ABSTRACT
Zinc resistance in livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) sequence type 398 (ST398) is primarily mediated by the czrC gene colocated with the mecA gene, encoding methicillin resistance, within the type V staphylococcal cassette chromosome mec (SCCmec) element. Because czrC and mecA are located within the same mobile genetic element, it has been suggested that the use of zinc in feed as an antidiarrheal agent has the potential to contribute to the emergence and spread of methicillin-resistant S. aureus (MRSA) in swine, through increased selection pressure to maintain the SCCmec element in isolates obtained from pigs. In this study, we report the prevalence of the czrC gene and phenotypic zinc resistance in U.S. swine-associated LA-MRSA ST5 isolates, MRSA ST5 isolates from humans with no swine contact, and U.S. swine-associated LA-MRSA ST398 isolates. We demonstrated that the prevalence of zinc resistance in U.S. swine-associated LA-MRSA ST5 isolates was significantly lower than the prevalence of zinc resistance in MRSA ST5 isolates from humans with no swine contact and swine-associated LA-MRSA ST398 isolates, as well as prevalences from previous reports describing zinc resistance in other LA-MRSA ST398 isolates. Collectively, our data suggest that selection pressure associated with zinc supplementation in feed is unlikely to have played a significant role in the emergence of LA-MRSA ST5 in the U.S. swine population. Additionally, our data indicate that zinc resistance is associated with the multilocus sequence type lineage, suggesting a potential link between the genetic lineage and the carriage of resistance determinants.
IMPORTANCE Our data suggest that coselection thought to be associated with the use of zinc in feed as an antimicrobial agent is not playing a role in the emergence of livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) ST5 in the U.S. swine population. Additionally, our data indicate that zinc resistance is more associated with the multilocus sequence type lineage, suggesting a potential link between the genetic lineage and the carriage of resistance markers. This information is important for public health professionals, veterinarians, producers, and consumers.
KEYWORDS: Staphylococcus aureus, zinc resistance
INTRODUCTION
Staphylococcus aureus commonly colonizes the skin and mucosal surfaces of mammalian and avian species and is present in the anterior nares of 20 to 30% of healthy humans (1). S. aureus is also a major opportunistic human pathogen with diverse clinical manifestations, ranging from mild skin and soft tissue infections to severe systemic infections and fatal sepsis. Prior to the availability of antibiotics, fatality rates for human cases of S. aureus bacteremia were estimated at 80% (2). Increased access to antibiotics has reduced the case fatality rate of S. aureus bacteremia to around 20 to 30% (2), but the capacity of S. aureus to acquire resistance to antibiotics has made multidrug-resistant strains a major public health concern (3).
Methicillin-resistant S. aureus (MRSA) was first reported in 1961 (4) and rapidly became endemic in hospitals (i.e., hospital-associated MRSA [HA-MRSA]) in many countries. During the 1990s, an increasing number of MRSA infections occurred in persons with no known risk factors for HA-MRSA infection (5). These infections developed in healthy members of the general community and were termed community-associated MRSA (CA-MRSA). S. aureus is considered a clonal organism, and genotypes associated with hospital infections typically differed from those associated with community infections, as well as varying geographically (6).
Although MRSA was first reported in food animals (dairy cattle) in 1972 (7), animal reservoirs were not considered to play a significant role in MRSA epidemiology until 2004, when an atypical MRSA variant was detected in three people in the Netherlands and was attributed to their residence on a swine farm (8). These initial isolates could not be typed by pulsed-field gel electrophoresis using SmaI restriction digestion, due to a variation in methylation by the type I restriction modification system. Multilocus sequence typing (MLST) revealed that these isolates belonged to a novel sequence type (ST), ST398 (9). This genotype was found to be widespread in the Dutch pig industry and to be present in other animal species, including cattle, poultry, and horses (1, 10, 11). Subsequent research revealed more complex epidemiology, and the predominant genotypes of MRSA found in swine vary geographically. In most Asian countries, ST9 variants are most common (12, 13); in the United States and Canada, both ST398 and ST5 MRSA variants appear to be relatively common, with ST9 MRSA being detected sporadically (14–17).
Resistance to tetracycline antibiotics has been almost universal in S. aureus isolates from pigs. Additionally, a prominent feature of livestock-associated MRSA (LA-MRSA) ST398 isolates from Europe and North America is the high prevalence (61 to 74%) of zinc resistance seen in swine-associated isolates (18–20), relative to isolates from veal calves (42%) or humans (48%) (19, 21). Zinc resistance in these MRSA isolates has been attributed to colocalization of the czrC gene (conferring zinc and cadmium resistance) on the type V staphylococcal cassette chromosome mec (SCCmec) element, which contains the mecA gene (conferring methicillin resistance). A strong correlation between phenotypic zinc resistance and the presence of czrC was reported previously, with 99% of MRSA ST398 isolates harboring czrC showing phenotypic zinc resistance and 96% of isolates exhibiting zinc resistance harboring the czrC gene (21). Dietary zinc supplementation at >2,400 ppm (compared with the minimum nutritional requirement of 100 to 165 ppm) for 5 to 10 days is commonly used in weaned pigs to control enteric disease. Since czrC and mecA are colocated on the SCCmec element, it has been suggested that the use of high concentrations of zinc in feed might have contributed to the emergence and spread of MRSA in swine, by increasing the selection pressure to maintain the SCCmec element in swine-associated ST398 isolates (20, 22–24).
While many reports detailing the prevalence of zinc resistance in LA-MRSA ST398 and ST9 isolates have been published, little to no information exists regarding the prevalence of zinc resistance in LA-MRSA ST5 isolates (21). Here we report the prevalence of zinc resistance in U.S. swine-associated LA-MRSA ST5 isolates and compare it with the prevalence in MRSA ST5 isolates obtained from humans with no swine contact and that in U.S. swine-associated LA-MRSA ST398 isolates, as well as that in previous studies reporting zinc resistance in LA-MRS ST398 isolates.
RESULTS
Prevalence of the czrC gene.
The czrC-specific PCR demonstrated that none of the tested swine-associated MRSA ST5 isolates (0/82 isolates) harbored the czrC gene (Table 1). In contrast, all LA-MRSA ST398 isolates (14/14 isolates) tested harbored the czrC gene. The prevalence of czrC in LA-MRSA ST5 isolates associated with swine was significantly lower than that in swine-associated LA-MRSA ST398 isolates (P < 0.0001) (Table 1) and also was lower than that reported for other LA-MRSA ST398 isolates (P < 0.0001) (21). Over one-fifth of MRSA ST5 isolates obtained from humans with no swine contact (16/73 isolates [22%]) contained the czrC gene (Table 1). The prevalence of the czrC gene among MRSA ST5 isolates obtained from humans with no swine contact was significantly higher than that among swine-associated ST5 isolates (P < 0.0001) (Table 1). Information on individual isolates is provided in Table S1 in the supplemental material.
TABLE 1.
Prevalence of phenotypic zinc chloride resistance and czrC presence in isolates from humans with no swine contact and swine-associated isolates
| Characteristic | No. of isolates/total no. of isolates (%) |
||
|---|---|---|---|
| MRSA ST5 from humans with no swine contact | LA-MRSA ST5 | LA-MRSA ST398 | |
| czrC prevalence | 16/73 (21.9)a | 0/82 (0) | 14/14 (100)a |
| Phenotypic zinc chloride resistance | 18/73 (24.7)a | 0/82 (0) | 14/14 (100)a |
Statistical significance (P < 0.0001), compared to LA-MRSA ST5.
Zinc chloride susceptibility testing.
Susceptibility testing revealed that no swine-associated MRSA ST5 isolates (0/82 isolates) were resistant to zinc chloride, while phenotypic resistance was seen for all LA-MRSA ST398 isolates (14/14 isolates). The prevalence of phenotypic resistance to zinc among MRSA ST5 isolates obtained from humans with no swine contact was 25% (18/73 isolates), greater than among swine-associated MRSA ST5 isolates (P < 0.0001) (Tables 1 and 2). Two MRSA ST5 isolates obtained from humans with no swine contact exhibited phenotypic resistance despite not harboring the czrC gene. Phenotypic zinc chloride resistance in the absence of czrC was reported previously for MRSA ST398 and non-ST398 isolates by Cavaco et al. (21), which indicates that an alternative mechanism for zinc resistance is also present in MRSA ST5 isolates.
TABLE 2.
Results from phenotypic zinc chloride resistance screen
| Isolate type | No. of isolates with susceptibility ofa: |
||||||
|---|---|---|---|---|---|---|---|
| 0.25 mM | 0.5 mM | 1 mM | 2 mM | 4 mM | 8 mM | 16 mM | |
| MRSA ST5 from humans with no swine contact (n = 73) | 6 | 0 | 20 | 29 | 14 | 4 | 0 |
| LA-MRSA ST5 (n = 82) | 0 | 20 | 53 | 9 | 0 | 0 | 0 |
| LA-MRSA ST398 (n = 14) | 0 | 0 | 0 | 0 | 14 | 0 | 0 |
Values of >2 mM indicate resistance.
SCCmec typing.
The swine-associated LA-MRSA ST5 isolates carried SCCmec type III (17/82 isolates [21%]) or type IV (42/82 isolates [51%]) or could not be typed using the primer sets published previously (23/82 isolates [28%]) (Table 3). Of the 23 untypeable isolates, 20 (24.4% of LA-MRSA ST5 isolates) carried a class D mec gene complex, which has not been assigned to a mec type, and 3 carried a class A mec gene complex without the traditional ccrA-ccrB gene combination. All LA-MRSA ST398 isolates tested harbored SCCmec type V (14/14 isolates). The MRSA ST5 isolates from humans with no known swine contact mostly carried SCCmec type II (69/73 isolates [95%]); the others were type IV (4/73 isolates [5.5%]).
TABLE 3.
SCCmec type and czrC gene prevalence among LA-MRSA ST5 and ST398 isolates and MRSA ST5 isolates from humans with no swine contact
| Isolate type | SCCmec type | czrC prevalence (no. of isolates/total no. of isolates) |
|---|---|---|
| LA-MRSA ST5 | III | 0/17 |
| IV | 0/42 | |
| Untypeablea | 0/23 | |
| MRSA ST5 from humans with no swine contact | II | 16/69 |
| IV | 0/4 | |
| LA-MRSA ST398 | V | 14/14 |
Isolates that are unable to be classified into an SCCmec type due to the presence of a ccr gene or mec complex unable to be determined using available primer sets or the presence of a ccr and mec complex combination not currently assigned an SCCmec type.
czrC localization.
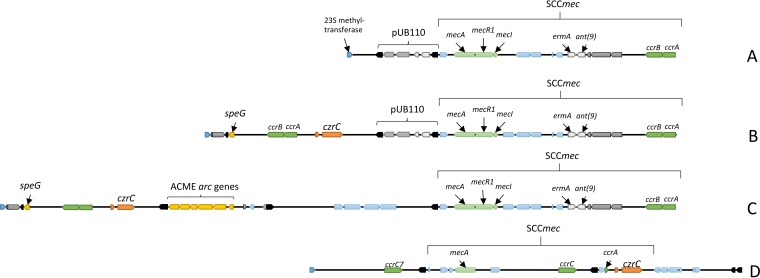
To determine the location of the czrC gene within the genomes of the 16 MRSA ST5 isolates obtained from humans with no swine contact that harbored the czrC gene, draft genome sequences were obtained, along with the complete genome sequences for two of the strains (UCI28 and UCI62) (25, 26). The gene content and organization of the SCCmec region and the surrounding mobile genetic elements for strains UCI28 and UCI62, along with strains Mu3 and S0385 for reference, are shown in Fig. 1. The SCCmec region for strains Mu3 and UCI28 contained pUB110 within the J3 region of the SCCmec element (Fig. 1A and B). Strain UCI28 and 12 other isolates (UCI3, UCI9, UCI19, UCI21, UCI28, UCI43, UCI45, UCI46, UCI48, UCI52, UCI56, and UCI64) harbored czrC downstream of speG and upstream of pUB110 and the SCCmec element (Fig. 1B) (25, 26). The nucleotide sequences containing speG and czrC located between the 23S methyltransferase and the second transposase were observed to be 100% identical between strains UCI28 and UCI62. The SCCmec elements of Mu3, UCI28, and UCI62 were observed to be 95.8% identical, with nucleotide differences being found in the J2 region. Isolate UCI62, as well as UCI11and UCI27, harbored czrC downstream of speG and upstream of the arginine catabolic mobile element (ACME) genes and the SCCmec element (Fig. 1C) (25, 26). The czrC gene for all 16 clinical isolates, even those lacking ACME, was located downstream of speG, a spermidine acetyltransferase that functions in the detoxification of spermidine and is often found within the ACME composite island. The location of the czrC gene within the SCCmec region of these isolates is different than the location of czrC in the LA-MRSA ST398 reference strain S0385, in which czrC is located downstream of the mecA gene within the type V SCCmec (Fig. 1D) (27).
FIG 1.
SCCmec region and surrounding mobile genetic elements for Mu3 (A), UCI28 (B), UCI62 (C), and S0385 (D). All regions start at the 23S methyltransferase (blue pentagons). Insertion sequences are depicted as solid black pentagons. The locations of the SCCmec elements, czrC, speG, and pUB110 are indicated, along with other previously annotated genes of interest. The SCCmec element of S0385 (D) is given as a reference for the location of czrC within the type V SCCmec element in LA-MRSA ST398.
DISCUSSION
The recent emergence of MRSA in livestock throughout the world has become a focal point in discussions regarding the role of antibiotic use in food animal production and the development of antibiotic-resistant clinical infections in humans. However, the mechanisms and factors responsible for this emergence, as well as the factors contributing to the geographical variations in genotypes of swine-associated MRSA found globally, are poorly understood. Although some causal role of antibiotic use in the emergence of LA-MRSA is hypothesized and may seem obvious, epidemiological evidence of such relationships has not been readily demonstrated (28). It is clear that other factors, including disinfectants and metals, may play selective roles in the emergence of particular MRSA clones in humans and animals (20, 29, 30).
In this study, zinc resistance mediated by the czrC gene was examined as a potential contributor to the prevalence of LA-MRSA ST5 on swine farms in the United States. A documented association exists between the presence of czrC and the mecA gene in LA-MRSA ST398 isolates obtained from swine farms, with swine-associated isolates having a higher prevalence of czrC than LA-MRSA ST398 isolates obtained from veal calves or humans (19, 21). The strong correlation (99%) reported between isolates harboring the czrC gene and phenotypic zinc resistance in LA-MRSA ST398 indicates that this gene orchestrates the predominant mechanism mediating zinc resistance in this lineage (21). The specific importance of the czrC gene and the physical link between mecA and czrC within the SCCmec element provide a mechanism by which dietary supplementation of zinc in swine rations could contribute to the persistence of methicillin resistance through coselection (19, 20, 22, 23). Evidence of the practical relevance of this mechanism comes from Denmark, where widespread use of zinc in weaned pig diets as an alternative to antibiotic therapy for controlling enteric disease followed the banning of antibiotics for growth promotion in 2000, approximately a decade before LA-MRSA ST398 became highly prevalent in the Danish swine industry (22).
Sequencing studies have demonstrated that the czrC gene is located within the type V and type VIII SCCmec elements of ST398 MRSA (19, 31). The majority of European LA-MRSA ST398 isolates investigated carried the type V SCCmec element containing the czrC gene downstream of the mecA gene (27), but none of the LA-MRSA ST5 isolates we examined carried the SCCmec type V element. All of the MRSA ST5 isolates from humans with no swine contact that carried the czrC gene contained the SCCmec type II element (Table 3), which has not been previously associated with the czrC gene. In those isolates, the czrC gene was located upstream of the SCCmec element and possibly transferred with the speG gene, which confers resistance to spermidine and a potential selective advantage for isolates colonizing and infecting humans (32). Importantly, the fact that none of the swine-associated LA-MRSA ST5 isolates harbored the SCCmec types seen in ST5 MRSA isolates from clinical infections provides further evidence that the animal and human reservoirs of ST5 MRSA appear to be phylogenetically distinct (33).
Previous reports examining czrC in LA-MRSA isolates indicated a higher prevalence of this gene in MRSA clonal complex 398 (CC398) isolates (72.5%), compared to all non-CC398 isolates evaluated (25.5%) (21). An absence of czrC in European and Asian LA-MRSA isolates of the CC5 and CC9 lineages has been reported, which is consistent with our results evaluating LA-MRSA ST5 isolates from the United States. Collectively, our results and previously published data indicate that the czrC gene has a lineage association and is prevalent in the ST398 lineage but is absent or rare among livestock-associated ST5 and ST9 lineages (21). An alternate explanation for the elevated prevalence of the czrC gene in the ST398 lineage is the selection pressure incurred with the use of elevated levels of zinc in feed. However, the prevalence of the czrC gene in non-ST398 LA-MRSA isolates from European swine was reported to be 30% in the tested isolates, while the phenotypic zinc resistance was reported to be 60% in the same isolates (21), arguing against selection pressure incurred with the use of elevated levels of zinc in feed being the sole factor controlling MRSA prevalence in swine, because the majority of these isolates lacked an SCCmec element carrying czrC. Although no national data concerning the use of zinc in swine rations exist, the practice is thought to be widespread in the United States (M. Tokach, personal communication). This appears not to have played the same role in propagating methicillin resistance in livestock isolates of S. aureus in the United States, as the majority of herds tested in recent studies were MRSA negative (14, 34).
Our results reported here, combined with previously reported results (21), open new avenues of research to be explored. First, the czrC gene has been identified in two methicillin-susceptible S. aureus ST398 isolates (18). The presence of this gene without the SCCmec element should be evaluated to determine whether czrC is a remnant from a previously methicillin-resistant isolate or whether the czrC gene has been integrated through a different mechanism. Both LA-MRSA isolates (21) and swine-associated methicillin-susceptible S. aureus isolates (J. Sun, R. S. Singer, S. J. Hau, T. L. Nicholson, and P. R. Davies, unpublished data) that show phenotypic zinc resistance without carrying the czrC gene have been identified. Such isolates should be screened for other mechanisms of zinc resistance, to determine the impact of other genes in conferring a resistant phenotype. Evaluation of the impact of czrC in non-ST398 LA-MRSA, specifically the ability of LA-MRSA ST5 isolates to acquire and to harbor czrC, and the impact of zinc in feed on the capability of LA-MRSA ST398 isolates to outcompete other lineages in swine also bears further investigation. Ultimately, zinc resistance in LA-MRSA is more complex than the presence or absence of czrC or the use of zinc in feed as an antimicrobial agent to combat disease in livestock. Further investigation is needed to determine the mechanisms leading to zinc resistance and to illuminate the impact of selective pressure on the emergence of particular MRSA clones in humans and animals.
Overall, the data reported here indicate that coselection associated with zinc supplementation in feed has not contributed to the persistence or prevalence of LA-MRSA ST5 in the U.S. swine population. This conclusion is contrary to theories surrounding the dissemination of LA-MRSA ST398 in Europe and, considering the presence of czrC in LA-MRSA ST398 isolates in the United States, indicates a potential link between the genetic lineage and the carriage of specific resistance markers, such as that seen for qacA resistance in CC22 in the hospital setting (30). Furthermore, the data reported here indicate that multiple mechanisms contribute to fitness and the ability of LA-MRSA ST5 and other lineages to compete and to persist in the nasal microbiota of pigs.
MATERIALS AND METHODS
Isolate acquisition.
Swine-associated LA-MRSA ST5 cultures were isolated from swine (n = 38), the environment within swine facilities (n = 26), and persons with short-term (n = 9) and long-term (n = 9) swine contact. These isolates were provided by Iowa State University and the University of Minnesota (14). Clinical isolates from humans with no swine contact were obtained from the University of California, Irvine (n = 64) (35), and the University of California, San Francisco (n = 9), hospitals servicing urban populations in Orange County (southern California) and the San Francisco area (northern California), respectively. Swine-associated LA-MRSA ST398 cultures obtained from Iowa State University were isolated from swine (n = 8) or the environment within swine facilities (n = 6) (14). Isolates were subjected to MLST and spa typing prior to acquisition (14, 35). Isolate sources and spa types are provided in Table S1 in the supplemental material.
Zinc susceptibility testing.
Zinc chloride MICs were determined by agar dilution, as described by Aarestrup and Hasman (36). Briefly, plates of Mueller-Hinton agar with an adjusted pH of 5.5 were supplemented with zinc chloride in 2-fold dilutions, with concentrations ranging from 0.25 to 16 mM. The isolate Salmonella enterica subsp. enterica serovar Typhimurium ATCC 14028 was used as a positive control, and S. aureus ATCC 29213 and ATCC 43300 were used as negative controls. A MIC value of >2 mM was used as the cutoff value to designate resistance, in accordance with previously published reports and the result for the positive control in this study (19, 21).
czrC PCR testing.
The presence of the czrC gene was determined by PCR using previously reported primers and protocols (19). Briefly, PCR was carried out in an MJ Research PCT-200 DNA Engine thermocycler (GMI, Ramsey, MN) using 50 ng of purified genomic DNA from the appropriate strains, the forward primer 5′-TAGCCACGATCATAGTCATG-3′, and the reverse primer 5′-ATCCTTGTTTTCCTTAGTGACTT-3′. Reaction mixtures included 0.4 μM primers, 1 U of AmpliTaq polymerase (Applied Biosystems, Foster City, CA), 2.5 μl of 10× buffer II (100 mM Tris-HCl [pH 8.3], 500 mM KCl), 2.5 mM MgCl2, and 200 μM deoxynucleoside triphosphates (dNTPs), in a final volume of 50 μl. Cycling conditions were 95°C for 2 min, 30 cycles of 95°C for 15 s, 52°C for 30 s, and 72°C for 1.5 min, and a final extension step of 72°C for 7 min. PCR products were run on a 1% agarose gel, stained with ethidium bromide, and visualized using UV light.
SCCmec typing.
SCCmec typing was completed using previously designed primer sets (Table 4) (37–42). Briefly, PCR was carried out in a MJ Research PCT-200 DNA Engine thermocycler (GMI) using 50 ng of purified genomic DNA from the appropriate strains; reaction mixtures included 0.4 μM primers, 1 U of AmpliTaq polymerase (Applied Biosystems), 2.5 μl of 10× buffer II (100 mM Tris-HCl [pH 8.3], 500 mM KCl), 2.5 mM MgCl2, and 200 μM dNTPs, in a final volume of 50 μl. PCR for the ccrA and ccrB genes was a multiplex reaction with cycling conditions of 95°C for 2 min, 10 cycles of 95°C for 15 s, 65°C for 30 s, and 72°C for 1.5 min, 25 cycles of 95°C for 15 s, 55°C for 30 s, and 72°C for 1.5 min, and a final extension step of 72°C for 7 min. PCR of the ccrC gene used cycling conditions of 95°C for 2 min, 30 cycles of 95°C for 15 s, 58°C for 30 s, and 72°C for 2 min, and a final extension step of 72°C for 7 min. PCR of the mec element genes was completed with cycling conditions of 95°C for 2 min, 30 cycles of 95°C for 15 s, 50°C for 30 s, and 72°C for 2 min, and a final extension step of 72°C for 7 min. PCR products were run on a 1% agarose gel, stained with ethidium bromide, and visualized using UV light.
TABLE 4.
Primer sets used for SCCmec typing of isolates
| Primer name | Nucleotide sequence | Expected product (bp) (forward primer used) | Source |
|---|---|---|---|
| ccrB-F | ATTGCCTTGATAATAGCCITCT | Ito et al. (37) | |
| ccrA1-R | AACCTATATCATCAATCAGTACGT | 694 (ccrB-F) | Ito et al. (37) |
| ccrA2-R | TAAAGGCATCAATGCACAAACACT | 937 (ccrB-F) | Ito et al. (37) |
| ccrA3-R | AGCTCAAAAGCAAGCAATAGAAT | 1,791 (ccrB-F) | Ito et al. (37) |
| ccrA4-R | GTATCAATGCACCAGAACTT | 1,287 (ccrB-F) | Kondo et al. (38) |
| ccrC-F | CGTCTATTACAAGATGTTAAGGATAAT | Kondo et al. (38) | |
| ccrC-R | CCTTTATAGACTGGATTATTCAAAATAT | 518 (ccrC-F) | Kondo et al. (38) |
| mecI-F | CAAGTGAATTGAAACCGCCT | Okuma et al. (39) | |
| mecI-R | CAAAAGGACTGGACTGGAGTCCAAA | 187 (mecI-F) | Okuma et al. (39) |
| mecR1-R | GTCTCCACGTTAATTCCATT | 1,920 (mecI-F) | Kobayashi et al. (40) |
| Class B-F | TATACCAAACCCGACAAC | Katayama et al. (41) | |
| IS1272-R | AACGCCACTCATAACATATGGAA | 1,996 (class B-F) | Okuma et al. (39) |
| Class C-F | AACGTTGTAACCACCCCAAGA | Hiramatsu et al. (42) | |
| IS431-R | TGAGGTTATTCAGATATTTCGATGT | 2,072 (class C-F) | Katayama et al. (41) |
Whole-genome sequencing and analysis.
Draft genome sequence data for 14 czrC-carrying isolates (UCI3, UCI9, UCI11, UCI19, UCI21, UCI24, UCI27, UCI43, UCI45, UCI46, UCI48, UCI52, UCI56, and UCI64) were generated using the Illumina MiSeq platform (Illumina, San Diego, CA) (25). Indexed libraries were generated and run on the MiSeq platform using the 500-cycle MiSeq v2 reagent kit. The data were assembled using MIRA 4.0.2 (http://mira-assembler.sourceforge.net/docs/DefinitiveGuideToMIRA.html) (43). Closed genomes were obtained for UCI28 and UCI62 as described previously (26). Briefly, genomic DNA was sequenced with a PacBio RSII instrument using a 10-kb insert library and one SMRT cell for each isolate. The data were assembled using PacBio SMRT Analysis 2.3.0 and CANU 1.3 software. The genomes were then polished and error corrected using Illumina MiSeq data and Broad Institute Pilon 1.18 software. Whole-genome sequence data were analyzed using Geneious 9.0.5 (Biomatters Ltd., Auckland, New Zealand). The SCCmec region was extracted from the closed genome sequences of Mu3 (GenBank accession number AP009324.1), UCI28 (GenBank accession number CP018768), UCI62 (GenBank accession number CP018766), and S0385 (GenBank accession number AM990992.1); these were compared visually in Geneious, using multiple sequence alignments to determine similarity. For the 14 draft genomes, the contig harboring the czrC gene was extracted and used for comparison. These regions were analyzed for similarity to UCI28 and UCI62 using multiple sequence alignments.
Statistical analysis.
Comparisons between isolates from humans with no swine contact and swine-associated isolates were completed using Fisher's exact test using GraphPad Prism (GraphPad Software, La Jolla, CA).
Accession number(s).
The whole-genome sequences for isolates UCI28 and UCI62 were deposited in DDBJ/ENA/GenBank with the following accession numbers: UCI28, CP018768 and CP018769; UCI62, CP018766 and CP018767 (26). The draft genome sequences obtained for 14 S. aureus ST5 isolates were deposited in DDBJ/ENA/GenBank with the following accession numbers: UCI3, LKYU00000000; UCI9, LKZA00000000; UCI11, LKZC00000000; UCI19, LKZK00000000; UCI21, LKZM00000000; UCI24, LKZP00000000; UCI27, LKZS00000000; UCI43, LLAI00000000; UCI45, LLAK00000000; UCI46, LLAL00000000; UCI48, LLAN00000000; UCI52, LLAR00000000; UCI56, LLAV00000000; UCI64, LLBD00000000 (25).
Supplementary Material
ACKNOWLEDGMENTS
We thank Danielle Fligg and Joann Kinyon of the bacteriology section of the Veterinary Diagnostic Laboratory at Iowa State University for their assistance in performing the zinc chloride susceptibility testing.
Funding was provided in part by the Iowa Pork Producers Association.
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00756-17.
REFERENCES
- 1.Monecke S, Coombs G, Shore AC, Coleman DC, Akpaka P, Borg M, Chow H, Ip M, Jatzwauk L, Jonas D, Kadlec K, Kearns A, Laurent F, O'Brien FG, Pearson J, Ruppelt A, Schwarz S, Scicluna E, Slickers P, Tan HL, Weber S, Ehricht R. 2011. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS One 6:e17936. doi: 10.1371/journal.pone.0017936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Hal SJ, Jensen SO, Vaska VL, Espedido BA, Paterson DL, Gosbell IB. 2012. Predictors of mortality in Staphylococcus aureus bacteremia. Clin Microbiol Rev 25:362–386. doi: 10.1128/CMR.05022-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeLeo FR, Chambers HF. 2009. Reemergence of antibiotic-resistant Staphylococcus aureus in the genomics era. J Clin Invest 119:2464–2474. doi: 10.1172/JCI38226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jevons M. 1961. “Celbenin”-resistant staphylococci. Br Med J 1:124–125. doi: 10.1136/bmj.1.5219.124-a. [DOI] [Google Scholar]
- 5.Diep BA, Otto M. 2008. The role of virulence determinants in community-associated MRSA pathogenesis. Trends Microbiol 16:361–369. doi: 10.1016/j.tim.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mediavilla JR, Chen L, Mathema B, Kreiswirth BN. 2012. Global epidemiology of community-associated methicillin resistant Staphylococcus aureus (CA-MRSA). Curr Opin Microbiol 15:588–595. doi: 10.1016/j.mib.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 7.Devriese LA, Van Damme LR, Fameree L. 1972. Methicillin (cloxacillin)-resistant Staphylococcus aureus strains isolated from bovine mastitis cases. Zentralbl Veterinarmed B 19:598–605. doi: 10.1111/j.1439-0450.1972.tb00439.x. [DOI] [PubMed] [Google Scholar]
- 8.Voss A, Loeffen F, Bakker J, Klaassen C, Wulf M. 2005. Methicillin-resistant Staphylococcus aureus in pig farming. Emerg Infect Dis 11:1965–1966. doi: 10.3201/eid1112.050428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uhlemann AC, Porcella SF, Trivedi S, Sullivan SB, Hafer C, Kennedy AD, Barbian KD, McCarthy AJ, Street C, Hirschberg DL, Lipkin WI, Lindsay JA, DeLeo FR, Lowy FD. 2012. Identification of a highly transmissible animal-independent Staphylococcus aureus ST398 clone with distinct genomic and cell adhesion properties. mBio 3:e00027-12. doi: 10.1128/mBio.00027-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Neeling AJ, van den Broek MJ, Spalburg EC, van Santen-Verheuvel MG, Dam-Deisz WD, Boshuizen HC, van de Giessen AW, van Duijkeren E, Huijsdens XW. 2007. High prevalence of methicillin resistant Staphylococcus aureus in pigs. Vet Microbiol 122:366–372. doi: 10.1016/j.vetmic.2007.01.027. [DOI] [PubMed] [Google Scholar]
- 11.van Duijkeren E, Hengeveld P, Zomer TP, Landman F, Bosch T, Haenen A, van de Giessen A. 2016. Transmission of MRSA between humans and animals on duck and turkey farms. J Antimicrob Chemother 71:58–62. doi: 10.1093/jac/dkv313. [DOI] [PubMed] [Google Scholar]
- 12.European Food Safety Authority. 2009. Scientific opinion of the Panel on Biological Hazards on a request from the European Commission on assessment of the public health significance of meticillin resistant Staphylococcus aureus (MRSA) in animals and foods. EFSA J 993:1–73. [Google Scholar]
- 13.Wagenaar JA, Yue H, Pritchard J, Broekhuizen-Stins M, Huijsdens X, Mevius DJ, Bosch T, Van Duijkeren E. 2009. Unexpected sequence types in livestock associated methicillin-resistant Staphylococcus aureus (MRSA): MRSA ST9 and a single locus variant of ST9 in pig farming in China. Vet Microbiol 139:405–409. doi: 10.1016/j.vetmic.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 14.Frana TS, Beahm AR, Hanson BM, Kinyon JM, Layman LL, Karriker LA, Ramirez A, Smith TC. 2013. Isolation and characterization of methicillin-resistant Staphylococcus aureus from pork farms and visiting veterinary students. PLoS One 8:e53738. doi: 10.1371/journal.pone.0053738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khanna T, Friendship R, Dewey C, Weese JS. 2008. Methicillin resistant Staphylococcus aureus colonization in pigs and pig farmers. Vet Microbiol 128:298–303. doi: 10.1016/j.vetmic.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 16.Smith TC, Pearson N. 2011. The emergence of Staphylococcus aureus ST398. Vector Borne Zoonotic Dis 11:327–339. doi: 10.1089/vbz.2010.0072. [DOI] [PubMed] [Google Scholar]
- 17.Molla B, Byrne M, Abley M, Mathews J, Jackson CR, Fedorka-Cray P, Sreevatsan S, Wang P, Gebreyes WA. 2012. Epidemiology and genotypic characteristics of methicillin-resistant Staphylococcus aureus strains of porcine origin. J Clin Microbiol 50:3687–3693. doi: 10.1128/JCM.01971-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Price LB, Stegger M, Hasman H, Aziz M, Larsen J, Andersen PS, Pearson T, Waters AE, Foster JT, Schupp J, Gillece J, Driebe E, Liu CM, Springer B, Zdovc I, Battisti A, Franco A, Zmudzki J, Schwarz S, Butaye P, Jouy E, Pomba C, Porrero MC, Ruimy R, Smith TC, Robinson DA, Weese JS, Arriola CS, Yu F, Laurent F, Keim P, Skov R, Aarestrup FM. 2012. Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. mBio 3:e00305-11. doi: 10.1128/mBio.00305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cavaco LM, Hasman H, Stegger M, Andersen PS, Skov R, Fluit AC, Ito T, Aarestrup FM. 2010. Cloning and occurrence of czrC, a gene conferring cadmium and zinc resistance in methicillin-resistant Staphylococcus aureus CC398 isolates. Antimicrob Agents Chemother 54:3605–3608. doi: 10.1128/AAC.00058-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Slifierz MJ, Friendship RM, Weese JS. 2015. Methicillin-resistant Staphylococcus aureus in commercial swine herds is associated with disinfectant and zinc usage. Appl Environ Microbiol 81:2690–2695. doi: 10.1128/AEM.00036-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cavaco LM, Hasman H, Aarestrup FM. 2011. Zinc resistance of Staphylococcus aureus of animal origin is strongly associated with methicillin resistance. Vet Microbiol 150:344–348. doi: 10.1016/j.vetmic.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 22.Aarestrup FM, Cavaco L, Hasman H. 2010. Decreased susceptibility to zinc chloride is associated with methicillin resistant Staphylococcus aureus CC398 in Danish swine. Vet Microbiol 142:455–457. doi: 10.1016/j.vetmic.2009.10.021. [DOI] [PubMed] [Google Scholar]
- 23.Slifierz MJ, Friendship R, Weese JS. 2015. Zinc oxide therapy increases prevalence and persistence of methicillin-resistant Staphylococcus aureus in pigs: a randomized controlled trial. Zoonoses Public Health 62:301–308. doi: 10.1111/zph.12150. [DOI] [PubMed] [Google Scholar]
- 24.Fairbrother JM, Nadeau E, Gyles CL. 2005. Escherichia coli in postweaning diarrhea in pigs: an update on bacterial types, pathogenesis, and prevention strategies. Anim Health Res Rev 6:17–39. doi: 10.1079/AHR2005105. [DOI] [PubMed] [Google Scholar]
- 25.Hau SJ, Bayles DO, Alt DP, Nicholson TL. 2017. Draft genome sequences of 14 Staphylococcus aureus sequence type 5 isolates from California, USA. Genome Announc 5:e00098-17. doi: 10.1128/genomeA.00098-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hau SJ, Bayles DO, Alt DP, Nicholson TL. 2017. Complete genome sequences of two Staphylococcus aureus sequence type 5 isolates from California, USA. Genome Announce 5:e00099-17. doi: 10.1128/genomeA.00099-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schijffelen MJ, Boel CH, van Strijp JA, Fluit AC. 2010. Whole genome analysis of a livestock-associated methicillin-resistant Staphylococcus aureus ST398 isolate from a case of human endocarditis. BMC Genomics 11:376. doi: 10.1186/1471-2164-11-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lassok B, Tenhagen BA. 2013. From pig to pork: methicillin-resistant Staphylococcus aureus in the pork production chain. J Food Prot 76:1095–1108. doi: 10.4315/0362-028X.JFP-12-341. [DOI] [PubMed] [Google Scholar]
- 29.Stegger M, Wirth T, Andersen PS, Skov RL, De Grassi A, Simoes PM, Tristan A, Petersen A, Aziz M, Kiil K, Cirkovic I, Udo EE, del Campo R, Vuopio-Varkila J, Ahmad N, Tokajian S, Peters G, Schaumburg F, Olsson-Liljequist B, Givskov M, Driebe EE, Vigh HE, Shittu A, Ramdani-Bougessa N, Rasigade JP, Price LB, Vandenesch F, Larsen AR, Laurent F. 2014. Origin and evolution of European community-acquired methicillin-resistant Staphylococcus aureus. mBio 5:e01044-14. doi: 10.1128/mBio.01044-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otter JA, Patel A, Cliff PR, Halligan EP, Tosas O, Edgeworth JD. 2013. Selection for qacA carriage in CC22, but not CC30, methicillin-resistant Staphylococcus aureus bloodstream infection isolates during a successful institutional infection control programme. J Antimicrob Chemother 68:992–999. doi: 10.1093/jac/dks500. [DOI] [PubMed] [Google Scholar]
- 31.Zhang K, McClure JA, Elsayed S, Conly JM. 2009. Novel staphylococcal cassette chromosome mec type, tentatively designated type VIII, harboring class A mec and type 4 ccr gene complexes in a Canadian epidemic strain of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 53:531–540. doi: 10.1128/AAC.01118-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Joshi GS, Spontak JS, Klapper DG, Richardson AR. 2011. Arginine catabolic mobile element encoded speG abrogates the unique hypersensitivity of Staphylococcus aureus to exogenous polyamines. Mol Microbiol 82:9–20. doi: 10.1111/j.1365-2958.2011.07809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hau SJ, Sun J, Davies PR, Frana TS, Nicholson TL. 2015. Comparative prevalence of immune evasion complex genes associated with β-hemolysin converting bacteriophages in MRSA ST5 isolates from swine, swine facilities, humans with swine contact, and humans with no swine contact. PLoS One 10:e0142832. doi: 10.1371/journal.pone.0142832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun J, Yang M, Sreevatsan S, Davies PR. 2015. Prevalence and characterization of Staphylococcus aureus in growing pigs in the USA. PLoS One 10:e0143670. doi: 10.1371/journal.pone.0143670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hudson LO, Murphy CR, Spratt BG, Enright MC, Elkins K, Nguyen C, Terpstra L, Gombosev A, Kim D, Hannah P, Mikhail L, Alexander R, Moore DF, Huang SS. 2013. Diversity of methicillin-resistant Staphylococcus aureus (MRSA) strains isolated from inpatients of 30 hospitals in Orange County, California. PLoS One 8:e62117. doi: 10.1371/journal.pone.0062117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aarestrup FM, Hasman H. 2004. Susceptibility of different bacterial species isolated from food animals to copper sulphate, zinc chloride and antimicrobial substances used for disinfection. Vet Microbiol 100:83–89. doi: 10.1016/j.vetmic.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 37.Ito T, Katayama Y, Asada K, Mori N, Tsutsumimoto K, Tiensasitorn C, Hiramatsu K. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 45:1323–1336. doi: 10.1128/AAC.45.5.1323-1336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kondo Y, Ito T, Ma XX, Watanabe S, Kreiswirth BN, Etienne J, Hiramatsu K. 2007. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother 51:264–274. doi: 10.1128/AAC.00165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okuma K, Iwakawa K, Turnidge JD, Grubb WB, Bell JM, O'Brien FG, Coombs GW, Pearman JW, Tenover FC, Kapi M, Tiensasitorn C, Ito T, Hiramatsu K. 2002. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J Clin Microbiol 40:4289–4294. doi: 10.1128/JCM.40.11.4289-4294.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kobayashi N, Taniguchi K, Kojima K, Urasawa S, Uehara N, Omizu Y, Kishi Y, Yagihashi A, Kurokawa I, Watanabe N. 1996. Genomic diversity of mec regulator genes in methicillin-resistant Staphylococcus aureus and Staphylococcus epidermidis. Epidemiol Infect 117:289–295. doi: 10.1017/S0950268800001461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katayama Y, Ito T, Hiramatsu K. 2001. Genetic organization of the chromosome region surrounding mecA in clinical staphylococcal strains: role of IS431-mediated mecI deletion in expression of resistance in mecA-carrying, low-level methicillin-resistant Staphylococcus haemolyticus. Antimicrob Agents Chemother 45:1955–1963. doi: 10.1128/AAC.45.7.1955-1963.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hiramatsu K, Asada K, Suzuki E, Okonogi K, Yokota T. 1992. Molecular cloning and nucleotide sequence determination of the regulator region of mecA gene in methicillin-resistant Staphylococcus aureus (MRSA). FEBS Lett 298:133–136. doi: 10.1016/0014-5793(92)80039-J. [DOI] [PubMed] [Google Scholar]
- 43.Chevreux B, Wetter T, Suhai S. 1999. Genome sequence assembly using trace signals and additional sequence information, p 45–46. In Computer science biology. Proceedings of the German Conference on Bioinformatics, GCB ’99. GCB, Hannover, Germany. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.