Significance
Bacterial migration, aggregation, and even host infection depend on the generation of mechanical force. Despite their biomedical importance, forces between bacteria and surfaces have not yet been measured during migration. We present a first study of bacterial cell–substrate traction using Myxococcus xanthus as a model organism. M. xanthus exhibits two common mechanisms of motility, namely, twitching and gliding. We find that these mechanisms lead to distinct patterns of traction during motion as an individual or in groups. Twitching leads to local, uncoordinated traction, and gliding in groups allows for collective emergence of directional traction. The forces produced by twitching or gliding of individual cells are significantly amplified when cells move in groups.
Keywords: Myxococcus xanthus, bacteria, traction force, twitching, gliding
Abstract
From colony formation in bacteria to wound healing and embryonic development in multicellular organisms, groups of living cells must often move collectively. Although considerable study has probed the biophysical mechanisms of how eukaryotic cells generate forces during migration, little such study has been devoted to bacteria, in particular with regard to the question of how bacteria generate and coordinate forces during collective motion. This question is addressed here using traction force microscopy. We study two distinct motility mechanisms of Myxococcus xanthus, namely, twitching and gliding. For twitching, powered by type-IV pilus retraction, we find that individual cells exert local traction in small hotspots with forces on the order of 50 pN. Twitching bacterial groups also produce traction hotspots, but with forces around 100 pN that fluctuate rapidly on timescales of <1.5 min. Gliding, the second motility mechanism, is driven by lateral transport of substrate adhesions. When cells are isolated, gliding produces low average traction on the order of 1 Pa. However, traction is amplified approximately fivefold in groups. Advancing protrusions of gliding cells push, on average, in the direction of motion. Together, these results show that the forces generated during twitching and gliding have complementary characters, and both forces have higher values when cells are in groups.
Many bacteria possess the ability to migrate over surfaces in large groups to facilitate such diverse phenomena as predation, aggregation, and biofilm formation. Research into the motility of microbes over the past few decades has made considerable progress toward an understanding of how single cells move, particularly the proteins involved, their regulation, and their ability to generate mechanical forces. However, the properties of the generated surface traction and the coordination of forces by multiple cells to produce coherent group motion remain unclear.
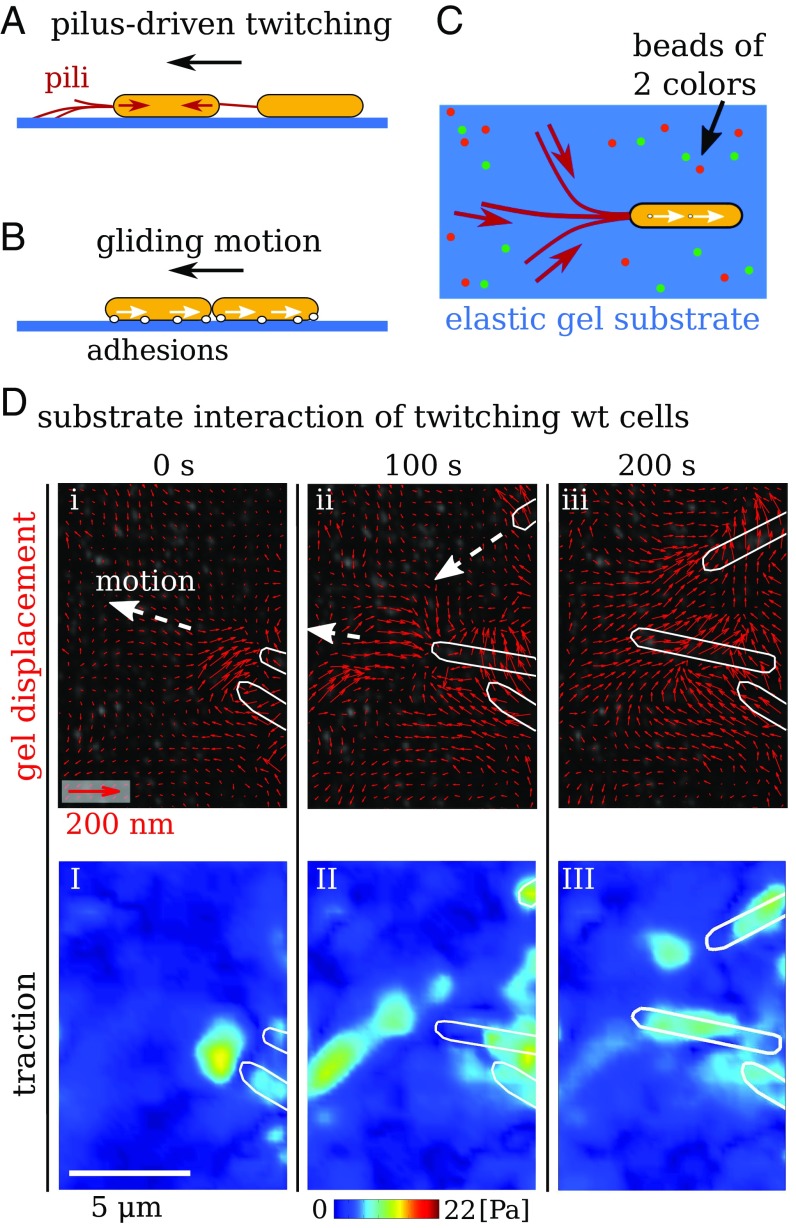
Myxococcus xanthus exhibits complex collective behaviors, including vegetative swarming, predation, and fruiting body formation (1). This organism is well characterized and uniquely suited for motility studies. It uses two migration machineries (2) to move in an intermittent forward–backward motion (3, 4) (Fig. 1 A and B). First, twitching, sometimes called social (S), motility (5, 6) is powered by the extension and retraction of type-IV pili, whereby extruded filaments adhere to the surface and filament retraction produces motility (7–9) (Fig. 1A). Pili also mediate cell–cell adhesion, and retraction has been shown to be triggered by polysaccharides on neighboring cells (10, 11). A second, genetically distinct, motility system (2, 12) is termed gliding, or adventurous (A), motility. Here, a gliding transducer complex (13) that spans the membranes and periplasm converts the transmembrane proton gradient into force (14, 15). Motion occurs through translation of substrate adhesion sites along the cell body (16, 17) (Fig. 1B).
Fig. 1.
Measurement of substrate traction resulting from the two migration machineries of M. xanthus. (A) Type-IV pilus extension–retraction cycles allow bacterial motion referred to as twitching motility or S motility. Pili can also mediate mechanical cell–cell coordination. (B) Gliding or A motility results from lateral translation of transmembrane complexes along the bacterium. Steric interactions allow directional coordination of gliding cells. (C) Top view of the setup for TFM. Cells are placed on a gel containing fluorescent marker beads of two colors. (D) Gel deformation and substrate traction resulting from motion of individual wild-type bacteria that can twitch and glide. White outlines show contours of bacteria. (i–iii) Red quivers show gel displacements. (I–III) Calculated traction magnitude.
Although many of the molecular details of these two systems are known, it is unclear if individual cells produce any measurable force during migration, or if and how groups of cells coordinate these forces. Inertia and hydrodynamic forces for these cells are negligible. For example, the drag force on a cell moving at a typical Myxococcus migration speed of is on the order of . Large traction forces will only occur if cells need to overcome friction with the surface or if the translation machinery itself has internal friction, similar to the situation for eukaryotic cells (18). Collective migration of bacteria within a contiguous group is even less understood. Could forces arise from a balance between cell–substrate and cell–cell interactions? Would this balance be local, or span larger distances within the group? Might one have “leader” cells at the advancing front of the group that exert forces locally to pull along those cells in the ranks behind? Or do all cells move forward from the back and push the advancing group forward? Finally, what are the timescales of force reorganization in groups? These mechanical aspects of bacterial migration have, to date, remained largely inaccessible to direct experimental measurement. In this study, we address such questions using a spatially resolved measurement of the cell–substrate stress resulting from twitching and gliding of M. xanthus using traction force microscopy (TFM) (18–21).
Results
For TFM, cells are placed on an elastic gel and imaged from above or below (Fig. 1C and SI Appendix, Fig. S1). Lateral cell–substrate forces during cell migration produce a deformation of the gel, allowing calculation of traction. A widely used elastic substrate for TFM is polyacrylamide (PAA), which has a broad range of tunable elasticity. Furthermore, nanometer-sized fluorescent beads can be embedded into the small gel pores, allowing tracking of gel displacement. PAA gels are prepared following established protocols (20, 22, 23), and elastic properties are characterized (see SI Appendix). Due to the weak bacterial force, it is necessary to use very soft gels with an elastic shear modulus of . We also use fluorescent marker beads of two colors, which increases the spatial resolution of TFM to around . To improve bacterial motility, gels are coated with chitosan, which has been previously used for studying M. xanthus (17, 24) (see SI Appendix). Time-lapse imaging of the fluorescent beads allows measurement of a spatiotemporally varying deformation field relative to the first frame of a sequence. This deformation field is then used to calculate a continuous field of relative traction that bacteria exert on to the substrate (20). The traction calculated in this way is measured relative to the possibly prestressed first frame of an imaging sequence. To obtain a reliable estimate of forces applied by pili, we use an alternative method relying on the assumption that pili produce point forces (20, 25) (see SI Appendix). To test if M. xanthus produces any measurable substrate forces during migration, we investigated wild-type cells with the ability to both twitch and glide. Fig. 1D shows representative results for the displacement field and traction maps that clearly demonstrate the presence of substrate forces below and ahead of migrating bacteria. For wild-type cells, the alternative modes of twitching and gliding lead to a high variability of traction in different experiments (SI Appendix, Fig. S2 A and B). A twitching- and gliding-deficient mutant pilA-aglQ (13, 17) produces no measurable traction (SI Appendix, Fig. S3).
Individual Twitching Cells Produce Small Hotspots of Traction.
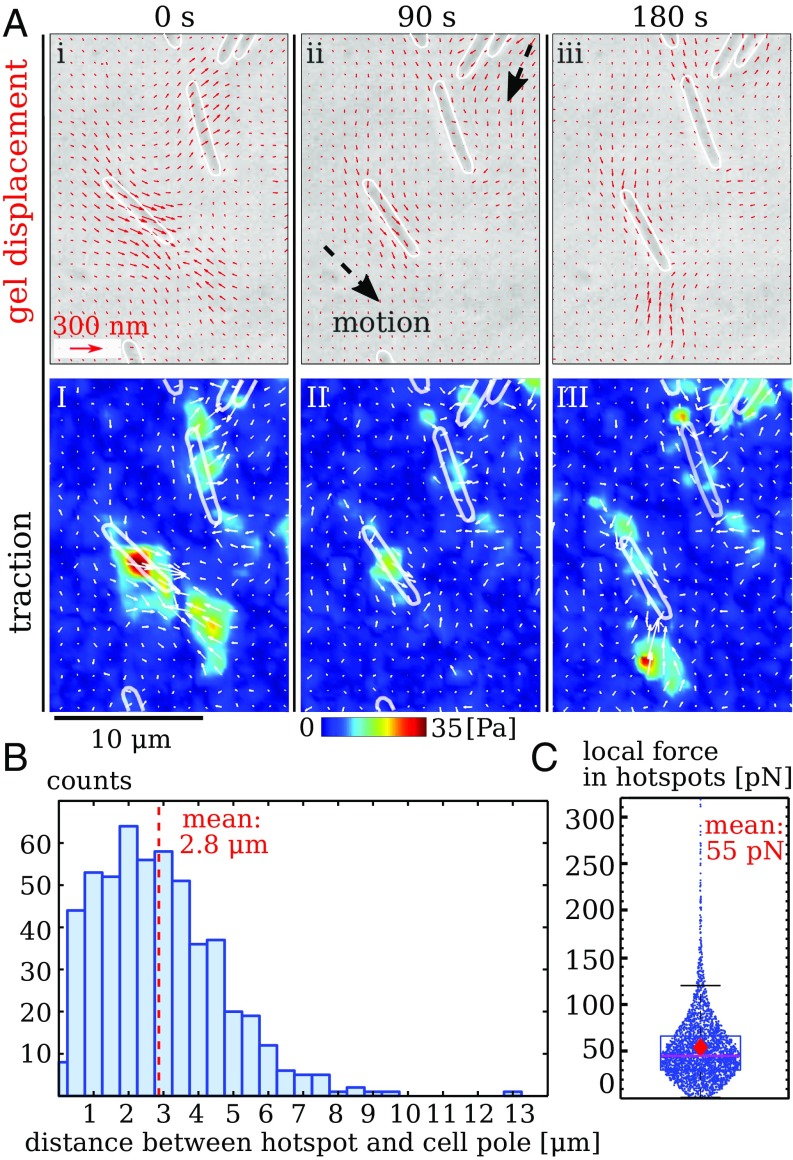
To isolate the different motility systems, we first probed twitching cells that lack the ability to glide due to a deletion of the aglQ gene (15). We observe localized areas of substrate deformation immediately in front of twitching cells, yielding bead displacements on the order of ; see Fig. 2A, I–III and Movie S1. The corresponding calculated traction is concentrated in hotspots, which have an apparent size on the order of due to resolution limitations. Time-lapse images in Fig. 2A, I–III demonstrate that the traction field is dynamic and changes on a timescale on the order of a minute. Among moving cells, not all show measurable traction at all times. If hotspots are present, we observe on average two to three of them, with as little as one and as many as six. Hotspots in front of cells mostly do not stretch all of the way to the cell bodies, which demonstrates that pili mostly engage the substrate at their tips. The distance between hotspots and the closest cell pole is, on average, but can be up to (Fig. 2B), consistent with electron microscopy data for pili lengths (26).
Fig. 2.
Twitching of individual, gliding-deficient bacteria that move by using their pili (aglQ). (A) Gel deformation and substrate traction at three time points. White outlines show contours of bacteria. (i–iii) Quiver plot of gel displacements. (I–III) Calculated traction magnitude. Note that hotspots appear in front of bacteria. (B) Distances between hotspots and the nearest cell pole. (C) Magnitude of overall force in individual hotspots as estimated by assuming point forces. Dots are individual measurements, box contains 25 to 75% of data around the median; diamond shows the mean. Data for B and C were collected from seven experiments with more than five cells each.
Twitching can lead to tug-of-war motion (27), where forces at different pili counteract each other. In our experiments, the random diffusive character of twitching motion (SI Appendix, Fig. S4) and the presence of counteracting forces are consistent with tug-of-war dynamics. However, these experiments do not distinguish between a model where counteracting forces result from pili pulling against other pili and a model where pili pull against passive resistance from the substrate interaction with the cell body. Note that the long range of pili allows bacteria to connect to each other even when they are seemingly far apart. These invisible mechanical links among cells render a detailed assessment of a force balance on the level of individual bacteria difficult. In addition, the force applied at individual hotspots cannot be estimated from local integration of the traction field, because undersampling suppresses those high-frequency spatial variations that affect the force magnitude strongest.
Nevertheless, the clear localization of traction in hotspots makes it possible to estimate the overall force corresponding to each hotspot by assuming that the force is applied only at one point (see SI Appendix). This approach yields an improved estimate of force magnitude for localized traction (20). For individual twitching bacteria, we find that the hotspots correspond, on average, to around , where almost all forces are smaller than (Fig. 2C). These numbers may be compared with data from optical tweezers. It has been reported in ref. 28 that retraction of M. xanthus type-IV pili stalls at maximum forces of . Pilus retraction speeds of up to /s were measured at low forces of . Motion of our bacteria is much slower than the maximum pilus retraction speed (see SI Appendix, Fig. S4), but forces measured at hotspots are consistent with the stall forces measured by optical tweezers.
Individual Gliding Cells Exert Very Little Traction.
To next investigate the motion of individual cells that do not use pili but move by the complementary gliding mechanism, we performed experiments using the twitching-deficient mutant pilA. SI Appendix, Fig. S6 shows typical results for bacteria that move individually without contacting each other. Here, substrate deformation is below , and very little overall traction is observed. Because displacements are close to the measurement precision, random noise is prominent. However, approximate colocalization of traction with bacteria implies that gliding bacteria do deform the substrate to some degree. Traction is localized not in front of the cells but beneath them. Estimated traction from individual gliding cells is on the order of times smaller than for twitching cells. Consequently, we conclude that gliding of individual cells is a low-friction process that hardly affects the environment mechanically.
Groups of Twitching Cells Exert Local, Fluctuating Traction.
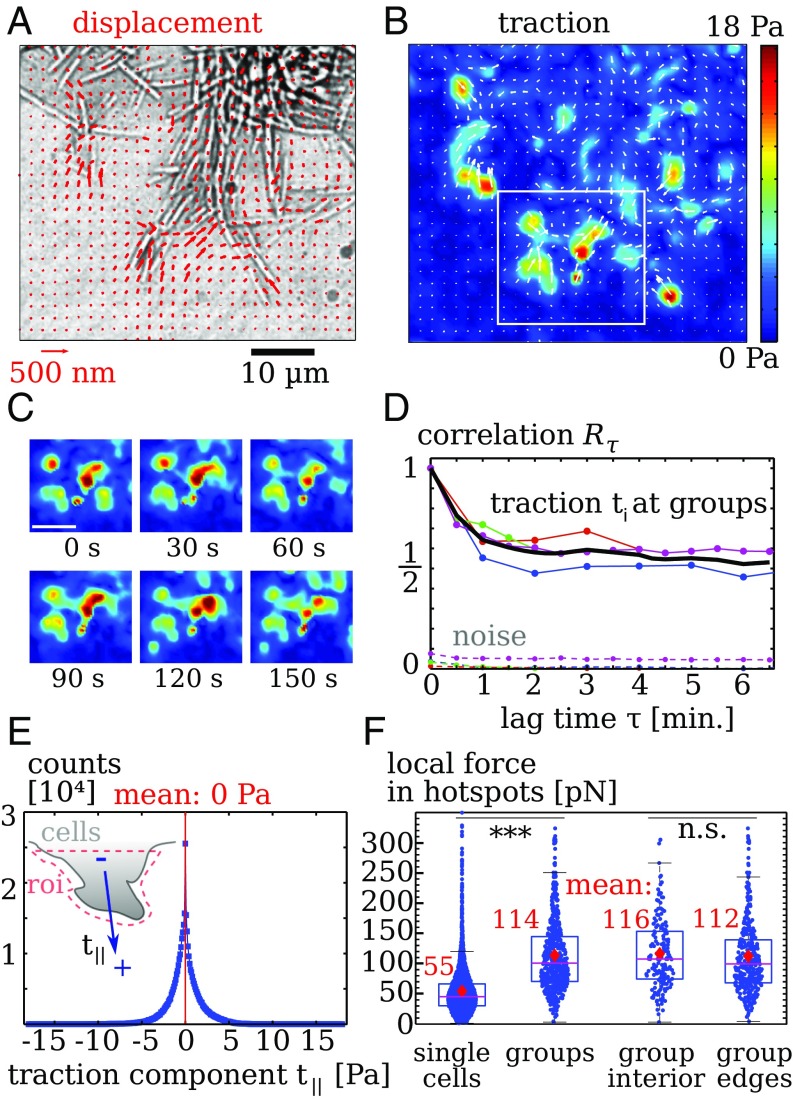
To investigate how groups of twitching cells distribute force while performing collective motility, we examined groups of twitching aglQ cells (15). Gliding-deficient mutants form slightly disorganized groups, where individual cells are not strongly aligned with each other. When deposited on a substrate, initial clumps of bacteria tend to spread out only slightly during the observation time. TFM analysis (Fig. 3 A and B) shows highly localized substrate forces in spots at the periphery of the group. The traction from the outermost spots points toward the cell group, as is expected from a pulling action of the pili. The snapshots of traction magnitude shown in Fig. 3C demonstrate a dynamical traction pattern.
Fig. 3.
Collective migration of twitching bacteria that are gliding-deficient (aglQ). (A) Gel displacements. Only every fourth measurement is displayed for clearer visibility. (B) Calculated traction showing hotspots. (C) Snapshots displaying evolution of of the traction pattern inside the region denoted by a white rectangle in B. (Scale bar: .) (D) The autocorrelation quantifies temporal fluctuations of traction. Upper lines denote data below cell groups and mean (black line). Lower, dotted lines denote traction noise measured in regions without cells. (E, Inset) Distribution of the traction component resulting from projection of on the main normal axis of an edge in a region of interest (roi). Data points: Tractions have a vanishing mean, showing that cell–substrate forces balance locally. Bin width is . (F) Force magnitude of hotspots. Single twitching cells produce significantly weaker hotspots than groups. In groups, forces at the edge are comparable to those measured below the cells. Dots are individual measurements. Stars indicate significantly different distributions with in a rank sum test. Data for cell groups in D and E were collected from four separate experiments with an overall of images taken at frame rates of 30 to 60 s.
To assess the traction dynamics quantitatively, we calculate its autocorrelation as , where is the traction vector at time at position with index . First, traction correlations are measured in the vicinity of the cells. Second, we also record correlations of traction noise far away from cells. Finally, correlations of real traction and noise are both normalized by the zero-lag correlation of real traction as . For long times, approaches a nonzero constant in our experiments (see SI Appendix).
In Fig. 3D, correlation data from four different experiments are shown where we distinguish between traction beneath cell groups and traction noise occurring away from cells. The noise correlation is clearly much smaller than the real signal. The correlations of traction decay very rapidly on timescales of , which demonstrates the presence of rapidly fluctuating traction. These fluctuations can also be observed in the raw data images, where rapid, local displacements of marker beads occur (Movies S2 and S3). The movies also demonstrate that individual cells move rapidly in a seemingly random fashion, whereas the group edge does not move much.
We next assess the traction orientation to see if cells at the group edge produce a net traction that pulls or pushes the group forward. As shown in Fig. 3E, Inset, we manually select regions of interest around the edges and record the traction components that are aligned with a main axis normal to the edge. Traction that pushes away from the group edge yields . We find a symmetric distribution of with a mean value of . Thus, groups of twitching aglQ cells do not exert coordinated traction.
Typical numbers of pili per M. xanthus bacterium have been reported to be around 4 to 10 (26), where, in some cells, up to pili were observed. Given the large number of potentially active pili in groups, it is not obvious that forces should be concentrated in the observed hotspots. However, if concentrated, the large number of available pili can produce strong forces on the order of nanonewtons (29), which is comparable to forces produced by much larger eukaryotes (30). Moreover, engaging the substrate with many pili simultaneously could potentially lead to very slow dynamics because motion would require detachment of many pili. To clarify this issue, we compare the absolute force magnitude of traction hotspots at groups with the magnitude of hotspots at individual cells (Fig. 3F). Although hotspots at individual cells have a magnitude of , we find, for hotspots at groups, a mean force of with an uncertainty approximately as large as the mean. Thus, measured forces are amplified in groups by about a factor of 2. Because these are smaller than the maximum stall force of (28), our result are still compatible with the notion that each traction hotspot in groups is caused only by one or a few pili. Furthermore, if many pili cooperated to produce one traction hotspot, weaker forces would be expected for the edge of groups where fewer pili are present. However, a comparison of force magnitude shows no significant differences for hotspots at group edges or below the group interior. Together, we find that, although groups of M. xanthus likely only use relatively few pili simultaneously to exert substrate forces, measured forces are considerably stronger in groups compared with single cells.
Gliding Groups Can Exert Persistent, Coordinated Force.
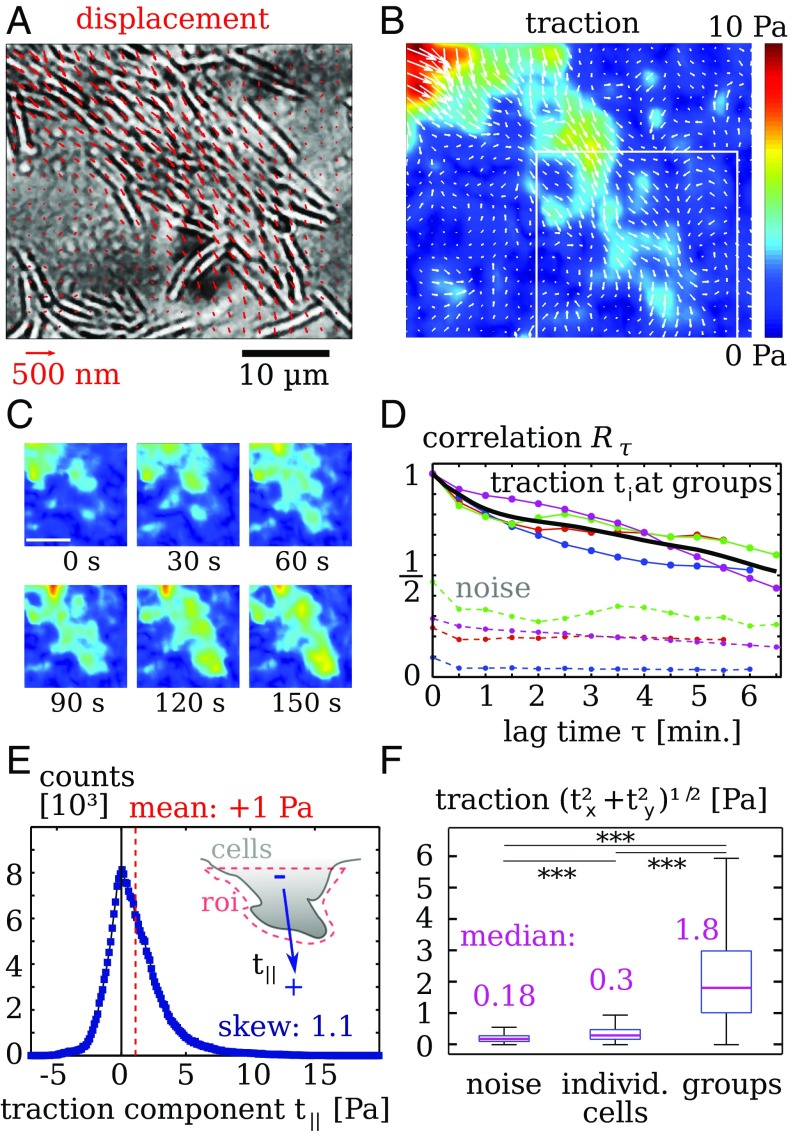
To next assess the collective mechanics of bacterial gliding, we probed groups of gliding pilA cells. When placed on the imaging substrate, clumps of bacteria present at the start of the experiment spread in a fingering fashion, where the fingers consist of closely packed bacteria that move parallel to each other. We find that, although gliding of individual cells does not produce much traction, gliding motion in groups leads to measurable forces (Fig. 4 A and B). Here, traction is distributed in diffuse patches underneath the moving group, and the traction magnitude is lower than in the presence of pili. Furthermore, the cell–substrate traction in the shown protrusion appears rather coordinated because the traction points in the direction of the advancing cells.
Fig. 4.
Collective migration of twitching-deficient strains (PilA). (A) Groups move with a finger-forming spreading pattern. Red quivers are gel displacements. Only every fourth measurement is displayed, for clearer visibility. (B) Calculated traction. Note that the traction points, on average, in the direction of motion. (C) Snapshots displaying evolution of of the traction pattern inside the region denoted by a white rectangle in B. (Scale bar: .) (D) Traction autocorrelation . Upper lines denote data from four cell groups with mean (black line). Lower, dotted lines denote traction noise measured in regions without cells. (E, Inset) Distribution of the traction component resulting from projection onto the average orientation of a fingering structure. Data points: Histogram bin width is . Data were recorded from areas where bacteria form a fingering structure. Positive values of the mean traction and the skewness demonstrate that twitching-deficient mutants in a growing finger tend to push the substrate in the direction of motion. (F) Bar plots of traction magnitude comparing noise away from cells, individual gliding PilA cells, and compact groups of PilA cells. Stars indicate significantly different distributions with in a rank sum test. Data for individual cells were recorded from overall cells in four experiments. Data for cell groups in D and E were collected from four experiments with image frames each.
The snapshots of traction magnitude shown in Fig. 4C illustrate that traction is dynamic, but changes appear less abrupt than for twitching cells. To assess the traction dynamics quantitatively, we calculate the correlation measure for the twitching-deficient mutants (Fig. 4C). Again, the correlation measure is normalized by the zero-lag autocorrelation below the cell groups in each movie . The contribution of measurement noise is here evidently stronger than in the case of twitching motion, due to the lower force magnitude. We find that the traction correlation does not show a rapid decay on short timescales as in Fig. 3D; instead, it decays over many minutes. Thus, gliding of groups causes traction variations that are slower than those resulting from pili.
Because the traction images of gliding groups in Fig. 4B and Fig. S7 suggest a “pushing” nature of the forces under advancing fingers, we quantitatively assess the directionality of forces in Fig. 4E. We manually select regions of interest around protruding fingers and record the traction components that are aligned with the protrusion direction. In contrast to the results from twitching cells, the distribution of is here asymmetric and heavy on the positive side, as quantified by a positive skew of . The pushing nature of cell–substrate traction below advancing fingers is corroborated by a positive distribution median of , where the hypothesis of a vanishing median is rejected with confidence by a sign test. These pushing forces necessarily require long-range load balance, where compression of cells at the rear end of the protruding finger balances pushing forces at the tip.
Finally, we compare the forces produced by gliding groups with forces from individual cells that are not touching each other. Because traction from gliding is distributed below the bacteria, we cannot use the assumption of discrete point forces to calculate absolute force values. Instead, we record the distribution of traction magnitude at every position either directly beneath individual cells or beneath densely packed groups. To obtain an estimate of the noise magnitude, traction magnitudes in areas without cells are also recorded. We find that individual gliding cells exert traction that is significantly above the noise threshold, but nevertheless quite weak, with a median below . Unexpectedly, we find that groups of gliding cells produce much higher traction than individual cells. The median of traction measured below gliding groups is more than 5 times higher than the median traction below individual cells, as shown in Fig. 4F. As a control, we also study a pilT mutant that has an impaired pilus retraction machinery (SI Appendix, Fig. S9). Similar to the gliding pilA strain, this strain produces low traction when moving individually. In groups, pilT mutants produce traction hotspots with an average force of , which is significantly weaker than for the twitching aglQ mutant. However, the presence of dynamic hotspots does suggest that the pilT strain can still use pili for transmission of forces that result either from the gliding machinery or from a pilT-independent retraction mechanism (28).
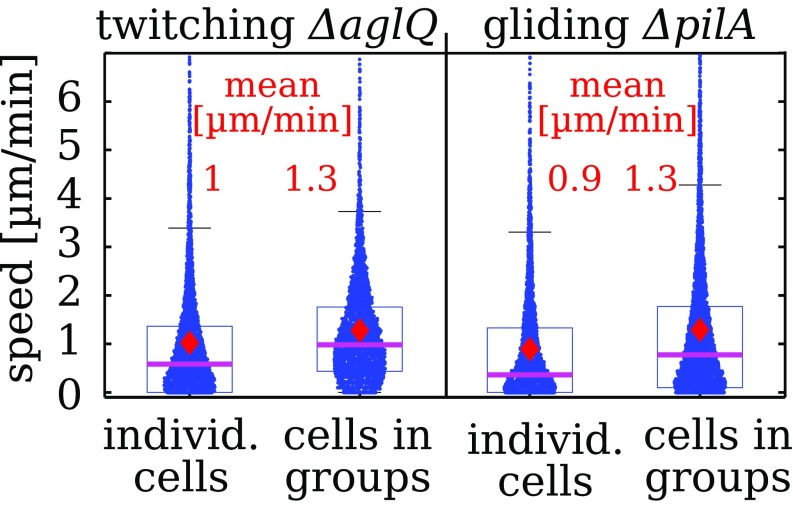
Cell Motion Is Faster in Groups.
To compare motility to traction generation, we track bacteria that move individually or in groups. For both twitching aglQ strains and gliding pilA strains, we find that the instantaneous speed is substantially larger in groups, as seen in Fig. 5 (Movies S4 and S5). Thus, higher measured traction in groups is accompanied by higher speed of individual cells. Because both motility mechanisms are associated with frequent direction changes on our chitosan-coated substrates, we test whether the motion has a diffusion-like character similar to other twitching bacteria (27, 31). For our observation timescale, the mean-square displacement of individually moving bacteria indicates a slightly superdiffusive motion of both twitching and gliding strains (SI Appendix, Figs. S4 and S5). In groups, twitching aglQ bacteria exhibit the most random motion patterns, with a velocity autocorrelation that decays within a minute timescale to zero. In contrast, the velocity autocorrelation of gliding pilA bacteria does not decay to zero entirely, which hints at limited persistence of motion, consistent with a superdiffusive random walk (SI Appendix, Figs. S4D and S5B).
Fig. 5.
Instantaneous speed of twitching and gliding strains is higher in groups. Data were obtained by manual tracking 150 to 170 cells in each condition. Data were obtained from seven experiments for groups of each kind and 13 experiments for individual cells of each kind.
Discussion
The presented data provide evidence for two very distinct patterns of force organization during the migration of M. xanthus. We find that pilus-driven twitching of individual cells results in counteracting substrate forces that are concentrated in hotspots with a force magnitude on the order of . In bacterial groups, the number of available pili per substrate area is high. Therefore, one might expect that the many pili in twitching groups produce a rather continuous traction pattern with coordinated directionality. However, we, instead, observe disorganized hotspots of traction, similar to what we observe for individual cells, albeit with a significantly amplified measured force magnitude around . The higher forces in groups are accompanied by faster, erratic twitching of cells. This collective increase in mechanical activity may originate from biochemical regulation (32, 33). However, higher forces in groups could also be caused by physical mechanisms, including cellular jamming that leads to higher resistance and thereby higher force generation in the retraction motors (28), and the collective action of pili. Traction and cell velocity in groups are both highly dynamic, and correlations decay on the timescale of a minute, whereas the groups as a whole hardly move over the time course of 10 to 20 min. We conclude that the forces generated by pili are not used efficiently for collective migration in our experiments. However, pili clearly provide anchoring to the substrate, and one might speculate about a sensory role that pilus retraction plays in allowing cells to probe their mechanical surroundings (34).
Gliding of M. xanthus is powered by elastically connected adhesion sites that are stationary with respect to the substrate and disassembled at the rear pole (17, 35). Our results for gliding mutants are almost the very reverse of those for twitching. Traction exerted by individually migrating cells is very low and often hardly measurable. Also, we rarely observe a build-up of traction at the rear ends of gliding cells. Such rear-end traction indicates that mechanical deadhesion limits motion and was found for the unrelated gliding of apicomplexa (36). In groups, gliding bacteria exert measurable traction that can push in the direction of motion. To produce this pushing traction, cells must transmit compressive force within a group. The amplified traction under gliding groups could result from contact-dependent biochemical mechanisms (37) or emerge from a mechanical cell–cell interaction because biochemically determined motion reversal of individual cells leads to stalling forces on other cells (38). In this picture, velocity variation produces an innately integrative mechanism for maintaining directional load while allowing group rearrangement.
Bacterial cell–substrate forces have been measured previously by using deformable micropillars as a substrate (29). In that work, forces up to result from parallel action of many pili bridging the voids between pillars. Our complementary assay is free of barriers to migration and allows a local force measurement. The spatial resolution of TFM results is limited by the density of measurements of the substrate deformation. Using fluorescent beads of two colors, substrate deformation can be measured approximately every , which is comparable to the bacterial thickness. Therefore, we expect that the real traction exerted by bacteria varies on a lengthscale comparable to, or shorter than, our measurement scale. We are then dealing with a spatially undersampled traction field, which is a problem that is routinely encountered in the context of TFM at eukaryotic adhesion sites. A consequence of undersampling of the displacement field is that the absolute traction magnitude is usually underestimated (20).
A number of challenging refinements of our methodology are desirable. First, accurate 3D tracking of beads in the substrate would possibly allow the assessment of vertical forces and allow for precise determination of the vertical distance between the beads and the bacteria. Such analysis was precluded, in our studies, by bacterial photodamage from the fluorescence excitation light. Second, the gel displacements are measured with respect to a prestressed state, because it proved difficult to recover the fully relaxed state after removal of bacteria. Although not essential for this study, it is generally desirable to obtain the relaxed state of the substrate. Third, comparison of TFM results with other migration assays would be facilitated if sufficiently soft and well-characterized agar-based TFM substrates were available.
Many facets of bacterial surface mechanics are yet poorly understood, for instance, substrate dependence of migration (39) or surface-dependent virulence of P. aeruginosa (34, 40). We have shown that the combination of traction measurement with genetic perturbations provides a viable tool for studying the emerging world of bacterial mechanics.
Note Added in Proof.
We would like to point interested readers to a recent preprint article on substrate forces during bacterial microcolony morphogenesis (41).
Supplementary Material
Acknowledgments
We thank S. Thutupalli for discussions and invaluable advice. We thank G. Laevsky and A. Perazzo for technical guidance. Tâm Mignot is thanked for providing bacterial strains. Work was supported by National Science Foundation Grants PHY-1401506 and MCB-1330288, German Research Foundation (DFG) Award Ko5239/1-1, and a German Academic Exchange Service (DAAD) fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. U.S.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1621469114/-/DCSupplemental.
References
- 1.Keane R, Berleman J. The predatory life cycle of Myxococcus xanthus. Microbiology. 2016;162:1–11. doi: 10.1099/mic.0.000208. [DOI] [PubMed] [Google Scholar]
- 2.Hodgkin J, Kaiser D. Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): Two gene systems control movement. Mol Gen Genet MGG. 1979;171:177–191. [Google Scholar]
- 3.Welch R, Kaiser D. Cell behavior in traveling wave patterns of Myxobacteria. Proc Natl Acad Sci USA. 2001;98:14907–14912. doi: 10.1073/pnas.261574598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mignot T, Merlie JP, Zusman DR. Regulated pole-to-pole oscillations of a bacterial gliding motility protein. Science. 2005;310:855–857. doi: 10.1126/science.1119052. [DOI] [PubMed] [Google Scholar]
- 5.Mattick JS. Type IV pili and twitching motility. Annu Rev Microbiol. 2002;56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- 6.Maier B, Wong GCL. How bacteria use type IV pili machinery on surfaces. Trends Microbiol. 2015;23:775–788. doi: 10.1016/j.tim.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 7.Merz AJ, So M, Sheetz MP. Pilus retraction powers bacterial twitching motility. Nature. 2000;407:98–102. doi: 10.1038/35024105. [DOI] [PubMed] [Google Scholar]
- 8.Skerker JM, Berg HC. Direct observation of extension and retraction of type IV pili. Proc Natl Acad Sci USA. 2001;98:6901–6904. doi: 10.1073/pnas.121171698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang Y-W, et al. Architecture of the type IVa pilus machine. Science. 2016;351:aad2001. doi: 10.1126/science.aad2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, et al. Extracellular polysaccharides mediate pilus retraction during social motility of Myxococcus xanthus. Proc Natl Acad Sci USA. 2003;100:5443–5448. doi: 10.1073/pnas.0836639100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Black WP, Xu Q, Yang Z. Type IV pili function upstream of the Dif chemotaxis pathway in Myxococcus xanthus EPS regulation. Mol Microbiol. 2006;61:447–456. doi: 10.1111/j.1365-2958.2006.05230.x. [DOI] [PubMed] [Google Scholar]
- 12.Nett M, König GM. The chemistry of gliding bacteria. Nat Prod Rep. 2007;24:1245–1261. doi: 10.1039/b612668p. [DOI] [PubMed] [Google Scholar]
- 13.Luciano J, et al. Emergence and modular evolution of a novel motility machinery in bacteria. PLoS Genet. 2011;7:e1002268. doi: 10.1371/journal.pgen.1002268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nan B, et al. Myxobacteria gliding motility requires cytoskeleton rotation powered by proton motive force. Proc Natl Acad Sci USA. 2011;108:2498–2503. doi: 10.1073/pnas.1018556108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun M, Wartel M, Cascales E, Shaevitz JW, Mignot T. Motor-driven intracellular transport powers bacterial gliding motility. Proc Natl Acad Sci USA. 2011;108:7559–7564. doi: 10.1073/pnas.1101101108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mignot T, Shaevitz JW, Hartzell PL, Zusman DR. Evidence that focal adhesion complexes power bacterial gliding motility. Science. 2007;315:853–856. doi: 10.1126/science.1137223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faure LM, et al. The mechanism of force transmission at bacterial focal adhesion complexes. Nature. 2016;539:530–535. doi: 10.1038/nature20121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oliver T, Jacobson K, Dembo M. Traction forces in locomoting cells. Cell Motil Cytoskeleton. 1995;31:225–240. doi: 10.1002/cm.970310306. [DOI] [PubMed] [Google Scholar]
- 19.Butler JP, Tolic-Norrelykke IM, Fabry B, Fredberg JJ. Traction fields, moments, and strain energy that cells exert on their surroundings. Am J Physiol Cell Physiol. 2002;282:C595–C605. doi: 10.1152/ajpcell.00270.2001. [DOI] [PubMed] [Google Scholar]
- 20.Sabass B, Gardel ML, Waterman CM, Schwarz US. High resolution traction force microscopy based on experimental and computational advances. Biophys J. 2008;94:207–220. doi: 10.1529/biophysj.107.113670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trepat X, et al. Physical forces during collective cell migration. Nat Phys. 2009;5:426–430. [Google Scholar]
- 22.Flanagan LA, Ju Y-E, Marg B, Osterfield M, Janmey PA. Neurite branching on deformable substrates. Neuroreport. 2002;13:2411–2415. doi: 10.1097/01.wnr.0000048003.96487.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plotnikov SV, Sabass B, Schwarz US, Waterman CM. High-resolution traction force microscopy. Methods Cell Biol. 2014;123:367–394. doi: 10.1016/B978-0-12-420138-5.00020-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ducret A, Valignat M-P, Mouhamar F, Mignot T, Theodoly O. Wet-surface–enhanced ellipsometric contrast microscopy identifies slime as a major adhesion factor during bacterial surface motility. Proc Nat Acad Sci USA. 2012;109:10036–10041. doi: 10.1073/pnas.1120979109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwarz US, et al. Calculation of forces at focal adhesions from elastic substrate data: The effect of localized force and the need for regularization. Biophys J. 2002;83:1380–1394. doi: 10.1016/S0006-3495(02)73909-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaiser D. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc Nat Acad Sci USA. 1979;76:5952–5956. doi: 10.1073/pnas.76.11.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marathe R, et al. Bacterial twitching motility is coordinated by a two-dimensional tug-of-war with directional memory. Nat Commun. 2014;5:3759. doi: 10.1038/ncomms4759. [DOI] [PubMed] [Google Scholar]
- 28.Clausen M, Jakovljevic V, Søgaard-Andersen L, Maier B. High-force generation is a conserved property of type IV pilus systems. J Bacteriol. 2009;191:4633–4638. doi: 10.1128/JB.00396-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biais N, Ladoux B, Higashi D, So M, Sheetz M. Cooperative retraction of bundled type IV pili enables nanonewton force generation. PLoS Biol. 2008;6:e87. doi: 10.1371/journal.pbio.0060087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balaban NQ, et al. Force and focal adhesion assembly: A close relationship studied using elastic micropatterned substrates. Nat Cell Biol. 2001;3:466–472. doi: 10.1038/35074532. [DOI] [PubMed] [Google Scholar]
- 31.Taktikos J, Stark H, Zaburdaev V. How the motility pattern of bacteria affects their dispersal and chemotaxis. PLoS One. 2013;8:e81936. doi: 10.1371/journal.pone.0081936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nudleman E, Wall D, Kaiser D. Cell-to-cell transfer of bacterial outer membrane lipoproteins. Science. 2005;309:125–127. doi: 10.1126/science.1112440. [DOI] [PubMed] [Google Scholar]
- 33.Konovalova A, Søgaard-Andersen L. Close encounters: Contact-dependent interactions in bacteria. Mol Microbiol. 2011;81:297–301. doi: 10.1111/j.1365-2958.2011.07711.x. [DOI] [PubMed] [Google Scholar]
- 34.Persat A, Inclan YF, Engel JN, Stone HA, Gitai Z. Type IV pili mechanochemically regulate virulence factors in Pseudomonas aeruginosa. Proc Nat Acad Sci USA. 2015;112:7563–7568. doi: 10.1073/pnas.1502025112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Treuner-Lange A, et al. The small G-protein MglA connects to the MreB actin cytoskeleton at bacterial focal adhesions. J Cell Biol. 2015;210:243–256. doi: 10.1083/jcb.201412047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Münter S, et al. Plasmodium sporozoite motility is modulated by the turnover of discrete adhesion sites. Cell Host Microbe. 2009;6:551–562. doi: 10.1016/j.chom.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 37.Jakobczak B, Keilberg D, Wuichet K, Søgaard-Andersen L. Contact-and protein transfer-dependent stimulation of assembly of the gliding motility machinery in Myxococcus xanthus. PLoS Genet. 2015;11:e1005341. doi: 10.1371/journal.pgen.1005341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang H, et al. Quantifying aggregation dynamics during Myxococcus xanthus development. J Bacteriol. 2011;193:5164–5170. doi: 10.1128/JB.05188-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi W, Zusman DR. The two motility systems of Myxococcus xanthus show different selective advantages on various surfaces. Proc Nat Acad Sci USA. 1993;90:3378–3382. doi: 10.1073/pnas.90.8.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siryaporn A, Kuchma SL, O’Toole GA, Gitai Z. Surface attachment induces Pseudomonas aeruginosa virulence. Proc Nat Acad Sci USA. 2014;111:16860–16865. doi: 10.1073/pnas.1415712111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duvernoy M-C, et al. 2017. Asymmetric adhesion of rod-shaped bacteria controls microcolony morphogenesis. bioRxiv: 10.1101/104679.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.