Significance
In the human organism, guanylate-binding proteins (GBPs) are involved in antimicrobial and antiviral activity, but the mechanism of GBPs’ action remains poorly understood. We have discovered that binding of a substrate molecule, GTP, to the enzyme triggers the release of an aforemasked lipid anchor, which results in GBP polymerization on the one hand and in the attachment of GBPs to lipid membranes on the other. Thus, membrane binding of GBPs competes with protein polymerization and, furthermore, leads to the membrane tethering, which could play a role in a clearance of engulfed pathogens from the cell. Altogether, our findings give deeper insights into GBPs’ molecular mechanism in the course of pathogen response.
Keywords: large GTPases, polymerization, membrane tethering, membrane binding, GBPs
Abstract
Dynamin-like proteins (DLPs) mediate various membrane fusion and fission processes within the cell, which often require the polymerization of DLPs. An IFN-inducible family of DLPs, the guanylate-binding proteins (GBPs), is involved in antimicrobial and antiviral responses within the cell. Human guanylate-binding protein 1 (hGBP1), the founding member of GBPs, is also engaged in the regulation of cell adhesion and migration. Here, we show how the GTPase cycle of farnesylated hGBP1 (hGBP1F) regulates its self-assembly and membrane interaction. Using vesicles of various sizes as a lipid bilayer model, we show GTP-dependent membrane binding of hGBP1F. In addition, we demonstrate nucleotide-dependent tethering ability of hGBP1F. Furthermore, we report nucleotide-dependent polymerization of hGBP1F, which competes with membrane binding of the protein. Our results show that hGBP1F acts as a nucleotide-controlled molecular switch by modulating the accessibility of its farnesyl moiety, which does not require any supportive proteins.
Human guanylate-binding protein 1 (hGBP1) is one of seven human guanylate-binding proteins (GBPs) and it is mostly activated in cells after stimulation with type II interferons (IFN-γ) (1). GBPs belong to the dynamin superfamily of large GTPases, also referred to as dynamin-like proteins (DLPs) (2, 3). Among the DLPs the GBPs are most closely related to atlastins, which mediate fusion of endoplasmic reticulum (ER) membranes (4). GBPs have been reported to be involved in the cellular immune responses against bacterial, viral, and protozoan pathogens (5). The antimicrobial function of GBPs has been linked to the ubiquitination and autophagic destruction of pathogen-containing inclusions, inflammasome activation, and the induction of pyroptosis (6, 7). The protein also shows antitumor and antiproliferative (8) effects and has actin remodeling activity (9). However, how the molecular mechanism of GBPs is linked to these diverse functions within the cell remains poorly understood.
In comparison with other DLPs, hGBP1 has a simpler domain architecture: it consists of the large GTPase (LG) domain at the N terminus and an elongated, purely α-helical moiety. The latter is subdivided in the middle domain and the C-terminal α12/13 domain (2, 10, 11). Patches of negatively and positively charged side chains on the LG and α12/13 domains, respectively, are responsible for a tight intramolecular interaction between these domains (12). Upon GTP hydrolysis, this contact is released and structural rearrangements can occur (13, 14).
In contrast to other GTPases, hGBP1 does not only catalyze the hydrolysis of GTP to GDP, but it is also able to catalyze a further phosphate cleavage step leading to the formation of GMP (15, 16). Whereas hGBP1 binds all three guanine nucleotides with similar low affinity (17), GTP binding results in the formation of homodimers of hGBP1 through an interface located on the LG domains (18). Moreover, the protein exhibits concentration-dependent self-activation for catalysis of GTP hydrolysis (11, 19). By formation of an LG domain dimer, an arginine residue is positioned toward the nucleotide pocket leading to enhancement of the catalytic activity (18). Reaction conditions that impair the formation of dimers, e.g., mutations, dilution to low hGBP1 concentrations, or immobilization in a dense packing, result in much slower GTP turnover and in less production of GMP (11, 19–21).
Most members of the dynamin superfamily are involved in processes of membrane reorganization, such as tubulation, fission, and fusion (10). The remodeling of the membrane requires close contact between the protein and the lipid bilayer. In the case of several GBPs, including hGBP1, this is mediated by isoprenylation of a C-terminal CaaX motif, which enables protein’s association to the membrane (22–24). HGBP1 itself is isoprenylated with a C15 farnesyl lipid moiety and in addition possesses a polybasic region directly adjacent to the CaaX motif of the protein, which could enhance membrane affinity. We were able to show that farnesylated hGBP1 (hGBP1F) interacts with membranes in the presence of GDP and aluminum fluoride (AlFx), which functions as a mimic of the transition state of GTP hydrolysis (25). In IFN-induced cells, hGBP1 is localized in cytosolic puncta, unidentified to date (26, 27). The addition of AlFx induces redistribution of hGBP1 to the Golgi complex in a farnesylation-dependent manner (26, 28). On the other hand, mutation of the contact between the LG domain and the α12/13 domain results in the localization of hGBP1 at the plasma membrane, whereas a protein mutant impaired in binding nucleotides displays a purely cytosolic staining (12, 27). In the case of murine GBPs, localization to the parasitophorous vacuole of Toxoplasma gondii depends on isoprenylation and a functional LG domain (29).
We addressed the mutual regulation of the membrane localization, the nucleotide-dependent oligomerization, and the GTPase activity of hGBP1 by using artificial, protein-free vesicles. Our results reveal that farnesylated hGBP1 can either polymerize into large ordered structures or engage in dynamic interactions with membranes, which results in membrane tethering. Both processes require the transient exposure of the farnesyl moiety to the solvent, which is regulated by GTP binding and GTP hydrolysis. Depletion of GTP or competition with GMP results in the disassembly of hGBP1 polymers and the release from membranes, respectively.
Results
Membrane Binding of hGBP1 Is Farnesylation Dependent and Nucleotide Controlled.
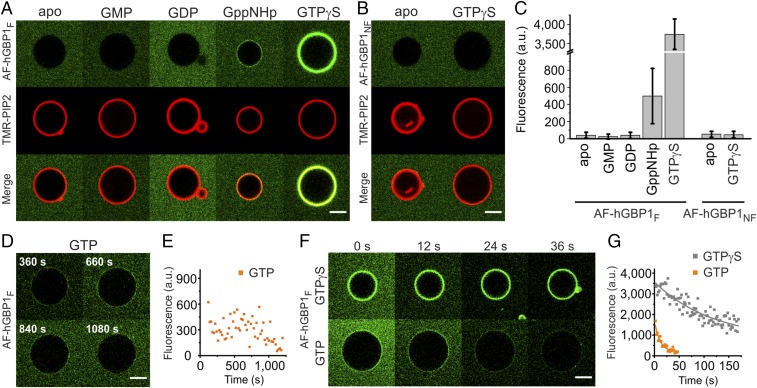
To observe the interaction of hGBP1 with membranes in real time, we investigated the behavior of hGBP1F and nonfarnesylated hGBP1 (hGBP1NF) in the presence of giant unilamellar vesicles (GUVs, 5–50 μm). GUVs were prepared from brain polar lipid (BPL) extract to represent membranes of the phagolysosomal pathway. First, we checked how the binding of different nucleotides might influence the interaction between hGBP1F and vesicles. Therefore, tetramethyl rhodamine-labeled GUVs were incubated with 2.5 μM Alexa Fluor 488-labeled hGBP1F (AF-hGBP1F) either in the absence of nucleotide (apo state) or in the presence of GMP, GDP, or the GTP analogs, GppNHp or GTPγS. No sign of AF-hGBP1F binding to GUVs was observed in the absence of nucleotide and in the presence of either GMP or GDP (Fig. 1 A and C). However, in the presence of GppNHp, a faint protein fluorescence was detectable on the surface of the GUVs (Fig. 1 A and C). Moreover, the presence of GTPγS led to a strong homogenous stain of AF-hGBP1F on the surface of GUVs, indicating that hGBP1 binding to membranes occurs in a GTP-dependent fashion (Fig. 1 A and C). In contrast, AF-hGBP1NF revealed no binding to GUVs—neither in the apo state nor in the presence of GTPγS (Fig. 1 B and C).
Fig. 1.
Interaction of hGBP1 with GUVs. (A) GUVs labeled with 1% TMR-PIP2 (red) in the presence of 2.5 µM AF-hGBP1F (green) and 400 µM corresponding nucleotides after 5 min of incubation. (B) Same as in A, but in the presence of AF-hGBP1NF. (C) Mean fluorescence intensities of AF-hGBP1 binding to GUVs (Methods) for different nucleotides. Statistics include the data for n = 25–80 GUVs under each experimental condition from at least two independent measurements. (D) Time-dependent dissociation of hGBP1F from a GUV in the presence of GTP monitored on a single vesicle. The experiment was started by addition of 1 mM GTP (t = 0 s) into a solution containing 2.5 µM AF-hGBP1F. (E) Numerical representation of D. (F) Time-lapse images of AF-hGBP1F dissociation from GUVs in the presence of 400 µM GTPγS (Upper) or 400 µM GTP (Lower) (see also Movie S1). (G) Numerical representation of F. (Scale bars, 5 µm.) a.u., arbitrary units.
To study the association of AF-hGBP1F with GUVs in the presence of the genuine substrate, we performed a similar set of experiments with GTP at its initial concentration of 1 mM. Due to the ability of hGBP1 to hydrolyze GTP rapidly, we followed the binding of AF-hGBP1F to GUVs over time. These experiments yielded initial binding of AF-hGBP1F to GUVs, which decayed over time due to depletion of GTP (Fig. 1 D and E). The decay was not due to photobleaching, as we used an oxygen scavenging solution in these experiments (Methods). Toward the end of each experiment (i.e., after 15–20 min) protein fluorescence was hardly detectable on the surface of GUVs and the background signal representing free AF-hGBP1F increased (Fig. 1D). We also observed the same behavior of AF-hGBP1F binding to GUVs in the presence of GTP when we monitored a pool of GUVs instead of a single vesicle (Fig. S1A). In this case, we disclosed different patterns of protein distribution on the membrane surface but altogether more than 70% of the total vesicle surface was covered by a faint, homogeneous stain, most likely representing a monolayer of AF-hGBP1F (Fig. S1 B and C–F).
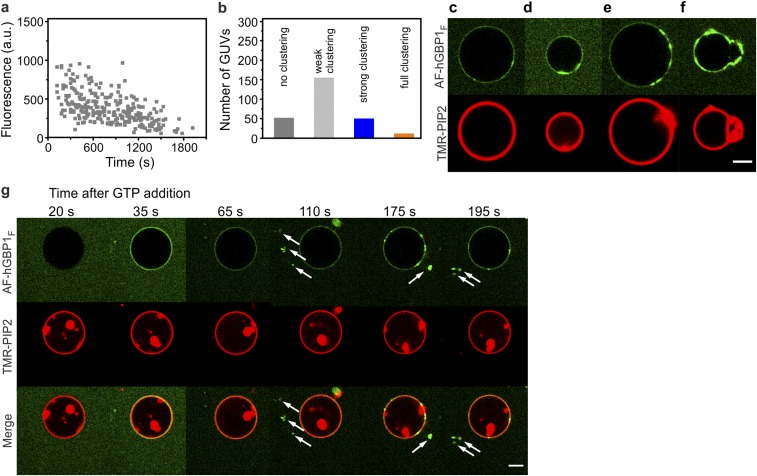
Fig. S1.
Binding of AF-hGBP1F to GUVs in the presence of GTP. (A) Referred to as the GUV pool, all GUVs in the chamber were screened one after another within a time slot of about 30 min. The experiment was started by addition of 1 mM GTP (t = 0 s) into 2.5 µM AF-hGBP1F premixed with GUVs. Time-dependent protein fluorescence was evaluated in homogeneous surface regions of GUVs. (B) Statistics of GUV phenotypes (as depicted in C–F) identified upon GTP-induced binding of AF-hGBP1F to a pool of vesicles. Representative images of AF-hGBP1F-covered GUVs illustrate four different phenotypes: (C) Homogeneous distribution of the protein on the GUV surface, protein amount within clusters was less than 5%. This phenotype accounted for ∼70% of all GUVs. (D) Weak clustering, protein amount within clusters was either tiny or it covered not more than 25% of GUV’s surface. (E) Strong clustering, clusters of AF-hGBP1F covered between 25% and 75% of GUV’s surface, and (F) full clustering, most of GUVs’ surface (>75%) was covered with protein clusters. (Scale bar, 5 µm.) (G) Time lapse of GTP-induced AF-hGBP1F binding to GUVs showed that initial binding was followed by a drop of background fluorescence and appearance of floating protein clusters (only visible in the protein channel of fluorescence and indicated with white arrows). (Scale bar, 5 µm.) a.u., arbitrary units.
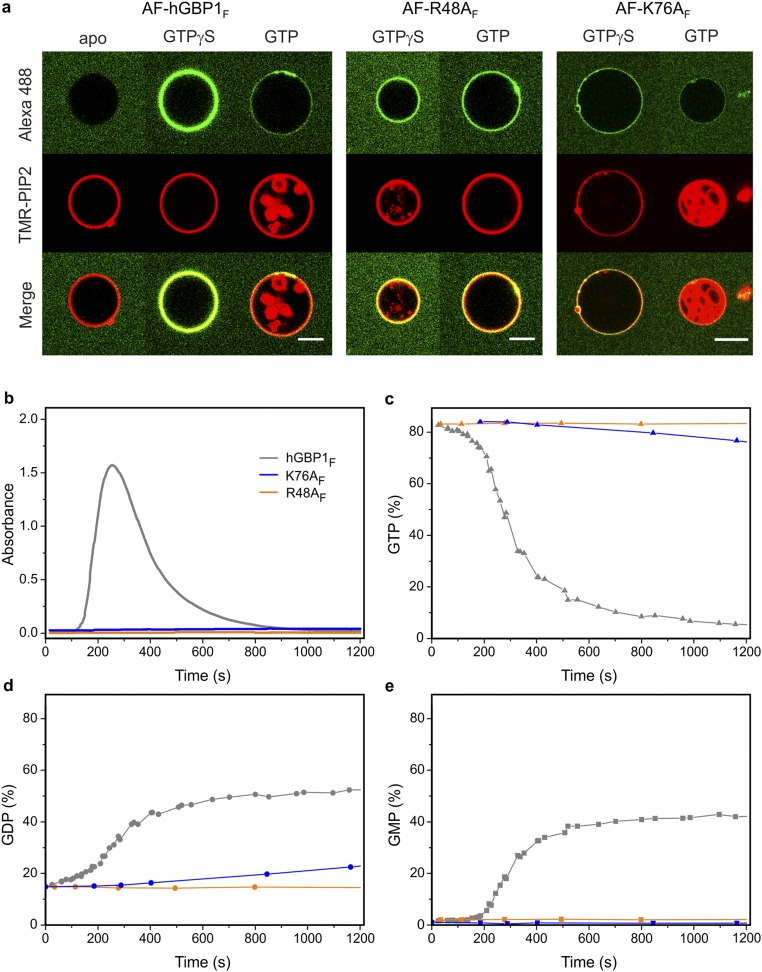
In addition to wild-type protein, we investigated GTPase-deficient and GDPase-deficient mutants of hGBP1 having R48A and K76A mutations (30), respectively, for their ability of membrane association with GUV-binding setup. Both Alexa-fluorescent R48A and K76A farnesylated mutants (AF-R48AF and AF-K76AF, respectively) bound to GUVs in the presence GTPγS and GTP, which was not the case in the absence of the nucleotide (Fig. S2A). Intriguingly, the degree of binding to GUVs of AF-R48AF in the presence of GTP was comparable to the presence of GTPγS, which was not the case for AF-hGBP1F and AF-K76AF (Fig. S2A).
Fig. S2.
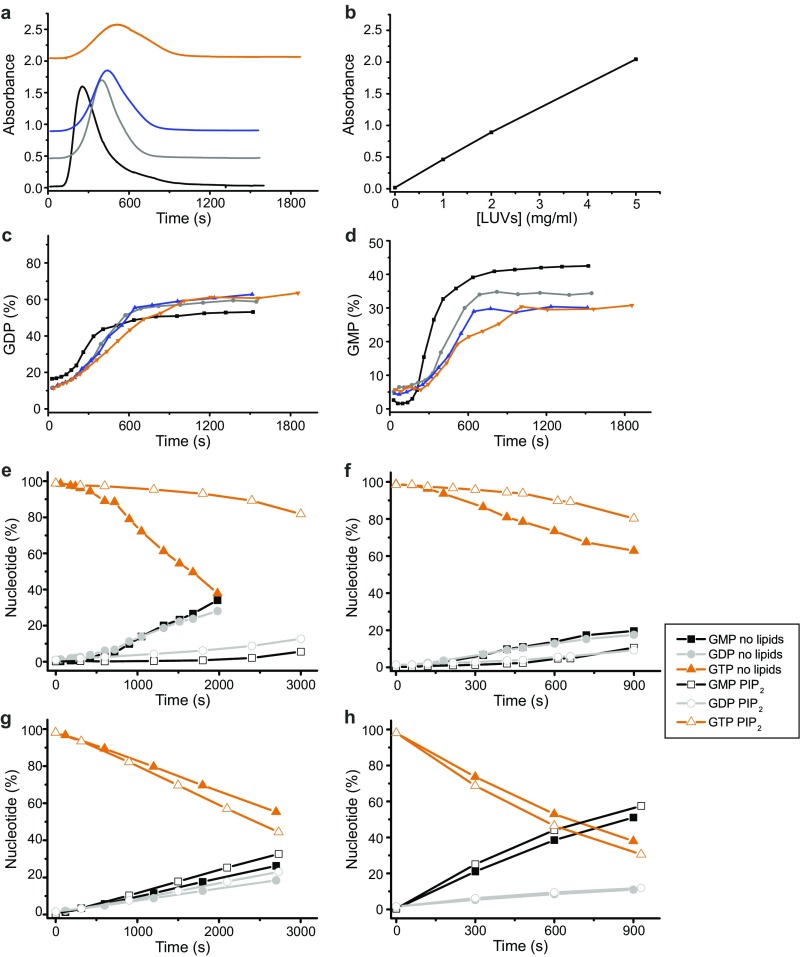
Membrane binding and polymerization properties of hGBP1 mutants. Farnesylated GTPase-deficient mutant R48AF (orange) and mutant K76AF (blue), which has a slower GTPase activity and a deficient second hydrolysis step leading to a lack of GMP formation, were investigated for their capability for GUV binding and polymerization. (A) Comparison of GTP- (10 min) and GTPγS-dependent GUV binding of Alexa-fluorescent R48AF (AF-R48AF) and K76AF (AF-K76AF) to AF-hGBP1F. Apo state of AF-hGBP1F is illustrated as representative for all mutants, which likewise failed to associate with membranes in the absence of nucleotide. (Scale bar, 5 µm.) (B–E) A total of 10 µM of hGBP1F wild type (gray), R48AF (orange), and K76AF (blue) were investigated for their capability to polymerize. Polymerization was triggered by addition of 1 mM GTP (t = 0 s) in the absence of liposomes. Time courses of absorbance (B) and the fractions of nucleotides GTP (C), GDP (D), and GMP (E) analyzed by HPLC at different time points along the turbidity assay.
In the previous experiments, GTP and AF-hGBP1F were mixed in a reaction tube before the mixture was transferred into the chamber. To see what happens directly after mixing the protein and GTP, we modified the previous experimental setup such that GTP was added into the chamber as a droplet only after AF-hGBP1F and GUVs were already preincubated. Under these experimental conditions, nucleotide spread in the chamber by diffusion. While recording a single GUV, we instantly observed binding of AF-hGBP1F comparable to the first experiment, which was followed by a sudden drop of the background fluorescence. Concomitantly, floating protein clusters became visible, suggesting that hGBP1F may be able to form polymers (Fig. S1G).
In the assay described above, the system remained at equilibrium conditions and it is difficult to conclude anything about the kinetics of the protein interaction with GUVs. To investigate the kinetics of AF-hGBP1F dissociation from GUVs, we performed an assay at nonequilibrium conditions. GUVs were incubated in the chamber with either GTP or GTPγS, and further, a short local injection of AF-hGBP1F into the vicinity of observed GUVs was performed from the injection pipette, containing 90 µM of protein. The presence of the corresponding nucleotide in the chamber resulted in instant binding of AF-hGBP1F to GUVs. Due to diffusion, the protein concentration in the vicinity of the observed GUV decreased, leading to dissociation of AF-hGBP1F from the GUV’s surface (Fig. 1F). Remarkably, AF-hGBP1F dissociation from GUVs occurred much faster in the presence of GTP compared with GTPγS (Fig. 1 F and G and Movie S1). Single exponential decays fitted to the recorded fluorescence of AF-hGBP1F on the GUVs (Fig. 1G) yielded observed rate constants (kobs) for dissociation of AF-hGBP1F from the membrane with values of 0.082 s−1 and 0.006 s−1 for experiments in the presence of GTP and GTPγS, respectively. Compared with Fig. 1 D and E, the decays were much faster, as they reflect predominantly the dissociation of the protein because reassociation was suppressed by the rapid diffusion of dissociated hGBP1 away from the GUV.
The comparison of GTP and GTPγS reveals a more than 10-fold faster dissociation rate of AF-hGBP1F from the surface of GUVs in the presence of GTP compared with its analog. It may be explained by the GTPase activity of AF-hGBP1F, involving structural changes including the C-terminal moiety. Of note, the very slow turnover of GTPγS bound to hGBP1F is 0.0001 s−1 (Fig. S3E), i.e., the membrane dissociation mentioned above is 60-fold faster. With the above-described dissociation assay, we also challenged farnesylated R48A, which showed almost no GTPase activity (Fig. S2C). Dissociation experiments with AF-R48AF revealed similar dissociation rates for the experiments performed with GTP and GTPγS (Movie S1), yielding dissociation rate constants of 0.012 s−1 and 0.014 s−1, respectively. These values are comparable to the wild-type protein in the presence of GTPγS, underlining the importance of GTPase activity for membrane dissociation.
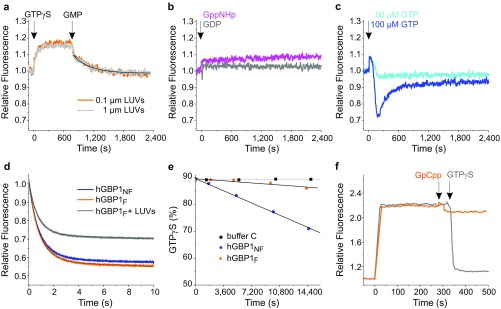
Fig. S3.
Liposome binding of hGBP1 induced by GTP analogs and their biochemical characterization. (A–C) Binding of AF-Q557CF to LUVs labeled with Rhodamine lipids. At time t = 0 s, indicated nucleotides (arrows) were added into a mixture of 0.5 mg/mL of LUVs and 2.5 µM of protein and an increase of fluorescence intensity reported liposome binding. (A) Addition of 200 µM GTPγS to LUVs of various sizes, 0.1 µm (orange) compared with 1 µm (gray). After ∼800 s, GTPγS was displaced with 5 mM of GMP, leading to dissociation of the protein from LUVs. Regardless of the LUV sizes, single exponential fits (black lines) yielded similar dissociation rate constants (koff = 0.0028 s−1 orange, koff = 0.0030 s−1 gray). (B) Addition of either 400 µM GppNHp (magenta) or GDP (gray) to 0.1 µm LUVs. (C) Addition of various concentrations of GTP to 0.1 µm LUVs. The fluorescence traces of GTP-dependent liposome binding were interfered with GTP-dependent polymerization events as demonstrated by different concentrations of the substrate (cyan, 50 µM GTP; blue, 100 µM GTP). (D) Farnesyl- and membrane-dependent dissociation kinetics of hGBP1 and GTP were investigated with hGBP1NF (blue) and hGBP1F either in the absence (orange) or presence of 2 mg/mL LUVs (gray) using the MANT-labeled GTP analog GTPγS (mGTPγS) (19). The preformed complex of 1 µM protein and 0.5 µM mGTPγS was rapidly mixed with 500 µM of nonlabeled GMP to trigger quasi-irreversible displacement of the GTP analog. The decrease of MANT fluorescence (λex = 366 nm, emission detected through 420-nm cutoff filter) reporting dissociation of mGTPγS was fitted with a single exponential equation (black lines) yielding similar dissociation rate constants under all tested conditions (koff = 1.18 s−1 blue, koff = 1.21 s−1 orange, and koff = 1.34 s−1 gray). (E) GTPγS hydrolysis catalyzed by hGBP1NF or hGBP1F was investigated with 500 µM GTPγS and 10 µM of protein. Kinetics of GTPγS turnover fitted with linear regression (solid lines) yielded specific activities of 0.037 min−1 (hGBP1NF) and 0.0058 min−1 (hGBP1F). In the absence of protein (buffer C only), the GTPγS concentration remained stable over time (dotted line). (F) Binding of the nonhydrolyzable GTP analog GpCpp to hGBP1F as tested by its ability to compete with mGTPγS used as fluorescent reporter of nucleotide binding (λex = 366 nm, λem = 435 nm). At t = 0 s, addition of 0.5 µM hGBP1F to 0.25 µM mGTPγS in the cuvette led to a fast and more than twofold increase of the fluorescence signal, indicating protein/nucleotide complex formation. Addition of 250 µM nonfluorescent GpCpp in contrast to GTPγS (arrows) did not suffice to displace the preformed protein/nucleotide complex, suggesting that hGBP1F does not bind GpCpp.
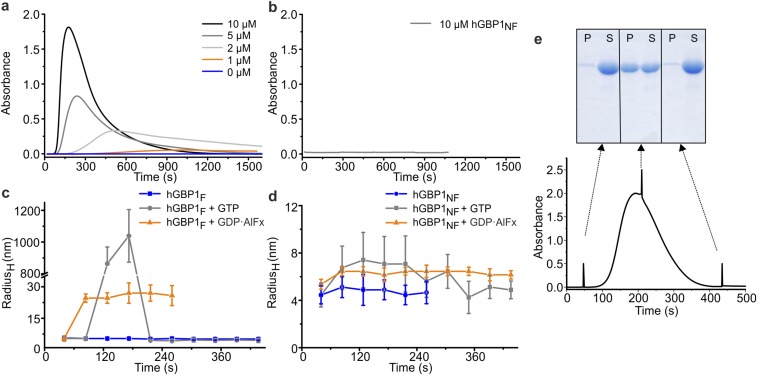
We used an independent FRET-based experimental setup to demonstrate nucleotide-dependent membrane binding of hGBP1F, in which we used large unilamellar vesicles (LUVs) of various sizes. LUVs, labeled with Rhodamine-PE lipids acting as acceptor fluorophore, were mixed with Alexa-488 labeled Q577C mutant of hGBP1 (AF-Q577CF) acting as donor fluorophore. After addition of GTPγS, the FRET efficiency increased reporting attachment of hGBP1F to the membrane. Moreover, the fluorescence value returned to its initial value after addition of a large molar excess of GMP (Fig. S3A). As nucleotide exchange is rapid (Fig. S3D), this finding is interpreted as dissociation of GMP-bound hGBP1F from the membrane with a rate constant of 0.003 s−1 (Fig. S3A). Intriguingly, both the increase of fluorescence and the dissociation rate are the same for small and large LUV diameters, respectively (Fig. S3A). With the same setting, the addition of GppNHp yielded a smaller and slower increase of fluorescence, whereas the addition of GDP led to only a very small jumpwise increase of fluorescence (Fig. S3B).
Electron Microscopy Analysis of hGBP1F.
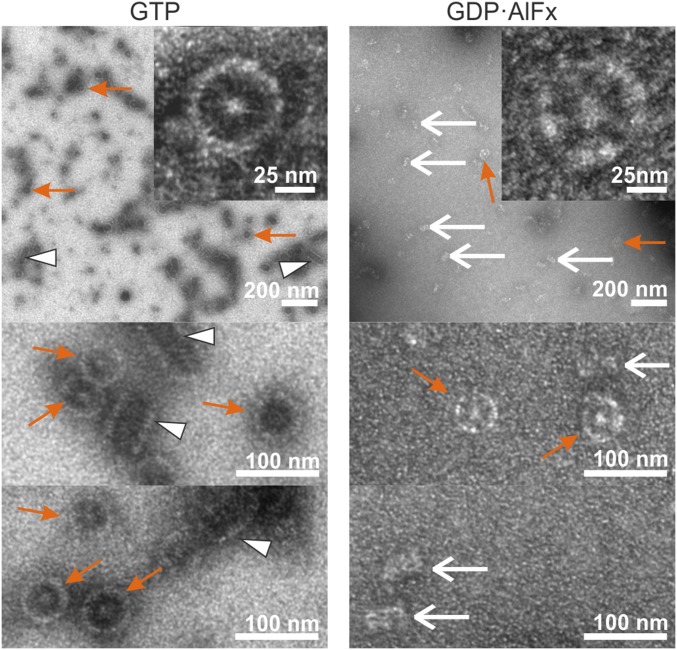
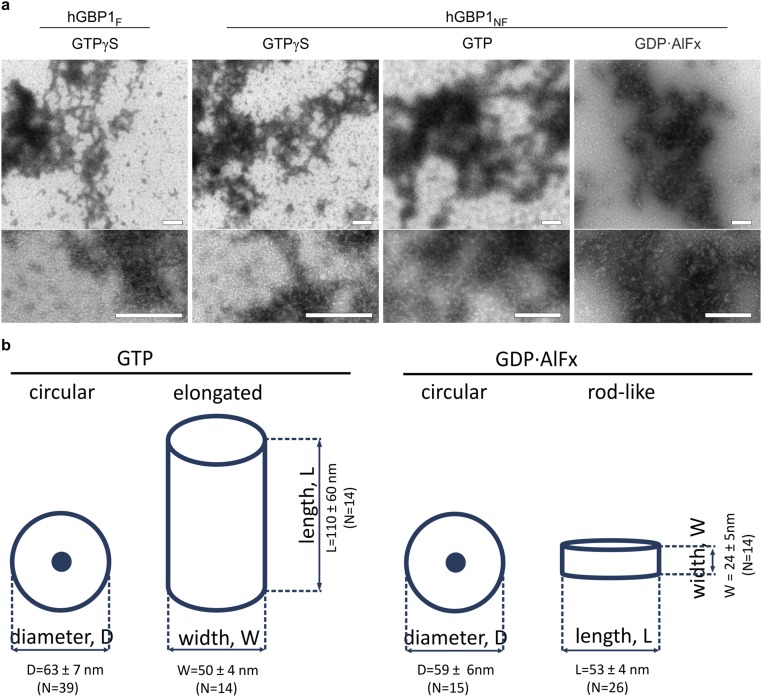
To gain insight into the architecture of the protein clusters formed by hGBP1F (Fig. S1G), we used negative stain electron microscopy (EM). In the presence of GTP, we observed a large number of circular structures with diameters around 60 nm (Fig. 2, orange arrows). The centers of these rings were always formed by a light dot. Besides these circular structures, we also observed some elongated structures of variable length between 60 nm and more than 200 nm and a width of about 50 nm (Fig. 2, white arrowheads). Circular structures with a white dot in the center and of a similar diameter were also observed for hGBP1F in the presence of GDP·AlFx. However, these GDP·AlFx-induced circular structures were lower in number than those observed in the presence of GTP (Fig. 2, orange arrows). Moreover, the elongated structures as observed with the natural substrate GTP were not formed in the presence of GDP·AlFx. Instead, we observed a large number of rod-like structures with a length of about 50 nm and a width of about 20 nm (Fig. 2, white arrows). These rods having a length similar to the ring diameter may therefore represent the same objects as the circular structures viewed from a different angle, in particular from the side view. Notably, the analog GTPγS did not induce any structured arrangement of hGBP1F at all (Fig. S4A), and hGBP1NF also did not yield any clusters irrespective of the nucleotide used (Fig. S4A). In summary, these data demonstrate that farnesylated hGBP1 is capable of forming polymeric structures in a nucleotide-dependent manner.
Fig. 2.
EM images of hGBP1. Representative images of hGBP1F in the presence of GTP or GDP·AlFx. Experimental mixture contained 10 μM of hGBP1F and 1 mM of GTP or 200 μM GDP, 300 μM AlCl3, and 10 mM NaF for samples with GDP·AlFx. Reactions were stopped after ∼10 min of incubation with GTP and after ∼20 min incubation with GDP·AlFx (Methods). Ordered polymer structures of hGBP1F in the presence of GTP or GDP·AlFx were identified during at least three independent measurements. Examples of observed ring-like structures are indicated with orange arrows, rods with white arrows, and elongated structures with white arrowheads. Scale bars are as indicated. The geometries of all identified polymer structures are quantified in Fig. S4B.
Fig. S4.
EM images of hGBP1. (A) Representative images of either hGBP1F or hGBP1NF incubated with the indicated nucleotides. Experimental mixture contained 10 μM of hGBP1F or hGBP1NF and 1 mM of corresponding nucleotide, except for samples with GDP·AlFx, which contained 200 μM GDP, 300 μM AlCl3, and 10 mM NaF. Reactions were stopped after ∼10 min (GTP) and after ∼20 min (GTPγS and GDP·AlFx). (Scale bars, 200 nm.) (B) Measures for the identified polymer structures (Fig. 2) of hGBP1F in the presence of indicated nucleotides as schematically drawn. The numbers of quantified structures (N in parentheses) as well as obtained mean values with SDs are indicated along each scheme.
Farnesylated hGBP1 Undergoes GTP-Dependent Reversible Polymerization.
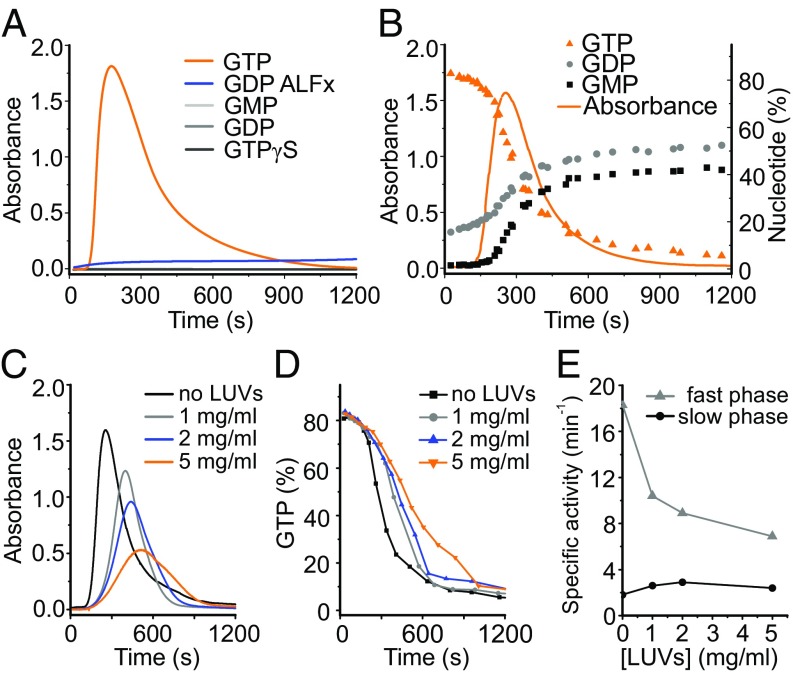
To further investigate the polymerization of hGBP1F, we followed the absorbance of protein samples at 350 nm upon injection of different nucleotides, which enabled us to detect turbidity as an indicator for polymerization of the protein. Injection of GTP induced a strong increase in turbidity of the hGBP1F solution, whereas this was not the case for hGBP1NF (Fig. 3A and Fig. S5B). Remarkably, turbidity appeared with a delay, peaked a few minutes after injection of GTP, and then decreased back to the baseline (Fig. 3A). The kinetics as well as the extent of hGBP1F polymerization depended on the protein concentration (Fig. S5A). The disappearance of turbidity was due to the complete dissolution of the polymers and not due to the sedimentation of large aggregates. This result was demonstrated by rapid centrifugation of samples taken (i) immediately after addition of GTP, (ii) at the time of peaking turbidity, and (iii) after turbidity had almost vanished and analysis by SDS/PAGE. Only for the second sample, hGBP1 was found in the pellet fraction but not for the first and the third samples (Fig. S5E). Using hGBP1F, the addition of GMP, GDP, or GTPγS did not trigger any polymerization of the protein (Fig. 3A). The addition of GDP·AlFx to hGBP1F revealed only a modest polymerization, which was not reversible (Fig. 3A). These observations are in good agreement with dynamic light scattering (DLS) experiments for hGBP1F and hGBP1NF. DLS experiments indicated reversible transient appearance of large polymers of hGBP1F in the presence of GTP hydrolysis and nonreversible appearance of relatively small polymers in the presence of GDP·AlFx (Fig. S5C) with an apparent hydrodynamic radius of about 30 nm, which corresponds reasonably well with the structures seen in the EM. Human GBP1NF was not able to polymerize at all, neither in the presence of GTP nor in the presence of GDP·AlFx (Fig. S5 B and D).
Fig. 3.
Regulation of GTPase activity of hGBP1F by polymerization and membrane interaction. (A) Absorbance signal of 10 μM hGBP1F after injection of nucleotides at t = 0 s. (B) Absorbance signal of 10 μM hGBP1F after injection of 1 mM GTP (t = 0 s) superimposed with nucleotide composition in the same solution. The plot includes data from three independent experiments; absorbance represents the average of all three datasets. (C) Absorbance signal of 10 μM hGBP1F after injection of 1 mM GTP in the presence of different concentrations of LUVs. The baseline of LUVs alone was subtracted from the raw data (Fig. S6 A and B). (D) GTP turnover from experiments described in C. (E) Enzymatic activity of 10 µM hGBP1F in the presence of different LUV concentrations. Specific activities of GTP hydrolysis in the slow and fast phases were calculated from the data presented in D.
Fig. S5.
Nucleotide-dependent polymerization of hGBP1F. (A and B) Absorbance of the sample with different concentrations of hGBP1F (A) and hGBP1NF (B) after addition of 1 mM GTP (t = 0 s). (C and D) Analysis of polymer formation of 20 µM hGBP1F (C) and hGBP1NF (D) by dynamic light scattering. The particle size, given as the hydrodynamic radius (RH) in nanometers without nucleotide (blue) and in the presence of 1 mM GTP (gray) or GDP·AlFx (orange) (300 µM AlCl3, 10 mM NaF, and 200 µM GDP) was monitored over 250–450 s at 25 °C. Farnesylated hGBP1 transiently forms very large polymers during GTP hydrolysis with a radius around 1,000 nm. In contrast, the addition of GDP·AlFx leads to an irreversible formation of polymers with an average RH of 25 nm. Nucleotide-free hGBP1NF shows a RH of 5 nm. Complexation with GDP·AlFx leads to a slight irreversible increase to 6.5 nm and addition of GTP to a transient increase to a maximum value of 7.2 nm. (E) Reversibility of GTP-induced hGBP1F polymers investigated by sedimentation. During a typical turbidity experiment (see A, 10 µM hGBP1F and 1 mM GTP added at t = 0 s), samples were taken at indicated time points (arrows), spun down, and the fraction of protein in pellet (P) and supernatant (S), respectively, was analyzed by SDS/PAGE (100-µL samples were taken, centrifuged at 30,000 × g, 4 °C and for 1 min. After removal of the supernatant, the pellet was resuspended in the same volume of buffer, and corresponding fractions of protein in pellet (P) and supernatant (S) were analyzed by SDS/PAGE. Considering that the proportion of protein in the pellet represents protein complexes large enough to sediment, e.g., polymers, SDS/PAGE analysis reflected the same observation as already monitored by time-trace of absorbance: The GTP-induced increase and decrease of the absorbance considered as reversible assembly and disassembly of hGBP1F polymers was accompanied by increase and decrease of protein share in the pellet.
To understand the link between polymerization of hGBP1F and its GTPase activity, we simultaneously measured the absorbance and analyzed the nucleotide composition in the same cuvette containing 10 μM hGBP1F after addition of 1 mM GTP (Fig. 3B). We observed a biphasic profile of GTPase activity of hGBP1F comprising an initial slow phase with a turnover number of 1.6 min−1 ± 0.6 min−1 followed by a fast phase with a turnover number of 15.8 min−1 ± 0.7 min−1. As could be seen from Fig. 3B, this acceleration of the catalytic hydrolysis reaction nicely correlated with the appearance of the turbidity. Moreover, there was an obvious delay in GMP production in the course of the experiment and the onset of GMP production coincided with the onset of turbidity as well as with the change from the slower to the faster turnover rate. We also checked the ability of R48AF and K76AF mutants of hGBP1 to polymerize using the turbidity assay. Intriguingly, both mutants did not show any increase of turbidity after mixing with GTP (Fig. S2B). As mentioned already, R48AF showed almost no GTPase activity. K76AF showed a reduced activity of GTP hydrolysis and almost no activity for the second step leading to GMP formation (Fig. S2 C–E).
We further questioned how the presence of liposomes alters the polymerization and GTPase activity of hGBP1F. To answer this question, we performed similar experiments in the presence of different concentrations of LUVs. Increasing amounts of LUVs resulted in a delayed and less pronounced polymerization of hGBP1F (Fig. 3C and Fig. S6A). Moreover, the presence of LUVs did not affect the first slow phase but did affect the second fast phase of GTP hydrolysis in two ways: with increasing LUV concentration, not only GTPase activity (Fig. 3 D and E) but also GMP formation decreased (Fig. S6D). Biphasic behavior of GTPase activity of hGBP1F was pronounced most remarkably at 25 °C, even for reduced protein concentrations (Fig. S6E). Nevertheless, at 37 °C this biphasic behavior could also be recognized in the absence of LUVs, and the presence of LUVs elongated the slow phase of enzymatic activity (Fig. S6F). In contrast, we did not observe biphasic behavior of catalytic activity for hGBP1NF, neither at 25 °C, nor at 37 °C (Fig. S6 G and H). The presence of LUVs did not alter the GTPase activity of hGBP1NF at either temperature (Fig. S6 G and H).
Fig. S6.
Polymerization and GTPase activity of hGBP1 under various lipid and temperature conditions. (A and B) Time courses of hGBP1F polymer absorbance (A) and the obtained background absorbance of liposomes as a function of lipid concentration (B). Experiments were performed with 10 µM hGBP1F in the absence of liposomes (black) and in the presence of 1 mg/mL (gray), 2 mg/mL (blue), and 5 mg/mL (orange) of final lipid concentration. Polymerization was triggered upon addition of 1 mM GTP (t = 0 s). (C and D) Analysis of hGBP1F catalyzed GTP hydrolysis products GDP (C) and GMP (D) for same experiments. (E–H) Nucleotide composition of the GTPase reaction performed either with 2 μM hGBP1F at 25 °C (E) and 37 °C (F) or with 2 μM hGBP1NF at 25 °C (G) and 37 °C (H) in the absence of liposomes (filled symbols) or in the presence of 0.67 mg/mL PIP2 liposomes (open symbols).
Having shown both GTP-dependent GUV binding and polymerization of hGBP1F, we used the FRET-based LUV binding assay described above. After addition of GTP to the mixture of LUVs and AF-Q577CF first, an increase of fluorescence intensity was observed which then changed to a decrease and after a few minutes came back to the initial value. This observation can be explained by rapid membrane attachment followed by polymerization of hGBP1F, which leads to decreased fluorescence intensity likely due to turbidity (Fig. S3C). Finally, after GTP is depleted, the initial situation of dissociated hGBP1F is reestablished; all this happened on time scales as observed in the turbidity assay reporting polymerization as shown in Fig. 3C.
Membrane-Bound hGBP1 Tethers GUVs in a Nucleotide-Dependent Manner.
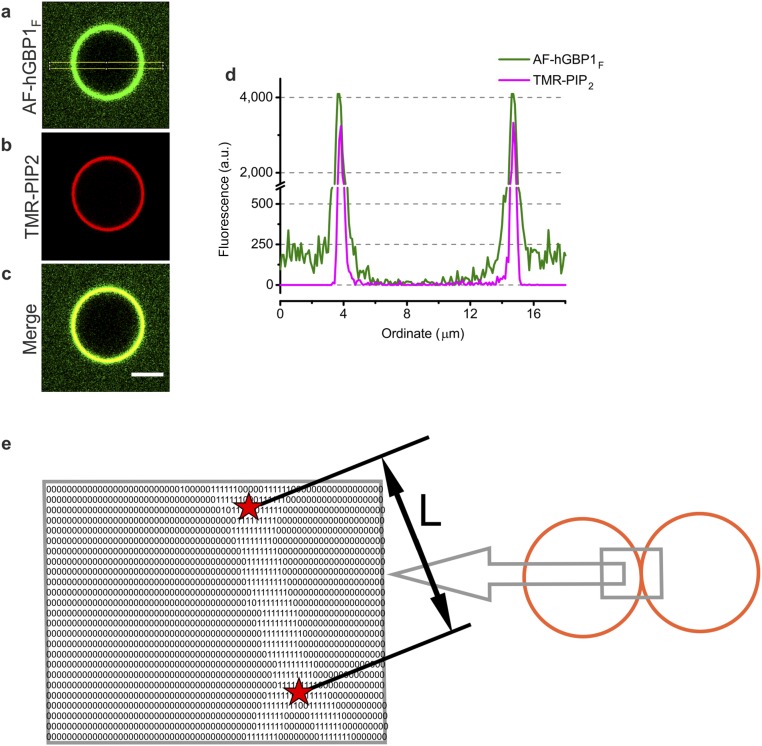
During GUV-binding experiments with AF-hGBP1F, we observed that GUVs spontaneously contacted each other. Notably, the contact area between those GUVs was larger in the presence of GTPγS compared with other nucleotides. To get insights into this phenomenon, we performed the following tethering assay: pairs of GUVs were gently brought into contact with each other in the presence of 22.5 µM of hGBP1F containing 11 mol% of AF-hGBP1F. Further, with the help of an injection pipette containing 400 µM of either GTPγS or GTP, the nucleotide was briefly injected (within 5–10 s) into the vicinity of the observed pair of GUVs. This event and subsequent rearrangements of the given GUV pair were tracked with confocal microscopy. Although the contact area gives the true measure for tethering, here, we exploited the contact length from confocal microscopy images instead (Fig. S7E and SI Methods), considering that an increase of the contact length implicates an increase of the contact area. Additionally to the contact length, we also quantified the fluorescence of membrane-bound protein for a given pair of GUVs.
Fig. S7.
Quantification of the fluorescence signal of membrane-bound hGBP1 and calculation of the contact length in the tethering assay. Typical images of GUVs for AF-hGBP1F in the presence of GTPγS for the protein channel of fluorescence (A), the lipid channel of fluorescence (B), or the merge of two fluorescence channels (C) as an example for quantification of the fluorescence signal of membrane-bound hGBP1. (Scale bar, 5 μm.) (D) Superimposed fluorescence intensity profiles across narrow equatorial rectangle as shown in B, taken from A and B. (E) Schematic representation of the image-processing algorithm for contact length calculation in tethering assay. The fluorescent signal from the lipid channel of fluorescence was set to the value of 1 when fluorescent intensity was higher than the threshold value and set to the value of 0 otherwise. The contact length (L) between the two vesicles is indicated. Points of membrane differentiation are indicated as red stars. The gray outlined arrow indicates that the two rectangles correspond to each other.
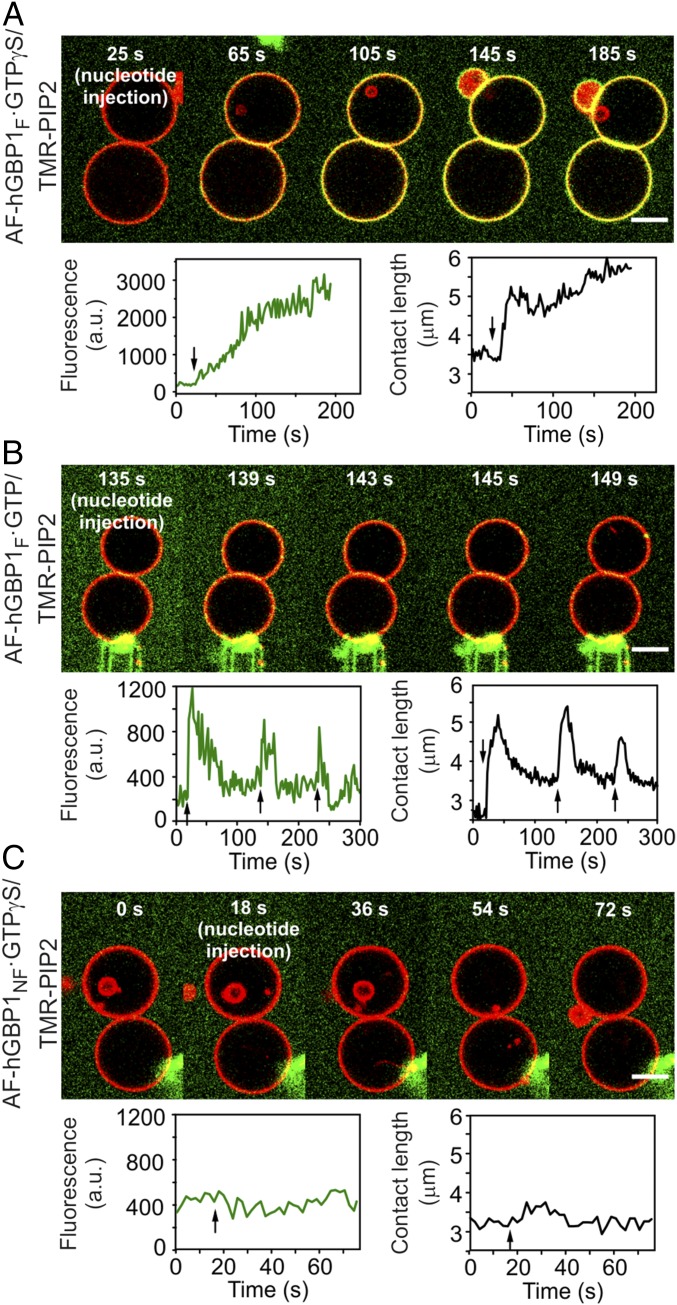
Strikingly, in the course of such experiments we noticed a different behavior for GTPγS compared with GTP. Upon injection of GTPγS we observed irreversible binding of AF-hGBP1F to GUVs and a nonreversible tethering effect of GUV pairs on a time scale of several minutes. After initial injection of the nucleotide, the contact length between pairs of GUVs increased until the maximum value was reached (Fig. 4A and Movie S2). In contrast, the binding of AF-hGBP1F and the tethering effect on GUVs was reversible upon injection of GTP and could be repeated several times for one pair of GUVs (Fig. 4B and Movie S3). Importantly, the change of the contact length between a given pair of GUVs correlated with the fluorescence signal of GUV-bound AF-hGBP1F resulting in a more dynamic system when the natural substrate GTP was supplied instead of the analog GTPγS (Fig. 4 A and B). As a control, tethering experiments with hGBP1NF did not reveal any binding of the protein or any increase of the contact length upon injection of GTPγS (Fig. 4C and Movie S4). The same was observed for hGBP1NF and injection of GTP (Movie S5). Generally, for experiments with hGBP1NF, GUVs that were initially brought into contact with each other had a tendency to separate from each other, rather than to adhere, upon injection of the nucleotide.
Fig. 4.
Tethering experiments on pairs of GUVs. (A) Time-lapse confocal images of GUV pairs (red) in the presence of hGBP1F (green) and GTPγS (Top) (see also Movie S2). Images were analyzed with respect to the fluorescence signal of the protein on the membrane of GUVs (Bottom Left) and contact length between a pair of GUVs (Bottom Right). Experiments were performed in the presence of 2.5 µM of labeled and 20 µM of nonlabeled protein. Arrows indicate time points of nucleotide injections. (B) Same as in A but with GTP (see also Movie S3). (C) Same as in A but with hGBP1NF (see also Movie S4). (Scale bars, 5 µm.) a.u., arbitrary units.
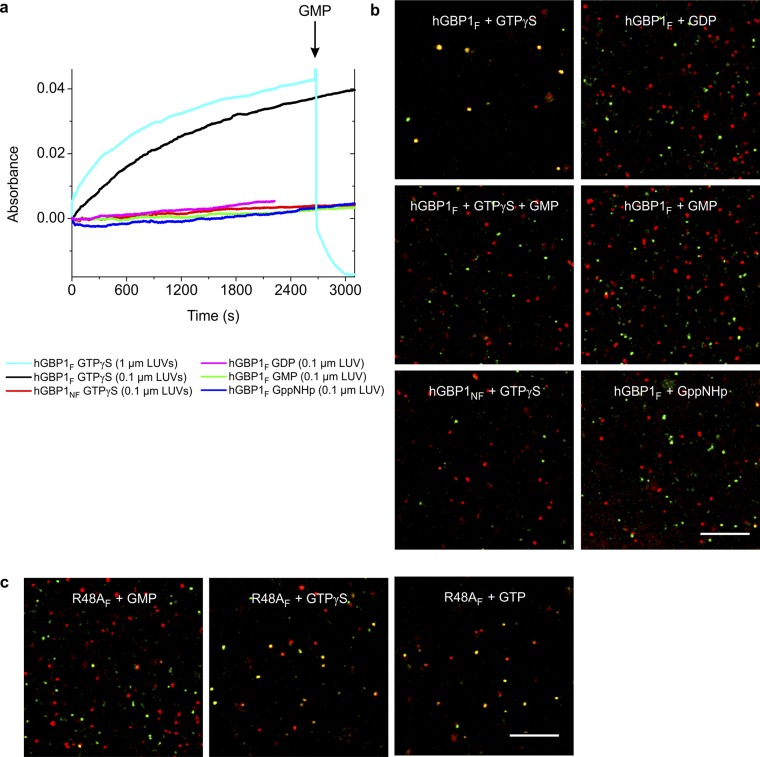
The notion that hGBP1F tethers vesicles in a GTP-dependent manner is supported by two additional experiments, as similarly performed on atlastin (31), which do not rely on initial contacting of the vesicles. First, LUVs of various sizes in the presence of hGBP1F showed an increase of turbidity after addition of GTPγS, which was not the case after addition of GMP, GDP, GppNHp, or when hGBP1NF was used instead (Fig. S8A). Moreover, the GTPγS-induced increase of turbidity was driven back by the addition of a large molar excess of GMP, which could be explained by disassembly of tethered vesicles (Fig. S8A). Second, green fluorescent LUVs and red fluorescent LUVs were mixed, showing separate spots under the microscope. After addition of GTPγS, the green and red spots disappeared as they merged, yielding yellow spots indicating colocalization, which we interpret as LUV tethering (Fig. S8B). Again, the addition of GMP could reverse this effect, leading to separate green and red spots. This is documented together with further control experiments in Fig. S8. There we show also weaker tethering mediated by the R48A variant in the presence of GTP and GTPγS, respectively (Fig. S8C).
Fig. S8.
Nucleotide-dependent membrane tethering mediated by hGBP1F. (A) HGBP1-mediated tethering followed by absorbance-based tethering assay. The increase in particle size was monitored by the absorbance at 350 nm. The initial absorbance value of 0.2 was subtracted. A total of 400 µM of the corresponding nucleotide was added at t = 0 s in the presence of 30 µM of the corresponding protein. Addition of GTPγS was done for LUVs of various sizes, 0.1 µm (black) compared with 1 µm (cyan). A total of 10 mM GMP was added after 45 min (arrow) to dissociate tethered LUVs. Reactions were done at 30 °C. (B) HGBP1-mediated tethering followed by visual tethering assay. LUVs, which also contained either ATTO-488 or Rhodamine-labeled lipids, respectively, were mixed at a 1:1 ratio (total lipid concentration, 0.8 mg/mL) in the presence of 30 µM of either hGBP1F or hGBP1NF. All samples were incubated with 400 µM of the corresponding nucleotide for 45 min at 30 °C. To dissociate tethered LUVs in the presence of GTPγS, one of the reaction mixtures with GTPγS was additionally supplied with GMP after 45 min (final concentration of 36 mM) and incubated for 10 more minutes at 30 °C. All samples were diluted, loaded into the observation chamber, and visualized by confocal microscopy. (Scale bar, 10 μm.) (C) As in B but with the GTPase-deficient farnesylated mutant R48AF.
HGBP1 Reveals Fast Membrane Dynamic in Cells.
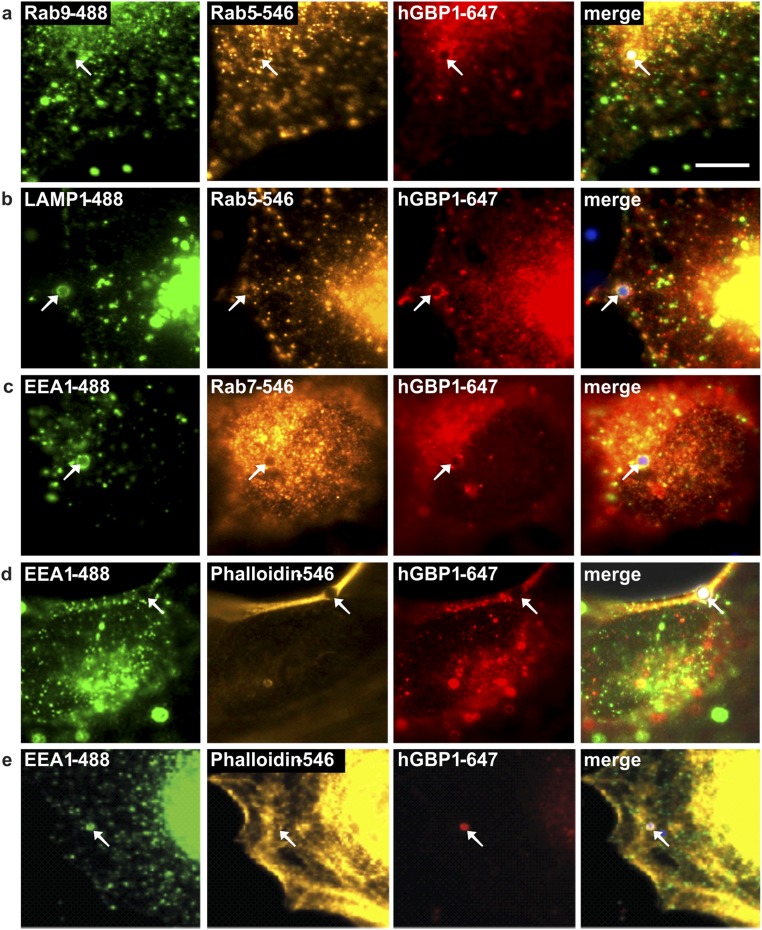
To compare the in vitro membrane dynamics to experiments in living cells we analyzed the recruitment of hGBP1 to cellular membranes in HeLa cells. In previous studies IFNγ-induced endogenous hGBP1 has been found in a punctate or vesicular cytoplasmic distribution throughout the whole cell (27). However, the exact assignment of the punctate structures to an intracellular membranous compartment has not been made. To probe for a phagolysosomal localization of hGBP1, we incubated IFNγ-induced HeLa cells with latex beads to induce phagocytosis (Fig. S9). Endogenous hGBP1 was recruited to these phagocytosed beads, although it was often not equally distributed around the entire beads but present in punctate structures. HGBP1 partially colocalized with marker proteins of the phagolysosomal pathway such as early endosomes (Rab5 and EEA1), late endosomes (Rab7), and lysosomes (LAMP1). These hGBP1+ puncta also colocalized with filamentous actin stained with fluorescently labeled phalloidin enclosing a phagocytosed bead.
Fig. S9.
Recruitment of endogenous hGBP1 to phagocytosed latex beads. HeLa cells were IFN-γ induced for 24 h and then incubated for 4 h with nonfluorescent (A and D) or blue-fluorescent (B, C, and E) latex beads with 2-µm (A–D) or 1-µm (E) diameter. The monoclonal 1B1-antibody against hGBP1 and several antibodies against distinct structures of the phagolysosomal pathway were used for costaining. (A) Rab9 (LE/TGN) and Rab5 (EE). (B) LAMP1 (lysosomes) and Rab5 (EE). (C) EEA1 (EE) and Rab7 (LE). (D and E) EEA1 (EE) and phalloidin (filamentous actin). Arrows mark engulfed latex beads within the cytoplasm. (Scale bar, 10 µm.) EE, early endosomes; LE, late endosomes; TGN, trans Golgi network.
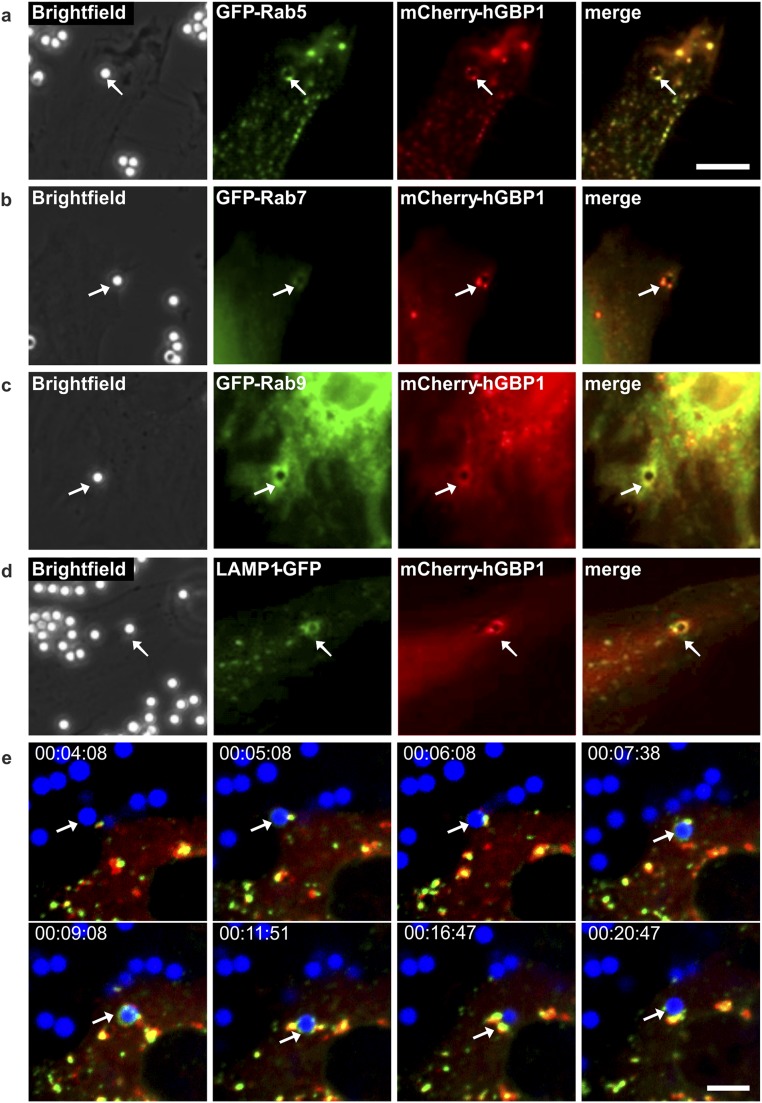
Similar events were observed upon transfection of unstimulated HeLa cells with fluorescent protein-tagged hGBP1 and marker proteins (Fig. S10). Recombinant mCherry-hGBP1 colocalized with GFP-Rab5, GFP-Rab7, GFP-Rab9, or LAMP1-GFP around bead-containing phagosomes (Fig. S10 and Movie S6). The colocalization of mCherry-hGBP1 with Rab+ vesicular structures was enhanced compared with untransfected cells.
Fig. S10.
Colocalization of recombinant hGBP1 and marker proteins around latex beads. Unstimulated HeLa cells were cotransfected with mCherry-hGBP1 as well as GFP-tagged marker proteins and incubated with 2 µm latex beads for 4 h to specify distinct stages of the phagocytic pathway. (A) GFP-Rab5. (B) GFP-Rab7. (C) GFP-Rab9. (D) LAMP1-GFP. (Scale bar, 5 µm.) Arrows mark engulfed latex beads within the cytoplasm. (E) Life-cell imaging of recruitment of hGBP1 and Rab5 to latex bead phagosomes. Uninduced HeLa cells transfected with mCherry-hGBP1 and GFP-Rab5 were incubated with blue fluorescent latex beads for 4 h and uptake of beads was followed by life-cell imaging. Arrows indicate a phagocytosed bead, which is attached to Rab+ (green) and hGBP1+ (red) punctate structures. See also Movie S6. Time scale, hours:minutes:seconds (h:min:s). (Scale bar, 10 µm.)
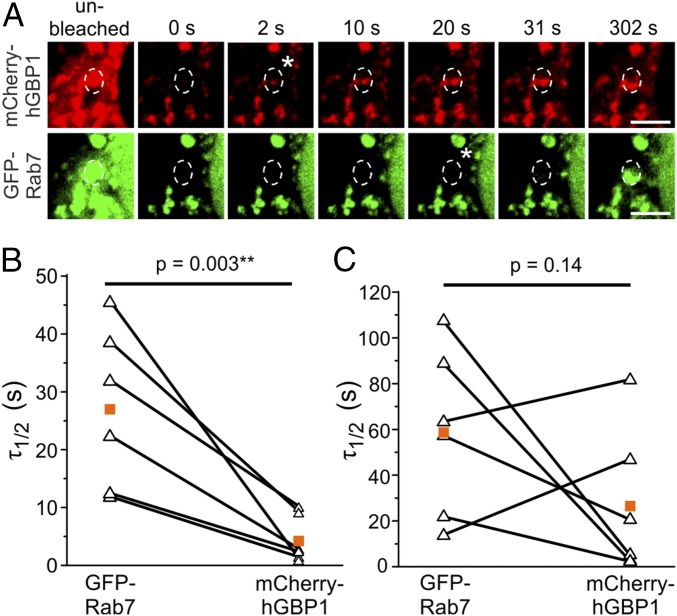
The dynamics of hGBP1 on cytosolic punctate structures and on bead-containing phagosomes (BCP) were determined by fluorescence recovery after photobleaching (FRAP). Before imaging, unstimulated HeLa cells cotransfected with mCherry-hGBP1 and GFP-tagged Rab7 were incubated with latex beads. After bleaching of a region of interest (ROI), the fluorescence bleached on BCPs or late endosomes positive for mCherry-hGBP1 and GFP-Rab7 recovered in all cases but usually not to the starting value. An exemplary time lapse for the recovery of mCherry-hGBP1 and GFP-Rab7 fluorescence on a vesicular structure is shown in Fig. 5. The signal for fluorescence of mCherry-hGBP1 reappeared quickly within 2 s, whereas the first faint GFP-Rab7 signal could only be detected after 20 s. It is striking that the overall fluorescence within the depicted cell decreased after bleaching of the ROI, which is most probably due to the long exposure with a high laser power and indicates a relatively fast exchange of the fluorescent components with the surrounding cytosol. Consequently, to normalize the data, an ROI was set outside of the observed cell additionally to the one in an unbleached cytoplasmic area inside the cell.
Fig. 5.
Fast recovery of mCherry-hGBP1 on GFP-Rab7+ structures. (A) FRAP: time lapse of mCherry-hGBP1 and GFP-Rab7 fluorescence recovery on a vesicle in cotransfected HeLa cells after bleaching. The first picture of each panel shows the region of interest (dashed circle) before bleaching. Asterisks mark first detection of recovering signals. (Scale bars, 10 μm.) (B and C) Data plots showing the half-lives of fluorescence recovery (τ1/2) of cotransfected mCherry-hGBP1 and GFP-Rab7 on vesicular structures (B) and BCP signals (C) after photobleaching. Values obtained for both protein constructs in the same measurement are connected by lines. Orange squares indicate the mean values.
Mean values of fluorescence recovery half-lives (τ1/2) are indicated in Fig. 5 B and C. On punctate vesicular structures, the signal for mCherry-hGBP1 recovers within a few seconds, which is significantly faster than the signal of GFP-Rab7, which recovers with a similar τ1/2 values in the range of 10–50 s as previously reported for human melanoma cells (32). On BCPs, the recovery of hGBP1 is slower and more variable and resembles that of GFP-Rab7. Because the entire vesicular structures or BCPs were bleached in each experiment, recovery of fluorescence could only be achieved by de novo delivery of fluorescent protein-tagged hGBP1 or Rab7 molecules, entering the bleached area from the surrounding parts of the cell and not by lateral diffusion. This indicates a very fast exchange of material between the membrane-bound pool of hGBP1 and the cytosol. In line with this notion is the finding that the fluorescence never recovered to more than 80% of the prebleach values, which can be explained by the long time needed to completely bleach the entire structures.
SI Methods
Protein Expression and Purification.
Synthesis of recombinant hGBP1 either in its nonfarnesylated (hGBP1NF) or farnesylated forms (hGBP1F) was performed in the E. coli strain BL21 (DE3) (Novagen). Therefore, cells were transformed with pQE80L-hGBP1 only (to obtain hGBP1NF), or according to an established method with both pQE80L-hGBP1 and pRSF-Duet-FTase α/β for coexpression of FTase, which catalyzes farnesylation of hGBP1 in prokaryotic cells (25). Cell cultures were supplemented with ampicillin (100 µg/mL) only (hGBP1NF) or with both ampicillin (100 µg/mL) and kanamycin (25 µg/mL) (hGBP1F) and grown at 37 °C to an OD600 of 0.6–0.8. Expression was induced with 100 µM isopropyl β-d-thiogalactopyranoside at 25 °C for 16–18 h. After harvesting (6,000 × g, 10 min, 4 °C), cells were frozen and stored at −80 °C. For protein extraction, cell pellets were resuspended in buffer A (50 mM Tris⋅HCl, pH 7.4, 500 mM NaCl, 5 mM MgCl2) containing 1 mM phenylmethylsulfonyl fluoride and disrupted by sonication on ice. Cell debris was removed by centrifugation at 25,000 × g, 4 °C for 1 h and soluble, N-terminally hexahistidine-tagged hGBP1 in the supernatant was purified by affinity chromatography: A 30-mL column with HisPur Cobalt Resin (Thermo Scientific) previously equilibrated with buffer A was loaded with supernatant. After washing of unbound protein with buffer A, specifically bound protein was eluted with an imidazole gradient from 10 mM to 150 mM. Fractions containing the target protein were precipitated with 3 M of (NH4)2SO4 and spun down at 7,000 × g and 4 °C for 15–20 min. Farnesylated and nonfarnesylated proteins were separated as previously described (25) using a Butyl Sepharose High-Performance Column (GE Healthcare) and a decreasing (NH4)2SO4 gradient from 1.2 M to 0 mM in 50 mM Tris⋅HCl, pH 7.9, and 5 mM MgCl2. A final step of gel filtration chromatography (Superdex 200 26/60, GE Healthcare) using buffer C (50 mM Tris⋅HCl, pH 7.9, 150 mM NaCl, 5 mM MgCl2) was performed to remove ammonium sulfate and to separate target monomeric protein from unspecifically formed protein complexes.
Expression and purification of hGBP1 mutants were performed as described for wild-type hGBP1. Both of the hGBP1 mutants R48A and K76A were already existing in pQE80L vectors, whereas mutation Q577C was introduced into pQE80L-hGBP1 using a QuikChange site-directed mutagenesis kit with KOD Hot Start DNA polymerase (Merck, Millipore) and the sense and antisense primers 5′-gcagaataatgaaaaatgagatatgcgatctccagacgaaaatgagac-3′ and 5′-cgtctcattttcgtctggagatcgcatatctcatttttcattattctg-3′, respectively. Oligonucleotides were purchased from MWG Eurofins Operon.
FRET-Based Tracking of Protein Binding to LUVs.
For monitoring hGBP1 binding to LUVs, the mutant Q577C in its farnesylated form (Q577CF) was used. Exploiting the additional cysteine residue at the C terminus, labeling with Alexa-488 dye was performed in a similar way as described in material and methods, yielding AF-Q577CF with a labeling efficiency of 300%. With this construct serving as donor and LUVs with Rhodamine-labeled lipids (Rhodamine-DPPE, Avanti) serving as acceptor, membrane binding of hGBP1 was monitored by FRET on the fluorescence spectrometer LS55 (Perkin-Elmer) (λex = 495 nm, λem = 590 nm, and slit width of 2.5/2.5 nm). AF-Q577CF was preincubated with LUVs (final lipid concentration, 0.5 mg/mL) and supplemented with the corresponding nucleotide. LUVs were made by extrusion using 0.1-µm membranes if not specified otherwise out of BPL containing 0.5% wt/wt Rhodamine lipids (16:0 Liss Rhod PE, 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (ammonium salt) (Avanti). All experiments were performed at 25 °C and in buffer C containing 1 mM MgCl2.
Quantification of Protein Binding to GUVs.
Fmembrane was quantified with the PlotProfile build-in command of ImageJ across a narrow equatorial rectangle (Fig. S7A). The width of the rectangle was kept constant for all calculations. The final Fmembrane value was taken as an average of two fluorescence maximums of the profile plot at the positions, which correspond to the opposite membranes of GUVs (Fig. S7D). Colocalization of GUV-bound protein with GUV’s membrane is seen from the merged picture of two fluorescent channels (Fig. S7C) and the fluorescence intensity profiles across a narrow equatorial rectangle taken for protein and lipid channels of fluorescence (Fig. S7D). For the dissociation and tethering assays, the fluorescence signal of the GUV-bound protein was quantified from the video recordings with the same ImageJ command in an automated way by using a javascript for ImageJ.
Quantification of the Contact Length Between a Pair of GUVs.
The contact length was calculated as a distance between two edges of shared segment of the membrane for a given pair of GUVs. Coordinates of the edges of the contact line were obtained by running the self-designed javascript in ImageJ in two steps. First, the picture from the lipid channel was transformed into a digital form as shown in Fig. S7E, using a threshold filter for recognizing the membrane. Second, the script was searching two points of membrane differentiation by a self-designed algorithm (Fig. S7E, red stars) and the contact length L was calculated as a distance between two points with known coordinates.
Absorbance-Based Tethering Assay.
The absorbance-based tethering assay used for atlastin studies (31) was modified such that the absorbance was monitored at 350 nm instead of 405 nm. GMP, GDP, GppNHp, or GTPγS was added at 400-µM concentration into the cuvette (Hellma Analytics) containing 0.5 mg/mL of LUVs and 30 µM of either hGBP1F or hGBP1NF. LUVs for these experiments were made by extrusion using 0.1-µm membranes if not specified otherwise out of BPL. For LUV preparation as well as for the reactions, buffer C1 (buffer C with 1 mM MgCl2) was used. Absorbance of the samples was measured on a Specord200 UV/Vis spectrophotometer (AnalytikJena), where the cuvette holder temperature was set to 30 °C.
Visual Tethering Assay.
Visual tethering assay was modified from the assay used for atlastin studies (31). A total of 30 µM of hGBP1F, hGBP1NF, or R48AF was premixed with liposomes containing ATTO-488-DOPE (ATTO-TEC) (0.5% wt/wt) or Rhodamine-DOPE (Avanti) (0.5% wt/wt) lipids. The vesicles were mixed 1:1 (final lipid concentration 0.8 mg/mL) in a reaction buffer of C1. After addition of the nucleotides, the reaction mixture was incubated for 45 min at 30 °C. Further, the samples of the reaction mixture (15 µL) were diluted 1:20 into buffer C1 and loaded into a newly assembled open homemade observation chamber (55). Fluorescent vesicles were visualized by confocal microscopy. ATTO-488 and Rhodamine dyes were excited with 488- and 561-nm lasers and their emissions were collected with a dichroic mirror (405/488/561) and 525/50-nm and 595/50-nm filters, respectively. Image collection was done using a Nikon Eclipse Ti inverted microscope (Nikon) and NIS software. Image brightness and contrast were adjusted across the entire image, and merged images showing both dyes were generated using Fiji (v1.51e). Fluorescent vesicles were made by extrusion using 0.1-µm membranes.
Fluorescence Microscopy.
For immunofluorescence microscopy, cells were fixed in PBS with 3% paraformaldehyde, permeabilized in PBS with 0.2% saponin (Sigma-Aldrich), and blocked in PBS, 3% BSA and 0.2% saponin. Primary and Alexa Fluor-labeled secondary antibodies (Life Technologies) were applied in blocking buffer. Coverslips were embedded in ProLong Gold Antifade (Life Technologies) and examined using a Zeiss Axioplan2 fluorescence microscope (Zeiss). Proteins and epitope tags were detected with the following antibodies against hGBP1 (1B1): Rab5 (3547, Cell Signaling), Rab7 (sc-10767, Santa Cruz Biotechnology), Rab9 (ALX-804–286, ENZO Life Sciences), EEA1 (610456, BD Biosciences), and Lamp1 (611042, BD Biosciences). For transient transfections, the GeneJuice reagent (Novagen) was used according to the manufacturer’s instructions.
Discussion
The guanylate-binding proteins function in innate immunity against bacteria, viruses, and protozoan pathogens (5–7). Furthermore, they are involved in the regulation of proliferation and migration of endothelial cells (8). For both functions, the ability to interact with cellular membranes is a prerequisite. In the case of hGBP1 and murine GBP2 (mGBP2), abolishing the C-terminal lipid modification results in biologically inactive proteins (26–28, 33). To understand the function and mechanism of hGBP1 on a molecular level, it is therefore of critical importance to study the protein in the lipid modified state.
Using GUVs and LUVs, we were able to study the binding of fluorescently labeled and farnesylated hGBP1 in real time, in particular in the presence of GTP. Most intriguingly, hGBP1F associated with membranes only in the presence of either GTP or its nonhydrolyzable analog GTPγS, but not in the absence of nucleotide or when the GTP hydrolysis products GDP or GMP were added (Fig. 1 and Fig. S3). Nonfarnesylated hGBP1 was not able to associate with GUVs, irrespective of the nucleotide supplied (Fig. 1B). The rate of association of hGBP1F to GUVs after addition of GTP occurs rapidly; most probably it is controlled by the rate of mixing and cannot be resolved accurately with these assays. The rate of hGBP1F dissociation from the GUV membrane could be defined more accurately: the dissociation of GTPγS-bound hGBP1F from GUVs occurred with a half-life of about 120 s. In the case of GTP-bound hGBP1F, the residence time is about 10 s, which is similar to the reciprocal rate constant of hGBP1-catalyzed GTP hydrolysis when bound to the membrane. Because the diffusion of a protein in the cytosol over a distance of a few micrometers is faster than a second, the dynamics of hGBP1 intermembrane traveling are most likely to occur in the range of 10 s, controlled by the rate of dissociation from the membrane. Our data suggest that GTP hydrolysis boosts dissociation of hGBP1F from the membrane. However, we have previously demonstrated the ability of hGBP1F to associate with liposomes only in the presence of GDP·AlFx but not in the presence of GTP (25). This disagreement to the present data is probably due to the dynamics of the membrane interaction of GTP-bound hGBP1F, which impairs the detection in a liposome cosedimentation assay. From our current study, we conclude that GTP binding is sufficient to trigger association of hGBP1F to the membrane and GTP hydrolysis is not required for that. Moreover, membrane binding of hGBP1F seems to not be sensitive to the membrane curvature as the assayed liposomes covered a wide range of diameter from 0.1 µm to 10 µm and did not show considerable differences in binding.
For other proteins carrying a membrane anchor like farnesyl, geranylgeranyl, palmitoyl, or myristoyl groups, in particular GTP-binding proteins, it is well known that their membrane localization is not permanent but rather dynamic, allowing the proteins to translocate to different membranes or even to the cytosol (34). Many examples of Ras-like proteins show nucleotide-dependent shuttling between various membrane compartments, e.g., the GTP-binding and hydrolysis-driven function of Rab proteins is the traveling between organelles and thereby directing the transport of cargo. These cycles of membrane binding and dissociation are facilitated by specific helper proteins [guanine-nucleotide dissociation inhibitors (GDIs)], which take up the isoprenyl anchor in a binding pocket to enable the GTP-binding protein to leave the membrane and travel through the cytosol (35). This mechanism is crucially different for hGBP1 and resembles that of N-terminally myristoylated Arf proteins (36). Human GBP1F attaches to the membrane upon GTP binding and leaves the membrane as a soluble protein when no more GTP is available, obviously not requiring any helper protein for positioning its farnesyl anchor inaccessible for other interaction partners (like the membrane) as is the case for small Ras-like Rho and Rab GTPases (35). Thus, we report that the accessibility of an isoprenyl anchor from a GTP-binding protein is exclusively controlled by the associated nucleotide. We term this mechanism nucleotide-dependent farnesyl switch of hGBP1.
In addition to farnesyl- and nucleotide-dependent membrane binding, we discovered the ability of farnesylated hGBP1 to polymerize in a GTP-dependent manner. Human GBP1F polymerized reversibly in the presence of GTP and irreversibly in the presence of GDP·AlFx. However, in the presence of GDP·AlFx, hGBP1F formed polymers to a much lower extent compared with GTP (Fig. 3). Despite the structural changes within the protein, which occur upon binding of GTPγS and which lead to the accessibility of the farnesyl moiety, binding of this nucleotide does not result in polymerization of hGBP1F as inferred from turbidity as well as from electron microscopy experiments (Fig. 2 and Fig. S4A). We suggest that only GTP hydrolysis is able to mediate conformational changes within the protein structure, which are sufficient for the assembly of hGBP1F and likely multiple rounds of GTP turnover are needed for efficient polymer formation. Considering the biphasic behavior of enzymatic activity, during the first slow phase (1.6 min−1) five GTP molecules can be estimated to be hydrolyzed per enzyme molecule but the second cleavage step resulting in GMP does not occur here. The second phase shows a roughly 10 times faster turnover of GTP, and additionally, the second cleavage step is observed yielding GMP. This biphasic behavior of hGBP1F is more pronounced at 25 °C but also present at 37 °C and has not been observed for nonfarnesylated hGBP1 (19) (Fig. S6), suggesting a principal difference between the way of action of farnesylated and nonfarnesylated hGBP1. The rate of the fast phase of hGBP1F is similar to the self-stimulated enzymatic activity of nonfarnesylated hGBP1 that is achieved by LG domain interactions at sufficiently high protein concentration. Also, the product ratio of GDP and GMP under these conditions for nonfarnesylated hGBP1 resembles the GDP/GMP ratio produced by farnesylated hGBP1 in this phase. In contrast, the slow phase of hGBP1F does not lead to detectable GMP production, similar to the observation with nonfarnesylated hGBP1 at low concentrations, where self-activation is not possible (16, 18). Furthermore, the observed onset of turbidity, accompanied by the change from slow to fast mode of hGBP1F activity, corresponds to a time of about five cycles of GTP hydrolysis in the slow phase. The strong but fully reversible polymerization of hGBP1F during GTP hydrolysis is highly reminiscent of the transient polymerization of murine Irga6 (37). The mutants R48A and K76A of hGBP1F, respectively, show no polymerization in the presence of GTP, underlining the importance of enzymatic activity (Fig. S2). Whereas R48A is almost completely impaired in GTPase activity, K76A showed a four- to fivefold reduced GTPase activity compared with the slow phase of wild type and almost no GMP production. As for this mutant, the second cleavage step is blocked but also the first step is slower than for the wild type; unfortunately, we cannot safely conclude as to the importance of the first and second cleavage steps for polymer formation. The absence of polymer formation of the K76A mutant can be due to the impaired second cleavage step of the nucleotide but it may also be explained by the smaller rate of the first step. For addressing the role of the second hydrolysis step within polymerization, we also considered GpCpp as a suitable GTP analog. However, hGBP1F did not bind GpCpp (Fig. S3F).
We have observed circular polymer structures of hGBP1F with a diameter of about 60 nm in the presence of both GTP and GDP·AlFx (Fig. 2). Similar ring-like structures have been reported for dynamin, human MxA, Dictyostelium discoideum dynamin A (DymA) in the absence of lipids (38–40), and for bacterial dynamin-like protein (BDLP) in the presence of lipids (41). DLPs involved in fission reaction can assemble into helical polymers even in the absence of nucleotides. In the case of hGBP1F, however, the circular structures are formed in the absence of lipids and in the presence of GDP·AlFx or GTP. The radius of these structures is strikingly similar to the hydrodynamic radius of hGBP1F in the presence of GDP·AlFx, observed by DLS and to the length of about 30 nm—a length that also applies to an hGBP1F molecule in a completely open and stretched conformation of the α12/13 domain. In this conformation, the hGBP1F molecule resembles a cone-like geometry. These observations and the analogy to other DLPs support our model shown in Fig. 6, where the LG domains reside outside and the farnesyl anchors in the center. With a width of about 4–5 nm for the LG domain, a full turn of the ring would comprise about 40–50 GBP molecules. Larger oligomers, which are observed only for GTP, may represent helices or stacks of circular structures leading to the elongated structures in Fig. 2.
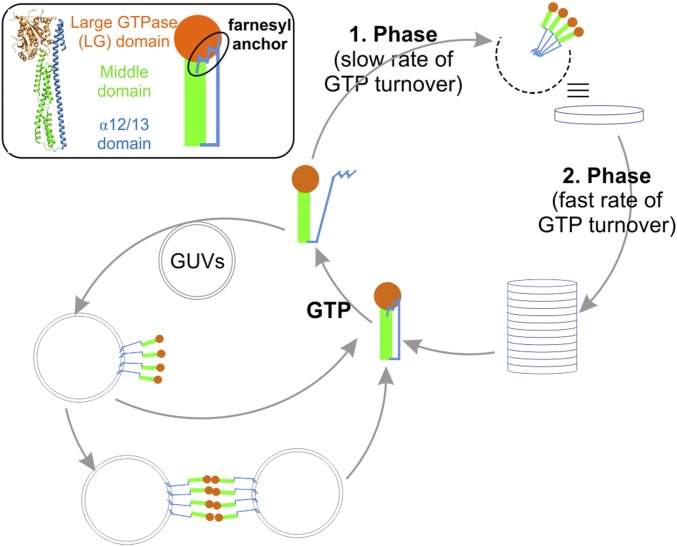
Fig. 6.
Model of hGBP1F function. After binding of GTP to hGBP1F and release of the farnesyl anchor, hGBP1F may follow two cycles: membrane binding with further membrane tethering (Left cycle) or polymerization by formation of a disk in a first phase accompanied by slow GTP hydrolysis followed by stacking of the circular structures to cylindrical structures enabling faster GTP turnover (Right cycle). See the text for more details. (Inset) With the subdomains highlighted in different colors, schematic representation of hGBP1 is according to the crystal structure of nonfarnesylated protein in the nucleotide-free state (Protein Data Bank ID code 1DG3). The farnesyl anchor at the C terminus is depicted as a blue zigzag line (ellipse).
Based on previous reports that nonfarnesylated hGBP1 undergoes major intramolecular opening and particularly releases the α12/13 domain upon GTP binding (12–14), we suggest that also farnesylated protein after mixing with GTP opens up in a similar way, thereby releasing the farnesyl anchor, which leads to polymerization of hGBP1F in an along-side orientation during the first phase. At the end of the first phase, this results in circular structures of the polymers with the farnesyl groups facing the center of the polymer and with the LG domain of the enzyme pointing outward as indicated in our model (Fig. 6). Up to this point, the LG domains do not form GTPase activating contacts to each other and hence provide only a low enzymatic activity. Now, with the change from the first to the second phase, we suggest that the circular structures grow or associate with each other stackwise, to form elongated, cylindrical structures becoming visible by turbidity. At the same time, the LG domain contacts established in the cylinder are responsible for the enhanced turnover rate and for the production of GMP, as both rate and product ratio are similar to LG domain dimer activity. Because the transition from the slow to the fast phase of GTP hydrolysis coincides with the appearance of large polymers, which can be detected by turbidity measurements, it is possible that the LG domains of hGBP1F in the circular structures are not able to form the activating dimer that was observed in the structure of the isolated LG domain (18), unless molecules from adjacent rings come into contact. Intriguingly, the presence of liposomes prevents the shift of the hydrolysis rate toward the fast mode (Fig. 3 D and E), also decreasing the formation of GMP (Fig. S6D). This observation can be rationalized by membrane insertion of hGBP1F at the expense of polymer formation: as in the phase of building up the circular structures by lateral assembly of hGBP1F, the protein apparently does not form activating LG domain contacts when bound to the membrane surface in cis. We have made a similar observation for hGBP1 artificially attached to a chip surface, which also resulted in decreased enzymatic activity (20). Considering a possible GTPase activating effect for hGBP1 dimers in trans—dimers bound to different membranes—this effect would be hardly detectable. The contact area between two associated vesicles is too small compared with the total vesicle surface, meaning there is only a small fraction of hGBP1 dimers in trans compared with the whole hGBP1 pool.
Recent studies on hGBP1 have reported a granular appearance of hGBP1 within the cell (27), which has also been observed for murine GBP2, an ortholog of hGBP1 (29). It has been suggested that the observed granular distribution of hGBP1 within the cell is based on binding of the protein to vesicular structures. We found hGBP1 partially colocalized with several different markers along the phagolysosomal pathway and in particular to phagosomes induced by latex beads (Figs. S9 and S10). Overexpression of marker proteins such as Rab5, Rab7, or Rab9 enhanced this colocalization. Other groups have already identified certain GBPs on phagocytosed latex bead (42, 43) and bacteria (44, 45). In some cases, we observed recruitment of hGBP1 very early during formation of phagocytic cups. In this context it is noteworthy to mention that a direct interaction between hGBP1 and actin has been recently shown (9). Several other groups have found that GBPs are involved in the control of cell proliferation and motility in both humans and mice by interfering with the integrin pathway (8). Along these lines, a role for hGBP1 in the maintenance of the barrier function of the colonic mucosa has been reported (8).
However, our data revealed that individual hGBP1 molecules are not stably membrane associated within the cell but rather in a constant dynamic exchange with the cytosolic pool. This observation is indicated by the fast recovery of hGBP1 in FRAP experiments on both punctate and phagosomal structures (Fig. 5), which is similar to the fast dynamics of the membrane interaction of hGBP1 we observed in vitro. We assume that at least those punctate structures that colocalize with Rab proteins are indeed vesicles. However, the ability of hGBP1F to polymerize in vitro, which we report in this study as well, implies that hGBP1 puncta that do not colocalize with phagolysosomal marker proteins could represent polymers of hGBP1. Similar to the current model for Mx proteins and the mitochondrial Drp1, such polymers of hGBP1 could serve as protein depots, ensuring a kind of buffer function for the rapid mobilization of the protein in immune response (46–48). Alternatively, such polymers could function as a scaffold for the assembly of other large immune-related complexes such as the inflammasome (5, 49, 50). At this point, we do not have data to judge the relative amounts of hGBP1 assembled in polymers or residing at membrane surfaces. High intracellular concentrations of hGBP1 are only present after stimulation by IFN-γ or other cytokines. Nevertheless, the enzymatic activity is crucial for polymer formation and at the same time it allows for a highly dynamic assembling process, making hGBP1 molecules available wherever in the cell and whenever they are needed (e.g., parasite combat).
Finally, we report membrane tethering features of hGBP1F, which are also strictly nucleotide controlled (Fig. 4). A number of DLPs are involved in the fusion events within the cell (4, 10, 51). The molecular mechanism of membrane tethering and fusion by dynamin-related GTPases is best understood for atlastins (31, 52). We suggest that hGBP1F might mediate the tethering effect through dimerization of LG domains similar to atlastins. Our previous results demonstrate the ability of hGBP1 to dimerize through its LG domains in the presence of nonhydrolyzable GTP analogs (11, 18). The observation of the vesicle tethering effect of hGBP1F in the presence of GTPγS implies that GTP hydrolysis is not required for the tethering activity. However, it is intriguing to see that GTP hydrolysis leads to a much faster increase of the contact area between the two vesicles. Along this line, after consumption of GTP, a relaxation back to a presumably nontensed situation of the two contacting membranes is observed. We did not observe any fusion events of GUVs mediated by hGBP1F, a finding also supported by reversible detachment of tethered LUVs upon GMP addition (Fig. S8). However, membrane fusion may require the aid of other proteins, which was not possible to elucidate with our experimental setup. Nevertheless, the observation of tethering underlines the functional similarity of GBPs with atlastins and also other dynamin family members as recently shown for mitofusin (53). Recent studies on the antibacterial effect of GBPs indicate that hGBP1 and 2 are recruited to bacterial inclusions of Chlamydia trachomatis and trigger their rerouting for lysosomal degradation (54). It is tempting to speculate that the tethering effect of hGBP1 and its localization along the phagolysosomal pathway, which we report here, could be the basis for the physiological function of the protein. The tethering of phagolysosomal membranes could result in an enhanced clearance of intracellular pathogens by a more efficient delivery to lysosomes. Similarly, hGBP1 could stimulate the internalization and down-regulation of surface receptors, such as certain integrins resulting in the described function in cell adhesion and motility (8). To test these hypotheses, the effect of tethering-impaired mutants of hGBP1 such as R48A could be tested with respect to membrane internalization, vesicular trafficking, and pathogen proliferation.
In conclusion, binding of farnesylated hGBP1 to membranes as well as the formation of hGBP1 polymers are strictly nucleotide controlled. Thus, the farnesyl anchor of hGBP1 is masked in the GDP- and the GMP-bound states, rendering the lipid-modified protein a soluble monomer in solution. As illustrated in our model in Fig. 6, binding of GTP leads to opening of the protein structure and to the release of the farnesyl anchor, enabling its immediate insertion into a membrane in the vicinity of the protein molecule (Fig. 6, Left cycle). Alternatively, the opening of hGBP1 and the concomitant availability of the farnesyl group and previously hidden protein surfaces facilitates polymerization of the enzyme, leading first to circular structures expanding to a cylindrical shape by stacking or helix formation in a second phase (Fig. 6, Right cycle). Hence, the nucleotide-controlled dynamic process of structural changes and release of the farnesyl anchor can result in membrane insertion followed by tethering processes (or more) on the one hand, and in the formation of well-defined polymers representing enzyme depots on the other.
Molecular switching between different nucleotide states by GTP binding and hydrolysis is the common feature for all GTPases, which is controlled by regulatory proteins. For hGBP1, which does not require any regulatory proteins for nucleotide exchange, we show that this switching controls the availability of the C-terminal farnesyl anchor, which is then used in a dual function, namely, membrane tethering and enzyme polymerization. In hGBP1 the combination of an assembly-stimulated GTPase reaction with a conformation-dependent release of the C-terminal farnesyl anchor in hGBP1 results in a unique molecular machinery—the farnesyl switch—which controls the membrane recruitment and polymerization activity of the protein.
Methods
Protein Expression, Purification, and Labeling.
Synthesis of recombinant hGBP1, either hGBP1NF or hGBP1F, was performed as previously described (25). Farnesylated as well as nonfarnesylated proteins were labeled with Alexa-488 C5 maleimide dye (Life Technologies). The reaction mixture was prepared in buffer C (50 mM Tris⋅HCl, pH 7.9, 5 mM MgCl2, 150 mM NaCl) by mixing 100 μM of protein with 400 μM of dye. After incubation on ice for 60 min, unreacted dye was removed via spin desalting columns (Thermo Scientific).
Preparation of Liposomes.
GUVs were electroformed as described before (55) from the mixture of 99% brain polar lipids extract (Avanti Polar Lipids) and 1% tetramethyl rhodamine phosphatidylinositol 4,5-bisphosphate (TMR-PIP2, Echelon) in 500 mM sucrose solution. LUVs were produced by extrusion through polycarbonate filters with a defined pore size of 0.1 µm, 0.20 µm, or 1 µm (Whatman) as previously described (25). The size of LUVs was confirmed by DLS.
Confocal Microscopy.
A newly assembled open homemade observation chamber was incubated with a casein solution (1 mg/mL) for 5 min to prevent adhesion of GUVs to the coverslip. After rinsing the chamber with buffer C (see above), the chamber was filled with 300 µL of experimental mixture prepared in buffer C, followed by injection of GUVs (3 µL). Protein and lipid fluorescence channels were observed by dual-color confocal microscopy (λ1 = 488 nm and λ2 = 543 nm) on a Nikon Eclipse Ti inverted microscope (Nikon). If required, GUVs were manipulated by micropipettes controlled by a motorized micromanipulator (MP-225, Sutter Instrument). Injection of either protein or nucleotide into the chamber was performed with an injection micropipette having typically a radius of 10 μm, controlled with a hydraulic micromanipulator (Narishige). All experiments were performed with identical laser power and gain settings as well as with the same illumination and detection settings. In dissociation and tethering assays, all experiments were recorded as fluorescent videos in the presence of oxygen scavenging mix [25 mM glucose, 0.37 mg/mL glucose oxidase (Sigma-Aldrich), 0.21 mg/mL catalase (Sigma-Aldrich), 5 mM DTT].
Image Analysis.
All fluorescent images were processed and analyzed with ImageJ software. Fluorescence signals of the protein on the membrane of GUVs, Fmembrane was quantified with the PlotProfile built-in command of ImageJ across narrow equatorial rectangles (Fig. S7 and SI Methods). The background fluorescence of unbound protein, Fbackground was measured in the vicinity of GUVs. The fluorescence intensity of the GUV-bound protein was denoted as I = (Fmembrane − Fbackground). For dissociation and tethering assays, fluorescence values of the GUV-bound protein were taken as Fmembrane. For the quantification of the tethering effect, the contact length was defined as a distance between two ends of a membrane segment shared by a pair of GUVs. The contact lengths were quantified from the images by a self-designed algorithm using a javascript for ImageJ (Fig. S7 and SI Methods).
Transfections and Fluorescence Microscopy.
HeLa B cells (European Collection of Authenticated Cell Cultures 85060701) were grown on coverslips in DMEM supplemented with 10% heat-inactivated calf serum (Sigma-Aldrich), penicillin (100 units/mL) (Gibco), streptomycin (100 mg/mL), and nonessential amino acids solution 10 mM (100×) (Gibco) at 37 °C with 5% CO2 in a humidified incubator. Expression of hGBP1 was induced with 200 units/mL of recombinant human IFN-γ (PeproTech) for at least 24 h. For transient transfections the GeneJuice reagent (Novagen) was used according to the manufacturer’s instructions.
Phagocytosis of latex beads was stimulated by starving in FCS-free DMEM for 2 h before adding of fluorescent or nonfluorescent latex beads (Life Technologies). After another 2 h, cells were either fixed for 20 min in 4% paraformaldehyde in PBS or analyzed by life cell imaging on a Nikon Eclipse Ti spinning disk confocal microscope. Images were analyzed with Volocity (Perkin-Elmer). After taking a reference picture of the unbleached cell, the ROI was bleached with 561-nm or 488-nm lasers with 100% laser power and a duration of 30 cycles (several seconds). Subsequently, cells were continuously imaged for 1 min and during the following 5 min, two pictures per minute were taken. In addition, one ROI of any fluorescent structure within the cell and another outside of the cell were imaged for normalization. Normalized curves were fitted with an exponential function yielding half-lives (τ1/2) of fluorescence recovery.
Absorbance-Based Polymerization Assay.
Absorbance-based measurements were performed in quartz precision cells (Hellma Analytics) on Specord200 UV/Vis spectrophotometer (AnalytikJena). The absorbance of the sample was measured at 350 nm in temperature-controlled cuvettes. Nucleotides were injected into the cuvette at final concentrations of 1 mM after initial preequilibration of the experimental mixture for 200 s. Experiments were performed either in buffer C (see above) or in buffer D (50 mM Tris⋅HCl, pH 8.0, 130 mM KCl, 2 mM MgCl2).
In the presence of GTP, we additionally monitored the nucleotide composition within the experimental mixture if required. For that monitoring, at defined time points, 5-μL samples were taken from the cuvettes and the GTPase reaction was immediately stopped by addition of 10 μL of 10% H3PO4, which was followed by neutralization with 30 μL of 0.77 M K2HPO4. Final samples were centrifuged for 2 min at 16,000 × g and further nucleotide composition was analyzed as described previously (19). Briefly, nucleotides were separated by reversed-phase HPLC using a Chromolith Performance RP-18 endcapped column (Merck) and monitored at 254 nm (MD-2010 Plus, Jasco). For quantifying the share of GMP, GDP, and GTP, peak areas corresponding to each nucleotide were integrated with the manufacturer’s software. For experiments in the presence of liposomes, LUVs were produced by extrusion through polycarbonate filters with a defined pore size of 0.1 µm (Whatman) as previously described (25).
Monitoring Polymerization of hGBP1 with Dynamic Light Scattering.
Dynamic light scattering of unmodified and farnesylated hGBP1 was determined with the Zetasizer Nano ZS (Malvern Instruments) under different conditions. A total of 20 µM of protein in buffer D (see above) in a UV microcuvette was placed into the cell holder and preincubated at 25 °C for 5 min. The measurement was started quickly after addition of 300 µM AlCl3, 10 mM NaF, and 200 µM GDP (GDP⋅AlFx) or 1 mM GTP in a total volume of 84 µL. The change of the particle size was monitored over 6–10 measurements, each consisting of four runs of 10 s each and with a delay of 4 s between the measurements. The hydrodynamic radius (RH) of the particles was determined with the manufacturer’s software.
Negative Stain Electron Microscopy.
Polymerization of hGBP1 was analyzed by negative stain electron microscopy. Pure protein (10 µM) in buffer D with 2 mM DTT was incubated at room temperature for 5–15 min in the presence of 200 µM GDP with 300 µM AlCl3 and 10 mM NaF or 1 mM GTP or GTPγS, respectively, or in the absence of nucleotide. Samples were bound to a Ni-300 grid with an incubation for 5 min. After removal of excess solution, negative staining was done two times for 2 min each with 2% uranyl acetate. Finally, preparations were examined in a Zeiss EM109 microscope (Zeiss).
Supplementary Material
Acknowledgments
We thank Klaus Boller and Regina Eberle for help with electron microscopy and Christiane Horst and Claudia Grundmann for excellent technical support. The research leading to these results has received funding from the European Commission’s Seventh Framework Program (FP7/2007-2013) under Grant TRANSPOL 264399; and the Deutsche Forschungsgemeinschaft with Grants HE 2679/6-1, PR 1090/3-1 within the priority program 1580, within the collaborative research center SFB635 and within the Cluster of Excellence RESOLV, EXC 1069.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1620959114/-/DCSupplemental.
References
- 1.Cheng YS, Colonno RJ, Yin FH. Interferon induction of fibroblast proteins with guanylate binding activity. J Biol Chem. 1983;258:7746–7750. [PubMed] [Google Scholar]
- 2.Praefcke GJK, McMahon HT. The dynamin superfamily: Universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol. 2004;5:133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- 3.Ferguson SM, De Camilli P. Dynamin, a membrane-remodelling GTPase. Nat Rev Mol Cell Biol. 2012;13:75–88. doi: 10.1038/nrm3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McNew JA, Sondermann H, Lee T, Stern M, Brandizzi F. GTP-dependent membrane fusion. Annu Rev Cell Dev Biol. 2013;29:529–550. doi: 10.1146/annurev-cellbio-101512-122328. [DOI] [PubMed] [Google Scholar]
- 5.Pilla-Moffett D, Barber MF, Taylor GA, Coers J. Interferon-inducible GTPases in host resistance, inflammation and disease. J Mol Biol. 2016;428:3495–3513. doi: 10.1016/j.jmb.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meunier E, Broz P. Interferon-inducible GTPases in cell autonomous and innate immunity. Cell Microbiol. 2016;18:168–180. doi: 10.1111/cmi.12546. [DOI] [PubMed] [Google Scholar]
- 7.Man SM, Karki R, Kanneganti T-D. Molecular mechanisms and functions of pyroptosis, inflammatory caspases and inflammasomes in infectious diseases. Immunol Rev. 2017;277:61–75. doi: 10.1111/imr.12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Britzen-Laurent N, Herrmann C, Naschberger E, Croner RS, Stürzl M. Pathophysiological role of guanylate-binding proteins in gastrointestinal diseases. World J Gastroenterol. 2016;22:6434–6443. doi: 10.3748/wjg.v22.i28.6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ostler N, et al. Gamma interferon-induced guanylate binding protein 1 is a novel actin cytoskeleton remodeling factor. Mol Cell Biol. 2014;34:196–209. doi: 10.1128/MCB.00664-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daumke O, Praefcke GJK. Invited review: Mechanisms of GTP hydrolysis and conformational transitions in the dynamin superfamily. Biopolymers. 2016;105:580–593. doi: 10.1002/bip.22855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prakash B, Praefcke GJK, Renault L, Wittinghofer A, Herrmann C. Structure of human guanylate-binding protein 1 representing a unique class of GTP-binding proteins. Nature. 2000;403:567–571. doi: 10.1038/35000617. [DOI] [PubMed] [Google Scholar]
- 12.Vöpel T, et al. Mechanism of GTPase-activity-induced self-assembly of human guanylate binding protein 1. J Mol Biol. 2010;400:63–70. doi: 10.1016/j.jmb.2010.04.053. [DOI] [PubMed] [Google Scholar]
- 13.Vöpel T, Kunzelmann S, Herrmann C. Nucleotide dependent cysteine reactivity of hGBP1 uncovers a domain movement during GTP hydrolysis. FEBS Lett. 2009;583:1923–1927. doi: 10.1016/j.febslet.2009.05.027. [DOI] [PubMed] [Google Scholar]
- 14.Vöpel T, et al. Triphosphate induced dimerization of human guanylate binding protein 1 involves association of the C-terminal helices: A joint double electron-electron resonance and FRET study. Biochemistry. 2014;53:4590–4600. doi: 10.1021/bi500524u. [DOI] [PubMed] [Google Scholar]
- 15.Schwemmle M, Staeheli P. The interferon-induced 67-kDa guanylate-binding protein (hGBP1) is a GTPase that converts GTP to GMP. J Biol Chem. 1994;269:11299–11305. [PubMed] [Google Scholar]
- 16.Kunzelmann S, Praefcke GJK, Herrmann C. Transient kinetic investigation of GTP hydrolysis catalyzed by interferon-gamma-induced hGBP1 (human guanylate binding protein 1) J Biol Chem. 2006;281:28627–28635. doi: 10.1074/jbc.M604911200. [DOI] [PubMed] [Google Scholar]
- 17.Praefcke GJK, Geyer M, Schwemmle M, Robert Kalbitzer H, Herrmann C. Nucleotide-binding characteristics of human guanylate-binding protein 1 (hGBP1) and identification of the third GTP-binding motif. J Mol Biol. 1999;292:321–332. doi: 10.1006/jmbi.1999.3062. [DOI] [PubMed] [Google Scholar]
- 18.Ghosh A, Praefcke GJK, Renault L, Wittinghofer A, Herrmann C. How guanylate-binding proteins achieve assembly-stimulated processive cleavage of GTP to GMP. Nature. 2006;440:101–104. doi: 10.1038/nature04510. [DOI] [PubMed] [Google Scholar]
- 19.Kunzelmann S, Praefcke GJK, Herrmann C. Nucleotide binding and self-stimulated GTPase activity of human guanylate-binding protein 1 (hGBP1) Methods Enzymol. 2005;404:512–527. doi: 10.1016/S0076-6879(05)04045-0. [DOI] [PubMed] [Google Scholar]
- 20.Syguda A, et al. Immobilization of biotinylated hGBP1 in a defined orientation on surfaces is crucial for uniform interaction with analyte proteins and catalytic activity. Langmuir. 2012;28:6411–6418. doi: 10.1021/la3008359. [DOI] [PubMed] [Google Scholar]
- 21.Abdullah N, Balakumari M, Sau AK. Dimerization and its role in GMP formation by human guanylate binding proteins. Biophys J. 2010;99:2235–2244. doi: 10.1016/j.bpj.2010.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olszewski MA, Gray J, Vestal DJ. In silico genomic analysis of the human and murine guanylate-binding protein (GBP) gene clusters. J Interferon Cytokine Res. 2006;26:328–352. doi: 10.1089/jir.2006.26.328. [DOI] [PubMed] [Google Scholar]
- 23.Stickney JT, Buss JE. Murine guanylate-binding protein: Incomplete geranylgeranyl isoprenoid modification of an interferon-gamma-inducible guanosine triphosphate-binding protein. Mol Biol Cell. 2000;11:2191–2200. doi: 10.1091/mbc.11.7.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nantais DE, Schwemmle M, Stickney JT, Vestal DJ, Buss JE. Prenylation of an interferon-gamma-induced GTP-binding protein: The human guanylate binding protein, huGBP1. J Leukoc Biol. 1996;60:423–431. doi: 10.1002/jlb.60.3.423. [DOI] [PubMed] [Google Scholar]
- 25.Fres JM, Müller S, Praefcke GJK. Purification of the CaaX-modified, dynamin-related large GTPase hGBP1 by coexpression with farnesyltransferase. J Lipid Res. 2010;51:2454–2459. doi: 10.1194/jlr.D005397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tripal P, et al. Unique features of different members of the human guanylate-binding protein family. J Interferon Cytokine Res. 2007;27:44–52. doi: 10.1089/jir.2007.0086. [DOI] [PubMed] [Google Scholar]
- 27.Britzen-Laurent N, et al. Intracellular trafficking of guanylate-binding proteins is regulated by heterodimerization in a hierarchical manner. PLoS One. 2010;5:e14246. doi: 10.1371/journal.pone.0014246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Modiano N, Lu YE, Cresswell P. Golgi targeting of human guanylate-binding protein-1 requires nucleotide binding, isoprenylation, and an IFN-gamma-inducible cofactor. Proc Natl Acad Sci USA. 2005;102:8680–8685. doi: 10.1073/pnas.0503227102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kravets E, et al. The GTPase activity of murine guanylate-binding protein 2 (mGBP2) controls the intracellular localization and recruitment to the parasitophorous vacuole of Toxoplasma gondii. J Biol Chem. 2012;287:27452–27466. doi: 10.1074/jbc.M112.379636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Praefcke GJK, et al. Identification of residues in the human guanylate-binding protein 1 critical for nucleotide binding and cooperative GTP hydrolysis. J Mol Biol. 2004;344:257–269. doi: 10.1016/j.jmb.2004.09.026. [DOI] [PubMed] [Google Scholar]
- 31.Liu TY, et al. Cis and trans interactions between atlastin molecules during membrane fusion. Proc Natl Acad Sci USA. 2015;112:E1851–E1860. doi: 10.1073/pnas.1504368112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jordens I, et al. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr Biol. 2001;11:1680–1685. doi: 10.1016/s0960-9822(01)00531-0. [DOI] [PubMed] [Google Scholar]
- 33.Kravets E, et al. Guanylate binding proteins (GBPs) directly attack T. gondii via supramolecular complexes. eLife. 2016;5:1–30. doi: 10.7554/eLife.11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmick M, Kraemer A, Bastiaens PIH. Ras moves to stay in place. Trends Cell Biol. 2015;25:190–197. doi: 10.1016/j.tcb.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 35.Goody RS, Rak A, Alexandrov K. The structural and mechanistic basis for recycling of Rab proteins between membrane compartments. Cell Mol Life Sci. 2005;62:1657–1670. doi: 10.1007/s00018-005-4486-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pasqualato S, Renault L, Cherfils J. Arf, Arl, Arp and Sar proteins: A family of GTP-binding proteins with a structural device for ‘front-back’ communication. EMBO Rep. 2002;3:1035–1041. doi: 10.1093/embo-reports/kvf221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uthaiah RC, Praefcke GJK, Howard JC, Herrmann C. IIGP1, an interferon-gamma-inducible 47-kDa GTPase of the mouse, showing cooperative enzymatic activity and GTP-dependent multimerization. J Biol Chem. 2003;278:29336–29343. doi: 10.1074/jbc.M211973200. [DOI] [PubMed] [Google Scholar]
- 38.Hinshaw JE, Schmid SL. Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature. 1995;374:190–192. doi: 10.1038/374190a0. [DOI] [PubMed] [Google Scholar]
- 39.Klockow B, et al. The dynamin A ring complex: Molecular organization and nucleotide-dependent conformational changes. EMBO J. 2002;21:240–250. doi: 10.1093/emboj/21.3.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haller O, Kochs G. Interferon-induced mx proteins: dynamin-like GTPases with antiviral activity. Traffic. 2002;3:710–717. doi: 10.1034/j.1600-0854.2002.31003.x. [DOI] [PubMed] [Google Scholar]
- 41.Low HH, Löwe J. A bacterial dynamin-like protein. Nature. 2006;444:766–769. doi: 10.1038/nature05312. [DOI] [PubMed] [Google Scholar]
- 42.Jutras I, et al. Modulation of the phagosome proteome by interferon-gamma. Mol Cell Proteomics. 2008;7:697–715. doi: 10.1074/mcp.M700267-MCP200. [DOI] [PubMed] [Google Scholar]
- 43.Trost M, et al. The phagosomal proteome in interferon-gamma-activated macrophages. Immunity. 2009;30:143–154. doi: 10.1016/j.immuni.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 44.Tietzel I, El-Haibi C, Carabeo RA. Human guanylate binding proteins potentiate the anti-chlamydia effects of interferon-gamma. PLoS One. 2009;4:e6499. doi: 10.1371/journal.pone.0006499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fiegl D, et al. Amphisomal route of MHC class I cross-presentation in bacteria-infected dendritic cells. J Immunol. 2013;190:2791–2806. doi: 10.4049/jimmunol.1202741. [DOI] [PubMed] [Google Scholar]
- 46.Patzina C, Haller O, Kochs G. Structural requirements for the antiviral activity of the human MxA protein against Thogoto and influenza A virus. J Biol Chem. 2014;289:6020–6027. doi: 10.1074/jbc.M113.543892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao S, et al. Structure of myxovirus resistance protein a reveals intra- and intermolecular domain interactions required for the antiviral function. Immunity. 2011;35:514–525. doi: 10.1016/j.immuni.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 48.Macdonald PJ, et al. A dimeric equilibrium intermediate nucleates Drp1 reassembly on mitochondrial membranes for fission. Mol Biol Cell. 2014;25:1905–1915. doi: 10.1091/mbc.E14-02-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shenoy AR, et al. GBP5 promotes NLRP3 inflammasome assembly and immunity in mammals. Science. 2012;336:481–485. doi: 10.1126/science.1217141. [DOI] [PubMed] [Google Scholar]
- 50.Finethy R, et al. Guanylate binding proteins enable rapid activation of canonical and noncanonical inflammasomes in Chlamydia-infected macrophages. Infect Immun. 2015;83:4740–4749. doi: 10.1128/IAI.00856-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Escobar-Henriques M, Anton F. Mechanistic perspective of mitochondrial fusion: Tubulation vs. fragmentation. Biochim Biophys Acta. 2013;1833:162–175. doi: 10.1016/j.bbamcr.2012.07.016. [DOI] [PubMed] [Google Scholar]
- 52.Hu J, Rapoport TA. Fusion of the endoplasmic reticulum by membrane-bound GTPases. Semin Cell Dev Biol. 2016;60:105–111. doi: 10.1016/j.semcdb.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 53.Qi Y, et al. Structures of human mitofusin 1 provide insight into mitochondrial tethering. J Cell Biol. 2016;215:621–629. doi: 10.1083/jcb.201609019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Al-Zeer MA, Al-Younes HM, Lauster D, Abu Lubad M, Meyer TF. Autophagy restricts Chlamydia trachomatis growth in human macrophages via IFNG-inducible guanylate binding proteins. Autophagy. 2013;9:50–62. doi: 10.4161/auto.22482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morlot S, et al. Membrane shape at the edge of the dynamin helix sets location and duration of the fission reaction. Cell. 2012;151:619–629. doi: 10.1016/j.cell.2012.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.