ABSTRACT
In our endeavor to improve the nitrogen fixation efficiency of a soil diazotroph that would be unaffected by synthetic nitrogenous fertilizers, we have deleted a part of the negative regulatory gene nifL and constitutively expressed the positive regulatory gene nifA in the chromosome of Azotobacter chroococcum CBD15, a strain isolated from the local field soil. No antibiotic resistance gene or other foreign gene was present in the chromosome of the engineered strain. Wheat seeds inoculated with this engineered strain, which we have named Azotobacter chroococcum HKD15, were tested for 3 years in pots and 1 year in the field. The yield of wheat was enhanced by ∼60% due to inoculation of seeds by A. chroococcum HKD15 in the absence of any urea application. Ammonium only marginally affected acetylene reduction by the engineered Azotobacter strain. When urea was also applied, the same wheat yield could be sustained by using seeds inoculated with A. chroococcum HKD15 and using ∼85 kg less urea (∼40 kg less nitrogen) than the usual ∼257 kg urea (∼120 kg nitrogen) per hectare. Wheat plants arising from the seeds inoculated with the engineered Azotobacter strain exhibited far superior overall performance, had much higher dry weight and nitrogen content, and assimilated molecular 15N much better. A nitrogen balance experiment also revealed much higher total nitrogen content. Indole-3-acetic acid (IAA) production by the wild type and that by the engineered strain were about the same. Inoculation of the wheat seeds with A. chroococcum HKD15 did not adversely affect the microbial population in the field rhizosphere soil.
IMPORTANCE Application of synthetic nitrogenous fertilizers is a standard agricultural practice to augment crop yield. Plants, however, utilize only a fraction of the applied fertilizers, while the unutilized fertilizers cause grave environmental problems. Wild-type soil diazotrophic microorganisms cannot replace synthetic nitrogenous fertilizers, as these reduce atmospheric nitrogen very inefficiently and almost none at all in the presence of added nitrogenous fertilizers. If the nitrogen-fixing ability of soil diazotrophs could be improved and sustained even in the presence of synthetic nitrogenous fertilizers, then a mixture of the bacteria and a reduced quantity of chemical nitrogenous fertilizers could be employed to obtain the same grain yield but at a much-reduced environmental cost. The engineered Azotobacter strain that we have reported here has considerably enhanced nitrogen fixation and excretion abilities and can replace ∼85 kg of urea per hectare but sustain the same wheat yield, if the seeds are inoculated with it before sowing.
KEYWORDS: Azotobacter, urea, wheat crop
INTRODUCTION
Inoculation of wheat seeds by different species of the soil diazotroph Azotobacter usually results in only about 8% to 10% wheat crop yield enhancement (1). It is, however, not clear if this enhancement is due to biologically reduced nitrogen or due to the plant growth substances influenced or elaborated by Azotobacter (2). On the other hand, application of 257 kg of urea (120 kg nitrogen) per hectare can enhance wheat grain yield by ∼100%. Production of any synthetic nitrogenous fertilizer, however, is highly energy intensive and results in ∼10-fold or higher emission of CO2 equivalent (3). Besides, only 30 to 40% of the applied urea is utilized by the wheat crop (4), and the rest pollutes groundwater and streams, rivers, and lakes. The situation can be severe. On 15 January 2017, taps ran dry in major parts of the city of Delhi, capital of India, because “dangerously high” levels of ammonia in raw water forced suspension of operations in two water treatment plants (5). Delhi receives most of its raw water from two neighboring states, both mainly agriculture oriented. The situation elsewhere may also be problematic. The pollution cost of chemical nitrogenous fertilizers for just the European Union has been estimated to be between €70 and €320 billion per year (6).
The positive regulator for all operons involved in the main nitrogen fixation pathway in Azotobacter is NifA. The negative regulator NifL, on activation by ammonia, interacts with NifA and inactivates it (7). In addition, the promoter of the nifLA operon is highly regulated (8). In order to achieve constitutive biological production of ammonia, even in the presence of chemically synthesized nitrogenous fertilizer, one has to inactivate or, better, delete nifL and simultaneously express nifA under a constitutive promoter. Unlike that in Klebsiella pneumoniae, the expression of the nifLA operon in Azotobacter vinelandii UW is not autogenously regulated (9).
The objective of the present study was to develop an improved Azotobacter strain from a local field isolate that would reduce nitrogen constitutively and to evaluate its ability to significantly enhance wheat crop yield. Though we had in our hands the engineered A. vinelandii UW (10), because of environmental concern we did not consider it prudent to apply to our field the bacterium whose parent was from Madison, WI. We also wanted to develop and test a general protocol by which in future the chromosome of other Azotobacter species isolated from fields of different and diverse areas of the world could be easily engineered by others in local laboratories to constitutively fix nitrogen. Again, because of environmental concern, we ensured that the mutant Azotobacter strain that we would use to inoculate wheat seeds would have neither any antibiotic resistance gene nor any other foreign gene. Here, we present the results obtained with Azotobacter chroococcum HKD15, a constitutive mutant (which can reduce nitrogen even in the presence of fixed nitrogen or chemical nitrogenous fertilizers) and a hyper-nitrogen fixer that has been engineered from the wild-type A. chroococcum CBD15, which was isolated from the fields of the Indian Agricultural Research Institute, New Delhi, India. This mutant strain, when used to inoculate wheat seeds, enhanced wheat grain yield by about 60% in the absence of any urea application. When urea was also applied, wheat grain yield could be sustained by applying ∼86 kg less urea than the usual ∼257 kg urea (∼120 kg nitrogen) per hectare. Such novel nitrogen-fixing strains can be of great environmental, agronomical, and economic significance. We strongly believe that the research work presented here is a viable and sustainable alternative to the nitrogen-reducing cereals which scientists pursued unsuccessfully for almost 4 decades.
RESULTS
The nifL gene of A. chroococcum CBD15.
Hybridization of the restriction subfragments of the ∼7.5-kb BamHI fragment of the genomic DNA of A. chroococcum CBD15 with nifL and nifA of A. vinelandii UW suggested that nifL was adjacent to nifA in A. chroococcum CBD15 (pCL6, Fig. 1A). The base sequence of nifL of A. chroococcum CBD15 (see Fig. S1 in the supplemental material) (GenBank accession no. KY781893) has 99% homology with the base sequence of nifL of A. vinelandii UW.
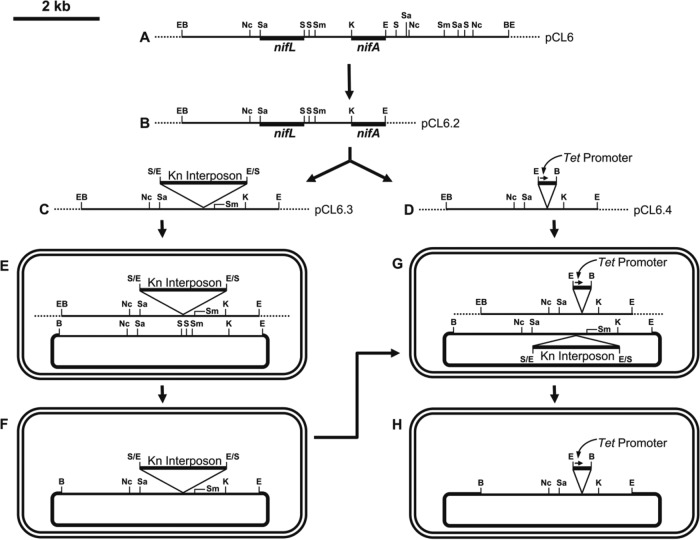
FIG 1.
Cloning steps involved for construction of engineered Azotobacter. (A) Partial restriction map of the ∼7.5-kb BamHI genomic fragment from A. chroococcum CBD15, which contains regions homologous to nifL and nifA of A. vinelandii UW, cloned in the BamHI site of pUC7. The construct has been designated pCL6. The restriction subfragments that hybridize with nifL and nifA of A. vinelandii UW are shown by thick lines. (B) The ∼4.8-kb EcoRI fragment from pCL6 containing the regions homologous to nifL and nifA of A. vinelandii UW was cloned in pUC7, and this construct has been designated pCL6.2. (C) The ∼2.0-kb EcoRI fragment from pHP45ΩKm containing the interposon ΩKm was inserted into the SalI sites of pCL6.2, and this construct has been designated pCL6.3. (D) The construct pCL6.2 was digested with SalI, and deletion of 1,112 bp was achieved by subsequent Bal 31 treatment; the 374-bp EcoRI-BamHI fragment from pBR322 containing the Tet promoter was cloned there in the correct orientation. This construct has been designated pCL6.4. (E) The construct pCL6.3 was introduced into A. chroococcum CBD15 by electroporation. (F) Insertion of the kanamycin interposon into the nifL gene in the genome of A. chroococcum CBD15, as a result of homologous recombination between pCL6.3 and the A. chroococcum CBD15 genome. (G) The construct pCL6.4 was introduced by electroporation into A. chroococcum CBD15, which already had the kanamycin interposon inserted into the nifL gene in its genome. (H) Replacement of the region of the nifL gene comprising 1,112 bp around the two SalI sites and the kanamycin interposon in the genome of A. chroococcum CBD15, as a result of homologous recombination with pCL6.4. Abbreviations: E, EcoRI; B, BamHI; Nc, NcoI; S, SalI; Sa, SacII; Sm, SmaI; K, KpnI; Kn, kanamycin.
Insertion of a constitutive promoter upstream of nifA after deletion in nifL in the chromosome of A. chroococcum.
Direct single-step insertion was ruled out because of the lack of an easy selection procedure. The strain with a deletion in nifL and the strain with subsequent insertion of the constitutive promoter are both likely to be nif+. A two-step approach was therefore chosen. In the first step, the kanamycin interposon ΩKm (11) was inserted into nifL (Fig. 1C, E, and F). The cassette KIXX (12, 13) was not used instead, as it would have provided a promoter to nifA downstream, and hence, the kanamycin-resistant A. chroococcum CBD15 would have been nif+. Insertion of ΩKm indeed resulted in a nif-minus derivative. This validated our assumption that nifL and nifA were in the same operon, nifL being proximal and nifA distal to the promoter, a structure similar to that in A. vinelandii UW. In the second step, the interposon was replaced by a deleted nifL containing the Tet promoter (Fig. 1D, G, and H) (the base sequence of the restriction fragment containing the Tet promoter is shown in Fig. S2), which resulted in a nif+ strain. This strain has been named Azotobacter chroococcum HKD15 (the base sequence of the nifL region is shown in Fig. S3).
Multiple copies of chromosomes in A. chroococcum CBD15.
The kanamycin-resistant cells obtained after the first step mentioned above, however, could also grow without ammonium acetate on Burk's nitrogen-free (BNF) medium and kanamycin. This could mean the presence of multiple copies of chromosomes in A. chroococcum CBD15, similar to A. vinelandii UW (14, 15), and incomplete segregation of chromosomes containing ΩKm. We continued subculturing on BNF medium containing kanamycin and ammonium acetate and examined nifL in the chromosome by PCR. Even after the sixth subculture, wild-type chromosomes containing no ΩKm were predominant compared to chromosomes containing ΩKm (lane 1, Fig. 2). A single PCR band representing nifL with the inserted ΩKm was visible only after the 18th subculture, with no sign of nifL devoid of any ΩKm (lane 2), and even with a large excess of PCR product (lane 3). The cells after 18 subcultures could no longer grow on BNF medium containing kanamycin in the absence of ammonium acetate. Nitrogen fixation had been abolished.
FIG 2.
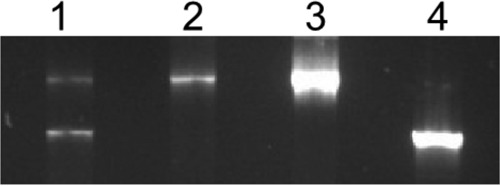
Photograph of agarose gel after electrophoresis of PCR products. Genomic DNA was isolated from kanamycin-resistant cells obtained after transformation by electroporation of A. chroococcum CBD15 with pCL6.3 (Fig. 1) and subcultured on BNF agar plus kanamycin and ammonium acetate. PCR of the nifL region was conducted by using the primers 5′-ACGAATCGATCCTCTCCAAC-3′ and 5′-TTCGCGATAGTCGGTGTCTAC-3′. The PCR products were electrophoretically separated on an agarose gel. Lane 1, PCR products of chromosomal DNA from cells subcultured six times; lane 2, PCR products of chromosomal DNA from cells subcultured 18 times; lane 3, large excess of PCR products of chromosomal DNA from cells subcultured 18 times; lane 4, PCR products of chromosomal DNA from A. chroococcum CBD15.
Similarly, after the second step, in which we attempted to replace ΩKm with a deleted nifL containing the Tet promoter, the cells could grow in the absence of ammonium acetate but were still resistant to kanamycin, again possibly because of incomplete segregation of the chromosomes containing the Tet promoter. Subculturing was continued 20 times on BNF medium without ammonium acetate and kanamycin. The cells then became sensitive to kanamycin and could grow without ammonium acetate.
Multiple copies of chromosomes were indeed found to be present in A. chroococcum CBD15. The DNA content of A. chroococcum CBD15 was determined to be 1.18 × 10−13 g per mid-exponential-phase cell when grown in a shake flask in BNF medium containing ammonium acetate. The size of the chromosome of A. chroococcum M4 was earlier estimated to be ∼5,300 kb (16). Assuming that the size of the chromosome of A. chroococcum CBD15 would be similar to that of A. chroococcum M4, we have calculated that each mid-exponential-phase cell of A. chroococcum CBD15 has ∼20 copies of the chromosome.
Confirmation of the deletion of a part of the nifL gene and the insertion of the Tet promoter there in the chromosome of A. chroococcum HKD15.
The deletion of a part of nifL and the insertion of the Tet promoter there in the chromosome of A. chroococcum HKD15 have been confirmed by PCR analysis (Fig. 3, top). Complete segregation of the Tet promoter containing copies of chromosomes was confirmed by the absence of any wild-type band at ∼1.4 kb (Fig. 3, bottom). Both primer sequences used for the PCR product shown in the bottom panel were chosen from the nifL sequence flanking the Tet promoter fragment, and only one band of ∼650 bp representing the Tet promoter-containing copies of chromosomes appeared.
FIG 3.
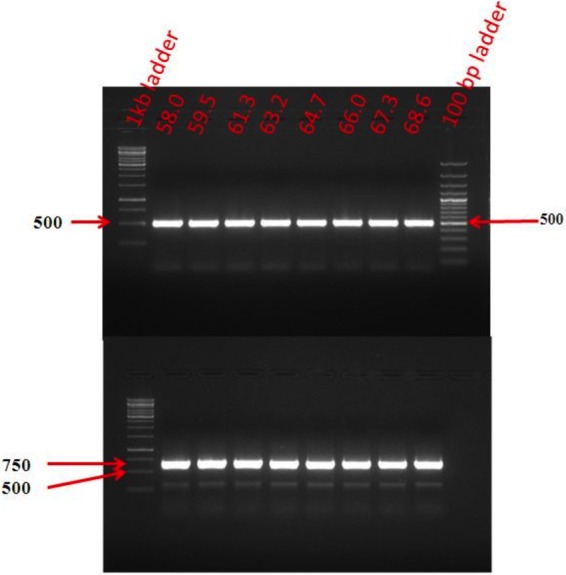
PCR analysis to check whether the DNA fragment containing the Tet promoter has indeed been integrated into the nifL gene in the genome of A. chroococcum HKD15. The PCR patterns have been obtained with genomic DNA of A. chroococcum HKD15. The annealing temperature (degrees Celsius) is shown on top of each lane. Primer sequences used for the reaction represented by the top panel were (i) 5′-CCGCACCATCACCGGCTACGGCAGC-3′ and (ii) 5′-GACGGGTGTGGTCGCCATGATCGCG-3′, the first one being from the 5′ end of the nifL sequence (see Fig. S1), while the second one was from the 3′ end of the Tet promoter fragment (see Fig. S2). Primer sequences used for reaction represented by the bottom panel were (i) 5′-CCGCACCATCACCGGCTACGGCAGC-3′ and (ii) 5′-GCTCGACGACGATCCGGCAGCCTTC-3′, the first one being from the 5′ end of the nifL sequence, while the second one was from the 3′ end (see Fig. S1).
Comparison of the DNA sequence of the nifL region of A. chroococcum HKD15 (Fig. S3) with that of nifL of A. chroococcum CBD15 revealed the deletion of 1,112 bases from nifL and the insertion of the 378-bp fragment containing the Tet promoter in the correct orientation. No PCR product was seen when primers (forward primer 5′-CAGCGAATTGCACGAACTGGAACA-3′; reverse primer 5′-TTGAGGTTGACCGGCATCTTGGA-3′) were chosen from the deleted region of nifL in DNA from A. chroococcum HKD15. The absence of a wild-type band at ∼700 bp was the final evidence of complete segregation of the deleted copies of the chromosome in A. chroococcum HKD15.
Fate of the NifL protein.
The nifL gene of A. chroococcum CBD15 has 1,560 bases excluding the chain-terminating codon; hence, the NifL protein has 520 amino acids. The deletion took place after the first 290 bases, and so only the first 96 amino acids of NifL remained. Altogether, 1,112 bases were deleted and 378 bases were added by the fragment containing the Tet promoter. Thus, a net 734 bases were lost. So, the bases downstream of the deletion are not in frame any more. Thus, all amino acids beyond the first 96 amino acids of A. chroococcum HKD15 NifL would be different from the amino acids of A. chroococcum CBD15 NifL. Study of the NifL-NifA interaction has revealed involvement of only the COOH-terminal half of NifL (17). NifA activity of A. chroococcum HKD15 would, therefore, not likely be affected even in the presence of fixed nitrogen.
Characteristics of the mutant Azotobacter.
Acetylene reduction by A. chroococcum HKD15 was more than 4 times that by A. chroococcum CBD15 (Table 1). Ammonium acetate reduced acetylene reduction by A. chroococcum CBD15 by ∼70% but reduced that by A. chroococcum HKD15 by only ∼15%. Ammonium excretion by A. chroococcum HKD15 was more than 8-fold that by A. chroococcum CBD15 (Table 2). KNO3 drastically reduced ammonium excretion by A. chroococcum CBD15, while that by A. chroococcum HKD15 was marginally affected. Production of indole acetic acid by A. chroococcum HKD15 was about the same as that by A. chroococcum CBD15 (Table S1).
TABLE 1.
Acetylene reduction by A. chroococcum CBD15 and A. chroococcum HKD15
| A. chroococcum strain | Ethylene produceda (nmol/mg protein/h) |
Inhibition by ammonium acetate (%) | |
|---|---|---|---|
| N-free growth medium | Growth medium containing 0.11% ammonium acetate | ||
| CBD15 | 755 (75) | 227 (13) | 70 |
| HKD15 | 3,153 (67) | 2,682 (126) | 15 |
Average from 3 experiments; standard deviations in parentheses.
TABLE 2.
Ammonium excretion by A. chroococcum CBD15 and A. chroococcum HKD15
| A. chroococcum strain | Ammonium excreteda (μg/mg protein) |
Inhibition of excretion (%) | |
|---|---|---|---|
| In absence of KNO3 | In presence of KNO3 (0.1%) | ||
| CBD15 | 31 (3) | 4 (1) | 87 |
| HKD15 | 267 (8) | 227 (4) | 15 |
Average from 3 experiments; standard deviations in parentheses.
Pot experiment with soil.
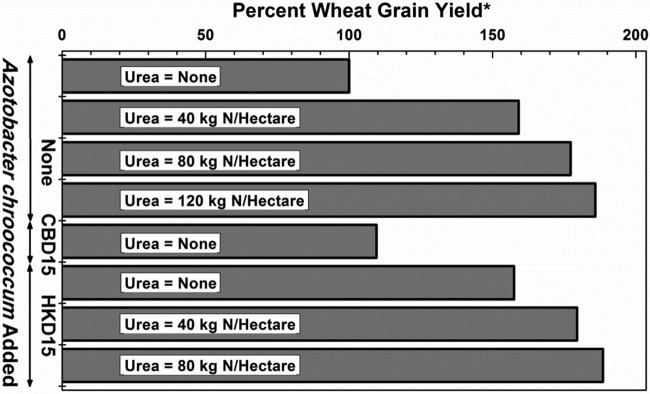
The average available nitrogen in the soil of the pots, as determined (18) in 12 random samples, was 1.56 × 10−4 kg N per kg soil (standard deviation, 0.145 × 10−4). Until about 2 months after sowing, the dry weight of wheat seedlings that arose from the seeds inoculated with A. chroococcum HKD15 but did not receive any urea application was comparable to the dry weight of seedlings that arose from seeds that were not inoculated but received 257 kg of urea (120 kg N) per hectare. Figure 4 shows the yield of wheat crop in the pot experiments. Inoculation of wheat seeds with A. chroococcum HKD15 enhanced wheat grain yield by ∼60% in the absence of any urea application. When urea was also applied, a savings of ∼86 kg urea (40 kg N) per hectare could be achieved on inoculation of wheat seeds with A. chroococcum HKD15.
FIG 4.
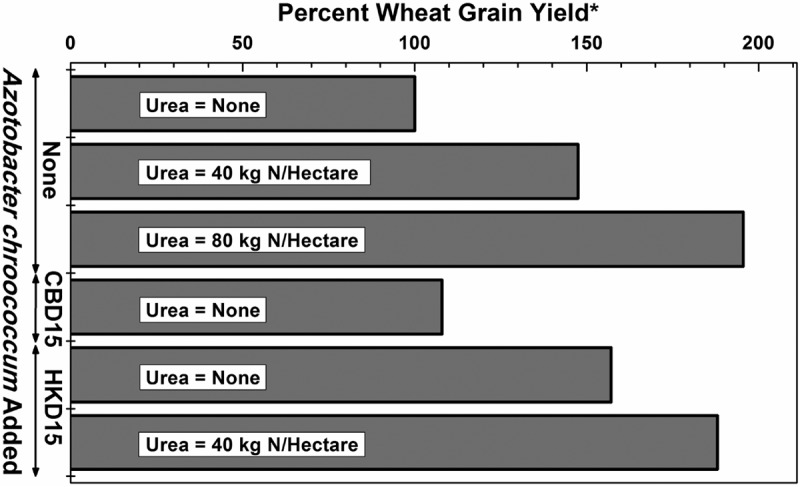
Effect of A. chroococcum on wheat (HD 2967) grain yield in pots. The pot experiment with soil was spread over 3 years, and 75 plants were cultivated per treatment per year. The amounts of urea mentioned are in kilograms of N per hectare. *, plants neither fertilized with urea nor inoculated with any bacteria were considered as the control (100%). The actual average wheat crop yield for this treatment per plant per year was 2.01 g. The standard errors of the means for the 1st, 2nd, and 3rd years are 0.05, 0.07, and 0.11, respectively. The critical differences (equivalent to least significant difference) at 5% for the 1st, 2nd, and 3rd years are 0.17, 0.20, and 0.31, respectively. The coefficients of variance for the first, second, and third years are 5.50, 6.90, and 8.10, respectively.
Field experiment.
Average available nitrogen in the soil of the field, as determined (18) in 9 random samples, was 1.64 × 10−4 kg N per kg soil (standard deviation, 0.157 × 10−4). Each plot was 1.5 m by 3.0 m. Three replicate plots were there for each treatment.
Did the mutant Azotobacter thrive in the field soil?
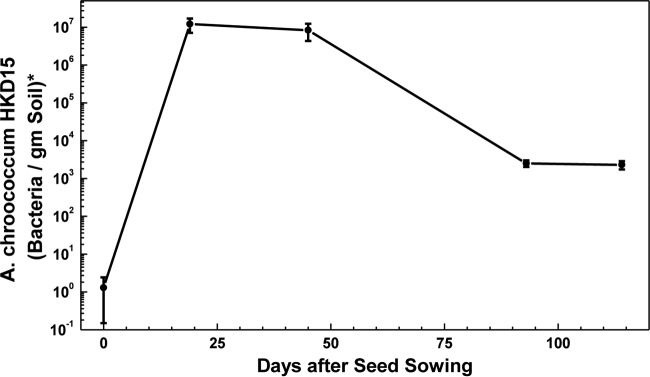
Because of our concern for the environment, we did not want to add to the field any bacteria containing any marker or any foreign gene. Hence, the number of A. chroococcum HKD15 cells in soil adhering to roots of wheat plants was determined by quantitative real-time PCR (19) and comparing the threshold cycle (CT) values of DNA from the soil with CT values obtained with DNA from a precounted number of A. chroococcum HKD15 cells. The forward primer chosen from the nifL region was 5′-AGGAAGTGCTCGGCAAGAACGAAT-3′, while the reverse primer from the inserted Tet promoter region was 5′-CGATGATAAGCTGTCAAACATGAG-3′. The results are presented in Fig. 5. We conclude from the data in Fig. 5 that A. chroococcum HKD15 cells did multiply rapidly until about 20 days after sowing, survived until at least 45 days after sowing, and continued subsequently to inhabit the rhizosphere but in a smaller number. The reduction after 45 days of sowing in the number of A. chroococcum HKD15 cells in the rhizosphere (Fig. 5) was possibly a consequence of various ecological factors, including other soil microorganisms that maintain the balance of the diverse microbial population in the soil. It was indeed necessary to inoculate the seeds every year with the engineered Azotobacter in order to be able to replace ∼85 kg urea and yet sustain the same wheat yield.
FIG 5.
Azotobacter chroococcum HKD15 cells (average of three field plots, five seedlings per field plot) in the rhizosphere of wheat seedlings. Values represent means ± standard deviations. *, g (dry weight).
Was the native population of microbes in the rhizosphere of the wheat plants in the field adversely affected by the inoculation of the seeds?
The population of bacteria, fungi, and actinomycetes in the rhizosphere of the wheat plants was determined periodically (Tables S2A, B, and C), but no adverse effect was observed. We concede that it might take several years for any untoward effect to be apparent, and many nonculturable microorganisms would be present in the soil.
Were the A. chroococcum HKD15 cells present in the rhizosphere of wheat plants in the field truly active?
The content of ammonium and nitrate nitrogen in the soil adhering to the roots of the wheat plants sown in three different locations in the field was determined. Data from seedlings arising out of wheat seeds not inoculated with any Azotobacter, inoculated with A. chroococcum CBD15, and inoculated with A. chroococcum HKD15 are presented in Fig. 6. It is obvious that the mutant strain was indeed active and was presumably fixing nitrogen.
FIG 6.
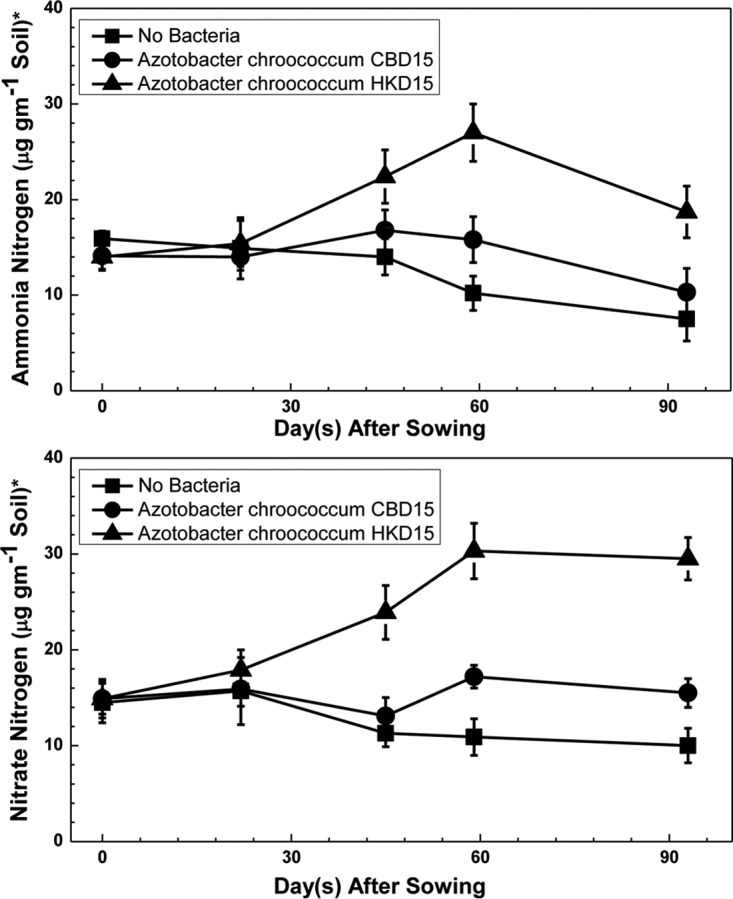
Ammonia (top) and nitrate (bottom) in rhizosphere postsowing in the absence of urea. ■, wheat seeds not inoculated (control); ●, wheat seeds inoculated with wild-type A. chroococcum CBD15; ▲, seeds inoculated with the engineered mutant, A. chroococcum HKD15. *, data presented are averages from three plots, five seedlings per plot.
Crop yield in field experiment.
Figure 7 shows the yield of the wheat crop in the field experiment. Inoculation of seeds with A. chroococcum HKD15 enhanced the yield of wheat grains by ∼60% in the absence of any urea application. When urea was also applied, a saving of 86 kg urea (40 kg N) per hectare was achieved on inoculation of the wheat seeds with A. chroococcum HKD15.
FIG 7.
Effect of inoculation of wheat HD 2967 with the engineered mutant, A. chroococcum HKD15, on yield of wheat grains in the field. The amounts of urea mentioned are in kilograms of N per hectare. Crop neither fertilized with urea nor inoculated with any bacteria was considered control (100%). *, the actual wheat crop yield (average of three plots) for this treatment was 291.0 g per m2. The critical difference (equivalent to least significant difference) at 5% was 69.2, the standard error of the mean was 23.1, and the coefficient of variance was 8.4.
Replacement of soil with vermiculite and perlite in the pot experiment.
We have presumed that the presence of enhanced ammonium and nitrate in the rhizosphere of the wheat plants in the field experiment and the enhanced crop yield from wheat seeds inoculated with A. chroococcum HKD15 in both pot and field experiments were because of enhanced nitrogen fixed by A. chroococcum HKD15. Soil, however, contains nitrogen that is unavailable to plants. Could A. chroococcum HKD15 be only making the unavailable nitrogen available to the wheat seedlings? A pot experiment was, therefore, conducted using vermiculite and perlite (2:1) instead of soil with Hoagland's solution (20) devoid of nitrogen as nutrient. Vermiculite and perlite contained no detectable nitrogen as determined by Kjeldahl digestion. The enhanced dry weight and nitrogen content of the root and shoot of wheat seedlings arising out of seeds inoculated with A. chroococcum HKD15, as shown in Table 3, must be because of enhanced nitrogen that was fixed.
TABLE 3.
Average dry biomass and nitrogen content of shoot and root of each wheat (HD 2967) seedling after 60 days of sowing in vermiculite-perlite in potsa
| Seed inoculant | Shoot biomass (mg) | Shoot nitrogen (mg/g) | Root biomass (mg) | Root nitrogen (mg/g) |
|---|---|---|---|---|
| None | 37.2 (12.5) | 4.7 (0.7) | 8.2 (2.0) | 3.3 (0.6) |
| CBD15 | 54.9 (13.9) | 7.7 (2.1) | 20.7 (4.2) | 4.9 (0.9) |
| HKD15 | 105.7 (16.3) | 14.8 (1.1) | 46.5 (16.6) | 10.2 (2.0) |
Average from four experiments, each experiment comprising four pots, each pot containing one seedling. Values in parentheses are standard deviations. The nutrient used was Hoagland's solution devoid of any nitrogen source.
Nitrogen balance experiment in pots.
The vermiculite-perlite experiment could not be continued until the maturity of the plants. Wheat seedlings that arose from seeds not inoculated with any Azotobacter started wilting ∼10 days postsowing. Even the ones that arose from seeds inoculated with A. chroococcum HKD15 did not survive beyond 70 days. The nitrogen fixed by A. chroococcum HKD15 could probably sustain the wheat seedlings but was not enough to nourish the full-grown plants. We wanted to conduct a nitrogen balance experiment with mature plants, so we replaced vermiculite-perlite with soil in the pots. Table 4 shows the results. We again conclude from the data in Table 4 that the enhanced combined total nitrogen content of soil plus those of inflorescence, shoot, and root after harvest of the wheat plants arising out of seeds inoculated with A. chroococcum HKD15 compared to the total nitrogen content of soil before sowing was because of enhanced nitrogen fixed by A. chroococcum HKD15. We, however, observed that the effect of inoculation of the wheat seeds with Azotobacter was less pronounced if soil was not autoclaved before sowing of the seeds. For example, the corresponding values in column vi of Table 4 were None, 1.302; A. chroococcum CBD15, 1.965; and A. chroococcum HKD15, 2.798, when soil was not autoclaved. We presume that this was because of the presence of other microorganisms in soil that interfered with the unhindered growth of Azotobacter. We had indeed seen a drop in the population of A. chroococcum HKD15 in the rhizosphere of the wheat plants after 45 days of sowing of the inoculated seeds (Fig. 5).
TABLE 4.
Nitrogen balancea
| Seed inoculant | Total nitrogen in: |
|||||
|---|---|---|---|---|---|---|
| (i) Soil before sowing any seed (g) | (ii) Inflorescence (mg) | (iii) Shoot (mg) | (iv) Root (mg) | (v) Soil after harvest (g) | (vi) Inflorescence, shoots, root, and soil (g) | |
| None | 1.410 (0.23) | 15 (1.3) | 27 (3.5) | 2 (0.29) | 1.36 (0.25) | 1.404 (0.26) |
| CBD15 | 1.410 (0.23) | 95 (6.4) | 93 (8.8) | 8 (0.44) | 2.772 (0.76) | 2.968 (0.77) |
| HKD15 | 1.410 (0.23) | 184 (9.8) | 199 (27.3) | 47 (6.1) | 3.906 (0.58) | 4.337 (0.62) |
Total nitrogen content of (i) soil (9 kg) before sowing of wheat HD 2967 seeds (the soil was autoclaved, nitrogen content was determined from 3 samples picked up at random, and then each of the 3 autoclaved pots was filled up with 9 kg of soil), (ii) inflorescences of 3 plants 135 days after sowing, (iii) shoots of 3 plants, (iv) roots of 3 plants, (v) soil after harvest of the plants 135 days after sowing, and (vi) inflorescences plus shoots plus roots plus soil. Values are averages (standard deviations). Each pot contained 3 wheat plants.
Experiment with 15N to conclusively prove that nitrogen fixed by Azotobacter was being utilized by the wheat plants.
Wheat seedlings in pots were exposed to air containing 15N gas, and 15N abundance in dried seedlings was determined. A small amount of 15N was present even in seedlings that arose from seeds which were not inoculated with any Azotobacter, possibly because of 15N fixed by nitrogen-fixing bacteria present in soil that was not autoclaved. Seedlings that arose from seeds inoculated with A. chroococcum CBD15 had twice as much 15N, while seedlings that arose from seeds that were inoculated with A. chroococcum HKD15 had four to six times as much 15N (Table 5).
TABLE 5.
Assimilation by wheat seedlings of 15N fixed by seed inoculant
| Seed inoculant |
15N abundance (% of total nitrogen) in: |
|
|---|---|---|
| Roots | Shoots | |
| None | 3.0 | 4.4 |
| A. chroococcum CBD15 | 6.8 | 8.2 |
| A. chroococcum HKD15 | 20.5 | 18.7 |
DISCUSSION
The presence of multiple chromosomes in A. vinelandii is a well-established fact (14, 15, 21–23). A. chroococcum CBD15, reported in this paper, has also revealed the presence of 20 copies of chromosomes at mid-exponential phase of its growth. Mutation of a gene in Azotobacter would, therefore, be inconsequential and liable to reversion, unless it is spread to all the chromosomes by culturing the cells for a large number of generations under selection pressure. Undefined mutants of Azotobacter which could reduce nitrogen even in the presence of fixed nitrogen (ammonium and nitrate), which were obtained in the past through standard mutagenesis techniques (24–31), were not of much practical use, as it was not recognized then that unless the mutation was transmitted to all the copies of the chromosome of Azotobacter, the mutant could not be stable.
The construction of the mutant in the present work has been carried out in two steps for ease of selection. The first step resulted in the insertion of the kanamycin interposon (11) in nifL, leaving nifA promoterless. The selection pressure was applied here by the addition of kanamycin to the culture medium along with ammonium. When the cells were cultured only 6 times, the persistence of the wild-type chromosomes was distinctly discernible. The cells needed culturing 18 times to achieve insertion of the kanamycin interposon into all the chromosomes. The second step has been aimed at the replacement of the kanamycin interposon by the Tet promoter in the correct orientation after deletion of a part of nifL. The selection pressure was applied here by omitting ammonium and also kanamycin, and the cells were cultured 20 times. The results presented in Fig. 2 confirmed that the kanamycin interposon has been replaced by the Tet promoter from all the chromosomes of A. chroococcum HKD15.
We had chosen a constitutive promoter in preference to an inducible promoter (cf. reference 32) because we realized that addition of the inducer to the field could be an environmental hazard. We also preferred to overexpress nifA and thus enhance nitrogen fixation so as to maximize the availability of ammonia, rather than reduce ammonia assimilation (cf. reference 32), as we appreciated that the two main products of ammonia assimilatory reactions, glutamine and glutamate, are both essential components of proteins.
In India, wheat is cultivated on ∼27 million hectares. If wheat seeds are inoculated everywhere in India with A. chroococcum HKD15, or similarly engineered Azotobacter bacteria derived from strains isolated from other wheat fields, there could be a savings of 2,100 million kg of urea per sowing season of wheat, assuming that the projected linear upscaling is valid.
Azotobacter has also been found to be beneficial for other cereal crops, vegetables, and cash crops (33). Engineered Azotobacter strains should be of use for these crops also. Not only in India but anywhere in the world where the agroclimatic conditions are conducive for Azotobacter to thrive, strains engineered in a similar way should be able to replace a good proportion of chemical nitrogenous fertilizer. The savings thus achieved would be not only in the cost of chemical nitrogenous fertilizers but, more importantly, in the cost of restoring the environment.
Fixed nitrogen is a $100 billion global industry annually, but environmental pollution by synthetic nitrogenous fertilizers remains a real problem. The Bill and Melinda Gates Foundation contributed $10 million to support research for producing nitrogen-fixing cereals (34). This is indeed a challenging task involving anywhere between 9 and 20 genes, taking care of the codon preference in the plant and the transcription of all the genes under plant promoters. It may be mentioned here that 9 nitrogen-fixing genes from Paenibacillus sp. strain WLY78, which was considered to be the minimum nitrogen fixation cascade, were only 10% active in Escherichia coli (35). Finally, even if the mission of producing nitrogen-fixing plants is accomplished, it may or may not be beneficial for the plants. The enzyme nitrogenase is extremely oxygen sensitive, and fixing nitrogen is metabolically expensive, which would be a heavy burden on plants. About 16 ATP molecules are needed to reduce one molecule of nitrogen (36). On the other hand, only 36 to 38 ATP molecules are released due to hydrolysis of one molecule of glucose (37). Cellulose is a universal backbone of plants, while starch is the most common stored nutrient in seeds and fruits. Both cellulose and starch are polymers of glucose. It will take about 1 mol of glucose (∼180 g) to reduce 2.4 mol (∼34 g) of nitrogen, which is equivalent to >5 g glucose per gram of reduced nitrogen. The energy requirement may be much higher when expressing nitrogenase in plants. This enormous energy load is likely to adversely affect the foliage and fruit development of plants.
Developing a biofertilizer with enhanced nitrogen-fixing ability, which is sustained even in the presence of synthetic nitrogenous fertilizers, is thus a practical and environmentally friendly answer to the growing demands for nitrogenous fertilizers for agriculture.
MATERIALS AND METHODS
Azotobacter chroococcum CBD15 was obtained from the Division of Microbiology, and the wheat variety HD 2967 was obtained from the Division of Genetics, Indian Agricultural Research Institute, New Delhi, India. HD 2967 has the following genealogy: ALDES/COC//URES/HD2160(M)//HD2278.
General methods.
Growth of bacteria, isolation of plasmids, isolation of genomic DNA, acetylene reduction assay, and labeling of DNA fragments were done as described earlier (38). Glucose in Burk's nitrogen-free (BNF) medium was replaced by 2% sucrose. DNA was estimated according to the method of Burton (39), indole acetic acid was determined according to the method of Hartmann et al. (40), and ammonium in culture medium was assayed according to the method of Chaney and Marbach (41). When both ammonium and nitrate were to be determined in soil, the method of Keeney and Nelson (42) was used. “Available nitrogen” in the soil of the field was determined by digestion of the soil samples with alkaline permanganate (18). Bacterial cells were counted in a hemocytometer.
Determination of base sequence of nifL of A. chroococcum CBD15.
The nifL region of A. chroococcum CBD15 was isolated by PCR using Phusion high-fidelity DNA polymerase and primers based on the sequence of nifL (9) of A. vinelandii UW. PCR was programmed with an initial denaturing at 94°C for 5 min followed by 30 cycles of denaturation at 94°C for 45 s, annealing at 64°C for 45 s, and extension at 70°C for 90 s and a final extension at 72°C for 10 min. The base sequence of the PCR product was determined commercially.
Construction of A. chroococcum HKD15.
A ∼7.5-kb BamHI fragment of A. chroococcum CBD15 genomic DNA hybridized with both nifL (9) and nifA (43) of A. vinelandii UW. This fragment was cloned in pUC7 and named pCL6 (partial restriction map shown in Fig. 1A). The ∼4.7-kb EcoRI fragment from pCL6, which contained the entire nifL gene and a major part of nifA, was cloned in pUC7 and was designated pCL6.2 (Fig. 1B). The plasmid pCL6.2 was digested with SalI, and the 5′ overhangs were filled up. The 5′ overhangs of the EcoRI fragment from pHP45ΩKm (11) were also filled up. These two DNA fragments were then ligated. E. coli DH5α was transformed, and kanamycin-resistant colonies were selected. The plasmid isolated from the kanamycin-resistant colonies was designated pCL6.3 (Fig. 1C). The construct pCL6.2 has a single BamHI site and a single SmaI site, while the EcoRI fragment containing ΩKm has two BamHI sites and three SmaI sites. These were used to confirm the insertion of ΩKm into nifL in pCL6.3.
The 5′ overhangs of the 381-bp EcoRI-BamHI fragment from pBR322 (base sequence in Fig. S1 in the supplemental material) containing the Tet promoter were filled up. The plasmid pCL6.2 was digested with SalI and treated with Bal 31 to obtain a deletion of ∼1 kb to ∼1.2 kb. The deleted DNA was consecutively treated with mung bean nuclease and Klenow fragment, along with all four deoxynucleoside triphosphates (dNTPs), and ligated with the DNA fragment from pBR322. We called the new construct pCL6.4 (Fig. 1D). The success of the ligation operation was confirmed by the appearance in pCL6.4 of a HindIII site and the disappearance of the SmaI site compared to pCL6.2.
The A. chroococcum CBD15 cells were transformed with pCL6.3 by electroporation, and the cells (Fig. 1E) were plated once on BNF agar containing ammonium acetate (0.11%). The cells were scraped off and plated again on BNF agar containing ammonium acetate and kanamycin (50 μg/ml). The kanamycin-resistant cells (Fig. 1F) were subcultured 20 times in the same medium and rechecked for their inability to grow on both BNF agar and BNF agar containing ammonium acetate and ampicillin (100 μg/ml).
The kanamycin-resistant derivative of A. chroococcum CBD15 (Fig. 1F) was next subjected to electroporation with pCL6.4, and the cells (Fig. 1G) were plated on BNF agar containing ammonium acetate. The cells were scraped off and plated again on BNF agar with no ammonium acetate and no kanamycin in it. The colonies were grown in BNF medium containing no kanamycin and no ammonium acetate, and the cells (Fig. 1H) were subcultured 20 times in the same medium and rechecked for their inability to grow on BNF medium containing ammonium acetate and kanamycin and also on BNF medium containing ammonium acetate and ampicillin.
PCR analysis.
PCR analysis was used to confirm the deletion of part of the nifL gene, the insertion of the Tet promoter there in the chromosome of A. chroococcum HKD15, and the achievement of complete segregation of the Tet promoter containing copies of the chromosomes. Taq DNA polymerase was used, and the operational steps were as follows: (i) initial denaturation for 4 min at 95°C; (ii) 30 cycles of amplification with 1 min of denaturation at 95°C, 1 min of annealing at 58°C to 68.5°C, and 1 min 30 s of extension at 72°C; and (iii) final extension for 10 min at 72°C.
Treatment of wheat seeds with bacteria.
Azotobacter strains were grown (38) to an A600 of 1.5, harvested, washed three times with sterile BNF medium containing sucrose but no ammonium acetate (38), and suspended in the washing medium but at 1/10 the volume of the culture. The concentrated bacterial suspension contained ∼1 × 1010 cells per ml. The wheat seeds were soaked in the concentrated bacterial suspension for 3 h at 25°C and air dried at 25°C. Seeds that were not intended to be treated with Azotobacter were treated exactly the same way in the same medium lacking bacteria. The number of Azotobacter cells adhering to each seed was on the order of 107.
Urea application.
Half of the desired dose of urea was applied immediately prior to sowing. The remaining urea was applied in two installments at successive intervals of 40 days.
Application of other fertilizers.
Potash was applied at 60 kg K per hectare and superphosphate was applied at 60 kg P per hectare in a single dose to all the pots and all the plots in the field a few days prior to sowing.
Pot experiments for crop yield.
Pot experiments were performed in a net house for 1 year at Jawaharlal Nehru University and for 2 years at the Division of Microbiology of the Indian Agricultural Research Institute, New Delhi. Each pot of 35-cm diameter contained 12 kg soil. Five plants were maintained per pot. Fifteen pots were used per treatment. The pots were distributed in the net house by randomized block design. The amount of fertilizer to be applied per pot was determined on the basis of the weight of soil per pot (12 kg), assuming that 1 hectare is equivalent to 2.24 × 106 kg of soil, which is approximately the weight of the upper 15 cm of the soil in 1 hectare (44).
Field experiment.
The field for confined field trials at the Indian Agricultural Research Institute, New Delhi, was used. Each plot was 1.2 m by 3.0 m, and the next plot was 3.0 m away in all directions. Three replicate plots were used for each treatment and distributed in the field by randomized block design. A 3.0-m-wide border of wheat plants was maintained on all four sides 3.0 m away from the experimental plots. Sowing of wheat seeds, 50 g per plot, was performed in six rows per plot occupying 1.2 m at the width of the plot. Each plot was notionally 3.0 m long, seeds falling over a little less or more. The exact length up to which the seeds fell was measured for each plot. The outer two rows of plants were discarded during harvesting, and crop yield was calculated on the basis of yield from the inner four rows of plants that occupied 0.8 m. One irrigation was applied prior to sowing. Six irrigations were applied after sowing. For determination of dry weight, wheat seedlings were uprooted immediately after an irrigation from a 20-cm length of each row chosen at random and washed free of all dust and soil, and the whole seedlings (shoot plus root) were dried at 100°C until they achieved a constant weight. Harvesting of the wheat crop was done manually 135 days after sowing.
Collection of soil samples adhering to roots of wheat plants.
Seeds treated with A. chroococcum were sown in the field in three different locations. Wheat seedlings were pulled out periodically and gently shaken to get rid of the loose soil from the roots, and the soil sticking to the roots was collected aseptically.
DNA isolation from soil samples.
DNA was isolated from 0.5 g soil adhering to roots of the wheat plants sown in three different locations by using the Zymo Research kit ZR soil microbe DNA MiniPrep (catalog no. D6001).
Determination of population of microbes in rhizosphere soil of wheat plants.
Soil adhering to roots of uprooted plants was suspended in normal saline and mixed thoroughly, the soil was allowed to sediment, and the supernatant suspension was plated on specific agar medium after appropriate dilution. Nutrient agar (45) was used for bacteria, Ken Knight's medium was used for Actinomycetes, and Martin's rose bengal medium (46) was used for fungi.
Determination of 15N assimilated by wheat seedlings.
Wheat seedlings in pots, 7 days after sowing, were exposed to 15N in a closed chamber. The gas volume of the chamber was 4.75 liters, and 1 liter of 15N was introduced by replacement of 1 liter of water. Since air has 78% nitrogen, there was 21% 15N in the total nitrogen in the chamber. The experiment was terminated after 20 days, and the roots and shoots were separately collected, dried at 100°C until they achieved a constant weight, and stored in a desiccator. The 15N abundance was determined in an isotope ratio mass spectrometer.
Supplementary Material
ACKNOWLEDGMENTS
Research grants from the Department of Biotechnology, Government of India, to H.K.D. and S.P. during the early stage of the work (engineering of A. chroococcum HKD15 and pot experiments) are acknowledged. S.G. and P.S. enjoyed fellowships from a research project funded by the Department of Biotechnology, Government of India. U.K.B. and M.S. enjoyed fellowships from the Council of Scientific and Industrial Research, India.
Thanks are due to Min Huang of the Hunan Agricultural University, China, for arranging 15N abundance determination in the Hauzhong Agricultural University, China. Thanks are also due to R. Srinivasan and S. R. Bhat for critical reading of the manuscript and valuable suggestions, to A. Singh for fruitful discussions and help, to I. Dasgupta and K. R. Koundal for help, and to R. S. Tomar, J. S. Bisht, and S. Srivastava for technical assistance.
H.K.D. conceived the work, designed the experiments, interpreted the results, and prepared the manuscript; U.K.B. assisted in preparing the manuscript and conducted the laboratory experiments; M.S., S.G., and P.S. conducted the laboratory experiments; S.P. performed the characterization of A. chroococcum HKD15, conducted the pot experiment for crop yield, and determined the microbial population in the rhizosphere soil; P.P.-S. assisted in preparing the manuscript and conducted the vermiculite-perlite experiment, the nitrogen balance experiment, and the 15N experiment; R.S.J. conducted the field experiment; R.Y. rendered valuable advice on the field experiment and edited the manuscript; D.R.B. helped in chemical analysis of soil; P.K.M. suggested the pot experiment using vermiculite and perlite and facilitated the work; and P.A.K., J.C.P., and K.A. facilitated the work.
The authors declare no conflict of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00590-17.
REFERENCES
- 1.Das HK. 1991. Biological nitrogen fixation in the context of Indian agriculture. Curr Sci 60:551–555. [Google Scholar]
- 2.Mrkovacki N, Milic V. 2001. Use of Azotobacter chroococcum as potentially useful in agricultural application. Ann Microbiol 51:145–158. [Google Scholar]
- 3.Zhang W-F, Dou Z-X, He P, Ju X-T, Powlson D, Chadwick D, Norse D, Lu Y-L, Zhang Y, Wu L, Chen X-P, Cassman KG, Zhang F-S. 2013. New technologies reduce greenhouse gas emissions from nitrogenous fertilizer in China. Proc Natl Acad Sci U S A 110:8375–8380. doi: 10.1073/pnas.1210447110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prasad R. 2009. Efficient fertilizer use—the key to food security and better environment. J Trop Agric 47:1–17. [Google Scholar]
- 5.Times News Network. 16 January 2017 Taps run dry in major part of city. High ammonia levels disrupt 2 plants; supply to remain affected today, p 3 The Times of India, New Delhi, India: http://timesofindia.indiatimes.com/city/delhi/taps-run-dry-in-major-parts-of-city/articleshow/56576079.cms?. [Google Scholar]
- 6.Sutton MA, Oenema O, Erisman JW, Leip A, van Grinsven H, Winiwarter W. 2011. Too much of a good thing. Nature 472:159–161. doi: 10.1038/472159a. [DOI] [PubMed] [Google Scholar]
- 7.Schmitz RA, Klopprogge K, Grabbe R. 2002. Regulation of nitrogen fixation in Klebsiella pneumoniae and Azotobacter vinelandii: NifL, transducing two environmental signals to the nif transcriptional activator NifA. J Mol Microbiol Biotechnol 4:235–242. [PubMed] [Google Scholar]
- 8.Mitra R, Das HK, Dixit A. 2005. Identification of a positive transcription regulatory element within the coding region of the nifLA operon in Azotobacter vinelandii. Appl Environ Microbiol 71:3716–3724. doi: 10.1128/AEM.71.7.3716-3724.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raina R, Bageshwar UK, Das HK. 1993. The Azotobacter vinelandii nifL-like gene: nucleotide sequence analysis and regulation of expression. Mol Gen Genet 237:400–406. [DOI] [PubMed] [Google Scholar]
- 10.Bageshwar UK. 1994. Studies on some nitrogen fixing genes of Azotobacter vinelandii, p 85–89. PhD thesis Department of Biosciences, Faculty of Natural Sciences, Jamia Milia Islamia, New Delhi, India. [Google Scholar]
- 11.Fellay R, Frey J, Krisch H. 1987. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designated for in vitro insertional mutagenesis of gram-negative bacteria. Gene 52:145–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- 12.Bali A, Blanco G, Hill S, Kennedy C. 1992. Excretion of ammonium by a nifL mutant of Azotobacter vinelandii fixing nitrogen. Appl Environ Microbiol 58:1711–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brewin B, Woodley P, Drummond M. 1999. The basis of ammonium release in nifL mutants in Azotobacter vinelandii. J Bacteriol 181:7356–7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagpal P, Jafri S, Reddy MA, Das HK. 1989. Multiple chromosomes of Azotobacter vinelandii. J Bacteriol 171:3133–3138. doi: 10.1128/jb.171.6.3133-3138.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manna AC, Das HK. 1997. Characterization and mutagenesis of the leucine biosynthetic genes of Azotobacter vinelandii: an analysis of the rarity of amino acid auxotrophs. Mol Gen Genet 254:207–217. doi: 10.1007/s004380050409. [DOI] [PubMed] [Google Scholar]
- 16.Manna AC, Das HK. 1994. The size of the chromosome of Azotobacter chroococcum. Microbiology 140:1237–1239. doi: 10.1099/13500872-140-5-1237. [DOI] [Google Scholar]
- 17.Martinez-Argudo I, Little R, Shearer N, Johnson P, Dixon R. 2004. The NifL-NifA system: a multidomain transcriptional regulatory couple that integrates environmental signals. J Bacteriol 186:601–610. doi: 10.1128/JB.186.3.601-610.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subbiah BV, Asija GL. 1956. A rapid procedure for the estimation of available nitrogen in soils. Curr Sci 25:259–260. [Google Scholar]
- 19.Zhang T, Fang HP. 2006. Applications of real time polymerase chain reaction for quantification of microorganisms in environmental samples. Appl Microbiol Biotechnol 70:281–289. doi: 10.1007/s00253-006-0333-6. [DOI] [PubMed] [Google Scholar]
- 20.Hoagland DR, Arnon DI. 1950. The water-culture method of growing plants without soil. California Agricultural Experiment Station circular 347. University of California, Berkeley, CA. [Google Scholar]
- 21.Sadoff HL, Berke E, Loperfido B. 1971. Physiological studies of encystment in Azotobacter vinelandii. J Bacteriol 105:185–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sadoff HL, Shimei B, Ellis S. 1979. Characterization of Azotobacter vinelandii deoxyribonucleic acid and folded chromosomes. J Bacteriol 138:871–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maldonado R, Jimenez J, Casadesus J. 1994. Changes of ploidy during the Azotobacter vinelandii growth cycle. J Bacteriol 176:3911–3919. doi: 10.1128/jb.176.13.3911-3919.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gordon JK, Brill WJ. 1972. Mutants that produce nitrogenase in the presence of ammonia. Proc Natl Acad Sci U S A 69:3501–3503. doi: 10.1073/pnas.69.12.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terzaghi BE. 1980. A method for the isolation of Azotobacter mutants derepressed for Nif. J Gen Microbiol 118:275–278. [Google Scholar]
- 26.Gordon JK, Jacobson MR. 1983. Isolation and characterization of Azotobacter vinelandii mutant strains with potential as bacterial fertilizer. Can J Microbiol 29:973–978. doi: 10.1139/m83-154. [DOI] [Google Scholar]
- 27.Narula N, Nijhawan DC, Lakshminarayana K, Kapoor RL, Verma OPS. 1991. Response of pearl millet (Pennisetum glaucum) to soil isolates and analogue resistant mutants of Azotobacter chroococcum. Indian J Agric Sci 61:484–487. [Google Scholar]
- 28.Jadhav AS, Shaikh AA, Harinarayana G. 1991. Response of rainfed pearl millet (Pennisetum glaucum) to inoculation with nitrogen-fixing bacteria. Indian J Agric Sci 61:268–271. [Google Scholar]
- 29.Lakshminarayana K, Narula N, Hooda IS, Faroda AS. 1992. Nitrogen economy in wheat (Triticum aestivum) through use of Azotobacter chroococcum. Indian J Agric Sci 62:75–76. [Google Scholar]
- 30.Kashyap LR. 1988. Azide resistance and role of various metabolites on Azotobacter growth. Curr Sci 57:1012–1014. [Google Scholar]
- 31.Narula N, Kukreja K, Suneja S, Lakshminarayana K. 1999. Ammonia excretion by ethylenediamine resistant (EDAR) mutants of Azotobacter chroococcum. Indian J Microbiol 39:93–97. [Google Scholar]
- 32.Ambrosio R, Ortiz-Marquez JC, Curatti L. 2017. Metabolic engineering of a diazotrophic bacterium improves ammonium release and biofertilization of plants and microalgae. Metab Eng 40:59–68. doi: 10.1016/j.ymben.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 33.Lakshminarayana K. 1993. Influence of Azotobacter on nitrogen nutrition of plants and crop productivity. Proc Indian Natl Sci Acad B 59:303–308. [Google Scholar]
- 34.Beatty PH, Good AG. 2011. Future prospects of cereals that fix nitrogen. Science 333:416–417. doi: 10.1126/science.1209467. [DOI] [PubMed] [Google Scholar]
- 35.Wang L, Zhang L, Liu Z, Zhao D, Liu X, Zhang B, Xie J, Hong Y, Li P, Chen S, Dixon R, Li J. 2013. A minimal nitrogen fixation gene cluster from Paenibacillus sp. WLY78 enables expression of active nitrogenase in Escherichia coli. PLoS Genet 9:e1003865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hill S. 1976. The apparent ATP requirement for nitrogen fixation in growing Klebsiella pneumoniae. J Gen Microbiol 95:297–312. doi: 10.1099/00221287-95-2-297. [DOI] [PubMed] [Google Scholar]
- 37.Rich PR. 2003. The molecular machinery of Keilin's respiratory chain. Biochem Soc Trans 3:1095–1105. doi: 10.1042/bst0311095. [DOI] [PubMed] [Google Scholar]
- 38.Raina R, Reddy MA, Ghosal D, Das HK. 1988. Characterization of the gene for the Fe-protein of the vanadium dependent alternative nitrogenase of Azotobacter vinelandii and construction of a Tn5 mutant. Mol Gen Genet 214:121–127. doi: 10.1007/BF00340189. [DOI] [PubMed] [Google Scholar]
- 39.Burton K. 1968. Determination of DNA concentration with diphenylamine. Methods Enzymol 12B:163–166. doi: 10.1016/0076-6879(67)12127-7. [DOI] [Google Scholar]
- 40.Hartmann A, Singh M, Klingmuller W. 1983. Isolation and characterization of Azospirillum mutant excreting high amounts of indole acetic acid. Can J Microbiol 29:916–923. doi: 10.1139/m83-147. [DOI] [Google Scholar]
- 41.Chaney AL, Marbach EP. 1962. Modified reagents for determination of urea and ammonia. Clin Chem 8:131–132. [PubMed] [Google Scholar]
- 42.Keeney DR, Nelson DW. 1982. Nitrogen-inorganic forms, p 643–698. In Page AL, Miller RH, Keeney DR (ed), Methods of soil analysis. Part 2. American Society of Agronomy, Madison, WI. [Google Scholar]
- 43.Reddy MA, Das HK. 1998. Cloning of a positive regulatory element involved in nitrogen fixation in Azotobacter vinelandii. J Genet 67:121–127. doi: 10.1007/BF02927792. [DOI] [Google Scholar]
- 44.Brady NC, Weil RR. 2002. The nature and properties of soils, p 960. Prentice Hall, Upper Saddle River, NJ. [Google Scholar]
- 45.Subba Rao NS. 1988. Biofertilizers in agriculture, p 198–199. Oxford & IBH Publishing Company, Delhi, India. [Google Scholar]
- 46.Martin JP. 1950. Use of acid, rose bengal and streptomycin in the plate method for estimating soil fungi. Soil Sci 69:215–232. doi: 10.1097/00010694-195003000-00006. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.