ABSTRACT
Streptococcus mutans is the primary etiological agent of dental caries and causes tooth decay by forming a firmly attached biofilm on tooth surfaces. Biofilm formation is induced by the presence of sucrose, which is a substrate for the synthesis of extracellular polysaccharides but not in the presence of oligosaccharides. Nonetheless, in this study, we found that raffinose, which is an oligosaccharide with an intestinal regulatory function and antiallergic effect, induced biofilm formation by S. mutans in a mixed culture with sucrose, which was at concentrations less than those required to induce biofilm formation directly. We analyzed the possible mechanism behind the small requirement for sucrose for biofilm formation in the presence of raffinose. Our results suggested that sucrose contributed to an increase in bacterial cell surface hydrophobicity and biofilm formation. Next, we examined how the effects of raffinose interacted with the effects of sucrose for biofilm formation. We showed that the presence of raffinose induced fructan synthesis by fructosyltransferase and aggregated extracellular DNA (eDNA, which is probably genomic DNA released from dead cells) into the biofilm. eDNA seemed to be important for biofilm formation, because the degradation of DNA by DNase I resulted in a significant reduction in biofilm formation. When assessing the role of fructan in biofilm formation, we found that fructan enhanced eDNA-dependent cell aggregation. Therefore, our results show that raffinose and sucrose have cooperative effects and that this induction of biofilm formation depends on supportive elements that mainly consist of eDNA and fructan.
IMPORTANCE The sucrose-dependent mechanism of biofilm formation in Streptococcus mutans has been studied extensively. Nonetheless, the effects of carbohydrates other than sucrose are inadequately understood. Our findings concerning raffinose advance the understanding of the mechanism underlying the joint effects of sucrose and other carbohydrates on biofilm formation. Since raffinose has been reported to have positive effects on enterobacterial flora, research on the effects of raffinose on the oral flora are required prior to its use as a beneficial sugar for human health. Here, we showed that raffinose induced biofilm formation by S. mutans in low concentrations of sucrose. The induction of biofilm formation generally generates negative effects on the oral flora. Therefore, we believe that this finding will aid in the development of more effective oral care techniques to maintain oral flora health.
KEYWORDS: Streptococcus mutans, biofilms, extracellular DNA, fructan, raffinose, small amount of sucrose
INTRODUCTION
Streptococcus mutans is an etiological agent of dental caries and infective endocarditis. This bacterium manifests virulence by forming a biofilm. The major traits of virulence are acidogenicity, aciduricity, and firm attachment of the biofilm (1). The biofilm formation mechanisms of S. mutans have been categorized as sucrose dependent and sucrose independent (2). The presence of sucrose is essential for polysaccharide-dependent biofilm formation, because all polysaccharide synthases conduct enzymatic reactions using sucrose as the substrate. S. mutans possesses 3 types of glucosyltransferases (Gtf-I, Gtf-SI, and Gtf-S, encoded by gtfB, gtfC, and gtfD, respectively) and a fructosyltransferase (Ftf, encoded by sacB). Gtf-I, which is absorbed on the bacterial cell surface, synthesizes insoluble glucans rich in 1,3-bonds and contributes to microcolony formation (3, 4). Gtf-SI, which is bound to a solid surface, synthesizes a mixture of soluble 1,6-bonds and insoluble glucans and acts as the attachment site for bacteria (4, 5). Gtf-S produces soluble glucans, which serve as primers for Gtf-I activity (6, 7). Fructan is synthesized by Ftf from sucrose and serves as a nutrient source for biofilm cells (8). Although fructan has been shown to contribute to biofilm formation by interacting with glucan (9), the role of fructan itself in biofilm formation is not fully understood.
In addition to the participation of these polysaccharides, extracellular DNA (eDNA) is involved in biofilm formation via a sucrose-independent mechanism. eDNA is believed to be genomic DNA released from dead cells and has important functions as an attachment factor for surfaces and an adhesive factor among bacteria during the initial stage of biofilm formation (10, 11). The degradation of eDNA by the addition of DNase I results in a significant decrease in biofilm formation (12). The emergence of dead cells (autolysis) is regulated by cell density-dependent gene expression (quorum sensing [QS]). In S. mutans, Streptococcus pneumoniae, and Streptococcus gordonii, autolysis leading to eDNA production is regulated by the expression of a secretory peptide pheromone (competence-stimulating peptide [CSP]) and by the recognition of CSP by a two-component signal transduction system (13–16). This QS system simultaneously controls genetic competence, bacteriocin production, and stress tolerance and contributes to the formation of a mature biofilm (17–19).
Conversely, the effects of sugars other than sucrose on biofilm formation are poorly understood, although S. mutans can metabolize many types of sugars, including oligosaccharides (20). We focused on raffinose, which is one oligosaccharide metabolized by S. mutans but is an indigestible oligosaccharide for humans (Fig. 1A). The sweetness of raffinose is approximately 20% of the sweetness of sucrose. Raffinose is known to be a sugar beneficial for human health due to its positive effects on the intestinal bacterial flora. Indeed, the continuous consumption of raffinose leads to a significant increase in the Bifidobacterium population and a significant decrease in the Clostridium and Bacteroides populations in healthy human intestines (21). In the oral cavity, raffinose has been reported to serve as the substrate for Ftf of Streptococcus salivarius, leading to the accumulation of extracellular polysaccharides (22). Nonetheless, the involvement of raffinose in S. mutans biofilm formation is unknown. In this study, we found that the presence of raffinose induced biofilm formation by S. mutans, and we investigated the underlying mechanism to gain a better understanding of biofilm formation and ultimately to prevent and treat bacterial infections.
FIG 1.
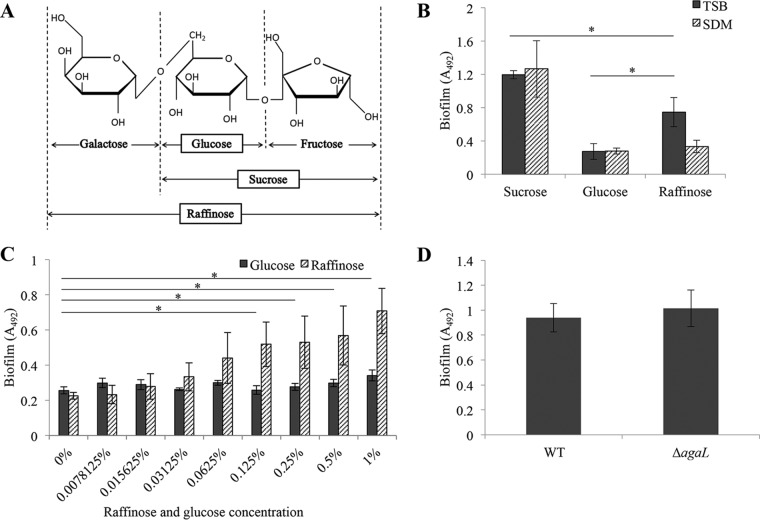
The stimulatory effect of raffinose on biofilm formation by S. mutans. (A) Structural comparison of sugars used in this study. (B) Biofilm formation assay in TSB or SDM supplemented with 0.25% (wt/vol) sucrose, glucose, or raffinose. We used sucrose and glucose as biofilm-inducible and non-biofilm-inducible controls, respectively. (C) The impact of a low concentration of sucrose and the stimulatory effect of raffinose on biofilm formation. To evaluate the effects of a low concentration of sucrose, we used SDM with 0.002% sucrose, which is not sufficient to induce a biofilm, as a base medium. We tested whether raffinose has a stimulatory effect of biofilm by adding various concentrations of raffinose to cell culture. Glucose was used as a negative control. (D) Comparison of the biomass of a biofilm formed by WT and ΔagaL mutant strains. Biofilm biomasses were comparable in the WT and ΔagaL mutant strains. The data indicate means ± standard deviations (SD) of the results from three independent experiments. The asterisks indicate a significant difference between two groups (Student's t test; P < 0.05).
RESULTS
The stimulatory effect of raffinose on biofilm formation.
We tested whether biofilm formation was induced by the presence of raffinose using several media. In this study, we used sucrose and glucose as the biofilm-inducing and non-biofilm-inducing substrates, respectively. As shown in Fig. 1B and in Fig. S1 in the supplemental material, biofilm formation was significantly increased in the tryptic soy broth with raffinose (TSBr) medium compared to TSB with glucose (TSBg), which served as a negative control for biofilm formation. In contrast, the biomass of the biofilm was significantly lower in TSBr than in TSB with sucrose (TSBs), which was used as a positive control. This result suggested that raffinose had the ability to induce biofilm formation by S. mutans. Conversely, biofilm formation was not induced when the base medium was changed to semidefined medium (SDM), brain heart infusion (BHI) agar, or Todd Hewitt broth (THB) (Fig. 1B and S2A and B). These results suggested that some components of TSB were necessary for the induction of the biofilm in the presence of raffinose. To identify the reason why a biofilm was induced in TSBr, we focused on the presence of sucrose in TSB. TSB consists of the components derived from tryptone and soy and contains approximately 0.0075% (wt/vol) sucrose, which is not sufficient for biofilm induction (data not shown). To evaluate the effect of raffinose at the low sucrose concentration, a biofilm formation assay was conducted in SDM supplemented with 0.002% (wt/vol) sucrose, which was a more severe condition in terms of sucrose availability, and with various raffinose concentrations. As shown in Fig. 1C, the biofilm biomass was significantly increased when a raffinose concentration greater than 0.125% was added to SDM supplemented with 0.002% (wt/vol) sucrose as the base substrate. As a control, the addition of glucose did not induce biofilm formation. This result suggested that raffinose had a stimulatory effect on biofilm formation. Therefore, raffinose was not only a nutrient.
Raffinose incorporated by S. mutans is degraded to galactose and sucrose under the influence of α-galactosidase as a first step of metabolism (23). We tested whether the sucrose formed by α-galactosidase during this process induced biofilm formation in TSBr. Then, we constructed an isogenic deletion mutant of the agaL gene (ΔagaL mutant), which encodes α-galactosidase. The biofilm formed by the ΔagaL mutant was quantified to examine the effect of α-galactosidase on the biofilm in TSBr. The biofilm biomass produced by the ΔagaL mutant was almost equal to the biofilm biomass of the wild-type (WT) strain (Fig. 1D). This result suggested that the sucrose derived from endogenous (intracellular) raffinose did not contribute to biofilm formation. Collectively, these results showed that extracellular sucrose was important for biofilm formation in TSBr.
Quantitative analysis and contribution of eDNA to biofilm formation.
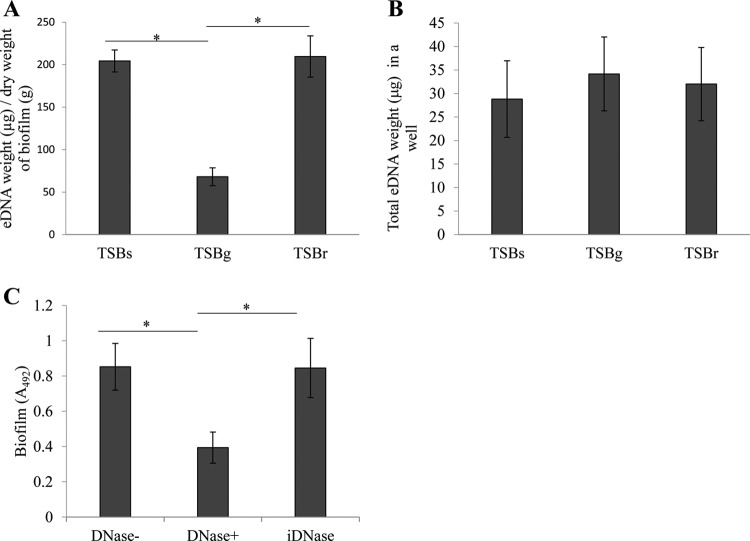
eDNA is a major component of biofilms in many bacteria, including S. mutans. We determined the amount of eDNA in the biofilm by isolating and calculating the eDNA weight per unit of dry weight of the biofilm. The amount of eDNA in the biofilm formed in TSBr was similar to the amount in the biofilm formed in TSBs but significantly greater than the amount in the biofilm formed in TSBg (Fig. 2A). The eDNA was confirmed to be derived from the genomic DNA of S. mutans UA159 using the random amplified polymorphic DNA (RAPD) method (Fig. S3). In contrast, the total eDNA included in the biofilm and culture supernatant did not change in any of the media (Fig. 2B). These results indicated that raffinose and sucrose, but not glucose, might induce the biofilm formation associated with eDNA. Next, to examine the contribution of eDNA to biofilm formation, we assessed the inhibitory effect of DNase I on biofilm formation. Biofilm formation was significantly but not completely inhibited by DNase I, whereas inhibition was not observed by heat-inactivated DNase I (Fig. 2C). These results indicated that eDNA was an important factor for biofilm formation in TSBr.
FIG 2.
Quantification of eDNA and the contribution of eDNA to biofilm formation. (A) Quantification of eDNA in biofilms. eDNA was isolated from a 20-h biofilm and quantified as the weight of eDNA per unit of dry weight of biofilm. The dry weights of biofilm obtained from TSBs, TSBg, and TSBr were 4.44 ± 0.3 μg, 2.49 ± 0.1 μg, and 2.80 ± 0.2 μg, respectively. (B) Quantification of total eDNA in biofilm and conditioned medium. Five hundred microliters of overnight culture was mixed with 4.5 ml of TSBs, TSBg, or TSBr in a 6-well culture plate. The biofilm formation was conducted for 20 h. These data indicate the total amount of eDNA detected from biofilm and 5 ml of culture supernatant. (C) Inhibitory effect of DNase I on biofilm formation. Taking into consideration the finding that eDNA contributes to the initial stage of the biofilm formation, we quantified biofilm formations in the 8-h biofilm. DNase I was added at a concentration of 50 U/ml (DNase+) before the incubation for biofilm formation. As a control, we also quantified biofilm formations in the absence of DNase I (DNase−) and heat-inactivated (100°C for 30 min) DNase I (iDNase). These data indicate mean ± SD of the results from three independent experiments. The asterisks indicate a significant difference between two groups (Student's t test, P < 0.05).
Quantitative analysis of polysaccharides.
Polysaccharides are important factors for the structural and functional stability of biofilms. In particular, insoluble polysaccharides are intimately implicated in firm attachment during biofilm formation in S. mutans and in microbial virulence in dental caries. In contrast, soluble polysaccharides are assumed to be indirect contributors to biofilms as the nutrient source for the biofilm cells. We determined whether the insoluble and soluble polysaccharides were synthesized by S. mutans in TSBr, because whether S. mutans possessed an enzyme capable of synthesizing a polysaccharide from raffinose was unknown. The amounts of insoluble polysaccharides synthesized were comparable in TSBr and TSBg; the absence of insoluble polysaccharide synthesis in TSBg was already known (Fig. 3A). The amount of soluble polysaccharides was increased in TSBr, and the detected amount was significantly greater than the amount formed in TSBs (Fig. 3A). Therefore, soluble polysaccharides were synthesized by S. mutans in TSBr. Because it is known that there are two types of soluble polysaccharides, glucan and fructan, synthesized by S. mutans, we attempted to determine whether the soluble polysaccharide(s) synthesized in TSBr was glucan and/or fructan. Using deletion mutants of gtfD (ΔgtfD) and sacB (ΔsacB), we performed a quantitative analysis of the soluble polysaccharides in TSBr. The soluble polysaccharide(s) was synthesized in TSBr by the Gtf-S ΔgtfD mutant but not by the Ftf ΔsacB mutant (Fig. 3B). These results indicated that the soluble polysaccharide synthesized in TSBr was most likely fructan.
FIG 3.
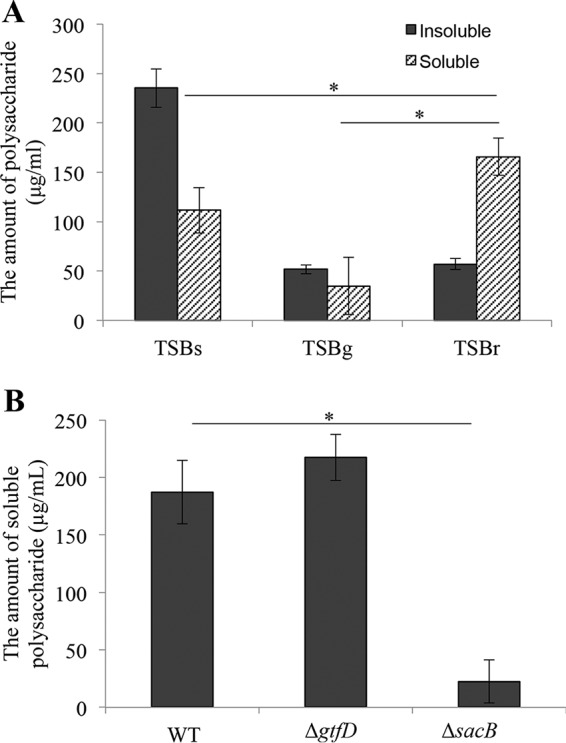
Quantification of polysaccharides. (A) Comparison of the soluble and insoluble polysaccharides synthesized in TSBs, TSBg, and TSBr. TSBs and TSBg serve as a positive control and negative control for the synthesis of polysaccharides, respectively. (B) Discrimination between glucan and fructan among the soluble polysaccharide synthesized in TSBr. We performed quantification of the soluble polysaccharide synthesized in TSBr using the ΔgtfD mutant, which cannot synthesize a soluble glucan, and the ΔsacB mutant, which cannot synthesize fructan. These data are presented as the means ± SD of the results from three independent experiments. The asterisks indicate a significant difference between two groups (Student's t test, P < 0.05).
Role of polysaccharides in biofilm formation.
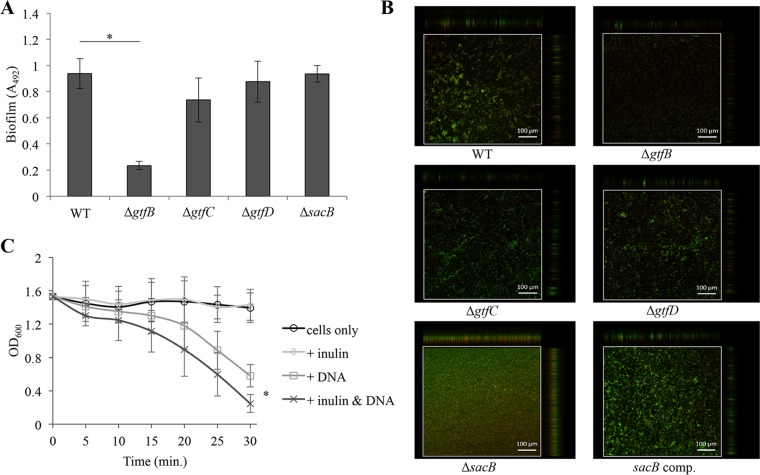
To evaluate the contribution of each enzyme involved in the synthesis of insoluble and soluble polysaccharides to biofilm formation, the ability of deletion mutants of polysaccharide synthase genes to form a biofilm in TSBr was compared with that of the WT strain. The biofilm biomass formed by the ΔsacB mutant was comparable to the biofilm biomass formed by the WT strain (Fig. 4A). Therefore, we assumed that the presence or absence of fructan was not associated with the biofilm biomass. However, the quality of the biofilm might be changed by a mutation in sacB. To analyze the biofilm quality, we examined the morphology of the biofilm following staining with the LIVE/DEAD BacLight bacterial viability kit using confocal laser scanning microscopy (CLSM). The cells attached on the surface as aggregates in the biofilm formed by the WT strain. In contrast, cell aggregation was not observed in the biofilm formed by the ΔsacB mutant, and the morphology of the biofilm was flat and largely stained by the LIVE/DEAD BacLight bacterial viability kit compared to the biofilm formed by the WT and ΔgtfD mutant strains (Fig. 4B). The defect in the biofilm morphology in the ΔsacB mutant was restored in the sacB-complemented strain (sacB comp.; Fig. 4B). Furthermore, the morphology was also restored when 200 μg/ml commercial inulin (a type of fructan synthesized by S. mutans) was added to TSBr (Fig. S4). Therefore, these results indicated that fructan was involved in the biofilm morphology without impacting the biofilm volume. Recently, the addition of levan (a type of fructan) to a DNA solution was reported to increase the viscosity of the solution (24). Ftf from S. mutans synthesizes inulin, which is a different type of fructan. Therefore, we tested the hypothesis that inulin facilitated cell aggregation by acting coordinately with the eDNA. We performed an aggregation assay of bacterial cells using genomic DNA from S. mutans UA159 and commercial inulin to mimic the main components of the extracellular matrix formed in TSBr. The addition of DNA alone or DNA and inulin in combination with the cell suspension led to cell aggregation. At 30 min, the extent of the aggregation was significantly higher when DNA and inulin were added to the cell suspension than with DNA alone (Fig. 4C). This result suggested that although fructan (inulin) alone did not lead to cell aggregation, fructan had a facilitative effect on cell aggregation in the presence of eDNA. Thus, fructan interacts with eDNA to aggregate bacterial cells and contribute to biofilm morphology in TSBr medium.
FIG 4.
The function of the polysaccharide synthase genes in biofilm formation in TSBr. (A) The amount of biofilm formed by the deletion mutants of the gene encoding polysaccharide synthase in TSBr. (B) CLSM analysis of the biofilm formed in TSBr. The cells in the biofilm were stained with the LIVE/DEAD BacLight bacterial viability kit for 30 min. Representative images from three independent experiments are presented. (C) An aggregation assay of the bacterial cells. The roles of eDNA and fructan in cell aggregation were examined by mimicking the extracellular matrix synthesized in TSBr. Five micrograms per milliliter genomic DNA of UA159 and/or 250 μg/ml commercial inulin was added to 3 ml of cell suspension, which was grown in BHI and adjusted to an OD600 of 1.5 with aggregation buffer. The data are presented as the means ± SD of the results from three independent experiments. The asterisks indicate a significant difference between two groups (Student's t test, P < 0.05).
Contribution of gtfB to biofilm formation.
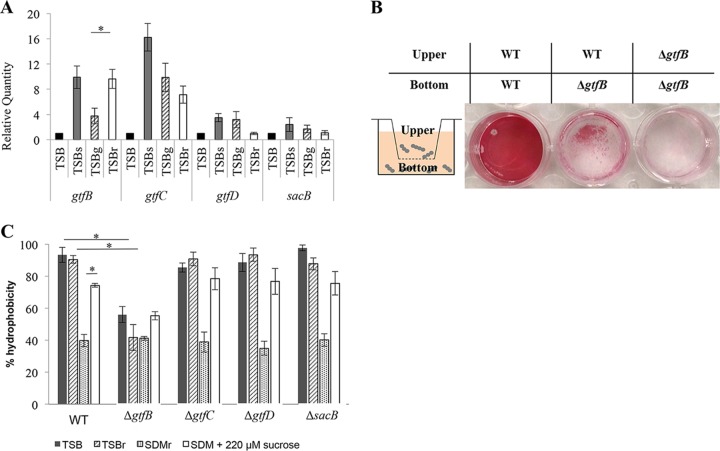
As shown in Fig. 4A and B, the deletion of gtfB, which encodes an enzyme involved in the synthesis of an insoluble glucan, resulted in a significant reduction in biofilm formation even though insoluble polysaccharides were synthesized ineffectively in TSBr (Fig. 3A). Additionally, the quantitative reverse transcription-PCR (RT-qPCR) results supported the importance of gtfB, because the gtfB expression was uniquely upregulated in the TSBr medium compared to TSBg (Fig. 5A). This result suggested that gtfB played a critical role in biofilm formation in TSBr. To clarify the involvement of gtfB in biofilm formation in TSBr, we performed a 2-compartment biofilm formation assay. Biofilm formation by the ΔgtfB mutant was partially complemented by supplying the secreted Gtf-I derived from the WT strain, which was inoculated into the upper compartment (Fig. 5B). gtfB has been reported to be adsorbed onto the bacterial cell surface and contribute to microcolony formation (4). Therefore, we focused on the differences in the activities on the cell surface between the WT and ΔgtfB mutant strains. We hypothesized that the presence or absence of gtfB would affect the hydrophobicity of the cell surface. When the cells were cultivated in TSBr, the hydrophobicity was lower in the ΔgtfB mutant than in the WT strain (Fig. 5C). In contrast, when the cells were cultivated in SDM with raffinose (SDMr), the hydrophobicities were equal among all of the strains (Fig. 5C). These results suggested that Gtf-I, which is encoded by gtfB, contributed to biofilm formation in TSBr by increasing the hydrophobicity of the cell surface. Furthermore, because the bacterial cell surface hydrophobicity did not change between cultivation in TSB and TSBr, TSB was important for the increase in bacterial hydrophobicity, and the presence of raffinose did not influence this process. Next, to identify which component(s) of TSB was involved in the increase in hydrophobicity, we examined the participation of sucrose, which was contained in TSB. When an amount equal (0.0075% [wt/vol]) to the amount of sucrose included in TSB was added to SDM, the bacterial cell surface hydrophobicity was significantly increased compared to SDM, but the level of hydrophobicity was lower than the level in TSB. These results suggested that components of TSB other than sucrose also contributed to this process. After upregulation of hydrophobicity on the cell surface, other responses associated with eDNA and fructan might have induced cell aggregation and biofilm formation. Therefore, gtfB contributed to cell aggregation and biofilm formation in TSBr by utilizing sucrose, and other components included in TSB increased the bacterial cell surface hydrophobicity and other responses associated with eDNA and fructan.
FIG 5.
The importance of gtfB for biofilm formation in TSBr. (A) Expression of genes which encode the polysaccharide synthases in a 20-h biofilm. This graph indicates the relative expression levels of the genes compared with the expression of each gene during cultivation in TSB. These data were normalized to the expression of the endogenous control (lactate dehydrogenase). (B) A two-compartment biofilm formation assay. This experiment revealed the importance of Gtf-I encoded by gtfB secreted by the WT cells inoculated into the upper compartment. The wells were separated by the hanging cell culture insert into the upper and bottom parts of the well. The biofilm formed on the bottom of the wells in TSBr was stained with a safranin solution. Representative images of three independent experiments are presented. (C) Comparison of the cell surface hydrophobicities between strains. The cells grown in TSB, TSBr, SDMr, or SDM supplemented with 0.0075% sucrose medium for 6 h were used. These data are presented as means ± SD of the results from three independent experiments. The asterisks indicate a significant difference between two strains (Student's t test, P < 0.05).
Biofilm formation on the sHA surface.
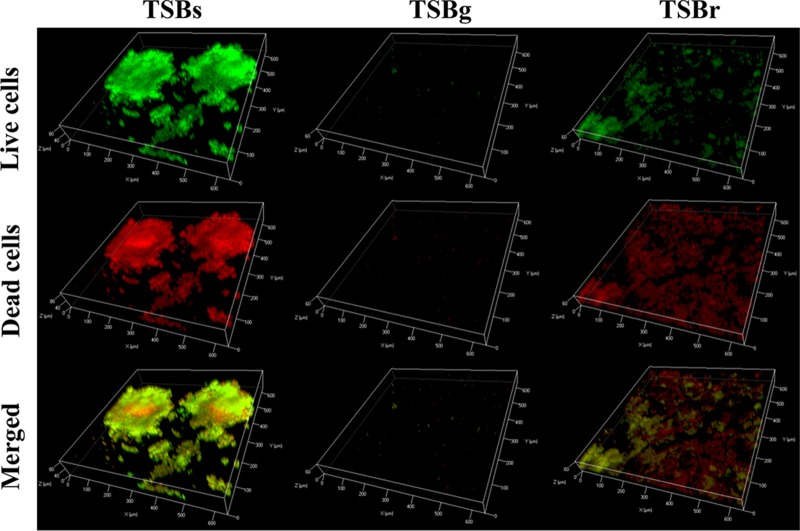
To confirm whether the biofilm formation in TSBr occurred on the tooth surface, biofilm formation by S. mutans was examined on the human saliva-coated hydroxyapatite disk (sHA) as an in vitro model of a smooth surface. We examined the cells in biofilms stained with the LIVE/DEAD BacLight bacterial viability kit by CLSM. Biofilm formation was observed on the sHA surface in TSBs and TSBr but not in TSBg. Small cell aggregates were scattered in TSBr on the sHA surface compared to those in TSBs (Fig. 6). The same result was obtained by examining safranin-stained biofilms (Fig. S5). In TSBr, colocalization of live cells (green) and dead cells (red) was observed in some small aggregates in the biofilm, whereas in other aggregates, dead cells were mainly observed on the entire sHA surface. In TSBs, colocalization of live and dead cells was observed in large aggregates in the biofilm. These results showed that the biofilm formed in TSBr was not simply a sediment on the bottom of the culture plate.
FIG 6.
CLSM examination of the biofilm formed on the sHA surfaces. Biofilm was formed on the sHA placed in a 24-well plate vertically and stained with the LIVE/DEAD BacLight bacterial viability kit. Live cells emitted green fluorescence, whereas the dead cells emitted red fluorescence. Representative three-dimensional (3D) images of three independent experiments are presented.
DISCUSSION
Our study revealed that biofilm formation by S. mutans was induced in TSBr but not in SDMr. Accordingly, the mechanism underlying biofilm formation in TSBr is divided into the participation of TSB and raffinose. Concerning the role of TSB, we propose that sucrose contained in TSB is important for increasing the hydrophobicity of the cell surface, which is mediated by the reaction of GtfI encoded by gtfB. Cell surface hydrophobicity has been reported to be important for microbial attachment and adhesion (25, 26). Indeed, the deletion of gtfB resulted in the reduction of hydrophobicity of the cell surface and decreased biofilm formation. Although the bacterial cell surface hydrophobicity of the ΔgtfB mutant was reduced by half compared to the WT level, the biofilm formation was dramatically decreased. According to these results, other effects were also predicted to be involved in the biofilm formation via the gtfB deletion. The importance of gtfB for biofilm formation was also suggested by RT-qPCR and the two-compartment biofilm formation assay results. Nonetheless, the effects of the gtfB deletion other than the decrease in hydrophobicity were not determined in this study.
The biofilm formation mechanism in the TSBr medium was different from the mechanism in TSBs. The biofilm formation results obtained for the ΔagaL mutant revealed that the induction of the biofilm was not mediated by sucrose produced after the incorporation of raffinose into the cells, because the biofilm biomass of the ΔagaL mutant was not changed. This result also implied the existence of a phenomenon that was uniquely caused by raffinose. This notion is supported by other reports which show that collective behaviors, such as biofilm formation and quorum sensing, are affected by the carbohydrate source (27, 28). Taken together, these results suggest that the biofilm formation in TSBr is induced by a mechanism whereby cell surface hydrophobicity is increased by the reaction of Gtf-I using sucrose included in TSB; then, the additional effects of raffinose occur.
Next, we explored the phenomena caused by the presence of raffinose and its involvement in biofilm formation. We revealed that eDNA was an important contributor to biofilm formation in TSBr. The amount of eDNA contained in the biofilm significantly increased in the biofilm formed in TSBr medium compared to TSBg medium. Nonetheless, the sum of the eDNA in the biofilm and conditioned medium did not change in TSBs, TSBg, and TSBr. These results indicate that raffinose is not involved in eDNA production but instead contributes to the aggregation of eDNA in the biofilm. Degradation of the eDNA by the addition of DNase I led to a significant decrease in biofilm formation. However, biofilm formation could not be completely inhibited by the addition of DNase I. Hence, eDNA is an important factor but not the only contributor. Furthermore, we showed that the eDNA detected in the biofilm formed in TSBr was likely to be genomic DNA released from dead cells based on the RAPD results, because the same band pattern was obtained for the eDNA and the genomic DNA used as the template for the PCR. This result agreed with other reports showing that the eDNA generated by cell lysis was important for biofilm formation by S. mutans (29).
We also revealed that a large amount of soluble polysaccharide was synthesized in the presence of raffinose and was identified as fructan. Ftf has been reported to synthesize fructan from raffinose in S. salivarius, Actinomyces viscosus, and Lactobacillus reuteri (30). Our results revealed that the Ftf of S. mutans also synthesized fructan from raffinose. The amount of soluble polysaccharide was significantly increased in TSBr compared to TSBg, whereas the expression of ftf was not upregulated in TSBr. Therefore, this increase in fructan can probably be attributed to the following mechanism: Ftf competes with glucosyltransferases for a limited supply of a substrate (sucrose) in TSBs but can synthesize fructan from the substrate (raffinose) in TSBr without competition with glucosyltransferases. This proposal is a reasonable explanation because the glucose moiety of raffinose is sandwiched between fructose and galactose residues, whereas the fructose moiety is available for polysaccharide synthesis because this moiety is exposed. However, the role of fructan in biofilm formation by S. mutans is ill defined even though fructan acts as an extracellular carbon source for the biofilm cells (31). In S. mutans V403, disruption of the ftf gene decreases cariogenic activity, but the capacity for adhesion to glass and rat tooth surfaces is not affected (32). In contrast, Wexler et al. showed that the number and severity of dental caries sites did not differ between UA159 and an isogenic mutant of the ftf gene (33). In contrast, Rozen et al. reported that fructan provided attachment sites for several cariogenic bacteria on sHA (34). Thus, the role of fructan in biofilm formation is complicated and unclear. In this study, we identified some of the functions of fructan in biofilm formation. During quantification of the biofilm, we found that deletion of the sacB gene did not apparently affect the biofilm biomass stained with safranin in TSBr. However, during qualitative observation of the biofilm, a flat form was represented in the biofilm of the ΔsacB mutant stained with the LIVE/DEAD BacLight bacterial viability kit. In contrast, aggregates were largely observed in the WT biofilm. These results indicated that the participation of fructan was associated with cell aggregates in the WT strain. Then, we focused on the role of fructan in the biofilm features. Levan, which is a type of fructan, was reported to enhance the viscosity of DNA solutions and was necessary for biofilm formation by Bacillus subtilis (24). Here, we showed that fructan from S. mutans facilitated cell aggregation when it interacted with eDNA. We believe that bacterial cells aggregate due to enhanced viscosity of the medium around the cells in the presence of eDNA and fructan.
Raffinose is considered a sugar beneficial for human health because it increases the population of Bifidobacterium spp. in the human gut and has antiallergic properties in rats (21, 35). Recently, raffinose was reported to inhibit the biofilm formation of Pseudomonas aeruginosa (36). In contrast, our study showed stimulatory effects of raffinose on biofilm formation by S. mutans in low concentrations of sucrose. Although the implications of this biofilm for cariogenicity were not studied here, we assumed that the induction of biofilm formation in the oral cavity negatively affected the maintenance of the healthy oral flora. Therefore, more detailed research into the effects of oligosaccharides, including raffinose, on bacteria is required prior to use as a beneficial sugar for human health. In addition to raffinose, we examined other trisaccharides, including melezitose, 1-kestose, and lactosucrose (Fig. S6). S. mutans could not increase in TSB supplemented with 0.25% (wt/vol) melezitose as a sugar source but could increase in TSB with 0.25% (wt/vol) 1-kestose or lactosucrose, compared with growth in TSB. Moreover, the biofilm formation of S. mutans was stimulated in these media. Whether biofilm formation in the presence of 1-kestose or lactosucrose was induced by a similar mechanism in the presence of raffinose is not clear. However, the combination of d-glucose and a terminal β-linked d-fructose, such as sucrose or raffinose, was involved in the structures of these trisaccharides, which induced biofilm formation. In the established consensus, carbohydrates, which possess a terminal β-linked d-fructose unit, are susceptible to the action of invertase (37). Invertase is an enzyme for the conversion of sucrose to glucose and fructose, and, like fructosyltransferase, it is important for the utilization of fructose (37, 38). However, melezitose, in lacking a terminal β-linked d-fructose unit, is insusceptible to invertase. S. mutans possesses invertase and shows enzyme activities to cleave sucrose and raffinose but not melezitose (39). Therefore, this combination of d-glucose and a terminal β-linked d-fructose in these trisaccharides may be key for growth and biofilm formation in collaboration with eDNA and fructan. Revealing the underlying mechanism of the formation of these biofilms is a future challenge.
In conclusion, we demonstrated that biofilm formation was induced by the cooperative effects of raffinose and sucrose. Our data indicate that raffinose works as a substrate for fructan synthesis by Ftf and that the low concentrations of sucrose contained in TSB contribute to an increase in cell surface hydrophobicity. Under these conditions, aggregates of S. mutans were induced by eDNA, which was released from dead cells, integrated into fructan, and adhered onto the surface of the plate or sHA. Our findings from this study will contribute to the understanding of the mechanism of S. mutans biofilm formation and human health.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains used in this study are listed in Table 1. S. mutans was grown in an aerobic atmosphere containing 5% CO2 (AnaeroPack-CO2; Mitsubishi Gas Chemical Company, Tokyo, Japan) in brain heart infusion (BHI) broth (Difco Laboratories, Detroit, MI) at 37°C prior to inoculation into culture plates that were used for biofilm formation. Escherichia coli DH5α was grown in Luria-Bertani (LB) broth (Affymetrix, Inc., Santa Clara, CA) or 1.5% agar at 37°C to be used for cloning and plasmid amplification.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant propertiesa | Source or reference |
|---|---|---|
| Strains | ||
| S. mutans | ||
| UA159 | Wild type; Erms Kans | 20 |
| ΔgtfB mutant | UA159::pSBB gtfB Ermr | This study |
| ΔgtfC mutant | UA159::pSBC gtfC Kanr | This study |
| ΔgtfD mutant | UA159::pSBD gtfD Kanr | This study |
| ΔsacB mutant | UA159 sacB Ermr | This study |
| ΔagaL mutant | UA159 agaL Ermr | This study |
| sacB comp. | ΔsacB mutant::pSMsacB::sacB Spcr | This study |
| E. coli | ||
| DH5α | Cloning host | TaKaRa |
| Plasmid | ||
| pUC19 | Cloning vector Ampr | 54 |
| pResEmMCS10 | Streptococcal integration plasmid Ermr | 42 |
| pDL276 | E. coli-Streptococcus shuttle vector; Kanr | 55 |
| pDL278 | E. coli-Streptococcus shuttle vector; Spcr | 56 |
| pSBB | pUC19 containing gtfB fragment; Ampr Kanr | This study |
| pSBC | pUC19 containing gtfC fragment; Ampr Ermr | This study |
| pSBD | pUC19 containing gtfD fragment; Ampr Kanr | This study |
| pSMsacB | pDL278::pldh, sacB; Spcr | This study |
Erms, erythromycin susceptibility; Kans, kanamycin susceptibility; Ermr, erythromycin resistance; Kanr, kanamycin resistance; Spcr, spectinomycin resistance; Ampr, ampicillin resistance.
Construction of mutants.
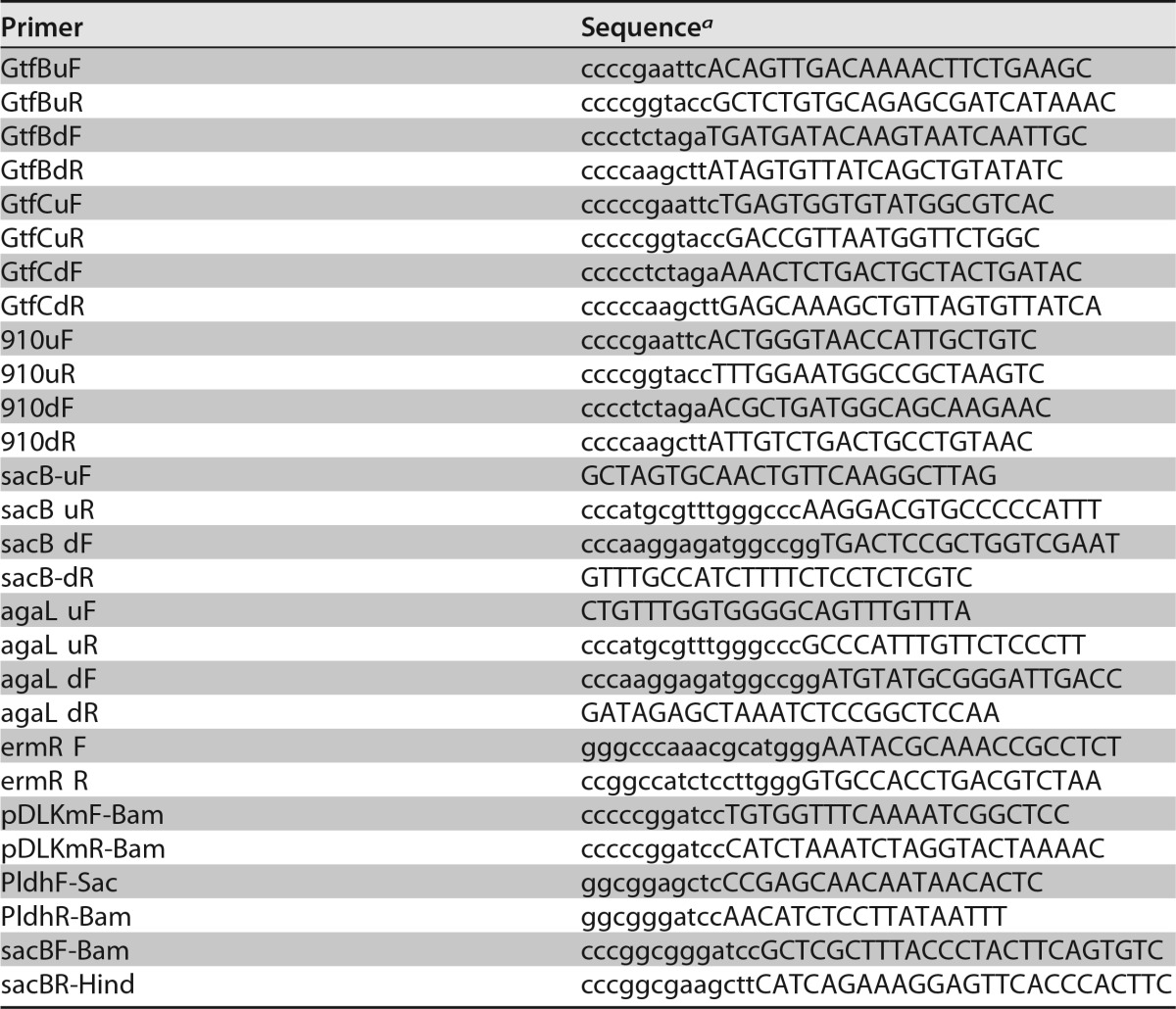
The primers used for the construction of bacterial mutants in this study are listed in Table 2. Sequence information was obtained from the KEGG (http://www.genome.jp/kegg/) and NCBI (http://www.ncbi.nlm.nih.gov/) databases. The ΔgtfB, ΔgtfC, and ΔgtfD mutants were created by inserting an erythromycin or kanamycin resistance gene using homologous recombination, as previously described (40). The ΔsacB and ΔagaL mutants were constructed by replacing the target gene with the erythromycin resistance gene using DNA fragments constructed by overlap extension PCR (41). The upstream and downstream regions of the target genes were amplified from the genomic DNA of S. mutans strain UA159, and the erythromycin resistance gene was obtained from pResEmMCS10 (42). The amplicons were ligated by overlap extension PCR and introduced into S. mutans UA159. The transformants were screened on Mitis Salivarius (MS) agar (Difco Laboratories, Detroit, MI) plates with erythromycin (10 μg/ml) or kanamycin (250 μg/ml). Insertion into the target region of the genomic DNA was confirmed by PCR and agarose gel electrophoresis.
TABLE 2.
Primers for mutant construction
aAdditional sequences which do not correspond to the sequences of the template DNA are shown with lowercase letters.
Construction of the complemented strain.
The primers used for the construction of the sacB-complemented strain are listed in Table 2. To construct a complemented strain constitutively expressing the sacB gene, a constitutive promoter (Pldh) and the sacB gene were amplified from the genomic DNA of S. mutans UA159 by PCR. The Pldh fragment was digested with SacI and BamHI, and the sacB gene fragment was digested with BamHI and HindIII. These fragments were inserted into the multicloning site of the pDL278 shuttle vector. To amplify the plasmid for sacB complementation, pSMsacB was introduced into E. coli DH5α competent cells and screened on LB plates with spectinomycin (50 μg/ml). The plasmid was extracted from the transformant, and the insertion of Pldh and the sacB gene into the target region of pDL278 was confirmed by PCR and agarose gel electrophoresis. Then, pSMsacB was introduced into competent S. mutans UA159 ΔsacB cells. The transformants were screened on MS plates with spectinomycin (200 μg/ml), and the complementation was confirmed by PCR and agarose gel electrophoresis.
Human saliva collection.
Whole-saliva samples from volunteers with good oral health were stimulated by chewing paraffin gum for 5 min, and the samples were collected in ice-chilled sterile bottles. After the saliva was clarified by centrifugation (10,000 × g, 10 min), the supernatant was passed through a 0.22-μm-pore-size filter (Millex-GP; Merck Millipore, Darmstadt, Germany) to remove debris. The sterilized saliva was used immediately for the biofilm formation assays.
Biofilm formation assays.
Biofilm formation was quantified using several medium types, including tryptic soy broth without dextrose (TSB; Difco Laboratories, Detroit, MI), TSB supplemented with 0.25% (wt/vol) sucrose (TSBs), glucose (TSBg), or raffinose (TSBr), and semidefined medium (SDM) supplemented with 0.25% (wt/vol) sucrose (SDMs), glucose (SDMg), or raffinose (SDMr) (43). The biofilm formation assays were performed as previously described (44). To evaluate biofilm formation by S. mutans, 20 μl of a bacterial cell suspension adjusted to an optical density at 600 nm (OD600) of 0.5 with phosphate-buffered saline (PBS) was mixed with 180 μl of each above-mentioned medium in a human saliva-coated 96-well microtiter plate (Sumitomo Bakelite, Tokyo, Japan). Biofilm formation was promoted at 37°C for 20 h in an aerobic atmosphere containing 5% CO2 under static conditions. After incubation, the medium was removed, and the wells were washed 2 times with distilled water (DW). The plate was desiccated, and the biofilm was stained with safranin (0.25% safranin and 0.5% ethanol in H2O; Muto Pure Chemicals, Tokyo, Japan) for 15 min. Then, the wells were washed 2 times with DW to remove the excess staining solution and were desiccated. Safranin was extracted from the biofilm with 100 μl of 70% (vol/vol) ethanol, and the absorbance at 492 nm (A492) was measured on a plate reader (Thermo Bioanalysis Japan, Tokyo, Japan).
Biofilm formation on a human saliva-coated hydroxyapatite disk (sHA, Cellyard pellet; Hoya Technosurgical Corporation, Tokyo, Japan) was evaluated in a 24-well culture plate (Corning Japan, Tokyo, Japan). The hydroxyapatite disk was coated with human saliva at 4°C overnight and placed vertically in a well of a 24-well culture plate (45). One hundred microliters of a bacterial cell suspension adjusted to an OD600 of 0.5 with PBS was mixed with 900 μl of medium in the 24-well culture plate. The culture plate was incubated at 37°C for 20 h in an aerobic atmosphere containing 5% CO2. After incubation, the medium was aspirated from the well, and the sHAs were washed 2 times with DW. Safranin staining of biofilms attached to the sHA surface was performed for 15 min. After that, these disks were washed two times with DW.
A 2-compartment biofilm formation assay was conducted in a 24-well culture plate using the Millicell hanging cell culture insert, with a pore size of 0.4 μm (Millipore, Billerica, MA). The bacterial cell suspensions were prepared using the same procedure described for the biofilm formation assay. One thousand eighty microliters of culture medium was added to the wells; then, the hanging cell culture inserts were placed into each well. Next, 60 μl of the cell suspensions adjusted to an OD600 of 0.5 with PBS was inoculated into the upper compartment and bottom. The plate was incubated at 37°C for 20 h in an aerobic atmosphere containing 5% CO2. Then, the hanging cell culture inserts and medium were removed, and the wells were washed 2 times with DW. The plate was air-dried, and the biofilm was stained with safranin for 15 min. The wells were washed 2 times with DW to remove excess staining solution and were desiccated. Biofilm formation on the bottom surface of the wells was assessed visually.
Quantitative analysis of eDNA.
Isolation and quantification of eDNA were performed as described elsewhere (46). Five hundred microliters of overnight culture was mixed with 4.5 ml of TSBs, TSBg, or TSBr in a 6-well culture plate (Iwaki, Tokyo, Japan). The plate was incubated at 37°C for 20 h in an aerobic atmosphere containing 5% CO2. The eDNA, which was contained in conditioned medium, was extracted with the cetyltrimethylammonium bromide (CTAB) method, as described elsewhere (47), after the samples were passed through a 0.22-μm-pore-size filter to remove cells. The eDNA contained in the biofilm was extracted with the following procedure. The biofilm was scraped from the bottom of the culture plate and air-dried at room temperature; then, the weight of the dried biofilm was measured. The biofilm samples were treated with 5 U/ml dextranase (Sigma Chemical, St. Louis, MO) at 40°C for 30 min. Next, the samples were treated with 5 μl/ml proteinase K (TaKaRa Bio, Inc., Shiga, Japan) at 37°C for 30 min. After these treatments, the samples were passed through a 0.22-μm-pore filter to remove the cells. The eDNA was isolated using the CTAB method. The eDNA concentration was measured by absorptiometry using a NanoDrop lite spectrophotometer (Thermo Fisher Scientific, Waltham, MA).
RAPD method.
A random amplified polymorphic DNA (RAPD) method was performed as described elsewhere, with slight modifications (48). Genomic DNA (gDNA) was isolated from S. mutans UA159 using the DNeasy blood and tissue kit. eDNA was isolated from the biofilm by the same procedure as in the quantification of eDNA. RAPD was carried out using the random primers that were published previously: OPA02 (5′-TGCCGAGCTG-3′) and OPA18 (5′-AGGTGACCGT-3′) (49, 50). The PCR was conducted using the PrimeSTAR premix (TaKaRa Bio, Inc., Shiga, Japan) under the following cycling conditions: 94°C for 1 min, 32°C for 1 min, and 72°C for 2 min for 40 cycles. RAPD products were separated by gel electrophoresis in 1% agarose and visualized with 0.2 μg/ml ethidium bromide.
Measurement of the inhibitory effect of DNase I on biofilm formation.
Fifty units per milliliter of DNase I (Roche Applied Science, Mannheim, Germany) was added to the medium prior to the biofilm formation assay. The preparation of the bacterial cell suspensions and the quantitation procedure were the same as those described for the biofilm formation assay. The incubation time was changed to 8 h, because eDNA was assumed to be an important contributor to the initial attachment, which was the primary step of biofilm formation. Inactivation of DNase I involved heating at 100°C for 30 min.
Aggregation assay.
An overnight culture of S. mutans grown in BHI was harvested by centrifugation and adjusted to an OD600 of 1.5 with aggregation buffer (1 mM Tris, 0.1 mM CaCl2, 0.1 mM MgCl2, 0.02% NaN3, and 0.15 M NaCl [pH 8.0]). We used genomic DNA isolated from S. mutans UA159 using the DNeasy blood and tissue kit (Qiagen, Venlo, The Netherlands) and inulin (Nacalai Tesque, Kyoto, Japan) to mimic the main components of the biofilm matrix formed in TSBr. Five micrograms per milliliter DNA and/or 250 μg/ml inulin were added to 3-ml cell suspensions. These samples were vortexed for 10 s and left to stand at room temperature. To assess cell aggregation, we measured the OD600 of the supernatant using a spectrophotometer (Beckman DU 530; Beckman Coulter, Brea, CA).
Quantitative analysis of polysaccharides.
Soluble and insoluble polysaccharides were isolated from the biofilm formed by the same procedure as in quantitative analysis of eDNA. After the biofilm formed in the 6-well plate, the culture supernatant was collected for quantification of soluble polysaccharides, and the biofilm attached to the bottom of the well was collected with a cell scraper for quantification of insoluble polysaccharides. We quantified the soluble and insoluble polysaccharides by the phenol-sulfuric acid method, as described elsewhere (46).
Confocal laser scanning microscopy.
Biofilms formed in a 96-well plate or on sHAs by the procedure described above were stained with the LIVE/DEAD BacLight bacterial viability kit (Molecular Probes, Leiden, The Netherlands). Complementation of the biofilm morphology was observed by adding 200 μg/ml commercial inulin to TSBr medium before biofilm formation. Syto 9 and propidium iodide (PI) stain bacteria with an intact membrane (live bacteria, green fluorescence) and bacteria with damaged membranes (dead bacteria, red fluorescence), respectively. After the stained biofilm was washed with DW 2 times to remove the excess dye, it was examined by confocal laser scanning microscopy (CLSM) (LSM 700; Carl Zeiss, Jena, Germany). Biofilm images were acquired using a Plan-Apochromat 10×/0.45 M27 objective lens, and a 4,880-nm laser line and a 555-nm laser line for excitation of Syto 9 and PI, respectively. The Zen software (Zeiss) was used for biofilm analysis.
RNA extraction and RT-qPCR.
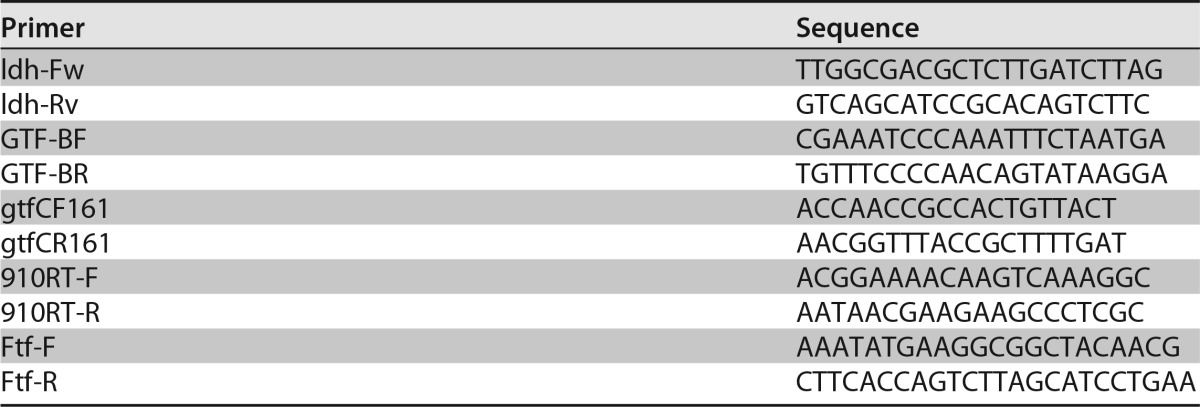
Total RNA was isolated from biofilm cells cultivated for 20 h in TSB, TSBs, TSBg, or TSBr (51). A biofilm was formed in a 6-well culture plate using the same procedure described for the quantification of eDNA. The biofilm was scraped from the bottom of the wells using a cell scraper. The samples were resuspended in cold PBS and sonicated at 2 W for 30 s to separate the extracellular matrix from the cells. The supernatant was removed after centrifugation (5,500 × g, 5 min), and these manipulations (washing and sonication) were repeated 2 times. The pellets were resuspended in 750 μl of NAES buffer (50 mM sodium acetate buffer, 10 mM EDTA, and 1% SDS [pH 5.0]). Then, the samples were transferred to a 2-ml tube containing 0.5 g of 0.5-mm-diameter glass beads. Next, 625 μl of acid phenol and 125 μl of chloroform were added and shaken on a tissue lyzer (Qiagen, Venlo, The Netherlands) for 3 min. After centrifugation (4°C, 14,000 × g, 5 min), the aqueous phase was transferred into a 1.5-ml Eppendorf tube, and 625 μl of acid phenol and 125 μl of chloroform were added (these manipulations were repeated one time). Next, 750 μl of phenol-chloroform-isoamyl alcohol (25:24:1) was added to the resultant sample solution. After centrifugation (4°C, 14,000 × g, 5 min), the aqueous phase was transferred into a 1.5-ml Eppendorf tube, and total RNA was precipitated with 2-propanol. cDNA synthesis was performed using the ReverTra Ace qPCR RT master mix with gDNA remover (Toyobo, Osaka, Japan) using 500 ng of total RNA. Quantitative reverse transcription-PCR (RT-qPCR) was conducted using the ABI Prism 7000 (Applied Biosystems, CA) with the Power SYBR Green PCR master mix (Applied Biosystems, Warrington, UK). We determined the expression level based on the relative quantitative comparative threshold cycle method (ΔΔCT). The cycling conditions were as follows: 2 min of initial denaturation at 95°C, and 40 cycles consisting of 95°C for 15 s and 60°C for 40 s. The primers for RT-qPCR are listed in Table 3. The expression levels were normalized to lactate dehydrogenase mRNA (endogenous control) (52).
TABLE 3.
Primers for real-time PCR
Hydrophobicity assay.
Hydrophobicity on the cell surface was measured by a hexadecane assay, as previously described (53). One hundred microliters of overnight culture was mixed with 4.9 ml of TSB, TSBr, SDMr, or SDM supplemented with 0.0075% (wt/vol) sucrose (which was the same concentration contained in TSB) in culture tubes. These tubes were next incubated at 37°C for 6 h in an aerobic atmosphere containing 5% CO2. The bacterial cells, which were collected by centrifugation (5,500 × g, 5 min) were washed with PBS and sonicated at 2 W for 15 s. These washing and sonication steps were repeated 3 times. After that, the bacterial cells were resuspended in PBS and adjusted to an OD600 of 1.0 in 14-ml polyethylene tubes. Next, 200 μl of n-hexadecane (Wako Pure Chemical Industries, Tokyo, Japan) was added to these samples, and the tubes were vortexed for 2 min. After these samples were incubated at room temperature for 10 min to allow for phase separation, the OD600 of the aqueous phase was measured. The percentage of cell surface hydrophobicity was calculated using the following formula: [1 − (OD600 after vortexing/OD600 before vortexing)] ×100.
Statistical analysis.
The statistical significance of differences between 2 groups was analyzed by Student's t test. All experiments in this study were repeated at least 3 times independently. Data with a P value of <0.05 were considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ryoma Nakao, Nozomu Obana, and Satoru Hirayama for their advice and valuable discussions. We also thank American Journal Experts for English language editing.
This work was supported in part by a grant-in-aid for the development of scientific research (grants 21390506, 24659821, and 16K11537) from the Ministry of Education, Science, and Culture of Japan.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00869-17.
REFERENCES
- 1.Loesche WJ. 1986. Role of Streptococcus mutans in human dental decay. Microbiol Rev 50:353–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krzyściak W, Jurczak A, Koœcielniak D, Bystrowska B, Skalniak A. 2014. The virulence of Streptococcus mutans and the ability to form biofilms. Eur J Clin Microbiol Infect Dis 33:499–515. doi: 10.1007/s10096-013-1993-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aoki H, Shiroza T, Hayakawa M, Sato S, Kuramitsu HK. 1986. Cloning of a Streptococcus mutans glucosyltransferase gene coding for insoluble glucan synthesis. Infect Immun 53:587–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiao J, Klein MI, Falsetta ML, Lu B, Delahunty CM, Yates JR III, Heydorn A, Koo H. 2012. The exopolysaccharide matrix modulates the interaction between 3D architecture and virulence of a mixed-species oral biofilm. PLoS Pathog 8:e1002623. doi: 10.1371/journal.ppat.1002623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanada N, Kuramitsu HK. 1988. Isolation and characterization of the Streptococcus mutans gtfC gene, coding for synthesis of both soluble and insoluble glucans. Infect Immun 56:1999–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanada N, Kuramitsu HK. 1989. Isolation and characterization of the Streptococcus mutans gtfD gene, coding for primer-dependent soluble glucan synthesis. Infect Immun 57:2079–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Critchley P, Wood JM, Saxton CA, Leach SA. 1967. The polymerisation of dietary sugars by dental plaque. Caries Res 1:112–129. doi: 10.1159/000259506. [DOI] [PubMed] [Google Scholar]
- 8.Burne RA. 1991. Oral ecological disaster: the role of short-term extracellular storage polysaccharides, p 351–364. In Bowen WH, Tabak LA (ed), Cariology for the nineties. University of Rochester Press, Rochester, NY. [Google Scholar]
- 9.Rozen R, Steinberg D, Bachrach G. 2004. Streptococcus mutans fructosyltransferase interactions with glucans. FEMS Microbiol Lett 232:39–43. doi: 10.1016/S0378-1097(04)00065-5. [DOI] [PubMed] [Google Scholar]
- 10.Das T, Sharma PK, Busscher HJ, van der Mei HC, Krom BP. 2010. Role of extracellular DNA in initial bacterial adhesion and surface aggregation. Appl Environ Microbiol 76:3405–3408. doi: 10.1128/AEM.03119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das T, Sehar S, Manefield M. 2013. The roles of extracellular DNA in the structural integrity of extracellular polymeric substance and bacterial biofilm development. Environ Microbiol Rep 5:778–786. doi: 10.1111/1758-2229.12085. [DOI] [PubMed] [Google Scholar]
- 12.Liao S, Klein MI, Heim KP, Fan Y, Bitoun JP, Ahn SJ, Burne RA, Koo H, Brady LJ, Wen ZT. 2014. Streptococcus mutans extracellular DNA is upregulated during growth in biofilms, actively released via membrane vesicles, and influenced by components of the protein secretion machinery. J Bacteriol 196:2355–2366. doi: 10.1128/JB.01493-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perry JA, Cvitkovitch DG, Lévesque CM. 2009. Cell death in Streptococcus mutans biofilms: a link between CSP and extracellular DNA. FEMS Microbiol Lett 299:261–266. doi: 10.1111/j.1574-6968.2009.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perry JA, Jones MB, Peterson SN, Cvitkovitch DG, Lévesque CM. 2009. Peptide alarmone signalling triggers an auto-active bacteriocin necessary for genetic competence. Mol Microbiol 72:905–917. doi: 10.1111/j.1365-2958.2009.06693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dagkessamanskaia A, Moscoso M, Hénard V, Guiral S, Overweg K, Reuter M, Martin B, Wells J, Claverys JP. 2004. Interconnection of competence, stress and CiaR regulons in Streptococcus pneumoniae: competence triggers stationary phase autolysis of ciaR mutant cells. Mol Microbiol 51:1071–1086. doi: 10.1111/j.1365-2958.2003.03892.x. [DOI] [PubMed] [Google Scholar]
- 16.Liu Y, Burne RA. 2011. The major autolysin of Streptococcus gordonii is subject to complex regulation and modulates stress tolerance, biofilm formation, and extracellular-DNA release. J Bacteriol 193:2826–2837. doi: 10.1128/JB.00056-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahn SJ, Wen ZT, Burne RA. 2006. Multilevel control of competence development and stress tolerance in Streptococcus mutans UA159. Infect Immun 74:1631–1642. doi: 10.1128/IAI.74.3.1631-1642.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reck M, Tomasch J, Wagner-Döbler I. 2015. The alternative sigma factor SigX controls bacteriocin synthesis and competence, the two quorum sensing regulated traits in Streptococcus mutans. PLoS Genet 11:e1005353. doi: 10.1371/journal.pgen.1005353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li YH, Tang N, Aspiras MB, Lau PC, Lee JH, Ellen RP, Cvitkovitch DG. 2002. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J Bacteriol 184:2699–2708. doi: 10.1128/JB.184.10.2699-2708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ajdić D, McShan WM, McLaughlin RE, Savić G, Chang J, Carson MB, Primeaux C, Tian R, Kenton S, Jia H, Lin S, Qian Y, Li S, Zhu H, Najar F, Lai H, White J, Roe BA, Ferretti JJ. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci U S A 99:14434–14439. doi: 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benno Y, Endo K, Shiragami N, Sayama K, Mitsuoka T. 1987. Effects of raffinose intake on human fecal microflora. Bifidobacteria Microflora 6:59–63. doi: 10.12938/bifidus1982.6.2_59. [DOI] [Google Scholar]
- 22.Song DD, Jacques NA. 1999. Purification and enzymic properties of the fructosyltransferase of Streptococcus salivarius ATCC 25975. Biochem J 341:285–291. doi: 10.1042/0264-6021:3410285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Russell RR, Aduse-Opoku J, Sutcliffe IC, Tao L, Ferretti JJ. 1992. A binding protein-dependent transport system in Streptococcus mutans responsible for multiple sugar metabolism. J Biol Chem 267:4631–4637. [PubMed] [Google Scholar]
- 24.Benigar E, Zupančič Valant A, Dogsa I, Sretenovic S, Stopar D, Jamnik A, Tomšič M. 2016. Structure and dynamics of a model polymer mixture mimicking a levan-based bacterial biofilm of Bacillus subtilis. Langmuir 32:8182–8194. doi: 10.1021/acs.langmuir.6b02041. [DOI] [PubMed] [Google Scholar]
- 25.Zita A, Hermansson M. 1997. Effects of bacterial cell surface structures and hydrophobicity on attachment to activated sludge flocs. Appl Environ Microbiol 63:1168–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goulter RM, Gentle IR, Dykes GA. 2009. Issues in determining factors influencing bacterial attachment: a review using the attachment of Escherichia coli to abiotic surfaces as an example. Lett Appl Microbiol 49:1–7. doi: 10.1111/j.1472-765X.2009.02591.x. [DOI] [PubMed] [Google Scholar]
- 27.Shields RC, Burne RA. 2016. Growth of Streptococcus mutans in biofilms alters peptide signaling at the sub-population level. Front Microbiol 7:1075. doi: 10.3389/fmicb.2016.01075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moye ZD, Son M, Rosa-Alberty AE, Zeng L, Ahn SJ, Hagen SJ, Burne RA. 2016. Effects of carbohydrate source on genetic competence in Streptococcus mutans. Appl Environ Microbiol 82:4821–4834. doi: 10.1128/AEM.01205-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petersen FC, Tao L, Scheie AA. 2005. DNA binding-uptake system: a link between cell-to-cell communication and biofilm formation. J Bacteriol 187:4392–4400. doi: 10.1128/JB.187.13.4392-4400.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Velázquez-Hernández ML, Baizabal-Aguirre VM, Bravo-Patiño A, Cajero-Juárez M, Chávez-Moctezuma MP, Valdez-Alarcón JJ. 2009. Microbial fructosyltransferases and the role of fructans. J Appl Microbiol 106:1763–1778. doi: 10.1111/j.1365-2672.2008.04120.x. [DOI] [PubMed] [Google Scholar]
- 31.Hamada S, Slade HD. 1980. Biology, immunology, and cariogenicity of Streptococcus mutans. Microbiol Rev 44:331–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schroeder VA, Michalek SM, Macrina FL. 1989. Biochemical characterization and evaluation of virulence of a fructosyltransferase-deficient mutant of Streptococcus mutans V403. Infect Immun 57:3560–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wexler DL, Penders JE, Bowen WH, Burne RA. 1992. Characteristics and cariogenicity of a fructanase-defective Streptococcus mutans strain. Infect Immun 60:3673–3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rozen R, Bachrach G, Bronshteyn M, Gedalia I, Steinberg D. 2001. The role of fructans on dental biofilm formation by Streptococcus sobrinus, Streptococcus mutans, Streptococcus gordonii and Actinomyces viscosus. FEMS Microbiol Lett 195:205–210. doi: 10.1111/j.1574-6968.2001.tb10522.x. [DOI] [PubMed] [Google Scholar]
- 35.Watanabe H, Sonoyama K, Watanabe J, Yamaguchi N, Kikuchi H, Nagura T, Aritsuka T, Fukumoto K, Kasai T. 2004. Reduction of allergic airway eosinophilia by dietary raffinose in Brown Norway rats. Br J Nutr 92:247–255. doi: 10.1079/BJN20041179. [DOI] [PubMed] [Google Scholar]
- 36.Kim HS, Cha E, Kim Y, Jeon YH, Olson BH, Byun Y, Park HD. 2016. Raffinose, a plant galactoside, inhibits Pseudomonas aeruginosa biofilm formation via binding to LecA and decreasing cellular cyclic diguanylate levels. Sci Rep 6:25318. doi: 10.1038/srep25318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kulshrestha S, Tyagi P, Sindhi V, Yadavilli KS. 2013. Invertase and its applications–a brief review. J Pharm Res 7:792–797. [Google Scholar]
- 38.Banguela A, Hernández L. 2006. Fructans: from natural sources to transgenic plants. Biotecnol Aplic 23:202–210. [Google Scholar]
- 39.Tanzer JM, Brown AT, McInerney MF. 1973. Identification, preliminary characterization, and evidence for regulation of invertase in Streptococcus mutans. J Bacteriol 116:192–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Narisawa N, Kawarai T, Suzuki N, Sato Y, Ochiai K, Ohnishi M, Watanabe H, Senpuku H. 2011. Competence-dependent endogenous DNA rearrangement and uptake of extracellular DNA give a natural variant of Streptococcus mutans without biofilm formation. J Bacteriol 193:5147–5154. doi: 10.1128/JB.05240-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Forloni M, Liu AY, Wajapeyee N. 2001. Methods for in vitro mutagenesis, p1080–1087. In Green MR, Sambrook J (ed). Molecular cloning: a laboratory manual, 3rd ed, vol 2 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 42.Shiroza T, Kuramitsu HK. 1993. Construction of a model secretion system for oral streptococci. Infect Immun 61:3745–3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Motegi M, Takagi Y, Yonezawa H, Hanada N, Terajima J, Watanabe H, Senpuku H. 2006. Assessment of genes associated with Streptococcus mutans biofilm morphology. Appl Environ Microbiol 72:6277–6287. doi: 10.1128/AEM.00614-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tamura S, Yonezawa H, Motegi M, Nakao R, Yoneda S, Watanabe H, Yamazaki T, Senpuku H. 2009. Inhibiting effects of Streptococcus salivarius on competence-stimulating peptide-dependent biofilm formation by Streptococcus mutans. Oral Microbiol Immunol 24:152–161. doi: 10.1111/j.1399-302X.2008.00489.x. [DOI] [PubMed] [Google Scholar]
- 45.Lemos JA, Abranches J, Koo H, Marquis RE, Burne RA. 2010. Protocols to study the physiology of oral biofilms. Methods Mol Biol 666:87–102. doi: 10.1007/978-1-60761-820-1_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kawarai T, Narisawa N, Suzuki Y, Nagasawa R, Senpuku H. 2016. Streptococcus mutans biofilm formation is dependent on extracellular DNA in primary low pH conditions. J Oral Biosci 58:55–61. doi: 10.1016/j.job.2015.12.004. [DOI] [Google Scholar]
- 47.Corinaldesi C, Danovaro R, Dell'Anno A. 2005. Simultaneous recovery of extracellular and intracellular DNA suitable for molecular studies from marine sediments. Appl Environ Microbiol 71:46–50. doi: 10.1128/AEM.71.1.46-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rose SJ, Babrak LM, Bermudez LE. 2015. Mycobacterium avium possesses extracellular DNA that contributes to biofilm formation, structural integrity, and tolerance to antibiotics. PLoS One 10:e0128772. doi: 10.1371/journal.pone.0128772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Y, Caufield PW, Emanuelsson IR, Thornqvist E. 2001. Differentiation of Streptococcus mutans and Streptococcus sobrinus via genotypic and phenotypic profiles from three different populations. Oral Microbiol Immunol 16:16–23. doi: 10.1034/j.1399-302x.2001.160103.x. [DOI] [PubMed] [Google Scholar]
- 50.Truong TL, Ménard C, Mouton C, Trahan L. 2000. Identification of mutans and other oral streptococci by random amplified polymorphic DNA analysis. J Med Microbiol 49:63–71. doi: 10.1099/0022-1317-49-1-63. [DOI] [PubMed] [Google Scholar]
- 51.Cury JA, Seils J, Koo H. 2008. Isolation and purification of total RNA from Streptococcus mutans in suspension cultures and biofilms. Braz Oral Res 22:216–222. doi: 10.1590/S1806-83242008000300005. [DOI] [PubMed] [Google Scholar]
- 52.Merritt J, Kreth J, Qi F, Sullivan R, Shi W. 2005. Non-disruptive, real-time analyses of the metabolic status and viability of Streptococcus mutans cells in response to antimicrobial treatments. J Microbiol Methods 61:161–170. doi: 10.1016/j.mimet.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 53.Nakao R, Ramstedt M, Wai SN, Uhlin BE. 2012. Enhanced biofilm formation by Escherichia coli LPS mutants defective in Hep biosynthesis. PLoS One 7:e51241. doi: 10.1371/journal.pone.0051241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vieira J, Messing J. 1987. Production of single-stranded plasmid DNA. Methods Enzymol 153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- 55.Dunny GM, Lee LN, LeBlanc DJ. 1991. Improved electroporation and cloning vector system for Gram-positive bacteria. Appl Environ Microbiol 57:1194–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.LeBlanc DJ, Lee LN, Abu-Al-Jaibat A. 1992. Molecular, genetic, and functional analysis of the basic replicon of pVA380-1, a plasmid of oral streptococcal origin. Plasmid 28:130–145. doi: 10.1016/0147-619X(92)90044-B. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.