Significance
G-protein–coupled receptors (GPCRs) can transmit external signals into the cytoplasm via the activation of G proteins. They also mediate G-protein–independent signaling that uses arrestin to activate an extracellular signal-regulated kinase (ERK) cascade. Using real-time optical measurements, we show that arrestin binds in two modes. Stable binding of arrestin to phosphorylated amino acids in the third intracellular loop of the muscarinic receptor upregulates ERK. Transient binding of arrestin to the unphosphorylated receptor downregulates ERK. Thus, the signaling bias of ERK can be determined by binding modes of arrestin to the receptors. Our results suggest a molecular mechanism for GPCR-arrestin-ERK coupling and pave the way for the development of new therapeutic approaches.
Keywords: arrestin, GPCR, ERK, muscarinic receptor, receptor kinase
Abstract
Binding of agonists to G-protein–coupled receptors (GPCRs) activates heterotrimeric G proteins and downstream signaling. Agonist-bound GPCRs are then phosphorylated by protein kinases and bound by arrestin to trigger desensitization and endocytosis. Arrestin plays another important signaling function. It recruits and regulates activity of an extracellular signal-regulated kinase (ERK) cascade. However, molecular details and timing of ERK activation remain fundamental unanswered questions that limit understanding of how arrestin-dependent GPCR signaling controls cell functions. Here we validate and model a system that tracks the dynamics of interactions of arrestin with receptors and of ERK activation using optical reporters. Our intermolecular FRET measurements in living cells are consistent with β-arrestin binding to M1 muscarinic acetylcholine receptors (M1Rs) in two different binding modes, transient and stable. The stable mode persists for minutes after agonist removal. The choice of mode is governed by phosphorylation on key residues in the third intracellular loop of the receptor. We detect a similar intramolecular conformational change in arrestin in either binding mode. It develops within seconds of arrestin binding to the M1 receptor, and it reverses within seconds of arrestin unbinding from the transient binding mode. Furthermore, we observed that, when stably bound to phosphorylated M1R, β-arrestin scaffolds and activates MEK-dependent ERK. In contrast, when transiently bound, β-arrestin reduces ERK activity via recruitment of a protein phosphatase. All this ERK signaling develops at the plasma membrane. In this scaffolding hypothesis, a shifting balance between the two arrestin binding modes determines the degree of ERK activation at the membrane.
Arrestin plays a fundamental role in the negative regulation of G-protein–coupled receptors (GPCRs) signaling because its binding to GPCRs hinders G-protein association with the receptors (1–5). Arrestin further recruits adaptor proteins and clathrin for the internalization and desensitization of the receptor (6). Additional functions have been suggested, with β-arrestin producing its own signaling through interactions with other effectors (7–9). For example, β-arrestin has binding sites for cRaf (MAPK kinase kinase), MEK (MAPK kinase), and ERK (MAPK) that mediate a signaling cascade for cell proliferation, cell migration, and actin dynamics after GPCR activation (10–13). Conventionally, MEK-dependent ERK signaling is described as involving GPCR–arrestin complexes on intracellular compartments such as endosomal vesicles (10, 14–16). However, whether similar complexes at the plasma membrane might trigger ERK signaling remains unknown. Electron microscope images of arrestin-receptor complexes together with modeling suggest that β-arrestin binds in two different modes, called transient binding and stable binding, to chimeric β2 adrenergic receptors (β2-AR) containing the C terminus of vasopressin type 2 receptors (3). X-ray crystal structures of rhodopsin–arrestin complexes also support such a concept (4).
From these observations, a common inference is that ERK activity is up-regulated while scaffolded on arrestin–GPCR complexes. An interesting alternative mechanism has been suggested recently, namely a briefly persistent conformational change of arrestin that allows it to continue to signal to ERK after dissociation from GPCRs (17)—a “memory effect” (16, 18). We studied the M1 muscarinic acetylcholine receptor (M1R), which couples to both Gq- and arrestin-dependent pathways, including ERK activation (19–22), and found strong evidence for two binding modes of arrestin to receptors that led to contrasting downstream signals. The M1R is a receptor for which desensitization and internalization are relatively weaker than for, for example, the β2-adrenergic receptor. Molecular interactions and conformational changes of arrestin upon M1R activation were monitored with intermolecular and intramolecular FRET and total internal reflection fluorescence microscopy. To isolate the arrestin effect on ERK activity, we used arrestin-biased mutations of the receptor.
Results
FRET Reveals Agonist-Induced Interactions of SNAP-Tagged M1R with Gq and β-Arrestin 2.
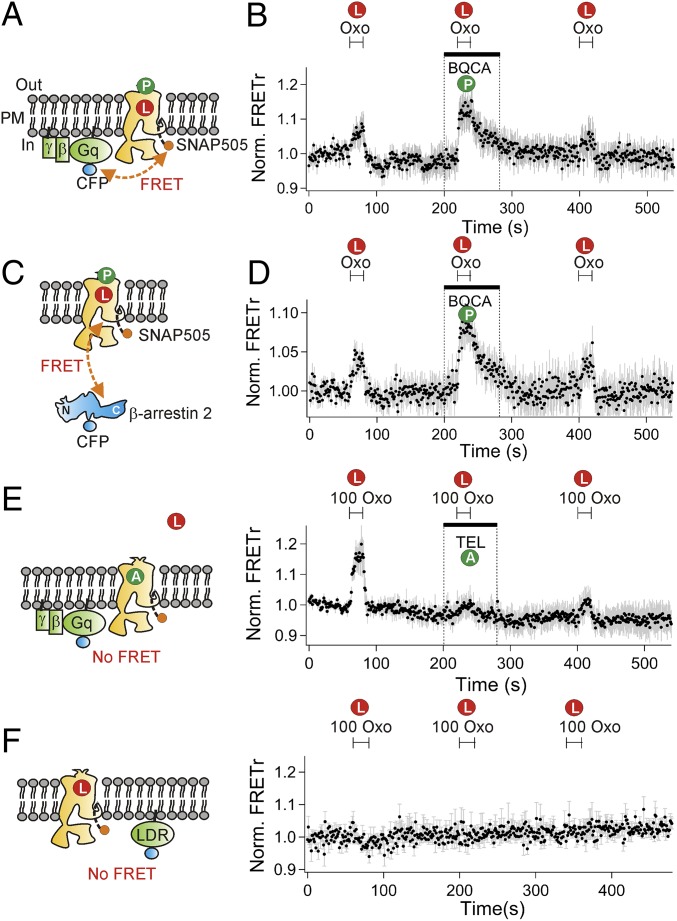
The formation of complexes between receptors and G proteins or β-arrestin was visualized by constructing fluorescent probes. The intracellular C terminus of the M1R was tagged with a SNAP tag (M1R-SNAP) (Fig. 1A). Because the SNAP tag can be labeled with different dyes, the same construct could be used in different spectral combinations in FRET and total internal reflection fluorescence (TIRF) imaging experiments. As a control, we tested whether the construct had normal function. tsA201 cells transiently expressing M1R-SNAP were treated with a cell-permeable SNAP505 dye, and the FRET interactions between Gq-CFP and M1R-SNAP505 (Fig. 1 A and B) or between β-arrestin 2-CFP and M1R-SNAP505 (Fig. 1 C and D) were monitored and expressed as FRET ratios (FRETr). Receptor-induced increases of FRETr using the muscarinic agonist oxotremorine (Oxo)-M (1 µM) verified the expected molecular interactions of Gq with M1R (Fig. 1B) and of β-arrestin 2 with M1R (Fig. 1D). In addition, benzyl quinolone carboxylic acid (BQCA), a specific allosteric modulator of M1R, further potentiated the coupling of Gq (Fig. 1B) and β-arrestin 2 (Fig. 1D) to the receptor as previously demonstrated (23). A competitive M1R antagonist, telenzepine (10 µM), almost completely abrogated the ability of a high concentration of oxotremorine (100 µM) to induce a FRETr increase (Fig. 1E, Left and Right), and there was no FRETr response when CFP was used to tag a nonspecific plasma-membrane–anchored LDR construct (5) instead of β-arrestin 2 (Fig. 1F, Left and Right). Thus, the FRETr signals reflect specific binding of Gq and β-arrestin 2 to the ligand-bound M1 receptor.
Fig. 1.
FRET reveals agonist-induced interactions of SNAP-tagged WT M1R with Gq-CFP and β-arrestin 2–CFP. Cell-permeable SNAP505 dye labeled the SNAP tag on the intracellular C terminus of M1R. P, L, and A denote the following: M1R potentiator (BQCA), Oxo, and antagonist (telenzepine), respectively. (A) Schematic for measuring FRET between Gq-CFP and M1R-SNAP505. PM indicates the plasma membrane. (B) Time course of FRETr. M1R agonist, Oxo (1 µM), increases FRET ratio (FRETr = SNAP505/CFP), and the allosteric modulator BQCA (30 µM) potentiates the response. FRETr was normalized to the value before the first agonist treatment. We conducted an average of five experiments (n = 5). Cells were pretreated with BQCA before the second application of oxotremorine as indicated by the bar. (C) Schematic for measuring the interaction between β-arrestin 2 CFP and M1R-SNAP505. (D) BQCA potentiates agonist-induced M1R and β-arrestin 2 interaction. n = 5. (E) M1R-specific antagonist telenzepine (TEL, 10 µM) inhibits the agonist-induced interaction between Gq-CFP and M1R-SNAP505. n = 5. (F) Absence of FRETr change between LDR-CFP and M1R-SNAP505. n = 3. In E and F, 100 µM Oxo was used.
Distinguishing Transient and Stable Binding Modes of β-Arrestin 2 to WT M1R.
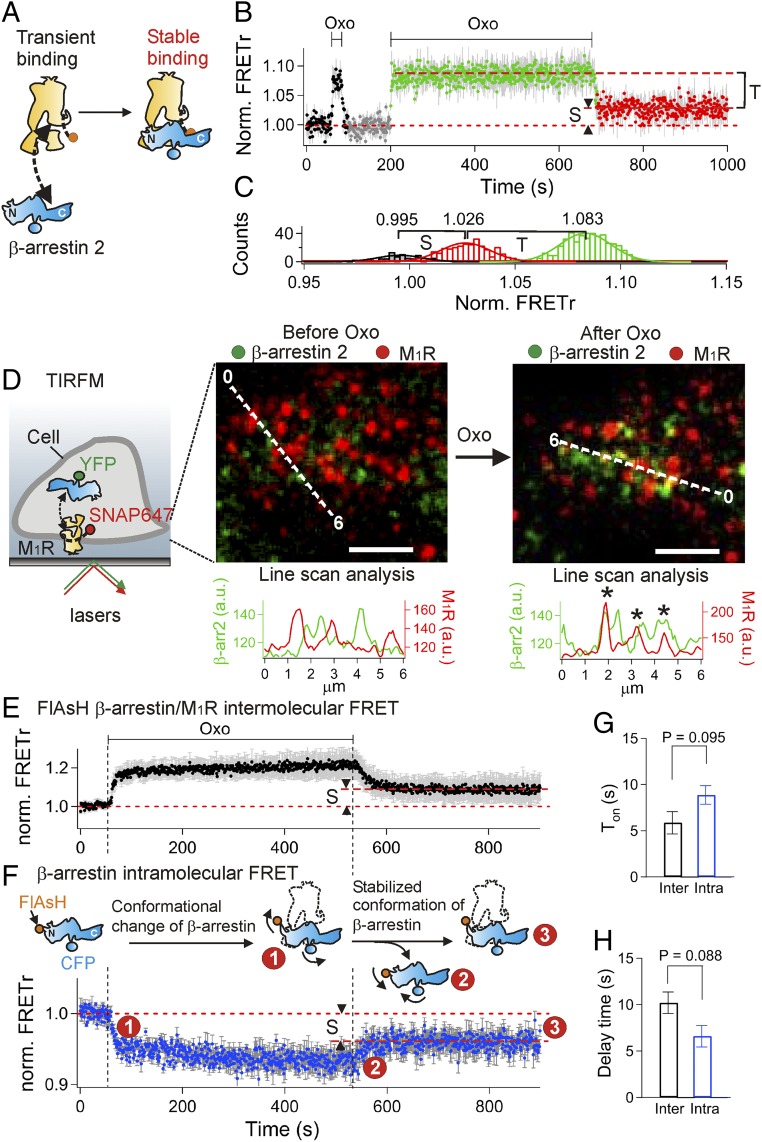
Two different modes of binding of arrestin to GPCRs have been suggested from structural studies with chimeric β2-AR (3) and rhodopsin (4). We also could distinguish two modes in our optical experiments in live cells (Fig. 2A). We monitored the binding of arrestin to the M1R using real-time FRET and TIRF microscopy at high sensitivity in live cells. Application of oxotremorine increased the FRETr interaction between arrestin and M1R within a second (Fig. 2B, τon = 1.2 ± 0.2 s). Following a short (20 s) agonist application, the arrestin-M1R FRETr increase was reversed within a few seconds (Fig. 2B, τoff = 4.3 ± 0.8 s) upon washout. In contrast, after an 8-min treatment, only about 67% of the FRETr increase reversed quickly and 33% persisted at least 6 min more after removal of the agonist (labeled “S” component in Fig. 2B). We denote this persistent component as reflecting a long-lasting “stable binding mode (S)” of the β-arrestin 2/M1R complex and the other, quickly reversed portion (∼67%), as reflecting a “transient binding mode (T).” Gaussian fitting to the all-points histogram of FRETr (Fig. 2C) indicated that the intermolecular FRETr level for the stable binding mode was significantly different from and intermediate between that in the basal state and the total binding seen while agonist was still present. Although the transition from transient to stable binding developed during the 8 min of receptor activation, there was no indication of an additional concomitant intermolecular FRETr change developing during that time. Apparently, the transient and stable complexes have strong and similar FRETr interactions. More conservatively, we could say that the product of the number of arrestin-bound receptors and the FRETr response from each does not change. One possibility is that, within the spatial resolution of FRET interactions (3–8 nm), there may have been no change in distance between the receptor and β-arrestin 2 labels during the switch from transient to stable binding. Increased recruitment and colocalization of β-arrestin with the M1R during agonist application could also be seen by TIRF microscopy (Fig. 2D).
Fig. 2.
Distinguishing transient and stable binding modes of β-arrestin to WT M1R. (A) Schematic for FRET between β-arrestin 2-CFP and M1R-SNAP505 and conversion of binding modes. (B) FRETr during a two-pulse protocol reveals two binding modes [transient (T) and stable (S)] of β-arrestin 2 to WT M1R (n = 4). The fraction of stable binding of arrestin was calculated as S/(T+S). (C) Distribution histogram of FRETr discriminates two binding modes of β-arrestin to M1R. The histograms were obtained from B. The colors match with the color in B for the individual states [gray, control; green, total binding (T+S); red, stable binding (S)]. (D) Fluorescence images at the plasma membrane measured with multicolor TIRF microscopy (TIRFM). Intensities of β-arrestin 2-YFP (green) and WT M1R-SNAP647 (red) were measured along the dashed scan lines and plotted below in arbitrary units (a.u.). Asterisks mark colocalization between M1R and β-arrestin 2 after 8 min in the presence of 100 µM Oxo. (Scale bar: 2 µm.) The basal YFP fluorescence was from near the plasma membrane in our TIRF mode. The contrast of the presented images was adjusted to promote visual comparison of two colors, whereas, for the line-scan analysis, the original images without contrast changes or background subtraction were used. (E) Intermolecular FRET between β-arrestin 2–CFP and M1R-SNAP505 (n = 5). (F) Intramolecular FRET (FlAsH/CFP) in β-arrestin 2 (n = 5). The red dashed horizontal lines indicate the portion of stabilized (S) β-arrestin 2 for intermolecular FRET (E) or for intramolecular FRET (F). Dashed vertical lines mark solution exchanges. The schematic reaction diagram is our interpretation of the conformational change and stabilization of β-arrestin 2 to M1R. (G) Average association time constant estimated from intermolecular FRET shown in E (black bar, Inter) or from intramolecular FRET shown in F (blue bar, Intra) upon agonist application. (H) Average delay time before the beginning of dissociation of β-arrestin from M1R begins (black bar, Inter) or the conformational change of β-arrestin (blue bar, Intra) after removal of agonist. All experiments in this figure used 100 µM Oxo.
Conformational Change of β-Arrestin 2.
Next, we monitored conformational changes of β-arrestin itself to explore a possible temporal relationship with the two modes of binding. Recent studies reported dynamic changes in the N and C domains of β-arrestin following activation of adrenergic and peptide receptors using deuterium exchange (3, 24) and interactions of optical probes (17, 25, 26). To study conformational changes of β-arrestin induced by the M1R in living cells, we measured intramolecular FRET between a FlAsH dye at residue 154 in the N domain of the β-arrestin 2 and CFP fused to the C terminus (FlAsH2–β-arrestin 2–CFP), as described (25, 27). Incorporating the FlAsH peptide sequence (CCPGCC) into β-arrestin 2 did not modify the persistence or magnitude of the stable binding mode to M1R (Fig. 2E). After labeling with FlAsH dye, the FlAsH–β-arrestin 2–CFP construct reported a clear intramolecular FRETr decrease from arrestin upon activation of the receptor (Fig. 2F). The recorded decrease suggests that the N domain and the C terminus of β-arrestin 2 move apart during binding to M1R. The onset kinetics of the conformational change (τon = 8.9 ± 1.0 s) were not significantly different from those of the β-arrestin-M1R intermolecular FRETr (τon = 5.9 ± 1.2 s, P = 0.095, Fig. 2G), indicating that β-arrestin changes its local structure quickly upon binding to the M1R. Was the conformational change of β-arrestin maintained even after dissociation from the GPCR? We followed the reversal of the arrestin intramolecular FRETr signal after agonist wash off. The recovery kinetics of the conformational signal showed a fast component and then a persistent plateau (Fig. 2F), closely resembling the fast unbinding and persistent plateau of β-arrestin interaction with the receptor (intermolecular FRETr) (Fig. 2E). The relative amplitudes of the persistent components were about the same (intramolecular FRETr: 54 ± 8% versus intermolecular FRETr: 45 ± 9%, P = 0.49; denoted by “S” in Fig. 2 E and F), and there was no clear difference of delay time after removal of agonist (intramolecular FRETr: 6.6 ± 1.4 s versus intermolecular FRETr: 10 ± 1.2 s, P = 0.088; Fig. 2H). Thus, we attribute the persistent component to arrestin molecules still bound in stable complexes with the receptor and the early fast-recovering component to arrestin molecules dissociating rapidly from transient complexes and losing their conformational change within seconds. This suggested that β-arrestin molecules retained an altered conformation as long as they remained bound to M1R and that arrestins released rapidly from transient complexes reverted quickly to their original state.
Contributions of Stable and Transient Binding of β-Arrestin 2 to ERK Activity.
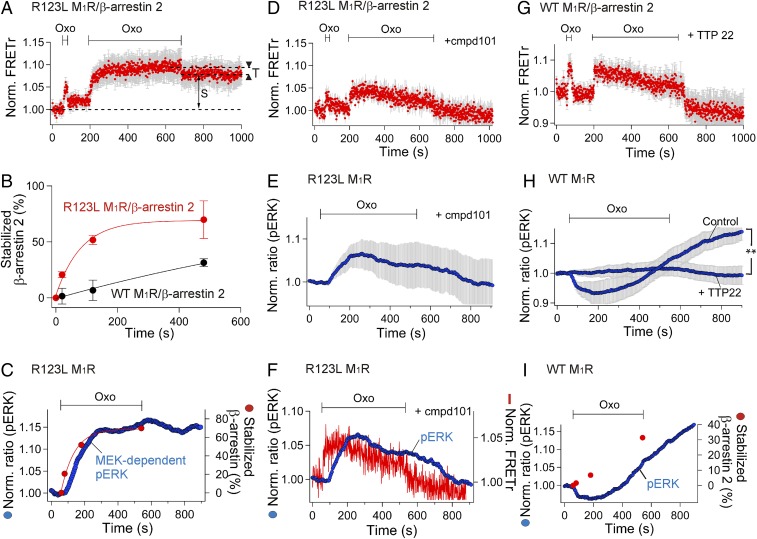
β-Arrestin interacts with members of the MAPK protein kinase cascade cRaf, MEK, and ERK (MAPK) (12, 27). We tested the contribution of β-arrestin–receptor binding modes to the activation by phosphorylation of the final member, ERK. Our hypothesis was that the stable mode of β-arrestin binding to the receptor mediates MEK and ERK activation. Because ERK can also be activated by G-protein–coupled signaling (28), we switched to a biased mutated M1R that binds β-arrestin but not G proteins. Arginine 123, implicated in G-protein binding, was mutated to leucine (R123L) in transmembrane segment 3 (22). We confirmed that the biased R123L M1R did not interact with Gq or activate phospholipase C (SI Appendix, Fig. S1). Interestingly, the stable interaction between β-arrestin and the receptor was facilitated, so even relatively short agonist treatments initiated some formation of stable complexes (Fig. 3 A and B and SI Appendix, Fig. S2). The development of stable binding was approximately five times faster than for WT M1R (Fig. 3B), and the percentage of stabilized arrestin rose to ∼70% by 8 min. This potentiation might be due to a variety of factors that we have not attempted to sort out. For example, stable arrestin binding is considered to require prior binding of a receptor kinase and phosphorylation of the receptor. Because the biased receptor does not interact productively with G proteins, both the receptor kinase-binding steps and the arrestin-binding steps could be facilitated by an absence of competition from G proteins (29).
Fig. 3.
Stable binding of β-arrestin and its contribution to ERK activity. (A) Development of stable binding monitored with intermolecular FRET (R123L M1R-SNAP505/β-arrestin 2–CFP). n = 8. (B) The stabilized fraction of β-arrestin 2 with R123L M1R (biased M1R) or WT M1R measured as a ratio of stabilized to total (transient+stable) FRETr change. Red and black symbols compare the values estimated with R123L M1R and WT M1R, respectively (n = 3–7 for each point). Error bars are the SEM. (C) Real-time monitoring of activity of ERK (pERK) using the ERK KTR probe. Blue and red symbols denote MEK-dependent ERK activity (SI Appendix, Fig. S3A) and stabilized β-arrestin 2 (B) on the R123L M1R, respectively. MEK-dependent pERK was evaluated from the net change of the ERK KTR ratio with or without U0126 (MEK1/2 blocker, see SI Appendix, Fig. S3A). (D–F) Preincubation (3 min) with GRK2/3 inhibitor, cmpd101, destabilized the β-arrestin 2 bound to R123L M1R (D, n = 7) and reduced ERK activity (E, n = 3 and n = 19). The two average traces are superimposed for comparison (F). (G and H) CK2 inhibitor, TTP22, destabilizes β-arrestin 2 WT/M1R interaction (G, n = 3) and decreased ERK activity (H, n = 4 and n = 37) compared with the control (n = 3 and n = 10; **P = 0.003). (I) Comparison of the time course of stabilization of β-arrestin to WT M1R (red) and ERK activity (blue). All experiments in this figure used 100 µM Oxo.
What happens to ERK? We measured the kinase activity of ERK functionally with the fluorescent probe ERK KTR that converts phosphorylation into a nucleocytoplasmic shuttling event (30). The probe contains ERK substrate, the phosphorylation of which turns off a nuclear localization signal and turns on a nuclear export signal. When phosphorylated by active ERK, the probe is exported from the nucleus, so the ratio of cytoplasmic to nuclear fluorescence intensity (Icyt/INuc) reflects ERK activity. We mention two subtleties in the use of the ERK KTR probe as an indicator of active pERK. One is temporal and one is spatial. The probe probably indicates ERK activity with a delay since the route to phosphorylation may be circuitous. One long scenario could involve pERK being imported into the nucleus, where it phosphorylates the probe, which then is exported to the cytoplasm giving the detected signal. The second subtlety is that the extent of phosphorylation of the probe is a dynamic indicator of two activities: the kinase activity of pERK opposed by protein phosphatase activity. It is possible that some signals from the probe might reflect changes in the relevant phosphatases rather than activation and deactivation of pERK. The real-time measurement with ERK KTR allows us to compare the time course of ERK activity with that of the β-arrestin–M1R interaction. As had been observed with the EGF receptor and the same probe (30), with this biased M1R, on average agonist induced a net increase of the ERK KTR ratio, significantly above the resting basal level—a “positive effect” (SI Appendix, Fig. S3A). With U0126, an inhibitor of MEK, the positive effect was abolished in most cells, on average being replaced by a “negative effect” (SI Appendix, Fig. S3A), so, as anticipated, the positive effect reflects phosphorylation of ERK by the upstream MAPK kinase MEK to yield active pERK, which in turn phosphorylates the probe.
The time course of the net MEK-mediated positive effect (SI Appendix, Fig. S3A, dashed line) was estimated by subtracting the averaged ERK activity response with the MEK inhibitor from the averaged control response. This time course matched well the kinetics of the stable binding of β-arrestin to the biased M1R (Fig. 3C) but with phosphorylation of the ERK probe lagging behind stable arrestin binding by ∼20 s. An increased ERK activity was sustained even after removal of the muscarinic agonist, suggesting that, as long as β-arrestin is stably bound, it continues to stimulate ERK phosphorylation. This activation of ERK took place without internalization of the biased receptor (SI Appendix, Fig. S4), suggesting that both stable binding of β-arrestin to M1R and the MEK-dependent ERK phosphorylation take place at the plasma membrane. Intriguingly, the mutations that produced the biased M1R not only removed G-protein interaction but also eliminated induction of the N domain to C-terminal intramolecular FRETr decrease in β-arrestin (SI Appendix, Fig. S5). Nevertheless, even in the absence of this arrestin conformational signal, ERK still became activated.
The receptor-stimulated probe phosphorylation by ERK was quite heterogeneous among individual cells (SI Appendix, Fig. S3B). The biased receptor resulted in a pure increase in some cells, a biphasic decrease and increase in some cells, and a pure decrease in other cells. The negative-tending trajectory, which became more obvious with the MEK inhibitor U0126 (SI Appendix, Fig. S3 A and C), implies a second receptor-stimulated mechanism that dephosphorylates pERK. Mechanisms underlying negative control of ERK signaling by M1R were not resolved in early biochemical studies (23, 31). However, in published coimmunoprecipitation experiments, activation of dopamine receptors recruited to β-arrestin the serine/threonine protein phosphatase PP2A that dephosphorylated Akt and pERK (32, 33), and, in mass-spectrometry experiments, activation of angiotensin II receptors recruited the protein phosphatases PP2Cα and PP2Cβ (and the MEK kinase MEKK1) to β-arrestin (7). To test for possible involvement of such phosphatases in the negative control of pERK by M1R, we first eliminated the positive effect with the MEK inhibitor. Applying the MEK inhibitor did not change the ERK KTR ratio of resting cells (SI Appendix, Fig. S6A), consistent with the idea that the kinase had not already been recruited at rest. However, when oxotremorine was added to the inhibitor-treated cells, there was a prominent loss of ERK activity that was significantly prevented by calyculin A, a blocker of serine/threonine protein phosphatases (SI Appendix, Fig. S6B), and was also prevented by knockdown of β-arrestin 1/2 by siRNA (SI Appendix, Fig. S6C) (5). Based on these results, we suggest that, in parallel with activation of ERK by phosphorylation, β-arrestin recruits phosphatases for ERK dephosphorylation during M1R stimulation—as previously reported with β-arrestin, Akt, and a dopamine receptor.
Regulation of β-Arrestin 2 Binding and ERK Activation by Protein Kinases.
The relation between stable binding of β-arrestin to M1R and the activation of ERK was probed further by modifying receptor phosphorylation. In general, β-arrestin binding is promoted by receptor-dependent phosphorylation of GPCRs through G-protein–coupled receptor kinases (GRKs) (34–37) or other protein kinases (5, 14, 38). For the M1R or M3R, both GRKs and casein kinases may participate in receptor phosphorylation (19, 35, 38). For the biased M1R, we first tried a GRK2/3 inhibitor, cmpd101. This inhibitor significantly reduced both the sustained levels of intermolecular FRETr between the biased receptor and β-arrestin 2 (Fig. 3D) and the ERK activity after washing agonist off (Fig. 3E). Again, the profiles of intermolecular FRETr and ERK activity looked similar but with ERK activity lagging ∼50 s behind (Fig. 3F). In contrast, with the WT M1R, the GRK2/3 blocker was without effect (SI Appendix, Fig. S7A), whereas the casein kinase 2 (CK2) blockers, TTP22 (Fig. 3G) and TBB (SI Appendix, Fig. S7B), completely prevented stable binding of β-arrestin to receptor. Also the activation of ERK was eliminated by the CK2 inhibitors (Fig. 3H and SI Appendix, Fig. S7C). Nevertheless, the transient binding mode still persisted, suggesting again that transient binding does not suffice to activate MEK/ERK signaling. With WT M1R, the development of stable β-arrestin binding and the development of ERK activity were slower than with biased M1R (Fig. 3 H and I).
Role of Phosphorylation on the Third Intracellular Loop of M1R for Stable β-Arrestin 2 Binding and ERK Activation.
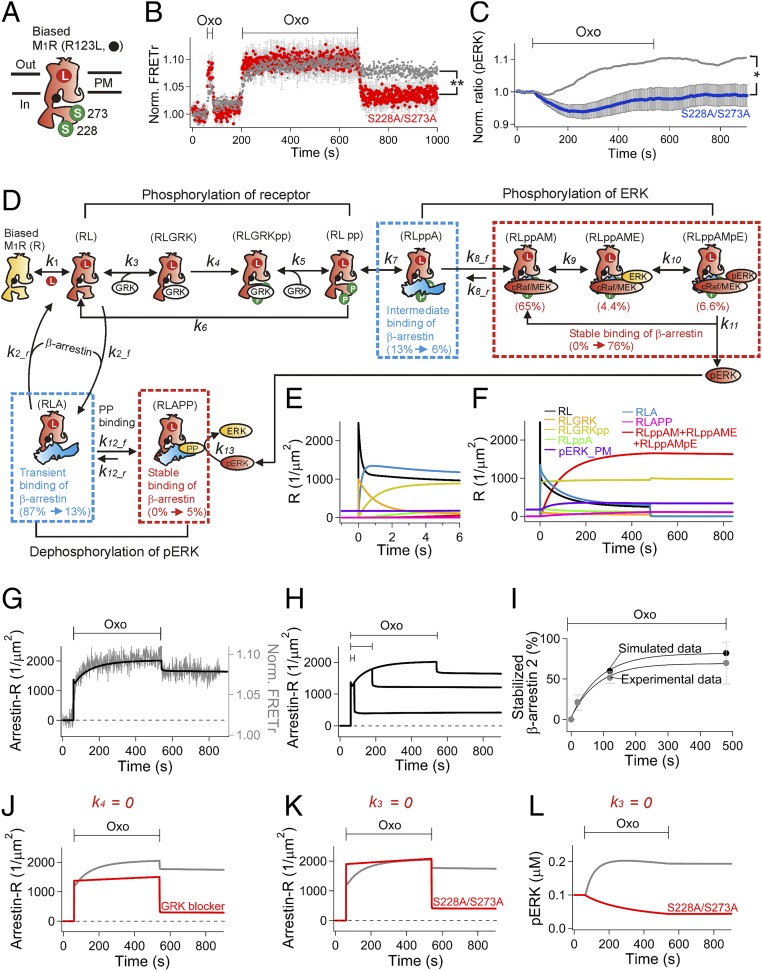
Finally, we asked whether phosphorylation of the receptor is needed for stable β-arrestin binding and for ERK activation. For this test, the biased M1R was first further mutated at two key target sites of phosphorylation on the third intracellular loop (39), S228 and S273, to nonphosphorylatable alanines (Fig. 4A). Mutating the biased receptor significantly reduced the stable binding of β-arrestin to the receptor (Fig. 4B) as well as ERK activation (Fig. 4C). As a negative control, we tested another mutant that had alanines replacing S451 and S457 in the C tail. This C-tail mutant showed normal stable binding of β-arrestin to the biased M1R and normal sustained ERK activity (SI Appendix, Fig. S8). These results support the concept that receptor phosphorylation controls stable β-arrestin binding and ERK activation (40) and that the two residues S228 and S273 in the intracellular loop are key, but residues S451 or S457 of the C tail are not. Additionally, based on the significant remaining negative effect on ERK activity (Fig. 4C), we suggest that protein phosphatases can still be recruited to arrestin that is transiently bound to the receptor.
Fig. 4.
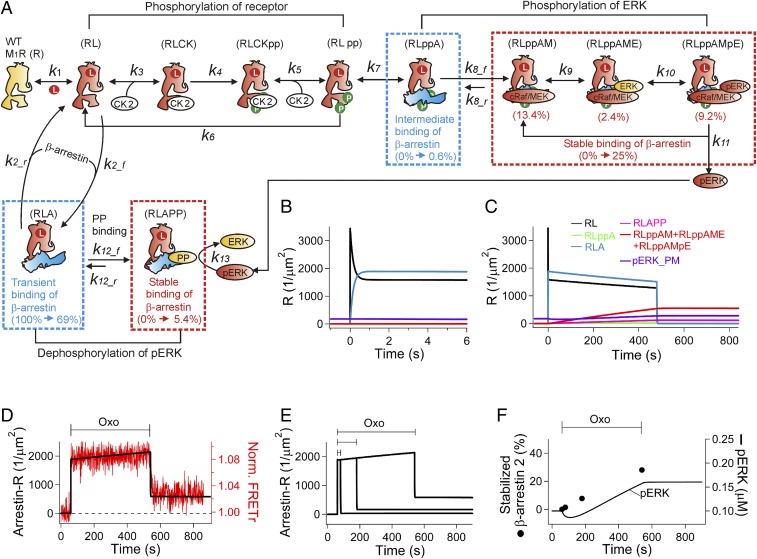
Role of phosphorylation on the third intracellular loop of the biased M1R for stable β-arrestin binding and ERK activation. (A) Nonphosphorylatable alanines replaced two serines (S228 and S273) in the biased M1R (R123L M1R). (B) Normalized FRETr between β-arrestin 2–CFP and R123L M1R-SNAP505 (red, n = 4). FRETr for biased M1R from Fig. 3A (gray) is presented for comparison. (C) Reduction of activated ERK in the phosphorylation-deficient biased M1R (n = 4, n = 52). The gray line indicates average pERK (SI Appendix, Fig. S3A, control) measured with the normal biased M1R. (D) Summary diagram of β-arrestin–dependent ERK activation for modeling the biased M1R. Arrestin binding to phosphorylated and nonphosphorylated receptor is indicated in the Upper and Lower rows, respectively. To describe the steady-state FRETr in control and mutant biased receptors after their activation (B), we assumed that GRK binds to the ligand-bound receptor in a manner competitive with arrestin. The relatively small pool of GRK (0.5 µM) competes with a high concentration of arrestin (15 µM). Formation of the stably bound complex with cRaf and MEK scaffold is >100 times slower than for forming the transiently and intermediately bound states. Therefore, this step, scaffolding cRaf/MEK to the arrestin-receptor complex, is a rate-limiting step for ERK activation. Abbreviations for the different states of the muscarinic receptor (R) are the following: ligand-bound receptor (RL), RL after phosphorylation at the key residues (RLpp), arrestin-bound phosphorylated receptor (RLppA), and RLppA complexed with cRaf/MEK (RLppAM). When ERK binds to RLppAM (RLppAME), MEK phosphorylates ERK (RLppAMpE). The arrestin also binds transiently to RL that lacks phosphorylation at the key residues to form RLA. Then PP produces a small pool of stably bound β-arrestin–M1R complex (RLAPP). Our simulation is able to describe time-dependent changes of each state. Change of percentile indicates the portion of the individual states before and after 8 min agonist treatment. The initial transient binding (87%) becomes significantly reduced to 13% after 8 min of agonist treatment because it gradually gives way to the stable binding mode. The final portions of stable binding to phosphorylated and nonphosphorylated receptor were 76% and 5%, respectively; i.e., the majority of stable binding of arrestin occurs with the phosphorylated receptor. (E and F) Simulated time courses of individual states from the model shown on two timescales. Ligand is added at 0 s and removed at 480 s. (G–L) Fitting experimental data with our mathematical model. (G) The FRET data for arrestin binding to the biased receptor (gray symbols in B) was simulated as the total number of arrestin-bound receptors (Arrestin-R, solid line). (H) Development of stable arrestin binding by different agonist treatments (Fig. 3A and SI Appendix, Fig. S2). (I) Time course of stable arrestin binding to the biased receptor compared with the experimental FRETr data (Fig. 3B). The error bars in the experimental data are SDs. (J–K) Simulated effect of the GRK blocker and mutations at the key phosphorylation sites on arrestin binding to the receptor. (J) The forward rate constant of phosphorylation (k4) was set to zero to mimic the effect of blocking GRK (Fig. 3D). (K) Mutation of phosphorylation sites changes the binding affinity of GRK to the ligand-bound receptor. The forward rate constant for binding of GRK (k3) was set to zero. The mutations accelerated arrestin binding to the receptor as measured with FRETr (red symbols in B) and as simulated (red line, K). (L) Simulated ERK phosphorylation in control or mutated (S228A/S273A) biased receptor when k3 equals to zero. **P = 0.01 and *P = 0.03 for B and C, respectively.
Kinetic Modeling of Arrestin-Dependent ERK Phosphorylation.
Our results gave sufficient quantitative kinetic information to develop a mathematical kinetic description to test our conclusions better. Perforce, the model must make specific assumptions and interpretations. Agreements of the model with the data demonstrate the self-consistency of these assumptions, but do not prove that some other assumptions would not have been consistent as well. We tried not to make unnecessarily complicated assumptions. We first propose a reaction scheme for the biased M1R, summarized in Fig. 4D. Starting at the far left in Fig. 4D is the resting inactive, unphosphorylated receptor, and proceeding along the top to the right, it binds ligand, is phosphorylated by GRK, binds arrestin, forms a scaffold for MEK and ERK, and finally ERK is phosphorylated. All these forms have bound ligand. These steps have been described qualitatively before.
After adjustments to fit our observations (see rate constants in SI Appendix, Table S1), our model can generate quantitative predictions for the time courses of each of these sequential steps. We start with an overall view (Fig. 4 E and F) and then proceed to simulations of individual experiments in the subsequent panels of Fig. 4. Inspection of Fig. 4 E and F suggests three timescales: subsecond, several seconds, and hundreds of seconds. The subsecond kinetics are not well determined as they are below the time resolution of our measurements. Fig. 4E shows that in the model the binding of ligand (k1) is subsecond and that the initial competitive binding of GRK (k3) or arrestin (k2) to RL is very fast, arbitrarily set at 1–2 s. Our live-cell measurements reveal that β-arrestin binds to the M1R in multiple kinetically distinguishable modes that have labels such as transient binding, intermediate binding, and stable binding in the diagram. The ligand binding step, the initial transient arrestin binding (Lower row in Fig. 4D), and the initial GRK binding all reverse in a few seconds when agonist is removed (Fig. 4F). These steps involve unphosphorylated receptors. Over the few seconds after agonist is applied, GRK phosphorylates receptors, and arrestin slowly begins to be recruited in a more stable complex. For simplicity, we diagrammed the reaction of GRK as two simultaneous phosphorylations forming RLGRKpp. This reaction, followed by loss of GRK and assembly of the arrestin-MEK-ERK scaffold, gradually depletes the initial species RL, RLA, and RLGRK over several hundred seconds, and active ERK (pERK) is formed in this long timescale. The early transient RLA complex recruits a protein phosphatase (PP) (k12) that can dephosphorylate pERK, reducing any tonic background ERK activity. The RLAPP complex is more stable and slower to reverse than RLA. Longer activation of M1R leads to MEK-dependent ERK signaling via the stable β-arrestin/phosphorylated-receptor complex. The simulation suggests that the rate-limiting step is recruitment of cRaf/MEK (k8) and that the most stable binding complexes are the cRaf/MEK-, cRaf/MEK/ERK-, and cRaf/MEK/pERK-bound states, ultimately involving ∼76% of the biased receptors.
The kinetic model in Fig. 4D for the biased M1R recapitulates our observations. It describes the initial rapid transient binding and slowly developing stable binding of arrestin to the receptor (Figs. 4 G–I and SI Appendix, Fig. S9). Blocking the phosphorylation step (k4) by GRK inhibition removes the stable binding (Fig. 4J). Setting the rate constant for the GRK-binding step (k3) to zero (Fig. 4K) mimics the effect of mutating two phosphorylation sites in Fig. 4B, consistent with the hypothesis that the double mutant decreases the binding affinity for GRK in addition to eliminating sites of phosphorylation. With k3 = 0, there is no stable arrestin binding along the upper pathway so that now the transient-binding lower pathway is augmented. The model also shows binding of protein phosphatase to the transient RLA form to yield RLAPP. Although involving only a small portion of the biased receptors (5%), this complex had to be made stable after the removal of the agonist to prevent the continued rise of the pERK level (Fig. 4L). The abundance of the RLAPP complex rose to 20% for the phosphorylation mutant (S228A/S273A) where its precursor the transient RLA form was augmented (Fig. 4K). In addition, our model reliably captures the interplay between positive regulation of ERK activity by the recruitment of MEK (SI Appendix, Fig. S3 D–F) and negative regulation by protein phosphatase (SI Appendix, Fig. S6 D–F). Taken together, the strength of ERK activity is controlled by a balance between the transient and stable branches of arrestin binding to M1R (Lower and Upper rows in Fig. 4D, respectively).
The kinetics of WT M1R were modeled in the same manner (Fig. 5A). However, GRK was replaced by CK2 as suggested by our experiments (Fig. 3 G–I) and by previous literature. GRK has a PIP2-binding pleckstrin homology domain that helps to anchor it at the plasma membrane (41). However, because the WT M1R couples to Gq, when the receptor is activated, PIP2 becomes depleted by phospholipase C (PLC), allowing the membrane pool of GRK to drift away. Thus, GRK becomes less available during strong activity of Gq-coupled receptors. Instead, cytosolic casein kinases phosphorylate WT M1R, albeit more slowly than GRK phosphorylates the biased receptor. Therefore, quite different kinetics and steady-state levels of stable arrestin binding (Fig. 3B) result for WT versus biased receptors (Fig. 5 B and C). Our WT M1R model described well the observed real-time FRET between receptor and arrestin (Fig. 5D), the time course of stabilization of arrestin depending on the duration of agonist application (Fig. 5 E and F, solid circles), and the delayed increase of pERK after significant dephosphorylation of pERK by protein phosphatase (Fig. 5F, line). The WT model did not reproduce the continued phosphorylation of ERK after washing off the agonist. Presumably, the amount of pERK is regulated by other G-protein–dependent mechanisms as described previously (27).
Fig. 5.
Simulation of WT M1R-arrestin signaling. (A) The model structure was the same as for the biased receptor, except that the receptor kinase is CK2 instead of GRK. The number of dissociated Gβγ (<50 molecules/µm2) from the activated WT M1R is estimated to be ∼100-fold lower than hydrolyzed PIs (PIP2 and PIP) and much lower than cytosolic arrestin. We did not include possible contributions of G proteins for competing with arrestin at active receptors. Rate constants for some of steps were different from those for the biased receptor (SI Appendix, Table S1), including a faster dephosphorylation of pERK by protein phosphatase complexed with RLA (RLAPP) to recapitulate the initial decreasing of pERK. (B and C) Simulated time courses of individual states from the model shown on two time scales. Some states that were too small to illustrate in the same scale were not included. (D–F) Simulation of arrestin-WT M1R interaction and ERK activity. (D) Simulation of the total number of arrestin-bound receptors (black line) compared with FRETr between WT M1R and arrestin (red). (E) Development of stable binding of arrestin to WT M1R. (F) Summary of stabilized arrestin (solid circles) and ERK phosphorylation (line).
Discussion
We have described the real-time kinetics of arrestin binding to the M1R followed by scaffolding of the MAPK pathway, and we have encapsulated them as a kinetic model. In some proposals for other GPCRs, β-arrestin first couples loosely to phosphorylated residues in the C terminus of an activated receptor and then more tightly to the core region of the receptor via the finger loop region of arrestin (3, 4). However, unlike rhodopsin and the β2-AR, M1R has only three potential phosphorylation sites in the C terminus, and, as we saw, mutating two of the serines there does not abrogate β-arrestin binding. Additionally, in a recent M1R crystal structure, the C-terminal tail seems poorly aligned to make an interface to β-arrestin 1 (42). Based on molecular modeling that compares the M1R C tail with that of PAR2 [SI Appendix, Fig. S10 A and B; see Materials and Methods for details of Iterative Threading Assembly Refinement (I-TASSER) modeling], we speculate that the third intracellular loop of M1R (missing in the crystal structure) crowds the C terminus. However, that loop apparently forms the interface region for arrestin binding and contains 12 serine/threonine residues. We have established two of these residues as key for phosphorylation, and their mutation weakens the stable binding of β-arrestin considerably. We postulate that arrestin binds (Upper Right in Figs. 4D and 5A) to phosphorylated sites in the third intracellular loop of M1R and then interacts with effector proteins such as cRaf, MEK, and ERK, further stabilizing the β-arrestin–M1R complex. With our optical measurements on the M1R, we found that stably bound β-arrestin forms a platform for the activation of the ERK signaling and that, upon subsequent dissociation of this complex, signaling from β-arrestin stops.
Conformational changes of arrestin after activation of the β2-AR may be important for signaling to downstream effectors (17, 26). It has been suggested that, like G proteins, arrestin might be able to dissociate from active β1-AR while still retaining an activated signaling conformation, a “memory effect” (16, 18). From our observations with the M1R, we can say that the recorded arrestin conformational change in the unliganded transient complex with receptor reverses very quickly after arrestin unbinds with no memory effect after transient binding. On the other hand, the arrestin conformational change in the activated stable complex is surprisingly long lasting. We attribute this to persistence of the complex itself for >400 s. Because there is little dissociation of the complex in that time, we cannot say whether the arrestin has an additional memory after dissociation from the stable complex with the M1R. Our broad supposition is that different receptors induce unique binding modes of arrestin, depending on the shape of the C tail and the size of the third intracellular loop of the active receptor, as well as the number and location of phosphorylation sites. Key factors for recruiting and stabilizing arrestin may be the alignment of the C-terminal tail of the receptor and how much the interface is screened by the third intracellular loop. Such factors are hard to compare among different receptors using a real-time FRET assay, but, according to our I-TASSER modeling, β-ARs have a less bulky third intracellular loop compared with muscarinic receptors. This situation leaves open the possibility that β-ARs may have different binding modes with arrestin that are released sooner and might give rise to memory effects.
So far, we have focused on the roles of specific receptor regions for recruiting arrestins. Unexpectedly, we found that the biased M1R without G-protein interaction did not induce an intramolecular FRET change of arrestin but still stimulated the MAPK pathway. At face value, this could mean that an arrestin conformational change is not needed for activation of ERK by the biased M1R; however, several other possibilities cannot be excluded. First, there may be a conformational change that is not captured by FRET between residue 154 and the C terminus of arrestin. Second, in line with a mega-complex model of receptor–G protein–arrestin (43), G proteins or their signaling may promote a conformational change of arrestin. This model proposed that the G protein and arrestin could be physically bound to the same receptor so that, for example, the G protein interacts with a core region of the receptor whereas arrestin hangs from the C-terminal tail of the receptor. Third, different GPCR kinases could generate distinct conformations of arrestin when they interact with active receptors.
GRKs are considered the primary kinases for phosphorylation of active rhodopsin and β-adrenergic receptors. One factor that aids receptor-induced plasma-membrane recruitment of GRK2/3 is binding to the Gβγ subunits dissociated from heterotrimeric G proteins (41), and another factor is binding to negatively charged phospholipids including the phosphatidylinositides PIP2 and PIP (41, 44). We found that the biased M1R, unable to activate Gq proteins or to stimulate PLC-dependent hydrolysis of PIP2, still appeared to be phosphorylated by GRKs (Fig. 3 D–F). In this case, basal PIP2 may suffice to localize GRKs. Although it generates diffusible Gβγ, the WT M1R is phosphorylated by CK2 instead of GRK2/3 possibly because it substantially reduces plasma membrane PIP2 and PIP with the strong stimuli that we use (20, 45).
What is the physiological significance of switching regulation of ERK activity by two modes of β-arrestin binding relying on receptor phosphorylation? We speculate that there is time- and intensity-dependent switching for different signals. For example, a short-term activation of muscarinic receptor at a cholinergic synapse can evoke Gq-coupled activation of phospholipase C and subsequent IP3- and diacylglycerol-dependent signaling. Evidently, transiently bound β-arrestin may also down-regulate any ongoing ERK signaling by ERK dephosphorylation (SI Appendix, Fig. S9). Longer activation of the receptor evokes stable binding of β-arrestin and augments ERK signaling, a molecular mechanism that could be involved in modulation of ion channels (46, 47) and formation of long-term memories (30, 39, 48). Evidently, by itself β-arrestin 1, but not β-arrestin 2, can act directly on plasma membrane ion channels (49).
Previous work with angiotensin II, vasopressin, and protease-activated receptors has suggested that phosphorylation of ERK follows internalization of complexed receptors via specific targeted endosomal vesicles (10, 14, 15). In contrast, our study of M1R supports the concept that significant MEK/ERK signaling starts at the plasma membrane without internalization of the receptor–β-arrestin complex (50). ERK activation at the cell surface could be physiologically significant for M1R (and β1 adrenergic receptors) that experience much less internalization compared with β2-AR (SI Appendix, Fig. S4) (18). Finally, in good agreement with barcode (51, 52) or multisite phosphorylation-threshold (53) hypotheses of phosphorylation of receptors, the extent and position of receptor phosphorylation by protein kinases controls downstream MEK/ERK signaling via the binding stability of β-arrestin to M1R. Altogether, our study provides a molecular explanation for differential signaling from a GPCR because it couples to MEK/ERK signaling and paves the way for the development of new therapeutic approaches.
Materials and Methods
Reagents and Solution.
Cell-permeable SNAP505 and SNAP647 dyes were from New England Biolabs. For labeling with SNAP505 dye, cells were incubated with 5 µM dye in a 37 °C CO2 incubator for 75–90 min, and then free dye was washed out three times with cell culture medium. For SNAP647 dye, the concentration was 0.3 µM, and the time was 5–10 min. Labeled cells were incubated in culture medium for at least 1 h before measurements were taken. M1Rs were cloned into the pSNAPf vector, putting the SNAP tag at the receptor C terminus. FlAsH-EDT2 [4′,5′-bis(1,3,2-dithioarsolan-2-yl)fluorescein-(1,2-ethanedithiol)2] dye and EDT (1, 2-ethanedithiol) were purchased from Santa Cruz Biotechnology. Gq-CFP and β-arrestin 2–CFP constructs were provided by Joachim Goedhart, University of Amsterdam, Amsterdam, The Netherlands, and Moritz Bünemann, University of Würzburg, Würzburg, Germany, respectively. The β-arrestin 2–CFP FlAsH2 construct, obtained from Carsten Hoffman, University of Würzburg, Würzburg, Germany, had the FlAsH peptide sequence (CCPGCC) between residues 154 and 155 in the N domain of β-arrestin (17). An EDT solution was used before FlAsH labeling with EDT2 dye to reduce nonspecific labeling of off-target proteins. First, 1 M EDT in dimethyl sulfoxide (DMSO) was prepared from the 9.5-M stock bottle (Santa Cruz Biotechnology) in the fume hood and diluted to 250 µM and further diluted to 10 µM in normal Ringer’s solution (see below). Cells were preincubated with 10 µM EDT and then treated with 0.5 µM FlAsH-EDT2 in the presence of 10 µM EDT for ∼90 min in the 37 °C CO2 incubator. The dye was washed out with Ringer’s solution containing 250 µM EDT for 5 min, and the cells were placed in the incubator for at least 1 h before the experiments. The ERK KTR vector (pLentiCMV Puro DEST ERKKTRClover), originally made by the Covert laboratory (29), was obtained from Addgene (#59150). TTP 22 and TBB were purchased from Tocris, and cmpd101 (GRK2/3 blocker) was from HelloBio. Oxo was from Sigma-Aldrich. The cell-permeable SNAP-cell star 505 (1 mM, SNAP505), SNAP-cell 647 (0.6 mM, SNAP647), and FlAsH-EDT2 (1 mM) were dissolved in DMSO. Oxotremorine was made as 10 mM of stock with distilled water. Stock solutions for TTP22 (100 mM), TBB (100 mM), calyculin A (20 µM), and cmpd101 (30 mM) were prepared in DMSO. Vigene Biosciences produced all mutants of the SNAP-tagged M1R using our WT M1R in pSNAPf vector. Standard external Ringer’s solution contained (in mM): 137.5 NaCl, 2.5 KCl, 2 CaCl2, 1 MgCl2, 10 glucose, and 10 Hepes (pH adjusted to 7.3 with NaOH). All experimental recordings were made at room temperature (22–24 °C).
Cell Culture and Transfection.
Human embryonic kidney 293-derived tsA201 cells were obtained from Sigma-Aldrich. The cells were cultured in DMEM containing glutamine (2 mM), sodium pyruvate, and glucose (25 mM) supplemented with 10% FBS, 1% penicillin, and streptomycin and subcultured every 3–4 d using 0.05% trypsin and EDTA. Cells were transfected with different amounts of cDNA depending on the type of experiment. For normal FRET experiments, we used 1 µg of cDNA for β-arrestins, G-protein subunits, and receptors. For FRET measurements between Gq and M1R, we transfected with equal amounts of Gq-CFP, Gβ1, and Gγ2, as well as M1R-SNAP constructs without β-arrestin constructs (20). For FRET between β-arrestin and M1R, cells were transfected with β-arrestins and M1R-SNAP without cDNA of G proteins. For confocal experiments, we used 0.3 µg cDNA for red fluorescent protein (RFP)-tagged PI(4,5)P2 sensitive pleckstrin homology domain probe (PH-RFP) and 1 µg cDNA for receptors. For TIRF experiments at high sensitivity, we used ∼20-fold less cDNA for receptors (TIRF Microscopy). X-tremegene 9 DNA transfection reagent (10 µL, Roche Applied Science) and cDNA were added to Opti-MEM solution (∼87 µL) sequentially. Cells at ∼75% confluency in a 35-mm culture dish were washed with Opti-MEM and supplemented with 0.5 mL Opti-MEM solution. Then the mixture of the transfection reagent and cDNAs (∼100 µL) was added into the Opti-MEM. Cells were incubated for 4–6 h. Finally, cells were treated with trypsin (0.05%, Sigma-Aldrich) and seeded onto small 5-mm glass chips for confocal and FRET experiments or 25-mm round coverslips for TIRF imaging. The glass chips and coverslips were coated with poly-l-ornithine (1 mg/mL, Sigma-Aldrich).
FRET.
The measurements were performed as described previously (5, 20). Briefly, epifluorescence photometry was used to measure the FRET between CFP and SNAP505 dye or between CFP and FlAsH dye. Emission light from CFP and SNAP505 (or FlAsH) was collected by photomultipliers in photon-counting mode using an inverted Nikon Diaphot microscope equipped with a 40× 1.3 N.A. oil-immersion objective. CFP was excited at 440 nm, and emissions for CFP and SNAP505 (or FlAsH) were collected at 480 nm and 535 nm, respectively. FRETr was taken as the ratio of SNAP505 (or FlAsH) emission divided by CFP emission after correction for background and bleedthrough. FRETr values were normalized to the control value before treatment with agonist to remove cell-to-cell variation. We collected data points every 1 s and used a local perfusion system that can exchange solutions within 1 s.
TIRF Microscopy.
For dual-color imaging in Fig. 2D, cells expressed SNAP-tagged WT M1R and β-arrestin 2 YFP. To label the SNAP tag, the cells were incubated with 0.3 µM SNAP647 dye for 5–10 min in the culture medium at 37 °C and then washed three times. The washing step is critical to remove the remaining dye from the cell interior. Cells were further incubated for at least 1 h before imaging. Photobleaching of the SNAP647 dye on the receptor was ∼15% within 1.5 s with a sampling rate of 33 frames/s. In the TIRF experiments, nonbleached receptors reentered the evanescent field from other regions of the cells during the intervals before and after treatment with agonist. To improve detection of YFP-tagged arrestin at near single-molecule sensitivity, we optimized several parameters: (i) to reduce the background signal, cDNA for arrestin (0.05 µg) was 20-fold less than used for ensemble FRET experiments (1 µg); (ii) to minimize the background signal collected from the cytosol, the angle of the TIRF excitation lasers was carefully adjusted in our home-built TIRF microscope (5); (iii) to reduce photobleaching of YFP-tagged arrestin, the power of the excitation laser was reduced, and the sampling time was kept at 30 ms to collect sufficient photons; and (iv) the high-gain mode was selected in the EMCCD camera (Andor). The cytosolic fluorescence was far brighter than that from the plasma membrane in epifluorescence mode, in agreement with previous observations that most arrestins are located in the cytosol. Most of the background signal bleached quickly after exposure to the laser light (5). TIRF images at high sensitivity were taken on a Nikon Eclipse Ti microscope with an APO TIRF 100×/1.49 N.A. objective (5). Separate lasers at 488 nm and 641 nm were used for the excitation of YFP and SNAP647 dye, respectively. Images were collected by the EMCCD cameras and a custom-built controller written in LabVIEW as described. Separate EMCCDs were used for simultaneously collecting emission from YFP and SNAP dye. Multifluorescence solid latex beads (0.5 µm, Polysciences) were used to correct position differences in the two EMCCD cameras in Image J (NIH). The images showed bright spots (puncta) of arrestin and receptor (Fig. 2D). The puncta were not further analyzed to determine whether they represented single molecules or small clusters of molecules.
Real-Time Imaging of ERK Activity.
The ERK KTR probe was used to measure phosphorylated ERK activity (30). The tsA201 cells were transiently transfected with the probe and M1R 1 d before use. The probe was excited at 488 nm using a Polychrome IV monochromator (Till Photonics), and the emission signal was recorded at 510 nm every 4 s using an EMCCD camera (Photometrics Technology) in an inverted Nikon TE2000 microscope equipped with a 20× objective lens and additional 1.5× magnification. For analysis, nuclear and the cytoplasmic regions-of-interest were identified from pseudocolor images. Background fluorescence measured in a cell-free area was subtracted.
Confocal Microscopy.
Confocal images were taken every 12 s for the PH-RFP probe to monitor PIP2 depletion by SNAP-tagged M1R with a Zeiss 710 confocal microscope (SI Appendix, Fig. S1C). Excitation and emission light were delivered and collected by a 40× oil immersion lens (1.3 N.A.) as previously (5).
Mathematical Modeling.
M1R- and β-arrestin–mediated cell signaling was simulated using a rule-based, deterministic, compartmental model in Virtual Cell version 6 (University of Connecticut). Reactions were represented as first-order chemical kinetic equations without any spatial diffusion of molecules. The kinetic model defines molecule pools by the states and reactions described in Figs. 4D and 5A with rate constants and initial conditions given in SI Appendix, Table S1. The numerical parameters were based on literature values where possible and adjusted manually for description of our data.
GPCR Structure Predicted with I-TASSER.
The I-TASSER server was developed by the Y. Zhang laboratory (University of Michigan). The structure of G-protein–coupled M1R and PAR2 were predicted using their server. The protocol for I-TASSER is detailed in their publication (54). In principle, I-TASSER predicts the structure of proteins of interest using an iterative threading assembly refinement. The predicted structure matches the 3D models with other known proteins and then finds the lowest energy level for the final structure. It has different features compared with homology modeling, which considers only similarity of the query structure with the known template structure. If similar structures are publically available, homology modeling would be more accurate. If not, the I-TASSER approaches can solve unknown domain or whole structures. For example, the crystal structure of the seven transmembrane domains of M1R was fully resolved (41), but the C tail and third intracellular loop of the receptor were not due to flexibility and truncation for insertion of a reference protein (T4 lysozyme), respectively. Therefore, in this study, we used I-TASSER modeling to complete the structure of M1R. The PAR1 and a new PAR2 structure are also available, but the C terminus tail and ICL3 regions are not included (55, 56). With I-TASSER, the c-score indicates the accuracy of the predictions and 90% accuracy of the protein fold is expected for a c-score > −1.5. The c-scores in our simulation were −1.59 and −0.77 for mouse M1R and human PAR2, respectively. Another important parameter is the TM score (0–1), which provides topological information. TM > 0.5 means a correct topology, and a TM score <0.17 suggests a random structure. TM scores were 0.51 and 0.62 for M1R and PAR2, respectively. The simulated results were rendered in PyMOL.
Statistics.
All statistical tests are performed using Student’s t test with Excel software (Microsoft), and we report average ± SEM in the main text and figure legends. P < 0.05 was considered statistically significant. N and n indicate independent experiments and the total number of single cells, respectively.
Supplementary Material
Acknowledgments
We thank Lea M. Miller for technical assistance; Drs. Martin Kruse and Jill B. Jensen in the B.H. lab for discussion; Dr. Carsten Hoffmann (University of Würzburg) for providing β-arrestin 2 CFP FlAsH2 construct; Dr. Joachim Goedhart (University of Amsterdam) for Gq-CFP; Dr. Charles L. Asbury for the use of his single-molecule microscopy system; Drs. Neil M. Nathanson, Charles Chavkin, Nephi Stella, and Chris Hague for insightful comments on the manuscript; and the Virtual Cell group at the University of Connecticut, especially Drs. Leslie M. Loew and James C. Schaff. The Virtual Cell group is supported by NIH Grant P41 GM103313. Our work was supported by NIH Grants R37-NS08174 (to B.H.) and R01-DK080840 (to D.-S.K.); a Research Fellowship from Brazilian Council for Science and Technology Development (CNPq) Brazil (to C.K.); and NIH Grants R01-GM099373 and 1S10RR026406 (to Charles L. Asbury).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1700331114/-/DCSupplemental.
References
- 1.Doan T, Mendez A, Detwiler PB, Chen J, Rieke F. Multiple phosphorylation sites confer reproducibility of the rod’s single-photon responses. Science. 2006;313:530–533. doi: 10.1126/science.1126612. [DOI] [PubMed] [Google Scholar]
- 2.Doan T, Azevedo AW, Hurley JB, Rieke F. Arrestin competition influences the kinetics and variability of the single-photon responses of mammalian rod photoreceptors. J Neurosci. 2009;29:11867–11879. doi: 10.1523/JNEUROSCI.0819-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shukla AK, et al. Visualization of arrestin recruitment by a G-protein-coupled receptor. Nature. 2014;512:218–222. doi: 10.1038/nature13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kang Y, et al. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature. 2015;523:561–567. doi: 10.1038/nature14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jung SR, et al. Contributions of protein kinases and β-arrestin to termination of protease-activated receptor 2 signaling. J Gen Physiol. 2016;147:255–271. doi: 10.1085/jgp.201511477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Posor Y, Eichhorn-Grünig M, Haucke V. Phosphoinositides in endocytosis. Biochim Biophys Acta. 2015;1851:794–804. doi: 10.1016/j.bbalip.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 7.Xiao K, et al. Functional specialization of β-arrestin interactions revealed by proteomic analysis. Proc Natl Acad Sci USA. 2007;104:12011–12016. doi: 10.1073/pnas.0704849104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shenoy SK, Lefkowitz RJ. β-Arrestin-mediated receptor trafficking and signal transduction. Trends Pharmacol Sci. 2011;32:521–533. doi: 10.1016/j.tips.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ostermaier MK, Peterhans C, Jaussi R, Deupi X, Standfuss J. Functional map of arrestin-1 at single amino acid resolution. Proc Natl Acad Sci USA. 2014;111:1825–1830. doi: 10.1073/pnas.1319402111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luttrell LM, et al. Activation and targeting of extracellular signal-regulated kinases by β-arrestin scaffolds. Proc Natl Acad Sci USA. 2001;98:2449–2454. doi: 10.1073/pnas.041604898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shenoy SK, Lefkowitz RJ. Seven-transmembrane receptor signaling through β-arrestin. Sci STKE. 2005;2005:cm10. doi: 10.1126/stke.2005/308/cm10. [DOI] [PubMed] [Google Scholar]
- 12.Song X, Coffa S, Fu H, Gurevich VV. How does arrestin assemble MAPKs into a signaling complex? J Biol Chem. 2009;284:685–695. doi: 10.1074/jbc.M806124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Min J, Defea K. β-arrestin-dependent actin reorganization: Bringing the right players together at the leading edge. Mol Pharmacol. 2011;80:760–768. doi: 10.1124/mol.111.072470. [DOI] [PubMed] [Google Scholar]
- 14.DeFea KA, et al. β-Arrestin-dependent endocytosis of proteinase-activated receptor 2 is required for intracellular targeting of activated ERK1/2. J Cell Biol. 2000;148:1267–1281. doi: 10.1083/jcb.148.6.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tohgo A, et al. The stability of the G protein-coupled receptor-β-arrestin interaction determines the mechanism and functional consequence of ERK activation. J Biol Chem. 2003;278:6258–6267. doi: 10.1074/jbc.M212231200. [DOI] [PubMed] [Google Scholar]
- 16.Ranjan R, Gupta P, Shukla AK. GPCR signaling: β-Arrestins kiss and remember. Curr Biol. 2016;26:R285–R288. doi: 10.1016/j.cub.2016.02.056. [DOI] [PubMed] [Google Scholar]
- 17.Nuber S, et al. β-Arrestin biosensors reveal a rapid, receptor-dependent activation/deactivation cycle. Nature. 2016;531:661–664. doi: 10.1038/nature17198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eichel K, Jullié D, von Zastrow M. β-Arrestin drives MAP kinase signalling from clathrin-coated structures after GPCR dissociation. Nat Cell Biol. 2016;18:303–310. doi: 10.1038/ncb3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Waugh MG, Challiss RA, Berstein G, Nahorski SR, Tobin AB. Agonist-induced desensitization and phosphorylation of m1-muscarinic receptors. Biochem J. 1999;338:175–183. [PMC free article] [PubMed] [Google Scholar]
- 20.Jensen JB, Lyssand JS, Hague C, Hille B. Fluorescence changes reveal kinetic steps of muscarinic receptor-mediated modulation of phosphoinositides and Kv7.2/7.3 K+ channels. J Gen Physiol. 2009;133:347–359. doi: 10.1085/jgp.200810075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falkenburger BH, Jensen JB, Hille B. Kinetics of M1 muscarinic receptor and G protein signaling to phospholipase C in living cells. J Gen Physiol. 2010;135:81–97. doi: 10.1085/jgp.200910344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeatman HR, et al. Allosteric modulation of M1 muscarinic acetylcholine receptor internalization and subcellular trafficking. J Biol Chem. 2014;289:15856–15866. doi: 10.1074/jbc.M113.536672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma L, et al. Selective activation of the M1 muscarinic acetylcholine receptor achieved by allosteric potentiation. Proc Natl Acad Sci USA. 2009;106:15950–15955. doi: 10.1073/pnas.0900903106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shukla AK, et al. Structure of active β-arrestin-1 bound to a G-protein-coupled receptor phosphopeptide. Nature. 2013;497:137–141. doi: 10.1038/nature12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shukla AK, et al. Distinct conformational changes in β-arrestin report biased agonism at seven-transmembrane receptors. Proc Natl Acad Sci USA. 2008;105:9988–9993. doi: 10.1073/pnas.0804246105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee MH, et al. The conformational signature of β-arrestin2 predicts its trafficking and signalling functions. Nature. 2016;531:665–668. doi: 10.1038/nature17154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lohse MJ, Hoffmann C. Arrestin interactions with G protein-coupled receptors. Handb Exp Pharmacol. 2014;219:15–56. doi: 10.1007/978-3-642-41199-1_2. [DOI] [PubMed] [Google Scholar]
- 28.Goldsmith ZG, Dhanasekaran DN. G protein regulation of MAPK networks. Oncogene. 2007;26:3122–3142. doi: 10.1038/sj.onc.1210407. [DOI] [PubMed] [Google Scholar]
- 29.Alvarez-Curto E, et al. Targeted elimination of G proteins and arrestins defines their specific contributions to both intensity and duration of G protein-coupled receptor signaling. J Biol Chem. 2016;291:27147–27159. doi: 10.1074/jbc.M116.754887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Regot S, Hughey JJ, Bajar BT, Carrasco S, Covert MW. High-sensitivity measurements of multiple kinase activities in live single cells. Cell. 2014;157:1724–1734. doi: 10.1016/j.cell.2014.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosenblum K, et al. The role of extracellular regulated kinases I/II in late-phase long-term potentiation. J Neurosci. 2002;22:5432–5441. doi: 10.1523/JNEUROSCI.22-13-05432.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beaulieu JM, et al. An Akt/β-arrestin 2/PP2A signaling complex mediates dopaminergic neurotransmission and behavior. Cell. 2005;122:261–273. doi: 10.1016/j.cell.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 33.Zhou B, Wang ZX, Zhao Y, Brautigan DL, Zhang ZY. The specificity of extracellular signal-regulated kinase 2 dephosphorylation by protein phosphatases. J Biol Chem. 2002;277:31818–31825. doi: 10.1074/jbc.M203969200. [DOI] [PubMed] [Google Scholar]
- 34.Pitcher JA, Freedman NJ, Lefkowitz RJ. G protein-coupled receptor kinases. Annu Rev Biochem. 1998;67:653–692. doi: 10.1146/annurev.biochem.67.1.653. [DOI] [PubMed] [Google Scholar]
- 35.Tsuga H, Okuno E, Kameyama K, Haga T. Sequestration of human muscarinic acetylcholine receptor hm1-hm5 subtypes: Effect of G protein-coupled receptor kinases GRK2, GRK4, GRK5 and GRK6. J Pharmacol Exp Ther. 1998;284:1218–1226. [PubMed] [Google Scholar]
- 36.Violin JD, Ren XR, Lefkowitz RJ. G-protein-coupled receptor kinase specificity for β-arrestin recruitment to the β2-adrenergic receptor revealed by fluorescence resonance energy transfer. J Biol Chem. 2006;281:20577–20588. doi: 10.1074/jbc.M513605200. [DOI] [PubMed] [Google Scholar]
- 37.Gurevich EV, Gainetdinov RR, Gurevich VV. G protein-coupled receptor kinases as regulators of dopamine receptor functions. Pharmacol Res. 2016;111:1–16. doi: 10.1016/j.phrs.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Torrecilla I, et al. Phosphorylation and regulation of a G protein-coupled receptor by protein kinase CK2. J Cell Biol. 2007;177:127–137. doi: 10.1083/jcb.200610018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Butcher AJ, et al. An antibody biosensor establishes the activation of the M1 muscarinic acetylcholine receptor during learning and memory. J Biol Chem. 2016;291:8862–8875. doi: 10.1074/jbc.M115.681726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nobles KN, et al. Distinct phosphorylation sites on the β(2)-adrenergic receptor establish a barcode that encodes differential functions of β-arrestin. Sci Signal. 2011;4:ra51. doi: 10.1126/scisignal.2001707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pitcher JA, Touhara K, Payne ES, Lefkowitz RJ. Pleckstrin homology domain-mediated membrane association and activation of the β-adrenergic receptor kinase requires coordinate interaction with Gβγ subunits and lipid. J Biol Chem. 1995;270:11707–11710. doi: 10.1074/jbc.270.20.11707. [DOI] [PubMed] [Google Scholar]
- 42.Thal DM, et al. Crystal structures of the M1 and M4 muscarinic acetylcholine receptors. Nature. 2016;531:335–340. doi: 10.1038/nature17188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomsen AR, et al. GPCR-G protein-β-arrestin super-complex mediates sustained G protein signaling. Cell. 2016;166:907–919. doi: 10.1016/j.cell.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Homan KT, Glukhova A, Tesmer JJ. Regulation of G protein-coupled receptor kinases by phospholipids. Curr Med Chem. 2013;20:39–46. [PubMed] [Google Scholar]
- 45.Traynor-Kaplan A, et al. Fatty-acyl chain profiles of cellular phosphoinositides. Biochim Biophys Acta. 2017;1862:513–522. doi: 10.1016/j.bbalip.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shui Z, Khan IA, Haga T, Benovic JL, Boyett MR. Control of the cardiac muscarinic K+ channel by β-arrestin 2. J Biol Chem. 2001;276:11691–11697. doi: 10.1074/jbc.M011007200. [DOI] [PubMed] [Google Scholar]
- 47.Yang S, et al. β-arrestin-dependent dopaminergic regulation of calcium channel activity in the axon initial segment. Cell Reports. 2016;16:1518–1526. doi: 10.1016/j.celrep.2016.06.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosenblum K, Futter M, Jones M, Hulme EC, Bliss TV. ERKI/II regulation by the muscarinic acetylcholine receptors in neurons. J Neurosci. 2000;20:977–985. doi: 10.1523/JNEUROSCI.20-03-00977.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu CH, et al. Arrestin-biased AT1R agonism induces acute catecholamine secretion through TRPC3 coupling. Nat Commun. 2017;8:14335. doi: 10.1038/ncomms14335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Budd DC, Rae A, Tobin AB. Activation of the mitogen-activated protein kinase pathway by a Gq/11-coupled muscarinic receptor is independent of receptor internalization. J Biol Chem. 1999;274:12355–12360. doi: 10.1074/jbc.274.18.12355. [DOI] [PubMed] [Google Scholar]
- 51.Reiter E, Ahn S, Shukla AK, Lefkowitz RJ. Molecular mechanism of β-arrestin-biased agonism at seven-transmembrane receptors. Annu Rev Pharmacol Toxicol. 2012;52:179–197. doi: 10.1146/annurev.pharmtox.010909.105800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Butcher AJ, et al. Differential G-protein-coupled receptor phosphorylation provides evidence for a signaling bar code. J Biol Chem. 2011;286:11506–11518. doi: 10.1074/jbc.M110.154526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lau EK, et al. Quantitative encoding of the effect of a partial agonist on individual opioid receptors by multisite phosphorylation and threshold detection. Sci Signal. 2011;4:ra52. doi: 10.1126/scisignal.2001748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roy A, Kucukural A, Zhang Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang C, et al. High-resolution crystal structure of human protease-activated receptor 1. Nature. 2012;492:387–392. doi: 10.1038/nature11701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheng RKY, et al. Structural insight into allosteric modulation of protease-activated receptor 2. Nature. 2017;545:112–115. doi: 10.1038/nature22309. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.