Significance
Fetal hemoglobin (HbF) expression is a tissue- and stage-specific marker of ontogeny in large mammals, which also has therapeutic importance for beta hemoglobinopathies. The heterochronic let-7 miRNAs, which regulate the time and sequence of stage-specific developmental events, have also been shown to regulate HbF in adult human erythroblasts. Here we provide a focused investigation of a let-7 target named “insulin-like growth-factor 2 mRNA-binding protein 1” (IGF2BP1), for its potential role in reactivating HbF in adult cells. IGF2BP1 overexpression caused robust increases of HbF and a reversal from the adult toward a fetal-like globin phenotype. IGF2BP1 effects are partially mediated by posttranscriptional regulation of the known HbF regulator BCL11A. These results suggest a novel mechanism for the regulation of BCL11A and HbF in humans.
Keywords: IGF2BP1, IMP1, let-7 targets, fetal hemoglobin, ontogeny
Abstract
Here we investigated in primary human erythroid tissues a downstream element of the heterochronic let-7 miRNA pathway, the insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1), for its potential to affect the hemoglobin profiles in human erythroblasts. Comparison of adult bone marrow to fetal liver lysates demonstrated developmental silencing in IGF2BP1. Erythroid-specific overexpression of IGF2BP1 caused a nearly complete and pancellular reversal of the adult pattern of hemoglobin expression toward a more fetal-like phenotype. The reprogramming of hemoglobin expression was achieved at the transcriptional level by increased gamma-globin combined with decreased beta-globin transcripts resulting in gamma-globin rising to 90% of total beta-like mRNA. Delta-globin mRNA was reduced to barely detectable levels. Alpha-globin levels were not significantly changed. Fetal hemoglobin achieved levels of 68.6 ± 3.9% in the IGF2BP1 overexpression samples compared with 5.0 ± 1.8% in donor matched transduction controls. In part, these changes were mediated by reduced protein expression of the transcription factor BCL11A. mRNA stability and polysome studies suggest IGF2BP1 mediates posttranscriptional loss of BCL11A. These results suggest a mechanism for chronoregulation of fetal and adult hemoglobin expression in humans.
Sickle-cell anemia and the beta-thalassemias are among the most common genetic diseases worldwide. They are caused by mutated or reduced expression of the adult-stage beta-globin gene (1, 2) and manifested when gene expression in the beta-globin locus sequentially switches from fetal to adult types (3) around the time of birth. It is predicted that maintaining fetal hemoglobin (HbF) at levels above 30% in circulating red cells should prevent the manifestation of most sickle-cell disease complications (4). Based on the therapeutic utility of HbF expression, considerable research has been directed toward understanding the fetal-to-adult hemoglobin transition. Over the last 50 years, understanding of globin gene epigenetics has significantly advanced, but the developmental timing mechanisms for hemoglobin switching remain vague (5, 6).
Multicellular organisms have networks of temporally regulated genes, known as heterochronic genes, responsible for the control of developmental timing (7). Heterochronic gene pathways were initially investigated in Caenorhabditis elegans, where the RNA-binding protein lin28 and its major target, the let-7 miRNA, were first described as having an important role in early development, including the regulation of the transition from larva to adulthood (8). During vertebrate development, the Lin28–let-7 axis is also functionally involved in the maternal-to-zygotic transition during early zebrafish embryogenesis, suggesting that the heterochronic Lin28–let-7 axis and its role in the larval- or fetal-to-adult transition is conserved across evolution (9). Based upon the evolutionary conservation of these heterochronic pathways, as well as the hemoglobin-switching phenomenon, we initially hypothesized that the heterochronic genes described in lower animals may contribute to developmental changes in hemoglobin expression (10).
In human reticulocytes, changes in the levels of the let-7 miRNAs were associated with the fetal-to-adult developmental transition (10). Experimentally, increased LIN28 or suppressed let-7 were shown to be developmentally regulated and both cause increased gamma-globin gene expression in cultured human adult erythroblasts (11). However, a more complete reversal of the fetal-to-adult hemoglobin profiles was not detected. We thus hypothesized that additional downstream elements of this RNA-binding regulatory cascade may be involved. Among the group of potential let-7 targets (12), IGF2BP1 and IGF2BP3 are generally silenced during mammalian development, whereas IGF2BP2 expression remains stable (12–15). Here we provide a focused examination of IGF2BP1 in human erythroid tissues as well as its potential to affect hemoglobin profiles in cultured human adult erythroblasts.
Results
IGF2BP1 Is Developmentally Regulated in Human Erythroid Tissues.
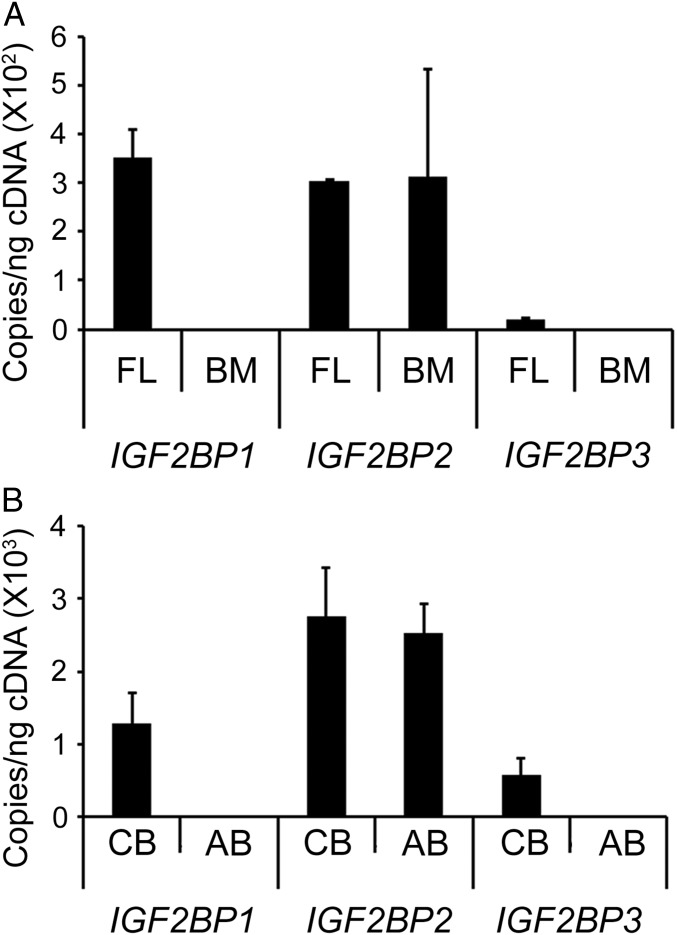
The IGF2BP family consists of three highly conserved RNA-binding proteins (IGF2BP1, IGF2BP2, and IGF2BP3) (16). IGF2BP1 and IGF2BP3 are expressed during early development and steadily decrease after birth in most tissues, but IGF2BP2 continues to be expressed in adult tissues (13, 14). To confirm their expression patterns in human erythroid tissues, we performed RT-qPCR in unsorted fetal liver, bone marrow, cord blood, and adult blood reticulocytes. We observed that both IGF2BP1 and IGF2BP3 are expressed in unsorted fetal liver but at background or undetectable levels in bone marrow samples, whereas IGF2BP2 is detected in both unsorted fetal liver and bone marrow samples (Fig. 1A). Among cultured primary erythroblasts, both IGF2BP1 and IGF2BP3 are expressed in cord blood erythroblasts but at background levels in adult blood erythroblasts. IGF2BP2 was detected at similar levels in both cord blood and adult blood erythroblasts (Fig. 1B). Compared with IGF2BP3, IGF2BP1 was detected at higher levels in each fetal tissue. Western blot analyses confirmed and further defined the higher level expression and developmental silencing of both proteins (Fig. 2A and Fig. S1A). Due to its high expression level in fetal tissues with reduced or absent expression in adult tissues, further studies were focused upon the effects of transgenic expression of IGF2BP1 in adult erythroblasts.
Fig. 1.
Expression pattern of IGF2BP1, IGF2BP2, and IGF2BP3 mRNAs in fetal and adult human tissues. IGF2BP1, IGF2BP2, and IGF2BP3 mRNA expression in (A) human fetal liver and adult bone marrow samples and (B) cultured cord blood and adult blood erythroblasts. Mean value ± SD is shown. AB, adult blood erythroblasts; BM, adult bone marrow; CB, cord blood erythroblasts; FL, fetal liver.
Fig. 2.
IGF2BP1 overexpression has cytoplasmic distribution and robust effects upon hemoglobin expression patterns in adult CD34(+) cells. (A) IGF2BP1 protein expression in Human Fetal Liver (FL) Protein Medley, adult bone marrow (BM), cord blood (CB), and adult blood (AB) erythroblasts. Beta-actin was used as a loading control. Molecular weight is shown in kilodaltons (kDa). IGF2BP1 overexpression (OE) was confirmed by (B) RT-qPCR, (C) Western blot, and (D–I) confocal analyses. RT-qPCR was performed at culture day 14. Mean value ± SD of three independent donors for each condition is shown. Western blot analysis confirmation of IGF2BP1 overexpression at culture day 14 in CD34(+) cells versus empty vector control (C) from three healthy adult donors (3 μg per lane). Beta-actin was used as a loading control. Molecular weight is shown in kilodaltons. Confocal analysis was performed at culture day 12 in IGF2BP1 overexpression cells versus empty vector control. Cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) (blue) and IGF2BP1 (green). HPLC analyses of hemoglobin from (J) empty vector control and (K) IGF2BP1 overexpression samples were performed at culture day 21. Datasets are representative of three independent donors for each condition. HbF and HbA peaks are labeled on each graph (y axis, mVolts; x axis, elution time in minutes). P value was calculated using two-tailed Student’s t test. *P < 0.05.
Fig. S1.
IGF2BP3 overexpression has moderate effects upon hemoglobin expression patterns. (A) IGF2BP3 protein expression in Human Fetal Liver (FL) Protein Medley, adult bone marrow (BM), cord blood (CB), and adult blood (AB) erythroblasts. Beta-actin was used as a loading control. Molecular weight is shown in kilodaltons (kDa). IGF2BP3 overexpression (OE) was confirmed by (B) RT-qPCR and (C) Western blot analyses. RT-qPCR was performed at culture day 14. Mean value ± SD of three independent donors for each condition is shown. Western blot confirmation of IGF2BP3 overexpression at culture day 14 in CD34(+) cells versus empty vector control (C) from three healthy adult donors is shown. Beta-actin was used as a loading control. Molecular weight is shown in kilodaltons. HPLC analyses of hemoglobin from (D) empty vector control and (E) IGF2BP3 overexpression samples were performed at culture day 21. Datasets are representative of three independent donors for each condition. HbF and HbA peaks are labeled on each graph (y axis, mVolts; x axis, elution time in minutes).
IGF2BP1 Overexpression Reverses Adult Erythroblasts to a More Fetal-Like Pattern of Globin Genes and Fetal Hemoglobin Expression.
To investigate the effects of IGF2BP1 expression in cultured adult erythroblasts, an erythroid-specific IGF2BP1 overexpression (IGF2BP1-OE) lentiviral vector was used in direct comparison with an empty vector control. Transductions were performed in CD34(+) cells from adult healthy volunteers cultured ex vivo in erythropoietin-supplemented serum-free media for 21 days. IGF2BP1 overexpression was confirmed by RT-qPCR and Western blot analyses at culture day 14 (Fig. 2 B and C). Confocal microscopy was performed on IGF2BP1-OE and empty vector control transductions and demonstrated that IGF2BP1 expression was predominantly cytoplasmic (Fig. 2 D–I).
Hemoglobin profiles (HPLC) were used to determine the effects of IGF2BP1 overexpression on HbF levels at culture day 21. Remarkably, IGF2BP1-OE caused a robust increase in the level of HbF [HbF/(HbF + HbA)] reaching 68.6 ± 3.9% in IGF2BP1-OE cells compared with 5.0 ± 1.8% in donor-matched control cells (P = 0.0004; Fig. 2 J and K). By comparison, IGF2BP3-OE caused only moderate effects on HbF levels [HbF/(HbF + HbA)] (18.6 ± 1.0% in IGF2BP3-OE cells versus 4.0 ± 2.1 in donor-matched control cells, P = 0.0021), but relatively low transgenic expression of IGF2BP3 was detected (Fig. S1 B–E).
IGF2BP1-related changes in the differentiation, maturation, and enucleation of the cultured erythroblasts were assessed by flow cytometry analysis of transferrin receptor (CD71) and glycophorin A (GPA) at culture days 14 (Fig. S2 A and B) and 21 (Fig. 3 A and B and Fig. S2 C and D). The reduced loss of surface CD71 in IGF2BP1-OE cells compared with empty vector control suggests altered maturation kinetics over the 21-day culture period. However, IGF2BP1-OE did not prevent hemoglobin accumulation or terminal maturation of the cells. Enucleation, as indicated by the lack of thiazole orange staining, was not significantly different in IGF2BP1-OE compared with empty vector control transduction (average enucleation: empty vector control, 31.5 ± 2.2; IGF2BP1-OE, 23.7 ± 8.4; P = 0.20). A representative donor is shown in Fig. 3 C and D. Finally, sorted IGF2BP1-OE enucleated cells were imaged and demonstrated equivalent cellular morphology compared with empty vector control samples (Fig. 3 E and F). Terminal maturation and enucleation did not cause a major reduction in HbF (Fig. S2 E and F), and a pancellular distribution of HbF was detected for IGF2BP1-OE cells (HbF stain: empty vector control, 67.6 ± 7.9; IGF2BP1-OE, 86.5 ± 3.0).
Fig. S2.
Quantitative analyses of the percentage of (A) day 14 CD71(+)/GPA(+), (B) day 14 CD71(+)/GPA(−), (C) day 21 CD71(+)/GPA(+), and (D) day 21 CD71(−)/GPA(+) measured by fluorescence-activated cell sorter analysis of control and IGF2BP1 overexpression in adult CD34(+) cells. Mean value ± SD of three independent donors for each condition is shown. P value was calculated using two-tailed Student's t test. *P < 0.05. IGF2BP1-OE increases in fetal hemoglobin are maintained after terminal differentiation. HPLC analyses of hemoglobin at culture day 21 from sorted IGF2BP1-OE (E) enucleated cells and (F) nucleated cells in adult CD34(+) cells. HbF and HbA peaks are labeled on each graph (y axis: mVolts; x axis: elution time in minutes). C, empty vector control; CD71, anti-transferrin receptor; GPA, anti-glycophorin A; OE, IGF2BP1 overexpression.
Fig. 3.
Effects of IGF2BP1 overexpression upon differentiation, maturation, enucleation, and cellular morphology of adult CD34(+) cells. Representative flow dot plots at culture day 21 stained for CD71 and GPA from (A) empty vector control cells and (B) IGF2BP1 overexpression. Thiazole orange staining was used to assess enucleation for (C) empty vector control and (D) IGF2BP1 overexpression at culture day 21. Sorted enucleated cells were imaged in (E) empty vector control and (F) IGF2BP1 overexpression after Wright–Giemsa staining. Datasets are representative of three independent donors for each condition. CD71, anti-transferrin receptor; GPA, anti-glycophorin A.
The effects of IGF2BP1-OE on the expression of globin genes were characterized by RT-qPCR analyses at culture day 14. The changes observed in the globin genes, including high-levels of gamma-globin mRNA accompanied by suppression of the adult delta- and beta-globin genes, reflected a more fetal-like pattern in the IGF2BP1-OE cells compared with control transductions (Fig. 4 A–H). IGF2BP1-OE caused a dominance (89.8% of all beta-like transcripts) of gamma-globin mRNA [control (empty vector) = 3.2E+06 ± 8.2E+05 copies per nanogram; IGF2BP1-OE = 2.0E+07 ± 5.9E+06 copies per nanogram; P = 0.046], whereas beta-globin mRNA decreased to minor levels of about 10.1% of all beta-like transcripts [control (empty vector) = 2.2E+07 ± 4.0E+06 copies per nanogram; IGF2BP1-OE = 2.2E+06 ± 6.2E+05 copies per nanogram; P = 0.019] compared with the empty vector control transductions. In addition, delta-globin mRNA was also reduced to 0.03% of all the beta-like transcripts in IGF2BP1-OE samples [control (empty vector) = 2.4E+05 ± 5.9E+04 copies per nanogram; IGF2BP1-OE = 7.4E+03 ± 1.8E+03 copies per nanogram; P = 0.020]. Importantly, the total output of transcripts from the beta- and alpha-gene clusters remained unchanged [beta-cluster total output: control (empty vector) = 2.5E+07 ± 1.0E+07 copies per nanogram; IGF2BP1-OE = 2.2E+07 ± 9.6E+06 copies per nanogram; P > 0.05; alpha-cluster total output: control (empty vector) = 1.4E+07 ± 6.8E+06 copies per nanogram; IGF2BP1-OE = 1.0E+07 ± 4.8E+06 copies per nanogram; P > 0.05].
Fig. 4.
IGF2BP1-OE causes a more fetal-like pattern of gene expression. IGF2BP1-OE and control transductions in adult CD34(+) cells were investigated for (A) epsilon-globin, (B) gamma-globin, (C) delta-globin, (D) beta-globin, (E) zeta-globin, (F) mu-globin, (G) alpha-globin, (H) theta-globin, (I) CA1, and (J) GCNT2. RT-qPCR analyses were performed at culture day 14. Open bars represent control transductions and black bars represent IGF2BP1-OE. Mean value ± SD of three independent donors for each condition is shown. P values were calculated using two-tailed Student’s t test. *P < 0.05. C, empty vector control transduction; OE, IGF2BP1 overexpression.
In addition to the globin genes, the human fetal-to-adult developmental transition is characterized by increased expression of the carbonic anhydrase I (CA1) gene and carbohydrate modification due to the increased expression of the GCNT2 gene (17–20). Interestingly, both CA1 and GCNT2 were significantly down-regulated in IGF2BP1-OE cells compared with empty vector control transductions (Fig. 4 I and J). Overall, these results demonstrate that IGF2BP1 regulates HbF and other markers of the fetal-to-adult developmental transition, suggesting that IGF2BP1 reverses adult erythroblasts toward a more fetal-like phenotype.
IGF2BP1 Overexpression Alters BCL11A and HMGA2 Protein Levels in Adult Erythroblasts.
Potential causes for the effects of IGF2BP1 upon developmentally related erythroid genes were explored. Initially, to identify mRNAs that potentially interact with IGF2BP1, IGF2BP1 immunoprecipitation was performed followed by RNA-Seq analysis of coprecipitating RNA. As predicted by the prevalence of the IGF2BP1-binding motif as well as earlier studies of mRNA binding in nonerythroid cells (21–23), a broad list of candidate genes was identified (Dataset S1). Due to the large number of potential binding partners in erythroid cells, additional studies were performed upon both mRNA and protein expression levels of IGF2BP1-binding partners. Three previously reported (13, 24, 25) IGF2BP1-binding partners were initially studied: c-MYC, IGF2, and beta-actin. In the primary erythroblasts, no significant changes were observed at their mRNA or protein levels in the IGF2BP1-OE samples compared with empty vector control transductions (Fig. S3). Of note, manipulation of IGF2BP1 in K562 erythroleukemia cells similarly had only minor effects on those genes (26).
Fig. S3.
IGF2BP1-OE has minor effects on selected known IGF2BP1 targets c-MYC, IGF2, and beta-actin. IGF2BP1-OE and control transductions were investigated for the mRNA levels of (A) c-MYC, (B) IGF2, and (C) beta-actin in adult CD34(+) cells. RT-qPCRs were performed at culture day 14. Open bars represent empty vector control and black bars represent IGF2BP1-OE. Mean value ± SD of three independent donors for each condition is shown. (D) Western analyses of c-MYC, IGF2, and beta-actin expression using protein extracts at culture day 14 of empty vector control and IGF2BP1 overexpression in adult CD34(+) cells. Alpha-tubulin was used as a loading control. Molecular weight is shown in kilodaltons (kDa). C, empty vector control; OE, IGF2BP1 overexpression.
Following these initial studies, the potential for IGF2BP1-OE to regulate the expression levels of the let-7 miRNAs or LIN28 in adult CD34(+) erythroblasts via a feedback loop was investigated. IGF2BP1-OE had little effect on the expression of the let-7 miRNAs (Fig. S4) and the LIN28A and LIN28B RNA binding proteins remained below detection limits.
Fig. S4.
IGF2BP1-OE has no major effects on the let-7 miRNAs. IGF2BP1-OE and control transductions in adult CD34(+) cells were investigated for (A) let-7a, (B) let-7b, (C) let-7c, (D) let-7d, (E) let-7e, (F) let-7f, (G) let-7g, (H) let-7i, and (I) miR-98. RT-qPCRs were performed at culture day 14. Open bars represent empty vector control and black bars represent IGF2BP1-OE. Mean value ± SD of three independent donors for each condition is shown. P value was calculated using two-tailed Student’s t test. *P < 0.05. C, empty vector control transduction; OE, IGF2BP1 overexpression.
In the context of fetal hemoglobin regulation, the effects of IGF2BP1-OE upon five erythroid-related transcription factors (BCL11A, HMGA2, ZBTB7A, KLF1, and SOX6) were investigated (27–29). IGF2BP1-OE caused little change in the mRNA levels of these genes with the exception of a small, but statistically significant decrease in ZBTB7A (Fig. 5 A–E). However, major down-regulation was detected in the protein level of BCL11A in IGF2BP1-OE compared with empty vector controls (Fig. 5F). HMGA2 demonstrated a doublet banding pattern reported elsewhere (30) with a marked increase in the intensity of the upper band after IGF2BP1-OE. ZBTB7A, KLF1, and SOX6 protein levels remained unchanged (Fig. 5 F and G). The mRNA and protein expression patterns of BCL11A and HMGA2 suggest that IGF2BP1 is a posttranscriptional regulator of both proteins. The posttranscriptional up-regulation of HMGA2 was expected because IGF2BP1 is a known competitor of let-7 for HMGA2 mRNA binding (31).
Fig. 5.
IGF2BP1-OE regulates BCL11A and HMGA2 in human adult erythroblasts. IGF2BP1-OE and control transductions in adult CD34(+) cells were investigated for the mRNA levels of (A) BCL11A, (B) HMGA2, (C) ZBTB7A, (D) KLF1, and (E) SOX6. RT-qPCRs were performed at culture day 14. Open bars represent empty vector control and black bars represent IGF2BP1-OE. Mean value ± SD of three independent donors for each condition is shown. P values were calculated using two-tailed Student’s t test. *P < 0.05. Western blot analyses of (F) BCL11A, HMGA2, and ZBTB7A and (G) KLF1 and SOX6 expression using protein extracts at culture day 14 of empty vector control and IGF2BP1 overexpressed CD34(+) cells are shown. Histone H3 and lamin B1 were used as loading controls. Molecular weight is shown in kilodaltons (kDa). C, empty vector control transduction; OE, IGF2BP1 overexpression.
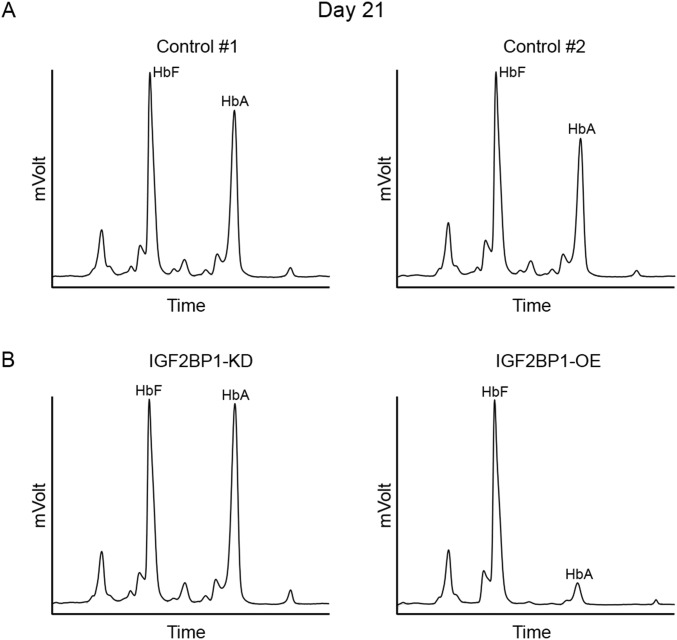
Because IGF2BP1 is expressed at near background levels in adult erythroblasts, IGF2BP1 knockdown (KD) studies were performed using fetal erythroblasts generated from cord blood CD34(+) cells. IGF2BP1 knockdown was confirmed by RT-qPCR and Western blot analyses at culture day 14 (Fig. S5). For comparison with the adult cell data above, IGF2BP1 overexpression studies were also performed in the human cord blood-derived erythroblasts (Figs. S6–S8). All studies were performed in triplicate. Neither knockdown, nor overexpression of IGF2BP1 in cord blood erythroblasts prevented the maturation or enucleation of the cells (Fig. S6). Moreover, IGF2BP1-KD had no significant effects in the HbF levels in cord blood erythroblasts (Fig. S7) (control, 49.5 ± 4.7%; IGF2BP1-KD, 47.0 ± 2.8%; P = 0.203). However, IGF2BP1-OE had an additive effect on the HbF levels of cord blood erythroblasts (control, 55.9 ± 2.7%; IGF2BP1-OE, 90.9 ± 1.7%; P = 0.001). Western analyses demonstrated that the IGF2BP1-OE effects upon BCL11A and HMGA2 were less pronounced in cord blood compared with adult CD34(+) cells. Consistent with its lack of HbF effects, IGF2BP1-KD had little effect upon the expression of BCL11A or ZBTB7A (Fig. S8). The expression of IGF2BP3 was slightly increased and LIN28B was maintained after IGF2BP1-KD (Fig. S8).
Fig. S5.
IGF2BP1 knockdown in cord blood erythroblasts was confirmed by (A) RT-qPCR and (B) Western blot analyses at culture day 14. Open bar represents empty vector control and black bar represents IGF2BP1 knockdown. Mean value ± SD of three independent donors for each condition is shown. P value was calculated using two-tailed Student’s t test. *P < 0.05. Beta-actin was used as a loading control. Molecular weight is shown in kilodaltons (kDa). C, empty vector control transduction; KD, IGF2BP1 knockdown.
Fig. S6.
Effects upon differentiation, maturation, and enucleation of IGF2BP1 knockdown or IGF2BP1 overexpression compared with each respective control transduction in cord blood CD34(+) cells. Representative flow dot plots at culture (A) day 14 and (B) day 21 stained for CD71 and GPA. (C) Thiazole orange staining was used to assess enucleation at culture day 21. Datasets are representative of three independent donors for each condition. CD71, anti-transferrin receptor; control 1, empty vector control transduction for knockdown vector; control 2, empty vector control transduction for overexpression vector; GPA, anti-glycophorin A; KD, knockdown; OE, overexpression.
Fig. S8.
Protein analyses of IGF2BP1 knockdown or overexpression in cord blood erythroblasts. Western images for IGF2BP1, IGF2BP3, LIN28B, BCL11A, HMGA2, and ZBTB7A expression using protein extracts at culture day 14. Lamin B1, histone H3, and beta-actin were used as loading controls. Molecular weight is shown in kilodaltons (kDa). Percentage of fetal hemoglobin (%HbF) shown corresponds to the representative HPLC profiles shown in Fig. S7. Control 1, empty vector control transduction for knockdown vector; control 2, empty vector control transduction for overexpression vector; KD, knockdown; OE, overexpression.
Fig. S7.
HPLC analyses of hemoglobin in cord blood CD34(+) cells at culture day 21. (A) Control transduction for knockdown vector (control 1) and control transduction for overexpression vector (control 2). (B) IGF2BP1 knockdown (KD) or IGF2BP1 overexpression (OE). Datasets are representative of triplicate experiments. HbF and HbA peaks are labeled on each graph (y axis, mVolts; x axis, elution time in minutes).
BCL11A Is Posttranscriptionally Regulated in IGF2BP1 Overexpressing Adult Cells.
Because increased BCL11A protein expression during human development is a major regulator of HbF (32), the potential for IGF2BP1-mediated, posttranscriptional regulation of BCL11A was pursued with great interest. The binding of IGF2BP1 to BCL11A mRNA was indicated by the significant enrichment of BCL11A transcripts compared with input and IgG control RNA samples after RNA immunoprecipitation (RIP) (Fig. 6A). Because IGF2BP1 may affect RNAs stability after binding (13), BCL11A mRNA decay was examined after treatment with the transcription inhibitor actinomycin D. As shown in Fig. 6B, BCL11A transcripts decayed at the same rate in IGF2BP1-OE samples compared with control transductions, suggesting that IGF2BP1 is not affecting the stability of BCL11A mRNA.
Fig. 6.
IGF2BP1 colocalizes with BCL11A in the ribosomes. RNA immunoprecipitation (RIP) using antibody against IGF2BP1 was performed to assess binding of RNAs to IGF2BP1 protein followed by RT-qPCR quantitation for (A) BCL11A transcripts. RIP was performed at culture day 14. Mean value ± SD of three independent donors for each condition is shown. (B) BCL11A stability analysis in IGF2BP1-OE versus empty vector control after actinomycin D treatment at culture day 14. Time course for RNA stability started by adding the transcription inhibitor actinomycin D (10 µg/mL) and cells were harvested at the indicated time points. Expression levels (percentage) were calculated considering the time point “0 h” as 100%. Mean value ± SD of two independent donors for each condition is shown. (C–H) Polysome profiling analysis of IGF2BP1-OE versus empty vector control at culture day 12. A representative polysome gradient (control versus IGF2BP1-OE) is shown in C. BCL11A mRNA was measured by RT-qPCR quantitation on the polysome fractions and mean value ± SD of two independent donors for each condition is shown in D. Western blot analysis of IGF2BP1 on representative polysome fractions from one representative donor is shown in E. Beta-globin (F), gamma-globin (G), and beta-globin + gamma-globin (H) mRNAs were measured by RT-qPCR quantitation on representative polysome profiling fractions. Results are shown from one representative donor. Molecular weight is shown in kilodaltons (kDa). P value was calculated using two-tailed Student’s t test. *P < 0.05. Experiments were performed in adult CD34(+) cells. BP1-RIP, IGF2BP1 RNA immunoprecipitation; IgG, immunoprecipitation with isotype control; Input, RNA sample before immunoprecipitation; OE, overexpression.
IGF2BP1 may also modulate mRNA translation (33); thus, polysome comparisons were made in IGF2BP1-OE and empty vector control transductions (Fig. 6 C–H). IGF2BP1-OE gradients were nearly identical to the control (Fig. 6C). The translational status of BCL11A was investigated, and demonstrated no differences in the polysome of IGF2BP1-OE compared with empty vector control (Fig. 6D). IGF2BP1 protein also localized with the RNAs in the polysomal fractions (Fig. 6E). The lack of differences between IGF2BP1-OE and control transductions upon the transcription, mRNA stability, and ribosomal loading of BCL11A protein expression suggests translational repression of BCL11A mediated by IGF2BP1.
In addition to whole-cell mRNA quantitation, beta-globin and gamma-globin mRNA levels were investigated in the polysome fractions to determine whether there were changes in the polysome-loaded profiles. In accordance with the RT-qPCR data (Fig. 4 B and D), a robust switch that included reduction of polysomal beta-globin, rather than a simple increase in gamma-globin, was observed. IGF2BP1-OE caused high levels of gamma-globin mRNA and low beta-globin levels on the highly translated fractions. In contrast, the empty vector control demonstrated high beta-globin and low gamma-globin levels on the same fractions (Fig. 6 F and G). Importantly, the total beta + gamma globin levels and polysome patterns remained unchanged (Fig. 6H). The switched globin pattern on the polysome profiles is consistent with the high levels of HbF produced.
Discussion
IGF2BP1 manifests highly regulated mRNA and protein expression patterns during human ontogeny with silencing during the transition from fetal to adult life (13–15). The developmental silencing of these RNA-binding proteins in vivo is significant because hemoglobin switching occurs during the same developmental period. The absence of IGF2BP1 expression in bone marrow was confirmed and studied further to demonstrate its presence in fetal liver and erythroblasts cultured from cord blood CD34(+) cells. Prior studies also validated developmental silencing by LIN28B and activation of let-7 in erythroid cells (10, 11). IGF2BPs function primarily in the binding and posttranscriptional regulation of multiple RNA targets (12, 21, 34–36), as a consequence having multiple biological functions, including pluripotency of stem cells and regulation of glucose metabolism (37, 38). Because let-7 miRNAs are downstream targets of LIN28B, and the IGF2BPs mRNAs are targets of let-7, a developmental regulatory pathway for fetal hemoglobin expression during ontogeny emerges. IGF2BP3 overexpression demonstrated weaker HbF effects, which will require further study, and could be a reflection of its lower expression levels. Alternatively, the HbF effects may have a structural basis because the two proteins have only a 73% similarity (13) and have been reported to exhibit distinct binding preferences (21).
Increased gamma-globin mRNA and suppressed adult delta- and beta-globin genes were identified in the adult cells, which are a pattern of changes predicted for a reversion to a more fetal-like phenotype. The switched pattern of gamma- and beta-globin mRNA was also detected in the polysome profiles. Several approaches were taken to understand the mechanism underlying the IGF2BP1-mediated changes in globin gene and protein expression. We observed that there was no significant change in the expression levels of the let-7 miRNAs, and no increases in the expression of LIN28. These results essentially exclude the interpretation that IGF2BP1 would act through a regulatory feedback loop in the let-7 cascade in adult cells. Studies of cord blood erythroblasts demonstrated no significant effects from IGF2BP1-KD, suggesting that lowering IGF2BP1 expression is not sufficient to reduce HbF levels in those cells. To identify candidate partners of interaction with IGF2BP1, we performed RNA immunoprecipitation followed by RIP-Seq analysis. An extensive list of mRNA-binding candidates was detected. However, IGF2BP1-OE modulation of proteins was more specific and clearly detected in BCL11A and HMGA2 levels (down- and up-regulation, respectively). HMGA2 is a known downstream target of let-7 and IGF2BP1 that has been recently associated with moderate increases in HbF levels in adult human erythroblasts (29).
IGF2BP1 acts in a tissue-specific manner to regulate the expression of proteins via multiple separate mechanisms, including increased mRNA decay, protein compartmentalization, or translational regulation (33, 39–43). Because an IGF2BP1-OE vector driven by the erythroid-specific SPTA1 promoter was designed for this study, stem cell reprogramming is not required for its erythroid effects. Also an oncofetal protein, IGF2BP1 is commonly detected in several tumors. Its effects upon growth are variable with speculation of both oncogenic as well as tumor-suppressive functionality (31, 44–46) based upon the cellular context of its expression. As shown here, overexpression of IGF2BP1 in the committed erythroblasts did not prevent their maturation or enucleation in ex vivo cultures.
Due to the importance of BCL11A on gamma-globin gene regulation (47), IGF2BP1 effects upon this transcription factor in adult cells were a primary focus of this study. We observed enrichment of BCL11A transcripts after RNA immunoprecipitation with IGF2BP1 antibody, but the broad list of potential binding partners identified by RIP-Seq reported here and elsewhere (21) suggest that better methods are needed to predict which mRNA species are specifically and functionally affected by IGF2BP1 binding. IGF2BP1 binding to BCL11A mRNA was reported previously to motifs located in both coding and noncoding regions (21). Informatics analysis suggests that BCL11A mRNA sequence (National Center for Biotechnology Information, NM_022893.3) contains several dozen IGF2BP1-binding motifs spread throughout the coding region and 3′ UTR, so potentially dynamic and complex IGF2BP1–BCL11A interactions may exist in the cells.
We further investigated the potential for IGF2BP1 to affect (i) the decay and (ii) the ribosomal loading and, therefore the translational status of BCL11A mRNA. Interestingly, IGF2BP1 did not affect the kinetics of BCL11A transcript decay as evidenced by no significant differences observed between IGF2BP1 overexpression cells compared with control transductions after actinomycin D treatment. In addition, IGF2BP1 also did not affect the ribosomal loading of BCL11A. These results confirm that IGF2BP1 regulates BCL11A most likely through posttranscriptional loss with a similar pattern of unchanged ribosomal loading that was identified in the cotranscriptional repression of the transcription factor lin-41 after binding of its RNA by the let-7a miRNA (48). However, the magnitude of gamma-globin induction (90% of total beta-like globin mRNA) as well as the reversal of gamma- and beta-globin in the polysome profiles suggests that BCL11A is not a sole globin effector of IGF2BP1.
Our findings suggest two paths for future research in this regard. First, in vivo models should be identified to determine whether RNA-binding regulatory cascades control mammalian developmental timing, including the timed transitions of hemoglobin during ontogeny. The notion that the let-7 cascade influences a general, rather than globin-specific timing mechanism is supported by regulation of two nonglobin genes, CA1 and GCNT2, which are regulated during the fetal-to-adult transition in vivo. Of note, developmental silencing of Igf2bp1 occurs during the transition from primitive to definitive erythropoiesis in mice (49). Because the HbF effects of IGF2BP1 were manifested in the setting of overexpression, murine or other experimental models are needed for the complete knockout of gene expression as well as gene rescue experiments to understand the physiological relevance of Igf2bp1 and other elements of the let-7 regulatory cascade during erythroid ontogeny. Second, these results may be further explored for their potential in developing novel therapeutic strategies for beta-hemoglobinopathies. Interestingly, 5-azacytidine reactivates IGF2BP1 expression (25). Because IGF2BP1 is a cytoplasmic protein, it also provides a potentially druggable route for posttranscriptional suppression of BCL11A in adult cells. Its globin- and nonglobin-related effects should be additionally studied for long-term effects on erythroblast growth and differentiation in vivo. The prevention or reversal of the disease course for patients with sickle-cell anemia and beta-thalassemia major are envisioned by safely achieving equivalent hemoglobin effects to those demonstrated here.
Methods
Cell Culture.
Written informed consent was obtained from all research subjects before participation in this study, and approval for the research protocol and consent documents using primary erythroblasts and peripheral blood samples was granted by the National Institute of Diabetes and Digestive and Kidney Diseases Institutional Intramural Review Board. CD34(+) cells from adult human healthy volunteers and human cord blood CD34(+) cells obtained from AllCells were studied ex vivo using a serum-free 21-day culture system as previously described (11).
Lentiviral Vector Construction, Production, and Transduction.
The empty lentiviral vector (control) with the human spectrin alpha gene (SPTA1) promoter was constructed as previously described (50). An erythroid-specific IGF2BP1 overexpression (IGF2BP1-OE) vector with human SPTA1 promoter encoding the IGF2BP1 ORF was produced by conventional RT-PCR with high-fidelity Taq from cultured cord blood CD34(+) mRNA with primers excluding the 5′ and 3′ UTR and containing XhoI and BamHI restriction sites for cloning as follows: 5′-ACCCTCGAGTATGAACAAGCTTTACATCGGCAACCTCAACGA-3′ and 5′-CCGGATCCTCACTTCCTCCGTGCCTGGGCCT-3′. The consensus let-7-binding sequence encoded in the 3′-UTR region of IGF2BP1 mRNA was excluded from the IGF2BP1-OE lentivirus transgene to prevent degradation of the transcripts in the adult CD34(+) cells. Human IGF2BP3 ORF was synthesized by Eurofins MWG Operon and cloned into SPTA1 promoter empty vector using XhoI and NotI restriction sites (IGF2BP3-OE). Lentivirus was packaged using HEK293T cells (Thermo Fisher Scientific) cotransfected with packaging helper virus plasmids (a gift from Dr. Derek Persons and Dr. Arthur Nienhuis, St. Jude Children’s Research Hospital, Memphis, TN) and the vector plasmid (SPTA1-empty vector control or SPTA1–IGF2BP1 overexpression vector or SPTA1–IGF2BP3 overexpression vector) as previously described (50–52). The human IGF2BP1 knockdown clone TRCN0000218799 as well as a nontargeting shRNA control vector (SHC002V) were purchased from Sigma-Aldrich. CD34(+) cells were transduced as previously described (50).
Flow Cytometry Analysis.
To evaluate the differentiation stages of maturation in adult or cord blood CD34(+) cells, antibodies directed against transferrin receptor (CD71) and glycophorin A (Life Technologies) were used on culture days 14 and 21 and analyzed using the BD FACSAria I flow cytometer (BD Biosciences) as previously described (53). Thiazole orange (Sigma-Aldrich) was used to assess enucleation on culture day 21. In adult CD34(+) cells, fetal hemoglobin was assessed with antibody directed against HbF (Life Technologies) at culture day 21.
Confocal Imaging.
For confocal microscopy, sorted erythroblasts at culture day 12 were fixed with 3.7% paraformaldehyde in PBS (Electron Microscopy Sciences) and permeabilized with 2% BSA/PBS containing 0.1% Triton X (Thermo Fisher Scientific). Cells were then incubated overnight at 4 °C with anti-human IGF2BP1 (1:500; 8482S, Cell Signaling) in 2% BSA/PBS solution, then for 1 h at 25 °C with anti-rabbit IgG (H+L) conjugated with CF-555 (1:500; Sigma) in 2% BSA/PBS. After secondary antibody incubation, slides were washed three times in PBS and once in distilled water, then mounted with ProLong Diamond Antifade Mountant with DAPI (Thermo Fisher Scientific). Laser scanned confocal images were obtained from Axio Observer Z1 (Carl Zeiss) and analyzed with Zen2012 software.
Cytospin Preparation and Wright–Giemsa Staining.
Cytospins were prepared by centrifugation of the cytoslides at 1,000 rpm for 2 min using the Shandon Cytospin 4 (Thermo Fisher Scientific). Cytoslides were then stained with Wright–Giemsa (Sigma-Aldrich) for 50 s and then washed twice in distilled water for 1 min.
Quantitative PCR for mRNAs.
Total RNA was isolated using miRNeasy mini kit with QIAzol (Qiagen) as previously described (29). cDNA was synthesized using SuperScript III reverse transcriptase (Thermo Fisher Scientific) as previously described (29, 54). Quantitative real-time PCR amplification was carried out in a 7900HT Fast Real-Time PCR System or 7500 Real Time PCR System (Thermo Fisher Scientific) using TaqMan Universal PCR Master Mix (Thermo Fisher Scientific) following manufacturer’s instructions. RT-qPCR assays and conditions were performed as previously described (11, 29, 55, 56). The following commercially available Assay-on-Demand gene expression products (Thermo Fisher Scientific) were used: IGF2BP1 (Hs00977566_m1), IGF2BP2 (Hs00538956_m1), IGF2BP3 (Hs01122560_g1), CA1 (Hs01100176_m1), GCNT2 (Hs00377334_m1), BCL11A (Hs00256254_m1), HMGA2 (Hs00971724_m1), ZBTB7A (Hs00792219_m1), KLF1 (Hs00610592_m1), SOX6 (Hs00264525_m1), c-MYC (Hs00153408_m1), IGF2 (Hs01005963_m1), and Beta-Actin (Hs99999903_m1). Absolute quantification for each gene was determined by comparison with a standard curve that was run in parallel with biological samples in all RT-qPCR reactions as previously described (29, 54). Reactions were performed in triplicate.
Quantitative PCR Analysis for the let-7 Family of miRNAs.
cDNA synthesis and RT-qPCR assays and conditions were performed as previously described for let-7a, let-7b, let-7c, let-7d, let-7e, let-7f, let-7g, let-7i, and miR-98 (10, 29). Known concentrations of a synthetic targeted mature miRNA oligonucleotide (1:10 serial dilutions, n = 6) were used for absolute quantification of each let-7 family in the biological samples as previously described (29). Reactions were performed in triplicate.
RNA-Binding Protein Immunoprecipitation.
RIP was performed on day 14 adult cells cultured from three independent donors transduced with IGF2BP1 lentiviral particles with antibody against IGF2BP1 (MBL International, RN007P) or rabbit IgG using RiboCluster Profiler RIP-Assay Kit (MBL International, RN1001) following manufacturer’s protocol. Briefly, 10–20 million cells were washed three times in cold PBS and lysed in 500 µL of lysis buffer supplemented with HALT Protease Inhibitor Mixture (Thermo Fisher Scientific/Pierce), 1.5 mM DTT (Life Technologies), and 200 units/mL RNAseOut (Life Technologies). Cell lysates were precleared using 50 µL of Dynabeads Protein A (Life Technologies) for 1 h at 4 °C. Antibody immobilized beads were prepared using 15 µg of IGF2BP1 antibody or rabbit IgG with 50 µL of Dynabeads Protein A for 1 h at 4 °C. Precleared cell lysates were then incubated with either IGF2BP1 Protein A beads or rabbit IgG Protein A beads at 4 °C overnight. A magnet was used to capture the antibody-immobilized beads and the supernatant was discarded. Beads were then washed four times with 1 mL of cold washing buffer supplemented with 1.5 mM of DTT. In the last wash, 100 µL was removed for Western blot analysis. RNA isolation was performed according to manufacturer’s protocol with overnight precipitation at −20 °C and reconstitution in 10 µL of RNase-free water.
RNA-Seq Analysis.
The RNA-Seq library was constructed from 1 µg of total RNA pooled from three donors after rRNA depletion using Ribo-Zero GOLD (Illumina). The Illumina TruSeq RNA Sample Prep V2 Kit was used according to manufacturer’s instructions except where noted. The cDNAs were fragmented to ∼275 bp using a Covaris E210. Amplification was performed using 10 cycles, which was optimized for the input amount and to minimize the chance of overamplification. The library was sequenced on one lane of a HiSeq2500 using Chemistry v4 to generate over 250 million 126-bp reads. Data were processed using RTA v1.18.64 and CASAVA 1.8.2. Sequence reads were aligned to hg19 reference build using STAR v2.5.1. QoRTs v1.0.1 (57) was run on the data using the Gencode v19 annotation release to get the gene counts. QoRTs also provided the reads supporting splice junctions. The number of reads supporting each junction was indicative of the presence of alternate transcripts as visualized in a University of California Santa Cruz bed track.
mRNA Decay Analysis.
RNA decay analysis was performed on culture day 14 adult cells from two independent donors transduced with the IGF2BP1 lentiviral vector or empty vector control transduction. Cells were directly harvested or treated with the transcription inhibitor actinomycin D (Sigma-Aldrich) at 10 μg/mL concentration and harvested at the indicated time points. DMSO was used as vehicle control. Total RNA isolation, cDNA synthesis, and RT-qPCR were performed as described above.
Polysome Profiling.
Analysis was performed on culture day 12 adult cells from two independent donors transduced with the IGF2BP1 lentiviral vector or empty vector control. Cells were incubated with cycloheximide (Santa Cruz Biotechnology) (100 μg/mL) in phase 2 media for 5 min and washed twice in cold PBS supplemented with cycloheximide (100 μg/mL) before lysis using 20 mM Tris⋅HCl (pH 8.5), 1.5 mM MgCl2, 140 mM KCl, 0.5 mM DTT, 0.5% Nonidet P-40, 200 units/mL ribonuclease inhibitor, and 0.1 mM cycloheximide as previously described (16). Sucrose gradient sedimentation analysis was performed by standard methods (58). Samples were loaded onto 10–50% gradients in 20 mM Tris⋅HCl (pH 8.0), 140 mM KCl, 5 mM MgCl2, 0.5 mM DTT, and 1 mg/mL cycloheximide buffer, and spun in a Beckman Coulter SW 41 Ti rotor at 40 krpm for 3 h at 4 °C. Gradients were mixed on a BioComp Gradient Master and analyzed using a Brandel BR-188 Density Gradient Fractionation System running at 1.5 mL/min and set to collect fractions of 0.5 mL. Data were recorded with a DataQ Instruments DI-155 and analyzed with Igor Pro-6.
HPLC Analysis.
Unsorted three million cultured cells or sorted thiazole orange negative (enucleated) and positive (nucleated) cells at day 21 were pelleted and lysed in distilled water with two repeat freeze/thaw cycles in a dry-ice ethanol bath. Cell debris was removed by filtration through Ultrafree-MC devices (Millipore). Hemoglobin content was analyzed for fetal globin (HbF) and adult globin (HbA) using a 20 × 4 mm PolyCAT A column (PolyLC Inc.) fitted to a Shimadzu Prominence HPLC System (Shimadzu Scientific Instruments). The hemoglobins were eluted during 15 min in 20 mM Bis-Tris, 2 mM KCN, pH 6.90 buffer with a gradient of 0–25% in a buffer consisting of 20 mM Bis-Tris, 2 mM KCN, 200 mM NaCl, pH 6.45. Variant Hemoglobin Control AF (Analytical Control Systems Inc.), which has values for HbA and HbF of ∼60% and 40%, respectively, was used as a standard control. Hemoglobin proteins were detected by absorbance measurements at 415 nm. The HbA and HbF peaks were quantitated using the LabSolutions software (Shimadzu Scientific Instruments). Total areas under the HbA and HbF peaks were used for ratio comparisons.
Western Analyses.
Cytoplasmic and nuclear extracts were prepared using the NE-PER Nuclear Protein Extraction Kit (Pierce Biotechnology) as previously described (11), and protein extracts were separated on a NuPAGE NOVEX 4–12% Bis-Tris or 12% Bolt Bis-Tris Plus gels as described previously (11). The nitrocellulose membranes were probed with the following antibodies: BCL11A (Abcam), IGF2BP1 (Cell Signaling Technology), IGF2BP3 (Abcam), LIN28B (Cell Signaling Technology), HMGA2 (GeneTex), c-Myc (Cell Signaling Technology), KLF1 (Abcam), SOX6 (Santa Cruz Biotechnology), ZBTB7A (Abcam), and IGF2 (NeoScientific). Histone H3 (Abcam), lamin B1 (Abcam), alpha-tubulin (ProSci Inc.), or beta-actin (Abcam) were used as loading controls. To assess IGF2BP1 expression in fetal liver and adult bone marrow, Human Fetal Liver Protein Medley (635342) was purchased from Clontech Laboratories, and the mononuclear cells from adult bone marrow were separated using Ficoll-Paque Premium (GE Healthcare Life Sciences). The amount of protein loaded was between 10 and 20 μg per lane, unless otherwise noted in the text.
Statistical Analysis.
Replicates are expressed as mean ± SD values and significance was calculated by two-tailed Student’s t test.
Supplementary Material
Acknowledgments
We thank Dr. Arthur Nienhuis and Dr. Derek Persons for their expertise, including material transfer of reagents for lentiviral production; Dr. Gerard Bouffard and the staff of the NIH Intramural Sequencing Center for DNA sequencing services and thoughtful discussions; and the Department of Transfusion Medicine for their expertise in obtaining CD34(+) cells. The Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases supported this work.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1609552114/-/DCSupplemental.
References
- 1.Weatherall DJ, Clegg JB. Recent developments in the molecular genetics of human hemoglobin. Cell. 1979;16:467–479. doi: 10.1016/0092-8674(79)90022-9. [DOI] [PubMed] [Google Scholar]
- 2.Kan YW, Dozy AM. Polymorphism of DNA sequence adjacent to human beta-globin structural gene: Relationship to sickle mutation. Proc Natl Acad Sci USA. 1978;75:5631–5635. doi: 10.1073/pnas.75.11.5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weatherall DJ, Wood WG, Jones RW, Clegg JB. The developmental genetics of human hemoglobin. Prog Clin Biol Res. 1985;191:3–25. [PubMed] [Google Scholar]
- 4.Steinberg MH, Chui DH, Dover GJ, Sebastiani P, Alsultan A. Fetal hemoglobin in sickle cell anemia: A glass half full? Blood. 2014;123:481–485. doi: 10.1182/blood-2013-09-528067. [DOI] [PubMed] [Google Scholar]
- 5.Wood WG, Bunch C, Kelly S, Gunn Y, Breckon G. Control of haemoglobin switching by a developmental clock? Nature. 1985;313:320–323. doi: 10.1038/313320a0. [DOI] [PubMed] [Google Scholar]
- 6.Bard H, Makowski EL, Meschia G, Battaglia FC. The relative rates of synthesis of hemoglobins A and F in immature red cells of newborn infants. Pediatrics. 1970;45:766–772. [PubMed] [Google Scholar]
- 7.Moss EG. Heterochronic genes and the nature of developmental time. Curr Biol. 2007;17:R425–R434. doi: 10.1016/j.cub.2007.03.043. [DOI] [PubMed] [Google Scholar]
- 8.Pasquinelli AE, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 9.Ouchi Y, Yamamoto J, Iwamoto T. The heterochronic genes lin-28a and lin-28b play an essential and evolutionarily conserved role in early zebrafish development. PLoS One. 2014;9:e88086. doi: 10.1371/journal.pone.0088086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noh SJ, et al. Let-7 microRNAs are developmentally regulated in circulating human erythroid cells. J Transl Med. 2009;7:98. doi: 10.1186/1479-5876-7-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee YT, et al. LIN28B-mediated expression of fetal hemoglobin and production of fetal-like erythrocytes from adult human erythroblasts ex vivo. Blood. 2013;122:1034–1041. doi: 10.1182/blood-2012-12-472308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyerinas B, et al. Identification of let-7-regulated oncofetal genes. Cancer Res. 2008;68:2587–2591. doi: 10.1158/0008-5472.CAN-08-0264. [DOI] [PubMed] [Google Scholar]
- 13.Bell JL, et al. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): Post-transcriptional drivers of cancer progression? Cell Mol Life Sci. 2013;70:2657–2675. doi: 10.1007/s00018-012-1186-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hansen TV, et al. Dwarfism and impaired gut development in insulin-like growth factor II mRNA-binding protein 1-deficient mice. Mol Cell Biol. 2004;24:4448–4464. doi: 10.1128/MCB.24.10.4448-4464.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leeds P, et al. Developmental regulation of CRD-BP, an RNA-binding protein that stabilizes c-myc mRNA in vitro. Oncogene. 1997;14:1279–1286. doi: 10.1038/sj.onc.1201093. [DOI] [PubMed] [Google Scholar]
- 16.Nielsen FC, Nielsen J, Kristensen MA, Koch G, Christiansen J. Cytoplasmic trafficking of IGF-II mRNA-binding protein by conserved KH domains. J Cell Sci. 2002;115:2087–2097. doi: 10.1242/jcs.115.10.2087. [DOI] [PubMed] [Google Scholar]
- 17.Brady HJ, et al. Expression of the human carbonic anhydrase I gene is activated late in fetal erythroid development and regulated by stage-specific trans-acting factors. Br J Haematol. 1990;76:135–142. doi: 10.1111/j.1365-2141.1990.tb07848.x. [DOI] [PubMed] [Google Scholar]
- 18.Marsh WL. Anti-i: A cold antibody defining the Ii relationship in human red cells. Br J Haematol. 1961;7:200–209. doi: 10.1111/j.1365-2141.1961.tb00329.x. [DOI] [PubMed] [Google Scholar]
- 19.Piller F, et al. Biosynthesis of blood group I antigens. Identification of a UDP-GlcNAc:GlcNAc beta 1-3Gal(-R) beta 1-6(GlcNAc to Gal) N-acetylglucosaminyltransferase in hog gastric mucosa. J Biol Chem. 1984;259:13385–13390. [PubMed] [Google Scholar]
- 20.Inaba N, et al. A novel I-branching beta-1,6-N-acetylglucosaminyltransferase involved in human blood group I antigen expression. Blood. 2003;101:2870–2876. doi: 10.1182/blood-2002-09-2838. [DOI] [PubMed] [Google Scholar]
- 21.Conway AE, et al. Enhanced CLIP uncovers IMP protein-RNA targets in human pluripotent stem cells important for cell adhesion and survival. Cell Reports. 2016;15:666–679. doi: 10.1016/j.celrep.2016.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jønson L, et al. Molecular composition of IMP1 ribonucleoprotein granules. Mol Cell Proteomics. 2007;6:798–811. doi: 10.1074/mcp.M600346-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.Nielsen FC, Nielsen J, Christiansen J. A family of IGF-II mRNA binding proteins (IMP) involved in RNA trafficking. Scand J Clin Lab Invest Suppl. 2001;234:93–99. [PubMed] [Google Scholar]
- 25.Ioannidis P, et al. CRD-BP/IMP1 expression characterizes cord blood CD34+ stem cells and affects c-myc and IGF-II expression in MCF-7 cancer cells. J Biol Chem. 2005;280:20086–20093. doi: 10.1074/jbc.M410036200. [DOI] [PubMed] [Google Scholar]
- 26.Liao B, et al. Targeted knockdown of the RNA-binding protein CRD-BP promotes cell proliferation via an insulin-like growth factor II-dependent pathway in human K562 leukemia cells. J Biol Chem. 2004;279:48716–48724. doi: 10.1074/jbc.M405853200. [DOI] [PubMed] [Google Scholar]
- 27.Sankaran VG, Orkin SH. The switch from fetal to adult hemoglobin. Cold Spring Harb Perspect Med. 2013;3:a011643. doi: 10.1101/cshperspect.a011643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith EC, Orkin SH. Hemoglobin genetics: Recent contributions of GWAS and gene editing. Hum Mol Genet. 2016;25:R99–R105. doi: 10.1093/hmg/ddw170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Vasconcellos JF, et al. HMGA2 moderately increases fetal hemoglobin expression in human adult erythroblasts. PLoS One. 2016;11:e0166928. doi: 10.1371/journal.pone.0166928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shell S, et al. Let-7 expression defines two differentiation stages of cancer. Proc Natl Acad Sci USA. 2007;104:11400–11405. doi: 10.1073/pnas.0704372104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Busch B, et al. The oncogenic triangle of HMGA2, LIN28B and IGF2BP1 antagonizes tumor-suppressive actions of the let-7 family. Nucleic Acids Res. 2016;44:3845–3864. doi: 10.1093/nar/gkw099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sankaran VG, et al. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science. 2008;322:1839–1842. doi: 10.1126/science.1165409. [DOI] [PubMed] [Google Scholar]
- 33.Hüttelmaier S, et al. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature. 2005;438:512–515. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- 34.Hafner M, et al. Identification of mRNAs bound and regulated by human LIN28 proteins and molecular requirements for RNA recognition. RNA. 2013;19:613–626. doi: 10.1261/rna.036491.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilbert ML, et al. LIN28 binds messenger RNAs at GGAGA motifs and regulates splicing factor abundance. Mol Cell. 2012;48:195–206. doi: 10.1016/j.molcel.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mizutani R, et al. Oncofetal protein IGF2BP3 facilitates the activity of proto-oncogene protein eIF4E through the destabilization of EIF4E-BP2 mRNA. Oncogene. 2016;35:3495–3502. doi: 10.1038/onc.2015.410. [DOI] [PubMed] [Google Scholar]
- 37.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 38.Frost RJ, Olson EN. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc Natl Acad Sci USA. 2011;108:21075–21080. doi: 10.1073/pnas.1118922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Farina KL, Huttelmaier S, Musunuru K, Darnell R, Singer RH. Two ZBP1 KH domains facilitate beta-actin mRNA localization, granule formation, and cytoskeletal attachment. J Cell Biol. 2003;160:77–87. doi: 10.1083/jcb.200206003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nielsen J, Kristensen MA, Willemoës M, Nielsen FC, Christiansen J. Sequential dimerization of human zipcode-binding protein IMP1 on RNA: A cooperative mechanism providing RNP stability. Nucleic Acids Res. 2004;32:4368–4376. doi: 10.1093/nar/gkh754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang HL, et al. Neurotrophin-induced transport of a beta-actin mRNP complex increases beta-actin levels and stimulates growth cone motility. Neuron. 2001;31:261–275. doi: 10.1016/s0896-6273(01)00357-9. [DOI] [PubMed] [Google Scholar]
- 42.Stöhr N, et al. ZBP1 regulates mRNA stability during cellular stress. J Cell Biol. 2006;175:527–534. doi: 10.1083/jcb.200608071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lemm I, Ross J. Regulation of c-myc mRNA decay by translational pausing in a coding region instability determinant. Mol Cell Biol. 2002;22:3959–3969. doi: 10.1128/MCB.22.12.3959-3969.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang W, et al. Identification and testing of a gene expression signature of invasive carcinoma cells within primary mammary tumors. Cancer Res. 2004;64:8585–8594. doi: 10.1158/0008-5472.CAN-04-1136. [DOI] [PubMed] [Google Scholar]
- 45.Wang G, et al. IMP1 suppresses breast tumor growth and metastasis through the regulation of its target mRNAs. Oncotarget. 2016;7:15690–15702. doi: 10.18632/oncotarget.7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang RJ, et al. MicroRNA-873 (miRNA-873) inhibits glioblastoma tumorigenesis and metastasis by suppressing the expression of IGF2BP1. J Biol Chem. 2015;290:8938–8948. doi: 10.1074/jbc.M114.624700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bauer DE, Orkin SH. Hemoglobin switching’s surprise: the versatile transcription factor BCL11A is a master repressor of fetal hemoglobin. Curr Opin Genet Dev. 2015;33:62–70. doi: 10.1016/j.gde.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nottrott S, Simard MJ, Richter JD. Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat Struct Mol Biol. 2006;13:1108–1114. doi: 10.1038/nsmb1173. [DOI] [PubMed] [Google Scholar]
- 49.Kingsley PD, et al. Ontogeny of erythroid gene expression. Blood. 2013;121:e5–e13. doi: 10.1182/blood-2012-04-422394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee YT, et al. Erythroid-specific expression of LIN28A is sufficient for robust gamma-globin gene and protein expression in adult erythroblasts. PLoS One. 2015;10:e0144977. doi: 10.1371/journal.pone.0144977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 52.Hanawa H, et al. Comparison of various envelope proteins for their ability to pseudotype lentiviral vectors and transduce primitive hematopoietic cells from human blood. Mol Ther. 2002;5:242–251. doi: 10.1006/mthe.2002.0549. [DOI] [PubMed] [Google Scholar]
- 53.Krivega I, et al. Inhibition of G9a methyltransferase stimulates fetal hemoglobin production by facilitating LCR/γ-globin looping. Blood. 2015;126:665–672. doi: 10.1182/blood-2015-02-629972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wojda U, Noel P, Miller JL. Fetal and adult hemoglobin production during adult erythropoiesis: Coordinate expression correlates with cell proliferation. Blood. 2002;99:3005–3013. [PubMed] [Google Scholar]
- 55.Sripichai O, et al. Cytokine-mediated increases in fetal hemoglobin are associated with globin gene histone modification and transcription factor reprogramming. Blood. 2009;114:2299–2306. doi: 10.1182/blood-2009-05-219386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goh SH, et al. A newly discovered human alpha-globin gene. Blood. 2005;106:1466–1472. doi: 10.1182/blood-2005-03-0948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hartley SW, Mullikin JC. QoRTs: A comprehensive toolset for quality control and data processing of RNA-Seq experiments. BMC Bioinformatics. 2015;16:224. doi: 10.1186/s12859-015-0670-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gandin V, et al. Polysome fractionation and analysis of mammalian translatomes on a genome-wide scale. J Vis Exp. 2014;87:e51455. doi: 10.3791/51455. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.