Abstract
Ankyrin-repeat family A protein (ANKRA) was originally cloned in mouse as an interacting protein to megalin, a member of low-density lipoprotein receptor superfamily. Here, we report that the isolation of rat ANKRA as a new binding partner for the α-subunit of rat large-conductance Ca2+-activated K+ channel (rSlo). We mapped the binding region of each protein by using yeast two-hybrid and in vitro binding assays. ANKRA expressed together with rSlo channels were colocalized near the plasma membrane and coimmunoprecipitated in transfected cells. We also showed that BKCa channel in rat cerebral cortex coprecipitated with rANKRA and colocalized in cultured rat hippocampal neuron. Although the coexpression of ANKRA did not affect the surface expression of rSlo, the gating kinetics of rSlo channel was significantly altered and the effects were highly dependent on the intracellular calcium. These results indicate that ANKRA could modulate the excitability of neurons by binding directly to endogenous BKCa channel and altering its gating kinetics in a calcium-dependent manner.
INTRODUCTION
Large-conductance calcium-activated potassium channels (BKCa channels) are ubiquitously distributed among tissues, except cardiac myocytes, and activated by membrane depolarization and increases of intracellular calcium concentration (Latorre et al., 1989; Toro et al., 1998). They play a key role in the control of neuronal excitability, hormone secretion, and smooth muscle tone (Robitaille et al., 1993; Nelson et al., 1995; Lingle et al., 1996; Fox et al., 1997). Thus, BKCa channels are considered to be molecular integrators of biochemical and electrical signals. In mammals, endogenous BKCa channel is thought as either homotetramer of a poreforming α-subunit encoded by a single gene, KCNMA1 (slo or slowpoke) (Butler et al., 1993) or an assembly of α-subunits and auxiliary β-subunits. Up to now, four different kinds of β-subunits (β1–β4) have been identified (Garcia-Calvo et al., 1994; Wallner et al., 1999; Behrens et al., 2000; Brenner et al., 2000; Meera et al., 2000; Weiger et al., 2000). Coexpression of β-subunits changes pharmacological properties, voltage dependence, and gating kinetics of BKCa channels (Orio et al., 2002).
Several proteins are known to associate with BKCa channel and modulate their functions. Two novel proteins, Slob and SLIP, were identified in Drosophila as the binding modulators of BKCa channels (Schopperle et al., 1998; Xia et al., 1998). Then, Src tyrosine kinase and protein kinase A (PKA) catalytic subunit also are found to interact with Drosophila BKCa channels (Wang et al., 1999). In neuronal cells, BKCa channels control calcium influxes and regulate the neurotransmitter release (Roberts et al., 1990; Sun et al., 1999), process that may require the colocalization with calcium channels at presynaptic sites (Issa and Hudspeth, 1994; Marrion and Tavalin, 1998; Hu et al., 2001; Orio et al., 2002). Recently, it was reported that β2 adrenergic receptor could simultaneously interact with both BKCa and L-type Ca2+ channels in vivo (Liu et al., 2004). Among intracellular signaling molecules in vertebrate, protein kinases and phosphatases, including PKA, protein kinase G, Src tyrosine kinase, Syk tyrosine kinase, focal adhesion kinase, and protein phosphatase 2A are most intensively studied as modulators of BKCa channels functions (Dworetzky et al., 1996; Sansom et al., 1997; Alioua et al., 1998; Ling et al., 2000; Alioua et al., 2002; Rezzonico et al., 2002, 2003; Tian et al., 2003). Recently, Syntaxin1A and β-catenine also have been reported as direct binding partners of BKCa channels. Syntaxin1A is reported to be associated with BKCa channel in rat brain and enhances its activity by changing gating kinetics at low calcium concentration (Ling et al., 2003). β-Catenine binds to the S10 segment of Slo channel protein and may provide a physical link between BKCa channel and voltage-gated calcium channel (Lesage et al., 2004).
In this study, we identified a new BKCa channel-binding protein, ANKRA, by using yeast two-hybrid screening and characterized its functional significance on channel activity. The expression pattern of rANKRA gene was investigated by reverse transcription-polymerase chain reaction (RT-PCR) analysis and well matched with that of slo gene. The critical regions for protein–protein interaction were mapped by yeast two-hybrid and in vitro binding assays. We found that the N-terminal part of rANKRA protein, including putative coiled-coil segment and the C-terminal segment of rSlo channel protein, are essential for binding. Coimmunoprecipitation experiment and immunofluorescence staining showed that the association of two proteins also occurs in transfected cells and native tissue. Finally, we investigated whether rANKRA protein affects the functional characteristics of rSlo channel. Coexpression of rANKRA resulted in enhanced activity of rSlo channels by increasing the activation rate and decreasing the deactivation rate in the range of micromolar calcium concentrations.
MATERIALS AND METHODS
Materials
Yeast two-hybrid constructs, epitope-tagged channel genes, and gene fusion constructs used the previously reported rat slo clone rslo27 (GenBank accession no. AF135265) (Ha et al., 2000). Plasmid vector pNBC-HA for in vitro transcription and translation reaction was modified from pGEM-HE vector (Liman et al., 1992). Briefly, HA-encoding oligonucleotide pairs (top, 5′-CCGGCCACCATGGCCTCCTACCCTTATGATGTGCCAGATTATGCCTCTGCCCCG-3′ and bottom, 5′-AGGCATAATCTGGCACATCATAAGGGTAGGAGGCCATGGTGG-3′) and multiple cloning site containing oligonucleotide pairs (top, 5′-GGGATCCGAATTCAATTGATCGATATGCATCTCGAG T-3′ and bottom, 5′-CTAGACTCGAGATGCATATCGATCAATTGAATTCGGATCCCCGGGGCAG-3′) were annealed in annealing buffer (50 mM Tris-Cl, pH 8.0, and 10 mM MgCl2) with a thermal ramp from 95 to 29°C >6 h. Two annealed oligonucleotide pairs were ligated simultaneously into pGEM-HE vector digested with XmaI and XbaI. pTracer-HA vector used in biochemical studies was similarly modified from pTracer-CMV (Invitrogen, Carlsbad, CA) as described above. Synthetic oligonucleotides were obtained from Cosmo Genentech (Seoul, Korea). The genotype of the Saccharomyces cerevisiae reporter strain AH109 is MATa, trp1-901, leu2-3, 112, ura3-52, his3-200, gal4Δ, gal80Δ, LYS2::GAL1UAS-GAL1TATA-HIS3, GAL2UAS-GAL2TATA-ADE2, URA3::MEL1UAS-MEL1TATA-lacZ. All chemicals were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise specified.
Yeast Two-Hybrid Screening
Before making bait constructs, conserved domains and predicted secondary structure of rSlo channel protein were analyzed. A construct for rSlo-C (corresponding to amino acids 951-1210) was generated by PCR by using a specific primer pair (sense, 5′-CGCGGATCCTCGCTAAGCCGGGCAAGT-3′ and antisense, 5′-ACGGGTCGACTCTGTAAACCATTTCTT T-3′) and subsequently subcloned into pGBK-T7, a Gal4 DNA-binding vector (BD Biosciences, San Jose, CA) by using standard techniques (Sambrook and Russell, 2001). Stable expression of bait proteins in a yeast strain AH109 was confirmed by immunoblotting by using mouse monoclonal antibody (mAb) directing GAL4 DNA-binding domain (Santa Cruz Biotechnology, Santa Cruz, CA). Library scale screening was performed on ∼3 million cotransformants according to the manufacturer's instructions. Rat brain cDNA library cloned in GAL4 activation-domain vector pACT2 (BD Biosciences) was cotransformed into yeast AH109 with bait clone by Li+-acetate method. Cotransformants were plated and selected on triple negative synthetic dropout media, SD/Trp-Leu-Adenine-, and grown colonies were further selected on quadruple negative SD/Trp-Leu-His-Adenine- media containing 1 mM 3-AT. To confirm the initial positives, clones survived on SD/Trp-Leu-His-Adenine- media were retransformed with bait rSlo C/pGBK-T7 or mock vector pGBK-T7. Subsequently, DNA sequences of confirmed positives were determined using dideoxy chain termination method.
For binding-site mapping experiment, further subdivided rSlo-C domain constructs rSlo-CN (corresponding to amino acids 951-1119) and rSlo-CC (1119–1210) were generated. Each subdomain bait construct was cotransformed with the positive clone or virgin pACT2 vector into yeast AH109. The growth and LacZ-based color appearance of each cotransformant on quadruple negative SD media containing X-α-Gal were monitored for examining specific interaction.
RNA Isolation and RT-PCR
Total RNAs were purified from isolated male Sprague Dawley rat tissues in TRI reagent (1 ml/100 mg of tissue; Molecular Research Center, Cincinnati, OH). Each tissue was homogenized using a glass-Teflon homogenizer and subsequent extracting steps were followed as described in product instruction. Complement DNA was synthesized from 5 μg of total RNA by priming of 1 μg of oligo(dT)12–18 (Promega, Madison, WI) by using M-MLV reverse transcriptase (Invitrogen) at 37°C for 1 h. To test whether rANKRA and rslo transcripts are expressed at particular tissues, gene-specific primers were used to generate 390-base pair cDNA fragments corresponding to nucleotides 332–721 of rANKRA (forward, 5′-TAAGGCACGTCTACACACC-3′ and reverse, 5′-CGTCAACTCCACAGTCTAGC-3′) and 783 base pairs to 2550–3332 of rslo, respectively. The primers for rslo were described in a previous report (Ha et al., 2000). To show minor variation in RNA preparations between different tissues, β-actin–specific primers used as internal control for RT-PCR reaction (forward, 5′-ACAGCTTCACCACCACAG-3′ and reverse, 5′-CGGATGTCAACGTCACAC-3′).
Expression of Channel Domains and In Vitro Binding Assay
rSlo-CN, -CC, and -C domains were subcloned into pGEX 4T-3 vectors (Amersham Biosciences UK, Little Chalfont, Buckinghamshire, United Kingdom). Fusion proteins were expressed in an Escherichia coli strain, BL21, by adding 0.5 mM isopropyl β-d-thiogalactoside and purified on glutathione Sepharose 4B (Amersham Biosciences UK) according to the manufacturer's instructions. After extensive washing steps, immobilized fusion proteins on glutathione Sepharose were directly used for in vitro binding assay without elution. For in vitro synthesis of rANKRA protein, full open reading frame (ORF) of rANKRA, partial domain N1, N2, N, C, and N2-C (corresponding to amino acids 1–313, 1–51, 52–150, 1–150, 151–313, and 52–313, respectively) were subcloned into pNBC-HA vector and synthesized with [35S]methionine (PerkinElmer Life and Analytical Sciences, Boston, MA) by using TNT coupled lysate system (Promega).
Three to 5 μg of glutathione S-transferase (GST) or GST-fused rSlo-C domains immobilized on Sepharose bead were incubated with 3 to 5 μl of 35S-labeled rANKRA proteins in binding buffer TBST-TBT (137 mM NaCl, 20 mM Tris-Cl, pH 7.6, 0.1% Tween 20, 100 μg/ml bovine serum albumin [BSA], and 0.5% Triton X-100), for 2–6 h at 4°C and washed three times with same buffer followed by additional three times with Tris-buffered saline (TBS)-T buffer (137 mM NaCl, 20 mM Tris-Cl, pH 7.6, and 0.1% Tween 20). After removal of washing buffer, protein complexes were eluted by boiling with 20–50 μl of 2× SDS sample buffer (100 mM Tris-Cl, pH 6.8, 4% SDS, 0.2% bromophenol blue, 20% glycerol, and 200 mM β-mercaptoethanol) and then separated by SDS-PAGE. Separated protein bands were detected by exposing dried gels on BioMax x-ray film (Eastman Kodak, Rochester, NY).
Cell Culture, Transient Transfection, and Preparation of Cell Lysate
Chinese hamster ovary (CHO)-K1 cells were cultured at 37°C in F-12K nutrient mixture, Kaighn's modification (Invitrogen) supplemented with 10% fetal bovine serum (FBS) (Invitrogen), in a humidified atmosphere containing 5% CO2. COS-7 and human embryonic kidney (HEK) 293 cells were maintained in DMEM with 10% FBS.
Subconfluent monolayer cells on six-wall plate were transfected in serum- and antibiotic-free media with 1.5 μg of total DNA by using 10 μl of PolyFect transfection reagent (QIAGEN, Valencia, CA) according to manufacturer's instructions. For the biochemical studies, rslo/pcDNA 3.1 and rANKRA/pTracer-HA were used. Transfection efficiencies were monitored by observing green fluorescence of GFP-Zeocin from pTracer vector using Olympus IX51 microscope. For immunofluorescence staining and electrophysiology, rslo::EGFP/pcDNA 3.1 and HA-rANKRA/pcDNA 3.1 were used.
CHO-K1 cells transfected transiently with rSlo, rANKRA, or both were gently lysed with Triton X-100 lysis buffer (10 mM Tris-Cl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 1 mM Na3VO4, 10% glycerol, 1% Triton X-100, and 1× protease inhibitor cocktail [Complete; Roche Diagnostics, Indianapolis, IN]) for 20 min on ice and then centrifuged at ∼16,000 × g for 10 min at 4°C for removing nuclei and any insoluble matrix.
Preparation of Anti-rANKRA Polyclonal Antibodies
The full-length rANKRA cDNA was subcloned in-frame into pGEX 4T-3 vector and overexpressed as GST fusion protein. The fusion protein was purified using glutathione Sepharose 4B and administrated to rabbits in series of three injections. After immunization, obtained serum was further purified by using GST::rANKRA affinity chromatography.
Preparation of Brain Membrane Fraction and Affinity Pull-Down Assay
Membrane-enriched fraction of cerebral cortex tissue from Sprague Dawley rats was prepared as described previously (Nadal et al., 2003). Briefly, isolated tissue was homogenized in 10 volumes of ice-cold sucrose buffer (0.32 M sucrose, 1 mM EDTA, 5 mM Tris-HCl, pH 7.4, and protease inhibitor cocktail). The homogenate was centrifuged at 1000 × g for 10 min, and the supernatants were centrifuged further at 27,000 × g for 40 min to yield a crude membrane pellet. The pellet was resuspended in TNE (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, and protease inhibitor cocktail) and solubilized with 1% Triton X-100 on ice. Insoluble material was removed by centrifugation at 20,000 × g for 30 min, and the supernatant was used for affinity pull-down assay. The solubilized membrane fraction (1 mg) was incubated overnight at 4°C with GST or GST::rANKRA protein in TNE buffer with 1% Triton X-100 and washed four times with same buffer. After eluting protein complexes with SDS-sample buffer, immunoblotting experiments were performed with anti-hSlo antibody.
Immunoprecipitation and Immunoblotting
After quantification of total proteins using Bradford method, 500 μg of soluble lysates from cortical membrane fraction or CHO-K1 cells were precleared with 50 μl of 50% protein A-Sepharose (Amersham Biosciences UK) by rotating for 2 h at 4°C. For immunoprecipitation of rSlo, 1 μg of mouse monoclonal anti-hSlo antibody (BD Transduction Laboratories, Lexington, KY) was added to the precleared samples and allowed to incubate for 4–12 h at 4°C and for rANKRA, 2.5 μg of purified anti-rANKRA antibody or 1.5 μg of rabbit polyclonal anti-HA antibody (Upstate Biotechnology, Chicago, IL) were used. The mixture was then incubated with 50 μl of 50% protein A-Sepharose for 2 h. Immobilized protein–antibody complexes on the bead were washed twice with lysis buffer and twice more with phosphate-buffered saline (PBS). Protein complexes were collected by centrifugation and eluted by 30–50 μl of 2× SDS-sample buffer at 37°C for 20 min.
For immunoblotting, immunoprecipitated samples or input representing 10% of materials used for immunoprecipitation were resolved on 6–12% SDS-acrylamide gel and then transferred on the polyvinylidene difluoride membranes (Amersham Biosciences UK). The blots were incubated with mouse anti-hSlo mAb or rabbit anti-HA antibody at 4°C overnight. After three washes with TBS-T, the blots were further incubated with appropriate horseradish peroxidase (HRP)-conjugated secondary antibodies, and immunoreactive protein bands were visualized by incubating membrane with ECL solution (Amersham Biosciences UK) and exposing on x-ray films.
Construction of GFP-fused cDNA Plasmid
To generate EGFP-fused rslo expression construct, termination codon removed rslo gene was amplified by PCR with primers (sense, 5′-ATAGGATCCATGAGCAATATCCACGCG-3′ and antisense, 5′-ATATGCGGCCGCTCTGTAAACCATTTC-3′) by using proofreading pfu polymerase. EGFP gene also was amplified in a separate PCR reaction (sense, 5′-AGTCGCGGCCGCCATGGTGAGCAAGGGC-3′ and antisense, 5′-GCGCTCGAGTTACTTGTACAGCTC-3′). After preparative digestion of each amplicon with BamHI/NotI or NotI/XhoI, gel-purified DNA fragments were ligated into pcDNA3.1(+) vector flanked by BamHI and XhoI. As a result, nine base pairs coding triple alanine (GCGGCCGCC) were added between rSlo and EGFP gene. To confirm the fusion gene construct, DNA sequencing was carried out using ABI 377 automatic DNA sequencer (PerkinElmer Life and Analytical Sciences).
Immunofluorescence Staining
The day after transfection, COS-7 cells were detached and transferred on gelatin-coated [0.2% (wt/vol)] Lab-Tak II chamber slide (Nalge Nunc International, Naperville, IL). After 24–36 h, cells were washed three times with PBS and then fixed with 4% paraformaldehyde in PBS for 10 min at room temperature. After washing in PBS, the cells were permeabilized with 0.5% Triton X-100 in PBS for 5 min at room temperature and then incubated for 1 h at room temperature, first with the rabbit polyclonal anti-HA primary antibodies diluted with PBS containing 2% BSA and 0.5% goat serum and then with Texas Red-conjugated donkey anti-rabbit IgG antibody (Jackson ImmunoResearch Laboratories, West Grove, PA). Thereafter, the chamber slide glasses were washed three times with PBS and mounted with Crystal/Mount (Biomeda, Forster, CA). The cells were viewed under a Leica DMIRD fluorescence microscope equipped with a 63× objective lens and appropriate filters.
Immunocytochemistry
Embryonic day 18 (E18) primary rat hippocampal neurons were prepared and maintained as described previously (Chang and De Camilli, 2001). Before antibody treatment, neurons were fixed with 4% paraformaldehyde in PBS containing 0.12 M sucrose and permeabilized in 0.25% Triton-X-100 for 5 min. The cultures were incubated with mouse anti-hSlo (2.5 ng/μl) and rabbit anti-rANKRA (2.0 ng/μl) in 2% BSA overnight at 4°C and subsequently labeled with fluorescein isothiocyanate-conjugated donkey anti-mouse secondary antibody (catalog no. 715-095-150; Jackson ImmunoResearch Laboratories) and Texas Red dye-conjugated donkey anti-rabbit secondary antibody (catalog no. 711-075-152; Jackson ImmunoResearch Laboratories) for 1 h at room temperature. Images were overlapped using Adobe Photoshop software.
Surface Biotinylation
After 2 d of transfection, biotinylation of cell surface proteins was performed as described in product instructions with minor modification. Briefly, cells grown on 35-mm plates were detached, washed three times with ice-cold PBS, and incubated 45 min with 1 mg/ml EZ-link Sulfo-NHS-LC-Biotin (Pierce Chemical, Rockford, IL) in PBS at 4°C with gentle inversions. The cells were washed twice with PBS-G (PBS with 100 mM glycine) and further incubated for 10 min with PBS-G to quench and to remove free biotin reagents. Then, cells were lysed in Triton X-100 lysis buffer and incubated on ice for 30 min. After centrifugation, supernatants were incubated with 50 μl of 50% slurry of Immobilized NeutrAvidin (Pierce Chemical) for 2 h at 4°C. Beads were collected by brief centrifugation, washed twice with lysis buffer, two more times with PBS, and then biotinylated proteins were eluted by boiling in 2× SDS-sample buffer. After probing biotinylated rSlo channel proteins with anti-hSlo antibody, relative intensities of protein bands were analyzed using GS-710 Image densitometer (Bio-Rad, Hercules, CA)
Electrophysiology
Ionic currents carried by rSlo::EGFP in the absence and presence of rANKRA were recorded from excised membrane patches of CHO-K1 cell in inside-out configuration by using an Axopatch 200B amplifier (Axon Instruments, Union City, CA). Recordings were performed at room temperature 24–72 h after transfection. Pipettes were prepared from thin-walled borosilicate glass (WPI, Sarasota, FL) and fire-polished to a resistance of 3 to 6 MΩ. Signals were filtered at 1 kHz by using a four-pole low-pass Bassel filter, digitized at a rate of 20 samples/ms by using Digidata 1200 (Axon Instruments), and stored in a personal computer. pClamp8 (Axon Instruments) software was used to control the amplifier and to acquire and analyze the data as described previously (Ha et al., 2004).
Pipette solutions contained 10 mM HEPES, 2 mM EGTA, 116 mM KOH, and 4 mM KCl. Intracellular solution for perfusing to internal face of excised patches was symmetrical to pipette solution except supplemented CaCl2. The amount of CaCl2 required to yield the desired concentration was calculated using stability constant for Ca-EGTA of 10.86 (log K), and the calculation included a pH adjustment (Martell and Smith, 1974). The pH of all recording solutions was adjusted to 7.2 with 2-[N-morpholino]ethanesulfonic acid. To analyze microscopic current, Origin 6.1 (OriginLab, Northampton, MA) was used.
RESULTS
An Ankyrin-Repeat-containing Protein, ANKRA, Was Identified as a Slo-Binding Protein
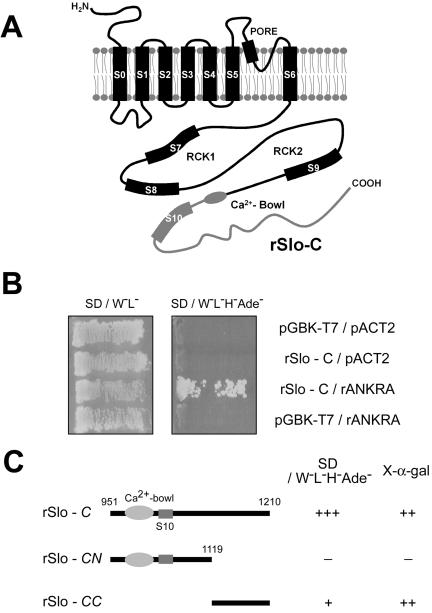
To identify proteins interacting with rat α-subunit of largeconductance calcium-activated potassium channel (rSlo), we screened rat brain cDNA library by using a C-terminal fragment (amino acids 951-1210) of rSlo channel as a bait (named rSlo-C, Figure 1A). Before preceding library screening, the stable expression of bait proteins in yeast was monitored by immunoblotting on transformed yeast lysate (our unpublished data). From ∼3 × 106 transformants, 184 positive clones maintained survival on selection media. Among those, we initially selected 38 clones to test direct interaction with rSlo channel by using in vitro binding assay. Finally, we have focused on a clone, which could encode rat ortholog of ankyrin-repeat family A (ANKRA) gene missing 5′ end 45 nucleotides independently found four times from the library screening. For cloning of full-length rANKRA gene, BLAST searches against dbEST (database for expressed sequence tags) were performed. One of the matched EST clones (dbEST ID 7079153) containing the full coding sequences was purchased from Research Genetics (Invitrogen), and its sequence was confirmed by sequencing. The nucleotide sequence of reconstituted rat ANKRA was deposited in the GenBank with accession no. AY566568. After reconstitution of full ORF in prey vector pACT2, yeast two-hybrid assay was performed using rSlo-C as bait (Figure 1B).
Figure 1.
Carboxy terminus of rSlo recruits an Ankyrin-repeat family A protein, rANKRA, in yeast two-hybrid screening. (A) Putative membrane topology of rSlo (α-subunit of large conductance Ca2+-activated potassium channel) protein. Cloned rSlo channel (AF135265) has seven membrane-spanning segments, including voltage-sensing S4, two regulating the conductance of K+ (RCK) domains, and “Calcium-Bowl” (a negatively charged region to be implicated in calcium binding). One-third of cytosolic C terminus (rSlo-C, amino acids 951-1210, gray line) including Calcium-Bowl was used to screen rat brain library (MATCHMAKER cDNA library; BD Biosciences). (B) Yeast, AH109 was transformed simultaneously with the two indicated plasmids, and cotransformants were selected on SD media lacking tryptophan and leucine. The interaction was monitored by histidine and adenine prototrophy. Yeasts only cotransformed with rSlo-C and rANKRA could survive on the SD/W-L-H-Ade- plate. (C) Using rSlo-C, -CN, and -CC domains as baits, region involved in recruiting rANKRA was investigated in yeast. Yeast cells harboring rSlo-C and rSlo-CC in bait vector were survived on selection media SD/W-L-H-Ade- containing X-α-gal (marked as +) and showed blue color with rANKRA in prey vector. Calcium-Bowl and S10 segment are indicated in molecular cartoon.
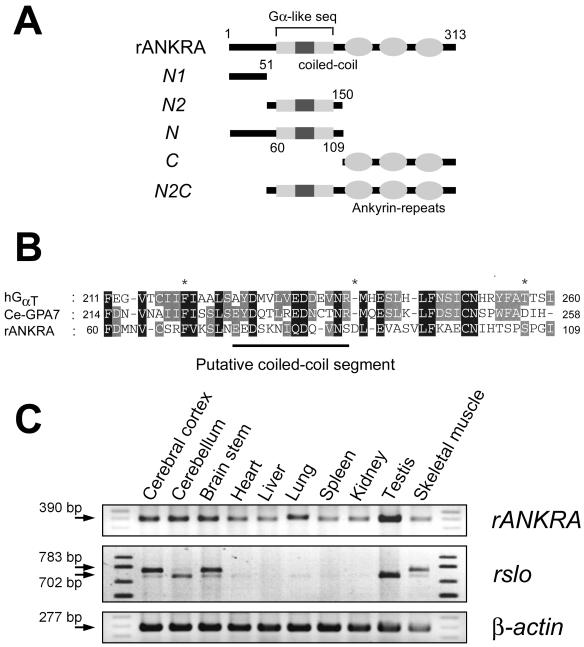
To obtain the clue for the evolutionary conserved function of rANKRA, we tried to find homologous sequences and known protein domains by using iterated psi-BLAST (National Center for Biotechnology Information, Bethesda, MD) and domain searching tools such as Pfam (Washington University, St. Louis, MO), and InterPro (European Bioinformatics Institute, Hinxton, Cambridge, United Kingdom). We were able to find a stretch of amino acid (amino acids corresponding 60–109) partially aligned with the α-subunit of trimeric G protein from psi-BLAST, which contains putative coiled-coil segment predicted from COILS (Lupas et al., 1991) but failed to detect any conserved domain except C-terminal three ankyrin-repeats (Figure 2, A and B).
Figure 2.
Domain structure of rANKRA and the expression profile of rANKRA and rslo transcripts in various tissues. (A) Cartoon for depicting domain structure and deletion construct of rANKRA. Carboxyl terminal three ankyrin-repeats were predicted from Pfam (Washington University), Gα-like sequence in the middle of ANKRA was found from psi-BLAST search, and putative coiled-coil segment was expected by algorithm COILS. Series of deletion mutants were generated based on domain structure and conserved amino acid sequences. (B) Amino acid sequence alignment of G protein α-like region (in A) in the middle of rANKRA. hGαT is a human rod-specific transducin α-subunit (gi 22027520) and Ce-GPA7 is a C. elegans G protein α-7 subunit (gi 17540930). A putative coiled-coil segment predicted by COIL (Lupas et al., 1991) is indicated as line. (C) Expression patterns of rANKRA (top) and rslo (bottom) were examined by RT-PCR. rANKRA transcripts were expressed ubiquitously in various tissues but more highly in brain subregions and testis (top). Although the expression of rslo was found in many tissues, the dominant expression was detected in brain, testis, and skeletal muscle. The alternative splicing variants of rslo (rslo0 and rslo27) were found in brain subregions and skeletal muscle. Although the rslo0 form (702 base pairs) was expressed highly in cerebellum, rslo27 (783 base pairs) was predominant in cerebral cortex and brain stem. β-actin transcripts were amplified for internal reaction control.
To determine the expression patterns of rANKRA, we performed RT-PCR analysis by using various tissues from rat (top, rANKRA, Figure 2C). Whereas the rANKRA transcripts were found in all tissues tested, higher level expressions were detected at brain subregions and testis. We also performed RT-PCR analysis on rslo by using identical RNA samples and found abundant expression of rslo transcripts in all brain subregions, testis, and skeletal muscle (Figure 2C, middle). The expression of rslo was detected in other tissues but only in low levels. Because our PCR primers were designed to flank the fifth splicing site of rslo gene, two different sizes of RT-PCR bands corresponding to rslo0 and rslo27 were expected (Ha et al., 2000). Although the rslo0 band (702 base pairs) was detected widely in all tissues tested, the splicing variant of rslo27 (783 base pairs) was observed predominantly in cerebral cortex, brain stem, and skeletal muscle. It is worth noticing that those tissues expressing rslo at high level also show an increased level of rANKRA messages.
N-Terminal Domain Confers Specific Binding on rANKRA Protein to the C-Terminal End of rSlo Channel
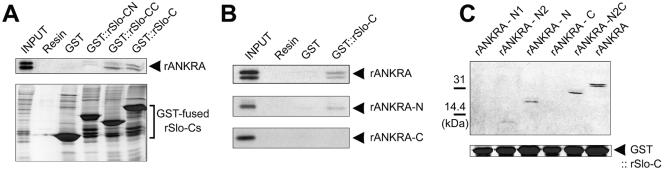
To map the interacting domains in rSlo and rANKRA, we examined the association of rSlo-C and rANKRA by using yeast two-hybrid and in vitro binding assay. At first, we tested possible interactions between full-length rANKRA and truncated rSlo-C proteins. In the consideration of amino acid sequence conservation between orthologues, rSlo-C was subdivided into N-terminal CN, which is highly conserved even with Drosophila and C. elegans orthologues, and C-terminal CC, specifically conserved among mammalian orthologues. Results from yeast two-hybrid assay indicated that rSlo-CC is involved in recruiting rANKRA protein (Figure 1C). To confirm the specific interaction between rSlo-C and rANKRA, we conducted binding assay in vitro. For the binding under cell-free condition, the rSlo-CN, rSlo-CC and rSlo-C domains were expressed in E. coli and purified as GST-fusion proteins. After affinity purification of GST fusion proteins, several distinct protein bands detected below each of expected molecular weight might be caused by proteolytic digestion of fusion proteins (Figure 3A, bottom). Radiolabeled rANKRA proteins were synthesized by in vitro transcription followed by translation using [35S]methionine. Synthesized rANKRA showed as two separated protein bands (Figure 3, A and B, top). Immunoblotting assay using anti-HA antibody and anti-ANKRA antibody revealed that low molecular weight protein was synthesized from second methionine after HA-epitope (our unpublished data). In accordance with yeast two-hybrid, rSlo-CC domain showed specific interaction with rANKRA (Figure 3A). To map the binding site within rANKRA, we tried to perform in vitro binding assay by using purified rSlo-C and biosected rANKRA-N (amino acids 1–150) or rANKRA-C domain (151–313). rANKRA-N domain showed specific association to rSlo-C domain compared with rANKRA-C domain (Figure 3B). None of rANKRA domains were associated to the negative controls, glutathione Sepharose resin or the immobilized GST protein (Figure 3B; our unpublished data). We then attempted to further localize interacting region within rANKRA-N domain by using a series of deletion mutants (Figure 2A). Despite of its weak affinity, 11 kDa of N2 domain (amino acids 52–150) itself is enough to interact with rSlo-C (Figure 3C). Only the constructs containing N2 region of 98 amino acids (N2, N, and N2C) were competent to interact with rSlo-C (Figure 3C).
Figure 3.
Mapping binding sites in rSlo and rANKRA by in vitro binding assay. (A) 35S-labeled rANKRA strongly interacted to immobilized GST::rSlo-CC and GST::rSlo-C but not to GST::rSlo-CN, the negative controls, glutathione Sepharose resin, and GST (top). Coomassie Blue staining image was shown as a control for pull-down processes (bottom). (B) N-terminal half of rANKRA can solely associate with rSlo-C. None of rANKRA domains showed specific binding to the Sepharose resin or GST proteins. (C) rANKRA-N2 (including Gα homologous part and putative coiled-coil segment, Figure 2B) is necessary for interaction with rSlo-C domain. Among rANKRA subdomains, N2 region containing domains such as N2, N, N2C, and whole region exhibited specific interactions with rSlo-C. Despite of its weak affinity, N2 region do interact alone with rSlo-C.
Interaction of rSlo and rANKRA in Transfected Cells
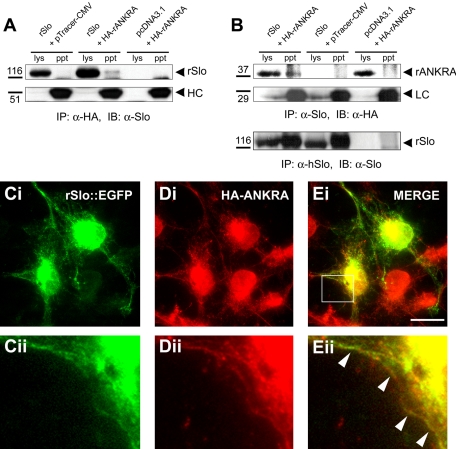
For the immunological assays on probing the rSlo channel protein, we used the commercial antibody raised against human ortholog. We initially tested cross-reactivity of hSlo antibody to rat Slo channel and the antibody recognized a specific band of approximately ∼130 kDa in rslo transfected cell lysate (our unpublished data). We then carried out coimmunoprecipitation experiments to test whether rSlo and rANKRA interact in vivo as full-length proteins. First, the HA-rANKRA protein was immunoprecipitated using rabbit polyclonal HA antibody, and the presence of rSlo proteins in the HA immunocomplex was detected by anti-hSlo antibody. A Slo channel immunoreactive band was only detected in the precipitate from cells cotransfected with rslo and rANKRA (Figure 4A). To confirm in vivo interaction between the rSlo channel and rANKRA, additional immunoprecipitation was performed in the reciprocal direction. We were also able to detect a HA-immunoreactive band corresponding to the size of HA-rANKRA in the Slo immunocomplex (Figure 4B). Together, we conclude that rSlo channels could associate with rANKRA proteins in intact cellular environments.
Figure 4.
Assemble of rSlo and rANKRA in transfected cells. (A) rSlo can be coimmunoprecipitated with HA-tagged rANKRA protein. Protein complexes were immunoprecipitated with rabbit polyclonal α-HA antibody from lysates of CHO-K1 cells transfected with rslo/pcDNA3.1 and pTracer-CMV, rslo/pcDNA3.1, and HA-tagged rANKRA/pTracer-CMV, or pcDNA3.1 and HA-tagged rANKRA/pTracer-CMV. Precipitated proteins were separated in 6% SDS-PAGE and probed with anti-hSlo antibody through immunoblotting. Slo-immunoreactive protein (∼130 kDa) was detected in the HA-immunoprecipitants. HC, IgG heavy chain. Relative positions of molecular standard are indicated (kilodaltons). (B) Proteins were reciprocally precipitated with mouse monoclonal α-hSlo antibody and probed with rabbit polyclonal HA antibody. Samples were separated in 12% SDS-PAGE. Immunoprecipitation procedures were validated by control experiments for detecting and precipitated rSlo protein and IgG light chain (LC). (C–E) Immunofluorescence microscopy of transiently transfected COS-7 cells by using rSlo::EGFP and HA-rANKRA proteins is shown. rSlo::EGFP channels were directly monitored under fluorescence microscope (Ci, green), and HA-rANKRA proteins were stained with Texas Red-conjugated secondary antibody (Jackson ImmunoResearch Laboratories) following rabbit anti-HA antibody (Di, red). The merged image was constructed using software Adobe Photoshop. Merged signals due to the colocalization of proteins are detected (white arrows) along the cell surface (Ei, yellow). Boxed regions marked in Ei were magnified for better visualization of colocalization (Cii, Dii, and Eii). Bar (C–E), 20 μm.
The previous biochemical experiments, GST-pull-down assays and coimmunoprecipitation experiments indicate that rSlo and rANKRA interact with each other in vitro and in transfected cells. Before examining the cellular localization of both proteins, an EGFP was fused in-frame to the carboxy terminus of the rSlo channel protein to facilitate detection of transfected cells and direct observation of its subcellular localizations without antibody staining. According to the previous report, the carboxyl terminal GFP-fused mouse Slo channel did not change the channel's own characteristics such as single-channel conductance, gating kinetics, calcium and voltage dependence of open probability, and charybdotoxin blockade (Myers et al., 1999). CHO-K1 cells were transfected with both rslo::EGFP and HA-rANKRA to perform an immunofluorescence study (Figure 4, C–E). In rSlo-transfected cells, punctuated green fluorescent signals of rSlo were visible along the plasma membrane, whereas much of signals are trapped in intracellular nets (Figure 4C). The staining patterns of HA-rANKRA showed diffused signals throughout the cytosol and somewhat punctuated near the plasma membrane (Figure 4D, red). In the merged image, both proteins displayed clear colocalizations near the cell surface (Figure 4E, white arrow). Thus, rSlo channel proteins can interact and colocalize with the rANKRA protein in cellular environment.
ANKRA Associates with Slo Channel in Native Tissue and Colocalizes in Cultured Neurons
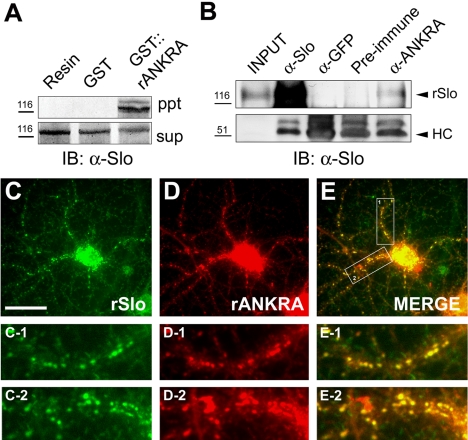
Although interaction between two proteins is valid in transfected cells, it still raises the question for the association of two proteins in native tissues. We first generated and purified GST-fused rANKRA proteins to examine possible association in native tissue. These fusion proteins were incubated with detergent-solubilized membrane proteins from rat cerebral cortex in affinity interaction GST pull-down assays. Immunoblotting analysis with an anti-hSlo antibody detected a protein of ∼130 kDa corresponding to rSlo channel (Figure 5A). To confirm specific association in brain lysate, we generated specific antibodies against full-length rANKRA protein. We used affinity-purified anti-rANKRA antibodies for characterization of rANKRA in vivo. In coimmunoprecipitation experiments on lysates of rat cerebral cortex, rANKRA antibodies coprecipitated rSlo (Figure 5B). These results consistently show that rANKRA associates with rSlo in native tissue.
Figure 5.
rANKRA associates and colocalizes with BKCa channel in vivo. (A) Affinity pull-down of rSlo protein from rat cerebral cortex tissue. Purified GST::rANKRA or GST protein immobilized on glutathione Sepharose 4B resin (bed volume, 20 μl) was incubated with 1 mg of membraneenriched fraction of rat cerebral cortical lysate and precipitated. After resolving proteins in 6% SDS-PAGE, protein bands were probed with goat anti-mouse IgG AP-conjugated antibody (Santa Cruz Biotechnology) after the treatment of mouse anti-hSlo antibody. Slo-immunoreactive bands were selectively isolated with GST::rANKRA compared with Sepharose resin only and GST protein (ppt). Sup represents 1/10 fraction of pull-down reaction. Relative positions of molecular standard are indicated (kilodaltons). (B) Coimmunoprecipitation of rSlo protein with rANKRA in brain lysate. Each of antibody against hSlo (1 μg), GFP (2.5 μg), rANKRA (2.5 μg), or preimmune serum (2 μl) was added to membraneenriched fraction of rat brain lysate (500 μg for anti-Slo and 1 mg for other antibodies) precleared with protein A/G resin. After resolving precipitated protein–antibody complexes in 8% SDS-PAGE, Slo-immunoreactive bands were probed with goat anti-mouse IgG HRP-conjugated antibody following α-hSlo antibody. Input corresponds to 1/10 fraction of immunoprecipitant. HC, IgG heavy chain. (C–E) ANKRA colocalizes with BKCa channel in cultured neurons. Mature rat hippocampal neurons (21 DIV) were double labeled for rSlo (green, C–C2) and for rANKRA (red, D–D2). Punctated colocalization patterns of rANKRA with rSlo along the axons were shown (yellow, E–E2). Boxed regions in E were magnified for better visualization of colocalization (C-1, D-1 and E-1; C-2, D-2 and E-2). Bar (C–E), 40 μm.
We then determined the subcellular location of rANKRA and its colocalization with rSlo in cultured hippocampal neurons (Figure 5, C–E). rANKRA-immunoreactivities were widely distributed in a punctated pattern along the neurites. Punctated structures were well colocalized with that of Slo-immunoreactivities in merged images (Figure 5E, E-1 and E-2). Collectively, we suggest that the endogenous ANKRA protein could interact with Slo channel in the isolated tissue and colocalize in culture neurons.
Surface Expression and Steady-State Activation Characteristics of rSlo Are Not Affected by the Coexpression of rANKRA
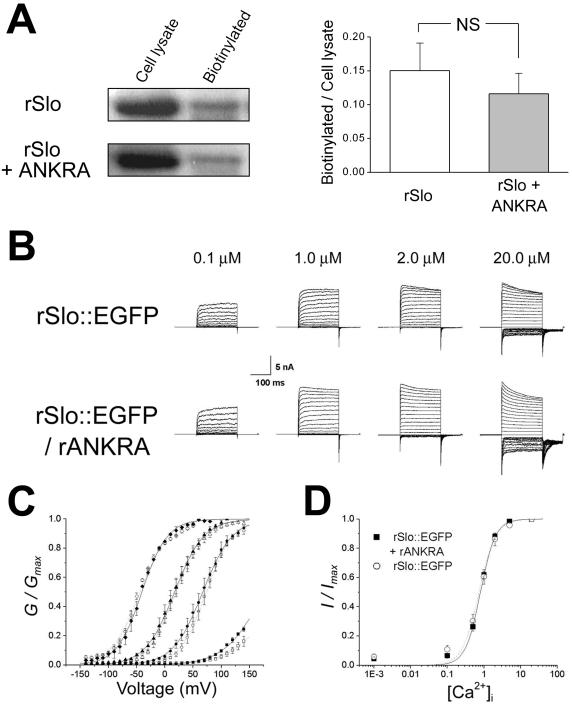
We also asked whether rANKRA affects the surface expression of rSlo protein. It is a pertinent question because the trapped rSlo proteins detected somewhat strongly in the intracellular compartments, such as seen in Figure 4E, could be due to the coexpression of rANKRA. We quantified the expression of rSlo in the absence and presence of rANKRA by using surface biotinylation experiments. Because there are four primary amines exposed to the extracellular space in rSlo channel, we used an amine-attacking biotinylation reagent, Sulfo-NHS-LC-Biotin, on both rslo-transfected and rslo/rANKRA-cotransfected CHO-K1 cells. The experimental results showed that the major portions of rSlo channels were found inside the cells independent of the expression of rANKRA protein (Figure 6A, left). The coexpression of rANKRA protein did not significantly affect the level of surface expression for rSlo channel proteins and the mean values of biotinylation faction were 0.15 and 0.12 for rSlo alone and rSlo with rANKRA, respectively (Figure 6B, right) (p >> 0.05, Student's t test). It is somewhat puzzling that >80% of translated rSlo channels are found in intracellular compartments. To investigate whether these observations are specific to expression system, we conducted surface biotinylation experiment on a different cell, HEK 293. The mean values of surface expression fractions were estimated as 0.14 for rSlo alone and 0.15 for rSlo with rANKRA in HEK 293 cells (our unpublished data). Thus, it might be concluded that the low level of surface expression is general nature in heterologous expression.
Figure 6.
Surface-expression and steady-state activation characteristics of rSlo were not affected by the coexpression of rANKRA. (A) Effects of rANKRA on the surface expression of rSlo channel. Cell lysates containing 60 μg of proteins and biotinylated proteins from equal amount of cell lysates were loaded and monitored using α-hSlo antibody (left). Quantification of membrane surface expression of rSlo. The biotinylated fractions of rSlo proteins were quantified by dividing their densitometric intensities of rSlo bands by those of total lysates. Each value represents the mean ± SE of three experiments from different cell batches (right). None of value differs from each other at the p < 0.05 level (paired Student's t test). (B) Representative macroscopic currents of rSlo::EGFP and rSlo::EGFP/rANKRA recorded in symmetrical 120 mM K+ solutions containing free [Ca2+]i of 0.1, 1.0, 2.0, and 20.0 μM, as indicated. For ionic currents, the potential was held at 0 mV, stepped to test potential ranging from –140 to +140 mV in 10-mV increments, and then stepped down to –100 mV to measure tail currents. Each current traces represent an average of three records in succession. (C) G-V relationships for rSlo::EGFP (open symbols) and rSlo::EGFP/rANKRA (closed symbols) were not changed significantly. Each symbol represents the value obtained from four different [Ca2+]i; 0.1 μM (square), 1.0 μM (circle), 2.0 μM (triangle), and 20.0 μM (diamond). Conductances (G) were measured at tail currents 1 ms after a tail step pulse and normalized by observed maximum conductances (Gmax). Data points were plotted the mean from four different patches for rSlo::EGFP or six for rSlo::EGFP/rANKRA. The lines in G-V represents the best fit of data points to Boltzmann equation G = Gmax/{1 + exp[(–V + V50) x zF/RT]}, where V50 is the half-activation voltage, z is the gating valence, F is Faraday's constant, R is the gas constant, and T is temperature (rSlo::EGFP, dotted line; rSlo::EGFP/rANKRA, solid line). (D) Calcium-dependent activation characteristics were not altered by the expression of rANKRA. Ca2+-dependent activation of rSlo::EGFP with or without rANKRA protein measured at +80 mV. Tail currents normalized by the maximum response evoked by 20 μM Ca2+ were plotted as a function of intracellular calcium concentration, and the data points were fitted with Hill equation, I = Imax ·[Ca 2+] in/(Kd App + [Ca2+]in). Each data point represents an average of several independent data sets (rSlo::EGFP, N = 4 and rSlo::EGFP/rANKRA, N = 5); error bars indicate SEM.
To investigate the effects of rANKRA interaction on the functional activity of rSlo channels, we characterized the channel currents evoked by rSlo in the absence or presence of rANKRA proteins. CHO-K1 cells were transiently transfected with rslo::EGFP or rslo::EGFP/rANKRA, and ionic currents from excised membrane patches were recorded in inside-out configuration after 1–3 d of transfection. Only those patches expressing currents greater than 4 nA at 100 mV were selected for analysis to avoid the fluctuation of ionic currents especially at low Ca2+. Representative current traces are shown for rSlo::EGFP and rSlo::EGFP/rANKRA (Figure 6B). To obtain relative conductance (G/Gmax) at each test potential, the tail currents at 1 ms after –100 mV tail pulse were normalized by maximum tail current. Then, G/Gmax values were plotted against test potentials for rSlo::EGFP and rSlo::EGFP/rANKRA (Figure 6C). The half-activation voltages (V1/2) for rSlo::EGFP channels were obtained from fitting data points to Boltzmann equation and determined as 175 ± 1.6 mV [at 0.1 μM intracellular calcium concentration ([Ca2+]i)], 67 ± 0.3 mV (at 1.0 μM [Ca2+]i), 17 ± 0.4 mV (at 2.0 μM [Ca2+]i), and –41 ± 0.6 mV (at 20 μM [Ca2+]i). V1/2 values for rSlo::EGFP/rANKRA were 204 ± 4.4 mV (at 0.1 μM [Ca2+]i), 73 ± 0.4 mV (at 1.0 μM [Ca2+]i), 19 ± 0.7 mV (at 2.0 μM [Ca2+]i), and –44 ± 1.1 mV (at 20 μM [Ca2+]i). Values for 0.1 μM calcium were extrapolated from fitted curve. Although conductance-voltage (G-V) relationships were shifted rightward at submicromolar concentration of calcium, e.g., ΔV1/2 of + 28.4 mV at 0.1 μM [Ca2+]i, the effect of rANKRA protein on the half-activation voltages became insignificant at micromolar Ca2+, e.g., V1/2 of + 6.4 mV at 1 μM. Steady-state activation profiles for different calcium concentration at 80 mV also were analyzed. Each value was fitted by Hill equation. Calculated Hill coefficients for rSlo::EGFP (1.92) and rSlo::EGFP/rANKRA (2.13) and apparent affinities were not much different to each other (Figure 6D).
Coexpression of rANKRA Modulates Gating Kinetics of rSlo Channels
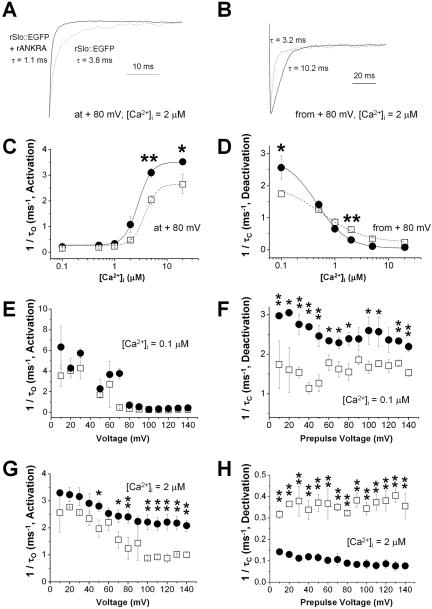
Although the steady-state activation characteristics (voltage- and Ca2+-dependent activation) of rSlo channel were not altered significantly by the coexpression of rANKRA, the effects of rANKRA were readily observed in opening and closing of macroscopic currents evoked by rSlo channels (Figure 6B). In the Figure 7, A and B, the trajectories of current activation and deactivation were compared between rSlo::EGFP (dotted line) and rSlo::EGFP/rANKRA (solid line). To understand quantitative effects of rANKRA on macroscopic gating characteristics of rSlo, we estimated the activation and deactivation time constants (τO and τC) by fitting each current trace to the single-exponential function. The activation rates (1/τO) of rSlo (open squares) and rSlo/rANKRA (closed circles) determined using +80-mV voltage step were plotted against intracellular calcium concentration (Figure 7A). As the [Ca2+]i increased, both rSlo::EGFP and rSlo::EGFP/rANKRA are activated faster. The effects of rANKRA proteins on the channel activation were highly dependent on intracellular Ca2+ concentration (Figure 7C). For the comparison at fixed calcium concentrations, the activation rates were plotted as function of test voltages. At submicromolar [Ca2+]i, no significant differences were found for activation rates between rSlo and rSlo/rANKRA (Figure 7E). However, at micromolar [Ca2+]i, increased activation rates by rANKRA are evident over all tested voltages (Figure 7G).
Figure 7.
Effects of rANKRA on activation and deactivation kinetics of macroscopic rSlo currents. (A) Exemplar activation currents of rSlo::EGFP in the absence (dotted line) and presence of rANKRA (solid line). Ionic currents were activated by a step pulse +80 mV from a holding potential of 0 mV in the presence of 2 μM intracellular Ca2+. Activated currents were normalized with their steadystate maximum currents for comparison. (B) Exemplar deactivation currents of rSlo::EGFP in the absence (dotted line) and presence of rANKRA (solid line). Ionic currents were deactivated by a step to –100 mV from a prepulse potential of +80 mV in the presence of 2 μM Ca2+ and the evoked tail currents were normalized with their maximum values. Activation, 1/τO, (C) and deactivation, 1/τC, (D) rates of rSlo::EGFP (open square and dotted line) and rSlo::EGFP/rANKRA (filled circle and solid line) were plotted as a function of [Ca2+]i. Time constants for activation (τO) and deactivation (τC) of macroscopic currents were obtained by fitting the individual current to single exponential function. Voltage dependence of activation rate was shown for two different [Ca2+]i, 0.1 μM (E) and 2 μM (G). Deactivation rate was plotted as a function of prepulse voltage. The biphasic effects of the rANKRA on the deactivation rate at different [Ca2+]i are shown for 0.1 μM (F) and 2 μM (H). Each data point represents an average of several independent data sets (rSlo::EGFP, N = 4 and rSlo::EGFP/rANKRA, N = 5); error bars indicate SEM. Compared values differed from each other at p < 0.05 level (*) or p < 0.01 level (**) are marked (paired Student's t test).
The effects of rANKRA on deactivation of macroscopic rSlo currents are more intriguing because the protein modulates the gating kinetics in a biphasic manner (Figure 7D). Although the deactivation rates of rSlo::EGFP/rANKRA are faster than that of rSlo::EGFP at submicromolar calcium concentrations, this effect is reversed at micromolar calcium range. This reversed effect of rANKRA on the deactivation kinetics of rSlo channel is more clearly seen in deactivation rate versus prepulse plots shown in Figure 7F (0.1 μM Ca2+) and H (2 μM Ca2+).
In summary, although the coexpression of rANKRA shows minor effect on the steady-state activation of rSlo::EGFP channel, the activation and deactivation kinetics of macroscopic channel currents are greatly affected and the gating effects are highly sensitive to the concentration of intracellular Ca2+.
DISCUSSION
In this report, we described the identification of a new interacting protein for BKCa channel, and its effects on the expression and function of the channel. To identify proteins directly binding to the channel and modulating its function, we launched a large-scale screening of rat brain cDNA library by using C-terminal region of rat BKCa channel α subunit rSlo as the baits. In designing the baits for yeast two-hybrid screening, we should consider domain architecture and possible structures of bulky cytosolic part of Slo. We initially generated three distinct bait constructs covering whole cytoplasmic domain of rSlo, rSlo-A of amino acids 394–655, rSlo-B of 656–950, and rSlo-C of 951-1210, based on the predicted secondary structures. Among them, rSlo-C covering from “Ca2+-Bowl” to the C-terminal end gave pertinent results from library screening. Recently, the cytoplasmic C terminus of BKCa channel was suggested to form a gating ring composed of eight RCK domains in analogy of the crystal structure of bacterial potassium channel MthK (Jiang et al., 2002). It is conceivable that the region outside of the tight gating structure of mammalian BKCa channel might be a good target for the binding of regulatory or auxiliary proteins and that the regions corresponding to two RCK domains (rSlo-A and rSlo-B) might gave much less initial candidates in this screening.
The identified rSlo-binding partner is 313 amino acids in length and the rat ortholog of ANKRA originally reported as a megalin-interacting protein in mouse kidney (Rader et al., 2000). The ankyrin-repeat domain is composed of ∼33 residues and has a general role as a molecular adapter. The motif has been recognized in >400 different proteins (Sedgwick and Smerdon, 1999). According to Rader et al. (2000), mANKRA interacts with a proline-rich motif of the megalin protein, a member of the low-density lipoprotein receptor superfamily (Farquhar et al., 1995), via ankyrin-repeats and C-terminal end (Rader et al., 2000). The anion exchanger, Na+-K+ ATPase, a voltage-dependent Na+ channel, a Na+/Ca2+-exchanger, Ca2+-release channel, and adhesion molecules have been reported to interact with proteins containing ankyrin domains (Bennett and Baines, 2001). We investigated the interaction site of rANKRA protein with rSlo channel by using several deletion constructs. Such experiments showed that N2 region including putative coiled-coil segment, not the C-terminal ankyrin repeats, of rANKRA is critical for interaction with the rSlo channel and rSlo C-terminal end, ∼90 amino acids in length, is required for direct binding (Figures 1C and 3). Interestingly, the C-terminal domain of rSlo localized to interact with rANKRA is conserved only among vertebrate orthologues and ANKRA proteins are not found in invertebrate genomes. The N2 region in rANKRA protein contains a stretch of amino acid well aligned with a part of switch region of G protein α-subunit, with which α-subunit interact with βγ subunit of trimetric G protein (Lambright et al., 1996). To address detailed binding mode between ANKRA and BKCa channel, more defined binding motifs within N2 on binding to rSlo should be addressed by mutagenesis and biochemical studies in the future.
We confirmed direct interaction between rSlo and rANKRA in several different ways both in vitro and in vivo. Initial screening and localization of binding domain by using yeast two-hybrid assay and in vitro binding assay unequivocally showed that the interaction between the rSlo C terminus and rANKRA (Figures 1 and 3). Reciprocal coimmunoprecipitation assays clearly indicated that the direct interaction between the two proteins in transfected cells (Figure 4, A and B). Subcellular location visualized by immunofluorescence also showed the colocalization of rSlo and rANKRA at the membrane surface in COS-7 cells (Figure 4, C–E). In addition to the evidences for protein–protein interaction in vitro and overexpression system, intrinsic association of rANKRA with rSlo was revealed by affinity pull-down assay and coimmunoprecipitation performed on rat brain lysate (Figure 5, A and B). Moreover, colocalization of Slo and ANKRA was clearly visualized in a punctated pattern along the neurites in mature hippocampal neurons by immunocytochemistry (21 DIV) (Figure 5, C–E). Localized distribution of rSlo in neurites is well matched to the concentrated mRNA localization in axonal compartment of hippocampus revealed by in situ hybridization (Knaus et al., 1996). This punctated pattern of colocalization strongly suggests that rANKRA be an authentic modulator or a scaffolding partner for BKCa channels in nervous system.
In addition, specific interactions between the two proteins were strongly supported by the functional effects of rANKRA on the activity of rSlo. We showed that the macroscopic gating kinetics of BKCa channel was markedly affected by coexpression with rANKRA (Figure 7). rANKRA not only changed in kinetic values of both activation and deactivation of rSlo but also affected them in a Ca2+-dependent manner. rANKRA increased the activation rate of macroscopic rSlo current when the intracellular Ca2+ raised above micromolar range. The effects of rANKRA were biphasic for the deactivation of rSlo: the increased deactivation rate under micromolar Ca2+ and the decreased rate above. As a result, the deactivation rate of rSlo decrease more than eightfold when intracellular calcium increases from 0.1 to 2 μM at 80 mV in the presence of rANKRA, whereas the identical condition give rise to the rate decrease of only 1.7-fold in the absence. Although rANKRA generally suppresses the activity of rSlo at low intracellular Ca2+, it potentiates the activity of rSlo by opening more quickly and closing more slowly when Ca2+ concentration is raised above micromolar level. Because the submembrane concentration of local calcium increases up to the tens of micromolar by bursts of action potential (Hille, 2001), ANKRA can magnify the effect of Ca2+ on the activities of BKCa channel and influence the excitability of bursting neurons by affecting the fast phase of afterhyperpolarization. This speculation is partly supported by the intimate colocalization patterns in neurites (Figure 5E). The effect of overexpressed ANKRA proteins on the gating properties of BKCa channel is reproducible in heterologous expression system despite of ubiquitous expression of ANKRA protein. These results could be generated if endogenous ANKRA proteins are not enough to occupy and modulate overexpressed BKCa channels. However, it remains to be investigated whether ANKRA constitutively interacts with BKCa channel as an auxiliary protein just like subunits or binds dynamically depending on the cellular environment. This question can be investigated using the targeted knockdown of endogenous form of ANKRA by siRNA.
Recently, multiple sequences in the carboxy terminus of Slo channel were suggested to be involved in surface expression and apical localization (Kwon and Guggino, 2004). A motif of six amino acid residues, DLIFCL, is required to exit from the endoplasmic reticulum, and another identified sequence, NAGQSRA, contains information necessary for apical sorting of BKCa channels. The determined sequences are located within Slo-CC domain identified as interacting domain with ANKRA. Interestingly, ANKRA was found to be enriched in the apical membrane fractions of rat kidney proximal tubule (Rader et al., 2000). Similarly, several experimental observations have suggested that spectrin and ankyrin have a role either in the formation of epithelia cell polarity or in maintenance of this polarity (Yeaman et al., 1999). Although cell surface expression level of rSlo channels was not significantly changed by coexpression of ANKRA in transfected cells (Figure 6, A and B), it might be possible that interaction between BKCa and ANKRA is important for the cellular sorting of BKCa channels in polarized cells rather than general surface expression. This possible role of ANKRA remained to be addressed in highly polarized cell with the information on the more defined interaction motifs in both proteins.
Knowing the fact that ANKRA expresses ubiquitously in all tissues tested and its mouse ortholog binds to another membrane protein in the kidney, we wonder whether this protein has more general roles in cellular physiology such as assembly and targeting of membrane proteins. ANKRA may act a multivalent protein-protein interacting adapter to the distinct membrane proteins and recruit specific modulating proteins near the target membrane proteins, including ion channels and membrane receptors. This idea is somewhat supported by the experimental evidences that ANKRA and its ortholog use different regions of the protein to interact their different partners, e.g., C-terminal ankyrin-repeat for megalin (Rader et al., 2000) and the Gα-like region for BKCa channel. Thus, it could even be imagined that ANKRA proteins recruit various signal mediators including membrane scaffolds, anchoring proteins, and effector molecules via ankyrin repeat domains to the BKCa channel and form a large cluster.
In summary, we identified ANKRA, a cellular protein containing ankyrin repeats, as an interacting protein for BKCa channel. The protein modulates the gating kinetics of BKCa channel in a Ca2+- and voltage-dependent manner in a heterologous system. Thus, we suggest that this protein, ANKRA, is a binding partner for BKCa channel in vivo and a potential modulator of cellular physiology such as neuroexcitability in central nervous system.
Acknowledgments
The authors are grateful to the members of Laboratory of Molecular Neurobiology at GIST for valuable comments and timely help throughout the work, to Dr. W. K. Song for providing facilities for neuron culture and imaging, and to C. Miller at Brandeis University for critical reading of the manuscript. This research was supported by research grants from the Ministry of Science and Technology of Korea (Korean Systems Biology Grant, M1–0309-00–0006 and 21C Frontier, 03K2201–00320) to C. –S. Park.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-06-0537) on December 22, 2004.
References
- Alioua, A., Mahajan, A., Nishimaru, K., Zarei, M. M., Stefani, E., and Toro, L. (2002). Coupling of c-Src to large conductance voltage- and Ca2+-activated K+ channels as a new mechanism of agonist-induced vasoconstriction. Proc. Natl. Acad. Sci. USA 99, 14560-14565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alioua, A., Tanaka, Y., Wallner, M., Hofmann, F., Ruth, P., Meera, P., and Toro, L. (1998). The large conductance, voltage-dependent, and calcium-sensitive K+ channel, Hslo, is a target of cGMP-dependent protein kinase phosphorylation in vivo. J. Biol. Chem. 273, 32950-32956. [DOI] [PubMed] [Google Scholar]
- Behrens, R., Nolting, A., Reimann, F., Schwarz, M., Waldschutz, R., and Pongs, O. (2000). hKCNMB3 and hKCNMB4, cloning and characterization of two members of the large-conductance calcium-activated potassium channel beta subunit family. FEBS Lett. 474, 99-106. [DOI] [PubMed] [Google Scholar]
- Bennett, V., and Baines, A. J. (2001). Spectrin and ankyrin-based pathways: metazoan inventions for integrating cells into tissues. Physiol. Rev. 81, 1353-1392. [DOI] [PubMed] [Google Scholar]
- Brenner, R., Jegla, T. J., Wickenden, A., Liu, Y., and Aldrich, R. W. (2000). Cloning and functional characterization of novel large conductance calcium-activated potassium channel beta subunits, hKCNMB3 and hKCNMB4. J. Biol. Chem. 275, 6453-6461. [DOI] [PubMed] [Google Scholar]
- Butler, A., Tsunoda, S., McCobb, D. P., Wei, A., and Salkoff, L. (1993). mSlo, a complex mouse gene encoding “maxi” calcium-activated potassium channels. Science 261, 221-224. [DOI] [PubMed] [Google Scholar]
- Chang, S., and De Camilli, P. (2001). Glutamate regulates actin-based motility in axonal filopodia. Nat. Neurosci. 4, 787-793. [DOI] [PubMed] [Google Scholar]
- Dworetzky, S. I., Boissard, C. G., Lum-Ragan, J. T., McKay, M. C., PostMunson, D. J., Trojnacki, J. T., Chang, C. P., and Gribkoff, V. K. (1996). Phenotypic alteration of a human BK (hSlo) channel by hSlobeta subunit coexpression: changes in blocker sensitivity, activation/relaxation and inactivation kinetics, and protein kinase A. modulation. J. Neurosci. 16, 4543-4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farquhar, M. G., Saito, A., Kerjaschki, D., and Orlando, R. A. (1995). The Heymann nephritis antigenic complex: megalin (gp330) and RAP. J. Am. Soc. Nephrol. 6, 35-47. [DOI] [PubMed] [Google Scholar]
- Fox, A. J., Barnes, P. J., Venkatesan, P., and Belvisi, M. G. (1997). Activation of large conductance potassium channels inhibits the afferent and efferent function of airway sensory nerves in the guinea pig. J. Clin. Investig. 99, 513-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Calvo, M., Knaus, H. G., McManus, O. B., Giangiacomo, K. M., Kaczorowski, G. J., and Garcia, M. L. (1994). Purification and reconstitution of the high-conductance, calcium-activated potassium channel from tracheal smooth muscle. J. Biol. Chem. 269, 676-682. [PubMed] [Google Scholar]
- Ha, T. S., Heo, M. S., and Park, C. S. (2004). Functional effects of auxiliary beta4-subunit on rat large-conductance Ca2+-activated K+ channel. Biophys. J. 86, 2871-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha, T. S., Jeong, S. Y., Cho, S. W., Jeon, H., Roh, G. S., Choi, W. S., and Park, C. S. (2000). Functional characteristics of two BKCa channel variants differentially expressed in rat brain tissues. Eur. J. Biochem. 267, 910-918. [DOI] [PubMed] [Google Scholar]
- Hille, B. (2001). Ion Channels of Excitable Membranes, Sunderland, MA: Sinauer Association.
- Hu, H., et al. (2001). Presynaptic Ca2+-activated K+ channels in glutamatergic hippocampal terminals and their role in spike repolarization and regulation of transmitter release. J. Neurosci. 21, 9585-9597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa, N. P., and Hudspeth, A. J. (1994). Clustering of Ca2+ channels and Ca2+-activated K+ channels at fluorescently labeled presynaptic active zones of hair cells. Proc. Natl. Acad. Sci. USA 91, 7578-7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, Y., Lee, A., Chen, J., Cadene, M., Chait, B. T., and MacKinnon, R. (2002). Crystal structure and mechanism of a calcium-gated potassium channel. Nature 417, 515-522. [DOI] [PubMed] [Google Scholar]
- Knaus, H. G., Schwarzer, C., Koch, R. O., Eberhart, A., Kaczorowski, G. J., Glossmann, H., Wunder, F., Pongs, O., Garcia, M. L., and Sperk, G. (1996). Distribution of high-conductance Ca2+-activated K+ channels in rat brain: targeting to axons and nerve terminals. J. Neurosci. 16, 955-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, S. H., and Guggino, W. B. (2004). Multiple sequences in the C terminus of MaxiK channels are involved in expression, movement to the cell surface, and apical localization. Proc. Natl. Acad. Sci. USA 101, 15237-15242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambright, D. G., Sondek, J., Bohm, A., Skiba, N. P., Hamm, H. E., and Sigler, P. B. (1996). The 2.0 A crystal structure of a heterotrimeric G protein. Nature 379, 311-319. [DOI] [PubMed] [Google Scholar]
- Latorre, R., Oberhauser, A., Labarca, P., and Alvarez, O. (1989). Varieties of calcium-activated potassium channels. Annu. Rev. Physiol. 51, 385-399. [DOI] [PubMed] [Google Scholar]
- Lesage, F., Hibino, H., and Hudspeth, A. J. (2004). Association of beta-catenin with the alpha-subunit of neuronal large-conductance Ca2+-activated K+ channels. Proc. Natl. Acad. Sci. USA 101, 671-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liman, E. R., Tytgat, J., and Hess, P. (1992). Subunit stoichiometry of a mammalian K+ channel determined by construction of multimeric cDNAs. Neuron 9, 861-871. [DOI] [PubMed] [Google Scholar]
- Ling, S., Sheng, J. Z., Braun, J. E., and Braun, A. P. (2003). Syntaxin 1A co-associates with native rat brain and cloned large conductance, calcium-activated potassium channels in situ. J. Physiol. 553, 65-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling, S., Woronuk, G., Sy, L., Lev, S., and Braun, A. P. (2000). Enhanced activity of a large conductance, calcium-sensitive K+ channel in the presence of Src tyrosine kinase. J. Biol. Chem. 275, 30683-30689. [DOI] [PubMed] [Google Scholar]
- Lingle, C. J., Solaro, C. R., Prakriya, M., and Ding, J. P. (1996). Calcium-activated potassium channels in adrenal chromaffin cells. Ion. Channels 4, 261-301. [DOI] [PubMed] [Google Scholar]
- Liu, G., Shi, J., Yang, L., Cao, L., Park, S. M., Cui, J., and Marx, S. O. (2004). Assembly of a Ca2+-dependent BK channel signaling complex by binding to beta2 adrenergic receptor. EMBO J. 23, 2196-2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas, A., Van Dyke, M., and Stock, J. (1991). Predicting coiled coils from protein sequences. Science 252, 1162-1164. [DOI] [PubMed] [Google Scholar]
- Marrion, N. V., and Tavalin, S. J. (1998). Selective activation of Ca2+-activated K+ channels by co-localized Ca2+ channels in hippocampal neurons. Nature 395, 900-905. [DOI] [PubMed] [Google Scholar]
- Martell, A. E., and Smith, R. M. (1974). Critical Stability Constant, New York: Plenum.
- Meera, P., Wallner, M., and Toro, L. (2000). A neuronal beta subunit (KC-NMB4) makes the large conductance, voltage- and Ca2+-activated K+ channel resistant to charybdotoxin and iberiotoxin. Proc. Natl. Acad. Sci. USA 97, 5562-5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers, M., Yang, J., and Stampe, P. (1999). Visualization and functional analysis of a maxi-K channel (mSlo) fused to green fluorescent protein (GFP). Electron. J. Biotechnol. 2, 140-151. [Google Scholar]
- Nadal, M. S., et al. (2003). The CD26-related dipeptidyl aminopeptidase-like protein DPPX is a critical component of neuronal A-type K+ channels. Neuron 37, 449-461. [DOI] [PubMed] [Google Scholar]
- Nelson, M. T., Cheng, H., Rubart, M., Santana, L. F., Bonev, A. D., Knot, H. J., and Lederer, W. J. (1995). Relaxation of arterial smooth muscle by calcium sparks. Science 270, 633-637. [DOI] [PubMed] [Google Scholar]
- Orio, P., Rojas, P., Ferreira, G., and Latorre, R. (2002). New disguises for an old channel: MaxiK channel beta-subunits. News Physiol. Sci. 17, 156-161. [DOI] [PubMed] [Google Scholar]
- Rader, K., Orlando, R. A., Lou, X., and Farquhar, M. G. (2000). Characterization of ANKRA, a novel ankyrin repeat protein that interacts with the cytoplasmic domain of megalin. J. Am. Soc. Nephrol. 11, 2167-2178. [DOI] [PubMed] [Google Scholar]
- Rezzonico, R., et al. (2003). Focal adhesion kinase pp125FAK interacts with the large conductance calcium-activated hSlo potassium channel in human osteoblasts: potential role in mechanotransduction. J. Bone Miner Res. 18, 1863-1871. [DOI] [PubMed] [Google Scholar]
- Rezzonico, R., Schmid-Alliana, A., Romey, G., Bourget-Ponzio, I., Breuil, V., Breittmayer, V., Tartare-Deckert, S., Rossi, B., and Schmid-Antomarchi, H. (2002). Prostaglandin E2 induces interaction between hSlo potassium channel and Syk tyrosine kinase in osteosarcoma cells. J. Bone Miner Res. 17, 869-878. [DOI] [PubMed] [Google Scholar]
- Roberts, W. M., Jacobs, R. A., and Hudspeth, A. J. (1990). Colocalization of ion channels involved in frequency selectivity and synaptic transmission at presynaptic active zones of hair cells. J. Neurosci. 10, 3664-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robitaille, R., Adler, E. M., and Charlton, M. P. (1993). Calcium channels and calcium-gated potassium channels at the frog neuromuscular junction. J. Physiol. 87, 15-24. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., and Russell, D. W. (2001). Molecular Cloning: A Laboratory Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Sansom, S. C., Stockand, J. D., Hall, D., and Williams, B. (1997). Regulation of large calcium-activated potassium channels by protein phosphatase 2A. J. Biol. Chem. 272, 9902-9906. [DOI] [PubMed] [Google Scholar]
- Schopperle, W. M., Holmqvist, M. H., Zhou, Y., Wang, J., Wang, Z., Griffith, L. C., Keselman, I., Kusinitz, F., Dagan, D., and Levitan, I. B. (1998). Slob, a novel protein that interacts with the Slowpoke calcium-dependent potassium channel. Neuron 20, 565-573. [DOI] [PubMed] [Google Scholar]
- Sedgwick, S. G., and Smerdon, S. J. (1999). The ankyrin repeat: a diversity of interactions on a common structural framework. Trends Biochem. Sci. 24, 311-316. [DOI] [PubMed] [Google Scholar]
- Sun, X. P., Schlichter, L. C., and Stanley, E. F. (1999). Single-channel properties of BK-type calcium-activated potassium channels at a cholinergic presynaptic nerve terminal. J. Physiol. 518, 639-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, L., Coghill, L. S., MacDonald, S. H., Armstrong, D. L., and Shipston, M. J. (2003). Leucine zipper domain targets cAMP-dependent protein kinase to mammalian BK channels. J. Biol. Chem. 278, 8669-8677. [DOI] [PubMed] [Google Scholar]
- Toro, L., Wallner, M., Meera, P., and Tanaka, Y. (1998). Maxi-KCa, a unique member of the voltage-gated K channel superfamily. News Physiol. Sci. 13, 112-117. [DOI] [PubMed] [Google Scholar]
- Wallner, M., Meera, P., and Toro, L. (1999). Molecular basis of fast inactivation in voltage and Ca2+-activated K+ channels: a transmembrane beta-subunit homolog. Proc. Natl. Acad. Sci. USA 96, 4137-4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Zhou, Y., Wen, H., and Levitan, I. B. (1999). Simultaneous binding of two protein kinases to a calcium-dependent potassium channel. J. Neurosci. 19, RC4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiger, T. M., et al. (2000). A novel nervous system beta subunit that down-regulates human large conductance calcium-dependent potassium channels. J. Neurosci. 20, 3563-3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, X., Hirschberg, B., Smolik, S., Forte, M., and Adelman, J. P. (1998). dSLo interacting protein 1, a novel protein that interacts with large-conductance calcium-activated potassium channels. J. Neurosci. 18, 2360-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeaman, C., Grindstaff, K. K., and Nelson, W. J. (1999). New perspectives on mechanisms involved in generating epithelial cell polarity. Physiol. Rev. 79, 73-98. [DOI] [PubMed] [Google Scholar]