Fisher Scientific, cat. no. S25239A)
2-mercaptoethanol (Sigma Aldrich, cat. no. 63689-25ML-F) !CAUTION. 2-mercaptoethanol is toxic. Use in chemical hood with gloves and seal before removal.
Glucose (Sigma Aldrich, cat. no. G8270)
EQUIPMENT
Tokuyasu Cryosectioning
-
Diamond trimming knife (Diatome, TrimTool 45, cat. no. TT-45)
!CAUTION Diamond knives are extremely sharp and fragile. Care must be taken not to touch the knife-edge to protect both user and the knife.
-
Cryo immuno diamond knife (Diatome, cat. no. DCIMM3530)
!CAUTION Diamond knives are extremely sharp and fragile. Care must be taken not to touch the knife-edge to protect both user and the knife.
Perfect loop (Electron Microscopy Sciences, cat. no. 70944)
Spin processor with vacuum chuck (Laurell cat no. WS-400-6NPP-LITE)
Aluminum cryo specimen pins (Electron Microscopy Sciences, cat. no. 70446)
Cell scraper (Research Products International Corp., cat. no. 162423)
Cryogenic vials (Sigma-Aldrich, cat. no. CLS431416)
Zeiss Merlin field-emission scanning electron microscope fitted with a focused ion beam gun (FEI) for 3D imaging (Zeiss, cat. no. 3495999001035000) or equivalent
Ultra-microtome (Leica, EM UC6)
Cryo-chamber for ultramicrotome (Leica, EM FC7)
Bench-top micro-centrifuge
Sputter–deposition system (Denton Explorer 14, Denton Vacuum)
Rotating platform (Electron Microscopy Sciences, cat. no. 61050-10)
Super-resolution fluorescence microscope. Can be custom-built or one of several commercially available models such as ELYRA (Zeiss), NSTORM (Nikon), Leica GSD (Leica), Vutara 352 (Bruker). Commercially available models all have differences in their abilities, including 3D capabilities.
Whole-cell mount
(Optional.) Critical point drying sample holder. We machined our own coverslip holder to look very similar to Tousimis, cat. no. 8767, but have an extended diameter to accommodate the 25 mm coverslip. Spare wavy washers (Tousimis, cat. no. 8767-01) should be used as coverslip spacers.
Two 100 ml beakers with a metal mesh shelf that will accommodate the critical point drying sample holder on top and a stir rod on the bottom
Magnetic stir plate
Sputter–deposition system (Denton Explorer 14, Denton Vacuum)
Rotating platform (Electron Microscopy Sciences, cat. no. 61050-10)
Zeiss Merlin field-emission scanning electron microscope fitted with a focused ion beam gun (FEI) for 3D imaging (Zeiss, cat. no. 3495999001035000) or equivalent
Super-resolution fluorescence microscope. Can be custom-built or one of several commercially available models such as ELYRA (Zeiss), NSTORM (Nikon), Leica GSD (Leica), Vutara 352 (Bruker). Commercially available models all have differences in their abilities, including 3D capabilities.
Platinum Replica Module
Branson Sonifier 450 Sonicator (VWR International 47727-492) with a 1/8″ tapered microtip (VWR International cat. no. 33996-163)
Critical point dryer (Tousimis Samdri cat. no. 795)
Freeze fracture system (Jeol JFD-V)
Glow discharge device (Pelco EasiGlow)
-
Diamond objective marker m25 thread (Leica 11505059) ^CRITICAL. You will most likely need to make an adaptor or spacer to fit this objective marker to your microscope.
We have machined a 14 mm spacer tapped with m25 to receive the marker and m25 threading to fit into our Nikon NSTORM turret.
Attofluor cell chamber (Life Technologies cat. no. A-7816), if using a commercial microscope stage. This is only for STORM and dSTORM. You may use any other coverslip chamber or slide but we find that this chamber has a useful upper lip where an additional coverslip can sit and seal in the STORM blinking buffer.
Platinum 5 mm Inoculation loop (Electron Microscopy Sciences cat. no. 62433-05)
Diamond scriber (Electron Microscopy Sciences cat. no. 62108-ST)
Single edge razor blade (Fisher Scientific, cat. no. 12-640)
Formvar and carbon coated 75 mesh copper TEM grids (Ted Pella cat. no. 01802-F)
Filter paper (Whatman cat. no. 1001-042)
Critical point drying sample holder. We machined our own coverslip holder that looks very similar to Tousimis cat. no. 8767 but has an extended diameter to accommodate the 25 mm coverslip. Spare wavy washers (Tousimis cat. no. 8767-01) should be used as coverslip spacers.
Two 100 ml beakers with a metal mesh shelf that will accommodate the critical point drying sample holder on top and a stir rod on the bottom
Magnetic stir plate
12-well plate (Corning cat. no. 3512)
Plastic bulb transfer pipettes (Fisher cat. no. 13-711-7M)
IMOD freeware software (http://bio3d.colorado.edu/imod/)
6-well cell culture plates (Fisher Sci. cat no. 08-772-1)
Jeol 1400 transmission electron microscope equipped with a high tilt specimen retainer (JEOL EM-21311HTR) and Serial EM Freeware
Upright light microscope equipped with 10× phase objective and camera. We use a Zeiss Axioplan 2 with a Zeiss A-Plan 10× / 0.25 NA Ph1 objective.
Super-resolution fluorescence microscope. Can be custom-built or one of several commercially available models such as ELYRA (Zeiss), NSTORM (Nikon), Leica GSD (Leica), Vutara 352 (Bruker). Commercially available models all have differences in their abilities, including 3D capabilities.
Resin embedding method
Heating plate with magnetic stirrer
Rotating platform (Electron Microscopy Sciences, cat. no. 61050-10)
Ultra-microtome (Leica, EM UC6)
Wohlwend HPF Compact 01 high-pressure freezer (Techno Trade)
HPF specimen carrier (TechnoTrade, Type A 0.1/0.2 mm, Type B flat)
Freeze substitution unit (Leica, EMAFS2)
Glow discharge unit (Ted Pella, cat. no. 91000 or Pelco EasiGlow)
Synaptek slot grid (Ted Pella, cat. no. 4514)
120-200 kV transmission electron microscope (FEI, Tecnai 20) and/or Zeiss Merlin field-emission scanning electron microscope (Zeiss, cat. no. 3495999001035000) or equivalent
30 ml Nalgene Teflon drop-dispensing bottle (Thermo Scientific, cat. no. 2414-0030)
Super-resolution fluorescence microscope. Can be custom-built or one of several commercially available models such as ELYRA (Zeiss), NSTORM (Nikon), Leica GSD (Leica), Vutara 352 (Bruker). Commercially available models all have differences in their abilities, including 3D capabilities.
Other
MATLAB (Mathworks) and Interactive Data Language (IDL), for running the PeakSelector software for fitting 3-dimensional interferometric datasets, for instance from iPALM
Biosafety cabinet
Chemical fume hood
PeakSelector (written in MATLAB and IDL). Available for license from HHMI. Please contact innovation@janelia.hhmi.org for licensing information.
Sample coating for SEM
Sputter–deposition system (Denton Explorer 14, Denton Vacuum)
Rotating platform (Electron Microscopy Sciences, cat. no. 61050-10)
REAGENT SETUP
20% (wt/vol) sucrose for Tokuyasu cryosectioning. Dissolve 20 g of sucrose in deionized water to a final total volume of 100 ml. Store at 4°C for up to 4 months.
Aldehyde fixative for Tokuyasu cryosectioning. Combine 3 ml distilled water, 2 ml 1 M phosphate buffer pH 7.4, 5 ml of 20% sucrose, 5 ml of 8% glutaraldehyde, and 5 ml of 16% paraformaldehyde. !CRITICAL Solution must be used the same day of preparation.
2.3 M Sucrose in phosphate buffer for Tokuyasu cryosectioning. Measure 100 ml water using a graduated cylinder and pour into 250 ml beaker. Mark meniscus; then pour out water. Add stir bar and then 80 g of sucrose to beaker. Add 10 ml of 1 M phosphate buffer pH 7.4 to beaker then fill to mark on beaker with distilled water. Stir until sucrose is dissolved then filter-sterilize using 0.22 μm vacuum filtration unit. Store at 4 °C for up to 4 months.
10% (wt/vol) gelatin for Tokuyasu cryosectioning. Combine 5 g of gelatin and 5 ml of 1 M phosphate buffer pH 7.4 in a 50 ml conical tube and bring to a final total volume of 50 ml with deionized water. Heat in a microwave until ~60°C (avoid boiling). Screw on cap and vortex until gelatin is dissolved. Centrifuge at 2000 g for 5 min at room temperature (20–22°C) and transfer to a clean container. Store at 4°C for up to 4 months.
50 mM glycine in PBS for Tokuyasu cryosectioning. Make a 0.1 M solution of glycine by dissolving 7.5 g of glycine in 600 ml of deionized water. Adjust the volume to 1 L. For a 50 mM glycine solution in PBS, mix 50 ml of 0.1 M glycine, 10 ml of 10× PBS pH 7.4, and 40 ml of deionized water. Store at 4°C for up to 12 months.
3% (wt/vol) uranyl acetate for Tokuyasu Method for two-dimensional electron microscopy. In an amber vial to protect the solution from light, dissolve 1.5 g uranyl acetate in deionized water to a final total volume of 50 ml. Mix until the powder is dissolved. Filter through a 0.45 μm filter and store protected from light at 4°C for up to 2 months.
2% (wt/vol) polyvinyl alcohol for Tokuyasu Method for two-dimensional electron microscopy. Dissolve 1 g of polyvinyl alcohol in deionized water to a final total volume of 50 ml. Mix well until the powder is dissolved. Filter through a 0.45 μm filter. Store at 4°C for up to 4 months.
0.6% (wt/vol) uranyl acetate in polyvinyl alcohol for Tokuyasu Method for two-dimensional electron microscopy. Mix 1 ml of 3% (wt/vol) uranyl acetate with 4 ml of 2% polyvinyl alcohol. Filter through a 0.45 μm filter prior to use. Solution should be made just prior to use.
5% (wt/vol) potassium ferrocyanide for Tokuyasu Method for two-dimensional electron microscopy. Dissolve 0.25 g of potassium ferrocyanide in 5 ml of deionized water. !CRITICAL Solution should be made just prior to use.
-
2% osmium reduced with potassium hexacyanoferrate (a.k.a. potassium ferrocyanide) for Tokuyasu Method for two-dimensional electron microscopy. Working in a hood, combine 0.32 ml distilled water, 0.4 ml of 1 M phosphate buffer pH 7.4, 1.28 ml of 5% potassium ferrocyanide, and 2 ml of 4% osmium tetroxide.
!CRITICAL Solution should be made just prior to use. !CAUTION Only use in a chemical safety cabinet.
Sato’s triple lead for Tokuyasu Method for two-dimensional electron microscopy. A mixture of 1% (wt/vol) lead nitrate, 1% lead citrate, 1% lead acetate and 2% sodium citrate in water. Add 0.1 g lead nitrate, 0.1 g lead citrate, 0.1 g lead acetate, and 0.2 g sodium citrate to 8.2 ml distilled water. Shake the mixture vigorously for 5 min, and sonicate for 30 seconds, then add in 1.8 ml of freshly made 4% sodium hydroxide (in distilled water). Store in a sealed container at 4°C for up to 1 year.
0.0075% (vol/vol) Sato’s triple lead in polyvinyl alcohol for Tokuyasu Method for two-dimensional electron microscopy. Mix 10 μl of 3% Sato’s lead with 4 ml of 2% polyvinyl alcohol. Solution should be made just prior to use.
5% (wt/vol) uranyl acetate for Tokuyasu Method for three-dimensional microscopy using FIB-SEM. In an amber vial to protect the solution from light, dissolve 2.5 g uranyl acetate in deionized water to a final total volume of 50 ml. Mix until the powder is dissolved. Filter through a 0.45 μm filter and store protected from light at 4°C for up to 2 months.
2% (wt/vol) methylcellulose for Tokuyasu Method for three-dimensional microscopy using FIB-SEM. Resuspend 4 g methylcellulose powder in 60°C deionized water with a final total volume of 50 ml. Resuspend methylcellulose powder in water by vortexing. Move the solution to 4°C for 3 d then centrifuge at 40,000 g for 1 hour at 4°C to pellet debris and undissolved methylcellulose. Store at 4°C for up to 4 months.
0.5% uranyl acetate in 1.8% methylcellulose for Tokuyasu Method for three-dimensional microscopy using FIB-SEM. Combine 0.2 ml of 5% uranyl acetate solution with 1.8 ml of 2% methylcellulose solution. Solution should be made just prior to use.
30 mM glycine for Whole Cell Mount. Make a 0.1 M solution of glycine by dissolving 7.5 g of glycine in 600 ml of deionized water. Adjust the volume to 1 L. For a 30 mM glycine solution in PBS, pH 7.4 combine 30 ml of 0.1 M glycine, 10 ml of 10× PBS pH 7.4, and 60 ml of deionized water. Store at 4°C for up to 12 months.
4% (wt/vol) paraformaldehyde and 0.2% (vol/vol) glutaraldehyde fixative in PBS for Whole Cell Mount. Mix 1 ml of 16% paraformaldehyde and 0.1 ml of 8% glutaraldehyde with 2.9 ml of phosphate buffered saline, pH 7.4. !CRITICAL Solution aliquots should be frozen at -20°C and freeze thawing should be minimized.
2% (wt/vol) glutaraldehyde in PBS for Whole Cell Mount. Mix 1 ml of 8% glutaraldehyde in 3 ml of phosphate buffered saline, pH 7.4 (makes 4 ml). !CRITICAL Solutions should be made prior to use in a chemical fume hood.
1% (wt/vol) osmium tetroxide for Whole Cell Mount. 1 ml of 4% OsO4 solution to 3 ml of distilled water (makes 4 ml). !CRITICAL Solutions should be made prior to use in a chemical fume hood.
5% (wt/vol) potassium ferrocyanide for Whole Cell Mount. Dissolve 0.25 g of potassium ferrocyanide in 5 ml of deionized water. !CRITICAL Solution should be made just prior to use.
1% osmium reduced with potassium hexacyanoferrate (a.k.a. potassium ferrocyanide) for Whole Cell Mount. Working in a hood, combine 1.32 ml distilled water, 0.4 ml of 1 M phosphate buffer pH 7.4, 1.28 ml of 5% potassium ferrocyanide, and 1 ml of 4% osmium tetroxide. !CRITICAL Solution should be made just prior to use. !CAUTION Only use in a chemical safety cabinet.
Ethanol solutions (15–95% vol/vol) for Whole Cell Mount. Dilute 200 proof ethanol in distilled water according to the desired percentage.
Molecular sieve dried ethanol for Whole Cell Mount. Add 200–500 sieve pellets into a fresh 200 proof ethanol bottle. Allow ethanol solution to dry for several days prior to use. !CRITICAL Ethanol solutions dried with molecular sieve should not be disturbed after incubation, pipette solution from top of liquid level without movement of sieve pellets (do not pour solution from bottle). Store dried ethanol at room temperature.
Hexamethyldisilazane (HMDS) solutions (25–75% vol/vol ethanol) for Whole Cell Mount. Molecular sieve dried ethanol is used to dilute the HMDS according to the desired percentage. Solutions should be made prior to use in a chemical fume hood.
Unroofing stabilization buffer for Platinum Replica Module In 1 L of water combine 5.22 g KCl (70 mM final conc.), 7.15 g HEPES (30 mM final conc.), 476 mg MgCl2 (5 mM final conc.), 1.14 g EGTA (3 mM final conc.), and bring to pH 7.4 with 5 M KOH. Store at room temperature (up to 6 months).
0.5% (wt/vol) Paraformaldehyde in PBS for unroofing. Solution diluted into unroofing stabilization buffer from freshly opened 16% ampule. Use immediately.
2% (wt/vol) Paraformaldehyde in PBS for post-unroofing fixation. Solution diluted into unroofing stabilization buffer from freshly opened 16% ampule. Use immediately.
2% (wt/vol) Glutaraldehyde in PBS for Platinum Replica Module. Mix 10 ml of 8% glutaraldehyde with 4 ml of 10× PBS and 26 ml of deionized water. Use immediately.
0.1% (wt/vol) tannic acid for Platinum Replica Module. 10 mg of solid tannic acid is added to 10 ml of water immediately prior to use.
0.1% (wt/vol) uranyl acetate for Platinum Replica Module. Dilute 1% (wt/vol) uranyl acetate stock solution 1:10 with deionized water. 1% ranyl acetate solution is made with 1 g of uranyl acetate in 100 ml of water and stirred in an amber vial to protect from light. May take minutes to hours to dissolve. Store protected from light at 4°C for up to 2 months.
4.9% (vol/vol) hydrofluoric acid for Platinum Replica Module. Dilute 49% hydrofluoric acid in water immediately prior to use in a polypropylene 15 ml conical tube. !CRITICAL Never use glass containers or pipettes to deal with hydrofluoric acid. Some polystyrenes may cause contamination. !CAUTION. Hydrofluoric acid is toxic. Use in chemical hood with gloves. Contact with skin requires immediate medical attention.
CLEM fixative for Resin-embedding. 4% (wt/vol) PFA, 0.2% glutaraldehyde in 0.1 M phosphate buffer (PB), pH 7.2. Mix 5 ml 16% PFA, 0.5 ml 8% glutaraldehyde, and 14.5 ml 0.1 M PB. Solution should be made just prior to use.
2% (wt/vol) agarose for Resin-embedding. Mix 2 g of agarose in 100 ml 0.1 M PB, and microwave to a clear liquid just prior to use.
1% (wt/vol) Osmium tetroxide for Resin-embedding. Dilute 4% osmium tetroxide to 1% osmium tetroxide in water. Can be stored in a fridge for months if sealed and double-bottled and wrapped in aluminum foil.
1% (wt/vol) Uranyl acetate (100 ml) for Resin-embedding. 1 g uranyl acetate in 100 ml water, mix well and filter through 0.2 μm filter. Store at 4°C for up to 1 month.
Sato’s triple lead for Resin-embedding. A mixture of 1% (wt/vol) lead nitrate, 1% (wt/vol) lead citrate, 1% (wt/vol) lead acetate and 2% (wt/vol) sodium citrate in water. Add 0.1 g lead nitrate, 0.1 g lead citrate, 0.1 g lead acetate, and 0.2 g sodium citrate to 8.2 ml distilled water. Shake the mixture vigorously for 5 min, and sonicate for 30 seconds, then add in 1.8 ml of freshly made 4% sodium hydroxide (in distilled water). Filter through 0.2 μm filter. Store at 4°C for up to 3 months.
GMA resin (20 ml) for Resin-embedding. Add 14 ml glycol methacrylate, 6 ml butyl methacrylate and 0.12 g benzoyl peroxide. Use glass pipet and glass vial. ! CRITICAL Sonicate mixture to clear liquid. Use immediately.
30% GMA for Resin-embedding. 30% (vol/vol) GMA in 95% ethanol. Use immediately.
70% GMA for Resin-embedding. 70% (vol/vol) GMA in 95% ethanol. Use immediately.
Freeze substitution medium for Resin-embedding. 96% (vol/vol) acetone, 3% (vol/vol) water, 0.1% (wt/vol) uranyl acetate, 1% (vol/vol) methanol. Add 10 mg uranyl acetate to 9.6 ml acetone and 0.1 ml methanol. Stir and sonicate until uranyl acetate dissolves. Then add 0.3 ml deionized H2O. Optionally include 0.5% OsO4 for improved ultrastructural preservation for OsO4-resistant probes such as mEos4. To add OsO4 while maintaining the concentrations of the other components, make 5% OsO4 in acetone by dissolving 0.1 g OsO4 in 2 ml acetone. For a 10 ml freeze substitution medium, dissolve 10 mg uranyl acetate in 8.6 ml acetone, 0.1 ml methanol and 1 ml 5% OsO4. After uranyl acetate is dissolved, add 0.3 ml deionized H2O. Aliquot the freeze substitution medium in cryo-vials and store in liquid nitrogen for up to 12 months.
1% pioloform for Resin-embedding. 1% (wt/vol) pioloform in 1,2-dichloroethane. Mix 0.2 g pioloform with 20 ml 1,2-dichloroethane in a fume hood. Stir until dissolved. Store at room temperature for up to 6 months.
1.2% hydrofluoric acid for Resin-embedding. Mix 0.5 ml 49% hydrofluoric acid with 19.5 ml water in 30 ml Teflon drop dispenser bottle. Store at room temperature indefinitely.
1 M K2HPO4. Dissolve 136 g of K2HPO4 in 800 ml deionized water. Bring to a final total volume of 1000 ml with deionized water. Store at room temperature indefinitely.
1 M KH2PO4. Dissolve 174 g of KH2PO4 in 800 ml deionized water. Bring to a final total volume of 1000 ml with deionized water. Store at room temperature indefinitely.
100 mM phosphate buffer, pH 7.4 For a 100 mM potassium phosphate buffer, combine 80.2 ml of 1 M K2HPO4, 19.8 ml of 1M KH2PO4, and 900 ml deionized water for a buffer with a final pH of 7.4. Store at room temperature indefinitely.
1× Phosphate buffered saline (PBS), pH 7.4. Dissolve 8 g of NaCl, 0.2 g of KCl, 1.44 g of Na2HPO4, and 0.24 g of KH2PO4 in 900 ml of deionized water. Adjust the pH to 7.4 with HCl. Add deionized water for a total final volume of 1 L. Sterilize by filtration through a 0.22 μm filter. Store at room temperature indefinitely.
10× Phosphate buffered saline (PBS), pH 7.4. Dissolve 80 g of NaCl, 2 g of KCl, 14.4 g of Na2HPO4, and 2.4 g of KH2PO4 in 800 ml of deionized water. Adjust the pH to 7.4 with HCl. Add deionized water for a total final volume of 1 L. Sterilize by filtration through a 0.22 μm filter. Store at room temperature indefinitely.
1% (wt/vol) bovine serum albumin. Dissolve 0.5 g of bovine serum albumin in 40 ml of deionized water. Add 5 ml of 1 M phosphate buffer, pH 7.4 then bring to a final volume of 50 ml with deionized water. Store at 4°C for up to 3 months.
4% (w/v) sodium hydroxide. Dissolve 4 g of sodium hydroxide in 80 mL of deionized water. Adjust final volume to 100 mL with deionized water. Make fresh just before use.
0.1% (wt/vol) Poly-L-lysine. For consistent results always make fresh 0.1% wt/vol solution of dry stock poly-L-lysine hydrochloride in deionized water.
10% wt/vol glucose in PBS for dSTORM buffer. Mix 3.5 g of glucose into 35 ml of PBS. Store at 4°C for up to 1 month.
80 mg/ml glucose oxidase for dSTORM buffer. Add 40 mg of glucose oxidase to a microcentrifuge tube and add 0.5 ml PBS. Make 25 μl aliquots to freeze at -20 °C for up to 3 months.
10 mg/ml catalase for dSTORM buffer. Add 10 mg of catalase to a microcentrifuge tube and add 1 ml of PBS. Make 10 μl aliquots and freeze at -20 °C for up to 3 months.
dSTORM buffer. To 2.5 ml of 10% glucose stock solution, add 25 μl 80 mg/ml glucose oxidase (0.8 mg/ml final conc.), 10 μl of 10 mg/ml Catalase (0.04 mg/ml final conc.), and 18.75 μl of 99% 2-mercaptoethanol. ! CAUTION. 2-mercaptoethanol is toxic. Use in chemical hood with gloves and seal before removal. !CRITICAL Must be made fresh within minutes of using and sealed from air after adding to the sample.
PROCEDURE
Sample Preparation. TIMING 2–7 d
-
1)
The following options are available depending on your desired imaging target and equipment availability. Perform your chosen option.
Step Method Imaging Target Specialized Equipment 1A Tokuyasu Cryosectioning Internal subcellular structures Ultramicrotome with a cryo attachment 1B Whole Cell Mount Extracellular surface structures Critical point dryer 1C Platinum replica of unroofed cells Cytosolic plasma membrane structures Sonicator, freeze fracture device 1D Resin embedding Internal cellular structures labeled with OsO4-resistant FPs 1E Resin embedding with high-pressure freezing and freeze substitution Internal cellular structures labeled with OsO4-resistant FPs High-pressure freezer and freeze-substitution device
A) Tokuyasu cryosectioning. -TIMING 2 d
Prepare tissue culture cells according to your particular study (i.e. transfected with an appropriate plasmid or subjected to a particular treatment). Flat, adherent cells such as 3T3 are typically appropriate (see Supplementary Note). Grow cells in normal tissue culture dishes.
Wash cells with PBS warmed to 37 °C (or cell-line appropriate temperature).
Add primary aldehyde fixative to cells and incubate at 37 °C for 15 min. !CRITICAL If performing small molecule or antibody labeling, or when using a sensitive fluorescent protein, a lower concentration of glutaraldehyde can be used.
Aspirate fixative solution, add fresh fixative, and incubate at room temperature for 1 hr.
If performing labeling with small molecule caged dyes, after aldehyde fixation, incubate the cells with 0.1% sodium borohydride solution in PBS for 7 min. Aspirate the sodium borohydride solution and permeabilize the cells with 0.1% saponin and 3% BSA in PBS for 15 min. Perform small molecule or antibody labeling according to desired protocol. !CRITICAL We found that using Triton X-100 detergent for permeabilization resulted in worse ultrastructure compared to saponin.
Wash twice for 5 mins each with PBS. Washes should be performed on platform shaker with gentle agitation.
Aspirate PBS and add 50 mM glycine in PBS. Incubate for 15 min at room temperature.
Aspirate previous solution and add 1% BSA solution in PBS. Using a Teflon cell scraper, scrape the cells from the tissue culture dish and place in a 50 ml conical tube or other appropriately sized tube.
Pellet the cells by centrifugation at 2000 g for 10 mins at room temperature.
Remove most of the supernatant, leaving behind ~0.5 ml covering the pellet. Resuspend the pelleted cells in the BSA solution and transfer to a 1.5 ml microcentrifuge tube.
Centrifuge at 2000 g for 5 mins at room temperature.
Put some solidified 10% gelatin into a microcentrifuge tube (can transfer chunks using a metal spatula). Warm gelatin in microcentrifuge tube until melted in a water bath set to 37°C, ~20 mins. Gelatin can also be melted in a microwave and kept molten in a 37°C water bath.
Also, warm the microcentrifuge tube containing the pellet in the water bath set to 37°C. This will prevent the gelatin from solidifying prematurely when added to the sample.
When the gelatin has melted, remove the supernatant covering the pellet and gently wash the pellet twice with the warm gelatin.
Add 1 ml of warm gelatin to the pellet containing microcentrifuge tube, and keeping the microcentrifuge tube in the warm water bath, resuspend the pellet in the gelatin. Mixing can be done using a syringe needle.
Once the pellet is resuspended in the gelatin, centrifuge at 16,000 g for 3 mins at room temperature.
Put the microcentrifuge tube on ice for 20 mins until the gelatin solidifies.
Use a razor blade to cut off the bottom of the tube just above the pellet and gently remove the gelatin containing the pellet. !CAUTION Take care when using a sharp razor blade. One can also try to dislodge and remove the pellet using a wooden stick without cutting the tube.
Place 1 ml of 2.3 M sucrose in 0.1 M phosphate buffer, pH 7.4 into a 1.5 ml microcentrifuge tube.
Place an aluminum plate on ice under a stereomicroscope. Place the gelatin embedded pellet on the plate and use a scalpel to cut away excess gelatin from the cell pellet (discard non-pellet-containing gelatin). Using a scalpel, cut the cell pellet into pyramidal pieces less than 1 mm3. !CAUTION Use caution when using a scalpel.
Using a metal spatula, transfer the small pieces of sample into the microcentrifuge tube containing the 2.3 M sucrose solution.
Place the microcentrifuge tube(s) on a rotator inside a 4°C refrigerator or cold room. Infiltrate the samples with sucrose by rotating overnight (12–16 hours).
The following day or after the incubation: Remove the sample pieces using a wooden applicator cut to have an angled flat edge, leaving a small amount of sucrose solution in contact with the sample.
Place a sample piece onto the center of an aluminum pin holder and freeze by plunging it into liquid nitrogen. The pins can then be placed in cryovials and stored until sectioning. PAUSE POINT Frozen samples can be stored indefinitely in liquid nitrogen until sectioning. Note that having a hole in the cryovial helps ensure the pins remain frozen in liquid nitrogen. It also reduces the probability of a cryovial exploding due to liquid nitrogen expansion.
Cut cryosections to required thickness. Various thicknesses can be cut based on the application and imaging mode. For two-dimensional imaging, sections with thicknesses of 100 nm give good results. For three-dimensional imaging, sections of 500 – 1000 nm may be used. For an in-depth protocol for cryosectioning see67,68
Place cryosections directly on the center of prepared coverslips (BOX 1). The methylcellulose-sucrose pick-up solution will automatically cover the section preventing dehydration. ?TROUBLESHOOTING PAUSEPOINT The sample should be stored at 4°C until just before imaging. It can be stored at 4°C for up to 24 hrs.
Just prior to imaging, place three large drops (~250 μl each) of PBS on a piece of Parafilm lying flat on a benchtop. Invert the coverslip, section side down onto the first drop and incubate for 2 mins to wash away residual methylcellulose/sucrose solution. Repeat for the other two PBS drops.
Place the inverted coverslip onto a drop of freshly prepared 0.5% sodium borohydride solution for 15 mins.
Wash twice for 2 mins on drops of PBS to remove sodium borohydride solution.
Proceed to fluorescence imaging (STEP 2).
B) Whole cell mount. -TIMING 2 – 3 d
Mark 18–25 mm coverslips at two locations to spatially distinguish a coordinate system for general referencing of the location of cellular specimens. Typically, a “north” and “east” direction marking is sufficient to locate cells of interest (BOX 2 and Figure 3a–b). ?TROUBLESHOOTING
Seed tissue culture cells to marked 18–25 mm coverslips using standard practices.
If required, transfect cells with plasmid DNA containing photoswitchable or photoactivatable probe fused to protein of interest. Incubate cell specimens for 18–36 hours to allow for protein fusion expression. Transfection should be performed using conditions and reagents suitable and optimized for your specific cell line and protein of interest (see Supplementary Note).
Rinse samples in PBS and fix using 4% paraformaldehyde and 0.2% glutaraldehyde for 30 minutes.
Quench residual fixative using 30 mM glycine in PBS solution for 15 minutes.
If only transfected FPs are used, proceed to PALM imaging (STEP 2). If immunolabeling native proteins or nucleic acids, prepare sample according to standard immunofluorescence protocols and optimize for each molecule of interest. If staining cellular structures with fluorescent dyes, follow manufacturer protocol. CRITICAL STEP: Many fluorescent labeling/staining protocols will have to be optimized for the highly sensitive nature of single molecule localization imaging.
C) Unroofed cells - TIMING 2 – 3 d
Place clean and dry Au nanoparticle coated coverslips (BOX 1) in a 6-well plate with the gold side up. Coat with whatever surface coating your cells prefer to stick to and grow well on. For HeLa cells we coat coverslips with 0.01% poly-L-lysine solution for 20 minutes. (Note that alternative coatings such as collagen or fibronectin could also work well.)
Seed adherent cultured cells onto the coverslip allowing room for growth to reach a final confluency of 80% for imaging (growth rate will be different for different cell lines).
Carry out transfection using conditions and reagents suitable and optimized for your specific cell line (see Supplementary Note). We use Lipofectamine 2000 or TurboFect according to the manufacturers’ instructions. If doing 3D localization, transfecting with myristoylated-PS-CFP2 to demark the membrane plane is useful for 3D alignment.
Fill two wells of a 6-well plate with stabilization buffer, one well with 4 ml of stabilization buffer containing 0.5% paraformaldehyde and one well with 4 ml of stabilization buffer containing 2% paraformaldehyde.
Prepare sonicator with 1/8″ tapered microtip that is in good condition and tightly secured to the horn of the sonicator. Place your 6-well plate immediately below the tip of the sonicator and set your sonicator to the appropriate settings. We use a single 400 ms pulse at the lowest output setting of our sonicator but the settings will need to be tested for your specific sonicator.
Take your cells from the incubator and immediately rinse for a minute in each well containing stabilization buffer.
Place the coverslip in the well containing 0.5% paraformaldehyde in stabilization buffer, bring the sonicator tip down about 5 mm above the coverslip (should be right below surface of liquid) and sonicate within 5 secs of the cells being in fixative. ?TROUBLESHOOTING
Move the coverslip to the well containing 2% paraformaldehyde in stabilization buffer and incubate for 20 mins.
If labeling your protein of interest with antibodies, use a typical antibody labeling procedure that will work for your specific antibodies. Permeabilization reagents like Triton-X should not be used, as the target is already accessible. ?TROUBLESHOOTING x) Image immediately (STEP 2) or store at 4 °C overnight. ^PAUSEPOINT. Do not store the cells for more than 24 hrs prior to imaging. Longer storage will result in sample degradation.
D) OsO4-resistant labels, GMA resin embedding, no high-pressure freezing - TIMING 5 – 7 d
Prepare tissue culture cells according to your particular study (e.g. transfected with an appropriate plasmid or subjected to a particular treatment). Flat, adherent cells such as 3T3 are typically appropriate (see Supplementary Note). Grow cells in a 35 mm tissue culture dish.
In a chemical fume hood, add 2 ml pre-warmed CLEM fixative to cells in the 35 mm tissue culture dish.
Place the dish on ice, blocking light, for 1 hr.
Wash with ice-cold 0.1 M PB three times for 3 mins on ice.
Pre-warm water bath to 42 °C.
Cut a piece of 2% agarose gel and microwave the gel to a clear liquid, and place in the 42 °C water bath.
Add 1 ml of 1% BSA to the primary-fixed cells and scrape the cells off the dish using a cell scraper.
Collect all cells in a 1.5 ml microcentrifuge tube and centrifuge at 3000 g for 3 min at room temperature.
Remove most of the supernatant, leaving enough to cover the cell pellet.
Place the microcentrifuge tube containing the cell pellet in the 42 °C water bath.
Add 1 ml clear 2% agarose liquid, stir gently in the water bath.
Centrifuge at 14,000 g for 2 min at room temperature and put on ice for 30 min to let the agarose solidify.
Use a scalpel to cut the agarose-embedded cell pellet into 0.5–1 mm3 pieces and store in 0.1 M PB. Samples can be stored in 0.1 M PB at 4 °C for up to 6 months.
Post-fix in desired concentration of OsO4 (e.g. 0.5% or 1%) in a fume hood for 1 hr, rotating, blocking light. !CAUTION Fumes of osmium tetroxide are toxic; open the reagent only in an appropriate fume hood.
Rinse with water three times for 15 mins to remove the excess OsO4, rotating and blocking light during washes.
In a fume hood, stain and stabilize in 1% UA for 1 hr. Protect from light.
Dehydrate in an ethanol series of 30%, 50%, 70%, 95% for 15 min each, rotating, blocking light.
Place sample in 95% ethanol in a freeze-substitution unit set to −20 °C for 1 hr.
Infiltrate with 30% GMA for at least 6 hrs at −20 °C. ^PAUSEPOINT Infiltration may go overnight up to 24 hrs.
Infiltrate with 70% GMA for at least 6 hrs at −20 °C. ^PAUSEPOINT Infiltration may go overnight up to 24 hrs.
Infiltrate with 100% GMA overnight at −20 °C. ^PAUSEPOINT Infiltration may go overnight up to 24 hrs.
Infiltrate with fresh 100% GMA two more times for at least 6 hours each time. ^PAUSEPOINT Each infiltration step may go overnight up to 24 hrs.
Add 0.15% (vol/vol) N,N-Dimethyl-p-toluidine to GMA. Pre-cool the GMA stock to −20 °C solution and quickly mix with N,N-Dimethyl-p-toluidine otherwise GMA will polymerize quickly and release heat that could denature fluorescent proteins. Let polymerization run at −20 °C for 24 – 48 hrs. Increase the temperature to 0 °C (5°C/hr). PAUSE POINT It is advisable to use freshly prepared samples due to potential decay of the fluorescent proteins. However, we had samples expressing mEos4 that retained fluorescence up to two years after preparation when stored at −20°C. ?TROUBLESHOOTING
Cut sections to required thickness using standard ultramicrotomy. Various thicknesses can be cut based on the application and imaging mode. For two-dimensional imaging, sections with thicknesses of 100 nm give good results. For three-dimensional imaging, sections of 500 – 1000 nm may be used. !CRITICAL STEP Allow the samples to stabilize at room temperature for 30 min prior to sectioning. Also, limit amount of exposure to light during sectioning to avoid bleaching fluorescent samples. ?TROUBLESHOOTING
If performing the “consecutive-section” approach where adjacent ultra-thin sections cut from resin are separately split between PALM imaging and EM fixation and imaging; cut one section and place it on the center of a coverslip then take the next or “consecutive” section and place it on a standard TEM grid. If performing the “same-section” approach, where a single resin-cut section is subjected to both PALM and TEM then place the section on a pioloform-coated coverslip (BOX 3). If performing the “same-section” approach with SEM then place the section on an ITO-coated coverslip with gold fiducials (BOX 1).
Proceed to fluorescence imaging (STEP 2) for sections on coverslips.
BOX 3. Pioloform coating of coverslips.
Clean coverslip by adding 0.5% HF on each side and hold over sink for 30 seconds. Rinse with deionized water and blow-dry with compressed air.
Attach a clean coverslip on a glass slide using double-sided tape in a way that it can be easily removed. Place the slide in the DiffSpin™ 2 Slide Spinner.
Put 100 μl of 1% pioloform onto the coverslip and immediately start the spinner at maximum speed. Spin for 1 min.
Glow discharge the coverslips: 25 mA, 10 sec.
Coat the coverslips with 100 μl 0.1% poly-L-lysine for 30 min.
Dilute 80 nm gold fiducials with water to 15% and sonicate for 10 min.
Rinse off poly-L-lysine with Milli-Q water and blow-dry the coverslips.
Put diluted fiducials on coverslips and incubate for 20 min.
Rinse with deionized water and blow-dry.
E) OsO4-resistant labels, High-pressure freezing and freeze substitution - TIMING 5 – 7 d
Prepare tissue culture cells according to your particular study (i.e. transfected with an appropriate plasmid or subjected to a particular treatment). Flat, adherent cells such as 3T3 are typically appropriate. Grow cells in a 35 mm tissue culture dish (see Supplementary Note).
Remove culture medium and quickly add 2 ml pre-warmed CLEM fixative to cells in the 35 mm tissue culture dish. Avoid exposure of cells to air.
Place dish on ice, blocking light, for 1 hr.
Wash with ice-cold 0.1 M PB three times for 3 mins on ice.
Coat the flat side of a Type B specimen carrier with hexadecene.
Add one drop of 1% BSA to the Petri dish. Scrape the cells off while keeping the dish on ice and collect in a 15 ml conical centrifuge tube.
Pellet cells by centrifuging at 3000 g for 10 min at 4 °C.
Wash the pellets with 20% BSA and resuspend in small volume (15 μl) of 20% BSA.
Add 0.8 μl cell suspension to the 0.1 mm-deep well of the Type A specimen carrier. Wet the edges of the well so no air bubbles will be trapped.
Take the hexadecene-coated Type B specimen carrier flat side-down with fine forceps. Touch the flat side against a piece of filter paper to remove excess hexadecene and place it over the 0.1/0.2 mm disc.
Load the carrier assembly into the sample holder and freeze in HPF machine per manufacturer’s instructions. Store samples in cryovials (labeled, and punctured in the cap and in the side wall to ensure liquid nitrogen access) in liquid nitrogen. PAUSE POINT Samples can be stored in liquid nitrogen indefinitely.
Under liquid nitrogen, force open the frozen sample carrier assembly with the tips of a pair of pre-cooled forceps. ?TROUBLESHOOTING
-
Transfer the Type A specimen carrier containing the sample to a 2 ml Nalgene cryovial containing 1 ml freeze substitution medium. Program the FS unit according to the following program.
Temp (°C) Temp. changing rate (°C/hr) Duration (hr) −140 - 1 −140 to −90 5 10 −90 - 31 −90 to −45 5 9 −45 - 5 −45 to −20 5 5 −20 - 2 −20 to −45 5 5 Wash with 95% ethanol six times over a period of 3 hrs at −45 °C.
Infiltrate with 30% GMA resin at −45 °C for at least 6 hrs to overnight.
Infiltrate with 70% GMA resin at −45 °C for at least 6 hrs to overnight.
Infiltrate with 100% GMA at −45 °C three times for 3 hrs each. The last infiltration can be done overnight.
Pre-cool the GMA stock solution to −45 °C, and add N,N-dimethyl-p-toluidine to a final concentration of 1.5μl/1ml GMA. Immediately return to −45 °C after quickly mixing.
Transfer the specimen carrier to an embedding capsule filled with the catalyzed GMA. Make sure that the sample side (0.1 mm-deep well) is facing up.
Run polymerization at −45 °C for 24 hours, and increase the temperature to 0 °C (5 °C/hr). PAUSE POINT It is advisable to use freshly prepared samples due to potential decay of the fluorescent proteins. However, we had samples expressing mEos4 that retained fluorescence up to two years after preparation when stored at −20 °C. ?TROUBLESHOOTING
Cut sections to required thickness using standard ultramicrotomy. Various thicknesses can be cut based on the application and imaging mode. For two-dimensional imaging, sections with thicknesses of 100 nm give good results. For three-dimensional imaging, sections of 500 – 1000 nm may be used. !CRITICAL STEP Allow the samples to stabilize at room temperature for 30 min prior to sectioning. Also, limit amount of exposure to light during sectioning to avoid bleaching fluorescent samples. ?TROUBLESHOOTING
If performing the “consecutive-section” approach where adjacent ultra-thin sections cut from resin are separately split between PALM imaging and EM fixation and imaging; cut one section and place it on the center of a coverslip then take the next or “consecutive” section and place it on a standard TEM grid. If performing the “same-section” approach, where a single resin-cut section is subjected to both PALM and TEM then place the section on a pioloform-coated coverslip (BOX 3). If performing the “same-section” approach with SEM then place the section on an ITO-coated coverslip with gold fiducials (BOX 1).
Proceed to fluorescence imaging (STEP 2) for sections on coverslips.
Super-resolution imaging TIMING 1 hr per area imaged
-
2)
Imaging proceeds in a similar manner for all samples at this point with two major variables. The first variable is whether a two-dimensional or three-dimensional imaging system with opposing objectives (e.g., iPALM) will be used. The second variable is whether endogenous fluorescent proteins (PALM) or dye-labeled antibodies (STORM or dSTORM) are used. If you are using a super-resolution microscope with a single objective, use the common holder for your system (step 2A). If using dual, opposing objectives (e.g., for iPALM) the sample will need to be sandwiched between two coverslips, as described in Step 2B. Identifying the same areas in both fluorescence and EM is made easier by either using a coordinate registration system or through the acquisition of low and high-magnification images of the sample using differential interference contrast (DIC) microscopy (See Box 2 and Figure 3). If you are not using a coordinate registration system, you can use a diamond objective marker to make a circle on the bottom of the coverslip around the region that you imaged. In the case that you will be making unroofed cell platinum replicas, we suggest you make a 4 mm circle with your region in the center (See Box 2 and Figure 5).
Figure 5.
Identifying imaged areas when using platinum replicas. (a) After fluorescence imaging, a 10× DIC map is used to find the cells imaged and a 4 mm circle is etched around the imaged region. (b) After coating the sample with platinum and carbon, 10× phase contrast of the circled region allows the cells to be re-found. (c) The replica is lifted onto a TEM grid where the cells should be found by comparing a 10× phase contrast image to the pre-lift phase contrast image of the replica. Images are snapshots taken to guide the correlative process (red pixels are saturated). The colored squares indicate cells that were imaged and located in all three steps. Red arrows indicate the changing orientation of the cellular landscape during each step. Adapted from22. Scale bars = 100 μm.
A) If using a super-resolution microscope with a single objective
-
i)
Mount sample in the common holder for your system as described in manufacturer instructions.
-
i) ii)
Carry out PALM or dSTORM imaging. Protocols for acquisition of SMLM images are microscope dependent and specifics will not be covered here. Please refer to the manufacturer’s instructions for your particular instrument. ?TROUBLESHOOTING iii) After SMLM imaging. Clean the oil off the coverslip carefully using a cotton swab and 70% ethanol or microscope objective cleaning fluid. Use caution not to disturb the sample or let it dry if using hydrated samples (i.e., cryosections, unroofed cells, or whole cells).
B) If using dual, opposing objectives
Obtain suitable coverslips. We typically use #1.5 Ø=25mm coverslip for the bottom, and #1.5 Ø=18mm coverslip for the top. This way it is easy to identify the top side, seal the sandwich, and pry the coverslip sandwich open after measurements.
Place small drops of epoxy near the outer edge of the glass coverslip to be used as top.
Place the top coverslip, with the epoxy drops down, onto the coverslip with the sample. The surface of the sample containing coverslip should be covered in approximately 100 μl of buffer (either PBS or dSTORM buffer). For dSTORM, make up a reducing buffer prior to imaging. Many different reducing buffers have been described25. See Materials for the STORM blinking buffer we typically use.
Press down firmly, but gently, on the top coverslip to create the sandwich.
Seal around the edge of the top coverslip using Vaseline. This can be done by ejecting Vaseline through a syringe needle.
Clean the top and bottom glass surface of the sample sandwich using a cotton swab and microscope objective cleaning fluid. Mount the sample onto the microscope.
Carry out PALM or dSTORM imaging. ?TROUBLESHOOTING
Take apart the coverslip sandwich by cleaning off the Vaseline, adding PBS around the sandwich to keep it wet, and using a very thin sheet of metal (we use a 0.0015″ thickness gauge) to slide between the two coverslips just around the edge to pry the top coverslip off.
Sample Preparation for EM Imaging
-
3)
For Tokuyasu cryosections, proceed to Steps 3A (for 2D SEM) or 3B (for FIB-SEM). For whole cells, proceed to Step 3C. For platinum replicas, proceed to Step 3D. For resin-embedded samples that will be imaged using TEM, proceed to Step 3E.
A) Staining for 2D SEM - TIMING 1 hr
Place the coverslip, section side up into a 35 mm dish and wash three times with 2 ml PBS for 1 min each at room temperature.
-
Aspirate PBS and add 2 ml of 2% OsO4/1.6% potassium ferrocyanide in 0.1 M phosphate buffer. Incubate for 15 mins at room temperature.
!CAUTION Osmium tetroxide is a very strong oxidizing fixative and a heavy metal toxin. It must be handled with extreme caution in a fume hood wearing protective clothing, gloves, and goggles.
Wash three times for 2 mins each with 2 ml deionized water.
Aspirate water and add 2 ml of 0.6% UA in 1.6% polyvinyl alcohol. Incubate for 15 mins at room temperature protected from light.
Wash three times for 3 secs with 2 ml of 2% polyvinyl alcohol.
Aspirate polyvinyl alcohol and add 2 ml of 0.0075% Sato’s triple lead in 2% polyvinyl alcohol for 15 mins at room temperature.
Wash three times for 3 secs with 2 ml of 2% polyvinyl alcohol.
Place coverslip on a spin processor with a vacuum chuck and spin at 5000 rpm for 10 secs.
Coat specimen with 5–10 nm of Au, Pt or Ir using sputter coating system following manufacturer protocol.
Remove coverslip and place in a desiccator until ready to image. The sample can be stored indefinitely. We have imaged samples 5 years after preparation with no apparent loss in sample quality.
Image by SEM. ?TROUBLESHOOTING
B) Staining for FIB-SEM - TIMING 15 mins
Place the coverslip, section side up into a 35 mm dish and wash three times with 2 ml PBS for 1 min each at room temperature.
Put three drops of 200 μl 0.5% UA in 1.8% methylcellulose on Parafilm that is placed on an aluminum block on ice.
Grabbing the coverslip with forceps, invert it so that the section side is down and place it on the first drop for 3 secs, transfer to the second drop for 3 secs and then transfer to the third drop. Incubate for 10 mins.
Grab the coverslip with forceps and drag the edge of the coverslip over Whatman 50 hardened paper until the methylcellulose solution is wicked away from the coverslip surface.
Once methylcellulose is dried, cover the sections with a small drop of cyanoacrylate.
Coat specimen with 5–10 nm of Au, Pt or Ir using sputter coating system following manufacturer protocol.
Image using FIB-SEM. ?TROUBLESHOOTING
C) Whole cell mount EM preparation - TIMING 3 – 6 hrs
Remove sample from fluorescence microscope and wash with PBS.
Post-fix the specimen with 2% glutaraldehyde in PBS buffer for 1–2 hrs. PAUSE POINT Fixation can be performed overnight (12 – 16 hrs) at 4 °C.
Rinse specimen with PBS and then with deionized water.
Apply 1% OsO4 in deionized water as secondary fixative for 1–2 hrs. Optional: Apply 1% OsO4 reduced with potassium ferrocyanide if additional membrane stabilization is required.
Rinse specimen thoroughly with deionized water to remove residual osmium salts.
Dehydrate the specimen with an increasing ethanol series beginning with 15% EtOH for 5 mins. Continue the process with 25%, 35%, 50%, 60%, 75%, 85%, 95%, and 100% EtOH, each for 5 mins. Repeat the 100% EtOH two additional times. Finally, dehydrate 3 times 5 mins with 100% EtOH that has been dried over molecular sieves.
Desiccate specimen using a CO2 critical point drying system according to the manufacturer’s protocol. CRITICAL STEP If you do not have access to a critical point drying system, hexamethyldisilizane (HMDS) can be used to dry the sample. To dehydrate with HMDS, perform a series of washes starting with 25% HMDS/75% dried EtOH for 5 mins. Continue the process by washing with 50% HMDS/50% dried EtOH and 75% HMDS/25% dried EtOH for 5 mins each. Dehydrate with 100% HMDS three times for 5 mins each and finally overlay the specimen with 50–100 μl of fresh HMDS and allow it to air dry slowly in the hood. !CAUTION HMDS wash solutions should be handled in a chemical fume hood. PAUSE POINT Sample can be stored desiccated under vacuum until SEM imaging can be performed. The sample can be stored up to 3 d.
Coat specimen with 5–10 nm of Au, Pt or Ir using sputter coating system following manufacturer protocol. CRITICAL Angular shadowing of the coating will aid in more complete coating of cellular structures and minimize charging effects during SEM imaging. The coated sample can be stored desiccated and under vacuum indefinitely.
Image by SEM. ?TROUBLESHOOTING
D) Platinum replica of unroofed cells - TIMING 3–5 hrs
Post-fix your specimen by placing it in 2% glutaraldehyde in PBS and incubating at 4 °C for 20 mins to 36 hrs.
Transfer coverslip to freshly made 0.1% tannic acid in deionized water for 20 mins at room temperature.
Rinse coverslips four times with deionized water.
Transfer the coverslip to 0.1% UA for 20 mins at room temperature. !CAUTION. UA is an alpha emitter and heavy metal toxin. UA is a cumulative toxin. Wear protective clothing, gloves and goggles.
Rinse coverslips four times with deionized water.
Place your critical point dryer holder in a water filled beaker that has a metal mesh shelf and a magnetic stir bar below the shelf.
Add your sample to the critical point dryer holder making sure that there are spacers that allow liquid to flow above and below your coverslip.
Move the sample into 15% EtOH in your other magnetic stir bar-containing beaker and stir for 5 mins to start the gradual dehydration process. Continue the process with 30%, 50%, 70%, 80%, 90%, 100% EtOH, each for 5 mins. Perform the 100% EtOH wash three times, rinsing out the beaker each time with 100% EtOH to remove residual water.
Place the sample holder into the critical point dryer in 100% EtOH and dry the sample according to the manufacturer’s instructions. In our Tousimis Samdri-795, we purge the chamber with liquid CO2 for 20 minutes.
Cut the sample to fit into the freeze fracturer using a diamond scriber to cut the coverslip around the etched circle region. It should be small enough to fit on your freeze fracturer stage. We use a lab marker to make dark dots on the bottom side of the coverslip making the region more visible. We then use a razor blade to guide our diamond scriber during cutting.
Place your sample on your freeze fracturer stage and use two pieces of ~1 mm2 double-stick tape to adhere the sample to a flat stage.
After placing your sample in the freeze fracturer, use the manufacturer’s instructions to rotary shadow (sputter-deposit platinum at an angle while rotating the sample) 2–4 nm of platinum/carbon onto your sample at a 17° angle (90° is no tilt). Then coat with carbon at 90° to stabilize the Pt/C coat (~5–10 nm).
Image your coverslip with a 10× phase contrast objective to be sure you can find the region that you previously imaged and you know what the region looks like after drying and coating (BOX 2 and Figure 5). You will use this image to help you find your cells of interest on your grid after the replica has been transferred.
Cut the sample again. Before lifting, the sample needs to be ~4 mm in diameter so you should be able to cut exactly around where the original diamond objective marking was. Cut with the diamond scriber again to assure that the sample is this size. It should fit completely within the 5 mm inoculation loop but not be so small that it can move around a lot within the loop. The region that you imaged should be directly in the center. It is possible to cut the sample down to this size the first time that you cut, which would make this step unnecessary. We prefer to cut twice because the larger piece gives room to handle the coverslip (attaching it to and pulling it off the double-stick tape with tweezers without scratching the region of interest).
Glow discharge TEM grids (75-mesh, with formvar and carbon coating) for one minute according to the manufacturer’s instructions.
If you are doing tomography, you should add 10 nm gold fiducials to the grids; these will enhance your tomography reconstruction. Dip your grids into a 10 nm gold solution or 10 nm gold conjugated antibody for two minutes, rinse with water, and place on filter paper to dry. You may need to optimize the density of gold labeling by changing the concentration of gold. If not doing tomography, you should not add 10 nm gold fiducial markers as it will add unnecessary spots to your image.
Place the grids on filter paper.
-
Place the cut sample, replica side up, on the surface of 5% hydrofluoric acid (HF) (diluted in pure water from 49% stock solution) in a 12-well plate. Wait for the glass coverslip to fall to the bottom of the well while the replica stays at the surface. Use a transfer pipette to replace the HF with water by successive dilutions while keeping the replica at the surface. ^CRITICAL. You must be delicate and avoid the replica touching the pipette or riding up the side of the well. Bringing the solution down to ~1.5 ml and adding ~5 ml of water 10–15 times has been sufficient for us. !CAUTION. Hydrofluoric acid is extremely toxic and should be used with gloves in a fume hood. Any contact with skin requires immediate medical attention.
?TROUBLESHOOTING
Lift the replica onto the grid by bringing a 4 mm Pt inoculation loop into the liquid and underneath the replica to lift it up out of the water. Bring the inoculation loop with replica down onto the grid and keep it there for 3 secs until the replica has stuck to the grid and the liquid has been soaked up by the filter paper.
Image the grid with a 10× phase contrast objective and find where the cells are on the grid (Box 3 and Figure 5). ^CRITICAL. There is a good chance that at least one or maybe two of the regions of interest have been obscured by a grid bar. You will not be able to correlate these cells.
-
Image by TEM montaging or perform electron tomography.
?TROUBLESHOOTING
E) Resin embedded samples preparation for TEM. TIMING 30 min
CRITICAL For plastic embedded sections, a same-section or consecutive-section approach can be taken. For the consecutive section approach, cut two consecutive sections, placing one on a TEM grid and the other on a coverslip for PALM. The section on the TEM grid can be stained as below, starting at step 3E step viii. For imaging the same section in both PALM and TEM, the section will need to be moved from the pioloform-coated coverslip (Box 3) to a TEM grid. This can be achieved by following all the steps in step 3E.
After PALM imaging, using a razor, score the fiducial side of the coverslip with a rectangular area surrounding the sections.
Place 1–2 drops 1.2% HF along one edge of the scored rectangle.
Tilt the coverslip slightly and let the HF seep under the pioloform film. ?TROUBLESHOOTING
Fill a glass jar with water. Once the HF solution has reached the other edge of the scored rectangle, dip the coverslip slowly into the water to float the pioloform membrane. ?TROUBLESHOOTING
Place a non-coated Synaptek slot grid on the floating film to capture the sections in the center of the opening slot. ?TROUBLESHOOTING
Lower a piece of Parafilm onto the floating film to pick up the grid.
After drying, carefully punch out the grid from the Parafilm with fine tweezers.
Place a drop of 1% UA on a piece of Parafilm in a Petri dish. Put the TEM grid on top of the 1% UA droplet, section side-down, for 10 min. Protect from light.
Pass the grid over 3 deionized water droplets for washing.
Dry the grid with filter paper by touching the edge of the grid.
Place a piece of Parafilm in a Petri dish, with a few NaOH pellets inside the dish.
Cover the Petri dish and wait 5 min for NaOH to absorb excess carbon dioxide from the air in the Petri dish.
Dispense a drop of Sato’s triple lead (filtered through 0.2 μm filter) on the Parafilm.
Place the grid on top of the lead droplet, section side-down. Put the lid back on the Petri dish and stain for 3 min.
Rinse quickly with deionized water.
Dry the grid with filter paper by touching the edge of the grid. The grid is now ready for TEM imaging.
Perform TEM imaging. ?TROUBLESHOOTING
Registering and transforming PALM and EM data sets. TIMING 1–2 h
-
4)
To register two-dimensional data sets follow Step 4A. To register three-dimensional data sets, follow Step 4B. Both processes are implemented in the PeakSelector routine. Step A can only be used when the same sample or section is imaged in both light and electron microscopy, i.e. not when consecutive sections are imaged. Consecutive sections have sufficiently different structures that they must be manually registered.)69.
Alternative methodologies for registration and alignment may also be used. Instead of small, near-point-emitter fiducials like Au nanoparticles, large feature-based correlation methods, such as autofluorescence correlation16 can be used. It should be noted that such methods have major drawbacks: (1) They require that LM and EM datasets have large-scale features that can be correlated, (2) Further, it requires specific knowledge of possible differences in these feature’s representation between LM and EM images (e.g. cytoplasmic labeling in LM, membrane labeling of the same structure in EM), and (3) They do not provide an easy way of quantifying the registration error, unlike the methods described in this protocol.
Methods based on Fourier ring correlation82 could be tested to see if they fit well into the user’s workflow. A fairly inclusive list of SMLM localization and deconvolution algorithms can be found at http://bigwww.epfl.ch/smlm/software.
A) Registering two-dimensional data sets
After PALM data has been collected and processed, determine the coordinate pairs { } of multiple reference fiducial points (Au nanoparticles).
Identify the same reference fiducial points in EM micrographs and determine their coordinate pairs { }.
Determine the transformation coefficients Kx, yij based on the corresponding reference sets { } and { } (Use POLYWARP function in IDL or CP2TFORM function in MATLAB).
Use the transformation coefficients Kx, yij, to calculate the new (transformed) PALM coordinates for reference fiducial points
- Calculate the registration errors for each fiducial point as:
Compare the determined registration errors εi for reference fiducial points with acceptable registration tolerance εtolerance (εtolerance = 20 nm is reasonable). If ⟦max {εi}⟧ ≥ εtolerance, exclude the point with maximum registration error from the reference sets { } and { } used to determine the transformation coefficients (assuming that the error is due to a fiducial object that is not a single dipole source). After this, repeat steps iii through vi. This will result in a new set of transformation coefficients Kx, yij with a new corresponding set of registration errors {εi}.
Once the condition ⟦max {εi}⟧ < εtolerance is met, use the transformation coefficients Kx, yij to calculate the new (transformed) PALM coordinates for all fluorescent localizations. ?TROUBLESHOOTING
- Determine the registration error as the average of the registration errors of all fiducial markers:
B) 3D )PALM and EM image registration
Identify the fiducial points.
Assuming that the fiducial points are Au nanoparticles on the surface of the coverslip, transform the entire 3D PALM data set so that all fiducial points lie in the same average plane of z=0. There may be small local variations between the vertical locations of fiducial points, so this transformation is not intended to put them exactly on the same plane. This transformation should be simple and global. We use a shear transformation, done with linear regression used initially to find a best-fit plane through all fiducial points. Next, shear transformation is applied, such that x- and y-coordinates remain unchanged, but z coordinates are changed so that the plane defined in linear regression coincides with the plane defined by condition z=0.
Perform 2D image registration of PALM and EM data sets leaving z coordinates unchanged.
Identify the features inside the volume of the sample, which can be detected in both PALM and EM data sets (ideally close to the top surface of the sample). These features may be fluorescently labeled proteins with locations easily identifiable in EM (such as clathrin or membrane proteins in the case of correlative iPALM-TEM of unroofed cells22), or random sparse fluorescence throughout the volume of the section (as in the case of iPALM-FIB-SEM of thick cryo-sections18).
Determine the scaling coefficient required to make these features overlap. In the example of iPALM-TEM of unroofed cells this means adjusting the scaling factor so that the membrane proteins overlap vertically with the Pt replica of the membrane. In the example of iPALM-FIB-SEM of thick cryosections the scaling is adjusted so that the random sparse fluorescence falls off at the same vertical location as the boundary of the section observed in the 3D EM data set. See Figure 4.
-
Apply the scaling coefficient to the entire 3D PALM data set.
?TROUBLESHOOTING
TIMING
Step 1A Tokuyasu cryosectioning. -TIMING 2 d
Step 1B Whole cell mount. -TIMING 2–3 d
Step 1C Unroofed cells - TIMING 2 – 3 d
Step 1D OsO4-resistant labels, GMA resin embedding, no high-pressure freezing - TIMING 5 – 7 d
Step 1E OsO4-resistant labels, High-pressure freezing and freeze substitution - TIMING 5 – 7 d
Step 2 Super-resolution imaging - TIMING 1 hr per area imaged
Step 3A Staining Tokuyasu cryosections for 2D SEM - TIMING 1 hr
Step 3B Staining Tokuyasu cryosections for FIB-SEM - TIMING 15 mins
Step 3C Whole cell mount EM preparation - TIMING 3 – 6 hrs
Step 3D Platinum replica of unroofed cells - TIMING 3–5 hrs
Step 3E Resin embedded samples preparation for TEM. TIMING 30 min
Step 4 Registering and transforming PALM and EM data sets. TIMING 1–2 h
?TROUBLESHOOTING
See Table 2 for troubleshooting guidance. Additional advice follows.
Table 2.
Troubleshooting
| Step | Problem | Possible Reason | Solution |
|---|---|---|---|
| 1.A.xxvi | Large folds in Tokuyasu cryosections | Not using correct cryosectioning equipment. | Use a sharp diamond knife designed for cryo-ultramicrotomy. Hold the loop perfectly steady when picking up the sections |
| 1.A.xxvi | Tokuyasu cryosections have large holes | Section retrieval solution not correct | Use a 1:1 methylcellulose:sucrose solution for section retrieval |
| 1.B.i | Loss of cardinal axis system on coverslip | The marking step is susceptible to removal by solvents used in the SEM sample preparation protocol. | Use marking method that is not susceptible to removal by solvents such as etch marking with a diamond knife. |
| 1.C.vii | Sonication blows away all of the cells | The cells weren’t adhered strongly enough | Try a stronger adhering surface. |
| Sonication was too strong | Decrease power on sonicator or length of time sonicating | ||
| None of the cells were unroofed | Cells were fixed too long prior to sonication | Sonicate more quickly after placing in fixative | |
| The sonication was not strong enough. | Sonicate stronger and closer, or make sure your sonicating tip is secured tighter to the sonicator horn, and is in good condition. | ||
| The unroofed cells are very broken up like shards of cells | Cells didn’t uniformly adhere to surface. | Treat the cells with poly-L-lysine and a hypo-osmotic shock prior to lifting, which will help the cells stick down more uniformly. Please see22. | |
| Intracellular structure is getting fixed together and tearing the cell apart. | Try sonicating in stabilization buffer without fixative and then quickly transferring to fixative. | ||
| 1.C.ix | The antibody labeling is not effective | One of the antibodies is bad. | Try a new antibody |
| The concentration has not been optimized | Try altering the concentration of antibodies | ||
| 1.D.xxiii 1.E.xx |
GMA not polymerizing at -45 °C | GMA resin without N,N-dimethyl-p-toludine was not completely removed prior to addition of catalyzed resin, resulting in the improper concentration of catalyst | Bring the temperature up to -30 °C or -20 °C |
| 1.D.xxiv 1.E.xxi |
GMA blocks are too soft for ultramicrotomy | GMA is not prepared correctly | Make sure instructions for preparation of GMA are followed correctly. Be especially careful that the catalyst is added at the correct concentration. |
| Blocks are too cold. | Warm up the block before cutting. | ||
| 1.E.xii | Frozen specimen carrier assembly cannot be split open | Specimen carrier was not coated with hexadecene | Ensure the flat disc is coated with hexadecene. |
| 2.A.iii 2.B.vii |
Little to no fluorescence in Tokuyasu cryosections | Fluorescent proteins are degraded. | Sections should be imaged by fluorescence within 24 hours after Tokuyasu cryosectioning. |
| 3.A.xi 3.B.vii |
Uneven contrast across Tokuyasu cryosections | Dehydration artifact. | Be sure sections are not exposed to air dehydration prior to final drying step. |
| 3.A.xi 3.B.vii |
Precipitate on Tokuyasu cryosections | Heavy metal stain precipitate | Make sure staining solutions are fresh. Filter UA and lead solutions just prior to use. Be sure salts are removed through water washes before UA/lead staining. |
| 3.C.ix | Membranes not preserved well (cracking or holes in membrane surface) as assessed by SEM imaging. | Sample not dehydrated properly | Extend the length of ethanol dehydration steps to 10 minutes each. Ensure that EtOH dried over molecular sieve is used in the final wash steps to eliminate any residual water remaining in the sample. |
| Fixation proceeded too long causing swelling and rupture. | Reduce fixation times with 2% glutaraldehyde and 1% OsO4 to 1 hour each. | ||
| Critical point drying step did not remove all EtOH from sample upon liquid CO2 exchange and prior to CO2 evaporation. | Proceed with additional exchanges and extend the mixing times between EtOH and liquid CO2. Subsequently extend the CO2 evaporation time by slowing the release rate during the evaporation routine. Follow manufacturer protocols for the specific critical point dryer used. | ||
| Cell samples too brittle for critical point drying. | Test the HMDS drying/desiccation procedure. | ||
| Not enough OsO4 to properly stabilize membranes. | Reduce OsO4 with potassium ferrocyanide prior to secondary fixation of specimen. | ||
| 3.D.xviii | The replica falls to the bottom of the well in the hydrofluoric acid. | The sample was placed upside down in the HF. | Check the orientation of the coverslip prior to placing on the HF. The cell surface generally appears rough and should be facing up. |
| The HF is contaminated from the plastic. | Check what types of plastics are coming into contact with the HF. Some polystyrenes will dissolve contaminants into your HF, making the surface tension lower. | ||
| The replica sticks to the wall of the 12-well plate and breaks. | Unknown | As soon as you see the replica approach the wall of the well, you should add more water to help it move away from the wall. The replicas should be constantly observed during this process. |
|
| 3.D.xix | The replica breaks into pieces when lifted out of solution or placed on the grid | The carbon layer is too thin. | Coat with more carbon to make the replica more stable. |
| 3.D.xxi | Sample looks melted in EM | There was trace water present during critical point drying | Make sure you are using 100% dry ethanol during dehydration. Open a new bottle before each dehydration or ensure that ethanol dried over a molecular sieve is used |
| The critical point dryer is not functioning properly. | Check to make sure critical-point dryer is operating at the appropriate temperatures and pressures and has enough CO2. | ||
| The sample dried out prior to critical point drying or became wet after critical point drying. | Be sure your sample never dries out prior to critical point drying, and gets rotary shadowed immediately after CPD to avoid rehydration from moisture in the air. | ||
| There are black spots around the grid bars in EM. | Hydrofluoric acid reacted with the grid. | Do more dilutions of the hydrofluoric acid prior to moving your replica onto the grid | |
| 3.E.iii | Hydrofluoric acid solution flows over sections | Excessive hydrofluoric acid used. | Use less hydrofluoric acid solution. Tilt the coverslip at a slight angle and let the hydrofluoric acid slowly seep under the pioloform membrane. |
| 3.E.iv | Some of the pioloform still adheres to the coverslip | Coverslip not scored cleanly. | When scoring the coverslip, use a ruler to help score a clean line. |
| 3.E.v | Pioloform film moves too much on water surface | Too much airflow in the room. | Minimize airflow in the room by working away from vents and limiting the movement of people and equipment. |
| 3.E.xvii | Dirty precipitate on grid | Precipitation of lead. | Lead reacts with carbon dioxide. Avoid airflow as much as possible during the lead staining procedure. Ensure grid is dry before touching the lead solution. |
| Poor sample morphology with artifacts and/or over-extraction in high-pressure frozen cells | High-pressure freezing was performed incorrectly | Do not expose cells to air at any point. Avoid air bubbles when placing Type B specimen carrier over Type A. After HPF, transfer the specimen carrier immediately to liquid nitrogen. After the sample is frozen with HPF, precool forceps in liquid nitrogen before touching the specimen carrier. Minimize temperature changes when changing media during washing and infiltration. |
|
| Section has too many wrinkles on the grid | Grids are hydrophobic | Use glow discharge to make grids hydrophilic | |
| 4.A.vii | Cellular specimen does not register to the corresponding fluorescent image with high fidelity, but Au fiducials are registered well and with a very small uncertainty. | The SEM sample preparation and dehydration procedure caused movement of the cellular specimen with respect to the coverslip (Au fiducial) surface. | Thick portions of the specimen often experience more movement on the nanometer scale upon desiccation. Choose regions of the cell to image by fluorescence that are thinner and less prone to movement/distortion upon desiccation. |
| 4.A.vii | Poor image registration or high residual uncertainty in SEM/fluorescence image alignment (based on fiducial alignment). | Au nanoparticle density is too low or non-reproducible | Choose region of interest during fluorescence image acquisition that is surrounded by many (>20) surface-exposed Au fiducials. Plating cells at lower density can help find unobstructed fields of view with detectable fiducials. Use new stock of Au nanoparticles. Au nanoparticles have a shelf life of a few months (80 nm nanospheres) to a few weeks (25 × 75 nm and 25 × 57 nm nanorods). Put a higher concentration of gold on your coverslips. Make sure the coverslips are cleaned properly. Do not recycle coverslips. Make fresh 0.1% poly-L-Lysine solution (shelf life ~ 1 week) so that Au nanoparticles adhere to coverslip. |
| All fiducials are confined to one region of the image, biasing the registration towards that region. | Choose cells of interest by fluorescence that have even (non-clustered) spatial distributions of fiducials. | ||
| 4.B.vi | Registration of FIB-SEM and iPALM data of Tokuyasu cryosections is not accurate | Shrinkage of sample | Tokuyasu cryosections are subject to significant shrinkage in the z-dimension during dehydration. Try using CPD or HMDS drying or place fiducials on top of the section. |
Fixation
All fixation procedures have the possibility to create artifacts in the sample. In general, cryofixation tends to have fewer artifacts than chemical fixation, but can still exhibit problems induced by freezing. Of chemical fixatives, aldehydes such as paraformaldehyde and glutaraldehyde generally provide the best sample preservation with the fewest artifacts, but can require optimization for each sample.
Make aldehydes fresh before each use as detailed in the protocols. Aldehydes polymerize over time, resulting in a loss of reactivity and an increase in size, such that they penetrate samples more slowly. Avoid using “ready-to-use” fixatives, as these can contain preservatives that inhibit polymerization. Instead, purchase “EM quality” glutaraldehyde, as it contains the highest fraction of monomers, which have optimal reactivity and sample penetration.
Temperature, pH and osmolarity should all be optimized to improve the quality of fixation. Fixation time is among the most important variable in sample quality and is easy to manipulate. In general, PFA fixation proceeds slowly and glutaraldehyde fixation more rapidly; conversely, PFA penetrates into the sample more quickly than the larger glutaraldehyde. A mixture of both of these typically provides a nice balance between fixing the sample quickly enough that its quality is maintained but slowly enough that sample damage such as oxidation doesn’t occur. Dehydration of under-fixed samples can frequently lead to protein precipitation, creating aggregates that can interfere with both LM and EM. In addition to cutting down on glutaraldehyde aromatization and subsequent autofluorescence generation (as discussed above), the use of aldehyde quenchers such as borohydrides also blocks free aldehydes, whose reactivity can become a problem in subsequent steps such as antibody addition.
Membrane blebs can be produced by cells as they die during fixation. The poor reactivity of aldehydes against lipids can lead to membrane motility such as vesiculation or the formation of “myelin figures” in samples. Use of membrane cross-linkers such as OsO4 can largely alleviate such potential artifacts. PFA can cross-link DNA, leading to chromatin alterations. In general, bright-field microscopy (such as DIC) can be a quick and effective way to diagnose fixation artifacts before proceeding to SMLM and EM. We find that optimization of the fixation protocol can largely alleviate concerns about these potential artifacts.
Fixation can also decrease the performance of fluorescent proteins. Optimization of fixation time can improve fluorescence preservation. Alternatively, specifically engineered fluorescent proteins such as mEos4 resist fluorescence loss better than unoptimized probes (but still show fluorescence loss). An easy way to optimize fixation conditions for fluorescent targets is to perform fixation of samples in a vessel (e.g., glass-bottom dish, chambered coverslip) in which the sample can be imaged and fluorescence monitored during the various steps of sample preparation.
Dehydration
Dehydration is a step common to all EM preservation techniques and will often destroy fluorescent proteins. Most of our sample preparation techniques avoid sample dehydration prior to fluorescence imaging (except for those using fixation-resistant proteins). Removal of water and other volatile liquids from samples can result in sample shrinkage and deformation. Overly rapid dehydration can lead to plasmolysis of cells, extraction of small organelles from the cell, and can also cause samples to become hard and brittle. On the other hand, the presence of residual dehydrating agents (particularly ethanol) can both cause blocks to become soft over time and can interfere with subsequent plastic resin polymerization steps. For an analysis of the artifacts that can be caused by dehydration and resin-embedding see83. If samples become noticeably disrupted during dehydration (which again, may be diagnosed by DIC imaging), these steps should be optimized for the sample. Both acetone and ethanol, and different concentration steps and incubation times should be tested on the sample.
Sectioning artifacts
The cutting of sections is also subject to artifact generation. Tears in the sample can decrease image quality and make alignment of torn halves problematic. Folds can obscure objects of interest and make alignment and interpretation difficult. Obtaining the services and advice of an expert in ultramicrotomy will be necessary to obtain good sections for imaging.
Anticipated Results
Correlation using gold nanoparticles
An example of the ability to identify gold in both fluorescence and EM channels can be seen in Figure 2.
Tokuyasu cryosectioning
The user should expect accurate localization of your fluorescently labeled protein or dye within a context-rich electron micrograph as seen in Figure 6a–c. In particular, membranes are well defined and visible in electron microscopy of Tokuyasu cryosections. If performing 3D CLEM, the fluorescent probes should be observable within a 3D volume (Figure 6 b,c). However, due to shrinkage of the sample one would expect significant collapse of the z height. This can be adjusted and appropriately aligned to give excellent 3D CLEM results.
Figure 6.
Examples of anticipated results. (a–c) Examples of CLEM using Tokuyasu cryosectioning adapted with permission from Kopek et al.18. NIH3T3 mouse embryonic fibroblast cells were transfected with a plasmid expressing the mitochondrial DNA-binding protein (mitochondrial transcription factor A (TFAM)) genetically fused to the paFP mEos2, rendered in red or magenta. (a) Example result for 2D CLEM imaging of mitochondrial nucleoids using the Tokuyasu protocol. (b,c) Example result for 3D CLEM of mitochondrial nucleoids using the Tokuyasu protocol with iPALM and FIB-SEM showing (b) an x–y plane and (c) a y–z plane sectioned through the volume as indicated by the marks in b. (d) Example results for whole-cell-mount method showing correlated iPALM and SEM of COS7 cells transfected with a plasmid expressing the HIV Gag protein fused to mEos2. (e) Platinum replica of an unroofed HeLa cell correlated with dSTORM of the Alexa-Fluor-647-labeled clathrin adaptor, AP-2. (f,g) Example of transmission electron micrograph (f) overlaid with a PALM (red/white) image (g) of NIH3T3 mouse embryonic fibroblast cells transfected with a plasmid expressing a mitochondrial matrix localized mEos4a. The sample was high-pressure frozen, freeze-substituted with 0.5% OsO4, and embedded in GMA resin. Scale bars, 500 nm.
Whole-cell Mount
If this protocol is performed properly, the user should expect two-dimensional correlative SEM and super-resolution point localization images detailing the topology of the specimen surface as well as the nanoscale distribution of the labeled molecule of interest as seen in Figure 6d21. The SEM/fluorescence images should be registered within <30 nm (often below 10 nm with sufficient fiducial coverage within the field of view) of one another, enabling a detailed topological analysis. Specimen shrinkage can occur during dehydration prior to SEM, which increases uncertainty in the final correlation (see Troubleshooting section regarding sample shrinkage issues for the Whole Cell Mount protocol).
Unroofed cells
In general, platinum replicas will show the complex architecture of the plasma membrane with its associated cytoskeleton, exocytic and endocytic structures, and membrane proteins as seen in Figure 6e. Clathrin coated pits can be clearly identified in TEM by their distinctive geometric basket patterns and caveolae appear as distinctive 80 nm-striped vesicles. Filaments of the actin cortex (appearing 8–12 nm in width) are also generally prominent features of these images.
When this correlative protocol is performed properly, you should be able to observe high quality PALM and platinum replicas. With smaller cells (15–20 μm) you should be able to correlate the entire adhering membrane. With larger cells, you should limit yourself to 20 μm regions for correlation to avoid large-scale imperfections in the film on the grid and EM image stitching. The final correlation should have 20-nm accuracy or better. If you are using antibodies for fluorescence labeling, there should be small (~20 nm) blobs in your EM that co-localize with your fluorescence localizations.
Resin-embedded samples
Both standard chemical fixation with aldehyde cross-linking and high-pressure freezing/freeze substitution (HPF-FS) should produce well-preserved cellular samples with good ultrastructure as seen in Figure 6f–g. Addition of OsO4 for samples with fluorescent labels resistant to this treatment (such as mEos420) should markedly improve ultrastructure preservation, including that of plasma and nuclear membranes, endoplasmic reticulum, Golgi apparatus and mitochondrial cristae. Chemical fixation is compatible with larger samples than HPF-FS, which typically requires samples smaller than 100 μm, in order to achieve uniform freezing without ice crystals.
The protocols described here should maintain fluorescence of the FP probes used, either targeted to fill specific organelles or the cytoplasm, or by fusion to specific proteins of interest. The user should expect good ultrastructure preservation, and the post-embedding use of aldehyde quenchers such as borohydride and cyanoborohydride should decrease autofluorescence without perturbing ultrastructure.
Supplementary Material
Acknowledgments
We would like to thank Richard Fetter and Wei-Ping Li of HHMI-Janelia Research Campus for assistance with electron microscopy. We would like to thank Matt Daniels and the NHLBI EM core facility for help with platinum replica TEM. U.S. Army Medical Research Institute of Infectious Disease (USAMRIID) Disclaimer: Opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the U.S. Army. M.G.P.-S., G.S., C.S.X., L.L.L. and H.F.H. were supported by the Howard Hughes Medical Institute. J.W.T. was supported by the Intramural Research Program of the NHLBI, US National Institutes of Health. This research was supported in part by funds from the Boettcher Foundation’s Webb-Waring Biomedical Research Awards program and The National Institute of Allergy and Infectious Disease, NIH_1R21AI127192-01 to S.B.vE.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing interests.
AUTHOR CONTRIBUTIONS
Tokuyasu protocol: B.G.K., G.S., C.S.X., H.F.H. Whole-mount protocol: S.B.v.E., G.S., H.F.H. Unroofing protocol: K.A.S., G.S., S.B.v.E., H.F.H., J.W.T. Resin embedding protocol: M.G.P-S., M.G.S., G.S., H.F.H., Y.W., L.L.L. iPALM/FIB-SEM protocol: C.S.X., G.S., H.F.H. All authors contributed to writing the paper.
Supplementary Note. Commonly Used Cell Lines and Plasmids
References
- 1.Shimomura O, Johnson FH, Saiga Y. Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J Cell Comp Physiol. 1962;59:223–239. doi: 10.1002/jcp.1030590302. [DOI] [PubMed] [Google Scholar]
- 2.Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 3.Shaner NC, Patterson GH, Davidson MW. Advances in fluorescent protein technology. J Cell Sci. 2007;120:4247–4260. doi: 10.1242/jcs.005801. [DOI] [PubMed] [Google Scholar]
- 4.Lippincott-Schwartz J, Manley S. Putting super-resolution fluorescence microscopy to work. Nat Methods. 2009;6:21–23. doi: 10.1038/nmeth.f.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 6.Hess ST, Girirajan TPK, Mason MD. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophysical Journal. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Westphal V, Hell SW. Nanoscale resolution in the focal plane of an optical microscope. Phys Rev Lett. 2005;94:143903. doi: 10.1103/PhysRevLett.94.143903. [DOI] [PubMed] [Google Scholar]
- 9.Gustafsson MG. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J Microsc. 2000;198:82–87. doi: 10.1046/j.1365-2818.2000.00710.x. [DOI] [PubMed] [Google Scholar]
- 10.Li D, et al. Extended-resolution structured illumination imaging of endocytic and cytoskeletal dynamics. Science. 2015;349:aab3500. doi: 10.1126/science.aab3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heilemann M, van de Linde S, Mukherjee A, Mukherjee A, Sauer M. Super-Resolution Imaging with Small Organic Fluorophores. Angew Chem Int Ed. 2009;48:6903–6908. doi: 10.1002/anie.200902073. [DOI] [PubMed] [Google Scholar]
- 12.Kim D, et al. Correlative Stochastic Optical Reconstruction Microscopy and Electron Microscopy. PLoS ONE. 2015;10:e0124581–20. doi: 10.1371/journal.pone.0124581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watanabe S, et al. Protein localization in electron micrographs using fluorescence nanoscopy. Nat Methods. 2011;8:80–84. doi: 10.1038/nmeth.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nanguneri S, Flottmann B, Horstmann H, Heilemann M, Kuner T. Three-dimensional, tomographic super-resolution fluorescence imaging of serially sectioned thick samples. PLoS ONE. 2012;7:e38098. doi: 10.1371/journal.pone.0038098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suleiman H, et al. Nanoscale protein architecture of the kidney glomerular basement membrane. eLife. 2013;2:e01149. doi: 10.7554/eLife.01149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perkovic M, et al. Correlative light- and electron microscopy with chemical tags. J Struct Biol. 2014;186:205–213. doi: 10.1016/j.jsb.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Löschberger A, Franke C, Krohne G, van de Linde S, Sauer M. Correlative super-resolution fluorescence and electron microscopy of the nuclear pore complex with molecular resolution. J Cell Sci. 2014;127:4351–4355. doi: 10.1242/jcs.156620. [DOI] [PubMed] [Google Scholar]
- 18.Kopek BG, Shtengel G, Xu CS, Clayton DA, Hess HF. Correlative 3D super resolution fluorescence and electron microscopy reveal the relationship of mitochondrial nucleoids to membranes. Proc Natl Acad Sci USA. 2012;109:6136–6141. doi: 10.1073/pnas.1121558109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kopek BG, Shtengel G, Grimm JB, Clayton DA, Hess HF. Correlative Photoactivated Localization and Scanning Electron Microscopy. PLoS ONE. 2013;8:e77209. doi: 10.1371/journal.pone.0077209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paez-Segala MG, et al. Fixation-resistant photoactivatable fluorescent proteins for CLEM. Nat Methods. 2015;12:215–218. doi: 10.1038/nmeth.3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Engelenburg SB, et al. Distribution of ESCRT machinery at HIV assembly sites reveals virus scaffolding of ESCRT subunits. Science. 2014;343:653–656. doi: 10.1126/science.1247786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sochacki KA, Shtengel G, Van Engelenburg SB, Hess HF, Taraska JW. Correlative super-resolution fluorescence and metal-replica transmission electron microscopy. Nat Methods. 2014;11:305–308. doi: 10.1038/nmeth.2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gould TJ, Verkhusha VV, Hess ST. Imaging biological structures with fluorescence photoactivation localization microscopy. Nat Protoc. 2009;4:291–308. doi: 10.1038/nprot.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shtengel G, et al. Imaging cellular ultrastructure by PALM, iPALM, and correlative iPALM-EM. Methods Cell Biol. 2014;123:273–294. doi: 10.1016/B978-0-12-420138-5.00015-X. [DOI] [PubMed] [Google Scholar]
- 25.van de Linde S, et al. Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nat Protoc. 2011;6:991–1009. doi: 10.1038/nprot.2011.336. [DOI] [PubMed] [Google Scholar]
- 26.Koga D, Kusumi S, Shodo R, Dan Y, Ushiki T. High-resolution imaging by scanning electron microscopy of semithin sections in correlation with light microscopy. Microscopy. 2015:dfv042. doi: 10.1093/jmicro/dfv042. [DOI] [PubMed] [Google Scholar]
- 27.Collman F, et al. Mapping synapses by conjugate light-electron array tomography. J Neurosci. 2015;35:5792–5807. doi: 10.1523/JNEUROSCI.4274-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Micheva KD, Smith SJ. Array Tomography: A New Tool for Imaging the Molecular Architecture and Ultrastructure of Neural Circuits. Neuron. 2007;55:25–36. doi: 10.1016/j.neuron.2007.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Egner A, et al. Fluorescence nanoscopy in whole cells by asynchronous localization of photoswitching emitters. Biophysical Journal. 2007;93:3285–3290. doi: 10.1529/biophysj.107.112201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dertinger T, Colyer R, Iyer G, Weiss S, Enderlein J. Fast, background-free, 3D super-resolution optical fluctuation imaging (SOFI) Proc Natl Acad Sci USA. 2009;106:22287–22292. doi: 10.1073/pnas.0907866106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rego EH, et al. Nonlinear structured-illumination microscopy with a photoswitchable protein reveals cellular structures at 50-nm resolution. Proc Natl Acad Sci USA. 2012;109:E135–43. doi: 10.1073/pnas.1107547108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bornfleth Satzler, Eils, Cremer High-precision distance measurements and volume-conserving segmentation of objects near and below the resolution limit in three-dimensional confocal fluorescence microscopy. J Microsc. 1998;189:118–136. [Google Scholar]
- 33.Van Oijen AM, Köhler J, Schmidt J, Müller M, Brakenhoff GJ. 3-Dimensional super-resolution by spectrally selective imaging. Chemical Physics Letters. 1998;292:183–187. [Google Scholar]
- 34.Lemmer P, et al. SPDM: light microscopy with single-molecule resolution at the nanoscale. Appl Phys B. 2008;93:1–12. [Google Scholar]
- 35.Hofmann M, Eggeling C, Jakobs S, Hell SW. Breaking the diffraction barrier in fluorescence microscopy at low light intensities by using reversibly photoswitchable proteins. Proc Natl Acad Sci USA. 2005;102:17565–17569. doi: 10.1073/pnas.0506010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fölling J, et al. Fluorescence nanoscopy by ground-state depletion and single-molecule return. Nat Methods. 2008;5:943–945. doi: 10.1038/nmeth.1257. [DOI] [PubMed] [Google Scholar]
- 37.Egner A, Jakobs S, Hell SW. Fast 100-nm resolution three-dimensional microscope reveals structural plasticity of mitochondria in live yeast. PNAS. 2002;99:3370–3375. doi: 10.1073/pnas.052545099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Egner A, Verrier S, Goroshkov A, Söling HD, Hell SW. 4Pi-microscopy of the Golgi apparatus in live mammalian cells. J Struct Biol. 2004;147:70–76. doi: 10.1016/j.jsb.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 39.Schrader M, Bahlmann K, Giese G, Hell SW. 4Pi-Confocal Imaging in Fixed Biological Specimens. Biophysical Journal. 1998;75:1659–1668. doi: 10.1016/S0006-3495(98)77608-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perinetti G, et al. Correlation of 4Pi and Electron Microscopy to Study Transport Through Single Golgi Stacks in Living Cells with Super Resolution. Traffic. 2009;10:379–391. doi: 10.1111/j.1600-0854.2008.00875.x. [DOI] [PubMed] [Google Scholar]
- 41.Schübbe S, Cavelius C, Schumann C, Koch M, Kraegeloh A. STED Microscopy to Monitor Agglomeration of Silica Particles Inside A549 Cells. Advanced Engineering Materials. 2010;12:417–422. [Google Scholar]
- 42.Chang YW, et al. Correlated cryogenic photoactivated localization microscopy and cryo-electron tomography. Nat Methods. 2014;11:737–739. doi: 10.1038/nmeth.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu B, et al. Three-dimensional super-resolution protein localization correlated with vitrified cellular context. Sci Rep. 2015;5:13017. doi: 10.1038/srep13017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaufmann R, et al. Super-resolution microscopy using standard fluorescent proteins in intact cells under cryo-conditions. Nano Lett. 2014;14:4171–4175. doi: 10.1021/nl501870p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson E, et al. Correlative in-resin super-resolution and electron microscopy using standard fluorescent proteins. Sci Rep. 2015;5:9583. doi: 10.1038/srep09583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ding B, Turgeon R, Parthasarathy MV. Substructure of freeze-substituted plasmodesmata. Protoplasma. 1992;169:28–41. [Google Scholar]
- 47.Los GV, et al. HaloTag: A Novel Protein Labeling Technology for Cell Imaging and Protein Analysis. ACS Chem Biol. 2008;3:373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 48.Keppler A, et al. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat Biotechnol. 2003;21:86–89. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- 49.Sharonov A, Hochstrasser RM. Wide-field subdiffraction imaging by accumulated binding of diffusing probes. Proc Natl Acad Sci USA. 2006;103:18911–18916. doi: 10.1073/pnas.0609643104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Endesfelder U. Advances in Correlative Single-Molecule Localization Microscopy and Electron Microscopy. NanoBioImaging. 2015;1:1–9. [Google Scholar]
- 51.de Boer P, Hoogenboom JP, Giepmans BNG. Correlated light and electron microscopy: ultrastructure lights up! Nat Methods. 2015;12:503–513. doi: 10.1038/nmeth.3400. [DOI] [PubMed] [Google Scholar]
- 52.Li J, Wang Y, Chiu SL, Cline HT. Membrane targeted horseradish peroxidase as a marker for correlative fluorescence and electron microscopy studies. Front Neural Circuits. 2010;4:6. doi: 10.3389/neuro.04.006.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Connolly CN, Futter CE, Gibson A, Hopkins CR, Cutler DF. Transport into and out of the Golgi complex studied by transfecting cells with cDNAs encoding horseradish peroxidase. J Cell Biol. 1994;127:641–652. doi: 10.1083/jcb.127.3.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lam SS, et al. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat Methods. 2014;12:51–54. doi: 10.1038/nmeth.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martell JD, et al. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat Biotechnol. 2012;30:1143–1148. doi: 10.1038/nbt.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shu X, et al. A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol. 2011;9:e1001041. doi: 10.1371/journal.pbio.1001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Viswanathan S, et al. High-performance probes for light and electron microscopy. Nat Methods. 2015;12:568–576. doi: 10.1038/nmeth.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mercogliano CP, DeRosier DJ. Concatenated metallothionein as a clonable gold label for electron microscopy. J Struct Biol. 2007;160:70–82. doi: 10.1016/j.jsb.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Q, Mercogliano CP, Löwe J. A ferritin-based label for cellular electron cryotomography. Structure. 2011;19:147–154. doi: 10.1016/j.str.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 60.Collins JS, Goldsmith TH. Spectral properties of fluorescence induced by glutaraldehyde fixation. J Histochem Cytochem. 1981;29:411–414. doi: 10.1177/29.3.6787116. [DOI] [PubMed] [Google Scholar]
- 61.Clancy B, Cauller LJ. Reduction of background autofluorescence in brain sections following immersion in sodium borohydride. J Neurosci Methods. 1998;83:97–102. doi: 10.1016/s0165-0270(98)00066-1. [DOI] [PubMed] [Google Scholar]
- 62.McKinney SA, Murphy CS, Hazelwood KL, Davidson MW, Looger LL. A bright and photostable photoconvertible fluorescent protein. Nat Methods. 2009;6:131–133. doi: 10.1038/nmeth.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chudakov DM, Lukyanov S, Lukyanov KA. Tracking intracellular protein movements using photoswitchable fluorescent proteins PS-CFP2 and Dendra2. Nat Protoc. 2007;2:2024–2032. doi: 10.1038/nprot.2007.291. [DOI] [PubMed] [Google Scholar]
- 64.Zhang M, et al. Rational design of true monomeric and bright photoactivatable fluorescent proteins. Nat Methods. 2012;9:727–729. doi: 10.1038/nmeth.2021. [DOI] [PubMed] [Google Scholar]
- 65.Wang S, Moffitt JR, Dempsey GT, Xie XS, Zhuang X. Characterization and development of photoactivatable fluorescent proteins for single-molecule-based super resolution imaging. Proc Natl Acad Sci USA. 2014;111:8452–8457. doi: 10.1073/pnas.1406593111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wysocki LM, et al. Facile and General Synthesis of Photoactivatable Xanthene Dyes. Angew Chem Int Ed. 2011;50:11206–11209. doi: 10.1002/anie.201104571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Slot JW, Slot JW, Geuze HJ, Geuze HJ. Cryosectioning and immunolabeling. Nat Protoc. 2007;2:2480–2491. doi: 10.1038/nprot.2007.365. [DOI] [PubMed] [Google Scholar]
- 68.Webster P, Webster A. Methods in Molecular Biology. Vol. 369. Humana Press; 2007. pp. 257–289. [DOI] [PubMed] [Google Scholar]
- 69.Shtengel G, et al. Interferometric fluorescent super-resolution microscopy resolves 3D cellular ultrastructure. Proc Natl Acad Sci USA. 2009;106:3125–3130. doi: 10.1073/pnas.0813131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Knott G, Marchman H, Wall D, Lich B. Serial section scanning electron microscopy of adult brain tissue using focused ion beam milling. J Neurosci. 2008;28:2959–2964. doi: 10.1523/JNEUROSCI.3189-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tokuyasu KT. Use of poly(vinylpyrrolidone) and poly(vinyl alcohol) for cryoultramicrotomy. Histochem J. 1989;21:163–171. doi: 10.1007/BF01007491. [DOI] [PubMed] [Google Scholar]
- 72.Larson DR, Johnson MC, Webb WW, Vogt VM. Visualization of retrovirus budding with correlated light and electron microscopy. Proc Natl Acad Sci USA. 2005;102:15453–15458. doi: 10.1073/pnas.0504812102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ryan US, Hart MA. Electron microscopy of endothelial cells in culture: II. Scanning electron microscopy and OTOTO impregnation method. Journal of Tissue Culture Methods. 1986;10:35–36. [Google Scholar]
- 74.Svitkina T. Methods in Molecular Biology. Vol. 586. Humana Press; 2009. pp. 187–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Collins A, Warrington A, Taylor KA, Svitkina T. Structural Organization of the Actin Cytoskeleton at Sites of Clathrin-Mediated Endocytosis. Current Biology. 2011;21:1167–1175. doi: 10.1016/j.cub.2011.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Heuser J. Quick-freeze, deep-etch preparation of samples for 3-D electron microscopy. Trends Biochem Sci. 1980;6:64–68. [Google Scholar]
- 77.Heuser J. The production of ‘cell cortices’ for light and electron microscopy. Traffic. 2000;1:545–552. doi: 10.1034/j.1600-0854.2000.010704.x. [DOI] [PubMed] [Google Scholar]
- 78.Caplan J, et al. Correlative Protein Localization in Yeast. 2013. [Google Scholar]
- 79.Kobs SF, Behrman EJ. Reactions of osmium tetroxide with imidazoles. Inorganica Chimica Acta. 1987;138:113–120. [Google Scholar]
- 80.Nielson AJ, Griffith WP. Tissue fixation by osmium tetroxide. A possible role for proteins. J Histochem Cytochem. 1979;27:997–999. doi: 10.1177/27.5.479559. [DOI] [PubMed] [Google Scholar]
- 81.Pertsinidis A, Zhang Y, Chu S. Subnanometre single-molecule localization, registration and distance measurements. Nature. 2010;466:647–651. doi: 10.1038/nature09163. [DOI] [PubMed] [Google Scholar]
- 82.Nieuwenhuizen RPJ, et al. Measuring image resolution in optical nanoscopy. Nat Methods. 2013;10:557–562. doi: 10.1038/nmeth.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mollenhauer HH. Artifacts caused by dehydration and epoxy embedding in transmission electron microscopy. Microscopy research and Technique. 1993;26:496–512. doi: 10.1002/jemt.1070260604. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.