ABSTRACT
Hydrogenotrophic methanogens typically require strictly anaerobic culturing conditions in glass tubes with overpressures of H2 and CO2 that are both time-consuming and costly. To increase the throughput for screening chemical compound libraries, 96-well microtiter plate methods for the growth of a marine (environmental) methanogen Methanococcus maripaludis strain S2 and the rumen methanogen Methanobrevibacter species AbM4 were developed. A number of key parameters (inoculum size, reducing agents for medium preparation, assay duration, inhibitor solvents, and culture volume) were optimized to achieve robust and reproducible growth in a high-throughput microtiter plate format. The method was validated using published methanogen inhibitors and statistically assessed for sensitivity and reproducibility. The Sigma-Aldrich LOPAC library containing 1,280 pharmacologically active compounds and an in-house natural product library (120 compounds) were screened against M. maripaludis as a proof of utility. This screen identified a number of bioactive compounds, and MIC values were confirmed for some of them against M. maripaludis and M. AbM4. The developed method provides a significant increase in throughput for screening compound libraries and can now be used to screen larger compound libraries to discover novel methanogen-specific inhibitors for the mitigation of ruminant methane emissions.
IMPORTANCE Methane emissions from ruminants are a significant contributor to global greenhouse gas emissions, and new technologies are required to control emissions in the agriculture technology (agritech) sector. The discovery of small-molecule inhibitors of methanogens using high-throughput phenotypic (growth) screening against compound libraries (synthetic and natural products) is an attractive avenue. However, phenotypic inhibitor screening is currently hindered by our inability to grow methanogens in a high-throughput format. We have developed, optimized, and validated a high-throughput 96-well microtiter plate assay for growing environmental and rumen methanogens. Using this platform, we identified several new inhibitors of methanogen growth, demonstrating the utility of this approach to fast track the development of methanogen-specific inhibitors for controlling ruminant methane emissions.
KEYWORDS: methanogen, greenhouse gas, Methanococcus maripaludis, high-throughput, rumen, Methanobrevibacter
INTRODUCTION
Methane emissions from ruminants are a significant contributor to global greenhouse gas emissions (1). In countries such as New Zealand, with a large pasture-based livestock sector, greenhouse gas emissions from agriculture represent approximately half of its total emissions (2). Methane is produced in the rumen principally by methanogens, a group of archaeal microorganisms. The methanogens that dominate the rumen belong to the Methanobacteriales and include the Methanobrevibacter ruminantium, Methanobrevibacter gottschalkii, and Methanosphaera clades (3, 4). Methanomassiliicoccaceae-affiliated species are also found (4). A number of technologies have been suggested for mitigating methane emissions (5, 6), including low-methane-emitting animals (7) and the use of special forages (8), phage or their lytic enzymes (5, 9), direct-fed microbials (10), vaccines (11), and inhibitors (12–15). Although some of these strategies have shown promise, not all directly target methanogens. Halogenated compounds (e.g., chloroform and bromochloromethane) are highly potent inhibitors of methanogenesis in ruminants (6, 16–21). However, these compounds are not considered appropriate for use in current animal husbandry due to environmental, human health, and animal welfare concerns. In addition, halogenated hydrocarbons (e.g., bromochloromethane) have potent ozone-depleting properties (22). Notwithstanding, there is still a significant potential for the discovery of narrow-spectrum methanogen-selective inhibitors that are more potent, more specific, and less toxic and that target methanogens and methane formation without negatively affecting animal productivity, consumers, or the environment (6, 23).
An early-stage drug development strategy that has undergone a recent resurgence in the discovery and development of small-molecule inhibitors of pathogenic microorganisms is phenotypic screening (24, 25). In phenotypic screening, a high-throughput platform using microtiter plates for the growth of the target bacterium is used to screen the toxicity of a compound library. Screening is an important prerequisite of this technology. Therefore, it has not been applied widely to microorganisms that have fastidious growth requirements, such as anaerobic bacteria (for an exception, see reference 26). Because hydrogenotrophic methanogens typically require H2 and CO2 overpressures in addition to strictly anaerobic conditions for growth, they represent additional challenges. Methanococcus maripaludis strain S2 is a well-characterized genetically tractable methanogen that can be grown in the absence of H2 and CO2 using formate (27–32). M. maripaludis grows quickly to high cell densities in contrast to slowly growing rumen isolates, such as Methanobrevibacter ruminantium M1, where cell densities are low (33, 34). Both methanogens are typically grown using anaerobic culturing techniques in 5-ml or greater culture volumes using appropriately sealed and pressurized glass tubes, which is incompatible with modern high-throughput screening techniques for drug discovery and phenotypic analysis (35). A microtiter plate method for performing antimicrobial peptide susceptibility testing has been reported for three different nonrumen methanogens (36). These methanogens were cultured with either methanol or H2-CO2 (36). To specifically perform high-throughput screening of large compound libraries, we sought to develop a microtiter plate method that did not require H2 overpressures for culturing rumen methanogens. This methodology was applied to the development of methanogen-specific inhibitors for controlling ruminant methane emissions.
Here, we report the culturing of marine and rumen methanogens in 96-well microtiter plates with methods that were optimized for robust growth, ease of use, and reproducibility. The methods were validated using published inhibitors, were statistically assessed for sensitivity and reproducibility, and were used to screen compound libraries as a proof-of-principle for their utility.
RESULTS AND DISCUSSION
Growth of methanogens in a microtiter plate format.
A previously published basal growth medium with formate (McF), with sodium sulfide as the reducing agent, was chosen for the growth of the fast-growing (2-h doubling time) marine methanogen M. maripaludis, eliminating the requirement for overpressurization with H2 (30). Using this McF medium, rapid progress was achieved in adapting the growth of M. maripaludis in 96-well microtiter plates (320-μl final volume). Cultures in the microtiter plates were inoculated with a 4% starter culture grown in Balch tubes (optical density at 600 nm [OD600] of 0.9). In initial experiments, plates were incubated at 37°C under two different anaerobic culturing conditions, namely, in an AGS AnaeroGen compact bag (Oxoid) that was sealed and kept either inside or outside the anaerobic chamber. An oxygen indicator (resazurin) in the medium enabled the detection of oxygen. For the cultures that were incubated outside the anaerobic chamber, anaerobic conditions lasted for approximately 40 h. Therefore, this method was not suitable for the experiments for optimizing growth conditions. Microtiter plates incubated in the anaerobic chamber reached a final optical density of 0.564 ± 0.083 after 5 days of growth (Table 1). Based on these results, further experiments were performed in the anaerobic chamber with a gas atmosphere of 5% H2, 5% CO2, and 90% N2 to maintain strictly anaerobic conditions in the microtiter plate format.
TABLE 1.
Growth of Methanococcus maripaludis strain S2 in 96-well microtiter plate format
| Replicate | Final OD600a |
||||
|---|---|---|---|---|---|
| Medium only | Inoculum only (4%) | DMSO (2%) | Monensin (1 μM) | BES (30 μM) | |
| R1 | 0 | 0.702 | 0.553 | 0.015 | 0.039 |
| R2 | −0.003 | 0.661 | 0.590 | 0.025 | 0.054 |
| R3 | −0.002 | 0.555 | 0.550 | 0.025 | 0.058 |
| R4 | −0.003 | 0.589 | 0.415 | 0.018 | 0.053 |
| R5 | −0.003 | 0.455 | 0.417 | 0.034 | 0.045 |
| R6 | −0.004 | 0.521 | 0.442 | 0.025 | 0.050 |
| R7 | −0.002 | 0.503 | 0.417 | 0.015 | 0.062 |
| R8 | −0.002 | 0.528 | 0.602 | 0.012 | 0.044 |
| Avg | −0.002 | 0.564 | 0.498 | 0.021 | 0.051 |
| SD | 0.001 | 0.083 | 0.083 | 0.007 | 0.008 |
Recorded after 5 days of incubation at 37°C using a 4% inoculum (approximate OD600 of 0.020) and McF medium containing 0.2% sodium sulfide (wt/vol) and 400 mM sodium formate. The Z′ for the assay was 0.64.
A range of inoculum sizes (1 to 10%) was tested, and the smallest size that gave consistently rapid and reproducible growth after 5 days (late exponential phase) was 4% (Table 1). Using this inoculum size, the concentration of sodium sulfide was optimized. Because volatile H2S was readily formed under these growth conditions, it was of special concern. Concentrations of 0.05%, 0.1%, 0.2%, and 0.3% (wt/vol) sodium sulfide were tested. On the basis of the growth rate and final optical density, the best concentration was 0.2% sodium sulfide (wt/vol), which is 4-fold higher than that used in sealed tubes (data not shown).
These adaptations of the standard growth conditions yielded reproducible growth of M. maripaludis in the 96-well microtiter plate format (Table 1). For instance, the average with standard deviation OD600 was 0.564 ± 0.083. The average with standard deviation OD600 for five biological replicates was 0.466 ± 0.094 (data not shown). Similarly, in the presence of the inhibitors monensin and 2-bromoethanesulfonic acid (BES), the values were 0.026 ± 0.017 and 0.036 ± 0.013, respectively (Table 1). Compound libraries are typically supplied with either dimethyl sulfoxide (DMSO) or some other organic solvent (e.g., ethanol) as the diluent. We tested the effects of 1% and 2% DMSO and 1% ethanol (Table 1 and data not shown). Neither solvent at these concentrations had a significant effect on the final optical density.
Methanobrevibacter sp. strain AbM4 is a slowly growing rumen isolate that grows without H2 in the presence of 20 mM methanol and 20 mM ethanol, the potential of which was indicated by Leahy et al. (37). The inoculum size and cysteine concentration were optimized for the growth of AbM4 in the 96-well microtiter plate format (320-μl final volume) using rumen fluid-based (RM02) medium. Inocula of 2.5%, 5%, and 10% (vol/vol) were evaluated after 4 days in the anaerobic chamber at 38°C (early-stationary-phase cultures). The mean absorbance values were 0.180, 0.396, and 0.421, respectively (Table 2). Based on the final OD600 reached and the number of population doublings achieved (>3), we chose a 5% inoculum for all further experiments. The reducing agents sodium sulfide (Na2S) at concentrations of 0.05%, 0.1%, and 0.2% (wt/vol) and cysteine at concentrations of 0.05%, 0.1%, and 0.5% (wt/vol) were checked for their ability to improve growth. The average absorbance values at 600 nm were 0.261, 0.406, and 0.368, respectively, for Na2S and 0.585, 0.503, and 0.394, respectively, for cysteine (data not shown). Thus, the highest growth was obtained with 0.05% cysteine, the standard concentration used for growth in Balch tubes. Thus, this concentration was used for all further experiments in 96-well microtiter plates. The effect of DMSO at concentrations of 0.5%, 1%, and 2% (vol/vol) was tested. Concentrations greater than 1% were inhibitory (Table 2 shows 2% DMSO). Lastly, the average with standard deviation of the growth yield (OD600) from five biological replicates in the 96-well format was 0.331 ± 0.056.
TABLE 2.
Growth of Methanobrevibacter species AbM4 in 96-well microtiter plate format
| Replicate | Final OD600a |
||||||
|---|---|---|---|---|---|---|---|
| Medium only | Inoculum only (2.5%) | Inoculum only (5%) | Inoculum only (10%) | DMSO (2%) | Monensin (1 μM) | BES (30 μM) | |
| R1 | 0 | 0.179 | 0.376 | 0.451 | 0.254 | 0.039 | 0.062 |
| R2 | −0.003 | 0.161 | 0.370 | 0.442 | 0.278 | 0.045 | 0.049 |
| R3 | −0.002 | 0.174 | 0.379 | 0.438 | 0.282 | 0.040 | 0.061 |
| R4 | −0.003 | 0.275 | 0.397 | 0.348 | 0.285 | 0.038 | 0.053 |
| R5 | −0.003 | 0.157 | 0.421 | 0.479 | 0.292 | 0.038 | 0.062 |
| R6 | −0.004 | 0.149 | 0.430 | 0.408 | 0.280 | 0.041 | 0.069 |
| R7 | −0.002 | 0.157 | 0.365 | 0.411 | 0.257 | 0.016 | 0.053 |
| R8 | −0.002 | 0.190 | 0.428 | 0.398 | 0.254 | 0.026 | 0.049 |
| Avg | −0.002 | 0.180 | 0.396 | 0.422 | 0.273 | 0.035 | 0.057 |
| SD | 0.001 | 0.041 | 0.027 | 0.040 | 0.015 | 0.010 | 0.007 |
Recorded after 4 days of incubation at 38°C using a 5% inoculum (approximate starting OD600 of 0.020) in medium containing reductant (0.05% cysteine [wt/vol]). The Z′ was 0.82.
Inhibition of methanogens in a microtiter plate format.
To further demonstrate that controlled inhibition of growth of M. maripaludis was achievable in our microtiter plate format, we tested the effects of two previously identified methanogen inhibitors on the growth of M. maripaludis (Table 1) and Methanobrevibacter sp. strain AbM4 (Table 2). Monensin, a sodium ionophore, and 2-bromoethanesulfonic acid (BES), an analogue of methyl-coenzyme M, are potent inhibitors of methanogens (34). Using monensin (1 μM) or BES (30 μM), nearly complete inhibition, i.e., >85% reduction of growth, was observed for both strains (Tables 1 and 2). The suitability of the assay for high-throughput screening was determined using the statistical parameter termed the Z-factor (38). The Z′ values for our microtiter plate screens were 0.64 for M. maripaludis and 0.82 for Methanobrevibacter sp. strain AbM4, indicating a high-quality assay exhibiting a wide separation between signal and background and low data variability.
Screening of compound libraries for new inhibitors of methanogens.
Using the microtiter plate format described above for M. maripaludis, the LOPAC 1280 library (Sigma-Aldrich, St. Louis, USA), comprising 1,280 biologically active compounds, was screened for inhibitors of M. maripaludis growth. The library was prepared as 1.0 mM stocks in dimethyl sulfoxide (DMSO) and assessed for inhibition of M. maripaludis growth at a final compound concentration of 20 μM. Each tested microplate contained control wells for DMSO (1% [vol/vol]) and positive inhibitor control wells (monensin and BES, 1 μM and 30 μM, respectively). The LOPAC screen was characterized by an average Z-factor of 0.67. Forty-one compounds were identified that caused ≥90% inhibition of growth after 5 days of incubation (see Table S1 in the supplemental material).
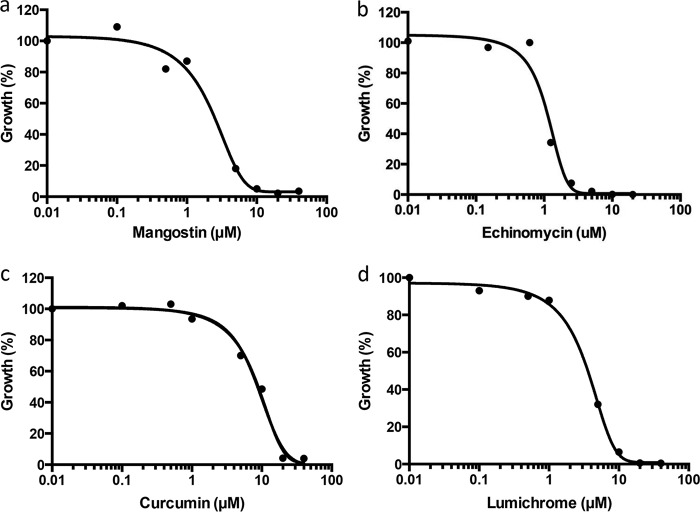
A second screen using M. maripaludis was performed with an in-house collection of 120 antibiotics and other natural products. Of the 120 compounds screened at final concentrations of 20 μM, 17 inhibited the growth yield of M. maripaludis by ≥90% (see Table S2). The screen was characterized by an average Z-factor of 0.78. These compounds were then screened at a range of concentrations from 0.02 to 20 μM to determine potency. Excluding previously reported inhibitors of methanogens (i.e., nigericin, valinomycin, and monensin) (Fig. 1) (39, 40), the most potent compounds identified in our natural product screen were mangostin (50% inhibitory concentration [IC50], 2.5 μM), lumichrome (IC50, 2.6 μM), echinomycin (IC50, 1.2 μM), and curcumin (IC50, 6.5 μM) (Fig. 2). A comparison between M. maripaludis and strain AbM4 revealed several potent inhibitors in common from screening natural product libraries, including nigericin, valinomycin, and echinomycin (inhibited growth at 2 μM). Others hits included daunorubicin hydrochloride, aristolochic acid, ellipticine, and actinomycin D, which showed some growth inhibition of AbM4 at 20 μM (data not shown). These data suggest that natural products have similar molecular targets in both methanogens, but the sensitivities differ between the two strains tested. The identity of the mode of action of these new inhibitors against methanogen growth is required to validate this proposal.
FIG 1.
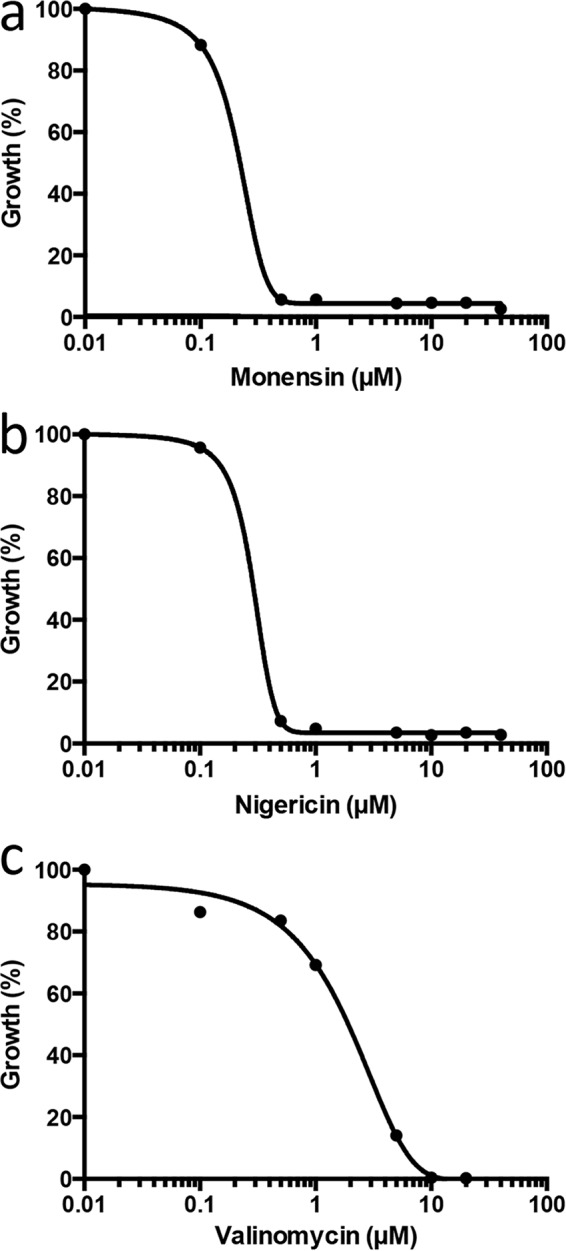
Determination of the median inhibitory concentration (IC50) values for monensin (0.20 μM) (a), nigericin (0.30 μM) (b), and valinomycin (1.3 μM) (c) on the growth (percentage) of M. maripaludis using the 96-well microtiter plate format. Growth was determined from OD600s recorded after 5 days of incubation at 37°C using a 4% inoculum and McF medium containing 0.2% sodium sulfide (wt/vol) and 400 mM sodium formate. IC50 values are the results from three independent experiments.
FIG 2.
Determination of the median inhibitory concentration (IC50) values for mangostin (2.5 μM) (a), echinomycin (1.2 μM) (b), curcumin (6.5 μM) (c), and lumichrome (2.6 μM) (d) on the growth (%) of M. maripaludis using the 96-well microtiter plate format. Growth was determined from OD600s recorded after 5 days of incubation at 37°C using a 4% inoculum and McF medium containing 0.2% sodium sulfide (wt/vol) and 400 mM sodium formate.
The inhibitors discovered in this study were identified through their ability to inhibit the growth of both methanogen species on either formate or ethanol and methanol, but there is reason to believe that core essential methanogen genes would also be essential for optimal growth on hydrogen/carbon dioxide (41), the major difference being the requirement for formate dehydrogenase or ethanol dehydrogenase under our growth conditions. The reduction in the total number of genes screened when using formate or methanol and ethanol is potentially quite small (1 to 2%). The increase in throughput capacity of the phenotypic screening methods (24, 25), which was our primary goal, outweighs the small loss in potential targets missed under hydrogenotrophic conditions.
In summary, we have developed, optimized, and validated a high-throughput microtiter plate assay for the growth of environmental and rumen methanogens that enables the rapid screening of compound libraries for the identification of novel methanogen-specific compounds. Using this platform, we identified several new inhibitors of methanogen growth, demonstrating the utility of this approach.
MATERIALS AND METHODS
Methanogen growth and microtiter plate development and screening.
Methanococcus maripaludis strain S2 was grown in basal medium at 37°C containing sodium formate (400 mM) (McF medium) essentially as described by Sarmiento et al. (30). For routine culture, strain S2 was grown in Balch tubes (15-ml working volume in 28-ml tubes). Balch tubes were pressurized to 15 lb/in2 with N2/CO2 (80:20 [vol/vol]) prior to autoclaving. The pH after autoclaving was 7.7 to 7.8. Prior to inoculation, 0.3 ml of 2.5% Na2S · 9H2O (wt/vol) was added per 15 ml of medium to ensure sufficient reducing conditions (final concentration, 0.05% Na2S · 9H2O). Methanobrevibacter species AbM4 was isolated in New Zealand (37, 42) and maintained using H2 and CO2 at 38°C in either Hungate tubes, Balch tubes, or serum vials using a 5% rumen fluid-based (RM02) medium (11). The medium was supplemented with a mixture of ethanol and methanol (both at 20 mM final concentration) which supported good growth and avoided the necessity for culturing with an overpressure of H2 and CO2. For storage of AbM4 cultures at −83°C, recovery was more reproducible when AbM4 was transferred to By+ medium using H2 and CO2 overpressures compared with that from storage in RM02 medium (both using 5% [vol/vol] DMSO for freezing) (43, 44). To recover cultures, the cultures were thawed and a 10% inoculum was transferred to new tubes of By+ medium and grown using H2 and CO2. The AbM4 inoculum that was used for screening was first adapted to growth with ethanol and methanol by at least two serial transfers from cultures grown in RM02 medium with H2 and CO2. For both methanogens, growth was measured by culture absorbance at 600 nm. Routine checks of culture purity were made using 16S rRNA PCR sequencing combined with fluorescence and phase-contrast microscopy.
The final optimized McF medium composition for growing M. maripaludis strain S2 in 96-well microtiter plates contained (per liter of ultrapure water; Milli-Q, Millipore, USA): glycyl glycine buffer (200 mM, pH 8.0), general salt solution (0.335 g/liter KCl, 2.25 g/liter MgCl · 6H2O, 3.45 g/liter MgSO4 · 9H2O, 0.5 g/liter NH4Cl, 0.14 g/liter CaCl2 · 2H2O, 0.14 g/liter K2HPO4, and 1.36 g/liter CH3COONa · 3H2O), trace minerals solution, pH 7.0 [15 mg/liter nitriloacetic acid, 1 mg/liter MnSO4, 1 mg/liter Fe(NH4)2(SO4) · 6H2O, 1 mg/liter CoCl2 · 6H2O, 1 mg/liter ZnSO4 · 7H2O, 0.1 mg/liter CuSO4 · 5H2O, 0.25 mg/liter NiCl2 · 6H2O, 2 mg/liter Na2SeO3, 1 mg/liter Na2MoO4 · 2H2O, and 1 mg/liter Na2WO4·2H2O], 1 mg/liter Fe(NH4)2(SO4) · 6H2O, 0.1% (wt/vol) resazurin, 27 g/liter HCOONa, 5 g/liter NaHCO3, and 500 mg/liter l-cysteine-HCl. The final optimized medium composition for growing Methanobrevibacter sp. AbM4 in 96-well microtiter plates was 5% rumen fluid-based (RM02) medium supplemented with a mixture of ethanol and methanol (both at 20 mM final concentration, 0.05% l-cysteine) (11).
For growth in 96-well microtiter plates (Corning Costar 96-well flat-bottom tissue culture), empty plates were preincubated in a Coy anaerobic chamber (gas mix, 4% H2, 5% CO2, and 91% N2) with <8 ppm O2 for at least 3 days to remove traces of O2. Test inhibitors were solubilized in dimethyl sulfoxide (DMSO) and placed in the preconditioned plates the day before the assay commenced in a sterile laminar flow hood (maximum final volume of DMSO was 1% [vol/vol] for AbM4 and 1 to 2% [vol/vol] for M. maripaludis). The plates were then sealed with a gas-permeable seal (Sigma AeraSeal film) and placed in an anaerobic chamber overnight to remove traces of O2 from the inhibitors. Cultures of M. maripaludis (Balch tube) or strain AbM4 (serum bottles) were used for inoculation in a final culture volume of 320 μl medium for 96-well plates. Optical density readings at 600 nm (OD600) (Flex Station 3 plate reader; Bio-strategy, USA) were performed after 5 days of incubation under anaerobic conditions. The assay performance was assessed by the statistical parameters Z and Z′, which take into account both data variability and signal window (38). The Z evaluates the performance of a high-throughput screening assay and Z′ is the characteristic parameter of the assay itself in the absence of test compounds (38). Statistical analysis was conducted in GraphPad Prism 6 using an unpaired t test (95% confidence, P < 0.05). The LOPAC1280 compound library was purchased from Sigma-Aldrich and screened in the 96-well format at a final concentration of 20 μM. An in-house collection of 120 natural products was screened at final concentrations of 20 μM.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the New Zealand Government to support the objectives of the Livestock Research Group of the Global Research Alliance on Agricultural Greenhouse Gases. Any view or opinion expressed does not necessarily represent the view of the Global Research Alliance.
We thank the Pastoral Greenhouse Gas Research Consortium (manager Mark Aspin), the Ministry for Primary Industries (Gerald Rys and Andrea Pickering), and the New Zealand Agricultural Greenhouse Gas Research Centre (Andy Reisinger and Harry Clark) for their ongoing support.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00396-17.
REFERENCES
- 1.Yusuf RO, Noor ZZ, Abba AH, Abu Hassan MA, Din MFM. 2012. Methane emission by sectors: a comprehensive review of emission sources and mitigation methods. Renew Sustain Energy Rev 16:5059–5070. doi: 10.1016/j.rser.2012.04.008. [DOI] [Google Scholar]
- 2.Clark H, Kelliher F, Pinares-Patino C. 2011. Reducing CH4 emissions from grazing ruminants in New Zealand: challenges and opportunities. Asian-Australas J Anim Sci 24:295–302. doi: 10.5713/ajas.2011.r.04. [DOI] [Google Scholar]
- 3.Janssen PH, Kirs M. 2008. Structure of the archaeal community of the rumen. Appl Environ Microbiol 74:3619–3625. doi: 10.1128/AEM.02812-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henderson G, Cox F, Ganesh S, Jonker A, Young W, Global Rumen Census Collaborators, Janssen PH. 2015. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci Rep 5:14567. doi: 10.1038/srep14567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buddle BM, Denis M, Attwood GT, Altermann E, Janssen PH, Ronimus RS, Pinares-Patino CS, Muetzel S, Neil Wedlock D. 2011. Strategies to reduce methane emissions from farmed ruminants grazing on pasture. Vet J 188:11–17. doi: 10.1016/j.tvjl.2010.02.019. [DOI] [PubMed] [Google Scholar]
- 6.Henderson G, Cook GM, Ronimus RS. 2016. Enzyme- and gene-based approaches for developing methanogen-specific compounds to control ruminant methane emissions: a review. Anim Prod Sci 2016:AN15757. doi: 10.1071/AN15757. [DOI] [Google Scholar]
- 7.Kittelmann S, Pinares-Patino CS, Seedorf H, Kirk MR, Ganesh S, McEwan JC, Janssen PH. 2014. Two different bacterial community types are linked with the low-methane emission trait in sheep. PLoS One 9:e103171. doi: 10.1371/journal.pone.0103171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun X, Henderson G, Cox F, Molano G, Harrison SJ, Luo D, Janssen PH, Pacheco D. 2015. Lambs fed fresh winter forage rape (Brassica napus L.) emit less methane than those fed perennial ryegrass (Lolium perenne L.), and possible mechanisms behind the difference. PLoS One 10:e0119697. doi: 10.1371/journal.pone.0119697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leahy SC, Kelly WJ, Altermann E, Ronimus RS, Yeoman CJ, Pacheco DM, Li D, Kong Z, McTavish S, Sang C, Lambie SC, Janssen PH, Dey D, Attwood GT. 2010. The genome sequence of the rumen methanogen Methanobrevibacter ruminantium reveals new possibilities for controlling ruminant methane emissions. PLoS One 5:e8926. doi: 10.1371/journal.pone.0008926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeyanathan J, Martin C, Morgavi DP. 2014. The use of direct-fed microbials for mitigation of ruminant methane emissions: a review. Animal 8:250–261. doi: 10.1017/S1751731113002085. [DOI] [PubMed] [Google Scholar]
- 11.Wedlock DN, Pedersen G, Denis M, Dey D, Janssen PH, Buddle BM. 2010. Development of a vaccine to mitigate greenhouse gas emissions in agriculture: vaccination of sheep with methanogen fractions induces antibodies that block methane production in vitro. N Z Vet J 58:29–36. doi: 10.1080/00480169.2010.65058. [DOI] [PubMed] [Google Scholar]
- 12.Haisan J, Sun Y, Guan LL, Beauchemin KA, Iwaasa A, Duval S, Barreda DR, Oba M. 2014. The effects of feeding 3-nitrooxypropanol on methane emissions and productivity of Holstein cows in mid lactation. J Dairy Sci 97:3110–3119. doi: 10.3168/jds.2013-7834. [DOI] [PubMed] [Google Scholar]
- 13.Martinez-Fernandez G, Abecia L, Arco A, Cantalapiedra-Hijar G, Martin-Garcia AI, Molina-Alcaide E, Kindermann M, Duval S, Yanez-Ruiz DR. 2014. Effects of ethyl-3-nitrooxy propionate and 3-nitrooxypropanol on ruminal fermentation, microbial abundance, and methane emissions in sheep. J Dairy Sci 97:3790–3799. doi: 10.3168/jds.2013-7398. [DOI] [PubMed] [Google Scholar]
- 14.Reynolds CK, Humphries DJ, Kirton P, Kindermann M, Duval S, Steinberg W. 2014. Effects of 3-nitrooxypropanol on methane emission, digestion, and energy and nitrogen balance of lactating dairy cows. J Dairy Sci 97:3777–3789. doi: 10.3168/jds.2013-7397. [DOI] [PubMed] [Google Scholar]
- 15.Hristov AN, Oh J, Giallongo F, Frederick TW, Harper MT, Weeks HL, Branco AF, Moate PJ, Deighton MH, Williams SR, Kindermann M, Duval S. 2015. An inhibitor persistently decreased enteric methane emission from dairy cows with no negative effect on milk production. Proc Natl Acad Sci U S A 112:10663–10668. doi: 10.1073/pnas.1504124112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denman SE, Tomkins N, McSweeney CS. 2007. Quantitation and diversity analysis of ruminal methanogenic populations in response to the antimethanogenic compound bromochloromethane. FEMS Microbiol Ecol 62:313–322. doi: 10.1111/j.1574-6941.2007.00394.x. [DOI] [PubMed] [Google Scholar]
- 17.Abecia L, Toral PG, Martin-Garcia AI, Martinez G, Tomkins NW, Molina-Alcaide E, Newbold CJ, Yanez-Ruiz DR. 2012. Effect of bromochloromethane on methane emission, rumen fermentation pattern, milk yield, and fatty acid profile in lactating dairy goats. J Dairy Sci 95:2027–2036. doi: 10.3168/jds.2011-4831. [DOI] [PubMed] [Google Scholar]
- 18.Abecia L, Waddams KE, Martinez-Fernandez G, Martin-Garcia AI, Ramos-Morales E, Newbold CJ, Yanez-Ruiz DR. 2014. An antimethanogenic nutritional intervention in early life of ruminants modifies ruminal colonization by Archaea. Archaea 2014:841463. doi: 10.1155/2014/841463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abecia L, Martin-Garcia AI, Martinez G, Newbold CJ, Yanez-Ruiz DR. 2013. Nutritional intervention in early life to manipulate rumen microbial colonization and methane output by kid goats postweaning. J Anim Sci 91:4832–4840. doi: 10.2527/jas.2012-6142. [DOI] [PubMed] [Google Scholar]
- 20.Knight T, Ronimus RS, Dey D, Tootill C, Naylor G, Evans P, Molano G, Smith A, Tavendale M, Pinares-Patino CS, Clark H. 2011. Chloroform decreases rumen methanogenesis and methanogen populations without altering rumen function in cattle. Anim Feed Sci Technol 166–167:101–112. doi: 10.1016/j.anifeedsci.2011.04.059. [DOI] [Google Scholar]
- 21.Mitsumori M, Shinkai T, Takenaka A, Enishi O, Higuchi K, Kobayashi Y, Nonaka I, Asanuma N, Denman SE, McSweeney CS. 2012. Responses in digestion, rumen fermentation and microbial populations to inhibition of methane formation by a halogenated methane analogue. Br J Nutr 108:482–491. doi: 10.1017/S0007114511005794. [DOI] [PubMed] [Google Scholar]
- 22.Rowland FS. 2006. Stratospheric ozone depletion. Philos Trans R Soc Lond B Biol Sci 361:769–790. doi: 10.1098/rstb.2005.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aung HL, Dey D, Janssen PH, Ronimus RS, Cook GM. 2015. A high-throughput screening assay for identification of inhibitors of the A1AO-ATP synthase of the rumen methanogen Methanobrevibacter ruminantium M1. J Microbiol Methods 110:15–17. doi: 10.1016/j.mimet.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 24.Macarron R, Banks MN, Bojanic D, Burns DJ, Cirovic DA, Garyantes T, Green DV, Hertzberg RP, Janzen WP, Paslay JW, Schopfer U, Sittampalam GS. 2011. Impact of high-throughput screening in biomedical research. Nat Rev Drug Discov 10:188–195. doi: 10.1038/nrd3368. [DOI] [PubMed] [Google Scholar]
- 25.Zheng W, Thorne N, McKew JC. 2013. Phenotypic screens as a renewed approach for drug discovery. Drug Discov Today 18:1067–1073. doi: 10.1016/j.drudis.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borglin S, Joyner D, Jacobsen J, Mukhopadhyay A, Hazen TC. 2009. Overcoming the anaerobic hurdle in phenotypic microarrays: generation and visualization of growth curve data for Desulfovibrio vulgaris Hildenborough. J Microbiol Methods 76:159–168. doi: 10.1016/j.mimet.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 27.Jones WJ, Paynter MJB, Gupta R. 1983. Characterization of Methanococcus maripaludis sp. nov., a new methanogen isolated from salt-marsh sediment. Arch Microbiol 135:91–97. doi: 10.1007/BF00408015. [DOI] [Google Scholar]
- 28.Whitman WB, Shieh J, Sohn S, Caras DS, Premachandran U. 1986. Isolation and characterization of 22 mesophilic methanococci. Syst Appl Microbiol 7:235–240. doi: 10.1016/S0723-2020(86)80012-1. [DOI] [Google Scholar]
- 29.Whitman WB, Tumbula DL, Yu JP, Kim W. 1997. Development of genetic approaches for the methane-producing archaebacterium Methanococcus maripaludis. Biofactors 6:37–46. doi: 10.1002/biof.5520060105. [DOI] [PubMed] [Google Scholar]
- 30.Sarmiento F, Leigh JA, Whitman WB. 2011. Genetic systems for hydrogenotrophic methanogens. Methods Enzymol 494:43–73. doi: 10.1016/B978-0-12-385112-3.00003-2. [DOI] [PubMed] [Google Scholar]
- 31.Hendrickson EL, Kaul R, Zhou Y, Bovee D, Chapman P, Chung J, Conway de Macario E, Dodsworth JA, Gillett W, Graham DE, Hackett M, Haydock AK, Kang A, Land ML, Levy R, Lie TJ, Major TA, Moore BC, Porat I, Palmeiri A, Rouse G, Saenphimmachak C, Soll D, Van Dien S, Wang T, Whitman WB, Xia Q, Zhang Y, Larimer FW, Olson MV, Leigh JA. 2004. Complete genome sequence of the genetically tractable hydrogenotrophic methanogen Methanococcus maripaludis. J Bacteriol 186:6956–6969. doi: 10.1128/JB.186.20.6956-6969.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haydock AK, Porat I, Whitman WB, Leigh JA. 2004. Continuous culture of Methanococcus maripaludis under defined nutrient conditions. FEMS Microbiol Lett 238:85–91. doi: 10.1016/j.femsle.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 33.Miller TL, Wolin MJ, Hongxue Z, Bryant MP. 1986. Characteristics of methanogens isolated from bovine rumen. Appl Environ Microbiol 51:201–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McMillan DG, Ferguson SA, Dey D, Schroder K, Aung HL, Carbone V, Attwood GT, Ronimus RS, Meier T, Janssen PH, Cook GM. 2011. A1Ao-ATP synthase of Methanobrevibacter ruminantium couples sodium ions for ATP synthesis under physiological conditions. J Biol Chem 286:39882–39892. doi: 10.1074/jbc.M111.281675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hungate RE. 1966. The rumen and its microbes. Academic Press, San Diego, CA. [Google Scholar]
- 36.Bang C, Schilhabel A, Weidenbach K, Kopp A, Goldmann T, Gutsmann T, Schmitz RA. 2012. Effects of antimicrobial peptides on methanogenic archaea. Antimicrob Agents Chemother 56:4123–4130. doi: 10.1128/AAC.00661-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leahy SC, Kelly WJ, Ronimus RS, Wedlock N, Altermann E, Attwood GT. 2013. Genome sequencing of rumen bacteria and archaea and its application to methane mitigation strategies. Animal 7 (Suppl 2):S235–S243. doi: 10.1017/S1751731113000700. [DOI] [PubMed] [Google Scholar]
- 38.Zhang JH, Chung TD, Oldenburg KR. 1999. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen 4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 39.Sauer FD, Mahadevan S, Erfle JD. 1980. Valinomycin inhibited methane synthesis in Methanobacterium thermoautotrophicum. Biochem Biophys Res Commun 95:715–721. doi: 10.1016/0006-291X(80)90844-X. [DOI] [PubMed] [Google Scholar]
- 40.Jarrell KF, Sprott GD. 1983. The effects of ionophores and metabolic inhibitors on methanogenesis and energy-related properties of Methanobacterium bryantii. Arch Biochem Biophys 225:33–41. doi: 10.1016/0003-9861(83)90004-8. [DOI] [PubMed] [Google Scholar]
- 41.Sarmiento F, Mrázek J, Whitman WB. 2013. Genome-scale analysis of gene function in the hydrogenotrophic methanogenic archaeon Methanococcus maripaludis. Proc Natl Acad Sci U S A 110:4726–4731. doi: 10.1073/pnas.1220225110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leahy SC, Kelly WJ, Li D, Li Y, Altermann E, Lambie SC, Cox F, Attwood GT. 2013. The complete genome sequence of Methanobrevibacter sp. AbM4. Stand Genomic Sci 8:215–227. doi: 10.4056/sigs.3977691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Joblin KN, Naylor GE, Williams AG. 1990. Effect of Methanobrevibacter smithii on xylanolytic activity of anaerobic ruminal fungi. Appl Environ Microbiol 56:2287–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Joblin KN. 2005. Methanogenic archaea, p 47–54. In Makkar HPS, McSweeney CS (ed), Methods in gut microbial ecology for ruminants, Springer Science+Business Media BV, Dordrecht, The Netherlands. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.