ABSTRACT
R-type tailocins are high-molecular-weight bacteriocins that resemble bacteriophage tails and are encoded within the genomes of many Pseudomonas species. In this study, analysis of the P. chlororaphis 30-84 R-tailocin gene cluster revealed that it contains the structural components to produce two R-tailocins of different ancestral origins. Two distinct R-tailocin populations differing in length were observed in UV-induced lysates of P. chlororaphis 30-84 via transmission electron microscopy. Mutants defective in the production of one or both R-tailocins demonstrated that the killing spectrum of each tailocin is limited to Pseudomonas species. The spectra of pseudomonads killed by the two R-tailocins differed, although a few Pseudomonas species were either killed by or insusceptible to both tailocins. Tailocin release was disrupted by deletion of the holin gene within the tailocin gene cluster, demonstrating that the lysis cassette is required for the release of both R-tailocins. The loss of functional tailocin production reduced the ability of P. chlororaphis 30-84 to compete with an R-tailocin-sensitive strain within biofilms and rhizosphere communities. Our study demonstrates that Pseudomonas species can produce more than one functional R-tailocin particle sharing the same lysis cassette but differing in their killing spectra. This study provides evidence for the role of R-tailocins as determinants of bacterial competition among plant-associated Pseudomonas in biofilms and the rhizosphere.
IMPORTANCE Recent studies have identified R-tailocin gene clusters potentially encoding more than one R-tailocin within the genomes of plant-associated Pseudomonas but have not demonstrated that more than one particle is produced or the ecological significance of the production of multiple R-tailocins. This study demonstrates for the first time that Pseudomonas strains can produce two distinct R-tailocins with different killing spectra, both of which contribute to bacterial competition between rhizosphere-associated bacteria. These results provide new insight into the previously uncharacterized role of R-tailocin production by plant-associated Pseudomonas species in bacterial population dynamics within surface-attached biofilms and on roots.
KEYWORDS: R-tailocin, bacterial competition, rhizosphere, Pseudomonas, bacteriocins, microbial ecology, rhizosphere-inhabiting microbes
INTRODUCTION
Bacteria produce a diversity of bacteriocins to kill closely related competitors (1). Tailocins are high-molecular-weight (HMW) bacteriocins produced by a variety of bacteria that resemble bacteriophage tails (2–5). Tailocin particles are protease resistant, thermolabile, and sedimentable by ultracentrifugation (2). These particles have been studied extensively for the opportunistic human pathogen Pseudomonas aeruginosa, strains of which are capable of producing either or both R-type and F-type tailocins (3). R-type tailocins are rigid rod-like particles with a contractile sheath and resemble T-even coliphage tails, whereas F-type tailocins are flexible sheathless particles that resemble lambda phage tails (6).
The P. aeruginosa R-tailocin particle is composed of a double hollow cylinder that consists of a sheath and core connected to a baseplate with six tail fibers (7, 8). In P. aeruginosa, R-tailocin synthesis is induced by the SOS response and yields 100 to 200 tailocins per cell (9). A lysis cassette similar to that utilized by lytic bacteriophage mediates the extracellular release of R-tailocin particles (6). The bacteriophage lysis cassette is composed of genes encoding the endolysin, holin, and the Rz and Rz1 proteins comprising the spanin complex (10). Some tailocin gene clusters encode all three lysis cassette components, while others, such as those in P. aeruginosa, generally do not encode the spanin complex (11). Endolysins are cytoplasmic proteins that catalyze the degradation of the cell wall peptidoglycan layer. Holins are small transmembrane proteins that accumulate in the inner membrane and permeabilize the membrane, which allows the endolysins to reach and degrade the peptidoglycan layer (12). Rz and Rz1 make the spanin complex, which is involved in disruption of the outer membrane and release of bacteriophage (13). Once released, R-type tailocin tail fibers interact with specific surface receptors (lipopolysaccharides) of susceptible bacterial competitors and insert a needle-like core through the cell wall. This depolarizes the membrane potential and stops macromolecular synthesis, leading to cell death (14, 15).
Host-killing range is determined by the specificity of the interaction between the tailocin particle and the target surface receptors and is generally limited to closely related species or strains. Producing strains are typically resistant to the R-tailocin they release (3, 16–19). Although host-killing ranges of some strains have been characterized, the extent to which R-tailocins contribute to the competitive dynamics of bacterial communities is less well understood. The importance of R-tailocin production and release in interstrain interactions was investigated for P. aeruginosa, and it was revealed that R-tailocin production results in a an advantage for the producing strain in mixed populations with susceptible strains in vitro as well as within human host tissues (20–22).
More recently, R-tailocins similar to those produced by P. aeruginosa have been identified in plant-associated Pseudomonas strains (11, 16, 17, 23, 24), suggesting that R-tailocin production may be an important competitive determinant of bacterial interactions for Pseudomonas inhabiting diverse environments, such as the plant microbiome. In the present study, we focused on Pseudomonas chlororaphis 30-84, a rhizosphere-colonizing strain selected for its ability to suppress take-all disease of wheat, caused by the fungal plant pathogen Gaeumannomyces graminis var. tritici. The production of phenazines by P. chlororaphis 30-84 is the primary mechanism of pathogen inhibition and contributes to rhizosphere persistence as well as biofilm development (25–28). Recently, production of the phenazine 2-hydroxy-phenazine-carboxylic acid (2-OH-PCA) by P. chlororaphis 30-84 was shown to promote biofilm formation by the release of extracellular DNA (eDNA) (29). Increased production of eDNA in the biofilm of P. chlororaphis 30-84 was due in part to cell autolysis as a result of the expression of a gene cluster with sequence similarity to R-tailocin gene clusters in P. aeruginosa (29).
Despite the identification of R-tailocin gene clusters in plant-associated Pseudomonas strains, they have received little attention beyond genomic comparison and killing spectrum studies. Previous comparison of the R-tailocin gene clusters among Pseudomonas strains revealed distinct evolutionary ancestries based on the protein sequences of the structural components that may be influenced by bacterial habitat (11, 16). Analysis of the P. chlororaphis 30-84 R-tailocin gene cluster suggested a unique evolutionary trajectory because it contains structural components with similarity to both P. aeruginosa and P. syringae, potentially representing a hybrid tailocin (16). Closer analysis, as described in this report, led us to hypothesize that the cluster may encode two functional R-tailocins sharing a single lysis cassette. If the two R-tailocins had different killing spectra, this could enhance the competitive ability of P. chlororaphis 30-84 strain over a broader spectrum of closely related species competing for similar ecological rhizosphere niches. The aims of this study were to (i) further characterize the P. chlororaphis 30-84 tailocin gene cluster in comparison to other plant-associated Pseudomonas species, (ii) determine whether P. chlororaphis 30-84 produces two functional R-tailocins as predicted from genome analysis and compare their killing spectra, (iii) determine the functionality of the lysis cassette in R-tailocin release, and (iv) evaluate the role of R-tailocin production in competition between rhizosphere-associated bacteria in surface-attached biofilms and on plant roots.
RESULTS
The P. chlororaphis 30-84 genome encodes two R-tailocin particles.
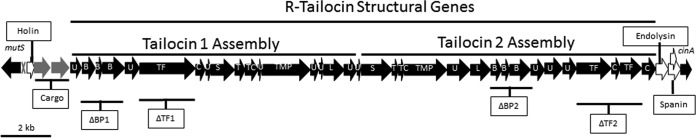
The tailocin gene cluster of P. chlororaphis 30-84, situated between the mutS (Pchl3084_1192) and the cinA (Pchl3084_1233) genes in the bacterial chromosome, is approximately 34 kb in length and consists of 40 putative open reading frames (ORFs) (Fig. 1). The cluster contains genes annotated as functioning in regulation, cell lysis and tailocin release, and tailocin particle assembly as well as cargo genes encoding proteins potentially carried with the tailocins (11, 23). At the beginning of the cluster, adjacent to mutS, is a gene with significant nucleotide sequence similarity to ptrR, the negative regulator of tailocin gene transcription in P. aeruginosa (11, 23). The cluster also contains a lysis cassette that consists of genes encoding a holin, an endolysin, and spanin complex involved in cell lysis and tailocin release. The genes in the lysis cassette flank the region containing the structural genes, with the holin gene upstream and the endolysin and spanin complex genes downstream of the structural genes. The region encoding the tailocin structural genes is nearly twice the size and has twice the number of ORFs as the genomic regions containing the R-tailocin structural genes previously described for other plant-associated pseudomonads (6, 11, 16, 17). This region can be divided into two smaller regions, each containing the genes required for the assembly of different R-tailocins (Fig. 1). The orders of the genes within the two R-tailocin assembly regions are different, as are the sequences of genes encoding proteins with similar functions. For example, the tail fiber and chaperone of the first tailocin are encoded by one gene each, whereas several genes encode the tail fibers and chaperones in the second cluster. The large cluster also includes two genes we refer to as cargo genes, Pchl3084_1195 and Pchl3084_1196, that are located immediately downstream of the holin gene and annotated as amidase domain-containing protein and the putidacin L1 bacteriocin, respectively (11, 16, 23).
FIG 1.
P. chlororaphis 30-84 R-tailocin gene cluster. (A) The R-tailocin gene cluster is situated between the mutS and cinA genes (flanking, black) within the P. chlororaphis 30-84 chromosome. This gene cluster contains 40 putative ORFs predicted to include a transcriptional regulator (gray X), a lysis cassette (white), cargo genes (gray), and R-tailocin structural genes (center, black). The lysis cassette is composed of genes encoding the holin protein and endolysin enzyme (hol and endo, respectively) and the spanin complex (rz and rz1, within the open reading frame of rz) and flanks the putative tailocin structural genes. The region including the R-tailocin structural genes is twice as large and contains twice as many genes as those observed in previously described clusters. This region can be divided into two subregions that contain all of the genes necessary to assemble two R-tailocin particles. The location of deletion mutations of the two tail fiber (ΔTF1 and ΔTF2) and the baseplate mutant (ΔBP1) are shown. Abbreviations: U, unknown; B, baseplate assembly; TF, tail fiber; C, tail fiber chaperone; S, sheath; T, tail tube; TC, tail assembly chaperone; TMP, tape measure protein; L, late control D protein.
R-tailocin gene clusters in P. chlororaphis 30-84 are evolutionarily distinct.
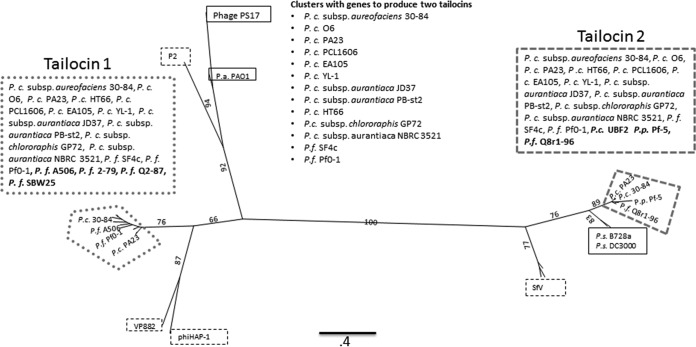
Examination of the tailocin gene clusters from the published genomes of other rhizosphere pseudomonads, including strains of P. chlororaphis, P. fluorescens, and P. protegens, revealed that several of these strains also had tailocin gene clusters predicted to encode more than one R-tailocin (Fig. 2). The tailocin gene clusters of all P. chlororaphis strains examined possess the genes predicted to produce two R-tailocins, with the exception of P. chlororaphis UBF2, which encodes one. In contrast, the gene clusters of a majority of P. fluorescens strains inspected encode just one R-tailocin, with the exception of P. fluorescens SF4c and P. fluorescens Pf0-1, which encode two. Phylogenetic trees were built from multiple amino acid sequence alignments of the predicted proteins encoding structural components, including the tail tube, sheath, and baseplate spike (Fig. 2). The phylogenetic trees obtained for each display similar topologies (see Fig. S1 to S3 in the supplemental material). Interestingly, for the strains that appear to encode two R-tailocins, components of one tailocin always cluster together and components of the other always cluster together, suggesting that if the genome encodes two tailocins, it is always the same two types. For the P. chlororaphis strains having two tailocins, the tailocin region designated tailocin 1 (Fig. 2) clusters with the regions in P. fluorescens strains A506, 2-79, SBW25, and Q287, each encoding one tailocin (Fig. 2). The R-tailocin 1 region sequences are most similar to the predicted amino acid sequences of Vibrio parahaemolyticus phage VP882 and Halomonas aquamarina phage phiHAP-1 (Fig. 2). Alternatively, the R-tailocin 2 region sequences cluster with the single tailocin sequences in P. chlororaphis UBF2, P. protegens Pf-5, and P. fluorescens Q8r1-96, as well as sequences in the phyllosphere pathogens P. syringae pv. syringae B728a and P. syringae pv. tomato DC3000. These are most similar to bacteriophage SfV amino acid sequences (Fig. 2).
FIG 2.
Phylogenetic analysis of tail tube proteins. The maximum likelihood tree was constructed from a multiple-sequence alignment (MUSCLE) of the amino acid sequence of tail tube protein homologs from Pseudomonas R-tailocin clusters as well as selected bacteriophage genomes (black dashed boxes). The two clusters containing the P. chlororaphis 30-84 tailocins are highlighted to indicate the similarity of each R-tailocin (1 and 2) to those produced by other P. chlororaphis and P. fluorescens strains. The tail tube sequences from the R-tailocin gene region designated tailocin 1 (circle, dashed gray boxes) cluster together and with some strains also encoding two R-tailocin particles (plain font) as well as some encoding only one R-tailocin particle (bold). The R-tailocin 1 tail tube sequences are most closely related to the bacteriophages VP882 and phiHAP-1 (black, dashed box). The tail tube sequences from the region designated tailocin 2 (dashed gray boxes) cluster together and with strains encoding two or one R-tailocin particle (standard or bold font, respectively) and are most closely related to bacteriophage SfV (black, dashed box). P. aeruginosa PAO1 and a few phyllosphere-colonizing P. syringae strains with tailocin clusters are shown for comparison (solid black box). Only selected strain names are shown on the tree, but all of the strains used to build the tree are listed in the boxes associated with the tree (for full annotation, see Fig. S1 to S3). The scale bar represents the number of amino acid substitutions per site. Bootstrap values (percentage of 1,000 replications) are at branches. Trees constructed using the sheath and baseplate spike sequences from these organisms are provided in Fig. S2 and S3. Abbreviations: P.c., P. chlororaphis; P.f., P. fluorescens; P.s., P. syringae.
P. chlororaphis 30-84 R-tailocin particles differ in length.
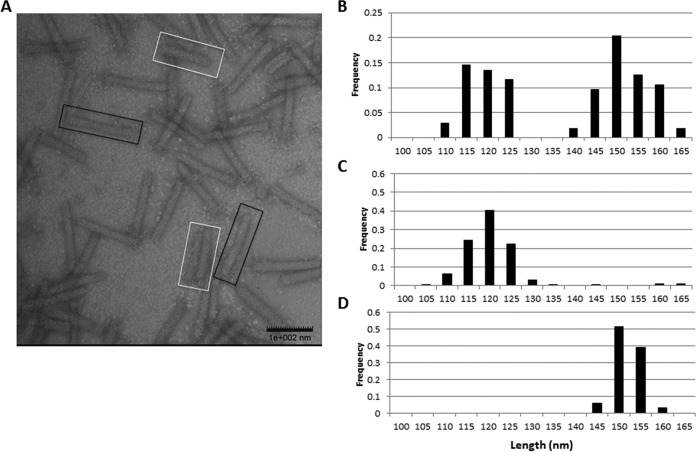
To characterize tailocin production in P. chlororaphis 30-84, an exponentially growing culture was exposed to UV light and the lysates were filter sterilized, concentrated by ultracentrifugation, and analyzed using transmission electron microscopy (TEM). The TEM micrograph revealed two distinct populations of rigid, rod-shaped, and capsidless bacteriophage tail-like particles that differed in length (Fig. 3A). Measurements of the lengths of R-tailocin particles using ImageJ confirmed that P. chlororaphis 30-84 produces two tailocin particles averaging 150 nm and 116 nm in length (Fig. 3B). The R-tailocin 1 cluster contains a larger (+97 amino acids [aa]) tape measure protein than the R-tailocin 2 cluster, which suggested that the R-tailocin 1 cluster encodes the larger R-tailocin particle. Consistent with this hypothesis, the smaller tailocin (118 ± 0.62 nm) and larger tailocin (149 ± 0.19 nm) were the only tailocins observed in the UV lysates of mutants in which baseplate genes of cluster 1 (ΔBP1) and cluster 2 (ΔBP2), respectively, were deleted (Fig. 1 and 3C and D).
FIG 3.
P. chlororaphis 30-84 produces two distinct populations of R-tailocin particles. (A) Observation of UV-induced lysates using transmission electron microscopy revealed two distinct populations of rigid, contractile bacteriophage tail-resembling particles that differ in length (white versus black boxes). (B) Frequency of particles measured in 5-nm categories. Measurement of the R-tailocin particles with ImageJ demonstrates that the R-tailocins differ in length, the longer being 149 ± 0.73 nm, compared to 116 ± 0.74 nm. (C) Frequency of particles produced by the ΔBP1 baseplate mutant measured in 5-nm categories. Measurement of the R-tailocin particles with ImageJ demonstrates that the shorter (149 ± 0.19 nm) R-tailocin is the only tailocin observed in the ΔBP1 UV lysates, indicating the tailocin 1 region encodes the larger tailocin. (D) Frequency of tailocins produced by the ΔBP2 baseplate mutant measured in 5-nm categories. Measurement of the R-tailocin particles with ImageJ demonstrates that the larger (149 ± 0.19) R-tailocin is the only tailocin observed in the ΔBP2 UV lysates, demonstrating the tailocin 2 region encodes the shorter tailocin.
P. chlororaphis 30-84 R-tailocin particles differ in killing spectra.
Tail fiber mutants with disruption of the assembly of the R-tailocin 1 or R-tailocin 2 particles (Fig. 1) or both R-tailocins were used to examine the killing spectrum of each R-tailocin particle produced by P. chlororaphis 30-84. In this experiment, the killing spectra of the mutants and complemented mutants (e.g., mutants having extra copies of the wild-type gene in trans) were compared. Concentrated UV-induced lysates were collected from the mutants and their complements, serially diluted, and spotted onto soft-agar overlays seeded with various target bacterial species or strains. Tailocins were considered the killing agent when clearing zones without the formation of plaques were observed. Among the strains tested, the host range of the tailocin particles was limited to Pseudomonas species (Table 1). The lysate of the tail fiber 1 mutant (ΔTF1) lost the ability to kill P. marginalis and P. fluorescens BL915. Complementation of the ΔTF1 mutant with a plasmid-borne copy of the gene and its chaperone restored the killing phenotype to the lysates, indicating that R-tailocin 1 targets P. marginalis and P. fluorescens BL915 (Table 1). The tail fiber 2 mutant (ΔTF2) lysate lost the ability to kill P. putida F1, P. syringae pv. tomato DC3000, P. syringae pv. phaseolicola, P. syringae pv. tomato A, P. aeruginosa PAK, and P. aureofaciens ATCC 13485. The ability to kill these strains was restored by complementation of ΔTF2, which demonstrates that R-tailocin 2 targets these strains (Table 1). Two strains, P. putida KT2440 and P. syringae pv. syringae B728a, were killed by both single-deletion mutants but not the double tail fiber mutant (ΔTF1/2), which suggests that these strains possess the receptor targeted by both tailocin particles. In fact, none of the strains tested were killed by the lysates collected from the double mutant, indicating that the tailocin particles are required for the killing activity (Table 1).
TABLE 1.
P. chlororaphis 30-84 R-tailocin killing spectruma
| Strain tested | WT | ΔTF1 | ΔTF1 (pTF1) | ΔTF2 | ΔTF2 (pTF2) | ΔTF1/2 |
|---|---|---|---|---|---|---|
| P. putida F1 | + | + | + | − | + | − |
| P. putida KT2440 | + | + | + | + | + | − |
| P. syringae pv. syringae B728a | + | + | + | + | + | − |
| P. syringae pv. tomato DC3000 | + | + | + | − | + | − |
| P. syringae pv. phaseolicola | + | + | + | − | + | − |
| P. syringae pv. tomato A | + | + | + | − | + | − |
| P. marginalis | + | − | + | + | + | − |
| P. fluorescens BL916 | + | − | + | + | + | − |
| P. aureofaciens ATCC 13485 | + | + | + | − | + | − |
| P. aeruginosa PAK | + | + | + | − | + | − |
| P. putida A514 | − | − | − | − | − | − |
| P. syringae pv. tabaci | − | − | − | − | − | − |
| P. protegens Pf5 | − | − | − | − | − | − |
| P. fluorescens 2-79 | − | − | − | − | − | − |
| P. aeruginosa PAO1 | − | − | − | − | − | − |
| P. fluorescens Q287 | − | − | − | − | − | − |
| P. fluorescens F113 | − | − | − | − | − | − |
| P. tolaasii | − | − | − | − | − | − |
| P. aureofaciens Z1B | − | − | − | − | − | − |
| E. coli DH5α | − | − | − | − | − | − |
| Pectobacterium carotovorum ATCC 15713 | − | − | − | − | − | − |
| Bacillus subtilis 613R | − | − | − | − | − | − |
| Bacillus megaterium | − | − | − | − | − | − |
| Agrobacterium tumefaciens | − | − | − | − | − | − |
| Erwinia amylovora | − | − | − | − | − | − |
Killing activity of UV lysates was determined using a soft-agar overlay assay; “+” and “−” indicate that test strains were killed or not killed, respectively, by concentrated lysates. Complemented strains are indicated as having a plasmid (in parentheses).
P. chlororaphis 30-84 hol and endo are involved in R-tailocin release.
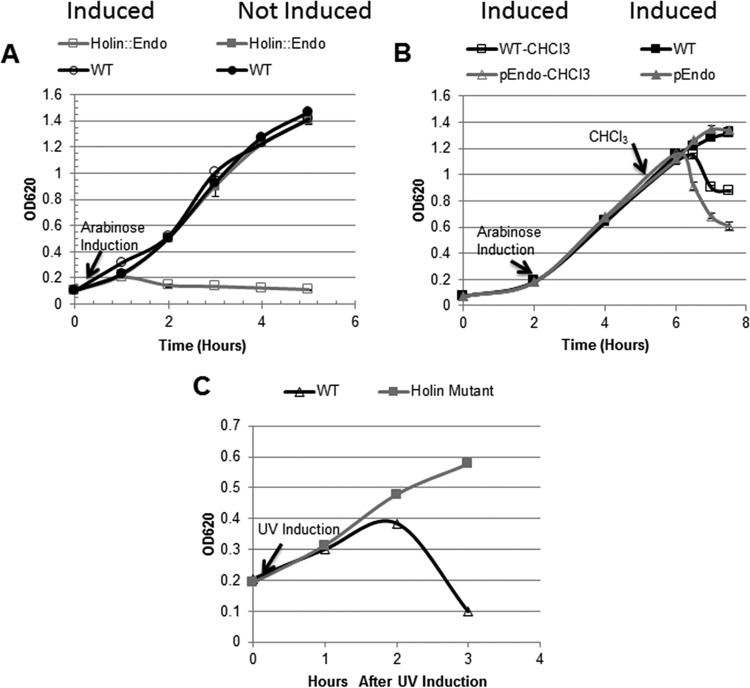
The hol and endo genes of the lysis cassette in the tailocin gene cluster were characterized in order to confirm their role in R-tailocin release. The two genes were cloned either together or separately downstream of an arabinose-inducible promoter in the vector pHERD20T. Coexpression of the hol and endo genes in P. chlororaphis 30-84 resulted in cell lysis (Fig. 4A), whereas the empty-vector controls and the noninduced cells did not lyse under these conditions. In contrast, expression of the endo gene alone did not alter growth following induction (Fig. 4B), indicating that without the activity of the holin protein, the endolysin is unable to reach and degrade the peptidoglycan layer. However, when cells expressing endo were treated with chloroform (1%), the cells rapidly lysed compared to cells expressing endo that were not treated with chloroform (Fig. 4B). Treatment of the wild type (having the empty vector) with chloroform also resulted in cell lysis but to a much lesser extent and at a lower rate, indicating the endolysin was important for rapid cell lysis. In this experiment, the chloroform substituted for the holin by permeabilizing the cytoplasmic membrane so that the endolysin could reach the peptidoglycan layer. These experiments were repeated with Escherichia coli and produced similar results (data not shown), indicating that the functionality of these proteins is not limited to Pseudomonas.
FIG 4.
Functionality of R-tailocin lysis cassette. (A) Wild-type P. chlororaphis 30-84 cells containing either the Hol::Endo plasmid or the empty vector were grown in AB + CAA and cell density was measured as OD620. Cultures were either induced or not induced with arabinose. Expression of the hol::endo construct resulted in cell lysis following arabinose induction (open squares), whereas noninduced cultures (closed squares) continued to grow at the same rate as the wild type containing the empty vector (open circles, induced; closed circles, noninduced). (B) Arabinose induced expression of endo without the hol gene did not result in altered growth patterns compared to that of the wild type with the empty vector (closed triangles and closed squares, respectively). Permeabilization of the cytoplasmic membrane with chloroform (1% CHCl3) resulted in rapid cell lysis (open triangles). Treatment of the wild type (having the empty vector) with chloroform also resulted in cell lysis, but to a lesser extent and at a lower rate (open squares). (C) The holin mutant does not lyse after UV induction (closed squares), whereas the wild type does (open triangles), indicating that the holin is required for cell lysis and release of the R-tailocin particles. Data are the means from nine biological replicates from three independent experiments, and error bars indicate standard errors.
A previously characterized hol mutant (29) was used to evaluate the role of holin in R-tailocin release. Consistent with the described role of holins in bacteriophage release (30), in our study the P. chlororaphis 30-84 holin deletion mutant did not lyse and continued to elongate hours after UV irradiation, whereas the wild-type cells lysed 2 h after induction (Fig. 3C). Induced cultures of the holin mutant were monitored for an additional 5 h and the cells still had not lysed. At 7 h after UV induction, 1% chloroform was added to the culture and the cells lysed rapidly, which yielded functional R-tailocins (data not shown). These observations indicated that UV-induced holin mutants do not lyse but endolysin and R-tailocin particles accumulate in the cytoplasm. These data confirm that the holin is an important component of the lysis cassette required for cell lysis and release of both R-tailocins produced by P. chlororaphis 30-84.
R-tailocin production bestows a competitive advantage in surface-attached biofilms.
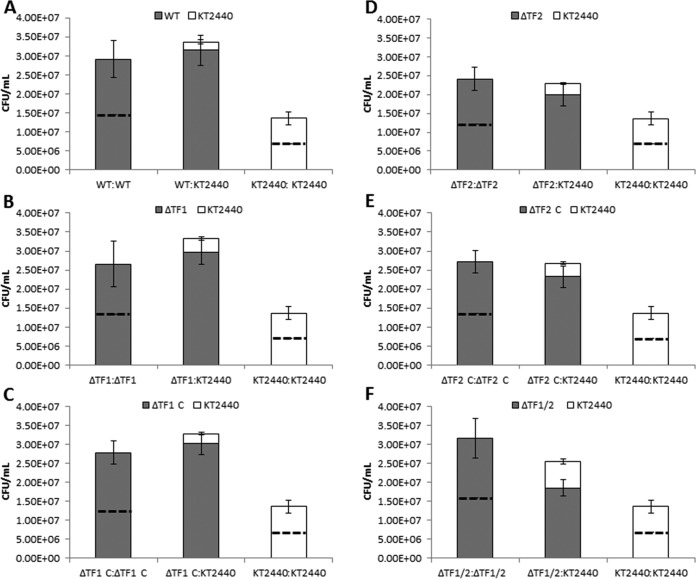
To determine whether R-tailocins produced by P. chlororaphis 30-84 contribute to the competitive fitness of P. chlororaphis 30-84 within mixed-species biofilms, a replacement series competition analysis was performed using wild-type or tail fiber mutant derivatives of P. chlororaphis 30-84 and P. putida KT2440 (sensitive to both R-tailocins). The replacement series consisted of a P. chlororaphis 30-84 strain and P. putida inoculated in different ratios (e.g., ratios 1:0, 0.5:0.5, and 0:1) maintaining a constant final cell density. Populations recovered from biofilms inoculated with either strain alone indicated the carrying capacity for each of the strains under these conditions. The carrying capacities of the P. chlororaphis 30-84 wild-type and tail fiber mutant populations were significantly greater than that of P. putida KT2440 but not significantly different from each other (Fig. 5). The similarity in the abilities of the wild type and tail fiber mutants to establish biofilm populations suggests that mutation to R-tailocin tail fibers does not influence the development of surface-attached biofilms under these conditions. Comparison of the final densities of each strain in the mixed population to their carrying capacities controls for differences between strains in carrying capacity and indicates their competitive fitness. Expectations are that in the absence of competition, each strain would attain a population size equivalent to 50% of its carrying capacity (since there was 50% less of it in the initial mixed inoculum than in the single-strain inoculum). However, in the surface-attached biofilms, wild-type P. chlororaphis 30-84 populations are significantly larger, whereas P. putida KT2440 populations are significantly smaller, than would be predicted in the absence of competition, indicating that the wild type has a competitive advantage over P. putida KT2440 in mixed surface-attached biofilms (Fig. 5A). Similarly, single tail fiber mutants ΔTF1 and ΔTF2 and their complements competitively excluded P. putida KT2440 in surface-attached biofilm populations (Fig. 5B to E). In contrast, the populations of the double mutant (ΔTF1/2) and P. putida KT2440 did not differ from the expected population sizes when mixed in a surface-attached biofilm (Fig. 5F), indicating that loss of both tailocins results in the loss of the competitive advantage of P. chlororaphis 30-84 over P. putida KT2440. Taken together, these data demonstrate that the production of at least one functional R-tailocin is required for P. chlororaphis 30-84 to have a competitive advantage over P. putida KT2440 in surface-attached biofilms.
FIG 5.
Role of R-tailocin in biofilm competition. Wild-type P. chlororaphis 30-84 (WT) and R-tailocin sensitive P. putida KT2440 (KT2440) or one of the P. chlororaphis 30-84 tail fiber mutants (ΔTF) and KT2440 were grown under biofilm conditions in AB minimal medium using a replacement series design (e.g., strains were introduced separately or in a 50:50 mixture, and all treatments had the same total cell density, 107 CFU/ml). Surface-attached biofilm populations were harvested after 48 h and quantified by dilution plating. Final population sizes of single strain treatments indicate the carrying capacity of each strain under the experimental conditions. Expectations are that in the absence of competition, each strain would attain a population size equivalent to 50% of its carrying capacity; this 50% expected population size is indicated by a line across the bars of each single strain treatment. (A) WT versus KT2440. In mixed surface-attached biofilm communities, WT populations are significantly larger, whereas KT2440 populations are significantly smaller, than expected, indicating that WT populations outcompeted KT2440. (B to E) Tail fiber mutants versus KT2440. In mixed surface-attached biofilm communities, ΔTF1, ΔTF1C, ΔTF2, and ΔTF2C populations are larger, whereas KT2440 populations are smaller, than expected, indicating that these populations outcompeted KT2440. (F) In mixed surface-attached biofilm communities, the double mutant ΔTF1/ΔTF2 and KT2440 population sizes are no different than the expected population sizes, indicating that the competitive advantage is lost with disruption of the tail fiber genes within both R-tailocin clusters. Data points represent the means from 15 biological replicates from four independent experiments, and error bars indicate standard errors.
R-tailocin production accords a competitive advantage in the rhizosphere.
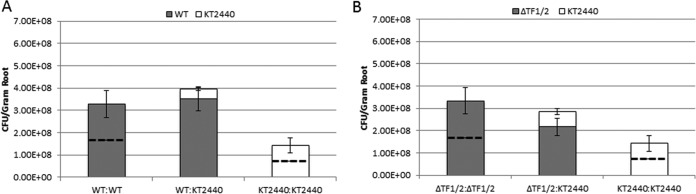
To determine whether the competitive advantage of R-tailocin production observed in mixed biofilms translates into a competitive advantage in the wheat rhizosphere, the replacement series experiment was repeated on wheat roots. The carrying capacities of wild-type P. chlororaphis 30-84 and ΔTF1/2 populations in the wheat rhizosphere were not significantly different (Fig. 6). This indicates that tailocin activity does not influence the ability of P. chlororaphis 30-84 to form rhizosphere populations in soil with reduced native microbial populations. In mixed populations, wild-type P. chlororaphis 30-84 populations were larger than predicted, whereas P. putida KT2440 populations were smaller than predicted, indicating that wild-type P. chlororaphis 30-84 was competitively excluding P. putida KT2440 in mixed-species rhizosphere communities. In mixed rhizosphere communities, the P. chlororaphis 30-84 double tail fiber mutant and P. putida KT2440 population sizes were similar to the expected population sizes, indicating that the benefit to P. chlororaphis 30-84 is diminished with the loss of tailocin activity.
FIG 6.
Role of R-tailocin release in rhizosphere competition. Wheat seeds (TAM304) were surface sterilized, pregerminated, and dip inoculated with each treatment (final concentration, 109 CFU/ml). After inoculation, seeds were planted in autoclaved wheat field soil, and rhizosphere populations were harvested 30 days after planting and quantified by dilution plating. (A) In mixed rhizosphere communities, WT populations reached the same final population size as the wild type inoculated alone and the KT2440 populations were smaller than the expected population size, suggesting that the WT benefits at the expense of KT2440 in mixed rhizosphere populations. (B) In mixed rhizosphere communities, the double tail fiber mutant and KT2440 population sizes were similar to the expected population sizes, indicating that the benefit to WT is lost with loss of tailocin activity. Similar to the case with the biofilm, there was no difference in the ability of the WT and the double tail fiber mutant to survive on plant roots in autoclaved soil. Data points represent the means from six biological replicates from one experiment, and error bars symbolize standard errors.
DISCUSSION
The P. chlororaphis 30-84 R-tailocin gene cluster contains twice as many structural genes as similar clusters in other plant-associated Pseudomonas species, such as P. fluorescens A506, P. putida BW11M1 (11, 23), P. protegens Pf-5 (24), and P. syringae pv. syringae B728a (16). Bioinformatic analysis revealed that the region potentially encodes the structural components to produce two R-tailocins that share a single lysis cassette. The two R-tailocin structural regions have distinct ancestries based on the maximum likelihood phylogenies showing that R-tailocin 1 and 2 are most closely related to the phages VP882/phiHAP-1 and SfV, respectively. Moreover, we found that the majority of the P. chlororaphis strains examined also potentially encode two R-tailocins with high amino acid similarity to R-tailocin 1 and R-tailocin 2 produced by P. chlororaphis 30-84. The speculation that some Pseudomonas species, including P. fluorescens SF4c, may produce more than one R-tailocin was reported previously (31); however, our study expands previous comparisons to include P. chlororaphis strains. Recent work categorized the R-tailocins produced by Pseudomonas species into four groups corresponding to evolutionary relationships and denoted by species complex of origin (11, 31). These groups include Rp1, Rp2, Rp3, and Rp4, represented by tailocins produced by P. aeruginosa, P. putida, P. fluorescens, and P. syringae, respectively. Using the classification system developed by Ghequire et al. (11), our results place R-tailocin 1 of the P. chlororaphis strains in the Rp3 group and R-tailocin 2 in the Rp4 group. Our study goes beyond bioinformatic characterization of the tailocin region by demonstrating that P. chlororaphis 30-84 produces two distinct, functional R-tailocin particles and suggests that pseudomonads encoding similar tailocin gene clusters also likely produce two different tailocins.
The R-tailocin gene cluster of P. chlororaphis 30-84 bears similarities to those of some plant-associated Pseudomonas species but differs in many ways from the gene clusters identified in P. aeruginosa and P. syringae. For example, the P. chlororaphis 30-84 R-tailocin gene cluster is integrated into the bacterial chromosome between mutS and cinA, which is similar to the insertion point observed in some plant-associated Pseudomonas species yet is different from the case with P. aeruginosa and P. syringae, in which the cluster is located between trpE and trpG (11, 16). The P. chlororaphis 30-84 R-tailocin lysis cassette encodes a holin, endolysin, and spanin module akin to those in other plant-associated pseudomonads; this spanin module is often missing from P. aeruginosa tailocin gene clusters (11). The P. chlororaphis 30-84 cluster also contains genes referred to as cargo genes in other plant-associated pseudomonads (11, 16). One of these (Pchl3084_1195) is predicted to encode an N-acetylmuramoyl-l-alanine amidase, potentially involved in peptidoglycan biosynthesis and hydrolysis, and its product has 96 to 98% amino acid sequence similarity to products of cargo genes in tailocin gene clusters in other plant-associated P. chlororaphis strains (e.g., PA23, O6, and 1606). The other gene (Pchl3084_1196) encodes a product with significant amino acid sequence similarity to the lectin-like bacteriocins LlpA1 (50%) and LlpA2 (49%) produced by P. protegens Pf-5 (32). Previous studies with P. putida suggested that the LlpA protein is physically associated with the R-tailocin particle produced by this strain (11). Further work is needed to determine whether there is a relationship between these proteins and R-tailocin activity.
The two P. chlororaphis 30-84 R-tailocins differ in size and killing spectrum.
TEM micrographs of concentrated lysates revealed the presence two populations of rigid, contractile capsidless bacteriophage tail-resembling particles that differ by length. Deletion of the genes required to assemble each of the R-tailocin baseplates confirmed that that R-tailocin regions 1 and 2 encode the larger and smaller particles, respectively.
Using specific tail fiber mutants, we demonstrated that the killing spectra of both R-tailocins were limited to Pseudomonas; however, within Pseudomonas species the killing spectra of the two tailocins differed. These findings suggest that the tailocins target different receptors. The finding that lysates collected from the double tail fiber mutants did not kill any of the strains screened indicated that the production of functional R-tailocin particles is required for killing activity. Our results demonstrate that the production of more than one functional R-tailocin particle increases the killing spectrum of the producing strain, thereby increasing strain competitiveness. Ongoing work seeks to identify the receptors targeted by each of the P. chlororaphis 30-84 R-tailocins.
R-tailocins confer a competitive advantage in biofilms and the rhizosphere.
Tailocin production was required for competitive survival against an R-tailocin-sensitive strain in surface-attached biofilm and rhizosphere populations. The use of a replacement series design was necessary to determine direct competition between strains, because P. putida KT2440 and P. chlororaphis 30-84 differed in their abilities to form in vitro biofilms and rhizosphere populations. We observed that wild-type populations had a strong competitive advantage over P. putida KT2440 in mixed biofilms and rhizosphere. However, a loss of functional R-tailocin production resulted in the loss of the competitive advantage over P. putida KT2440 in mixed biofilms and the rhizosphere. The importance of R-tailocins as determinants of interactions among closely related strains of P. aeruginosa was shown previously (20–22). Although the killing capacities of R-tailocins produced by plant-associated Pseudomonas strains have been described, to our knowledge this is the first demonstration that R-tailocins function in competition between plant-associated bacterial species in surface-attached biofilms or in rhizosphere communities.
Recent conceptualizations of root tip colonization provide a framework for relating root development and bacterial population dynamics (33). As bacteria are recruited from the soil to the root tip region, attach, and develop into microcolonies, the root environment changes as the root tip matures and develops. The simultaneous maturation of the many proliferating root tips creates a highly diverse environmental landscape composed of recruitment niches at the tips that evolve into mature root niches within a matter of days. The need for competitive strategies targeting closely related combatants likely changes as the root and root-inhabiting microbial populations develop. In future studies, regard for the dynamics of microbial populations in relation to root tip maturation may identify niches where R-tailocin production is most effective.
MATERIALS AND METHODS
Bacterial strains and media.
The bacterial strains and plasmids used in this study are described in Table 2. A spontaneous rifampin-resistant derivative of P. chlororaphis 30-84 was used in all studies. P. chlororaphis 30-84 and P. putida KT2440 were grown at 28°C in Luria-Bertani (LB) medium or AB minimal medium supplemented with 2% Casamino Acids (AB + CAA) (34). E. coli strains were grown at 37°C in LB medium. Antibiotics were used when appropriate at the following concentrations: for E. coli, kanamycin (Km) at 50 μg/ml, and for Pseudomonas, Km, rifampin (Rif), and gentamicin (Gm) at 50, 100, and 50 μg/ml, respectively. Cycloheximide (100 μg/ml) was used to inhibit fungal growth in the rhizosphere competition assay.
TABLE 2.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| P. chlororaphis 30-84 | Wild type; Rifr | W. W. Bockus |
| P. chlororaphis 30-84 ΔHolin | hol gene replaced with Kmr cassette; Kmr | 29 |
| P. chlororaphis 30-84 ΔTF1 | TF1 gene replaced with Kmr cassette | This study |
| P. chlororaphis 30-84 ΔTF2 | TF2 gene replaced with Tcr cassette | This study |
| P. chlororaphis 30-84 ΔTF1/2 | TF1 and TF2 genes replaced with Kmr and Tcr, respectively | This study |
| P. chlororaphis 30-84 ΔBP1 | BP1 genes replaced with Kmr cassette; Kmr | This study |
| P. chlororaphis 30-84 ΔBP2 | BP2 genes replaced with Tcr cassette; Tcr | This study |
| P. chlororaphis 30-84 ΔBP1/2 | BP1 and BP2 genes replaced with Kmr and Tcr, respectively | This study |
| Escherichia coli DH5α | F− recA1 endA1 hsdRl7 supE44 thi-J gyrA96 reLA1A (argF-lacZYA) 1169 + 801acZAM15/− | GIBCO-BRL |
| Escherichia coli HB101 | F− hsdS20(rB−mB−) supE44 recAl aral4proA2 lacYl galK2 rpsL20 xyl-5mtl-5I−; GIBCO-BRL | GIBCO-BRL |
| P. putida F1 | Rhizosphere associated | ATCC |
| P. putida KT2440 | Rhizosphere associated | D. C. Gross (Plant Pathology and Microbiology, TAMU) |
| P. syringae pv. tomato | Plant pathogen | 43 |
| P. syringae pv. phaseolicola N4SP | Plant pathogen | Teaching collectionsa |
| P. marginalis L | Plant pathogen | Teaching collections |
| P. syringae pv. syringae B728A | Plant pathogen | 44 |
| P. syringae pv. tomato DC3000 | Plant pathogen | 45 |
| P. aeruginosa PAK | Opportunistic human pathogen | 46 |
| P. aureofaciens ATCC 13485 | Rhizosphere associated | ATCC |
| P. fluorescens BL915 | Rhizosphere associated | 47 |
| P. syringae pv. tabaci | Plant pathogen | Teaching collections |
| P. protegens Pf-5 | Rhizosphere associated | 24 |
| P. fluorescens 2-79 | Rhizosphere associated | 25 |
| P. aeruginosa PAO1 | Opportunistic human pathogen | 48 |
| P. fluorescens Q287 | Rhizosphere associated | 49 |
| P. fluorescens F113 | Rhizosphere associated | 50 |
| P. tolaasii NCPPB 1116 | Fungal pathogen | 51 |
| P. aureofaciens Z1B | Rhizosphere associated | Pierson Lab |
| Bacillus megaterium | Rhizosphere associated | Teaching collections |
| Bacillus subtilis 613R | Rhizosphere associated | Teaching collections |
| Pectobacterium carotovorum ATCC 15713 | Plant pathogen | ATCC |
| Agrobacterium tumefaciens C58 | Plant pathogen | Teaching collections |
| Erwinia amylovora ea1189 | Plant pathogen | 52 |
| Plasmids | ||
| pEX18Ap | Apr | 38 |
| pUC4K | Kmr Apr aph | 53 |
| pKRP12 | Tcr | 54 |
| pUCP20Gm | Gmr | 39 |
| pHERD20TKm | pHERD20T derivative bearing Kmr cartridge; Kmr | This study |
| pHERD20TKmHE | pHERD20TKm containing 946-bp hol::endo fragment; Kmr | This study |
| pHERD20TKmE | pHERD20TKm containing 581-bp endo fragment; Kmr | This study |
| pTF1 | pUCP20Gm containing 3,092-bp tail fiber 1 and chaperone fragment; Gmr | This study |
| pTF2 | pUCP20Gm containing 4,141-bp tail fiber 2 and chaperone fragment; Gmr | This study |
University of Arizona School of Plant Sciences or Texas A&M University Department of Plant Pathology and Microbiology teaching collection.
Phylogenetic analysis.
The predicted amino acid sequences of the tail tube, sheath, and baseplate spike encoded within each tailocin region of P. choloraphis 30-84 were used to identify related gene clusters in other P. chlororaphis, P. fluorescens, and P. protegens strains as well as within a few bacteriophages using BLAST (Basic Local Alignment Search Tool). Sequence alignments and phylogenetic analyses were performed with MEGA7 (35). Amino acid alignments were performed with MUSCLE (MEGA7) and used to build maximum likelihood trees using the Jones, Taylor, and Thorton (JTT) substitution model. The resulting phylogenies were edited with FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/).
UV induction of P. chlororaphis 30-84 cultures.
UV irradiation was used to induce R-tailocin synthesis and cell lysis. Overnight LB cultures of P. chlororaphis 30-84 and the appropriate mutants were collected by centrifugation, washed with sterile water, resuspended in 20 ml of fresh LB (optical density at 620 nm [OD620] = 0.05), and grown at 28°C with shaking (200 rpm). When the cultures reached an OD620 of 0.5, they were centrifuged (7,500 × g) for 10 min at 4°C. The pellets were washed once and resuspended in 10 ml of 0.85% NaCl. The 10-ml suspensions in sterile petri plates were UV irradiated (400 μW/cm2/s) for 7 s with constant shaking of the plates to ensure even exposure to UV. The irradiated suspensions were transferred to foil-covered 500-ml flasks containing LB (final concentration, 1×; 50 ml). These cultures were grown at 28°C with shaking (200 rpm), and the optical density (620 nm) was monitored hourly. After cell lysis, the lysates were collected and filter sterilized to remove cellular debris.
TEM.
Transmission electron microscopy (TEM) was performed at the Microscopy and Imaging Center at Texas A&M University. UV-induced lysates were ultracentrifuged (2.5 h at 48,000 × g) and resuspended in 2.2 ml of λ buffer (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 8 mM MgSO4) to generate a 40× concentration for TEM analysis. Tailocins were collected by dipping carbon-coated mica into the R-tailocin sample for 1 min and staining with 2% aqueous uranyl acetate for 10 s. The floating carbon film was attached to the grid and excessive liquid was removed for analysis. Specimens were visualized on a JEOL 1200EX TEM operating at an acceleration voltage of 100 kV. Images were recorded at calibrated magnifications with a charge-coupled-device (CCD) camera, and measurements were acquired using ImageJ (36).
Soft-agar overlay spot assay.
Killing activity of UV lysates was gauged using a soft-agar overlay assay similar to one previously reported (37). Overnight cultures of P. putida KT2440 (and the strains listed in Table 1) were diluted to an OD620 of 0.05 and grown at 28°C with shaking (200 rpm) until the culture reached an OD620 of 0.4 to 0.5. Liquid LB containing 0.7% agar was kept at 55°C, and 4 ml was transferred to 5-ml plastic culture tubes (Falcon). Samples (100 μl) of the bacterial culture were added to the 4 ml of liquid LB top agar, vortexed, and poured onto an LB agar (1.5%) plate. Plates were allowed to solidify for 10 min before lysates were spotted. Lysates concentrated by ultracentrifugation and suspended in 5 ml of λ buffer were serially diluted and a concentration gradient of lysates was spotted (10 μl) on the plates in duplicate. Zones of growth inhibition without plaques, indicating that the killing agent is nonreplicative, signified tailocin activity.
Heterologous expression of hol and endo genes from P. chlororaphis 30-84.
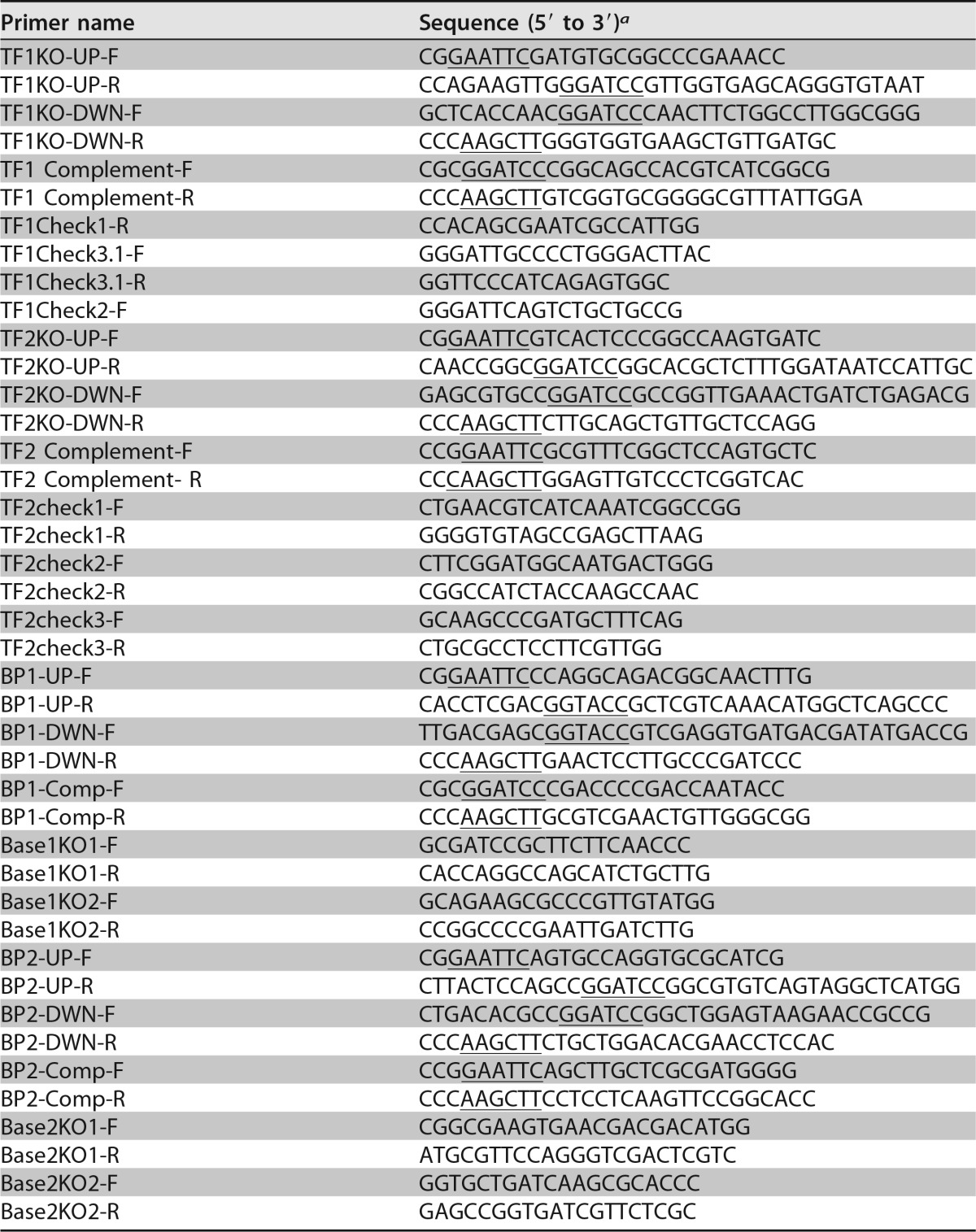
For gene expression studies in P. chlororaphis 30-84, a modified version of the expression vector pHERD20T (containing an arabinose-inducible promoter) was constructed by replacing the Apr cassette with the Kmr cassette of pUC4K (Table 2). The Kmr cassette was amplified using the primer set km-F/km-R (Table 3) and cloned into pHERD20T to create pHERD20TKm (Table 2). Primer pairs hol-F/hol-R and endo-F/endo-R (Table 3) were used to amplify DNA fragments containing the native ribosome-binding site and the complete coding sequence of the holin and endolysin genes from P. chlororaphis 30-84, respectively. The endolysin PCR fragment was cloned together with the holin fragment or separately into the vector pHERD20TKm, generating a plasmid expressing both hol and endo (pHERD20TKmHE) and a plasmid expressing only endo (pHERD20TKmE) (Table 2). Plasmids were introduced by triparental conjugation.
TABLE 3.
Primers used in this study
aUnderlined nucleotides indicate restriction enzyme sites at the end of primers.
Lysis cassette gene expression assays.
To test the functionality of the lysis cassette, P. chlororaphis 30-84 strains containing pHERD20TKmHE or the control plasmid pHERD20TKm were grown overnight in AB + CAA supplemented with the appropriate antibiotics. These overnight cultures were diluted to an OD620 of ∼0.1 and grown at 28°C with shaking. When the cultures reached an OD620 of ∼0.2, half of the cultures were induced by the addition of arabinose (final concentration, 1%). After induction, the optical density (620 nm) of the cultures was measured every hour to monitor cell lysis. To examine the role of the endolysin in cell lysis, P. chlororaphis 30-84 strains containing pHERD20TKmE or the control plasmid pHERD20TKm were grown and treated as in the previous experiment. However, after arabinose induction, when the cell cultures reached an OD620 of ∼1, chloroform was added to half of the cultures of cells (final concentration of 1% [vol/vol]) to permeabilize the cytoplasmic membrane, enabling the endolysin to reach the peptidoglycan layer. The optical density of the cultures was measured every half hour after chloroform addition to monitor cell lysis.
Deletion of tail fiber and baseplate genes.
P. chlororaphis 30-84 tail fiber mutants (ΔTF, which can no longer target susceptible strains) were generated using the suicide vector pEX18Ap (Table 2) as described by Hoang et al. (38). To generate ΔTF1, sequences (1,000 nucleotides [nt]) flanking the tail fiber gene of tailocin 1 (Pchl3084_1202) (Fig. 1) were amplified by two-step PCR using the primer pairs TF1KO-UP-F/TF1KO-UP-R and TF1KO-DWN-F/TF1KO-DWN-R (Table 3). Using the primer pair TF1KO-UP-F and TF1KO-DWN-R with the product of the previous PCR resulted in a construct that contained the upstream fragment separated from the downstream fragment by a BamHI restriction site. This fragment was ligated into the EcoRI-to-HindIII site in the pEX18Ap multiple-cloning region (38). The kanamycin resistance cassette with its promoter was then digested from a pUC4K (Table 2) template with the BamHI enzyme and ligated between the upstream and downstream fragments of the tail fiber genes in pEX18Ap. The final construct was electroporated into P. chlororaphis 30-84 and transformants were plated onto LB amended with Km as performed previously. Double-crossover mutants were obtained by counterselection with LB amended with Km and 6% sucrose and confirmed using PCR primers specific to the internal regions of the tail fiber coding sequence (Table 3). Similarly, to generate ΔTF2, the sequences (1,500 nt) flanking both tail fiber genes of tailocin 2 (Pchl3084_1227-1230) (Fig. 1) were amplified by two-step PCR using the primer pairs TF2KO-UP-F/TF2KO-UP-R and TF2KO-DWN-F/TF2KO-DWN-R, followed by amplification using the primer pair TF2KO-UP-F and TF2KO-DWN-R (Table 3). This fragment was ligated into pEX18Ap at the EcoRI-to-HindIII site (38). The tetracycline resistance cassette with its promoter was digested from the pKRP13 template with the BamHI enzyme and ligated between the upstream and downstream fragments of the tail fiber genes in pEX18Ap. The final construct was electroporated into P. chlororaphis 30-84, and mutants were selected using the appropriate antibiotics, as described above. A double mutant, ΔTF1/ΔTF2, was generated by creating a tail fiber 2 deletion in a tail fiber 1 mutant (ΔTF1) and confirmed with the primers used to confirm tail fiber 1 and 2 mutants (Table 3). P. chlororaphis 30-84 baseplate mutants (ΔBP, which no longer produces a tailocin particle) were generated as described above. The ΔBP1 mutant was generated by replacement of three genes required for baseplate assembly, Pchl3084_1198 to Pchl3084_1120 (Fig. 1), using a kanamycin antibiotic resistance marker and the primer set BP1-UP-F, BP1-UP-R, BP1-DWN-F, and BP1-DWN-R (Table 3). Similarly, ΔBP2 was generated by replacement of three genes required for baseplate assembly, Pchl3084_1221 to Pchl3084_1123, with a tetracycline antibiotic resistance marker using the primer set BP2-UP-F, BP2-UP-R, BP2-DWN-F, and BP2-DWN-R (Table 3). All mutants were confirmed using PCR primers specific to the internal regions of the target gene coding sequence (Table 3).
Complementation of tail fiber and baseplate mutants.
The deletion of the tail fiber coding sequences removed the ribosome-binding site of the genes downstream of both tail fiber deletions (Pchl3084_1203 and Pchl3084_1227 to Pchl3084_1230) (Fig. 1). Both of these genes (Pchl3084_1204 and Pchl3084_1231) are predicted to encode a chaperone that is required for the attachment of tail fibers to the tailocin baseplate. To ensure proper expression of both of these genes in the mutant background, the coding sequence of the tail fibers and their chaperones were PCR amplified with the primers TF1Comp-F and TF1Comp-R and TF2Comp-F and TF2Comp-R (Table 3). The resulting fragments were digested with the appropriate restriction enzymes and cloned into the expression vector pUCP20Gm (39). A similar method was used to complement the ΔBP1 mutant using the primers BP1-Comp-F and BP1-Comp-R (Table 3). The final constructions were introduced into the respective tail fiber or baseplate mutants by electroporation.
Biofilm replacement series analysis.
A replacement series analysis was used to examine whether R-tailocin production confers a competitive advantage to P. chlororaphis 30-84 in mixed biofilms with P. putida KT2440 (40, 41). The replacement series consisted of different ratios of the two target strains in the starting inoculum, with a final cell density of 107 CFU/ml for all treatments. Treatments consisted of 100% P. chlororaphis 30-84 (wild type, ΔTF1, ΔTF1 pTF1, ΔTF2, ΔTF2 pTF2, and ΔTF1/2), 100% P. putida KT2440, or a 50:50 mixture of P. chlororaphis 30-84 strains and P. putida KT2440. The replacement series was used to infer competition by comparing the observed population sizes of each strain in mixed populations to the predicted sizes based on the carrying capacity observed for the single strain treatments (e.g., rather than from the relative proportion of each strain in the final mixture). Total population density and ratios within the inoculum were confirmed by dilution plating. Biofilm cultures were grown as described previously (42). Briefly, bacterial strains were grown overnight in AB + CAA at 28°C with agitation (200 rpm), harvested, washed, and resuspended in fresh medium before creating the single strain or mixed starting cultures. Replacement series treatments (3 ml) were added to 15-ml polypropylene tubes, and the tubes were incubated at 28°C without shaking. After 48 h, the liquid and loosely adherent cells were removed by pipetting. The surface-attached biofilm was washed three times with sterile water. After washing, 5 ml of phosphate-buffered saline (pH 7.4) was added and the tubes were sonicated (3 times for 10 s) and vortexed (3 times for 10 s) to remove surfaced-adhered bacteria. Population sizes were quantified by dilution plating. Serial dilutions were plated on LB agar, where P. chlororaphis 30-84 colonies were easily differentiated from P. putida KT2440 colonies without antibiotic selection by phenazine pigment production and differences in colony morphology. In this experiment, all strains carried a plasmid: either one or both of the tail fiber complements or the empty vector control pUCP20GM. Since no antibiotics were used in the competition assay, to confirm the stability of the plasmids in the complemented strains, dilutions were plated on media with and without gentamicin. No significant differences in CFU were found, indicating that the plasmid was stable over the 48-h assay (data not shown).
Rhizosphere replacement series analysis.
A smaller replacement series analysis was conducted to examine the role of R-tailocins in P. chlororaphis 30-84 (wild type versus ΔTF1/2) competition with P. putida KT2440 in mixed rhizosphere populations. This replacement series consisted of the same population ratios as described for the biofilm replacement series, but the total cell density of the treatments used to inoculate wheat seeds was 109 CFU/ml. The inoculum for the different treatments was prepared as described above, and final cell densities and ratios were confirmed via serial dilution plating. Wheat seeds (TAM304) were surface disinfected by incubation in 70% ethanol for 10 min, incubated in 90% commercial bleach (1 min), and then washed with sterile water (5 times for 1 min). Disinfected seeds were pregerminated on germination paper for 48 h, and then the seedlings were suspended in the bacterial inoculum for 10 min. The inoculated seedlings were sown in autoclaved (45 min twice, with a 24-h break) wheat field soil (Bushland, TX) and grown for 30 days (dark [8 h] and light [16 h] cycle at 27 ± 2°C). The rhizosphere bacterial populations were estimated as described previously (40). Briefly, the whole root system of each plant was collected, placed in 5 ml of phosphate-buffered saline (pH 7.4), and sonicated (10 s) and vortexed (10-s pulses) three times. Serial dilutions were plated on LB agar supplemented with cycloheximide, and colonies were differentiated as described above (e.g., without antibiotic selection). The roots were dried for 48 h in a 65°C oven and populations were standardized to dry root weight.
Statistical analysis.
Comparisons of observed and expected surface-attached biofilm populations were analyzed statistically using Student's t test. P values less than 0.05 were considered significant.
Supplementary Material
ACKNOWLEDGMENTS
We thank Carlos Gonzalez and lab members Tram Le and Guichun Yao for assistance with the transmission electron microscope and guidance on bacteriophage analyses. We thank Dennis Gross for providing strains and helpful discussions. We are grateful to Dongping Wang, Panatda Saenkham, Huiqiao Pan, and Tessa Ries for helpful discussions throughout the project.
Research was funded by support from E. Pierson and the Department of Plant Pathology and Microbiology.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00706-17.
REFERENCES
- 1.Riley MA, Gordon DM. 1999. The ecological role of bacteriocins in bacterial competition. Trends Microbiol 7:129–133. doi: 10.1016/S0966-842X(99)01459-6. [DOI] [PubMed] [Google Scholar]
- 2.Bradley DE. 1967. Ultrastructure of bacteriophage and bacteriocins. Bacteriol Rev 31:230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michel-Briand Y, Baysse C. 2002. The pyocins of Pseudomonas aeruginosa. Biochimie 84:499–510. doi: 10.1016/S0300-9084(02)01422-0. [DOI] [PubMed] [Google Scholar]
- 4.Gebhart D, Williams SR, Bishop-Lilly KA, Govoni GR, Willner KM, Butani A, Sozhamannan S, Martin D, Fortier L-C, Scholl D. 2012. Novel high-molecular-weight, R-type bacteriocins of Clostridium difficile. J Bacteriol 194:6240–6247. doi: 10.1128/JB.01272-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zink R, Loessner MJ, Scherer S. 1995. Charaterization of cryptic prophages (monocins) in Listeria and sequence analysis of a holin/endolysin gene. Microbiology 141:2577–2584. doi: 10.1099/13500872-141-10-2577. [DOI] [PubMed] [Google Scholar]
- 6.Nakayama K, Takashima K, Ishihara H, Shinomiya T, Kageyama M, Kanaya S, Ohnishi M, Murata T, Mori H, Hayashi T. 2000. The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol Microbiol 38:213–231. doi: 10.1046/j.1365-2958.2000.02135.x. [DOI] [PubMed] [Google Scholar]
- 7.Ishii S-I, Nishi Y, Egami F. 1965. The fine structure of a pyocin. J Mol Biol 13:428–431. doi: 10.1016/S0022-2836(65)80107-3. [DOI] [PubMed] [Google Scholar]
- 8.Higerd TB, Baechler CA, Berk RS. 1969. Morphological studies on relaxed and contracted forms of purified pyocin particles. J Bacteriol 98:1378–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsui H, Sano Y, Ishihara H, Shinomiya T. 1993. Regulation of pyocin genes in Pseudomonas aeruginosa by positive (prtN) and negative (prtR) regulatory genes. J Bacteriol 175:1257–1263. doi: 10.1128/jb.175.5.1257-1263.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang N. 2006. Lysis timing and bacteriophage fitness. Genetics 172:17–26. doi: 10.1534/genetics.105.045922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghequire MG, Dillen Y, Lambrichts I, Proost P, Wattiez R, De Mot R. 2015. Different ancestries of R tailocins in rhizospheric Pseudomonas isolates. Genome Biol Evol 7:2810–2828. doi: 10.1093/gbe/evv184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang I-N, Smith DL, Young R. 2000. Holins: the protein clocks of bacteriophage infections. Annu Rev Microbiol 54:799–825. doi: 10.1146/annurev.micro.54.1.799. [DOI] [PubMed] [Google Scholar]
- 13.Berry J, Rajaure M, Pang T, Young R. 2012. The spanin complex is essential for lambda lysis. J Bacteriol 194:5667–5674. doi: 10.1128/JB.01245-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaziro Y, Tanaka M. 1965. Studies on the mode of action of pyocin. J Biochem 58:357–363. doi: 10.1093/oxfordjournals.jbchem.a128212. [DOI] [PubMed] [Google Scholar]
- 15.Uratani Y, Hoshino T. 1984. Pyocin R1 inhibits active transport in Pseudomonas aeruginosa and depolarizes membrane potential. J Bacteriol 157:632–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hockett KL, Renner T, Baltrus DA. 2015. Independent co-option of a tailed bacteriophage into a killing complex in Pseudomonas. mBio 6:e00452-15. doi: 10.1128/mBio.00452-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischer S, Godino A, Quesada JM, Cordero P, Jofré E, Mori G, Espinosa-Urgel M. 2012. Characterization of a phage-like pyocin from the plant growth-promoting rhizobacterium Pseudomonas fluorescens SF4c. Microbiology 158:1493–1503. doi: 10.1099/mic.0.056002-0. [DOI] [PubMed] [Google Scholar]
- 18.Ohsumi M, Shinomiya T, Kageyama M. 1980. Comparative study on R-type pyocins of Pseudomonas aeruginosa. J Biochem 87:1119–1126. [PubMed] [Google Scholar]
- 19.Williams SR, Gebhart D, Martin DW, Scholl D. 2008. Retargeting R-type pyocins to generate novel bactericidal protein complexes. Appl Environ Microbiol 74:3868–3876. doi: 10.1128/AEM.00141-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oliveira NM, Martinez-Garcia E, Xavier J, Durham WM, Kolter R, Kim W, Foster KR. 2015. Biofilm formation as a response to ecological competition. PLoS Biol 13:e1002191. doi: 10.1371/journal.pbio.1002191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Köhler T, Donner V, van Delden C. 2010. Lipopolysaccharide as shield and receptor for R-pyocin-mediated killing in Pseudomonas aeruginosa. J Bacteriol 192:1921–1928. doi: 10.1128/JB.01459-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heo Y-J, Chung I-Y, Choi KB, Cho Y-H. 2007. R-type pyocin is required for competitive growth advantage between Pseudomonas aeruginosa strains. J Microbiol Biotechnol 17:180–185. [PubMed] [Google Scholar]
- 23.Ghequire MG, De Mot R. 2014. Ribosomally encoded antibacterial proteins and peptides from Pseudomonas. FEMS Microbiol Rev 38:523–568. doi: 10.1111/1574-6976.12079. [DOI] [PubMed] [Google Scholar]
- 24.Mavrodi DV, Loper JE, Paulsen IT, Thomashow LS. 2009. Mobile genetic elements in the genome of the beneficial rhizobacterium Pseudomonas fluorescens Pf-5. BMC Microbiol 9:8. doi: 10.1186/1471-2180-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mazzola M, Cook RJ, Thomashow LS, Weller D, Pierson L III. 1992. Contribution of phenazine antibiotic biosynthesis to the ecological competence of fluorescent pseudomonads in soil habitats. Appl Environ Microbiol 58:2616–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maddula VK, Zhang Z, Pierson E, Pierson L III. 2006. Quorum sensing and phenazines are involved in biofilm formation by Pseudomonas chlororaphis (aureofaciens) strain 30-84. Microb Ecol 52:289–301. doi: 10.1007/s00248-006-9064-6. [DOI] [PubMed] [Google Scholar]
- 27.Maddula V, Pierson E, Pierson L. 2008. Altering the ratio of phenazines in Pseudomonas chlororaphis (aureofaciens) strain 30-84: effects on biofilm formation and pathogen inhibition. J Bacteriol 190:2759–2766. doi: 10.1128/JB.01587-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pierson LS III, Thomashow LS. 1992. Cloning and heterologous expression of the phenazine biosynthetic locus from Pseudomonas aureofaciens 30-84. Mol Plant Microbe Interact 5:330–339. doi: 10.1094/MPMI-5-330. [DOI] [PubMed] [Google Scholar]
- 29.Wang D, Yu JM, Dorosky RJ, Pierson LS III, Pierson EA. 2016. The phenazine 2-hydroxy-phenazine-1-carboxylic acid promotes extracellular DNA release and has broad transcriptomic consequences in Pseudomonas chlororaphis 30-84. PLoS One 11:e0148003. doi: 10.1371/journal.pone.0148003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Young R. 2002. Bacteriophage holins: deadly diversity. J Mol Microbiol Biotechnol 4:21–36. [PubMed] [Google Scholar]
- 31.Ghequire MG, De Mot R. 2015. The tailocin tale: peeling off phage tails. Trends Microbiol 23:587–590. doi: 10.1016/j.tim.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 32.Parret AH, Temmerman K, De Mot R. 2005. Novel lectin-like bacteriocins of biocontrol strain Pseudomonas fluorescens Pf-5. Appl Environ Microbiol 71:5197–5207. doi: 10.1128/AEM.71.9.5197-5207.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dupuy LX, Silk WK. 2016. Mechanisms of early microbial establishment on growing root surfaces. Vadose Zone J 15:1–13. doi: 10.2136/vzj2015.06.0094. [DOI] [Google Scholar]
- 34.Pierson L, Keppenne VD, Wood DW. 1994. Phenazine antibiotic biosynthesis in Pseudomonas aureofaciens 30-84 is regulated by PhzR in response to cell density. J Bacteriol 176:3966–3974. doi: 10.1128/jb.176.13.3966-3974.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.National Institutes of Health. 2012. ImageJ. National Institutes of Health, Bethesda, MD. [Google Scholar]
- 37.Hockett KL, Baltrus DA. 2017. Use of the soft-agar overlay technique to screen for bacterially produced inhibitory compounds. J Vis Exp 2017(119):e55064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. doi: 10.1016/S0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 39.Chiang P, Burrows LL. 2003. Biofilm formation by hyperpiliated mutants of Pseudomonas aeruginosa. J Bacteriol 185:2374–2378. doi: 10.1128/JB.185.7.2374-2378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chancey ST, Wood DW, Pierson EA, Pierson LS. 2002. Survival of GacS/GacA mutants of the biological control bacterium Pseudomonas aureofaciens 30-84 in the wheat rhizosphere. Appl Environ Microbiol 68:3308–3314. doi: 10.1128/AEM.68.7.3308-3314.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Driscoll WW, Pepper JW, Pierson LS, Pierson EA. 2011. Spontaneous Gac mutants of Pseudomonas biological control strains: cheaters or mutualists? Appl Environ Microbiol 77:7227–7235. doi: 10.1128/AEM.00679-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang D, Dorosky RJ, Han CS, Lo C-C, Dichosa AE, Chain PS, Yu JM, Pierson LS, Pierson EA. 2015. Adaptation genomics of a small-colony variant in a Pseudomonas chlororaphis 30-84 biofilm. Appl Environ Microbiol 81:890–899. doi: 10.1128/AEM.02617-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He P, Warren RF, Zhao T, Shan L, Zhu L, Tang X, Zhou J-M. 2001. Overexpression of Pti5 in tomato potentiates pathogen-induced defense gene expression and enhances disease resistance to Pseudomonas syringae pv. tomato. Mol Plant Microbe Interact 14:1453–1457. doi: 10.1094/MPMI.2001.14.12.1453. [DOI] [PubMed] [Google Scholar]
- 44.Thakur PB, Vaughn-Diaz VL, Greenwald JW, Gross DC. 2013. Characterization of five ECF sigma factors in the genome of Pseudomonas syringae pv. syringae B728a. PLoS One 8:e58846. doi: 10.1371/journal.pone.0058846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Petnicki-Ocwieja T, Schneider DJ, Tam VC, Chancey ST, Shan L, Jamir Y, Schechter LM, Janes MD, Buell CR, Tang X. 2002. Genomewide identification of proteins secreted by the Hrp type III protein secretion system of Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci U S A 99:7652–7657. doi: 10.1073/pnas.112183899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strom MS, Lory S. 1986. Cloning and expression of the pilin gene of Pseudomonas aeruginosa PAK in Escherichia coli. J Bacteriol 165:367–372. doi: 10.1128/jb.165.2.367-372.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hill D, Stein J, Torkewitz N, Morse A, Howell C, Pachlatko J, Becker J, Ligon J. 1994. Cloning of genes involved in the synthesis of pyrrolnitrin from Pseudomonas fluorescens and role of pyrrolnitrin synthesis in biological control of plant disease. Appl Environ Microbiol 60:78–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vincent MN, Harrison L, Brackin J, Kovacevich P, Mukerji P, Weller D, Pierson E. 1991. Genetic analysis of the antifungal activity of a soilborne Pseudomonas aureofaciens strain. Appl Environ Microbiol 57:2928–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shanahan P, O'Sullivan DJ, Simpson P, Glennon JD, O'Gara F. 1992. Isolation of 2, 4-diacetylphloroglucinol from a fluorescent pseudomonad and investigation of physiological parameters influencing its production. Appl Environ Microbiol 58:353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wong W, Preece T. 1979. Identification of Pseudomonas tolaasi: the white line in agar and mushroom tissue block rapid pitting tests. J Appl Bacteriol 47:401–407. doi: 10.1111/j.1365-2672.1979.tb01200.x. [DOI] [Google Scholar]
- 52.Wang D, Korban SS, Zhao Y. 2009. The Rcs phosphorelay system is essential for pathogenicity in Erwinia amylovora. Mol Plant Pathol 10:277–290. doi: 10.1111/j.1364-3703.2008.00531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grindley NDF, Joyce CM. 1981. Analysis of the structure and function of the kanamycin-resistance transposon Tn903. Cold Spring Harbor symposia on quantitative biology: Movable genetic elements, vol 45 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [DOI] [PubMed] [Google Scholar]
- 54.Reece KS, Phillips GJ. 1995. New plasmids carrying antibiotic-resistance cassettes. Gene 165:141–142. doi: 10.1016/0378-1119(95)00529-F. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.