ABSTRACT
Calderihabitans maritimus KKC1 is a thermophilic, hydrogenogenic carboxydotroph isolated from a submerged marine caldera. Here, we describe the de novo sequencing and feature analysis of the C. maritimus KKC1 genome. Genome-based phylogenetic analysis confirmed that C. maritimus KKC1 was most closely related to the genus Moorella, which includes well-studied acetogenic members. Comparative genomic analysis revealed that, like Moorella, C. maritimus KKC1 retained both the CO2-reducing Wood-Ljungdahl pathway and energy-converting hydrogenase-based module activated by reduced ferredoxin, but it lacked the HydABC and NfnAB electron-bifurcating enzymes and pyruvate:ferredoxin oxidoreductase required for ferredoxin reduction for acetogenic growth. Furthermore, C. maritimus KKC1 harbored six genes encoding CooS, a catalytic subunit of the anaerobic CO dehydrogenase that can reduce ferredoxin via CO oxidation, whereas Moorella possessed only two CooS genes. Our analysis revealed that three cooS genes formed known gene clusters in other microorganisms, i.e., cooS-acetyl coenzyme A (acetyl-CoA) synthase (which contained a frameshift mutation), cooS–energy-converting hydrogenase, and cooF-cooS-FAD-NAD oxidoreductase, while the other three had novel genomic contexts. Sequence composition analysis indicated that these cooS genes likely evolved from a common ancestor. Collectively, these data suggest that C. maritimus KKC1 may be highly dependent on CO as a low-potential electron donor to directly reduce ferredoxin and may be more suited to carboxydotrophic growth compared to the acetogenic growth observed in Moorella, which show adaptation at a thermodynamic limit.
IMPORTANCE Calderihabitans maritimus KKC1 and members of the genus Moorella are phylogenetically related but physiologically distinct. The former is a hydrogenogenic carboxydotroph that can grow on carbon monoxide (CO) with H2 production, whereas the latter include acetogenic bacteria that grow on H2 plus CO2 with acetate production. Both species may require reduced ferredoxin as an actual “energy equivalent,” but ferredoxin is a low-potential electron carrier and requires a high-energy substrate as an electron donor for reduction. Comparative genomic analysis revealed that C. maritimus KKC1 lacked specific electron-bifurcating enzymes and possessed six CO dehydrogenases, unlike Moorella species. This suggests that C. maritimus KKC1 may be more dependent on CO, a strong electron donor that can directly reduce ferredoxin via CO dehydrogenase, and may exhibit a survival strategy different from that of acetogenic Moorella, which solves the energetic barrier associated with endergonic reduction of ferredoxin with hydrogen.
KEYWORDS: Wood-Ljungdahl pathway, acetogen, carbon monoxide dehydrogenase, carboxydotroph, electron bifurcation, genome, hydrogenogen
INTRODUCTION
Carbon monoxide (CO) is a potent electron donor (1) that can serve as an energy and carbon source for thermophilic carboxydotrophs (CO-oxidizing microbes) (2). CO utilization requires specific carbon monoxide dehydrogenases (CODHs) to catalyze the reversible reaction CO + H2O ⇔ CO2 + 2H+ + 2e− (3, 4). CODHs from anaerobic microbes possess a nickel-containing reaction center (Ni-CODHs) (5, 6), whereas aerobic-type CODHs contain a highly conserved molybdenum-based active site. From thermodynamic considerations, CO oxidation can be coupled to the reduction of most redox-active cofactors (7). A number of diverse physiological anaerobic carboxydotrophs and CO oxidizers have been described, such as acetogens, methanogens, sulfate reducers, iron reducers, and hydrogenogens (8), many of which possess multiple Ni-CODH genes (9–11). Ni-CODHs are subdivided into the Cdh type, almost all of which are found in archaea, and the CooS type, which are more frequent in bacteria (10). While CooS-type CODHs contain one [Ni-Fe-S] cluster (C cluster) and two [4Fe-4S] clusters (B and D clusters), Cdh types harbor two additional [4Fe-4S] clusters (E and F clusters) (12) and generally show relatively low homology to CooS-type CODHs.
The functions of CODHs have often been predicted from other genes located in close proximity to CODH genes (genomic context) (10). CODHs can be divided into four functional groups according to their genomic context (10): (i) within an acetyl coenzyme A (acetyl-CoA) synthase (ACS) gene cluster, (ii) adjacent to an energy-converting hydrogenase (ECH) gene cluster, (iii) adjacent to a ferredoxin-like electron transfer Fe-S protein (CooF) gene but not an ECH gene cluster, and (iv) other than types I to iii. CODHs in category i form CODH/ACS complexes that catalyze the reduction of CO2 to CO and acetyl-CoA synthesis in the final step of the Wood-Ljungdahl pathway (3). These complexes are widespread in CO-oxidizing and non-CO-oxidizing anaerobes that employ the Wood-Ljungdahl pathway, such as acetogens (13, 14), methanogens (15–17), sulfate reducers (18, 19), and thermophilic hydrogenogenic carboxydotrophs (6). Cdh-type CODHs fall exclusively in the type i category, while type ii CODH genes cluster with those encoding ECH, whose presence is therefore considered a feature of hydrogenogenic carboxydotrophs that oxidize CO to produce CO2 and hydrogen gas (CO + H2O → CO2 + H2) (11). ECH is a membrane-associated, H2-evolving enzyme that requires CooF or ferredoxin as an electron donor and stores energy by proton translocation (20). Two types of cooS-ECH gene clusters are known in bacteria. One corresponds to the coo (CO-oxidizing) gene cluster found in Carboxydothermus hydrogenoformans and Rhodospirillum rubrum (9, 21) and includes CO-induced hydrogenase genes. The second is found in Caldanaerobacter subterraneus subspecies (22) and clusters with the hyf/hyc-type ECH genes long known as the hydrogenase module of formate hydrogen lyase complexes. Moreover, cooS genes of type iii are believed to encode an Ni-CODH responsible for generating electrons during CO oxidation and transferring them to CooF, which in turn relays them to various redox reactions. Members of group iv are “lone” cooS genes, in that they are not found in a genomic context with known CO metabolism-related genes.
Some thermophilic hydrogenogenic carboxydotrophs, such as C. hydrogenoformans and C. subterraneus subsp. pacificus, can propagate on high concentrations of CO as the sole carbon and energy source (7). Thermophilic hydrogenogenic carboxydotrophs have been studied extensively as models of CO metabolism, and a genomic study revealed that C. hydrogenoformans possessed five distinct cooS genes, one each of types i, ii, and iv and two of type iii (9). In contrast, the genome of C. subterraneus subsp. pacificus includes only one CooS gene cluster of type ii (22). As mentioned above, type ii cooS gene clusters in both organisms are distinct even though they exhibit physiology similar to that of thermophilic hydrogenogenic carboxydotrophs. Thus, the presence of highly divergent CooS gene cluster combinations prompts fundamental questions on their function, evolution, and origin.
Here, we describe the de novo sequencing and feature analysis of the Calderihabitans maritimus KKC1 genome. C. maritimus KKC1 is a hydrogenogenic carboxydotrophic thermophile isolated from a sediment core sample taken from a submerged marine caldera (23). According to 16S rRNA phylogenetic analysis, C. maritimus KKC1 belongs to the family Thermoanaerobacteraceae and the phylum Firmicutes and is most closely related to members of the genus Moorella (23). Although Moorella stamsii and Moorella thermoacetica strain AMP are reported to be hydrogenogenic carboxydotrophs like C. maritimus KKC1 (24, 25), most Moorella strains are known homoacetogens. M. thermoacetica is the type species for the genus and is a well-known model of acetogenic bacteria that can grow autotrophically using H2 plus CO2 or CO to produce acetate via the Wood-Ljungdahl pathway (1, 26). We compared the overall genomic features of C. maritimus KKC1 (a hydrogenogenic carboxydotroph) to those of acetogenic M. thermoacetica ATCC 39073 and Moorella perchloratireducens An10 and analyzed CODH gene clusters to gain insight into the physiological and phylogenetic differences between C. maritimus KKC1 and Moorella groups.
RESULTS
General features of the C. maritimus KKC1 genome and subsequent phylogenetic analysis.
Overall, draft assemblies of the C. maritimus KKC1 genome yielded 223 contigs with an average GC content of 47%. The draft genome was approximately 3.1 Mbp, and a total of 3,509 coding sequences (CDSs) were identified (Table 1). C. maritimus KKC1 possessed a single copy of 16S and 23S rRNA genes, two 5S rRNA genes (each of which was on different contigs), and a total of 48 tRNA genes coding for all 20 amino acids.
TABLE 1.
General features of the genomes from C. maritimus KKC1, M. thermoacetica, and M. perchloratireducens
| Parameter | Value for: |
||
|---|---|---|---|
| C. maritimus KKC1 | M. thermoacetica ATCC 39073 (NC_007644.1) | M. perchloratireducens An10 (2506520025) | |
| Genome size (bp) | 3,064,849 | 2,628,784 | 3,307,499 |
| G+C content (%) | 47 | 55.8 | 53.8 |
| No. of: | |||
| CDSs | 3,509 | 2,463 | 3,349 |
| rRNAs | 4 | 3 | 3 |
| tRNAs | 48 | 51 | 52 |
| No. (%) of genes in COG | 2,287 (65.2) | 1,953 (79.3) | 2,518 (75.2) |
| No. of contigs | 223 | 1 | 133 |
| Source | This study | RefSeq | IMG |
C. maritimus KKC1 can grow heterotrophically on pyruvate, lactate, fumarate, glucose, fructose, and mannose with thiosulfate as an electron acceptor under an N2 atmosphere (23). Metabolic pathways predicted by Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis revealed that the C. maritimus KKC1 genome encoded a complete glycolytic pathway and an incomplete tricarboxylic acid (TCA) cycle that lacked citrate synthase (present in Moorella species) and malate dehydrogenase (see Fig. S2 in the supplemental material). It also possessed one gene encoding a NAD-dependent malic enzyme (EC 1.1.1.38) (Fig. S2), which is responsible for linking the TCA cycle to glycolysis by catalyzing the interconversion of malate and pyruvate (27). In addition, C. maritimus KKC1 maintained the fructose utilization pathway driven by the phosphoenolpyruvate-dependent phosphotransferase system (28) and l-lactate dehydrogenase (Fig. S2). Therefore, we suggest that when C. maritimus KKC1 utilizes lactate, fumarate, and fructose, these compounds are converted into pyruvate. Pathways for mannose metabolism were not predicted by our analysis of the C. maritimus KKC1 genome; hence, the underlying mechanism remains unclear. C. maritimus KKC1 utilized the Wood-Ljungdahl pathway for autotrophy, but genes encoding key enzymes for other known carbon fixation pathways, such as RuBisCO and 4-hydroxybutyryl-CoA dehydratase, were not found.
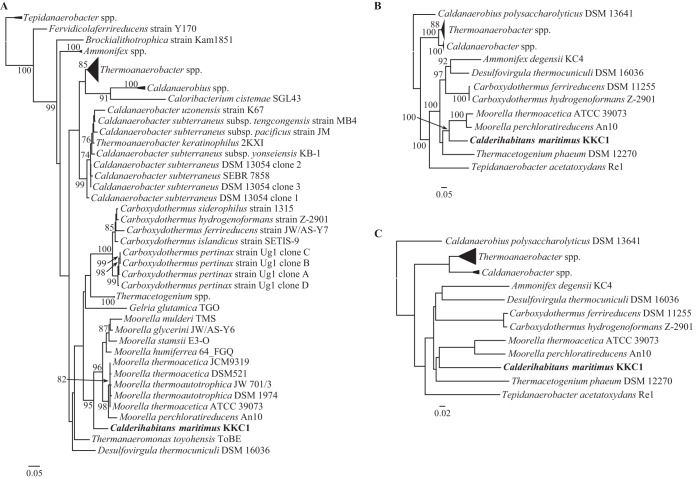
As reported previously, the C. maritimus KKC1 is most closely related to Moorella species on the basis of 16S rRNA phylogenetic analysis (23) (Fig. 1A). The C. maritimus KKC1 genome showed a relatively low GC content (47%) compared to those of M. thermoacetica and M. perchloratireducens (55.8 and 53.8%, respectively). We conducted phylogenetic analyses based on five housekeeping genes and the genomic similarity score (GSS), which confirmed that Moorella species were the most closely related to C. maritimus KKC1 (Fig. 1B and C). In both phylogenetic trees, the sister group of the C. maritimus KKC1 and Moorella clades included known hydrogenogenic carboxydotrophs, such as Carboxydothermus. This was particularly true of the maximum-likelihood (ML) tree of housekeeping genes, as indicated by the high bootstrap replica value.
FIG 1.
Phylogenetic reconstruction of Thermoanaerobacteraceae. (A) Maximum-likelihood (ML) phylogenetic analysis using 16S rRNA. (B) ML phylogenetic analysis of five concatenated housekeeping genes. Only bootstrap support values (out of 100 runs) greater than or equal to 70 are shown in both panels A and B. (C) Genomic similarity score (GSS) distance matrix plotted as a neighbor-joining tree. Calderihabitans maritimus KKC1 is indicated in bold font.
Genomic comparison between C. maritimus KKC1 and Moorella species.
Recent studies revealed that M. thermoacetica is an “ECH-acetogen” (29) that utilizes two metabolic modules, the CO2-reducing Wood-Ljungdahl pathway and the energy-conserving ECH-based module energized by reduced ferredoxin. M. thermoacetica possesses HydABC and NfnAB, which catalyzes the endergonic reduction of low-potential ferredoxin with H2 by flavin-based electron bifurcation (29). It also possesses pyruvate:ferredoxin oxidoreductases (PFORs) or CODHs that generate reduced ferredoxin, an actual “energy equivalent,” by pyruvate or CO oxidation, respectively (29, 30), and can utilize versatile energy sources in acetogenic growth (31). M. perchloratireducens can grow on CO, methanol, pyruvate, glucose, fructose, cellobiose, mannose, xylose, and pectin, but no growth is observed on H2 plus CO2 (32). The products from substrate utilization are acetate, CO2, and H2. On the other hand, C. maritimus KKC1 is a hydrogenogenic carboxydotroph that can grow on CO with production of H2 in a medium containing ferric citrate (10 mg/liter) as the only organic compound. Acetogenic growth on H2 plus CO2 has not been observed in C. maritimus KKC1. It can grow heterotrophically on pyruvate, lactate, fumarate, glucose, fructose, and mannose with thiosulfate as an electron acceptor under an N2 atmosphere but not without any electron acceptors (23). We performed a functional classification of open reading frames (ORFs) from C. maritimus KKC1, M. thermoacetica, and M. perchloratireducens by BLAST search against clusters of orthologous groups (COGs) (see Fig. S1 in the supplemental material). The number of ORFs assigned to COG categories related to central metabolic pathways (C, energy production and conversion; E, amino acid transport and metabolism; G, carbohydrate transport and metabolism) varied substantially between C. maritimus KKC1 and Moorella species, as described below.
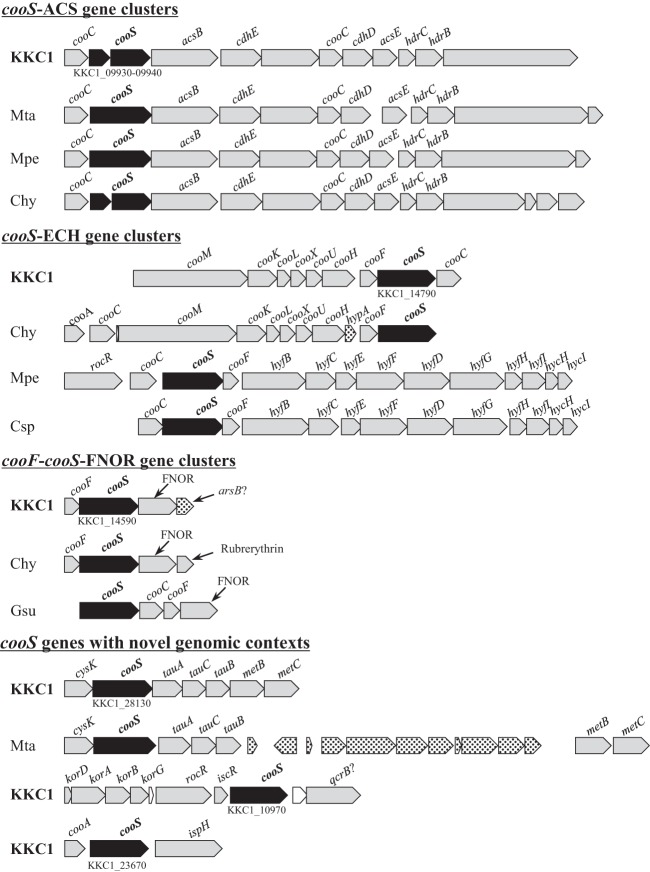
While each Moorella species possessed one hyc/hyf-type ECH gene cluster, the C. maritimus KKC1 genome contained two ECH complexes: one coo-type ECH (forming cooS-ECH) and one hyc/hyf-type ECH clustered with a formate dehydrogenase gene (fdoG). The structures of the hyc/hyf-type ECH gene clusters from C. maritimus KKC1, M. thermoacetica, and M. perchloratireducens were very similar, but only the hyf/hyc-type ECH from M. perchloratireducens lacked a formate dehydrogenase gene and clustered with cooS (Fig. 2; see Fig. S5 in the supplemental material). The C. maritimus KKC1 genome conserved a complete Wood-Ljungdahl pathway, like M. thermoacetica (although cooS within the ACS gene cluster was frameshifted, as mentioned below), while M. perchloratireducens lacked formate dehydrogenase (Fdh), which catalyzes the first CO2 fixation step in the Wood-Ljungdahl pathway (Fig. S2). Unlike for Moorella species, no HydABC and NfnAB homologs were found in the C. maritimus KKC1 genome, consistent with its failure of acetogenic growth on H2 and CO2 (23). The authentic PFOR of M. thermoacetica is encoded in Moth_0064 and contains three domains, the α, γ, and β subunits, annotated as COG0674, COG1014, and COG1013, respectively (31). A Moth_0064 homolog was found in the M. perchloratireducens genome but not in that of C. maritimus KKC1 (see Table S1 in the supplemental material). Although C. maritimus KKC1 possessed six sets of genes annotated as COG0674, COG1014, and COG1013, these were more similar to 2-oxoglutarate (α-ketoglutarate):ferredoxin oxidoreductase (KFOR) than to PFOR, according to KEGG orthology annotation.
FIG 2.
Schematic representation of CooS gene clusters from C. maritimus KKC1, Moorella thermoacetica, Moorella perchloratireducens, Caldanaerobacter subterraneus subsp. pacificus, and Carboxydothermus hydrogenoformans. KKC1, C. maritimus; Mta, M. thermoacetica; Mpe, M. perchloratireducens; Csp, C. subterraneus subsp. pacificus; Chy, C. hydrogenoformans. Black, cooS; dots, inserted genes; gray, other functional proteins.
Remarkably, C. maritimus KKC1 harbored six CooS genes with conserved residues linked to metal clusters in its genome (3, 5) (see Fig. S3 in the supplemental material). Functional types of the six CooS genes were affiliated to each of types i, ii, and iii and three of type iv, although cooS within the cooS-ACS type i was frameshifted. We also detected the simultaneous transcription of all six CooS genes in C. maritimus KKC1 during carboxydotrophic growth by reverse transcription-PCR (RT-PCR) (data not shown). Moreover, both Moorella species possessed only two cooS clusters (Fig. 2), one engaged in the Wood-Ljungdahl pathway (i.e., type i CODH). The other cooS clusters were types iv and ii in M. thermoacetica and M. perchloratireducens, respectively. We discuss the details of the CooSs in the following sections.
Genomic contexts of the six CooS genes in C. maritimus KKC1.
Of the six CooS genes from C. maritimus KKC1, three presented already-known genomic contexts in other microorganisms: the type i cooS-ACS, type ii cooS-ECH, and type iii cooF-cooS-FNOR gene clusters. These are almost identical to the cooS-I, III, and IV clusters of C. hydrogenoformans, respectively (9), but with some variation (Fig. 2). The sequence of the cooS gene within the cooS-ACS gene cluster in C. maritimus KKC1 was split into two ORFs (KKC1_09930 and KKC1_09940) owing to a frameshift. The cooS-ECH gene cluster of C. maritimus KKC1 was a coo type. However, unlike in C. hydrogenoformans, a homolog of cooA, encoding a heme-containing regulator of the coo operon, was not found upstream of the ECH gene cluster (cooMKLXUH) (Fig. 2). The cooS-IV gene (type iii) from C. hydrogenoformans forms an operon with cooF and the genes encoding FAD-NAD oxidoreductase (FNOR) and rubrerythrin-like protein, which is thought to play a role in reactive oxygen species detoxification (9). In C. maritimus KKC1, cooS (KKC1_14590) in the cooF-cooS-FNOR gene cluster lacked a gene encoding rubrerythrin (Fig. 2). Similar gene clusters consisting of sequential genes putatively coding for CooS, CooF, and FNOR have been found in some sulfate reducers (e.g., Geobacter sulfurreducens), thermophilic fermenting bacteria, and Clostridium species (33) (Fig. 2).
The other three cooS genes of C. maritimus KKC1 (KKC1_28130, KKC1_10970, and KKC1_23670) were found in novel genomic contexts. The cooS gene in KKC1_28130 was associated with those encoding cysteine synthase A (CysK), a putative ABC transport system with domains similar to those of TauABC, cystathionine γ-synthase (MetB), and β-lyase (MetC) (Fig. 2). The genomic context of KKC1_28130 was similar to that of type iv cooS genes found in M. thermoacetica (Fig. 2). CysK catalyzes the formation of l-cysteine and acetate from O-acetyl-l-serine and sulfide (34). TauABC is required for the utilization of taurine as an organic sulfur source when inorganic sulfur is not available (35). MetB and MetC catalyze consecutive trans-sulfuration reactions in the biosynthesis of methionine (36). Three copies of cysK and two copies of TauABC genes were found in the genome of C. maritimus KKC1, whereas metBC was found only in the proximity of KKC1_28130.
The cooS gene in KKC1_10970 clustered with those encoding the KFOR δ, α, β, and γ subunits (KorDABG; KKC1_10900 to KKC1_10930), which are one of the six sets of genes putatively encoding KFOR as described above (Table S1), and two putative transcriptional regulators (RocR and IscR; KKC1_10940 and KKC1_10950). KFOR is a TCA cycle-related enzyme that catalyzes the oxidative decarboxylation of 2-oxoglutarate and the reverse reaction (succinyl-CoA carboxylation) in autotrophic bacteria that fix CO2 by the reductive TCA (RTCA) cycle (37, 38).
The cooS gene in KKC1_23670 clustered with those encoding CooA (KKC1_23660) and 4-hydroxy-3-methylbut-2-enyl diphosphate reductase (IspH; KKC1_23680). In the C. maritimus KKC1 genome, KKC1_23660 was the sole cooA homolog that conserves the His-82 residue (the axial ligands of the Fe[III] and Fe[II] hemes) in CooA from C. hydrogenoformans (39). When searching the upstream regions of CooS genes, we identified CooA-binding sites (5′-TGTCA-N6-CGACA) previously reported in R. rubrum (40), 95 bp and 85 bp upstream of the CooA gene (KKC1_23660) and cooS-ECH gene cluster (KKC1_14720-800), respectively. IspH catalyzes the terminal step of the nonmevalonate route, a biosynthetic pathway for isopentenyl diphosphate and dimethylallyl diphosphate, which are universal precursors for all isoprenoids or terpenes (e.g., steroids and carotenoids) in living organisms (41, 42). In particular, quinones in the electron transport chain, such as ubiquinone and menaquinone, or polyprenols, including the carbohydrate carrier bactoprenol from eubacteria, represent ubiquitous bacterial isoprenoids (43).
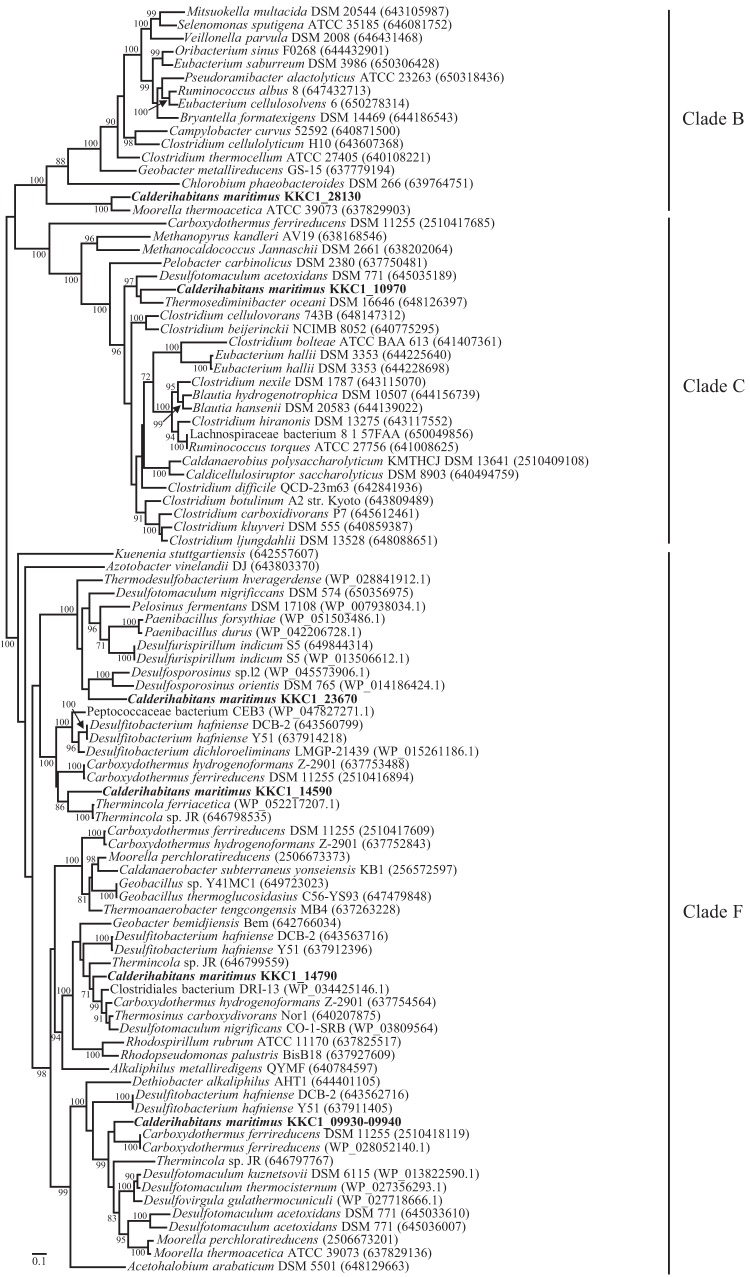
Phylogenetic analysis of CooSs.
Comprehensive phylogenetic analysis of CODH genes revealed the presence of six distinct clades (10). Following previously described criteria (10), CooSs encoded in KKC1_09930-40 (from cooS-ACS), KKC1_14790 (from cooS-ECH), KKC1_14590 (from cooF-cooS-FNOR), and KKC1_23670 (in the proximity of cooA and ispH) were classified as clade F (Fig. 3). In contrast, CooSs encoded in KKC1_28130 (in the proximity of cysK, tauACB, and metBC) and KKC1_10970 (in the proximity of korDABG) were classified as clade B and clade C, respectively.
FIG 3.
ML phylogenetic tree of CooSs. CooSs from C. maritimus KKC1 are indicated in bold font. Only bootstrap support values (out of 100 runs) equal to or greater than 70 are shown.
Clade F CooSs encoded within the cooS-ACS, cooS-ECH, and cooF-cooS-FNOR gene clusters from C. maritimus KKC1 showed 71%, 82%, and 68% identity with respect to their counterparts in Desulfotomaculum kuznetsovii (WP_013822590.1), Thermosinus carboxydivorans (WP_007288856.1), and Thermincola potens (WP_013120796.1), respectively, and formed subclades with each one from C. hydrogenoformans. However, the CooS encoded in KKC1_23670 did not form a subclade with Thermoanaerobacteraceae and instead formed a subclade together with Thermodesulfobacterium (phylum Thermodesulfobacteria), Desulfotomaculum, Desulfosporosinus (order Clostridiales, phylum Firmicutes), Desulfurispirillum (phylum Chrysiogenetes), Paenibacillus (class Bacilli, phylum Firmicutes), and Pelosinus (class Negativicutes, phylum Firmicutes). The clade B CooS (encoded in KKC1_28130) was phylogenetically close to the type iv cooS of M. thermoacetica (76% identity), which presented a similar genomic context (Fig. 2), forming the most deeply branched members of clade B (Fig. 3) (10). Clade C CooS (encoded in KKC1_10970) had 70% identity with counterparts from Desulfotomaculum acetoxidans (WP_015758381.1). Both CooSs were phylogenetically distinct from those from members of Thermoanaerobacteraceae.
Type i CooS genes from M. thermoacetica and M. perchloratireducens clustered in the same subclade in clade F, which is constituted with only type i CooS genes (Fig. 3). The type ii CooS gene (Integrated Microbial Genomes [IMG] Gene ID 2506673373) clustered with the hyf/hyc-type ECH gene cluster in M. perchloratireducens was phylogenetically distinct from those of M. thermoacetica or C. maritimus KKC1 but formed the same subclade with type iii cooS-II (clade F) from C. hydrogenoformans (Fig. 3).
Horizontal gene transfer analysis of six CooSs from C. maritimus KKC1.
To determine whether cooS was obtained by horizontal gene transfer, we performed a simple test for sequence composition (see Fig. S4 in the supplemental material). In this test, we calculated Euclidean distances between CDS tetranucleotide frequencies and the whole genome and evaluated the significance of distances of CooSs. As a general rule, horizontally transferred DNA fragments exhibit the oligonucleotide composition of the species they are derived from, and the screening of local variations of oligonucleotide composition along genomes is expected to reveal regions of interest where horizontally transferred genes might be located (44). A study predicted that the average proportion of horizontally transferred genes per genome was ∼12% of all CDSs, ranging from 0.5% to 25% depending on the prokaryotic lineage (11% in Bacillus subtilis 168 [Firmicutes]) (45). Therefore, we used 75% (corresponding to a distance of 0.03024) as a loose threshold for the detection of horizontally transferred CooSs. Accordingly, the distances of four cooS genes, KKC1_14790 (from cooS-ECH), KKC1_28130 (in the proximity of cysK, tauACB, and metBC), KKC1_10970 (in the proximity of korDABG), and KKC1_23670 (in the proximity of cooA and ispH), to the C. maritimus KKC1 genome were from 0.02004 to 0.02277, whereas the other two cooS genes, KKC1_09930-40 (from cooS-ACS) and KKC1_14590 (from cooF-cooS-FNOR), showed slightly higher values (0.0290 and 0.0279, respectively). However, distance values for all cooS genes from C. maritimus KKC1 were below the threshold, suggesting that all cooS genes descended from a common ancestor.
DISCUSSION
The similar branching pattern observed by phylogenetic analyses of 16S rRNA, five housekeeping genes, and GSS (Fig. 1) indicates that the thermophilic, hydrogenogenic carboxydotroph C. maritimus KKC1 and members of the genus Moorella, one of the most studied groups of acetogenic bacteria, evolved from a common ancestor. Both M. thermoacetica and C. maritimus KKC1 possessed the CO2-reducing Wood-Ljungdahl pathway and the energy-conserving ECH-based module energized by reduced ferredoxin. However, in contrast to M. thermoacetica, C. maritimus KKC1 lacked the electron-bifurcating enzymes HydABC and NfnAB. HydABC couples the simultaneous endergonic reduction of ferredoxin with H2 to the exergonic reduction of NAD+ with H2 (29), and NfnAB catalyzes the reduction of two NADP+ molecules with one NADH and one reduced ferredoxin to generate two NADPH molecules, which are required for the reduction of CO2 to acetate in M. thermoacetica (46). Because the potential of CO is lower than that of ferredoxin, reduction of ferredoxin by oxidation of CO may not need electron bifurcation in C. maritimus KKC1. The frameshift mutation in cooS within cooS-ACS, which has been reported in C. hydrogenoformans (9), was also found in C. maritimus KKC1. Even so, C. maritimus KKC1 is known to produce a small amount of acetate during hydrogenogenic growth under a CO atmosphere (23). According to a study by Svetlitchnyi and colleagues (6) which suggests that CooS may be unnecessary for operation of the Wood-Ljungdahl pathway, the frameshift mutation in cooS (KKC1_09930-40) within the cooS-ACS gene cluster might not affect pathway function.
M. thermoacetica is able to catalyze the near-stoichiometric conversion of glucose to 3 mol of acetate using PFOR, which couples the glycolytic pathway to the Wood-Ljungdahl pathway (1, 47). In contrast, C. maritimus KKC1 cannot grow on glucose (and other organic compounds) without electron acceptors, but it can grow with electron acceptors such as thiosulfate, resulting in a small amount of acetate (23). Genomic analysis of C. maritimus KKC1 revealed that it lacks genes encoding authentic PFOR, which is conserved across Moorella species. From a thermodynamic perspective, acetogenesis from glucose is less effective in supporting growth than anaerobic respiration using electron acceptors except for CO2 (1). Therefore, it is assumed that the lack of authentic PFOR in C. maritimus KKC1 might result in a survival strategy different from that of M. thermoacetica, which can thrive where no electron acceptors (except for CO2) are available. The small production of acetate during heterotrophic growth with thiosulfate by C. maritimus KKC1 might be explained by the presence of six gene sets encoding putative KFORs, because KFOR shows significant similarity with PFOR and some KFORs show broad specificity for pyruvate and 2-oxoglutarate (48). In this case, why C. maritimus KKC1 cannot grow acetogenically on glucose without electron acceptors using KFORs is unknown. One possibility is that the reaction efficiency of KFOR is lower than that of PFOR in oxidation of pyruvate, but further studies are needed to understand the mechanism of heterotrophic growth of C. maritimus KKC1.
The highest number of cooS genes ever reported in a single genome is five (cooS-I to -V) in C. hydrogenoformans (10). Thus, C. maritimus KKC1 harboring six CooS genes (five cooS genes conserving all residues linked to metal clusters [49] and one frameshifted cooS within the cooS-ACS gene cluster) possessed the most CooS genes of microbes with sequenced genomes. As described above, the simultaneous transcription of five cooS genes in C. maritimus KKC1 during carboxydotrophic growth was observed, and all might contribute to its CO metabolism. Three of the six cooS genes formed cooS-ACS (type i), cooS-ECH (type ii), and cooF-cooS-FNOR (type iii) gene clusters. On the other hand, the other three type iv cooS genes (KKC1_28130, KKC1_10970, and KKC1_23670) exhibit an uncharacterized genomic context. Although the cooS gene in KKC1_23670 clustered with a CooA homolog, KKC1_28130 and KKC1_10970 were not flanked by any genes with obvious roles in CO-related processes. However, the genomic context of cooS in KKC1_10970 is interesting, because the KFOR encoded upstream of KKC1_10970 is a redox enzyme that requires ferredoxin and produces (or consumes) CO2. Therefore, an interaction between CooS and KFOR could result in a novel CO fixation pathway where CooS oxidizes CO to produce CO2 and reduced ferredoxin, which could then be used to produce 2-oxoglutarate by KFOR. Sequence composition analysis of the six cooS genes from C. maritimus KKC1 showed that their distances to the whole genome were not exceedingly high (see Fig. S4 in the supplemental material), suggesting that they could descend from a common ancestor.
M. perchloratireducens is phylogenetically and physiologically similar to M. thermoacetica but cannot grow acetogenically on H2 plus CO2, unlike M. thermoacetica (32). This might be explained by the lack of formate dehydrogenase (Fdh), which fixes CO2 to formate in the first step of the Wood-Ljungdahl pathway. Although hydrogenogenic carboxydotrophic growth has never been reported, M. perchloratireducens possessed a cooS-hyf/hyc-type ECH gene cluster that might form from replacement of the fdoG-hycB by cooC-cooS-cooF in hyf/hyc-type ECH gene clusters conserved in M. thermoacetica and C. maritimus KKC1 (see Fig. S5 in the supplemental material). It appears that the assembly of the CooS gene and hyf/hyc-type ECH gene cluster might occur in the course of M. perchloratireducens evolution to efficiently generate energy by CO oxidation and proton translocation with hydrogen production. In addition, the origin of the CooS gene from cooS-hyf/hyc-type ECH might be the same as for type iii cooS-II from C. hydrogenoformans, implying that the common ancestor of Moorella species and C. maritimus KKC1 may have harbored a cooS-II homolog, which might have been lost by C. maritimus KKC1 and M. thermoacetica during evolution, while M. perchloratireducens retained it in the cooS-hyf/hyc-type ECH. Nevertheless, further studies, including genomic analysis of hydrogenogenic Moorella, such as M. stamsii and the M. thermoacetica strain AMP, are needed to understand the complex evolution of CooS genes and the emergence of acetogen and hydrogenogen within Moorella bacteria.
In conclusion, de novo genome sequencing and analysis of the hydrogenogenic carboxydotroph C. maritimus KKC1 revealed genomic contents largely different from that of acetogenic Moorella species, despite their phylogenetic similarity. Both species utilize energy-converting ECH-based modules that require the low-potential electron carrier ferredoxin. The lack of bifurcating enzymes and authentic PFOR and the presence of six copies of cooS genes in the C. maritimus KKC1 genome suggested that the organism may be highly dependent on CO as an electron donor, which can directly reduce ferredoxin, and more adaptive to carboxydotrophic growth than the acetogenic growth observed in Moorella species. In other words, the C. maritimus KKC1 genome might reveal its survival strategy of reliance on the energy-rich substrate CO, whereas the genomes of Moorella species show an adaptation at the thermodynamic limit (29). Thus, C. maritimus KKC1 may serve as a model for understanding the evolution and adaptation of CO metabolism.
MATERIALS AND METHODS
Bacterial strains, genome sequencing, and assembly.
C. maritimus KKC1 was isolated and maintained in our laboratory at 65°C in hypotonic artificial seawater (hASW) medium under a 100% CO atmosphere (23). Genomic DNA was extracted by the NaOH method as previously described (23) and sequenced by Fasmac Co. Ltd., (Kanagawa, Japan) using MiSeq, NexteraXT, and TruSeq DNA sample preparation kits (Illumina, San Diego, CA, USA). We obtained 4,553,796150-bp paired-end reads; those displaying a Phred score above Q20 for 80% of the bases were quality filtered using the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). This yielded 2,835,116 reads, which were then assembled with Velvet 1.2.10 (50).
ORF prediction and annotation.
To predict ORFs in the C. maritimus KKC1 genome, we employed Glimmer 3.02 (51), which uses Markov's interpolated models, and GeneMarkS 4.29 (52), followed by a manual curation process. After the ORFs were determined, protein sequences were further analyzed by BLASTP searches against nonredundant protein sequences in the National Center for Biotechnology Information (NCBI), KEGG, and COG databases (53). tRNA and rRNA were predicted using tRNA Scan-SE 1.3.1 (54) and RNAmmer 1.2 (55), respectively.
Phylogenetic analysis based on 16S rRNA, housekeeping genes, and GSS.
We retrieved the 16S rRNA gene sequences of the Thermoanaerobacteraceae family from the Reference Sequence Database in NCBI (Ref_seq). The sequences were aligned using MUSCLE 3.8.31 (56), and gap positions were removed automatically using trimAL1.4 (57). Phylogenetic reconstructions were performed by the ML method using PhyML3.1 (58) and visualized with MEGA 6.06 (59). Robustness of the topology of the phylogenetic trees was evaluated by bootstrap analysis based on 100 runs.
For genome-wide phylogenetic analysis, we collected 31 publicly available genomes of Thermoanaerobacteraceae members: from NCBI, Ammonifex degensii KC4 (NC_013385), Caldanaerobacter subterraneus subsp. tengcongensis MB4 (NC_003869), Carboxydothermus hydrogenoformans Z-2901 (NC_007503), Moorella thermoacetica ATCC 39073 (NC_007644), Tepidanaerobacter acetatoxydans Re1 2011 (NC_015519), Tepidanaerobacter acetatoxydans Re1 2013 (NC_019954), Thermoacetogenium phaeum DSM 12270 (NC_018870), Thermoanaerobacter brockii subsp. finnii Ako-1 (NC_014964), Thermoanaerobacter italicus Ab9 (NC_013921), Thermoanaerobacter mathranii subsp. mathranii A3 (NC_014209), Thermoanaerobacter pseudethanolicus ATCC 33223 (NC_010321), Thermoanaerobacter sp. X513 (NC_014538), Thermoanaerobacter sp. X514 (NC_010320), and Thermoanaerobacter wiegelii Rt8.B1 (NC_015958); from Integrated Microbial Genomes (IMG): Caldanaerobacter subterraneus subsp. pacificus DSM 12653 (647533123), Caldanaerobacter subterraneus subsp. yonseiensis KB-1 (2563367176), Caldanaerobius polysaccharolyticus DSM 13641 (2510065085), Carboxydothermus ferrireducens DSM 11255 (2510065088), Desulfovirgula thermocuniculi DSM 16036 (2524023160), Moorella perchloratireducens An10 (2506520025), Moorella thermoacetica Y72 (2582580993), Thermoanaerobacter ethanolicus CCSD1 (645058764), Thermoanaerobacter ethanolicus JW 200 (2503538027), Thermoanaerobacter indiensis BSB-33 (2517287027), Thermoanaerobacter kivui DSM 2030 (2576861811), Thermoanaerobacter siderophilus SR4 (2509276025), Thermoanaerobacter sp. strain A7A, Thermoanaerobacter sp. strain X561 (645058760), Thermoanaerobacter thermocopriae JCM 7501 (2546825535), and Thermoanaerobacter thermohydrosulfuricus WC1 (2517572224) (60).
We retrieved the amino acid sequences corresponding to the genes for ribosome recycling factor (frr), transcription elongation factor (nusA), 50S ribosomal protein L2 (rplB), 50S ribosomal protein L27 (rpmA), and elongation factor Ts (tsf) from the genomes of the Thermoanaerobacteraceae species listed above. The sequences were aligned and trimmed as described above. Concatenated alignments of five genes were then used to build an ML tree using PhyML (bootstrap = 100).
To compute the similarity between genomes of Thermoanaerobacteraceae, we calculated the corresponding genomic similarity score (GSS) (61). This measurement is based on the sum of bit scores of shared orthologs. These are determined by the all-versus-all BLASTP search using protein sets and are normalized against the sum of bit scores of the compared genes against themselves (self-bit score). We used protein sets for each genome with coverage of 70% of both genes, with an E value of 1 × e−5 at an effective database size of 107. The GSS ranged from 0 to 1, and the maximum score was obtained when two proteomes were identical. The neighbor-joining tree was built using a GSS distance matrix (62).
Phylogenetic analysis of CooS genes.
We retrieved CooS amino acid sequences by BLASTP searches against Ref_seq proteins. We also used some sequences of the Ni-CODH phylogenetic tree from Techtmann et al. (10) as references. The sequences were aligned and trimmed, and used to build an ML tree (bootstrap = 100) as described above.
Horizontal gene transfer analysis of CooS genes.
We calculated tetranucleotide frequencies of CDSs with the length of greater than or equal to 500 bp from C. maritimus KKC1 and its genome (all contigs were catenated, and sequence gaps “N” were removed). Sequences were extended with their reverse complements. The observed frequencies of all 256 possible tetranucleotides were computed for these sequences. We calculated Euclidean distances of tetranucleotide frequencies of CDSs to that of whole genome and evaluated the significance of distances of CooSs.
Accession number(s).
The draft genome sequence generated in this study has been deposited in the DNA Data Bank of Japan (DDBJ) database under accession numbers BDGJ01000001 to BDGJ01000223.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Koichiro Nakano and Kazuto Takasaki of FASMAC Co. Ltd. for the preparation of genome libraries and sequencing of C. maritimus KKC1. We thank Stephen M. Techtmann for kindly providing information on the CooS gene sequence that greatly improved our knowledge of CooS and anaerobic carboxydotrophic microbes. Part of the computational analysis was completed at the Super System Institute for Chemical Research, Kyoto University. We also thank Takashi Daifuku for technical assistance with genomic analysis and anonymous reviewers for critical reviews of the manuscript.
We declare that we have no conflicts of interest.
This work was supported by Grants-in-Aid for Scientific Research (A) 20248023, (A) 25252038, and (S) 16H06381 from The Ministry of Education, Culture, Sports, Science and Technology (MEXT) and by a Grant-in-Aid from the Japan Society for the Promotion of Science (JSPS) Fellows (16J11269).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00832-17.
REFERENCES
- 1.Drake HL, Daniel SL. 2004. Physiology of the thermophilic acetogen Moorella thermoacetica. Res Microbiol 155:869–883. doi: 10.1016/j.resmic.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 2.Techtmann SM, Colman AS, Robb FT. 2009. “That which does not kill us only makes us stronger”: the role of carbon monoxide in thermophilic microbial consortia. Environ Microbiol 11:1027–1037. doi: 10.1111/j.1462-2920.2009.01865.x. [DOI] [PubMed] [Google Scholar]
- 3.Ragsdale SW. 2004. Life with carbon monoxide. Crit Rev Biochem Mol Biol 39:165–195. doi: 10.1080/10409230490496577. [DOI] [PubMed] [Google Scholar]
- 4.Ferry JG. 1995. CO dehydrogenase. Annu Rev Microbiol 49:305–333. doi: 10.1146/annurev.mi.49.100195.001513. [DOI] [PubMed] [Google Scholar]
- 5.Dobbek H, Svetlitchnyi V, Gremer L, Huber R, Meyer O. 2001. Crystal structure of a carbon monoxide dehydrogenase reveals a [Ni-4Fe-5S] cluster. Science 293:1281–1285. doi: 10.1126/science.1061500. [DOI] [PubMed] [Google Scholar]
- 6.Svetlitchnyi V, Dobbek H, Meyer-Klaucke W, Meins T, Thiele B, Römer P, Huber R, Meyer O. 2004. A functional Ni-Ni-[4Fe-4S] cluster in the monomeric acetyl-CoA synthase from Carboxydothermus hydrogenoformans. Proc Natl Acad Sci U S A 101:446–451. doi: 10.1073/pnas.0304262101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oelgeschläger E, Rother M. 2008. Carbon monoxide-dependent energy metabolism in anaerobic bacteria and archaea. Arch Microbiol 190:257–269. doi: 10.1007/s00203-008-0382-6. [DOI] [PubMed] [Google Scholar]
- 8.Sokolova TG, Henstra AM, Sipma J, Parshina SN, Stams AJM, Lebedinsky AV. 2009. Diversity and ecophysiological features of thermophilic carboxydotrophic anaerobes. FEMS Microbiol Ecol 68:131–141. doi: 10.1111/j.1574-6941.2009.00663.x. [DOI] [PubMed] [Google Scholar]
- 9.Wu M, Ren Q, Durkin S, Daugherty SC, Brinkac LM, Dodson RJ, Madupu R, Sullivan SA, Kolonay JF, Nelson WC, Tallon LJ, Jones KM, Ulrich LE, Gonzalez JM, Zhulin IB, Robb FT, Eisen JA. 2005. Life in hot carbon monoxide: the complete genome sequence of Carboxydothermus hydrogenoformans Z-2901. PLoS Genet 1:e65. doi: 10.1371/journal.pgen.0010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Techtmann SM, Lebedinsky AV, Colman AS, Sokolova TG, Woyke T, Goodwin L, Robb FT. 2012. Evidence for horizontal gene transfer of anaerobic carbon monoxide dehydrogenases. Front Microbiol 3:132. doi: 10.3389/fmicb.2012.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sokolova T, Lebedinsky A. 2013. CO-oxidizing anaerobic thermophilic prokaryotes, p 203–231. In Satyanarayana T, Littlechild J, Kawarabayasi Y (ed), Thermophilic microbes in environmental and industrial biotechnology. Springer, Houten, Netherlands. [Google Scholar]
- 12.Gencic S, Duin EC, Grahame DA. 2010. Tight coupling of partial reactions in the acetyl-CoA decarbonylase/synthase (ACDS) multienzyme complex from Methanosarcina thermophila: acetyl C-C bond fragmentation at the A cluster promoted by protein conformational changes. J Biol Chem 285:15450–15463. doi: 10.1074/jbc.M109.080994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drake HL, Küsel K, Matthies C. 2002. Ecological consequences of the phylogenetic and physiological diversities of acetogens. Antonie Van Leeuwenhoek 81:203–213. doi: 10.1023/A:1020514617738. [DOI] [PubMed] [Google Scholar]
- 14.Ragsdale SW. 1997. The Eastern and Western branches of the Wood/Ljungdahl pathway: how the East and West were won. Biofactors 6:3–11. doi: 10.1002/biof.5520060102. [DOI] [PubMed] [Google Scholar]
- 15.Ferry JG. 1999. Enzymology of one-carbon metabolism in methanogenic pathways. FEMS Microbiol Rev 23:13–38. doi: 10.1111/j.1574-6976.1999.tb00390.x. [DOI] [PubMed] [Google Scholar]
- 16.Stupperich E, Hammel KE, Fuchs G, Thauer RK. 1983. Carbon monoxide fixation into the carboxyl group of acetyl coenzyme A during autotrophic growth of Methanobacterium. FEBS Lett 152:21–23. doi: 10.1016/0014-5793(83)80473-6. [DOI] [PubMed] [Google Scholar]
- 17.Ladapo J, Whitman WB. 1990. Method for isolation of auxotrophs in the methanogenic archaebacteria: role of the acetyl-CoA pathway of autotrophic CO2 fixation in Methanococcus maripaludis. Proc Natl Acad Sci U S A 87:5598–5602. doi: 10.1073/pnas.87.15.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schauder R, Preuß A, Jetten M, Fuchs G. 1988. Oxidative and reductive acetyl CoA/carbon monoxide dehydrogenase pathway in Desulfobacterium autotrophicum. Arch Microbiol 151:84–89. doi: 10.1007/BF00444674. [DOI] [Google Scholar]
- 19.Spormann AM, Thauer RK. 1988. Anaerobic acetate oxidation to CO2 by Desulfotomaculum acetoxidans. Demonstration of enzymes required for the operation of an oxidative acetyl-CoA/carbon monoxide dehydrogenase pathway. Arch Microbiol 150:374–380. [Google Scholar]
- 20.Vignais PM, Billoud B. 2007. Occurrence, classification, and biological function of hydrogenases: an overview. Chem Rev 107:4206–4272. doi: 10.1021/cr050196r. [DOI] [PubMed] [Google Scholar]
- 21.Fox JD, Yiping HE, Shelver D, Roberts GP, Ludden PW. 1996. Characterization of the region encoding the CO-induced hydrogenase of Rhodospirillum rubrum. J Bacteriol 178:6200–6208. doi: 10.1128/jb.178.21.6200-6208.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sant'Anna FH, Lebedinsky AV, Sokolova TG, Robb FT, Gonzalez JM. 2015. Analysis of three genomes within the thermophilic bacterial species Caldanaerobacter subterraneus with a focus on carbon monoxide dehydrogenase evolution and hydrolase diversity. BMC Genomics 16:757. doi: 10.1186/s12864-015-1955-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoneda Y, Yoshida T, Yasuda H, Imada C, Sako Y. 2013. A thermophilic, hydrogenogenic and carboxydotrophic bacterium, Calderihabitans maritimus gen. nov., sp. nov., from a marine sediment core of an undersea caldera. Int J Syst Evol Microbiol 63:3602–3608. doi: 10.1099/ijs.0.050468-0. [DOI] [PubMed] [Google Scholar]
- 24.Alves JI, van Gelder AH, Alves MM, Sousa DZ, Plugge CM. 2013. Moorella stamsii sp. nov., a new anaerobic thermophilic hydrogenogenic carboxydotroph isolated from digester sludge. Int J Syst Evol Microbiol 63:4072–4076. doi: 10.1099/ijs.0.050369-0. [DOI] [PubMed] [Google Scholar]
- 25.Jiang B, Henstra A-M, Paulo PL, Balk M, van Doesburg W, Stams AJM. 2009. Atypical one-carbon metabolism of an acetogenic and hydrogenogenic Moorella thermoacetica strain. Arch Microbiol 191:123–131. doi: 10.1007/s00203-008-0435-x. [DOI] [PubMed] [Google Scholar]
- 26.Fontaine FE, Peterson WH, McCoy E, Johnson MJ, Ritter GJ. 1942. A new type of glucose fermentation by Clostridium thermoaceticum. J Bacteriol 43:701–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyer FM, Stülke J. 2013. Malate metabolism in Bacillus subtilis: distinct roles for three classes of malate-oxidizing enzymes. FEMS Microbiol Lett 339:17–22. doi: 10.1111/1574-6968.12041. [DOI] [PubMed] [Google Scholar]
- 28.Fraenkel DG, Vinopal RT. 1973. Carbohydrate metabolism in bacteria. Annu Rev Microbiol 27:69–100. doi: 10.1146/annurev.mi.27.100173.000441. [DOI] [Google Scholar]
- 29.Schuchmann K, Müller V. 2014. Autotrophy at the thermodynamic limit of life: a model for energy conservation in acetogenic bacteria. Nat Rev Microbiol 12:809–821. doi: 10.1038/nrmicro3365. [DOI] [PubMed] [Google Scholar]
- 30.Diender M, Stams AJM, Sousa DZ. 2015. Pathways and bioenergetics of anaerobic carbon monoxide fermentation. Front Microbiol 6:1275. doi: 10.3389/fmicb.2015.01275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pierce E, Xie G, Barabote RD, Saunders E, Han CS, Detter JC, Richardson P, Brettin TS, Das A, Ljungdahl LG, Ragsdale SW. 2008. The complete genome sequence of Moorella thermoacetica (f. Clostridium thermoaceticum). Environ Microbiol 10:2550–2573. doi: 10.1111/j.1462-2920.2008.01679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balk M, van Gelder T, Weelink SA, Stams AJM. 2008. (Per)chlorate reduction by the thermophilic bacterium Moorella perchloratireducens sp. nov., isolated from underground gas storage. Appl Environ Microbiol 74:403–409. doi: 10.1128/AEM.01743-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geelhoed JS, Henstra AM, Stams AJM. 2016. Carboxydotrophic growth of Geobacter sulfurreducens. Appl Microbiol Biotechnol 100:997–1007. doi: 10.1007/s00253-015-7033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mino K, Ishikawa K. 2003. Characterization of a novel thermostable O-acetylserine sulfhydrylase from Aeropyrum pernix K1. J Bacteriol 185:2277–2284. doi: 10.1128/JB.185.7.2277-2284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van der Ploeg JR, Weiss MA, Saller E, Nashimoto H, Saito N, Kertesz MA, Leisinger T. 1996. Identification of sulfate starvation-regulated genes in Escherichia coli: a gene cluster involved in the utilization of taurine as a sulfur source. J Bacteriol 178:5438–5446. doi: 10.1128/jb.178.18.5438-5446.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lill R, Mühlenhoff U. 2005. Iron-sulfur-protein biogenesis in eukaryotes. Trends Biochem Sci 30:133–141. doi: 10.1016/j.tibs.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 37.Shiba H, Kawasumi T, Igarashi Y, Kodama T, Minoda Y. 1985. The CO2 assimilation via the reductive tricarboxylic acid cycle in an obligately autotrophic, aerobic hydrogen-oxidizing bacterium, Hydrogenobacter thermophilus. Arch Microbiol 141:198–203. doi: 10.1007/BF00408058. [DOI] [Google Scholar]
- 38.Shiba H, Kawasumi T, Igarashi Y, Kodama T, Minoda Y. 2014. The deficient carbohydrate metabolic pathways and the incomplete tricarboxylic acid cycle in an obligately autotrophic hydrogen-oxidizing bacterium. Agric Biol Chem 46:2341–2345. doi: 10.1080/00021369.1982.10865433. [DOI] [Google Scholar]
- 39.Inagaki S, Masuda C, Akaishi T, Nakajima H, Yoshioka S, Ohta T, Pal B, Kitagawa T, Aono S. 2005. Spectroscopic and redox properties of a CooA homologue from Carboxydothermus hydrogenoformans. J Biol Chem 280:3269–3274. doi: 10.1074/jbc.M409884200. [DOI] [PubMed] [Google Scholar]
- 40.He Y, Shelver D, Kerby RL, Roberts GP. 1996. Characterization of a CO-responsive transcriptional activator from Rhodospirillum rubrum. J Biol Chem 271:120–123. doi: 10.1074/jbc.271.1.120. [DOI] [PubMed] [Google Scholar]
- 41.Adam P, Hecht S, Eisenreich W, Kaiser J, Grawert T, Arigoni D, Bacher A, Rohdich F. 2002. Biosynthesis of terpenes: studies on 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase. Proc Natl Acad Sci U S A 99:12108–12113. doi: 10.1073/pnas.182412599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seemann M, Tse Sum Bui B, Wolff M, Miginiac-Maslow M, Rohmer M. 2006. Isoprenoid biosynthesis in plant chloroplasts via the MEP pathway: direct thylakoid/ferredoxin-dependent photoreduction of GcpE/IspG. FEBS Lett 580:1547–1552. doi: 10.1016/j.febslet.2006.01.082. [DOI] [PubMed] [Google Scholar]
- 43.Rohmer M. 1999. The discovery of a mevalonate-independent pathway for isoprenoid biosynthesis in bacteria, algae and higher plants. Nat Prod Rep 16:565–574. doi: 10.1039/a709175c. [DOI] [PubMed] [Google Scholar]
- 44.Dufraigne C, Fertil B, Lespinats S, Giron A, Deschavanne P. 2005. Detection and characterization of horizontal transfers in prokaryotes using genomic signature. Nucleic Acids Res 33:e6. doi: 10.1093/nar/gni004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakamura Y, Itoh T, Matsuda H, Gojobori T. 2004. Biased biological functions of horizontally transferred genes in prokaryotic genomes. Nat Genet 36:760–766. doi: 10.1038/ng1381. [DOI] [PubMed] [Google Scholar]
- 46.Huang H, Wang S, Moll J, Thauer RK. 2012. Electron bifurcation involved in the energy metabolism of the acetogenic bacterium Moorella thermoacetica growing on glucose or H2 plus CO2. J Bacteriol 194:3689–3699. doi: 10.1128/JB.00385-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Furdui C, Ragsdale SW. 2000. The role of pyruvate ferredoxin oxidoreductase in pyruvate synthesis during autotrophic growth by the Wood-Ljungdahl pathway. J Biol Chem 275:28494–28499. doi: 10.1074/jbc.M003291200. [DOI] [PubMed] [Google Scholar]
- 48.Fukuda E, Wakagi T. 2002. Substrate recognition by 2-oxoacid:ferredoxin oxidoreductase from Sulfolobus sp. strain 7. Biochim Biophys Acta 1597:74–80. doi: 10.1016/S0167-4838(02)00280-7. [DOI] [PubMed] [Google Scholar]
- 49.Inoue T, Takao K, Yoshida T, Wada K, Daifuku T, Yoneda Y, Fukuyama K, Sako Y. 2013. Cysteine 295 indirectly affects Ni coordination of carbon monoxide dehydrogenase-II C-cluster. Biochem Biophys Res Commun 441:13–17. doi: 10.1016/j.bbrc.2013.09.143. [DOI] [PubMed] [Google Scholar]
- 50.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Delcher AL, Bratke KA, Powers EC, Salzberg SL. 2007. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Besemer J, Lomsadze A, Borodovsky M. 2001. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res 29:2607–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tatusov RL, Natale DA, Garkavtsev IV, Tatusova TA, Shankavaram UT, Rao BS, Kiryutin B, Galperin MY, Fedorova ND, Koonin EV. 2001. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res 29:22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res 33:W686–W689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lagesen K, Hallin P, Rødland EA, Staerfeldt H-H, Rognes T, Ussery DW. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 59.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Markowitz VM, Korzeniewski F, Palaniappan K, Szeto E, Werner G, Padki A, Zhao X, Dubchak I, Hugenholtz P, Anderson I, Lykidis A, Mavromatis K, Ivanova N, Kyrpides NC. 2006. The integrated microbial genomes (IMG) system. Nucleic Acids Res 34:D344–348. doi: 10.1093/nar/gkj024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moreno-Hagelsieb G, Janga SC. 2008. Operons and the effect of genome redundancy in deciphering functional relationships using phylogenetic profiles. Proteins 70:344–352. doi: 10.1002/prot.21564. [DOI] [PubMed] [Google Scholar]
- 62.Alcaraz LD, Moreno-Hagelsieb G, Eguiarte LE, Souza V, Herrera-Estrella L, Olmedo G. 2010. Understanding the evolutionary relationships and major traits of Bacillus through comparative genomics. BMC Genomics 11:332. doi: 10.1186/1471-2164-11-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.