ABSTRACT
Feed efficiency (FE) is critical in pig production for both economic and environmental reasons. As the intestinal microbiota plays an important role in energy harvest, it is likely to influence FE. Therefore, our aim was to characterize the intestinal microbiota of pigs ranked as low, medium, and high residual feed intake ([RFI] a metric for FE), where genetic, nutritional, and management effects were minimized, to explore a possible link between the intestinal microbiota and FE. Eighty-one pigs were ranked according to RFI between weaning and day 126 postweaning, and 32 were selected as the extremes in RFI (12 low, 10 medium, and 10 high). Intestinal microbiota diversity, composition, and predicted functionality were assessed by 16S rRNA gene sequencing. Although no differences in microbial diversity were found, some RFI-associated compositional differences were revealed, principally among members of Firmicutes, predominantly in feces at slaughter (albeit mainly for low-abundance taxa). In particular, microbes associated with a leaner and healthier host (e.g., Christensenellaceae, Oscillibacter, and Cellulosilyticum) were enriched in low RFI (more feed-efficient) pigs. Differences were also observed in the ileum of low RFI pigs; most notably, Nocardiaceae (Rhodococcus) were less abundant. Predictive functional analysis suggested improved metabolic capabilities in these animals, especially within the ileal microbiota. Higher ileal isobutyric acid concentrations were also found in low RFI pigs. Overall, the differences observed within the intestinal microbiota of low RFI pigs compared with that of their high RFI counterparts, albeit relatively subtle, suggest a possible link between the intestinal microbiota and FE in pigs.
IMPORTANCE This study is one of the first to show that differences in intestinal microbiota composition, albeit subtle, may partly explain improved feed efficiency (FE) in low residual feed intake (RFI) pigs. One of the main findings is that, although microbial diversity did not differ among animals of varying FE, specific intestinal microbes could potentially be linked with porcine FE. However, as the factors impacting FE are still not fully understood, intestinal microbiota composition may not be a major factor determining differences in FE. Nonetheless, this work has provided a potential set of microbial biomarkers for FE in pigs. Although culturability could be a limiting factor and intervention studies are required, these taxa could potentially be targeted in the future to manipulate the intestinal microbiome so as to improve FE in pigs. If successful, this has the potential to reduce both production costs and the environmental impact of pig production.
KEYWORDS: swine, residual feed intake, feces, ileum, cecum
INTRODUCTION
Feed accounts for ∼70% of the total cost of producing a pig (1). Therefore, improving feed efficiency (FE) will increase profitability while also reducing the environmental impact of pig production (2). The porcine intestinal microbiota is considered an important “organ” with a crucial role to play in nutrient processing and the harvesting of ingested energy (3–5). Therefore, it is plausible to suggest that the porcine intestinal microbiota could potentially be targeted to improve FE. Indeed, porcine metabolism is impacted by the complex interplay between the resident intestinal microbes, their metabolites, e.g., volatile fatty acids (VFAs), and enterocyte function (6–9). Microbial mechanisms of potential relevance to FE include positive feedback between certain microbes and mucin production, goblet cells along the villi, and upregulation of butyric acid production (10, 11). Interestingly, studies in cattle have shown differences in the intestinal microbiota in animals differing in FE (12, 13). However, very few studies to date have explored the possible link between the intestinal microbiota and FE in pigs.
In recent years, the pig microbiome has become the focus of much attention (14–18). The porcine intestinal microbiota is dominated at the phylum level by Firmicutes, Bacteroidetes, and Proteobacteria (3, 4, 19, 20). Differences within the intestinal microbiota have explained variability in body weight in pigs; for example, at the phylum level, Firmicutes and Planctomycetes have been found at higher relative abundance in heavier pigs, while Bacteroidetes were more abundant in lighter pigs (21). In the same study, body weight-associated differences were found at the genus level (21), and a study by Mach et al. showed that Prevotella was positively correlated with body weight (22). Intestinal microbiota composition has also been shown to vary between lean and obese pigs, with an increased abundance of Firmicutes found in obese pigs (15). In addition, bacterial diversity within the intestinal tract was found to be higher in pigs with heavier body weights and improved growth rates (3, 21). However, to our knowledge, only one study to date has investigated the association between FE in pigs and the intestinal microbiota (14). It demonstrated an increased abundance of Lactobacillus in the cecum of more feed-efficient pigs; however, only Firmicutes, Bacteroidetes, Bacteroides, Lactobacillus, and Enterobacteriaceae were measured (by quantitative PCR). Pigs with better FE also tended to have higher concentrations of total VFAs in the cecum and butyric acid in the colon, which may be explained by differences in microbial composition and function (14, 23).
Characterizing the intestinal microbiota of highly feed-efficient pigs could help to define an “optimal” microbial profile for improved FE. Shifts in microbial community structure associated with FE might suggest opportunities to modulate the intestinal microbiota composition to improve FE. In particular, the enrichment of specific microbes, supported by beneficial functionality, could pinpoint prospective microbial biomarkers for FE within the porcine intestinal microbiota. Optimization of the microbiota could then potentially be achieved through the use of these specific bacterial taxa as probiotics or, alternatively, by increasing their abundance via the use of prebiotics or other dietary supplements (10, 24, 25) or by fecal microbiota transplantation (26).
Therefore, in this study, we investigated the hypothesis that the composition and potential functionality of the intestinal microbiota are linked with FE in pigs. The objective was to determine if there were any differences in microbial diversity and/or relative abundance of bacterial taxa, at the phylum, family and genus levels within the fecal microbiota throughout the life of the pig and in the ileal and cecal microbiota at slaughter (at ∼166 days of age), in pigs ranked based on residual feed intake ([RFI] a metric for FE).
RESULTS
Growth performance of pigs ranked by residual feed intake.
The mean growth performance data recorded for pigs between weaning and day 126 postweaning (pw) are presented according to RFI rank in Table 1. Selected pigs in the low, medium, and high RFI ranks had distinct RFI values, with RFI reduced by 127 g/day for pigs ranked as low compared with that of pigs ranked as high RFI (P < 0.001). Pigs with a low RFI were the most feed efficient, as indicated by a reduction in the average daily feed intake (ADFI) of 219 g/day (P < 0.001), but showed an improvement in feed conversion efficiency (FCE) of 0.12 g/g (P < 0.01) compared with that of high RFI pigs. However, medium RFI pigs had statistically similar ADFI and FCE to both the high and low RFI ranks. No differences between RFI ranks were observed for average daily gain ([ADG] P > 0.05) or for any of the carcass quality measures (Table 1) or organ weights (P > 0.05) (see Table S2 in the supplemental material).
TABLE 1.
Effect of ranking pigs by RFI (between weaning and day 126 postweaning) on growth performance parameters and carcass traits
| Parametera | High RFI (n = 10) | Medium RFI (n = 10) | Low RFI (n = 12) | SEMb | P value |
|---|---|---|---|---|---|
| RFI (g/day) | 76.0c | 6.0d | −51.0e | 15.40 | <0.001 |
| ADG (g/day) | 910 | 877 | 855 | 28.4 | 0.38 |
| ADFI (g/day) | 1,850c | 1,732c,d | 1,631d | 51.2 | <0.01 |
| FCE (g/g) | 1.91c | 1.86c,d | 1.79d | 0.025 | <0.01 |
| Slaughter wt (kg) | 150.3 | 147.2 | 141.0 | 2.50 | 0.51 |
| Carcass cold wt (kg) | 113.4 | 113.1 | 108.1 | 3.59 | 0.48 |
| Kill out (%) | 79.2 | 78.9 | 77.9 | 0.53 | 0.19 |
| Muscle depth (mm) | 61.7 | 61.0 | 63.2 | 1.78 | 0.66 |
| Fat (mm) | 17.2 | 17.9 | 16.4 | 0.79 | 0.56 |
| Lean meat (%) | 54.9 | 54.1 | 55.5 | 0.69 | 0.49 |
RFI, residual feed intake; ADG, average daily gain; ADFI, average daily feed intake; FCE, feed conversion efficiency.
Least-squares means and pooled standard errors of the means are presented.
Within each row, values that do not share a common superscript are significantly different (P ≤ 0.05).
Salivary cortisol and ileal histology in pigs ranked by residual feed intake.
Cortisol concentrations were measured, as cortisol has been suggested as a biomarker for FE, with animals that were more feed efficient having lower serum concentrations (27). However, in this study, salivary cortisol concentrations measured in pigs at the end of the finishing period were unaffected by RFI rank (P > 0.05) (see Table S2).
As intestinal structure is another factor that can potentially influence FE, ileal histology measurements were determined in this study (i.e., villus height and width, crypt depth, villus height-to-crypt depth ratio, and the number of goblet cells) (see Table S3). However, only the numbers of goblet cells were affected by RFI rank, with low RFI pigs having 7.5 fewer goblet cells per villus and 0.02 fewer goblet cells/μm of villus height compared with those of high RFI pigs (P < 0.05) (Table S3).
Microbial load and diversity in pigs ranked by residual feed intake.
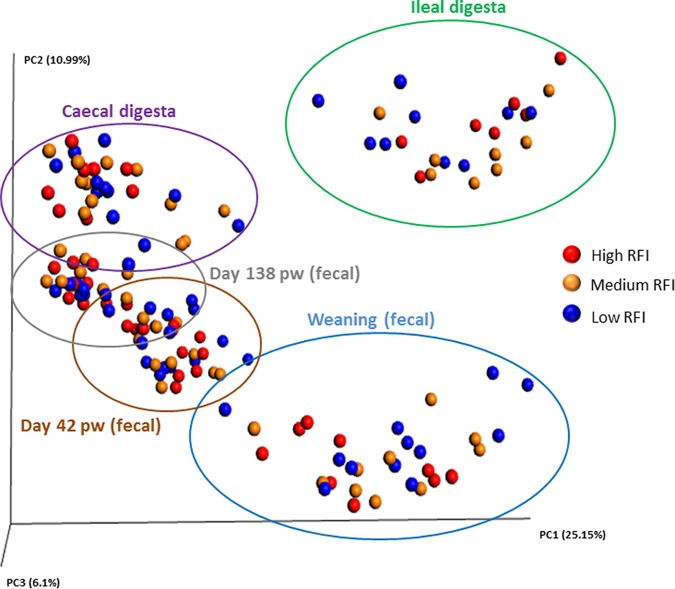
The total bacterial loads were quantified in all fecal and digesta samples. No differences were observed between high and low RFI pigs in the feces at any time point or in the ileal and cecal digesta collected at slaughter (P > 0.10) (see Table S4). Likewise, no significant differences for any of the indices of α diversity measured, i.e., richness based on rare operational taxonomic units ([OTUs] Chao1) or richness and evenness (Shannon and Simpson) were observed between RFI ranks (see Fig. S1). Furthermore, β diversity analyses showed that samples did not cluster based on RFI rank, but clustering on the basis of sample type was observed, with ileal samples distinctly different from fecal and cecal samples (Fig. 1). Fecal samples also clustered according to age, with the greatest variance detected for weaning samples. No sex-associated differences were observed for intestinal microbial diversity (data not shown).
FIG 1.
Principal-coordinate analysis (PCoA) plot (based on OTUs) according to residual feed intake (RFI) rank and sample type (n = 150). Low RFI: feces (n = 36) and digesta (cecal, n = 12; ileal, n = 9); medium RFI: feces (n = 30) and digesta (ceca, n = 10; ileal, n = 9); high RFI: feces (n = 30) and digesta (cecal, n = 8; ileal, n = 6). Plot is based on the unweighted UniFrac distances. The amount of variance is depicted by the percentages in parentheses on each axis. Ellipses denote clustering according to fecal sample time points and intestinal location.
Intestinal microbiota composition in pigs ranked by residual feed intake.
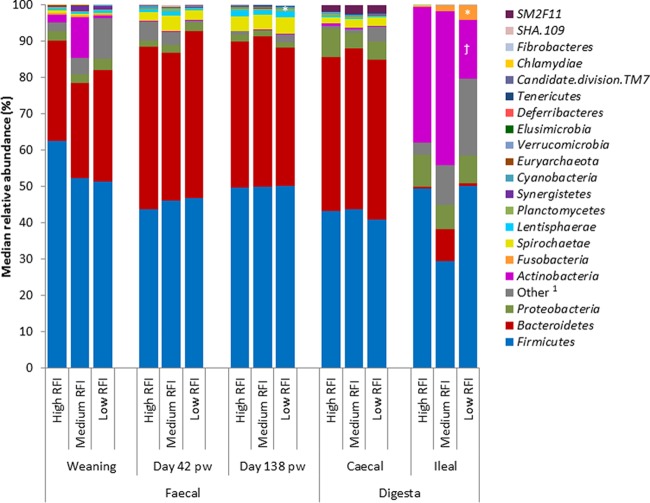
From a taxonomic perspective, 21 phyla, 161 families, and 295 genera were identified across all pig fecal/intestinal samples. Phylum profiles differed depending on sample type (Fig. 2). For example, Firmicutes and Bacteroidetes were the most abundant phyla in the feces and cecal digesta. However, a distinct profile was observed in the ileum, where Firmicutes and Actinobacteria predominated. Other general observations at the phylum level included the fact that Proteobacteria were relatively more abundant in the cecal and ileal digesta than in the feces. Also, Spirochaetes increased in relative abundance in the feces as the pigs aged, and were present in the cecal but not ileal digesta. However, no sex-associated differences were observed for intestinal microbial composition (data not shown).
FIG 2.
Median relative abundances (%) of bacterial phyla present in pigs ranked by residual feed intake (RFI) across all fecal time points (n = 96) and both intestinal locations (n = 54). 1, no blast hits/uncultured; *, significant difference (P ≤ 0.05) (candidate division TM7 in the feces at day 138 postweaning and Fusobacteria in the ileum); †, tendency toward significant differences (Actinobacteria in the ileum; P = 0.06) observed between high and low RFI pigs within each sample type.
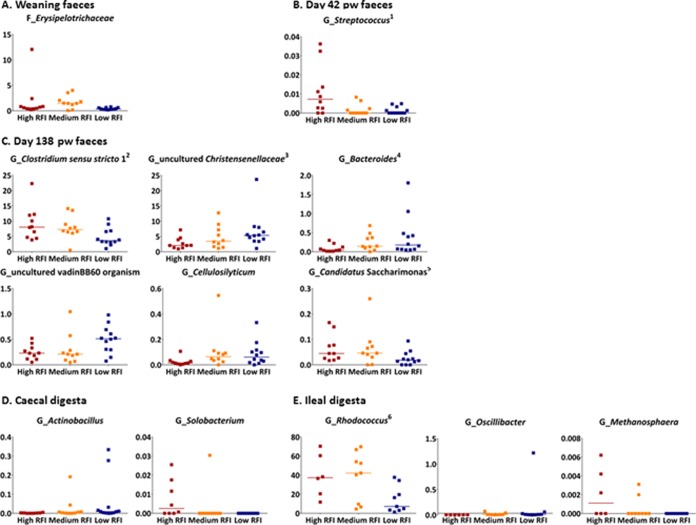
Certain taxa were identified as differentially abundant according to RFI rank (Fig. 2 and 3). Differences between low and high RFI pigs were detected for two phyla, candidate division TM7, which were 2.5-fold lower in relative abundance in the feces from low RFI pigs at day 138 pw (P < 0.05), and Fusobacteria, which were 14-fold higher in abundance in the ilea of low RFI pigs (P < 0.05); Actinobacteria also tended to be almost 3-fold lower in abundance in the ilea of low RFI pigs (P = 0.06) (Fig. 2). Six bacterial families and 12 genera also differed in relative abundances between high and low RFI pigs (P < 0.05), the details of which are outlined below, with five of the genus-level differences reflecting differences at the family level (i.e., relative abundances were identical) (Fig. 3). Relative abundance differences were observed across all sample types, but mostly in the feces at day 138 pw (Fig. 3C). Biological variation in microbial composition occurred between individuals, as evidenced by outliers in the relative abundance plots (Fig. 3), although only four pigs (one low RFI and three high RFI) had outlying data for more than one taxon (but not for all taxa).
FIG 3.
Median relative abundance (%) of microbial taxa found to be differentially abundant between pigs ranked with low and high residual feed intake (RFI) (P < 0.05) in feces at weaning (n = 32) (A), feces at day 42 pw (n = 32) (B), feces at day 138 pw (n = 32) (C), cecal digesta (n = 30) (D), and ileal digesta (n = 24) (E). Low RFI: feces (n = 36) and digesta (cecal, n = 12; ileal, n = 9); medium RFI: feces (n = 30) and digesta (cecal, n = 10; ileal, n = 9); high RFI: feces (n = 30) and digesta (cecal, n = 8; ileal, n = 6). F, family; G, genus. Horizontal lines in the plots indicate median values of the distributions. The Fusobacteria phylum was also differentially abundant, and this is illustrated in Fig. 2. Some genus-level differences shown in the plots reflect differences at a higher taxonomic level which are not shown here as follows: 1, Streptococcaceae family; 2, Clostridiaceae family; 3, Christensenellaceae family; 4, Bacteroidaceae family; 5, candidate division TM7 phylum; 6, Nocardiaceae family. Some of the animals for which the highest variance from the median values of the taxa distribution was seen had outlying data for more than one taxon, i.e., one low RFI pig had higher relative abundances than the median values for uncultured Christensenellaceae, Actinobacillus, and Oscillibacter and three high RFI pigs had higher relative abundances than the median value for Rhodococcus and Methanosphaera.
At the family level, Erysipelotrichaceae were 2-fold lower in the feces collected at weaning and Streptococcaceae (Streptococcus spp.) were 1-fold lower in the feces collected at day 42 pw in low RFI pigs compared with that in high RFI pigs (P < 0.05) (Fig. 3A and B). At day 138 pw (Fig. 3C), differences occurred mainly within the Firmicutes phylum, and mostly for members of Clostridiales. Within this order, the median relative abundance of the genus Clostridium sensu stricto 1 (belonging to the Clostridiaceae family) was 2-fold lower and abundances of an uncultured member of the vadinBB60, an uncultured genus from the Christensenellaceae family, and the Cellulosilyticum genus were respectively 1-fold, 2.5-fold, and 6-fold higher in low RFI pigs than in their high RFI counterparts (P < 0.05). Within the Bacteroidetes phylum, Bacteroides spp. were 4.5-fold higher in low versus high RFI pigs while the genus “Candidatus Saccharimonas” was 2-fold lower in low RFI pigs than in high RFI pigs (P < 0.05) (Fig. 3C), with the latter accounting for the difference observed at the phylum level (i.e., relative abundances were identical). In the cecum, low RFI pigs had a >3-fold lower abundance of Solobacterium spp. but a 4.5-fold higher abundance of Actinobacillus spp. (P < 0.05) (Fig. 3D).
In the ilea of low RFI pigs, the tendency for a lower relative abundance of Actinobacteria (Fig. 2) was reflected by a concomitantly lower relative abundance (3-fold) of the Nocardiaceae family and the Rhodococcus genus (P < 0.05) (Fig. 3E). Of all the taxa that differed between low and high RFI pigs, these were at the highest relative abundances. The genus Methanosphaera from Archaea was also lower in the ilea of low RFI pigs. However, low RFI pigs had a higher abundance of Oscillibacter spp. (from Clostridiales), although one pig in the low RFI rank appears to have skewed the data (P < 0.05) (Fig. 3E).
The RFI-associated differences outlined above were generally mirrored at the OTU level (see Fig. S2 and Table S5). However, there were some discrepancies. For example, at weaning and day 42 pw, no differences between RFI ranks were found (Table S5). Furthermore, in the ileum, Treponema berlinense from Spirochaetaceae was found at a higher relative abundance in low RFI pigs, and in the cecum, an uncultured Clostridiales bacterium from Ruminococcaceae was present at a lower relative abundance in low RFI pigs, while Actinobacillus porcinus was higher in the low RFI pigs (Table S5).
Eighty percent of the OTUs in the feces collected at weaning were common between high and low RFI pigs; 85% were common at day 42 pw, and 82% at day 138 pw in the feces and in the cecal digesta, and 66% in the ileal digesta. On the other hand, a number of OTUs (belonging to 17 phyla) were found exclusively in either low or high RFI ranked pigs (Fig. S2). Low RFI pigs harbored more of these OTUs than the high RFI pigs, particularly in the ileum, where 60 OTUs were found to be exclusive to low RFI pigs versus 28 in their high RFI counterparts. Some of the OTUs exclusively found in low RFI pigs represent potentially beneficial microbes, for example, Akkermansia found in the feces at weaning and in the ileum, Bifidobacterium in the feces at day 138 pw, uncultured bacteria from Prevotellaceae in the feces at weaning and day 138 pw, Mucispirillum in the feces at day 42 pw and in the cecum, and Butyricimonas in the ileum. Most of the RFI-specific OTUs were members of Firmicutes, especially, the uncultured microorganisms and those from the Clostridiales order.
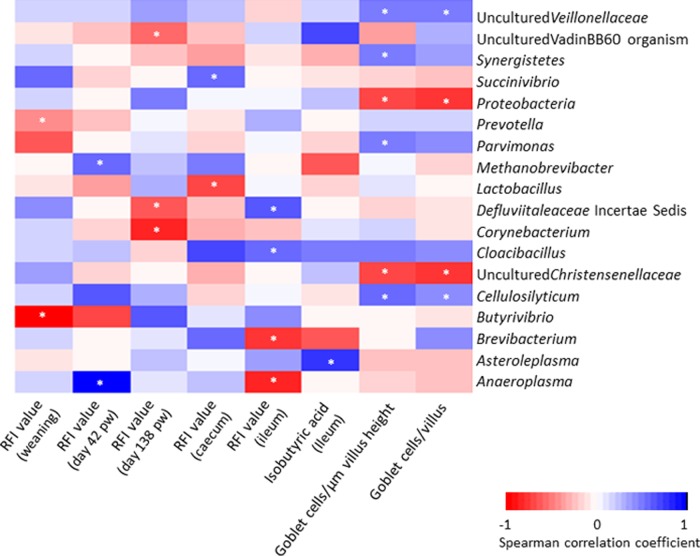
Intestinal microbiota correlations with RFI value.
A correlation analysis was performed between the intestinal microbiota composition, at the phylum and genus levels, and RFI value, and although no significant RFI-associated correlations were found at the phylum level, 13 genera correlated with RFI value (P < 0.05) (Fig. 4), including eight with low RFI (negative correlation) and five with high RFI (positive correlation) across the different sample types. An uncultured organism of the vadinBB60 family was correlated with a low RFI value, i.e., with better FE, in the feces at day 138 pw (P < 0.05) and was the only RFI-correlated genus previously identified as RFI-associated from the relative abundance data (Fig. 3C). Butyrivibrio, from which two uncultured OTUs were found exclusively in the feces from low RFI pigs at day 42 pw, was strongly correlated with a low RFI value at weaning (P < 0.05). Furthermore, Prevotella in the feces collected at weaning, Corynebacterium and Defluviitaleaceae incertae sedis in the feces collected prior to slaughter (day 138 pw), Lactobacillus in the cecal digesta, and Brevibacterium and Anaeroplasma in the ileal digesta were also correlated with low RFI values, although not differing in relative abundances between high and low RFI pigs. On the contrary, Anaeroplasma was strongly correlated with a high RFI value in the feces on day 42 pw, and other weaker correlations were found with microbes in the cecum and ileum.
FIG 4.
Heatmap showing Spearman correlations between bacterial taxa and physiological measures in pigs ranked by residual feed intake (RFI). Low RFI: feces (n = 36) and digesta (cecal, n = 12; ileal, n = 9); medium RFI: feces (n = 30) and digesta (cecal, n = 10; ileal, n = 9); high RFI: feces (n = 30) and digesta (cecal, n = 8; ileal, n = 6). pw, postweaning. Correlations were examined between bacterial taxa (at both the phylum and genus levels) and physiological measures found to be significantly different between low and high RFI pigs. *, P ≤ 0.05.
Predictive functional analysis of the intestinal microbiota of pigs ranked by residual feed intake.
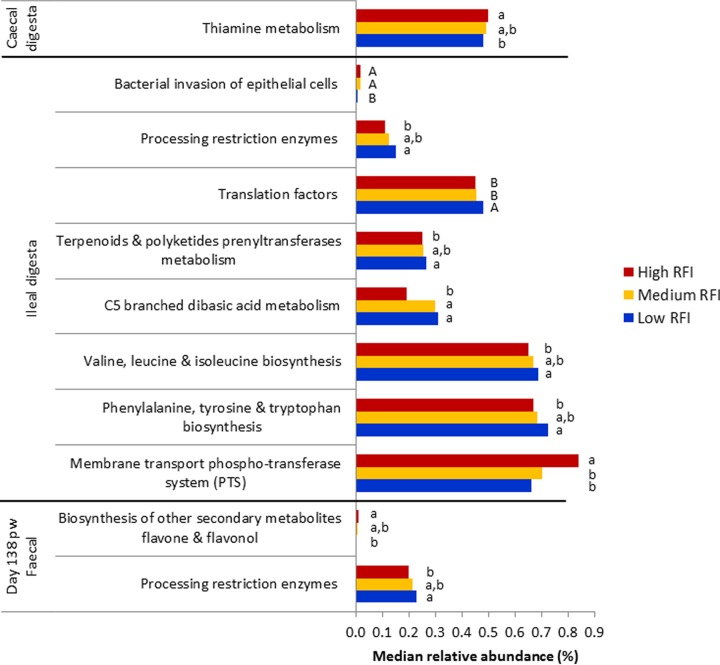
PICRUSt was employed to gain some insight into the functional capacity of the intestinal microbiota (28) of the pigs in this study and to explore any potential links with FE. Between 47 and 93% of the sequences were taxonomically assigned in the Greengenes database with 97% homology. Most of the predicted pathways identified were at very low median relative abundances (0.001% to 0.99%) (Fig. 5). Nine predicted microbial pathways differed significantly in abundances between low and high RFI pigs, including two in the feces at day 138 pw and one in the cecal digesta, but most (i.e., six) were in the ileal digesta, where another two pathways also tended to be different (P ≤ 0.10). The differentially abundant predicted pathways at the highest relative abundance were related to metabolic function in the ilea of low RFI pigs. These were enriched in low RFI pigs and included pathways involved in the biosynthesis of amino acids (phenylalanine, tyrosine, tryptophan, valine, leucine, and isoleucine) and the metabolism of C5-branched dibasic acid, terpenoids, and polyketides, as well as restriction enzyme processing. Higher restriction enzyme activity in the fecal microbiota of low RFI pigs on day 138 pw was also inferred. Furthermore, translation factors tended to be higher in relative abundance in low than in high RFI pigs (P = 0.06). Contrary to this, some of the bacterial pathways inferred were at lower relative abundances in low RFI pigs, for example, those involved in the biosynthesis of secondary metabolites in the feces at day 138 pw, thiamine metabolism in the cecum, and the phosphotransferase system (PTS) in the ileum. A pathway involved in bacterial invasion of epithelial cells also tended to be less abundant in the ilea of low RFI pigs (P = 0.08).
FIG 5.
Comparison of predicted functional pathways for the fecal and intestinal microbiota of pigs ranked by residual feed intake (RFI). Low RFI: feces (n = 12) and digesta (cecal, n = 12; ileal, n = 9); medium RFI: feces (n = 10) and digesta (cecal, n = 10; ileal, n = 9); high RFI: feces (n = 10) and digesta (cecal, n = 8; ileal, n = 6). Pathways are from the KEGG database and level 3 pathways are presented. Only 11 of 23 differences between all RFI ranks are shown, as 6 were pathways present at <0.001% median relative abundance and another 6 were differences observed for medium RFI pigs. Within each pathway, bars that do not share lowercase letters (a, b, and c) are significantly different (P ≤ 0.05), whereas those that do not share uppercase letters (A, B, and C) tended to be different (P ≤ 0.10).
Volatile fatty acid concentrations in the feces and digesta of pigs ranked by residual feed intake.
Volatile fatty acid concentrations measured in the feces collected throughout the lifetime of the pigs and in the cecal and ileal digesta collected at slaughter are presented in Tables S6 and S7 in the supplemental material. Only one difference was found between low and high RFI pigs. Low RFI pigs had a 2.3-fold higher concentration of isobutyric acid in the ileal digesta compared with that in high RFI pigs (P < 0.05) (Table S7).
Correlations between microbial composition and physiological traits in pigs ranked by residual feed intake.
Correlations were examined between bacterial taxa (at both the phylum and genus levels) and the physiological measures found to differ significantly between low and high RFI pigs (Fig. 4). In the ileum, the concentration of isobutyric acid was positively correlated with the relative abundance of Asteroleplasma. The number of goblet cells in the ileum (both per villus and per μm villus height) was negatively correlated with Proteobacteria and an uncultured bacterium from Christensenellaceae (P < 0.05). The phylum Synergistetes, an uncultured genus from Veillonellaceae, and the genera Cellulosilyticum and Parvimonas were positively correlated with goblet cell number per μm villus height (P < 0.05). Of these, both the uncultured member of Veillonellaceae and the Cellulosilyticum genus were also positively correlated with the numbers of goblet cells per villus (P < 0.05).
DISCUSSION
The advent of high-throughput sequencing has facilitated comprehensive profiling of the resident bacteria in the digestive tracts of pigs (17, 22, 29, 30). However, this study is one of the first to exploit this technology to examine the intestinal microbiota among pigs of varying FE. The metric used for FE was RFI, and ranking pigs according to this measure was particularly useful, as it allowed the selection of pigs that consume less feed to achieve the same weight gain, in agreement with previous findings for pigs divergent in RFI (14, 31). To minimize the variability in FE due to external factors, pigs were ranked by RFI within litter (to control for genetic influences), and all pigs were subjected to the same management, environmental, and nutritional conditions. When considering reasons why pigs may differ in FE, it is interesting to note that the energy-related physiological parameters measured in this study were not associated with RFI. For example, there was no difference in stress levels, as determined by salivary cortisol concentrations, despite serum cortisol having previously been suggested as a biomarker for FE in cattle (27). Furthermore, low RFI pigs tended to have higher plasma cortisol concentrations in a previous study (32), albeit no differences in salivary concentrations were found, in agreement with our findings. In addition, there were no differences in carcass weight, leanness, or organ weights among animals divergent in RFI in this study, although previous work has found that low RFI pigs have lower back fat (33). However, there likely are unmeasured attributes contributing to FE.
When looking at the intestinal microbiota, similar to previous findings in cattle, the overall intestinal bacterial diversity did not cluster by RFI rank, but rather, RFI-associated variations in community membership were detected (12, 34). Interestingly, in our study, microbial diversity, as well as composition, was impacted by both age and intestinal site. Furthermore, in feces taken at three time points, clusters converged with age, indicating that the intestinal microbiota became more homogenous among pigs over time. In agreement with results from previous studies (4, 20, 22), the core phyla within the fecal and cecal microbiota were Firmicutes and Bacteroidetes. However, the ileal microbiota composition differed from that previously found in pigs (35), with Actinobacteria replacing Bacteroidetes as the second most abundant phylum.
The hypothesis that the composition and potential functionality of the intestinal microbiota are linked with FE in pigs was supported by the differences in fecal/intestinal bacterial profiles found between RFI ranks throughout the lifetime of the pigs. However, these differences can be considered subtle, as of all the taxa detected, relatively few differed, and most that did were present at low relative abundances (<2%). Nonetheless, these taxa may still influence FE, as ultimately, it is the complex interplay within the intestinal community that would have the most influence on host homeostasis and FE. On the other hand, we cannot disregard the fact that biological variation in microbial composition between pigs could account for some of the differences found. It may also be that FE is influencing the intestinal microbiota, meaning that pigs with low RFI are more feed efficient for a number of reasons, and because of this, they have a somewhat different microbiota; but, further studies are needed to elucidate such causality. Furthermore, as sex influences FE in pigs, one might expect differences in the microbiota profiles due to sex, as previously reported (36). However, no association with sex was observed in this study.
As outlined above, most of the differences in the composition and predicted functionality of the fecal and intestinal microbiota observed between RFI ranks were subtle. Several members of the Clostridiales order previously associated with carbohydrate degradation and better metabolic efficiency (i.e., a leaner phenotype in humans and less fatness in pigs) were enriched in low RFI pigs in the feces immediately preslaughter, e.g., uncultured Christensenellaceae and Cellulosilyticum (35, 37, 38). Additionally, OTUs exclusively found in low RFI (more feed-efficient) pigs are from this order, including those that were higher in abundance in these pigs, for example, an unknown genus belonging to the vadinBB60 family. Previously, an unclassified genus belonging to this family was increased in relative abundance in rats fed a high-fat diet for 4 weeks, i.e., during the preobese state, indicating its possible role in metabolism (40).
At weaning, the butyrate producer, Butyrivibrio, was strongly correlated with low RFI, which could also be linked to an enhanced ability to ferment complex carbohydrates (41). Moreover, Prevotella, a member of Bacteroidetes considered another key microbe capable of fermenting complex carbohydrates (42, 43), was correlated with a low RFI value at weaning. The fact that this was observed at weaning may be due to the introduction of a cereal-based diet and its likely role in enhancing the growth rate postweaning (22). These findings indicate that pigs that are more feed efficient are likely to have an intestinal microbiota that is more competent in terms of digesting the carbohydrate component of the diet. Some of these microbes could also have a role in influencing ileal morphology as indicated by the negative correlation between an uncultured Christensenellaceae OTU (enriched in low RFI pigs) and the number of goblet cells per μm of villus height (lower in low RFI pigs). Furthermore, increases in members of Bacteroidetes have been reported as a driver for leaner phenotypes (35, 44–46), which was substantiated in this study by an enrichment of Bacteroides spp. in low RFI pigs. On the contrary, some other members that may have a role in carbohydrate utilization, such as the genus “Candidatus Saccharimonas” (47) or Methanosphaera, were less abundant in low RFI pigs, albeit they were present at very low median relative abundances. Actinobacillus was present at a higher abundance in the ceca of low RFI pigs, but despite this, some species potentially pathogenic to pigs (OTUs of A. pleuropneumoniae, A. porcitonsillarum, and A. rossii) were found, albeit at very low abundances, in the ilea of both low and high RFI pigs. Potentially undesirable bacteria that were less abundant in pigs that were more feed efficient included the Erysipelotrichaceae family, associated with intestinal inflammation in humans (48). In addition, genera with a possible negative effect on FE, as they contain potentially pathogenic members, e.g., Streptococcus and Solobacterium (49, 50), were relatively less abundant in the feces from pigs that were more feed efficient at day 42 pw and in the cecum at slaughter, respectively, albeit they were at low relative abundances in all groups.
The ileal microbiota was notable by virtue of the number of unique FE-associated OTUs harbored and by the fact that the bacterial metabolite isobutyric acid was found at a higher concentration in the ilea of pigs that were more feed efficient, albeit at relatively low concentrations in both groups. Isobutyric acid is an end product of protein fermentation, and increased concentrations could be indicative of better utilization of dietary protein by the microbiota (51). Furthermore, the predicted higher relative abundance of the valine, leucine, and isoleucine biosynthesis pathway in low RFI pigs may be linked to the higher concentrations of isobutyric acid in the ilea of these animals, as isobutyric acid is the end product of the microbial deamination of valine (52). On the other hand, it could also mean that these pigs are less efficient at digesting protein, leaving more available for microbial fermentation. This is especially noteworthy as it occurred in the ileum. However, poorer protein utilization would not be expected in pigs that are more feed efficient. With regard to the ileal microbiota, the most notable difference in relative abundance across RFI-ranked pigs occurred for members of Rhodococcus, a genus containing species known to cause disease in pigs (specifically, infections of the submaxillar and mesenteric lymph nodes [53, 54]), which were substantially lower in the low RFI pigs. This provides evidence that the microbiota of pigs that are more feed efficient could potentially be “healthier.” In addition, the high relative abundance of this genus is remarkable, as to our knowledge, it has not previously been reported as abundant in pigs.
In addition, ileal histology is important when considering FE. For example, longer villi and shorter crypts enhance absorptive capacity (55–57). However, while we found no differences in these parameters between pigs of varying FE, we did find fewer goblet cells along the villi of pigs that were more feed efficient. This suggests reduced mucin secretion in these animals, perhaps indicating increased nutrient absorptive capacity, as excess mucin can act as a physical barrier to absorption (58). It may also indicate less diversion of energy away from growth, as the animals are producing less mucin. Both of these hypotheses may help to explain the better FE in these animals. In agreement with the potentially lower mucin production in low RFI pigs, the Clostridium sensu stricto 1 genus, less abundant in the feces of these animals, is a mucin promoter (10), while Mucispirillum, an opportunistic mucin degrader previously found to play a role in active colitis in murine models, was more abundant in the cecum (59). Within the Clostridiales order, butyric acid-producing bacteria and mucin degraders, for which a greater number of OTUs were found in low RFI pigs, have been associated with improved gastrointestinal health in humans and animals, including pigs (60, 61), likely through increased mucin production in the colon, which enhances epithelial barrier function (11). It is noteworthy that Akkermansia, among other OTUs exclusively found in the ilea of pigs that are more feed efficient, is also linked with mucin degradation and can indicate better/healthier intestinal function, as it has been inversely correlated with metabolic disorders and intestinal inflammation (62, 63). The involvement of different metabolic pathways predicted to be either more or less relatively abundant in pigs that are more feed efficient could further justify differences in the host phenotype. For instance, genes encoding the PTS, a bacterial sugar transport system, were predicted to be relatively more abundant in the small intestine of pigs with poor FE. This could be linked with a higher bacterial energy uptake, leaving less sugar available for growth of the animal (35, 38, 64).
In conclusion, this study has revealed FE-related compositional differences within the intestinal microbiota throughout the life of the pig, but mostly at the end of the finishing period, suggesting that the intestinal microbiota has a possible link with FE in pigs. Specifically, a higher relative abundance of potentially beneficial bacteria, most notably members of Clostridiales and Bacteroidetes, and a lower relative abundance of potentially undesirable bacteria, such as Rhodococcus and Erysipelotrichaceae, were found in animals that were more feed efficient. However, it should be noted that many of the FE-associated compositional differences were relatively subtle, occurring for taxa present at low relative abundances. Nonetheless, the differentially abundant intestinal taxa identified could potentially be exploited as biomarkers for FE or manipulated by dietary means to improve FE. Although, when examined at the genus and OTU levels, some members of these taxa were uncultured, advances in culturing techniques may facilitate their exploitation in the future. However, additional research is needed to investigate the reliability of the FE-associated microbial taxa identified here, i.e., across batches of pigs/rearing environments. Furthermore, intervention studies are required to confirm the insights provided so as to improve FE in pigs.
MATERIALS AND METHODS
Ethical approval.
The pig study was approved by the animal ethics committees of Teagasc (TAEC9/2013) and Waterford Institute of Technology (13/CLS/02) and performed according to European Union regulations outlining the minimum standards for the protection of pigs (91/630/EEC) and concerning the protection of animals kept for farming purposes (98/58/EC). An experimental license (number AE1932/P004) was obtained from the Irish Health Products Regulatory Authority (HPRA).
Animal management and sample collection.
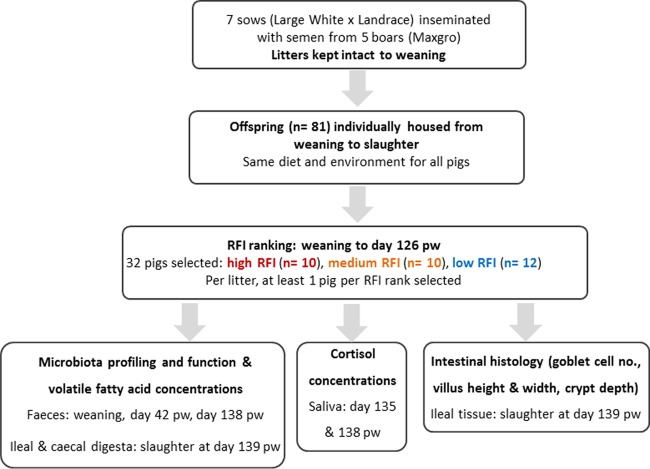
A schematic illustration depicting animal management, selection, and sample collection is shown in Fig. 6. Multiparous F1 sows (Large White × Landrace; Hermitage Genetics, Kilkenny, Ireland) were selected at weaning and randomly inseminated using semen from one of five Hylean Maxgro boars (Hermitage Genetics). At farrowing, piglets were tagged for identification purposes and weighed. Litters were kept intact between farrowing and weaning, but when necessary for welfare reasons, surplus/nonthriving pigs were fostered by nontrial sows. At weaning, 7 litters comprising male and female pigs, each with 11 to 12 pigs, were selected, so that a total of 81 pigs (44 males and 37 females) were blocked by litter ancestry and randomly assigned to individual weaner pens (1.2 m by 0.9 m) with plastic slats (Faroex, Manitoba, Canada) and solid plastic dividers between pens. On day 42 pw, pigs were transferred to individual finisher pens (1.81 m by 1.18 m, fully slatted with solid plastic panel partitions), where they remained until the end of the study. Feed was available ad libitum as dry pellets via stainless steel dry feed hoppers, 30 cm in length (O'Donovan Engineering, County Cork, Ireland). The ingredients and chemical compositions of the diets are shown in Table S1 in the supplemental material. Any pigs treated with antibiotics were removed from the study.
FIG 6.
Flow chart depicting animal management, selection, and sample collection. RFI, residual feed intake; pw, postweaning.
Individual fecal samples were collected following rectal stimulation at weaning and at days 42 pw and 138 pw (day prior to slaughter at the end of the finishing period), and were immediately snap-frozen in liquid nitrogen and stored at −80°C for microbiota and VFA analyses.
Individual body weights and feed disappearance were manually recorded every 2 weeks from weaning up to day 126 pw and used to calculate performance indicators (ADFI, ADG, and FCE). Ultrasonic back fat and muscle depth measurements were recorded using a Piglog 105 ultrasound scanner (Carometec, Herley, Denmark) on the same day as weighing, between days 42 and 126 pw. Back fat and muscle depth were measured between the 3rd and 4th last lumbar vertebrae, 7 cm from the midline for the calculation of lean meat content.
On day 126 pw, extremes for RFI (the metric used for FE in this study) were selected on the basis of measurements calculated from weaning for each pig. Residual feed intake measures the difference between actual and expected feed intake, where the expected feed intake is based on live weight, rate of gain, body fat, and muscle content of the individual pig (31). It was calculated as the residuals from a least-squares multiple-regression model of ADFI on ADG, metabolic live weight, sex, and all relevant two-way interactions, as well as the effects of back fat and muscle depth. Pigs were ranked, within each litter, by RFI (low, medium, and high, where low RFI pigs are the most feed efficient), so that a minimum of two standard deviations in RFI existed between the means of the low and the high RFI pigs within the litter. Thirty-two pigs were selected (low RFI, n = 12; medium RFI, n = 10; and high RFI, n = 10), and samples from these ranked pigs were used in all subsequent analyses.
As outlined above, pigs were tested between weaning and day 126 pw (to represent the normal productive life of pigs in Ireland). However, the mean live weight at day 126 pw was ∼129 kg, which was higher than originally predicted for this age. Following the test period, the selection of extremes in RFI was undertaken as outlined above, and, as this took time, all pigs were slaughtered 2 weeks later, on day 139 pw (corresponding to ∼166 days of age) by CO2 stunning followed by exsanguination. The hot carcass weight was recorded immediately following slaughter and was multiplied by 0.98 to obtain the cold carcass weight. Kill-out percentage was calculated as carcass weight/body weight at slaughter × 100. Back fat and muscle depth were measured at 6 cm from the edge of the split back at the third and fourth last ribs using a Hennessy grading probe (Hennessy and Chong, Auckland, New Zealand). Lean meat yield was estimated according to the following formula: lean meat yield = 60.30 − 0.847 X1 + 0.147 X2, where X1 is the back fat depth (mm) and X2 is the muscle depth (mm). Immediately after slaughter, the hearts, kidneys, livers, and lungs were collected and trimmed of fat, any blood clots were removed, and the organs were blotted dry and weighed. The stomachs were emptied of contents, flushed with water, and blotted dry before being weighed. Digesta samples were collected from the 32 selected pigs from the terminal ileum (15 cm proximal to the ileocecal junction) and from the terminal tip of the cecum. Samples were immediately snap-frozen in liquid nitrogen and stored at −80°C for subsequent microbiota and VFA analyses. Ileal tissue samples (∼3 cm sections) were collected, rinsed in phosphate-buffered saline (PBS), and placed in NOTOXhisto fixative (Scientific Device Laboratory, Des Plaines, IL, USA) on a shaker for 48 h. The samples were then stored at room temperature until histological analysis. After sampling, the small intestines were emptied of contents, flushed with water, trimmed of connective tissue, blotted dry, and weighed.
Salivary cortisol analysis.
Cortisol concentrations were determined from saliva samples collected in the days prior to slaughter (days 135 and 138 pw) in duplicate using a high sensitivity enzyme-linked immunosorbent assay (ELISA) kit (Salimetrics Europe Ltd., Suffolk, UK) according to the manufacturer's instructions.
Histological analysis of ileal tissue.
Fixed ileal tissue samples were dehydrated through a graded alcohol series, cleared with a Sub-X clearing agent (Surgipath, Richmond, IL, USA), and embedded in paraffin wax. Tissue samples were sliced using a microtome (Leica, Wetzlar, Germany), mounted on microscope slides, and stained with hematoxylin and eosin (Sigma-Aldrich, St. Louis, MO, USA) for histological analysis. Ten villi per slide were examined for villus height, villus width, crypt depth, and the number of goblet cells under a light microscope at ×400 magnification.
16S rRNA gene amplicon sequencing of fecal and intestinal microbiota.
Total DNA was extracted from fecal, ileal, and cecal samples using the QIAamp DNA stool minikit (Qiagen, Crawley, United Kingdom) according to the manufacturer's instructions, apart from adding a bead beating step after sample addition to the InhibitEX buffer and increasing the lysis temperature to 95°C to increase the DNA yield (65).
Microbial profiling was performed using high-throughput sequencing of the V3-V4 region of the 16S rRNA gene (paired-end reads of 300 bp or 250 bp) on an Illumina MiSeq platform according to the standard Illumina protocol, except that the PCR mix volume was doubled in the first PCR step, and 30 cycles were used instead of 25 (66). Any samples with less than 40,000 postquality reads were removed from the analysis. Raw sequences were merged using Flash (with a minimum overlap of 30 bp and a minimum read length of 460 bp) and quality checked using the split libraries script (with default parameters) from the QIIME package version 1.9.1. Reads were clustered into OTUs using de novo picking, with a 97% sequence identity threshold, and chimeras and singletons were removed with the 64-bit version of USEARCH (version 7) (67). Subsequently, OTUs were aligned to the SILVA rRNA specific database (version 111) to assign taxonomy, and a phylogenetic tree was generated within QIIME. Alpha diversity indices, i.e., Chao1 (which measures richness based on rare OTUs) and Shannon and Simpson (which measure richness and evenness), and β diversity analyses were also calculated within QIIME, again using a rarefaction level of 97% identity. Principal-coordinate analysis (PCoA) plots, based on unweighted UniFrac distances, were visualized using EMPeror v0.9.3-dev. Subsequent downstream images were generated with the R package Phyloseq (68).
Prediction of microbial function.
Phylogenetic Investigation of Communities by Reconstruction of Unobserved Species (PICRUSt), a tool that employs the 16S rRNA gene as a marker (28) using the 13_5 version of the Greengenes database for taxonomy and OTU assignments, was used to predict the functionality of the fecal/intestinal microbiota of the low, medium, and high RFI pigs. The prediction of functions was inferred based on Kyoto Encyclopedia of Genes and Genomes (KEGG) annotations for level 3 pathways. Pathways for which the relative abundance was <0.001% were dismissed.
Total bacterial quantification using quantitative PCR.
Quantification of the 16S rRNA gene was performed by quantitative PCR (qPCR) for all fecal and digesta samples collected in this study. A standard curve was prepared using 10-fold serial dilutions (109 to 102 copies of 16S rRNA gene/μl cloned into the pTOPO plasmid). The plasmid was first linearized and purified and the number of copies of the plasmid determined. Reactions for standards and samples were run in triplicate on a LightCycler 480 (Roche, Mannheim, Germany) using the following conditions: denaturation at 95°C for 3 min, followed by 45 amplification cycles of 95°C for 10 s, 60°C for 20 s, and 72°C for 1 s. The reactions were performed in a final volumes of 20 μl using 10 μl of Kapa SYBR fast mastermix (Kapa Biosystems, London, UK), 0.4 μM of each primer (16S rRNA forward, ACTCCTACGGGAGGCAGCAG and 16S rRNA reverse, ATTACCGCGGCTGCTGG), 7.2 μl of water, and 1 μl of DNA. Averages were calculated and the values were then converted to number of copies/μl of total DNA extracted.
Volatile fatty acid analyses of fecal and intestinal digesta samples.
Volatile fatty acid concentrations were measured in triplicate in fecal, ileal, and cecal digesta samples. Approximately 8 g of sample was weighed and the pH was recorded, and the samples were diluted with 5% trichloroacetic acid ([TCA] 2.5 × the weight of the sample) and centrifuged at 1,800 × g for 10 min at 4°C. A 1.5-ml volume of the resultant supernatant and 1.5 ml internal standard were mixed gently and filtered through a 0.45-μm-pore-size Whatman filter (VWR International Ltd., Dublin, Ireland) into a labeled 8-mm amber GC vial (Antech Solutions Ltd., Waterford, Ireland). Extracts were stored at −80°C until analysis by gas chromatography, as previously described (25, 69).
Statistical analysis.
Residual feed intake was calculated between weaning and day 126 pw as the residuals from a least-squares multiple-regression model of ADFI on ADG, with metabolic live weight, sex, and all relevant two-way interactions, as well as the effects of back-fat and muscle depth, using PROC GLM in SAS 9.3 (SAS Institute Inc., Cary, NC).
Growth performance parameters (weight, ADG, ADFI, and FCE) were analyzed using a fixed-effects linear model with sex, RFI rank, time period (biweekly weight and feed intake recordings; repeated measures), and a two-way interaction between RFI rank and time period considered fixed effects. Body weight at weaning (initial weight) was included as a covariate in the analysis. “Sow” was used as the random effect, and a repeated measures model was used to describe correlations between time periods. Physiological parameters measured at only one time point (i.e., ileal histology and salivary cortisol concentrations assessed at and prior to slaughter, respectively) were analyzed using a mixed linear model also, with the aforementioned fixed effects included in the model. Body weight at slaughter was used as a covariate in the analysis of organs (liver, lungs, heart, and kidney), stomach, and small intestine weights, and carcass cold weight was used as a covariate for carcass traits (muscle depth and fat and lean meat percentages). The full model was fitted using the Mixed procedure of SAS 9.3. Detailed comparisons of means were carried out using a Tukey's correction for multiplicity to adjust P values for the pairwise comparisons using t tests. Residual checks were made to ensure that the assumptions of the analysis were met.
Statistical differences for microbiota compositions between high, medium, and low RFI pigs were calculated in R using the SILVA 16S specific database (version 111) and were estimated using the Kruskal-Wallis test for independent samples and the Wilcoxon-Rank test for paired samples. Corrections for multiple comparisons were made using the Benjamini-Hochberg method. The qPCR data (following log10 transformation) and VFA concentrations were analyzed using a generalized linear mixed model (PROC GLIMMIX) in SAS, with the same fixed effects used in the model, as described above (feces; repeated measures analysis).
For all data, only significant differences between high and low RFI pigs are discussed, and for microbial composition data, only those significantly different bacterial taxa which were present at >0.001% median relative abundance are discussed.
Spearman rank-order correlations were performed between physiological measures found to be significantly different between low and high RFI pigs (i.e., ileal isobutyric acid concentrations and ileal goblet cell numbers) and for RFI values and taxonomic relative abundances at the phylum and genus levels for each sample type. Correlations were calculated using the PROC CORR procedure in SAS 9.3, and multiple comparisons were corrected for using the Stepdown Bonferroni test. A heatmap showing correlations was produced in R (Heatmap3 package).
Accession number(s).
The 16S rRNA gene sequence data were deposited in the European Nucleotide Archive (ENA) under the study accession number PRJEB19324.
Supplementary Material
ACKNOWLEDGMENTS
We thank the farm staff in the Pig Development Department at Teagasc, Moorepark, for assistance with pig management, as well as work placement students and technicians assisting with the pig study and laboratory work. We also thank Joseph Cassidy at University College Dublin for preparation of the ileal histology slides and Gwynneth Halley, who was supported by the Society for Applied Microbiology to work on this study.
P.G.L., G.E.G., D.B., and P.D.C. conceived the study and, together with B.U.M.-Z., E.M., and P.V., designed the experiment. P.G.L., P.D.C., and G.E.G. directed the study. P.V. supplied semen and breeding values for boars used in the study. U.M.M, T.R., and S.G.B. conducted the animal study. U.M.M., S.G.B., P.G.L., T.R., and G.E.G. collected intestinal samples. S.G.B. and D.B. analyzed and ranked the pigs by RFI. U.M.M., T.C., M.L.P., F.C., and S.G.B. performed laboratory analyses. Bioinformatics and microbiota statistical analyses were performed by O.O.S. U.M.M. statistically analyzed animal growth performance and physiological data. T.C. interpreted the microbial data together with U.M.M., G.E.G., and P.G.L. U.M.M. and T.C. wrote and edited the manuscript, and G.E.G., P.D.C., D.B., F.C., O.O.S., and P.G.L. revised the manuscript. All authors read and approved the final version of the manuscript.
The research leading to these results was funded by the European Union's Seventh Framework Programme (ECO-FCE project no. 311794) for research, technological development, and demonstration independently of any commercial input, financial or otherwise. U.M.M. is funded by the Teagasc Walsh Fellowship programme.
None of the authors had a financial or personal conflict of interest in regard to this study.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00380-17.
REFERENCES
- 1.Teagasc Pig Development Department. 2015. Pig herd performance report 2015. Teagasc, Oak Park, Carlow, Ireland. [Google Scholar]
- 2.Rotz CA. 2004. Management to reduce nitrogen losses in animal production. J Anim Sci 82(E Suppl):E119–E137. [DOI] [PubMed] [Google Scholar]
- 3.Ramayo-Caldas Y, Mach N, Lepage P, Levenez F, Denis C, Lemonnier G, Leplat JJ, Billon Y, Berri M, Dore J, Rogel-Gaillard C, Estelle J. 2016. Phylogenetic network analysis applied to pig gut microbiota identifies an ecosystem structure linked with growth traits. ISME J 10:2973–2977. doi: 10.1038/ismej.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiao L, Estellé J, Kiilerich P, Ramayo-Caldas Y, Xia Z, Feng Q, Liang S, Pedersen AØ, Kjeldsen NJ, Liu C, Maguin E, Doré J, Pons N, Le Chatelier E, Prifti E, Li J, Jia H, Liu X, Xu X, Ehrlich SD, Madsen L, Kristiansen K, Rogel-Gaillard C, Wang J. 2016. A reference gene catalogue of the pig gut microbiome. Nat Microbiol 2016:16161. doi: 10.1038/nmicrobiol.2016.161. [DOI] [PubMed] [Google Scholar]
- 5.Fouhse JM, Zijlstra RT, Willing BP. 2016. The role of gut microbiota in the health and disease of pigs. Anim Front 6:30–36. doi: 10.2527/af.2016-0031. [DOI] [Google Scholar]
- 6.Willing BP, Van Kessel AG. 2010. Host pathways for recognition: establishing gastrointestinal microbiota as relevant in animal health and nutrition. Livest Sci 133:82–91. doi: 10.1016/j.livsci.2010.06.031. [DOI] [Google Scholar]
- 7.Kennelly J, Aherne F, Sauer W. 1981. Volatile fatty acid production in the hindgut of swine. Can J Anim Sci 61:349–361. doi: 10.4141/cjas81-043. [DOI] [Google Scholar]
- 8.Liu Y. 2015. Fatty acids, inflammation and intestinal health in pigs. J Anim Sci Biotechnol 6:41. doi: 10.1186/s40104-015-0040-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shirkey T, Siggers R, Goldade B, Marshall J, Drew M, Laarveld B, Van Kessel A. 2006. Effects of commensal bacteria on intestinal morphology and expression of proinflammatory cytokines in the gnotobiotic pig. Exp Biol Med (Maywood) 231:1333–1345. [DOI] [PubMed] [Google Scholar]
- 10.Wlodarska M, Willing BP, Bravo DM, Finlay BB. 2015. Phytonutrient diet supplementation promotes beneficial Clostridia species and intestinal mucus secretion resulting in protection against enteric infection. Sci Rep 5:9253. doi: 10.1038/srep09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burger-van Paassen N, Vincent A, Puiman PJ, van der Sluis M, Bouma J, Boehm G, van Goudoever JB, van Seuningen I, Renes IB. 2009. The regulation of intestinal mucin MUC2 expression by short-chain fatty acids: implications for epithelial protection. Biochem J 420:211–219. doi: 10.1042/BJ20082222. [DOI] [PubMed] [Google Scholar]
- 12.Myer PR, Smith TPL, Wells JE, Kuehn LA, Freetly HC. 2015. Rumen microbiome from steers differing in feed efficiency. PLoS One 10:e0129174. doi: 10.1371/journal.pone.0129174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carberry CA, Waters SM, Kenny DA, Creevey CJ. 2014. Rumen methanogenic genotypes differ in abundance according to host residual feed intake phenotype and diet type. Appl Environ Microbiol 80:586–594. doi: 10.1128/AEM.03131-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vigors S, O'Doherty JV, Kelly AK, O'Shea CJ, Sweeney T. 2016. The effect of divergence in feed efficiency on the intestinal microbiota and the intestinal immune response in both unchallenged and lipopolysaccharide challenged ileal and colonic explants. PLoS One 11:e0148145. doi: 10.1371/journal.pone.0148145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pedersen R, Andersen AD, Mølbak L, Stagsted J, Boye M. 2013. Changes in the gut microbiota of cloned and non-cloned control pigs during development of obesity: gut microbiota during development of obesity in cloned pigs. BMC Microbiol 13:30. doi: 10.1186/1471-2180-13-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frese SA, Parker K, Calvert CC, Mills DA. 2015. Diet shapes the gut microbiome of pigs during nursing and weaning. Microbiome 3:28. doi: 10.1186/s40168-015-0091-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buzoianu SG, Walsh MC, Rea MC, O'Sullivan O, Crispie F, Cotter PD, Ross RP, Gardiner GE, Lawlor PG. 2012. The effect of feeding Bt MON810 maize to pigs for 110 days on intestinal microbiota. PLoS One 7:e33668. doi: 10.1371/journal.pone.0033668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Metzler-Zebeli BU, Schmitz-Esser S, Mann E, Grüll D, Molnar T, Zebeli Q. 2015. Adaptation of the cecal bacterial microbiome of growing pigs in response to resistant starch type 4. Appl Environ Microbiol 81:8489–8499. doi: 10.1128/AEM.02756-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim HB, Isaacson RE. 2015. The pig gut microbial diversity: understanding the pig gut microbial ecology through the next generation high throughput sequencing. Vet Microbiol 177:242–251. doi: 10.1016/j.vetmic.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 20.Zhao W, Wang Y, Liu S, Huang J, Zhai Z, He C, Ding J, Wang J, Wang H, Fan W, Zhao J, Meng H. 2015. The dynamic distribution of porcine microbiota across different ages and gastrointestinal tract segments. PLoS One 10:e0117441. doi: 10.1371/journal.pone.0117441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han GG, Lee J-Y, Jin G-D, Park J, Choi YH, Chae BJ, Kim EB, Choi Y-J. 2016. Evaluating the association between body weight and the intestinal microbiota of weaned piglets via 16S rRNA sequencing. Vet Microbiol 196:55–62. doi: 10.1016/j.vetmic.2016.10.020. [DOI] [PubMed] [Google Scholar]
- 22.Mach N, Berri M, Estellé J, Levenez F, Lemonnier G, Denis C, Leplat J-J, Chevaleyre C, Billon Y, Doré J, Rogel-Gaillard C, Lepage P. 2015. Early-life establishment of the swine gut microbiome and impact on host phenotypes. Environ Microbiol Rep 7:554–569. doi: 10.1111/1758-2229.12285. [DOI] [PubMed] [Google Scholar]
- 23.Vigors S, Sweeney T, O'Shea CJ, Kelly AK, O'Doherty JV. 2016. Pigs that are divergent in feed efficiency, differ in intestinal enzyme and nutrient transporter gene expression, nutrient digestibility and microbial activity. Animal 10:1848–1855. doi: 10.1017/S1751731116000847. [DOI] [PubMed] [Google Scholar]
- 24.Hou C, Liu H, Zhang J, Zhang S, Yang F, Zeng X, Thacker PA, Zhang G, Qiao S. 2015. Intestinal microbiota succession and immunomodulatory consequences after introduction of Lactobacillus reuteri I5007 in neonatal piglets. PLoS One 10:e0119505. doi: 10.1371/journal.pone.0119505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prieto ML, O'Sullivan L, Tan SP, McLoughlin P, Hughes H, O'Donovan O, Rea MC, Kent RM, Cassidy JP, Gardiner GE, Lawlor PG. 2014. Evaluation of the efficacy and safety of a marine-derived Bacillus strain for use as an in-feed probiotic for newly weaned pigs. PLoS One 9:e88599. doi: 10.1371/journal.pone.0088599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Vos WM. 2013. Fame and future of faecal transplantations–developing next-generation therapies with synthetic microbiomes. Microb Biotechnol 6:316–325. doi: 10.1111/1751-7915.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richardson E, Herd R, Archer J, Arthur P. 2004. Metabolic differences in Angus steers divergently selected for residual feed intake. Aust J Exp Agric 44:441–452. doi: 10.1071/EA02219. [DOI] [Google Scholar]
- 28.Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C. 2013. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buzoianu SG, Walsh MC, Rea MC, Quigley L, O'Sullivan O, Cotter PD, Ross RP, Gardiner GE, Lawlor PG. 2013. Sequence-based analysis of the intestinal microbiota of sows and their offspring fed genetically modified maize expressing a truncated form of Bacillus thuringiensis Cry1ab protein (Bt maize). Appl Environ Microbiol 79:7735–7744. doi: 10.1128/AEM.02937-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim J, Nguyen SG, Guevarra RB, Lee I, Unno T. 2015. Analysis of swine fecal microbiota at various growth stages. Arch Microbiol 197:753–759. doi: 10.1007/s00203-015-1108-1. [DOI] [PubMed] [Google Scholar]
- 31.Patience JF, Rossoni-Serão MC, Gutiérrez NA. 2015. A review of feed efficiency in swine: biology and application. J Anim Sci Biotechnol 6:33. doi: 10.1186/s40104-015-0031-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lefaucheur L, Lebret B, Ecolan P, Louveau I, Damon M, Prunier A, Billon Y, Sellier P, Gilbert H. 2011. Muscle characteristics and meat quality traits are affected by divergent selection on residual feed intake in pigs. J Anim Sci 89:996–1010. doi: 10.2527/jas.2010-3493. [DOI] [PubMed] [Google Scholar]
- 33.Cai W, Casey D, Dekkers J. 2008. Selection response and genetic parameters for residual feed intake in Yorkshire swine. J Anim Sci 86:287–298. doi: 10.2527/jas.2007-0396. [DOI] [PubMed] [Google Scholar]
- 34.Jewell KA, McCormick CA, Odt CL, Weimer PJ, Suen G. 2015. Ruminal bacterial community composition in dairy cows is dynamic over the course of two lactations and correlates with feed efficiency. Appl Environ Microbiol 81:4697–4710. doi: 10.1128/AEM.00720-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang H, Huang X, Fang S, Xin W, Huang L, Chen C. 2016. Uncovering the composition of microbial community structure and metagenomics among three gut locations in pigs with distinct fatness. Sci Rep 6:27427. doi: 10.1038/srep27427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou Z, Zheng W, Shang W, Du H, Li G, Yao W. 2015. How host gender affects the bacterial community in pig feces and its correlation to skatole production. Ann Microbiol 65:2379–2386. doi: 10.1007/s13213-015-1079-0. [DOI] [Google Scholar]
- 37.Miller DA, Suen G, Bruce D, Copeland A, Cheng JF, Detter C, Goodwin LA, Han CS, Hauser LJ, Land ML, Lapidus A, Lucas S, Meincke L, Pitluck S, Tapia R, Teshima H, Woyke T, Fox BG, Angert ER, Currie CR. 2011. Complete genome sequence of the cellulose-degrading bacterium Cellulosilyticum lentocellum. J Bacteriol 193:2357–2358. doi: 10.1128/JB.00239-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, Beaumont M, Van Treuren W, Knight R, Bell JT, Spector TD, Clark AG, Ley RE. 2014. Human genetics shape the gut microbiome. Cell 159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reference deleted.
- 40.Lin H, An Y, Hao F, Wang Y, Tang H. 2016. Correlations of fecal metabonomic and microbiomic changes induced by high-fat diet in the pre-obesity state. Sci Rep 6:21618. doi: 10.1038/srep21618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hespell RB, Wolf R, Bothast RJ. 1987. Fermentation of xylans by Butyrivibrio fibrisolvens and other ruminal bacteria. Appl Environ Microbiol 53:2849–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Umu ÖCO, Frank JA, Fangel JU, Oostindjer M, da Silva CS, Bolhuis EJ, Bosch G, Willats WGT, Pope PB, Diep DB. 2015. Resistant starch diet induces change in the swine microbiome and a predominance of beneficial bacterial populations. Microbiome 3:16. doi: 10.1186/s40168-015-0078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. 2008. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Microbiol 6:121–131. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- 44.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. 2009. A core gut microbiome in obese and lean twins. Nature 457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Torok VA, Hughes RJ, Mikkelsen LL, Perez-Maldonado R, Balding K, MacAlpine R, Percy NJ, Ophel-Keller K. 2011. Identification and characterization of potential performance-related gut microbiotas in broiler chickens across various feeding trials. Appl Environ Microbiol 77:5868–5878. doi: 10.1128/AEM.00165-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. 2011. Linking long-term dietary patterns with gut microbial enterotypes. Science 334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niu Q, Li P, Hao S, Zhang Y, Kim SW, Li H, Ma X, Gao S, He L, Wu W, Huang X, Hua J, Zhou B, Huang R. 2015. Dynamic distribution of the gut microbiota and the relationship with apparent crude fiber digestibility and growth stages in pigs. Sci Rep 5:9938. doi: 10.1038/srep09938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaakoush NO. 2015. Insights into the role of Erysipelotrichaceae in the human host. Front Cell Infect Microbiol 5:84. doi: 10.3389/fcimb.2015.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goyette-Desjardins G, Auger J-P, Xu J, Segura M, Gottschalk M. 2014. Streptococcus suis, an important pig pathogen and emerging zoonotic agent-an update on the worldwide distribution based on serotyping and sequence typing. Emerg Microbes Infect 3:e45. doi: 10.1038/emi.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pedersen RM, Holt HM, Justesen US. 2011. Solobacterium moorei bacteremia: identification, antimicrobial susceptibility, and clinical characteristics. J Clin Microbiol 49:2766–2768. doi: 10.1128/JCM.02525-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walsh AM, Sweeney T, Bahar B, Flynn B, O'Doherty JV. 2013. The effects of supplementing varying molecular weights of chitooligosaccharide on performance, selected microbial populations and nutrient digestibility in the weaned pig. Animal 7:571–579. doi: 10.1017/S1751731112001759. [DOI] [PubMed] [Google Scholar]
- 52.Zarling EJ, Ruchim MA. 1987. Protein origin of the volatile fatty acids isobutyrate and isovalerate in human stool. J Lab Clin Med 109:566–570. [PubMed] [Google Scholar]
- 53.Lara GH, Ribeiro MG, Leite CQ, Paes AC, Guazzelli A, da Silva AV, Santos AC, Listoni FJ. 2011. Occurrence of Mycobacterium spp. and other pathogens in lymph nodes of slaughtered swine and wild boars (Sus scrofa). Res Vet Sci 90:185–188. doi: 10.1016/j.rvsc.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 54.Komijn RE, Wisselink HJ, Rijsman VM, Stockhofe-Zurwieden N, Bakker D, van Zijderveld FG, Eger T, Wagenaar JA, Putirulan FF, Urlings BA. 2007. Granulomatous lesions in lymph nodes of slaughter pigs bacteriologically negative for Mycobacterium avium subsp. avium and positive for Rhodococcus equi. Vet Microbiol 120:352–357. doi: 10.1016/j.vetmic.2006.10.031. [DOI] [PubMed] [Google Scholar]
- 55.Chen H, Mao XB, Che LQ, Yu B, He J, Yu J, Han GQ, Huang ZQ, Zheng P, Chen DW. 2014. Impact of fiber types on gut microbiota, gut environment and gut function in fattening pigs. Anim Feed Sci Technol 195:101–111. doi: 10.1016/j.anifeedsci.2014.06.002. [DOI] [Google Scholar]
- 56.Pluske JR, Pethick DW, Hopwood DE, Hampson DJ. 2002. Nutritional influences on some major enteric bacterial diseases of pigs. Nutr Res Rev 15:333–371. doi: 10.1079/NRR200242. [DOI] [PubMed] [Google Scholar]
- 57.Lalles JP, Bosi P, Smidt H, Stokes CR. 2007. Nutritional management of gut health in pigs around weaning. Proc Nutr Soc 66:260–268. doi: 10.1017/S0029665107005484. [DOI] [PubMed] [Google Scholar]
- 58.Montagne L, Piel C, Lalles JP. 2004. Effect of diet on mucin kinetics and composition: nutrition and health implications. Nutr Rev 62:105–114. doi: 10.1111/j.1753-4887.2004.tb00031.x. [DOI] [PubMed] [Google Scholar]
- 59.Rooks MG, Veiga P, Wardwell-Scott LH, Tickle T, Segata N, Michaud M, Gallini CA, Beal C, van Hylckama-Vlieg JET, Ballal SA, Morgan XC, Glickman JN, Gevers D, Huttenhower C, Garrett WS. 2014. Gut microbiome composition and function in experimental colitis during active disease and treatment-induced remission. ISME J 8:1403–1417. doi: 10.1038/ismej.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Berni Canani R, Sangwan N, Stefka AT, Nocerino R, Paparo L, Aitoro R, Calignano A, Khan AA, Gilbert JA, Nagler CR. 2016. Lactobacillus rhamnosus GG-supplemented formula expands butyrate-producing bacterial strains in food allergic infants. ISME J 10:742–750. doi: 10.1038/ismej.2015.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Levine UY, Looft T, Allen HK, Stanton TB. 2013. Butyrate-producing bacteria, including mucin degraders, from the swine intestinal tract. Appl Environ Microbiol 79:3879–3881. doi: 10.1128/AEM.00589-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, de Vos WM, Cani PD. 2013. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A 110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schneeberger M, Everard A, Gómez-Valadés AG, Matamoros S, Ramírez S, Delzenne NM, Gomis R, Claret M, Cani PD. 2015. Akkermansia muciniphila inversely correlates with the onset of inflammation, altered adipose tissue metabolism and metabolic disorders during obesity in mice. Sci Rep 5:16643. doi: 10.1038/srep16643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Deutscher J, Francke C, Postma PW. 2006. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev 70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Buzoianu SG, Walsh MC, Rea MC, O'Sullivan O, Cotter PD, Ross RP, Gardiner GE, Lawlor PG. 2012. High-throughput sequence-based analysis of the intestinal microbiota of weanling pigs fed genetically modified MON810 maize expressing Bacillus thuringiensis Cry1Ab (Bt Maize) for 31 days. Appl Environ Microbiol 78:4217–4224. doi: 10.1128/AEM.00307-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fouhy F, Deane J, Rea MC, O'Sullivan O, Ross RP, O'Callaghan G, Plant BJ, Stanton C. 2015. The effects of freezing on faecal microbiota as determined using MiSeq sequencing and culture-based investigations. PLoS One 10:e0119355. doi: 10.1371/journal.pone.0119355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 68.McMurdie PJ, Holmes S. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lynch MB, O'Shea CJ, Sweeney T, Callan JJ, O'Doherty JV. 2008. Effect of crude protein concentration and sugar-beet pulp on nutrient digestibility, nitrogen excretion, intestinal fermentation and manure ammonia and odour emissions from finisher pigs. Animal 2:425–434. doi: 10.1017/S1751731107001267. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.