Significance
Definitive mesoderm (e.g., muscle cells) evolved in the ancestor of the bilateria but is not present in their sister group, Cnidaria. Forward transcriptomics and gene knockdown in the anthozoan Nematostella vectensis show that both cWnt and BMP 2/4 signaling pathways reciprocally regulate components of the endomesodermal gene regulatory network (GRN) during development to set up distinct regional territories prior to the onset of gastrulation. Furthermore, the conserved “kernel” of the bilaterian heart mesoderm GRN is operational in the endomesoderm of the anthozoan N. vectensis. This GRN fails to have a “lockdown” feedback loop found in bilaterian GRNs that may help explain the highly regenerative potential of adult cnidarian endomesoderm, suggesting that the endoderm and mesoderm arose from the bifunctional endomesoderm.
Keywords: cell signaling, cell fate, evodevo, heart kernel, gene regulatory network
Abstract
Gastrulation was arguably the key evolutionary innovation that enabled metazoan diversification, leading to the formation of distinct germ layers and specialized tissues. Differential gene expression specifying cell fate is governed by the inputs of intracellular and/or extracellular signals. Beta-catenin/Tcf and the TGF-beta bone morphogenetic protein (BMP) provide critical molecular signaling inputs during germ layer specification in bilaterian metazoans, but there has been no direct experimental evidence for a specific role for BMP signaling during endomesoderm specification in the early branching metazoan Nematostella vectensis (an anthozoan cnidarian). Using forward transcriptomics, we show that beta-catenin/Tcf signaling and BMP2/4 signaling provide differential inputs into the cnidarian endomesodermal gene regulatory network (GRN) at the onset of gastrulation (24 h postfertilization) in N. vectensis. Surprisingly, beta-catenin/Tcf signaling and BMP2/4 signaling regulate a subset of common downstream target genes in the GRN in opposite ways, leading to the spatial and temporal differentiation of fields of cells in the developing embryo. Thus, we show that regulatory interactions between beta-catenin/Tcf signaling and BMP2/4 signaling are required for the specification and determination of different embryonic regions and the patterning of the oral–aboral axis in Nematostella. We also show functionally that the conserved “kernel” of the bilaterian heart mesoderm GRN is operational in N. vectensis, which reinforces the hypothesis that the endoderm and mesoderm in triploblastic bilaterians evolved from the bifunctional endomesoderm (gastrodermis) of a diploblastic ancestor, and that slow rhythmic contractions might have been one of the earliest functions of mesodermal tissue.
Gastrulation is the morphogenetic event in metazoan embryonic development that leads to germ layer specification and organismal axial patterning (1). The complex cell signaling events that regulate both morphogenetic and cell-type specification is controlled temporally and spatially by complex networks of hierarchical regulatory inputs and outputs referred to as gene regulatory networks (GRNs) (1–5). During germ layer specification in triplobloblastic bilaterians with three germ layers (ectoderm, mesoderm, and endoderm), early endomesoderm (the embryonic precursor of both endoderm and mesoderm) induction is followed by the segregation of endodermal (e.g., lining of the gut and its derivatives) from mesodermal (e.g., muscle, connective tissue, kidney, somatic gonad, and coelomic lining) derivatives. Generally accepted to have evolved at the base of the bilaterian lineage, the presence of a mesoderm has played a crucial role in the evolution of complex animal body plans (5, 6). GRNs regulating initial embryonic endomesoderm specification are relatively well known in a variety of model bilaterian systems (7–12). Canonical Wnt (cWnt) signaling (13–17) and BMP signaling (18–21) play crucial regulatory roles during axis patterning and endomesoderm specification in bilaterians.
To characterize the molecular basis for the evolution of distinct mesodermal and endodermal tissues from an ancestral bifunctional endomesoderm, we studied the development of the diploblastic anthozoan cnidarian, Nematostella vectensis. Cnidarians are the sister group to the triploblastic bilaterians (Fig. 1B) and although anthozoans possess clear signs of bilaterality (22), their adult body plan contains only an outer epidermis and an inner bifunctional gastrodermis that lines the gastric cavity and has both absorptive and contractile properties (Fig. 1A). As in bilaterians, cWnt signaling plays a critical role in regulating gastrulation and germ layer specification in N. vectensis (Fig. 1A). A protein called beta-catenin becomes localized to the nucleus in the blastomeres of the animal hemisphere, the location of presumptive endomesoderm formation, through the function of Disheveled, a key cytoplasmic component of the Wnt signaling pathway. Stabilization of beta-catenin, along with its coactivator TCF activates downstream target genes of the endomesodermal GRN (23–26) (Fig. 1A). Based on the expression of downstream target genes of cWnt signaling, the animal hemisphere contains at least four different domains: the central domain, the central ring, the central domain + ring, and the external ring before the onset of gastrulation (23–25) (Fig. 1C). Although BMP2/4 signaling in Nematostella has been studied during the formation of the directive axis (26–29) and implicated in a role in endomesoderm specification (26–28) (Fig. 1A), there has been no direct experimental evidence for a specific role played by BMP signaling during endomesoderm specification or for interactions with cWnt signaling.
Fig. 1.
Gastrulation and germ layer specification in Nematostella. (A) cWnt signaling provides inputs into the endomesodermal GRN in Nematostella to activate endomesodermal gene expression in the animal half blastomeres. BMP2/4 expession during early and later development in Nematostella suggests that it might be playing different regulatory roles during development. (B) Phylogenetic relationships among major metazoan lineages showing the position of Cnidaria as the sister taxon to all bilaterians (61). (C) At 24 hpf (late blastula stage) the animal hemisphere contains at least four domains defined by differential gene expression (25).
Results
NvBMP2/4 Is Required for Normal Gastrulation Movements and Germ Layer Segregation in Nematostella.
NvBMP2/4 shows localized expression in the animal hemisphere from early in development, indicating a potential role in gastrulation and germ layer segregation (27, 28). To test this hypothesis, we injected a translation-blocking NvBMP2/4 antisense morpholino (NvBMP2/4-MO) (26) into uncleaved zygotes to knockdown NvBMP2/4 protein levels in developing Nematostella embryos and collected midgastrula-stage embryos [48 h postfertilization (hpf)] and analyzed them using scanning confocal microscopy. Results of this analysis showed that both NvBMP2/4-MO–injected embryos and control-MO–injected embryos had undergone the initial stages of gastrulation morphogenesis normally, but the NvBMP2/4 morphant embryos were unable to form an organized epithelial endodermal layer (Fig. 2A). Both control and morphant embryos showed normal development of the ectoderm (Fig. 2A). This phenotype was highly reproducible and was similar to the phenotype seen in embryos overexpressing a dominant negative (dn) form of NvDishevelled that inhibits Wnt/beta-catenin signaling (24, 30). This similarity in phenotypes pointed to a potential role for NvBMP2/4 in germ layer specification in Nematostella embryos through an interaction with canonical Wnt signaling.
Fig. 2.
Differential inputs from cWnt and BMP2/4 signaling. (A) Effect of NvBMP2/4 knockdown and NvTcf knockdown on germ layer specification in Nematostella embryos. (A′) Control-MO–injected embryo at 48 hpf. (B′) NvBMP2/4-MO–injected embryo at 48 hpf. Embryos are stained with phalloidin (green) and propidium iodide (red). Note how disorganized the internal issue is and the lack of endomesodermal epithelialization. (B) Effects on gene expression after BMP MO injection (E′–H′ and M′–P′) compared with control embryos (A′–D′ and I′–L′) analyzed by in situ hybridization. Insets show oral views of the embryo. (C) Venn diagram depicting unique and shared sets of down-regulated and up-regulated genes between dnTCF-injected embryos vs. dextran-injected control embryos and BMP2/4 morpholino-injected embryos vs. dextran-injected control embryos at 24 hpf. (D) Model GRN circuit that would potentially result in separating the central ring domain into an inner and an outer ring. (E) At 24 hpf, the animal hemisphere contains at least four domains defined by differential gene expression as demonstrated by Röttinger et al. (25): the central domain, the central ring, the central ring + ring domain, and the external ring. Based on the model proposed in this study, integrated inputs from canonical Wnt signaling and BMP2/4 signaling splits the central ring domain into two: an outer central ring coexpressing NvTcf and NvBMP2/4 and an inner central ring expressing NvBra, NvFoxA, and NvWntA.
cWnt and BMP2/4 Signaling Provides Reciprocal Inputs to the Cnidarian Endomesodermal GRN.
We then specifically investigated the roles of both cWnt signaling and BMP2/4 signaling in endomesodermal patterning at the onset of gastrulation in Nematostella (24 hpf) to determine whether these two pathways are providing differential inputs to the cnidarian endomesodermal GRN, and if so, whether the interaction between these two signaling pathways provides any insight into the evolutionary origins of distinct mesodermal and endodermal tissues. We used RNA-sequencing (RNA-seq) to identify differential inputs into the GRN through beta-catenin/TCF (cWnt) signaling and BMP2/4 signaling in the developing Nematostella embryo. The dataset consisted of three different transcriptomes with three biological replicates: one from NvTcf-deficient embryos (by injection of a dominant negative mRNA form of NvTcf that lacks the transactivation domain, one from BMP2/4-deficient embryos (by injection of antisense morpholino-injected embryos), and one from dextran-injected control embryos. This analysis identified a set of 271 genes that were up-regulated in embryos in which beta-catenin/TCF signaling was inhibited (Table S1) and another set of 227 genes that were down-regulated in the same embryos (Table S2). In embryos with reduced BMP2/4 signaling, 147 genes showed increased expression levels (Table S3), whereas 133 genes were down-regulated, compared with control embryos (Table S4). Interestingly, there was no overlap between genes being up-regulated or down-regulated, between embryos with decreased cWnt signaling compared with decreased BMP2/4 signaling. Instead, a subset of genes was differentially regulated reciprocally by Tcf signaling and BMP2/4 signaling (up-regulated by Tcf and down-regulated by BMP2/4 and vice versa) (Fig. 2C). Eighty-six genes showed increased expression levels in cWnt signaling-deficient embryos and showed decreased expression levels in BMP2/4 signaling-deficient embryos (Table S5). Another 57 genes were down-regulated in cWnt signaling-deficient embryos while being up-regulated in BMP2/4 signaling-deficient embryos (Table S6). Furthermore, some of the differentially regulated genes are already known to play a critical role in specifying the oral–aboral axis, highlighting a crucial early function of BMP2/4 signaling before a role in the specification of the directive axis during later development (26, 31, 32) and confirming potential roles suggested by previous in situ expression patterns (26–28). Thus, these results demonstrate that beta-catenin/TCF and BMP2/4 signaling interact to provide additional spatial patterning of the endomesoderm (Fig. 2).
Integration of Inputs from Beta-Catenin/Tcf Signaling and BMP2/4 Signaling Plays a Role in Defining Specialized Domains in the Developing Nematostella Embryo.
Unlike the case for bilaterians, endomesoderm is specified at the animal pole of the developing Nematostella embryos and beta-catenin/TCF signaling plays a crucial role in the specification and segregation of germ layers in these embryos (23–25, 30). Based on differential gene expression, the animal hemisphere of the 24-hpf Nematostella embryo has been divided into at least four domains (Fig. 1C): a central domain, a central ring (where both NvBMP2/4 and NvTcf are expressed), central domain plus central ring, and an external ring (25). Using RNA-seq, we determined that genes were being regulated in opposite ways through beta-catenin/TCF signaling and BMP2/4 signaling in all four expression domains in the animal hemisphere. In the external ring, NvChordin and NvWnt4 were up-regulated in NvBMP2/4 morphant embryos, whereas they were down-regulated in TCF-deficient embryos. Central ring genes NvFoxA, NvWntA, and NvBra were also up-regulated in BMP2/4 morphant embryos, whereas NvTcf, another central ring gene, was down-regulated. In contrast, embryos deficient in beta-catenin/Tcf signaling showed decreased NvFoxA, NvWntA, and NvBra expression and increased NvTcf expression. NvAxin, which is expressed in the central domain and the central ring, was up-regulated in NvBMP2/4 morphant embryos but down-regulated in TCF-deficient embryos. Central domain gene NvEhand showed increased levels of expression in beta-catenin/Tcf signaling-deficient embryos, whereas it was down-regulated in BMP2/4 morphant embryos (Fig. 2B). Reduced expression levels of Chordin, NvFoxA, NvBra, NvAxin, and NvWnt 4 were also seen in a separate study (25), which analyzed the expression of these genes in embryos by overexpressing a dominant negative version of the TCF protein using whole mount in situ hybridization (WMISH). Differential gene expression observed in NvBMP2/4 morphant embryos were confirmed with WMISH experiments in this study (Fig. 2B). Our results from transcriptome analysis and validated by in situ hybridization experiments indicate a crucial role for combinatorial signaling events resulting from the integration of beta-catenin/Tcf signaling and BMP2/4 signaling in defining specialized domains after the initial, broad specification of endomesoderm in the animal hemisphere by the activation of canonical Wnt signaling.
The Evolutionarily Conserved “Kernel” of the Bilaterian Heart Mesoderm GRN Is Operational in Nematostella.
Within the differentially regulated set of genes (Tables S5 and S6), we identified several key genes expressed in bilaterian mesoderm, including key components of the bilaterian heart-field specification GRN kernel (Fig. 3B) identified by Davidson and Erwin (4). The transcription factors NvNkx2.5 and NvEhand were up-regulated in embryos deficient in cWnt signaling but were down-regulated in embryos with reduced BMP2/4 signaling (Fig. 3A). Nkx2.5 (a homolog of Drosophila gene Tinman) (33) is part of the core regulatory network regulating bilaterian heart development and is expressed in cardiac progenitor cells of all vertebrates and functions as a target of inductive signals that initiate cardiogenesis (34–39). Hand genes are also part of the GRN kernel regulating heart development and regulate ventricular growth (33, 40, 41). Activation of the bilaterian heart GRN kernel requires signaling input from BMP/Dpp signaling, leading to the subsequent activation of downstream target genes (4, 33, 42–44). Therefore, the down-regulation of NvNkx2.5, the Nematostella homolog of Drosophila Tinman (45) and NvEhand in the developing gastrodermis of BMP2/4-deficient Nematostella embryos is surprisingly similar to what has been observed during cardiac mesoderm differentiation in bilaterian embryos and suggests the presence of an ancestral heart-field specification GRN kernel in the bifunctional endomesoderm of Nematostella. GRN kernels, or subcircuits regulating early specification, such as the heart-field specification GRN kernel, are evolutionarily more constrained compared with subcircuits responsible for subsequent fine-scale patterning and terminal differentiation (4). Therefore, identification of an ancestral heart-field specification GRN kernel in a nonbilaterian diploblastic cnidarian would provide evidence for the conservation of these GRN kernels across eumetazoans.
Fig. 3.
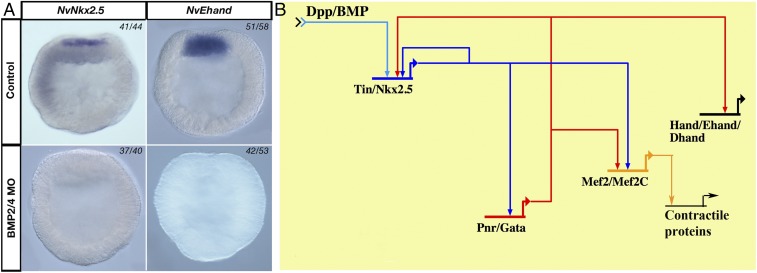
BMP2/4-regulated components of heart-field specification GRN kernel. (A) BMP2/4-deficient embryos lack NvNkx2.5 and NvEhand expression compared with control-MO–injected embryos at 24 hpf. (B) Potential heart-field specification GRN kernel assembled from experimental data from Drosophila and vertebrates [adapted from Davidson and Erwin (4)].
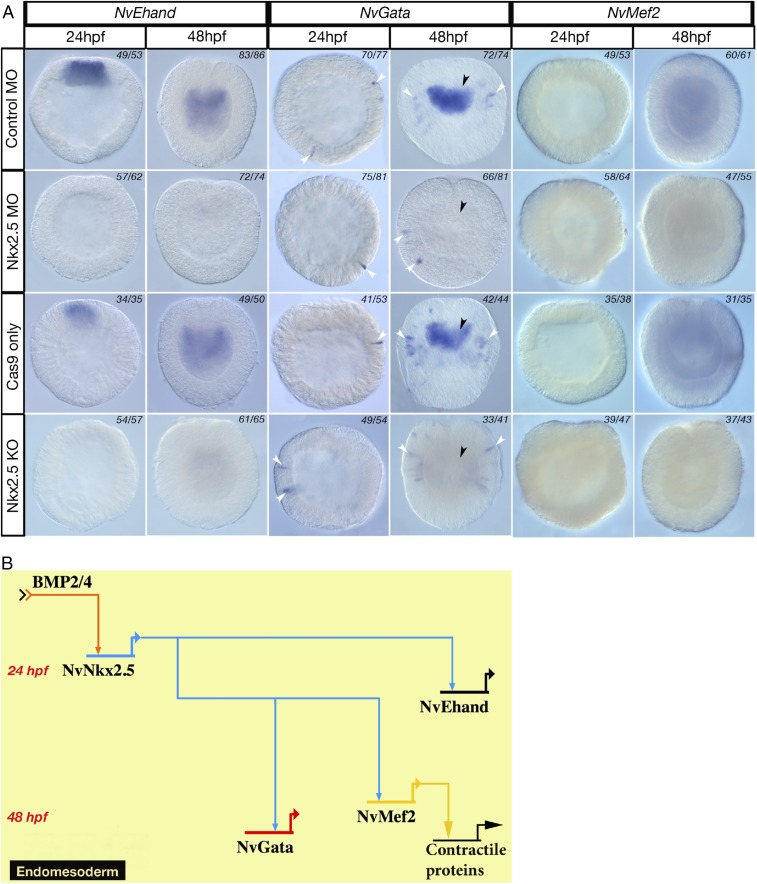
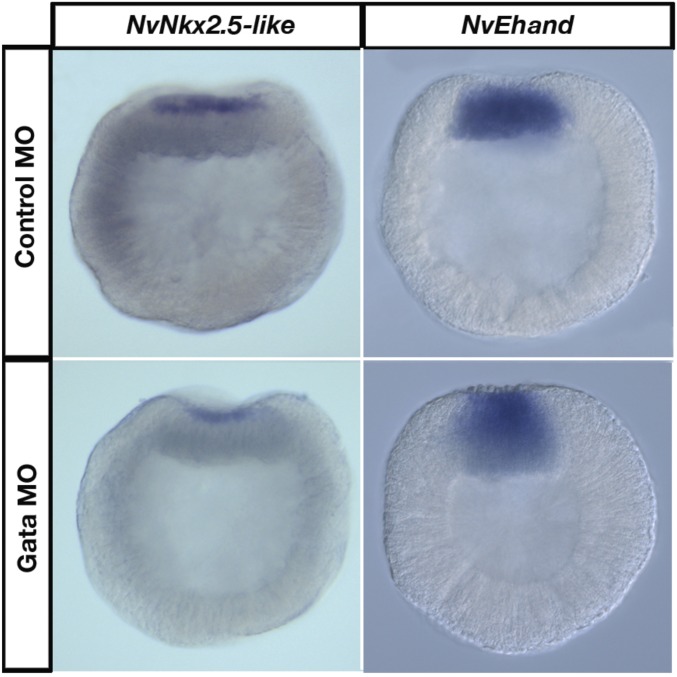
To determine whether the architecture of the putative Nematostella heart-field specification GRN is similar to bilaterians, we investigated the regulatory links within the GRN kernel by systematically knocking down transcription factors in the kernel, using CRSPR/Cas9 genome editing and antisense morpholinos (Figs. S1 and S2). In NvNkx2.5-deficient embryos, no NvEhand expression was seen at the animal pole at 24 hpf, confirming that NvEhand required regulatory input from NvNkx2.5 for its activation (Fig. 4A). By 48 hpf, all components of the GRN kernel are expressed in the endomesoderm. NvGata expression is pan-endomesodermal (6), whereas the endomesoderm-specific slice variant of NvMef2 is also expressed pan-endomesodermally at 48 hpf (46). Endomesodermal expression of NvGata at 48 hpf is completely lost in NvNkx2.5-deficient embryos in both NvNkx2.5 morpholino injection and CRISPR/Cas9. Interestingly NvGata also has a “salt-and-pepper” expression in the ectoderm starting at 24 hpf, and this ectodermal expression remained unchanged following NvNkx2.5 morpholino injection or CRISPR/Cas9-mediated NvNkx2.5 knockout. The fact that both morpholino and CRISPR/Cas9 treatments give the same phenotype, and that only endodermal expression is missing, confirms the specificity of the heart kernel to only endomesodermal tissue (Fig. 4A). Thus, by 48 hpf, components of the bilaterian heart-field specification GRN kernel are expressed in the endomesodermal gastrodermis of Nematostella and, as in bilaterians, the entire circuit is activated by BMP2/4, which activates NvNkx2.5, which in turn activates the downstream components NvEhand, NvGata, and NvMef2 (Fig. 4B). In the bilaterian heart-field specification GRN kernel, in addition to the BMP2/4- and Nkx2.5-dependent activation of downstream components, there are additional feedback loops, including autoregulation of Nkx2.5 by itself and positive feedback from Gata, which activates Nkx2.5, Ehand, and Mef2 (4). To test whether there is any feedback from the downstream components in to the GRN, we knocked down NvGata through morpholino-mediated gene knockdown. There were no detectable changes in the expression of any of the component genes in the endoderm (Fig. S3), suggesting that the fine-scale regulation of feedback mechanisms seen in bilaterians is not yet present, or has been lost, in the Nematostella GRN kernel.
Fig. S1.
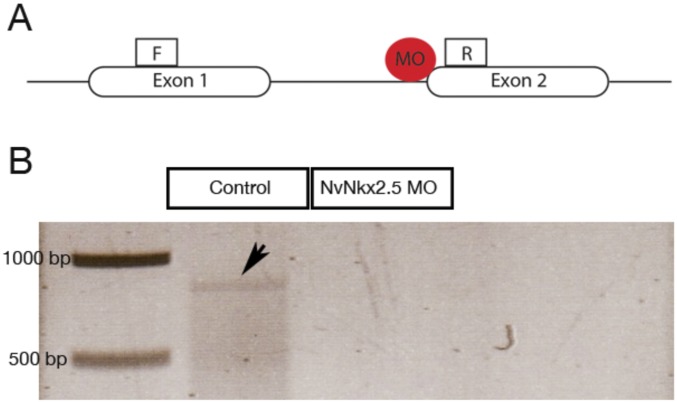
Morpholino mediated knock down and CRISPR/Cas9 mediated knock out of NvNkx2.5 in Nematostella. (A) A splice blocking anti-sense morpholino oligo was designed targeting the splice site between intron1 and exon2. (B) PCR showing an amplified DNA fragment of 880bp from cDNA from control embryos but absent in cDNA from NvNkx2.5 MO injected embryos. NvNkx2.5 gene deletion efficiency was determined by amplifying the NvNkx2.5 gene fragment.
Fig. S2.
CRISPR/Cas9 mediated knock out of NvNkx2.5 in Nematostella. (A) gRNA sequences and primers used for the PCR assay are shown on the genomic sequence of NvNkx 2.5. (B) Note the truncated fragments of NvNkx2.5 with a smaller size than the wild type resulting from CRISPR/Cas9 mediated mutagenesis. (C) NvNkx2.5 knockout embryos (Cas9 and gRNAs) lack NvNkx2.5 expression compared with control embryos (Cas9 only) embryos at 24 hpf.
Fig. 4.
Ancestral heart-field specification GRN kernel is present in Nematostella endoderm. (A) Components of the bilaterian heart-field specification GRN, NvEhand, and NvGata are lost in the endoderm (black arrowheads) and NvMef2 of NvNkx2.5-deficient embryos compared with their normal expression in control-MO–injected embryos. Ectodermal expression of NvGata is not affected by knockdown of NvNkx2.5 (white arrowheads). (B) An ancestral heart-field specification GRN kernel is present in Nematostella where NvNkx2.5 is activated by BMP2/4, which in turn activates NvEhand, NvGata, and NvMef2 in the endoderm. Note the absence of the Gata lockdown feedback loop.
Fig. S3.
NvGata does not provide regulatory inputs into the heart-field specification GRN kernel in Nematostella. NvNkx2.5 and NvEhand expression is not affected in NvGata MO-injected embryos compared with control MO-injected embryos at 24 hpf.
Discussion
BMP2/4 Signaling Plays a Key Role in Germ Layer Segregation in Nematostella.
The first provisional GRN-regulating endomesoderm specification in Nematostella was developed by Röttinger et al., using inputs from canonical Wnt signaling at the onset of gastrulation at 24 hpf (25). Here we provide evidence that BMP signaling provides additional inputs to the endomesodermal GRN based on NvBMP2/4 function in the animal hemisphere before gastrulation. These findings are supported by other studies on BMP function during development of Nematostella, where it was suggested that BMP signaling might be playing a role in endomesoderm specification based on the gene expression phenotypes seen in embryos with altered BMP signaling (26–28, 32). Two of these studies (26, 32) are mainly focused on BMP function at later stages of development postgastrulation and the findings have provided compelling evidence for a role for BMP signaling in patterning the directive axis in Nematostella (26, 32). However, given the highly dynamic temporal and spatial expression patterns of BMP2/4 and other components of the pathway, BMP signaling has the potential of playing multiple roles in regulating different developmental processes (27). The current study confirms that BMP2/4 is involved in endomesoderm specification before its role in patterning the directive axis based on the abnormal endomesoderm development and loss of endomesodermal marker genes (e.g., NvSnail, NvEhand, and NvOtxc) in BMP2/4 morphant embryos (Fig. 2B). Therefore, results of this study suggest that BMP2/4 signaling plays a critical role in further patterning endomesoderm in specific specialized tissues in Nematsotella after the initial, broad specification of endomesoderm through cWnt signaling. They further indicate a more specific role for BMP2/4 signaling in regulating “mesodermal” genes (NvSnail, NvEhand, and NvOtxC) in the bifunctional endomesoderm. However, the specific mechanisms of BMP signaling-mediated regulation of initial endomesoderm specification in Nematostella requires further investigation. Another aspect of this regulation that needs further investigation is the differential regulation of NvSnail by cWnt signaling and BMP2/4 signaling. Röttinger et al. (25) showed that knockdown of NvTcf by a dominant negative approach does not affect NvSnail expression in the developing embryo but the knockdown of NvBMP2/4 does result in the loss of NvSnail expression (Fig. 2B). However, NvSnail was not detected in the NvBMP2/4 knockdown RNA-seq dataset, likely because it is expressed in a small number of cells or its transcripts are not expressed at high levels in the cells that express it.
Reciprocal Signaling Events Resulting from Integration of cWnt and BMP2/4 Signaling Fine Tune Endomesodermal Patterning of the Early Nematostella Embryo.
This study shows that canonical Wnt signaling and BMP2/4 signaling differentially regulate a subset of genes at 24 hpf in developing Nematostella embryos and these include some key genes that are important in specifying different expression domains in the animal hemisphere. This differential regulation of components of these different domains shows how canonical Wnt signaling and BMP2/4 signaling integrate with one another to define distinct gene expression domains. At 24 hpf, the animal hemisphere of the developing Nematostella embryo contains at least four domains defined by differential gene expression (25): the central domain, the central ring, the central ring + central domain, and the external ring. The model proposed in this study suggests that there are at least five domains based on gene expression (at 24 hpf) because the integrated inputs from canonical Wnt signaling and BMP2/4 signaling produce two ring-like domains: an outer central ring coexpressing NvTcf and NvBMP2/4 and an inner central ring expressing NvBra, NvFoxA, and NvWntA. NvTcf expression in developing Nematostella embryos is dynamic, with uniform expression in early cleavage stages and the expression getting progressively restricted to the animal hemisphere such that by 24 hpf, it is expressed in a ring-like domain (the “central ring”) around the blastopore (25, 47). Based on the transcriptome data, canonical Wnt signaling down-regulates NvTcf expression and the progressive repression of NvTcf expression in the animal hemisphere, leading to the activation of canonical Wnt signaling at around 8 hpf. This initial wave of canonical Wnt signaling activates NvBMP2/4 in the animal hemisphere (25). However, after this initial activation, NvBMP2/4 up-regulates NvTcf expression and at 24 hpf, both NvBMP2/4 and NvTcf have overlapping ring-like expression domains (Fig. 2 B and E). The later expression of NvBMP2/4 appears to be independent of canonical Wnt signaling, thus reciprocally regulating only a subset of genes in this domain. This continued expression of NvTcf in the outer central ring overlaps with NvBMP2/4 expression driven by BMP signaling and results in the inhibition of canonical Wnt signaling in this domain (Fig. 2E). These interactions result in the inhibition of other target genes of canonical Wnt signaling, including NvFoxA, NvWntA, and NvBra, limiting them to the inner central ring, and NvChordin and NvWnt 4, limiting these genes to the external central ring. Thus, these two signaling pathways subfunctionalize the outer ring to pharyngeal (inner central ring) and oral (outer central ring) domains. Studies on lineage differentiation potential in mouse embryonic stem cells have shown that canonical Wnt signaling regulates differentiation of stem cells through the down-regulation of Tcf3 (48). A similar mechanism could be operating here to inhibit canonical Wnt signaling in a specific domain in the animal hemisphere; however, further experiments are needed to confirm this idea.
Mesodermal Gene Expression in Nematostella and the Origins of Triploblasty.
The formation of mesodermal derivatives was a critical event in metazoan evolution, leading to the evolution of more complex body plans in bilaterians. Generally believed to be a bilaterian invention, presence of a mesodermal germ layer has led to the division of metazoans into two main groups: the triploblasts with three germ layers, an ectoderm, an endoderm and a mesoderm; and the diploblasts, with only an ectoderm and an endoderm (6). However, investigations aimed at understanding the evolutionary origins of triploblasty has led to the discovery of “mesodermal” gene expression in the gastrodermis (endoderm) of the diploblastic nonbilaterian Nematostella (6). These findings support the hypothesis that the mesoderm and endoderm of bilaterians evolved from the bifunctional endomesoderm of diploblasts (8). This idea is further supported by the mechanisms by which endoderm and mesoderm are specified in bilaterians, where initial endomesoderm specification is followed by the segregation of distinct endodermal and mesodermal tissues (12, 49). The mechanisms of endomesoderm specification in Nematostella show remarkable similarities to bilaterians from the signaling pathways involved to the wiring of the endomesodermal GRN (23–25, 30). However, the mechanisms of segregating endomesoderm to discreet endodermal and mesodermal tissues is understood in only a few systems (12). Segregation of endomesoderm and further differentiation of the endoderm and the mesoderm involve the function of multiple signaling pathways, including cWnt, Notch, FGF, and BMP (12, 18, 50–53). Some of these bilaterian mechanisms are still being characterized but to understand the evolutionary origins of these mechanisms, the function of pathways involved in these processes has to be looked at in diploblastic animals with a bifunctional endomesoderm, and Nematostella appears to be an ideal candidate for further study.
GRN Kernels and Terminal Differentiation of Germ Layers.
The presence of an ancestral heart-field specification GRN kernel in the diploblastic cnidarian Nematostella provides valuable evolutionary insights into the origin of mesoderm. Diploblastic nonbilaterians lack a distinct mesodermal tissue even though the genome possesses a complex set of bilterian mesodermal orthologs (54) that are expressed in the gastrodermis (6), and cnidarian polyps have myoepithelial cells in the gastrodermis (55, 56). The Nematostella muscular system is divided into a body column and a tentacle system. The body column contains three morphologically and functionally distinct muscle groups that are endomesodermal in origin and are used to generate peristaltic movements along the oral–aboral axis to mix the contents of the gut (55, 56). The presence of the heart-field specification GRN kernel in the Nematostella gastrodermis suggests that it might be regulating the specification of smooth myoepithelial-like muscle cells in the gastrodermis that function to produce slow contractions, similar to those seen in smooth muscles by calcium-dependent phosphorylation of the myosin regulatory light chain and the phosphorylation of caldesmon on the actin filaments, also present in Nematostella (57). Nematostella also possess slow rectifying K+ channels characteristic of slow rhythmic smooth muscle contraction expressed in the gastrodermis (58, 59), which is consistent with the specification of myoepithelial cells with smooth muscle characteristics. These data support the hypothesis that an ancestral heart-field specification GRN kernel as seen in the myoepithelial cells in Nematostella regulated the specification of slow contracting smooth muscles that was later co-opted to regulate cardiac muscle in bilaterians. This likely corresponded with the evolution of novel contractile proteins and additional regulatory inputs into the GRN. For example, the absence of troponin in Nematostella, a key regulatory protein in powerful cardiac muscle contraction and relaxation in vertebrates (57, 60), suggests that this core kernel was modified to give rise to novel tissue types later in bilaterian evolution. It is also of interest that we detected evolutionary differences in the architecture of the heart kernel GRN. Nematostella appears to lack the Gata “lockdown” feedback loop (compare Fig. 3B to Fig. 4B) found in other bilaterian animals studied, which presumably maintains the stability of heart tissue cell fate. One fundamental difference between most bilaterian heart tissue cells and the endomesoderm in cnidarians like Nematostella is the dramatic ability of adult cells to regenerate, which appears to have been lost in many bilaterian taxa. It will be of interest to investigate the GRN “logic” in other cell types between regenerating and nonregenerating species to determine whether there are potential patterns in regulatory structure that can help explain the differences in regenerative ability.
There is strong evidence, both embryological and molecular, to suggest that the bifunctional endomesoderm of cnidarians gave rise to both the endoderm and the mesoderm in triploblastic bilaterians (58). Therefore, the discovery of an ancestral mesodermal GRN kernel regulating the expression of mesodermal genes during the specification and differentiation of the gastrodermis of Nematostella further reinforces the hypothesis that the endoderm and mesoderm in triploblastic bilaterians evolved from the bifunctional endomesoderm of a diploblastic ancestor.
SI Materials and Methods
Culture and Spawning and Microinjection of Nematostella vectensis.
Nematostella embryos were cultured at the Whitney Laboratory for Marine Bioscience, University of Florida. Males and females were kept in separate bowls in one-third (1/3×) seawater, and the adults were spawned by putting them in a light cycle as described by Hand and Uhlinger (62). The gelatinous mass around the eggs was removed using 4% l-cysteine followed by five washes of 1/3× seawater. Microinjections were carried out as described earlier (63). The fertilized/microinjected eggs were cultured in filtered 1/3× seawater at 17 °C.
Morpholino Antisense Oligonucleotide Sequences.
The NvBMP2/4 morpholino sequences have been reported previously (26): NVNkx2.5 MO (5′–3′): CCATTGACACTGAAAAGTTACAAGG; NvGata MO (5′–3′): ACTGTTTCCATTGAGATCACAGGGA; and standard control MO (5′–3′): CCTCTTACCTCAgTTACAATTTATA.
A 2-mM stock solution of each morpholino was diluted in autoclaved miliQ water to a final concentration of 1 mM in the microinjection mix.
CRISPR/Cas9-Mediated Mutagenesis.
Single-guide RNAs for CRISPR/Cas9-mediated mutagenesis of Nematostella NvNkx2.5 were synthezied using the following oligos as described by Dang et al. (64): NvNkx2.5 GR1, ATGTCAATCAATACCAATT; NvNkx2.5 GR2, GTTAAAGCAACGCGAATA; NvNkx2.5 GR3, GCCTGCCTCTTTGTAGTT; NvNkx2.5 GR4, AAAAGCGACTCTAGCCAAG; NvNkx2.5 GR5, AATGCATATCTTGGCTTTC; NvNkx2.5 GR6, GAGAACAAAGCGCGTGCTG; NvNkx2.5 GR7, AGATGGAGAGACGTGCTTC; and NvNkx2.5 GR8, GACAAGACCTGTTCGTGAG.
Cas9 protein was obtained from PNA Bio. A total of 1.25 μg/μL single-guide RNA mix and 1 μg/μL Cas9 protein were injected into fertilized eggs. Control embryos were injected with Ca9 protein only. DNA was prepared from individual polyps and PCR was carried out using the following oligo sequences to identify deletions in the genome: NvNkx2_5 gDNA F1AATGTTTCTGTCTTGCCTTGCGAG, NvNkx2_5 gDNA F2 GTGTCCCGTCTAACAGCCTGAG.
Whole Mount RNA in Situ Hybridization, Actin, and Nuclear Staining.
Whole mount in situ hybridization of Nematostella eggs and embryos was carried out as previously described (6). Digoxigenin-labeled RNA probes were synthesized using the MegaScript Transcription Kit (Ambion). Hybridization of the DIG-labeled RNA probe (1 ng/L) was carried out at 60 °C. Visualization of the labeled probe was carried out using NBT/BCIP as substrate for the alkaline phosphatase-conjugated anti-DIG antibody (Roche Applied Bioscience). In situ hybridizations were performed as previously described (6). Bodipy FL Phallacidin (Molecular Probes/Invitrogen, B607) and propidium iodide (Sigma, 81845) were used to stain F-actin and cell nuclei, respectively, as described previously (65) to analyze embryonic morphology.
In situ hybridization images were taken on a Zeiss AxioScop 2 mounted with an Axiocam camera triggered by Axiovision software (Carl Zeiss). Confocal images of stained embryos were taken using a Zeiss LSM710 microscope running the LSM ZEN software (Cal Zeiss).
RNA-Seq and Reference Mapping.
All nine samples were sequenced in a single lane on the Illumina HiSEq 2500 at the University of Texas Austin Genomic Sequencing and Analysis Facility. Reads were single-end 50 bp. Read quality was checked with Fastqc (https://www.bioinformatics.babraham.ac.uk/projects/) and the first 12 bases were trimmed using the FASTX toolkit along with the removal of any sequences with phred scores below 33. FastqMCF was used to remove any adaptor contamination and homopolymers. The reads were mapped to the edited Nematostella reference transcriptome from ref. 66, which is the genomic filtered model set with the exclusion of mitochondrial and ribosomal RNA sequences. Using Bowtie 2.2.0 beta3 with –verysensitive-local and -a flags. Counts were generated using the custom script from ref. 66. Differential gene expression was found by using EdgeR version edgeR_3.8.5 (67). One count per million in three or more samples was kept to filter low number reads, and the reads were normalized to the trimmed mean of M-values. Testing for differential expression was done using the generalized linear model approach and genes with adjusted P values below 0.05 were considered differentially expressed.
Supplementary Material
Acknowledgments
This work is supported by National Institutes of Health Grant GM093116.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1701607114/-/DCSupplemental.
References
- 1.Martindale MQ. The evolution of metazoan axial properties. Nat Rev Genet. 2005;6:917–927. doi: 10.1038/nrg1725. [DOI] [PubMed] [Google Scholar]
- 2.Levine M, Davidson EH. Gene regulatory networks for development. Proc Natl Acad Sci USA. 2005;102:4936–4942. doi: 10.1073/pnas.0408031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lyons DC, Kaltenbach SL, McClay DR. Morphogenesis in sea urchin embryos: Linking cellular events to gene regulatory network states. Wiley Interdiscip Rev Dev Biol. 2012;1:231–252. doi: 10.1002/wdev.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davidson EH, Erwin DH. Gene regulatory networks and the evolution of animal body plans. Science. 2006;311:796–800. doi: 10.1126/science.1113832. [DOI] [PubMed] [Google Scholar]
- 5.Pérez-Pomares JM, Muñoz-Chápuli R. Epithelial-mesenchymal transitions: A mesodermal cell strategy for evolutive innovation in Metazoans. Anat Rec. 2002;268:343–351. doi: 10.1002/ar.10165. [DOI] [PubMed] [Google Scholar]
- 6.Martindale MQ, Pang K, Finnerty JR. Investigating the origins of triploblasty: ‘Mesodermal’ gene expression in a diploblastic animal, the sea anemone Nematostella vectensis (phylum, Cnidaria; class, Anthozoa) Development. 2004;131:2463–2474. doi: 10.1242/dev.01119. [DOI] [PubMed] [Google Scholar]
- 7.Kimelman D, Griffin KJ. Vertebrate mesendoderm induction and patterning. Curr Opin Genet Dev. 2000;10:350–356. doi: 10.1016/s0959-437x(00)00095-2. [DOI] [PubMed] [Google Scholar]
- 8.Rodaway A, Patient R. Mesendoderm: An ancient germ layer? Cell. 2001;105:169–172. doi: 10.1016/s0092-8674(01)00307-5. [DOI] [PubMed] [Google Scholar]
- 9.Kikuchi Y, et al. Notch signaling can regulate endoderm formation in zebrafish. Dev Dyn. 2004;229:756–762. doi: 10.1002/dvdy.10483. [DOI] [PubMed] [Google Scholar]
- 10.Revinski DR, Paganelli AR, Carrasco AE, López SL. Delta-Notch signaling is involved in the segregation of the three germ layers in Xenopus laevis. Dev Biol. 2010;339:477–492. doi: 10.1016/j.ydbio.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 11.Hinman VF, Davidson EH. Evolutionary plasticity of developmental gene regulatory network architecture. Proc Natl Acad Sci USA. 2007;104:19404–19409. doi: 10.1073/pnas.0709994104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sethi AJ, Wikramanayake RM, Angerer RC, Range RC, Angerer LM. Sequential signaling crosstalk regulates endomesoderm segregation in sea urchin embryos. Science. 2012;335:590–593. doi: 10.1126/science.1212867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imai K, Takada N, Satoh N, Satou Y. (beta)-catenin mediates the specification of endoderm cells in ascidian embryos. Development. 2000;127:3009–3020. doi: 10.1242/dev.127.14.3009. [DOI] [PubMed] [Google Scholar]
- 14.Henry JQ, Perry KJ, Wever J, Seaver E, Martindale MQ. β-catenin is required for the establishment of vegetal embryonic fates in the nemertean, Cerebratulus lacteus. Dev Biol. 2008;317:368–379. doi: 10.1016/j.ydbio.2008.02.042. [DOI] [PubMed] [Google Scholar]
- 15.Schneider S, Steinbeisser H, Warga RM, Hausen P. β-catenin translocation into nuclei demarcates the dorsalizing centers in frog and fish embryos. Mech Dev. 1996;57:191–198. doi: 10.1016/0925-4773(96)00546-1. [DOI] [PubMed] [Google Scholar]
- 16.Logan CY, Miller JR, Ferkowicz MJ, McClay DR. Nuclear beta-catenin is required to specify vegetal cell fates in the sea urchin embryo. Development. 1999;126:345–357. doi: 10.1242/dev.126.2.345. [DOI] [PubMed] [Google Scholar]
- 17.Martindale MQ, Lee PN. The development of form: Causes and consequences of developmental reprogramming associated with rapid body plan evolution in the bilaterian radiation. Biol Theory. 2013;8:253–264. [Google Scholar]
- 18.Hoppler S, Moon RT. BMP-2/-4 and Wnt-8 cooperatively pattern the Xenopus mesoderm. Mech Dev. 1998;71:119–129. doi: 10.1016/s0925-4773(98)00004-5. [DOI] [PubMed] [Google Scholar]
- 19.Duboc V, et al. Nodal and BMP2/4 pattern the mesoderm and endoderm during development of the sea urchin embryo. Development. 2010;137:223–235. doi: 10.1242/dev.042531. [DOI] [PubMed] [Google Scholar]
- 20.Nikaido M, Tada M, Saji T, Ueno N. Conservation of BMP signaling in zebrafish mesoderm patterning. Mech Dev. 1997;61:75–88. doi: 10.1016/s0925-4773(96)00625-9. [DOI] [PubMed] [Google Scholar]
- 21.Shi Y, Katsev S, Cai C, Evans S. BMP signaling is required for heart formation in vertebrates. Dev Biol. 2000;224:226–237. doi: 10.1006/dbio.2000.9802. [DOI] [PubMed] [Google Scholar]
- 22.Finnerty JR, Pang K, Burton P, Paulson D, Martindale MQ. Origins of bilateral symmetry: Hox and dpp expression in a sea anemone. Science. 2004;304:1335–1337. doi: 10.1126/science.1091946. [DOI] [PubMed] [Google Scholar]
- 23.Wikramanayake AH, et al. An ancient role for nuclear β-catenin in the evolution of axial polarity and germ layer segregation. Nature. 2003;426:446–450. doi: 10.1038/nature02113. [DOI] [PubMed] [Google Scholar]
- 24.Lee PN, Kumburegama S, Marlow HQ, Martindale MQ, Wikramanayake AH. Asymmetric developmental potential along the animal-vegetal axis in the anthozoan cnidarian, Nematostella vectensis, is mediated by Dishevelled. Dev Biol. 2007;310:169–186. doi: 10.1016/j.ydbio.2007.05.040. [DOI] [PubMed] [Google Scholar]
- 25.Röttinger E, Dahlin P, Martindale MQ. A framework for the establishment of a cnidarian gene regulatory network for “endomesoderm” specification: The inputs of ß-catenin/TCF signaling. PLoS Genet. 2012;8:e1003164. doi: 10.1371/journal.pgen.1003164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saina M, Genikhovich G, Renfer E, Technau U. BMPs and chordin regulate patterning of the directive axis in a sea anemone. Proc Natl Acad Sci USA. 2009;106:18592–18597. doi: 10.1073/pnas.0900151106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matus DQ, et al. Molecular evidence for deep evolutionary roots of bilaterality in animal development. Proc Natl Acad Sci USA. 2006;103:11195–11200. doi: 10.1073/pnas.0601257103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matus DQ, Thomsen GH, Martindale MQ. Dorso/ventral genes are asymmetrically expressed and involved in germ-layer demarcation during cnidarian gastrulation. Curr Biol. 2006;16:499–505. doi: 10.1016/j.cub.2006.01.052. [DOI] [PubMed] [Google Scholar]
- 29.Leclère L, Rentzsch F. RGM regulates BMP-mediated secondary axis formation in the sea anemone Nematostella vectensis. Cell Reports. 2014;9:1921–1930. doi: 10.1016/j.celrep.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 30.Kumburegama S, Wijesena N, Xu R, Wikramanayake AH. Strabismus-mediated primary archenteron invagination is uncoupled from Wnt/β-catenin-dependent endoderm cell fate specification in Nematostella vectensis (Anthozoa, Cnidaria): Implications for the evolution of gastrulation. Evodevo. 2011;2:2. doi: 10.1186/2041-9139-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rentzsch F, et al. Asymmetric expression of the BMP antagonists chordin and gremlin in the sea anemone Nematostella vectensis: Implications for the evolution of axial patterning. Dev Biol. 2006;296:375–387. doi: 10.1016/j.ydbio.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 32.Genikhovich G, et al. Axis patterning by BMPs: Cnidarian network reveals evolutionary constraints. Cell Reports. 2015;10:1646–1654. doi: 10.1016/j.celrep.2015.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holland ND, Venkatesh TV, Holland LZ, Jacobs DK, Bodmer R. AmphiNk2-tin, an amphioxus homeobox gene expressed in myocardial progenitors: Insights into evolution of the vertebrate heart. Dev Biol. 2003;255:128–137. doi: 10.1016/s0012-1606(02)00050-7. [DOI] [PubMed] [Google Scholar]
- 35.Harvey RP. NK-2 homeobox genes and heart development. Dev Biol. 1996;178:203–216. doi: 10.1006/dbio.1996.0212. [DOI] [PubMed] [Google Scholar]
- 36.Cripps RM, Olson EN. Control of cardiac development by an evolutionarily conserved transcriptional network. Dev Biol. 2002;246:14–28. doi: 10.1006/dbio.2002.0666. [DOI] [PubMed] [Google Scholar]
- 37.Satou Y, Imai KS, Satoh N. The ascidian Mesp gene specifies heart precursor cells. Development. 2004;131:2533–2541. doi: 10.1242/dev.01145. [DOI] [PubMed] [Google Scholar]
- 38.Davidson B, Levine M. Evolutionary origins of the vertebrate heart: Specification of the cardiac lineage in Ciona intestinalis. Proc Natl Acad Sci USA. 2003;100:11469–11473. doi: 10.1073/pnas.1634991100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet. 2005;6:826–835. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- 40.Srivastava D, Olson EN. A genetic blueprint for cardiac development. Nature. 2000;407:221–226. doi: 10.1038/35025190. [DOI] [PubMed] [Google Scholar]
- 41.Brand T. Heart development: Molecular insights into cardiac specification and early morphogenesis. Dev Biol. 2003;258:1–19. doi: 10.1016/s0012-1606(03)00112-x. [DOI] [PubMed] [Google Scholar]
- 42.Reim I, Frasch M. The Dorsocross T-box genes are key components of the regulatory network controlling early cardiogenesis in Drosophila. Development. 2005;132:4911–4925. doi: 10.1242/dev.02077. [DOI] [PubMed] [Google Scholar]
- 43.Oliveri P, Davidson EH. Gene regulatory network controlling embryonic specification in the sea urchin. Curr Opin Genet Dev. 2004;14:351–360. doi: 10.1016/j.gde.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 44.Putnam NH, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- 45.Chourrout D, et al. Minimal ProtoHox cluster inferred from bilaterian and cnidarian Hox complements. Nature. 2006;442:684–687. doi: 10.1038/nature04863. [DOI] [PubMed] [Google Scholar]
- 46.Genikhovich G, Technau U. Complex functions of Mef2 splice variants in the differentiation of endoderm and of a neuronal cell type in a sea anemone. Development. 2011;138:4911–4919. doi: 10.1242/dev.068122. [DOI] [PubMed] [Google Scholar]
- 47.Lee PN, Pang K, Matus DQ, Martindale MQ. A WNT of things to come: Evolution of Wnt signaling and polarity in cnidarians. Semin Cell Dev Biol. 2006;17:157–167. doi: 10.1016/j.semcdb.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 48.Atlasi Y, et al. Wnt signaling regulates the lineage differentiation potential of mouse embryonic stem cells through Tcf3 down-regulation. PLoS Genet. 2013;9:e1003424. doi: 10.1371/journal.pgen.1003424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peter IS, Davidson EH. Evolution of gene regulatory networks controlling body plan development. Cell. 2011;144:970–985. doi: 10.1016/j.cell.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ciruna B, Rossant J. FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Dev Cell. 2001;1:37–49. doi: 10.1016/s1534-5807(01)00017-x. [DOI] [PubMed] [Google Scholar]
- 51.Green SA, Norris RP, Terasaki M, Lowe CJ. FGF signaling induces mesoderm in the hemichordate Saccoglossus kowalevskii. Development. 2013;140:1024–1033. doi: 10.1242/dev.083790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carmena A, Murugasu-Oei B, Menon D, Jiménez F, Chia W. Inscuteable and numb mediate asymmetric muscle progenitor cell divisions during Drosophila myogenesis. Genes Dev. 1998;12:304–315. doi: 10.1101/gad.12.3.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Halfon MS, Carmena A, Baylies M, Michelson AM. A molecular basis for competence and for the specificity of Ras signaling in mesoderm development. Dev Biol. 2000;222:270. [Google Scholar]
- 54.Jahnel SM, Walzl M, Technau U. Development and epithelial organisation of muscle cells in the sea anemone Nematostella vectensis. Front Zool. 2014;11:44. doi: 10.1186/1742-9994-11-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frank P, Bleakney JS. Histology and sexual reproduction of the anemone Nematostella vectensis Stephenson 1935. J Nat Hist. 1976;10:441–449. [Google Scholar]
- 56.Renfer E, Amon-Hassenzahl A, Steinmetz PRH, Technau U. A muscle-specific transgenic reporter line of the sea anemone, Nematostella vectensis. Proc Natl Acad Sci USA. 2010;107:104–108. doi: 10.1073/pnas.0909148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martinson AS, et al. Functional evolution of Erg potassium channel gating reveals an ancient origin for IKr. Proc Natl Acad Sci USA. 2014;111:5712–5717. doi: 10.1073/pnas.1321716111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li X, et al. Bimodal regulation of an Elk subfamily K+ channel by phosphatidylinositol 4,5-bisphosphate. J Gen Physiol. 2015;146:357–374. doi: 10.1085/jgp.201511491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Layland J, Solaro RJ, Shah AM. Regulation of cardiac contractile function by troponin I phosphorylation. Cardiovasc Res. 2005;66:12–21. doi: 10.1016/j.cardiores.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 60.Technau U, Steele RE. Evolutionary crossroads in developmental biology: Cnidaria. Development. 2011;138:1447–1458. doi: 10.1242/dev.048959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dunn CW, et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452:745–749. doi: 10.1038/nature06614. [DOI] [PubMed] [Google Scholar]
- 62.Hand C, Uhlinger KR. The culture, sexual and asexual reproduction, and growth of the sea anemone Nematostella vectensis. Biol Bull. 1992;182:169–176. doi: 10.2307/1542110. [DOI] [PubMed] [Google Scholar]
- 63.Layden MJ, Röttinger E, Wolenski FS, Gilmore TD, Martindale MQ. Microinjection of mRNA or morpholinos for reverse genetic analysis in the starlet sea anemone, Nematostella vectensis. Nature Protoc. 2013;8:924–934. doi: 10.1038/nprot.2013.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dang Y, et al. Optimizing sgRNA structure to improve CRISPR-Cas9 knockout efficiency. Genome Biol. 2015;16:280. doi: 10.1186/s13059-015-0846-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Magie CR, Daly M, Martindale MQ. Gastrulation in the cnidarian Nematostella vectensis occurs via invagination not ingression. Dev Biol. 2007;305:483–497. doi: 10.1016/j.ydbio.2007.02.044. [DOI] [PubMed] [Google Scholar]
- 66.Helm RR, Siebert S, Tulin S, Smith J, Dunn CW. Characterization of differential transcript abundance through time during Nematostella vectensis development. BMC Genomics. 2013;14:266. doi: 10.1186/1471-2164-14-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.